Abstract
The availability of acyl-Coenzyme A (acyl-CoA) thioester compounds affects numerous cellular functions including autophagy, lipid oxidation and synthesis, and post-translational modifications. Consequently, the acyl-CoA level changes tend to be associated with other metabolic alterations that regulate these critical cellular functions. Despite their biological importance, this class of metabolites remains difficult to detect and quantify using current analytical methods. Here we show a universal method for metabolomics that allows for the detection of an expansive set of acyl-CoA compounds and hundreds of other cellular metabolites. We apply this method to profile the dynamics of acyl-CoA compounds and corresponding alterations in metabolism across the metabolic network in response to high fat feeding in mice. We identified targeted metabolites (>50) and untargeted features (>1000) with significant changes (FDR < 0.05) in response to diet. A substantial extent of this metabolic remodeling exhibited correlated changes in acyl-CoA metabolism with acyl-carnitine metabolism and other features of the metabolic network that together can lead to the discovery of biomarkers of acyl-CoA metabolism. These findings show a robust acyl-CoA profiling method and identify coordinated changes of acyl-CoA metabolism in response to nutritional stress.
Thioester compounds containing acyl-coenzyme A (acyl-CoA)1 are key metabolites in intermediary metabolism. The most prominent of which is acetyl-CoA whose levels regulate critical cellular processes such as energy metabolism, protein acetylation, lipid synthesis and catabolism, and even autophagy (1–4). Other acyl-CoA compounds are also increasingly appreciated as playing important roles in diverse cellular processes (5–8). These compounds are generated from multiple pathways, such as glycolysis, the citric acid cycle (TCA cycle), beta-oxidation, and branched chain amino acid catabolism. As the acyl group carrier, acyl-CoA can partake in chemical reactions on proteins including histones resulting in mediation of chromatin biology. Therefore, considerable effort has been spent on developing methods for acyl-CoA and corresponding acyl protein modification measurements (9–17). Liquid chromatography coupled to mass spectrometry (LC-MS) is the most frequently used method for small molecule analysis in large part because of superior sensitivity. Moreover, LC-MS analysis can handle a broad range of complex biological mixtures and the analysis is relatively easier compared with many other methods, such as NMR, scintillation counting, and UV detection.
Reversed phase LC coupled to a triple quadrupole mass spectrometer has been frequently used as for targeted measurements of specific acyl-CoA compounds, because acyl-CoA compounds undergo a common fragmentation, the neutral loss of adenosine diphosphate, which is the basis of multiple reaction monitoring for acyl-CoA measurements. Especially when stable isotope labeled acyl-CoA standards are used, this method has shown high accuracy and precision (11, 14). However, these methods involve several laborious steps of sample purification and enrichment before LC-MS analysis, such as solid phase extraction, which in addition to often being time- and cost-prohibitive, can also result in poor sensitivity and accuracy because of imperfect metabolite recovery. Moreover, reversed phase ion-paired chromatography coupled to high-resolution MS has also been used for short, medium, and long chain acyl-CoA identification or quantification with the help of stable isotope labeled standards (10, 13). However, these methods were also developed with limited coverage of metabolites, and the quantitative capacity without using stable isotope labeled standards was not evaluated.
We therefore developed a novel method for sensitive, rapid, and quantitative acyl-CoA profiling, with a compatible sample preparation procedure that has been previously shown for polar metabolite analysis (18). The method involves LC-MS using reversed phase chromatographic separation coupled to a high-resolution Orbitrap mass spectrometer with label free quantitation. With a single liquid extraction from a few milligrams of tissue, followed by three separate chromatography methods, a broad coverage of metabolites is achieved, which is especially valuable when sample availability is limited.
To show the utility of our approach, we considered the alterations in the metabolic network that accompany high fat (HF) feeding. Conditions of high fat feeding that induce nutritional stress are shown to induce global changes in enzymes in metabolism (19, 20), but a comprehensive assessment of the global alterations in metabolism that remains include possible remodeling of acyl-CoA metabolism remain unknown. We reasoned that under such a condition, a dynamic response involving alterations in acyl-CoA levels along with the rest of the metabolome may be observed. This remodeling could also be associated with acyl-carnitine metabolism that often serves as both a readout of acyl-CoA metabolism and other features of metabolism status. Propionyl-CoA that is mainly generated from branched chain amino acid (BCAAs) catabolism and has been implicated in contributing to insulin resistance (21, 22), exhibits large changes. We applied our method to understand the metabolic changes that accompany HF feeding in mouse liver (23). We identify acyl-CoA compounds with dramatic changes after administration of a HF diet. Hierarchical clustering and principle component analysis (PCA) of metabolites measured in liver tissue show further diet-dependent metabolic profiling changes. Moreover, measurements of acyl-carnitine compounds have been used to reflect acyl-CoA levels (24), but the correlation between these two species has not been studied. Our method with coverage of both acyl-carnitine and acyl-CoA enabled us to evaluate acyl-carnitine as a biomarker of acyl-CoA status. In turn, we were able to confirm many relationships between acyl-CoA and acyl-carnitine levels but also discovered several unexpected relationships as well.
EXPERIMENTAL PROCEDURES
Reagents
RPMI 1640 medium was purchased from Cellgro. Fetal Bovine Serum (FBS), penicillin, and streptomycin were purchased from Hyclone Laboratories, Logan, UT. Optima ammonium acetate, ammonium hydroxide, Optima LC-MS grade formic acid, acetonitrile, methanol, and water were purchased from Fisher Scientific, Agawam, MA. U-13C-glucose was obtained from Cambridge Isotope Laboratories, Tewksbury, MA. CoA and acyl-CoA thioester standards were purchased from Sigma-Aldrich, St Louis, MO, except suberyl-CoA was synthesized in house. Inorganic salts were purchased from BDH Chemicals, VWR International, West Chester, PA.
Mouse Models
Wild-type (WT) C57/B6 (CD45.2+; 000664) mice were purchased from the Jackson Laboratory, Bar Harbor, ME and bred at Cornell University. Mice were housed in a pathogen-free facility. All animal procedures were approved by the Cornell Institutional Animal Care and Use Committee. Six-week-old male mice were fed with either standard chow (SC), 13% fat, 67% carbohydrate, and 20% protein from Harlan Teklad, Madison, WI (2914) or high fat diet (HF), 60% fat, 20% carbohydrate, and 20% protein from Research Diets Inc, New Brunswick, NJ. (D12492) for 12 weeks. Mice were fasted for 4 h before they were sacrificed by cervical dislocation and tissues were snap frozen in liquid nitrogen. Frozen tissues were stored at −80 °C for further analysis (23).
Cell Culture
HCT116 cells were provided as a generous gift from Dr. Lewis Cantley's laboratory, Weill Cornell Medical College, NY. HCT116 cells were first cultured in 6-well plate with full growth medium, which contains RPMI 1640, 10% FBS, 100 U/ml penicillin, and 100 μg/ml streptomycin. Cells were grown in 37 °C incubator with 5% CO2.
HPLC Method
Ultimate 3000 UHPLC (Dionex) is coupled to Q Exactive-Mass spectrometer (QE-MS, Thermo Scientific, San Jose, CA) for metabolite separation and detection. For acyl-CoA analysis, a reversed phase liquid chromatography (RPLC) method was used. For short to medium chain acyl-CoA compounds, a Luna C18 column (100 × 2.0 mm i.d., 3 μm; Phenomenex, Torrance, CA) is employed with mobile phase A: water with 5 mm ammonium acetate (pH = 6.8), and mobile phase B: methanol. Linear gradient is: 0 min, 2% B; 1.5 min, 2% B; 3 min, 15% B; 5.5 min, 95%B; 14.5 min, 95%B; 15 min, 2%B, and 20 min, 2% B. For medium to long chain acyl-CoA compounds, a Luna C18 column (100 × 2.0 mm i.d., 3 μm; Phenomenex) is employed with mobile phase A: water with 10 mm ammonium acetate (pH = 8.5, adjusted with ammonium hydroxide), and mobile phase B: acetonitrile. Linear gradient is: 0 min, 20% B; 1.5 min, 20% B; 5 min, 95% B; 14.5 min, 95%B; 15 min, 20%B, and 20 min, 20% B. Flow rate is 0.2 ml/min. Column temperature: room temperature. For additional polar metabolite analysis, a hydrophilic interaction chromatography method (HILIC) with an Xbridge amide column (100 × 2.1 mm i.d., 3.5 μm; Waters, Milford, MA) is used for compound separation at room temperature. The mobile phase and gradient information were described previously (18).
MS Method
The Q Exactive mass spectrometer (QE-MS) is equipped with a HESI probe, and the relevant parameters are as listed: heater temperature, 120 °C; sheath gas, 30; auxiliary gas, 10; sweep gas, 3; spray voltage, 3.6 kV for positive mode and 2.5 kV for negative mode. Capillary temperature was set at 320 °C, and S-lens was 55. A full scan range was set at 60 to 900 (m/z) when coupled with the HILIC method, or 100 to 1500 (m/z) when coupled with the RPLC method. The resolution was set at 70,000 (at m/z 200). The maximum injection time (max IT) was 200 ms. Automated gain control (AGC) was targeted at 3 × 106 ions. For targeted MS/MS analysis, the isolation width of the precursor ion was set at 1.5 (m/z), high energy collision dissociation (HCD) was 35%, and max IT is 100 ms. The resolution and AGC were 35,000 and 200,000, respectively.
Metabolite Extraction
When cultured cells were used, the procedure was similar as described in previous study (18). Basically, the media were quickly removed when cells reach 80% confluence, and the plates were placed on top of dry ice. One milliliter of extraction solvent was immediately added, and the plates were then transferred to the −80 °C freezer. The plates were left for 15 min and then cells were scraped into extraction solvent on dry ice. The whole solution was centrifuged with the speed of 20,000 × g at 4 °C for 10 min. When liver tissue was used, the tissue sample was first homogenized in liquid nitrogen and then 5 to 10 mg was weighed in a new Eppendorf tube. Ice cold extraction solvent (250 μl) was added to tissue sample, and mixer was used to further break down tissue chunk and form an even suspension. After incubation on ice for additional 10 min, the tissue extract mix was centrifuged with the speed of 20,000 × g at 4 °C for 10 min. Here, cell extracts were prepared from three wells to make triplicates. Six extraction solvents, 80% methanol/water (“80MeOH”), 80% methanol/water with 5% formic acid (“80MeOH_5FA”), methanol/acetonitrile/water (2:2:1, v/v, “MeOH_ACN”) with or without 5% formic acid (“MeOH_ACN_5FA”) were tested. Ten percent of the supernatant from tissue extract was transferred to a new tube for LC-QE-MS analysis using a HILIC method, whereas the rest was split into two tubes for CoA analysis using a reversed phase liquid chromatography (RPLC) method (one tube as backup). All samples were dried in a vacuum concentrator (SpeedVac) and stored at −80 °C as a dry pellet to preserve the stability.
Evaluation of Acyl-CoA Stability
Freshly prepared acyl-CoA standard solution was diluted with different solvents, including water (named as “Water”), 50 mm ammonium acetate at pH 4.0 (named as “AQ-pH4.0,” adjusted with acetic acid), 50 mm ammonium acetate at pH 6.8 (“AQ-pH6.8”), 50% methanol/water (“50%Water”), 50% pH 4.0/methanol (“50%AQ-pH4.0”), and 50% pH 6.8/methanol (“50%AQ-pH6.8”). The final concentration of every acyl-CoA is 1 μm. These six samples were placed in the LC autosampler at 4 °C, and an injection of 5 μl was done every 8 h, ending up with six injections over 2 days. To test the acyl-CoA stability in liver extract, dry liver extract was dissolved in AQ-pH6.8 solvent, and an injection of 5 μl from the same vial was done after 0, 3, 6, 9, 12, and 24 h after reconstitution.
Sample Preparation for LC-MS Analysis
Samples (as a dry pellet) are reconstituted into solvent in batches (20–30 samples per batch), and analyzed as soon as possible once they are dissolved into solvent. For short to medium chain acyl-CoA analysis, cell or liver extract was dissolved into 30 μl of 50 mm ammonium acetate, pH 6.8. For medium to long chain acyl-CoA analysis, the extract was dissolved into the same ammonium acetate buffer with 20% acetonitrile. For polar metabolite analysis, water was added to dissolve the pellet of tissue extract and after centrifugation, the same volume of methanol/acetonitrile (1:1, v/v) was added to dilute water to minimize the sample solvent effect on HILIC chromatography. Samples were centrifuged at 20,000 × g at 4 °C for 3 min and the supernatant was transferred to LC vials. The injection volume was 7 μl for CoA analysis, which is equivalent to a metabolite extract from 1 mg tissue injected on the column. The injection volume was 5 μl for HILIC chromatography, which is equivalent to a metabolite extract of 160 μg tissue injected on the column. To evaluate the dynamic range of current LC-QE-MS setup, a serial dilution of the sample was done to make different concentrations of samples.
Preparation of 13C Labeled Extract and Evaluation of Tissue Extraction Method
Escherichia coli strain NCM3722 (E. coli) was a generous gift from Joshua Rabinowitz's group (Princeton University). The E. coli culture and metabolite extraction method were modified from a previous studies (25, 26). Briefly, E. coli was grown in a 2 liter flask containing minimal liquid medium with 13C-glucose overnight, and the supernatant was removed by centrifugation at 1000 × g 4 °C for 5 min, followed by adding extraction solvent, acetonitrile/methanol/water (2:2:1, v/v) with 0.1 N formic acid. The flask was kept at −20 °C for 30 min, and the solvent was centrifuged at 6000 × g at 4 °C for 20min. The supernatant was neutralized by addition of 0.1 N ammonium hydroxide before drying in the vacuum concentrator. The dry pellet was further purified by using a C18 solid phase extraction cartridge (Sigma) (27). Ten, 30, 90, and 180 μl of final eluent were transferred to a new Eppendorf tube and dried under vacuum. Here, 10 μl of E. coli extract is roughly equivalent to 1 ml of original E. coli in liquid culture after overnight growth. Finally, the 13C labeled E. coli pellets were reconstituted in 20 μl water and spiked into 4 mg liver extract. The rest of the liver extraction procedure was the same as described above. The final pellet was dissolved in 20 μl ammonium acetate buffer (50 mm, pH 6.8) and 7 μl was injected to LC-QE-MS.
Peak Extraction
Raw data collected from LC-QE-MS were processed on Thermo Scientific software Sieve 2.0. Peak alignment and detection were performed according to manufacturer protocols. For a targeted metabolomics analysis, a frame seed including 194 metabolites was used for targeted metabolites analysis with data collected in positive mode, whereas a frame seed of 263 metabolites was used for negative mode, m/z width is set at 10 ppm as previously described (18). A similar method, including standard compound comparison, MS/MS fragmentation pattern, isotope ratio comparisons, was used to build a frame seed containing acyl-CoA. For the untargeted metabolomics analysis, the following parameter values were used to extract untargeted features (pairs of m/z, putatively assigned chemical composition, and RT): background signal to noise ratio, 3; minimum ion count, 1 × 105; minimum scans across peak, 5; m/z step, 10 ppm.
Statistic Analysis
To assess the linear range, the data were filtered as follows: for each metabolite in the targeted list, if the lowest integrated mass spectrometer signal (MS intensity) in all of the samples is less than 103 and meanwhile the highest signal is less than 104, then this metabolite is considered as below the detection limit and will be excluded for further data analysis; if the lowest signal is less than 103, but the highest signal is more than 104, then a value of 103 is imputed. For acyl-CoA profiling, this noise threshold is set at 100, whereas for untargeted metabolomics, the threshold is 5000. p value (student's t test, two tailed) was adjusted with False Discovery Rate (FDR) correction. The data filtering process, linear regression, t test, PCA, and elastic net regression (lambda = 0.5) were performed in the R computing language (http://www.r-project.org/). Hierarchical clustering and heat map were done with the software Gene-e (Broad Institute, http://www.broadinstitute.org/cancer/software/GENE-E/download.html). Pathway analysis using the KEGG pathway database (http://www.genome.jp/kegg/) was carried out with Metaboanalyst package (http://www.metaboanalyst.ca/MetaboAnalyst/faces/Home.jsp).
RESULTS
Simultaneous Determination of Acyl-CoA Thioesters and Other Metabolites
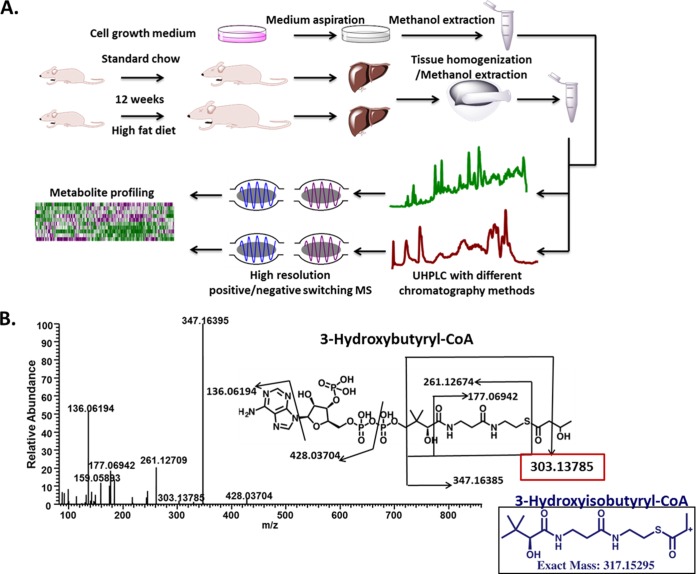
A high throughput method for the simultaneous measurement of acyl-CoA compounds and the polar complement of the metabolome was developed, and applied to both cultured cells and tissue samples (Fig. 1A). The detailed sample preparation procedure from cultured cells was discussed in a previous study (21) but the method was extended to consider tissue extraction. Cell or tissue extracts were separated by three chromatography methods comprising one normal and two reversed phase chromatography methods followed by high-resolution mass analysis with positive and negative mode switching coupled to an Orbitrap mass analyzer. With improved ion optics and faster scanning speeds, we hypothesized that the resulting enhanced sensitivity could allow for robust profiling of acyl-CoA compounds. First for metabolite assignments, a targeted MS/MS was performed (Fig. 1B and Table I), and the major fragment ions are listed in Table I. A representative MS/MS spectra and major fragment ion structure assignment of 3-hydroxybutyryl-CoA is shown in Fig. 1B.
Fig. 1.
Workflow. A, Workflow of acyl-CoA thioesters and other metabolite analysis. Cold methanol was used to extract acyl-CoA or other metabolites after cell medium aspiration or tissue homogenization. The colorectal cancer cell line HCT116 was used as cell line model, whereas liver from standard chow or high fat diet fed mice was used as tissue samples. Cell or tissue extracts were split into two and analyzed by three different liquid chromatography (LC) methods coupled to one Orbitrap based positive and negative switching mode high-resolution mass spectrometer (HRMS, LC-QE-MS). Finally, the raw data from these two methods were combined to give both acyl-CoA and other metabolites profiling. B, MS/MS spectrum and fragment interpretation of 3-hydroxybutyryl-CoA detected from liver extract. Fig. 1 was made with software Chembiodraw 13.0.
Table I. Summary of measured acyl-CoA compound. Acyl-CoA compound names are listed in the first column. BT: Butyryl; IBT: Isobutyryl; VAL: Valeryl; IVAL: Isovaleryl; HMG: 3-Hydroxymethylglutaryl; HBT: 3-Hydroxybutyryl; SUC: Succinyl; MML: Methylmalonyl. The number of carbons (C) in the name indicates the number of carbons in acyl group, while the number after “C:” indicates the number of double bonds. R represents the CoA moiety, and the structures of acyl groups are provided in column 2. Retention time (RT) of each acyl-CoA compound is shown in column 3. The mass error (ppm) of acyl-CoA detected from standard (STD), HCT116 cell, and liver samples is listed in column 4 to 6. The major MS/MS fragments generated at a collision energy of 35% and resolution of 35000 from acyl-CoA detected from the liver sample are listed in column 7. NA means standard is not available, and ND means acyl-CoA is not detected in HCT116 (2 × 105 cells) or liver extract (1 mg) with current method setup.
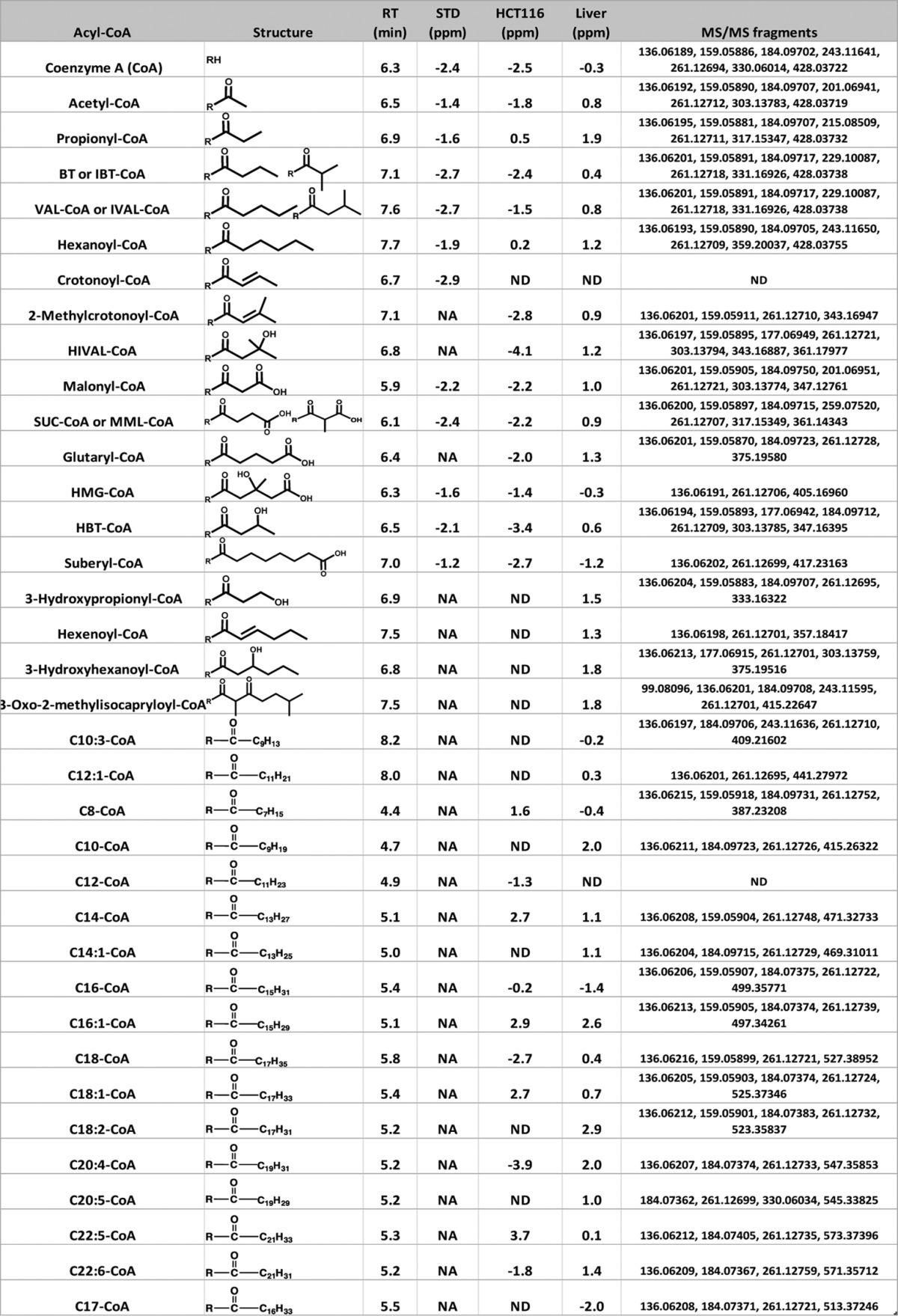
The common fragments shared by each acyl-CoA compound are those generated from the breakdown of the CoA moiety. The most commonly observed cleavage pattern was the neutral loss of adenosine diphosphate. When there is a functional group attached to the acyl group, such as a hydroxyl, carboxylate, the carbon bond next to these function groups is weakened, and the cleavage of the C–C bond in the acyl group facilitates structure elucidation. For example, we are able to distinguish 3-hydroxybutyryl-CoA (HBT-CoA) and 3-hydroxyisobutyryl-CoA (HIBT-CoA) from the characteristic fragment ion, m/z = 303.13758, which is generated by cleavage between C2 and C3 in 3-hydroxybutyryl-CoA, but may not be readily generated from 3-hydroxyisobutyryl-CoA (cleavage between C2 and C3 generates m/z = 317.15295, Fig. 1B). The measured acyl-CoA names, including abbreviation names, acyl group structures, retention time (RT) and mass error of every acyl-CoA detected from standards (STD), HCT116 cells and liver are summarized in Table I. Unresolvable isomers, such as butyryl-or isobutyryl-CoA (BT or IBT-CoA), valeryl or isovaleryl-CoA (VAL or IVAL-CoA) are grouped together. Acyl groups with unsaturated double bonds are named by the number of carbon in the acyl group and number of double bonds.
Method Optimization and Evaluation
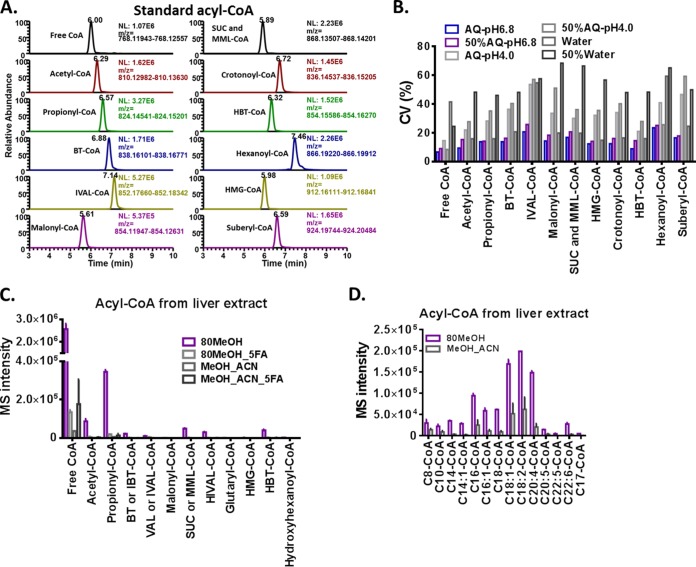
Twelve acyl-CoA thioester standards including free CoA were used to optimize conditions. As shown in Fig. 2A, around 1 ng of each standard was retained on the column and yielded retention times between 5 to 8 min. Most were well separated except for succinyl and methylmalonyl-CoA (SUC and MML-CoA). Another challenge is the stability of each compound, so we tested compound stability in different sample solvents by making standard solutions (final concentration: 1 μm). Then we injected from the same vial three times per day over a course of 2 days. The coefficient of variation (CV) of the integrated MS intensity of these six injections was calculated for each acyl-CoA compound. As shown in Fig. 2B, ammonium acetate buffered solvent at neutral pH stabilized most of the acyl-CoA compounds, whereas the addition of methanol did not affect the stability. Extracts obtained from cells or liver tissue were prepared in duplicate with different extraction conditions. Free CoA and acyl-CoA compounds exhibited highest MS intensities with 80% methanol extraction, while there is very poor or no signal for most of the acyl-CoA compounds when formic acid or acetonitrile is present in the extraction solvent. When 80% methanol is used as the extraction solvent, 26 CoA and acyl-CoA compounds are measured from milligram level of liver tissue, and 24 CoA compounds are detected from a cell extract of 2 × 105 HCT116 cells (Fig. 2C and supplemental Fig. S4). The number of acyl-CoA compounds detected and corresponding measured MS intensities depend on the amount of liver used. Eighteen short to medium chain acyl-CoA and 14 medium to long chain acyl-CoA compounds are detected from an extract of 2 mg or 8 mg of liver, respectively, and this number decreases as the amount of liver decreases (supplemental Fig. S1A). Meanwhile, the corresponding MS intensity decreases linearly when the concentration decreases from 2000 to 0.6 μg or 8000 to 500 μg (supplemental Fig. S1B).
Fig. 2.
Acyl-CoA extraction method optimization. LC-QE-MS condition for acyl-CoA analysis was optimized based on 12 acyl-CoA standards. A, Extracted ion chromatography of CoA standards with m/z window of 6 ppm (± 3ppm). B, Coefficient variation (CV) of every acyl-CoA standard in different solvents at 4 °C over 48 h. CV was calculated based on the MS intensities of six injections over 48 h. C, MS intensities of short chain acyl-CoA thioester detected in liver tissue (0.25 mg) using different extraction solvents. Samples were prepared in duplicate, and the error bars show standard deviation. D, MS intensities of medium to long chain acyl-CoA detected in liver tissue (1.25 mg) using different extraction solvents. Samples were prepared in duplicate, and the error bar shows standard derivation. The abbreviation of solvent names in Fig. 2B, 2C, and 2D are described in the experimental procedures section.
The degradation of acyl-CoA compounds in the liver extract was evaluated (supplemental Fig. S2). A liver extract in ammonium acetate buffer (pH 6.8) was stored at 4 °C and injected to LC-MS six times over 24 h and these endogenous acyl-CoA compounds degraded at different rates in solution. Free CoA, acetyl-CoA, propionyl-CoA, HBT-CoA, C8, C10, and C18:1-CoA degraded by 30% or less after 24 h, whereas hexanoyl-CoA, suberyl-CoA, 3-oxo-2-methylisocaprylonyl-CoA, and C14:1-CoA degraded by more than 90% of their starting values after 24 h. Most of these acyl-CoAs have less than 30% degradation in solution after 9 h at 4 °C, except for SUC/MML-CoA and C14:1-CoA, with a 32 and 44% degradation, respectively. Therefore, our label free method requires samples to be analyzed as soon as possible after reconstitution. Even with potential degradation during sample preparation, this label free method nevertheless results in a linear increase of MS intensity when increasing amounts of E. coli extract (containing 13C-acetyl-CoA) was added to liver tissue in the very beginning extraction step (supplemental Fig. S1C). Addition of acid to the extract although conceivably stabilizing the acyl-CoA compounds by eliminating base-catalyzed hydrolysis, the MS signals decreased more than 10 times when formic acid was added to the extraction solvent (Fig. 2C). Addition of sulfosalicylic acid to the sample also led to complete elimination of the signal. The reason is likely because of the presence of additional ions in the extraction solvent that result in ion suppression.
Acyl-CoA Profiling in Response to Differences in Diet
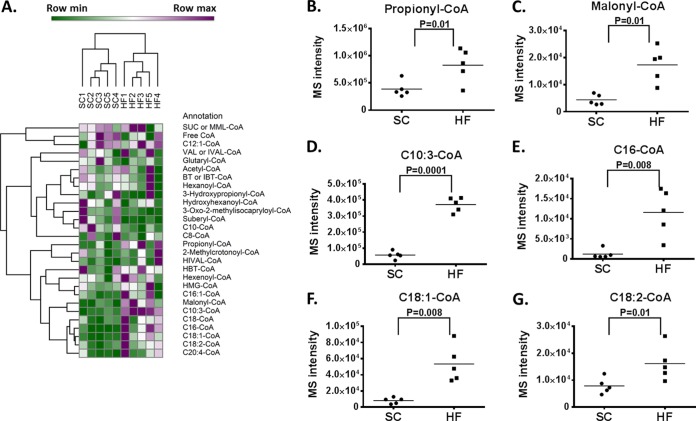
Six week-old mice were fed with either standard chow (SC, 13% fat) or a high fat diet (HF, 63% fat) for 12 weeks, and by that time, the HF mice gained weight and developed insulin resistance as has been described (18). Because lipid metabolism involves acyl-CoA as an intermediate, we reasoned that there could exist remodeling of the acyl-CoA profile under these conditions. We measured the set of acyl-CoA compounds across these mice and carried out unsupervised hierarchical clustering. The 10 liver samples are clustered based on the relative MS intensity of the acyl-CoA compound detected and clearly, there are two major clusters, corresponding to SC and HF fed mice liver samples (Fig. 3A). Within these acyl-CoA compounds, propionyl, malonyl, and C10:3, C16, C18:1, C18:2-CoA significantly (p = 0.01, 0.01, 0.0001, 0.008, 0.008, and 0.01, respectively, student t test, two-tailed) increased in HF fed mice (Fig. 3B to G). Together these findings show remodeling of acyl-CoA metabolism in response to HF feeding.
Fig. 3.
Acyl-CoA profiling in standard chow and high fat diet fed mouse liver. Five standard chow (SC) and five high fat diet (HF) fed mouse liver samples were used for acyl-CoA and other metabolites analysis. A, Hierarchical clustering of 10 samples based on relative MS intensity of acyl-CoA compounds. B to G, Dot plots of four acyl-CoA compounds with large changes in HF samples compared with SC liver samples.
Metabolomics in Response to Feeding
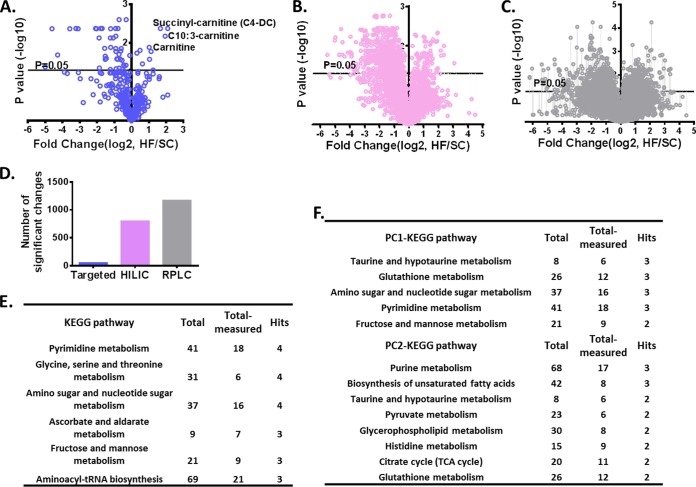
To investigate the global changes that occur in metabolism alongside changes in acyl-CoA metabolism, a polar metabolomics analysis was next carried out as previously described (21). Targeted metabolite analysis was performed on the LC-QE-MS raw data, and a volcano plot was computed based on the fold change (HF/SC) and corresponding p value (FDR adjusted) of relative MS intensities of detected metabolites in 10 mouse liver samples (Fig. 4A). Meanwhile, there were an additional 5391 features (pairs of m/z, putatively assigned chemical composition, and RT) detected together with these targeted metabolites when the HILIC method was coupled to the QE-MS, and 8396 features recorded together with acyl-CoA thioesters when the RPLC method was applied. p value, and fold change of individual feature are plotted in Fig. 4B and C. Fifty-one out of 395 targeted metabolites, 800 out of 5391 untargeted features (HILIC), and 1173 out of 8396 features (RPLC) show significant changes in HF diet samples (Fig. 4D). Pathway analysis of targeted metabolites exhibiting the largest variations (fold change of the integrated MS intensity greater than 1.5 or less than 0.5, HF/SC) is shown in Fig. 4E. Principal Components Analysis (PCA) was conducted with these 83 metabolites. The top 50 values (absolute values of loadings) in PC (principle component) 1 and PC2 used for KEGG pathway analysis, and the results are listed in Fig. 4F. The PC1 corresponds to an overall shift in taurine and hypotaurine metabolism, glutathione metabolism, amino sugar, and nucleotide sugar metabolism, and so on. The PC2 corresponds to changes in purine metabolism, biosynthesis of unsaturated fatty acids pathways among others. Together this analysis reveals coordinated changes in metabolism in a defined set of metabolic pathways that occur in response to differences in diet. These pathways involve several metabolic processes that have been lesser known to be affected by differences in diet.
Fig. 4.
Metabolites profiling in SC and HF diet fed mouse liver. A, Volcano plot of SC and HF fed mouse liver samples based on relative MS intensities of targeted metabolites measured by HILIC method. B, and C, Volcano plot of untargeted features from the HILIC method and reversed phase LC method (RPLC), respectively. For A, B, and C, x axis, intensity ratio (HF/SC, log2); y axis, p value (-log10). p value is adjusted using false discovery rate. D, summary of significant changes (FDR<0.05) generated from targeted HILIC, untargeted HILIC and untargeted RPLC method. E, KEGG pathway analysis of metabolites measured in SC and HF samples. The first column shows KEGG pathways, the second column shows the total number of metabolites, the third column lists the number of metabolites measured in liver samples, and the last column shows the number of metabolites with significant changes. F, Principal components and pathway analysis. The first column shows KEGG pathway names, and second column lists the number of metabolites (hits) within the pathway with top 50 loadings in PC1 or PC2. PCA was done based on 83 metabolites with fold change (HF/SC) either larger than 1.5 or less than 0.5.
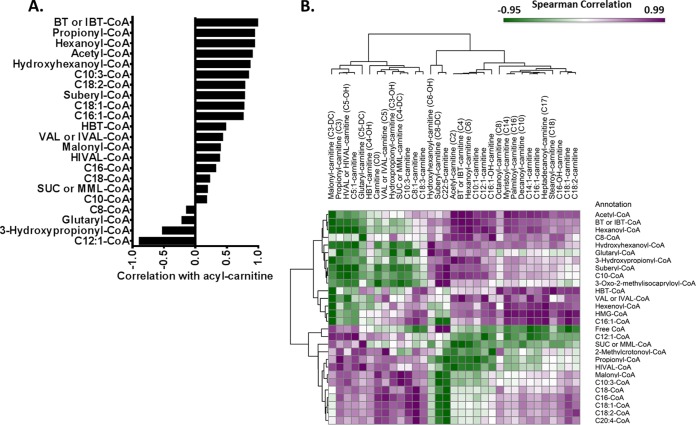
Correlation of acyl-CoA with acyl-carnitine
Given acyl-carnitines are often the carriers of the acyl moiety, acyl-carnitine metabolite levels are thought to correlate with the levels of the corresponding acyl-CoA metabolite. We investigated the generality of this assumption by comparing acyl-carnitine and acyl-CoA levels. Spearman correlation coefficients of each pair of acyl-CoA and acyl-carnitine (22 pairs) is plotted in Fig. 5A. There is a strong positive correlation between these two species, with some exceptions, which might be because of undistinguished isomers or relative low signal in some samples. Hierarchical clustering based on the spearman correlation of 28 acyl-CoA (including free CoA) and 33 acyl-carnitine (including free carnitine) compound is plotted in Fig. 5B. These results reveal an immediate interpretation of CoA and carnitine metabolism. The level of each long chain acyl-carnitine tends to be coordinately regulated with acetyl-CoA levels as well as other CoA compounds involved in lipid metabolism. This finding reveals highly coordinated albeit not general levels of CoA and acyl-CoA metabolism that serve as signatures and markers of several processes involved in lipid metabolism and metabolic status.
Fig. 5.
Correlation of acyl-CoA and acyl-carnitine. A, Spearman correlation of 22 pairs of acyl-CoA and acyl-carnitine sharing the same acyl group in 10 liver samples. B, Hierarchical clustering based on spearman correlations of 28 acyl-CoA and 33 acyl-carnitine compounds. The number of carbons (C) indicates the number of C in the acyl group. DC represents dicarboxylic acid in the acyl group. OH means a hydroxyl group attached to the acyl group. The number after “C:” indicates the number of double bonds.
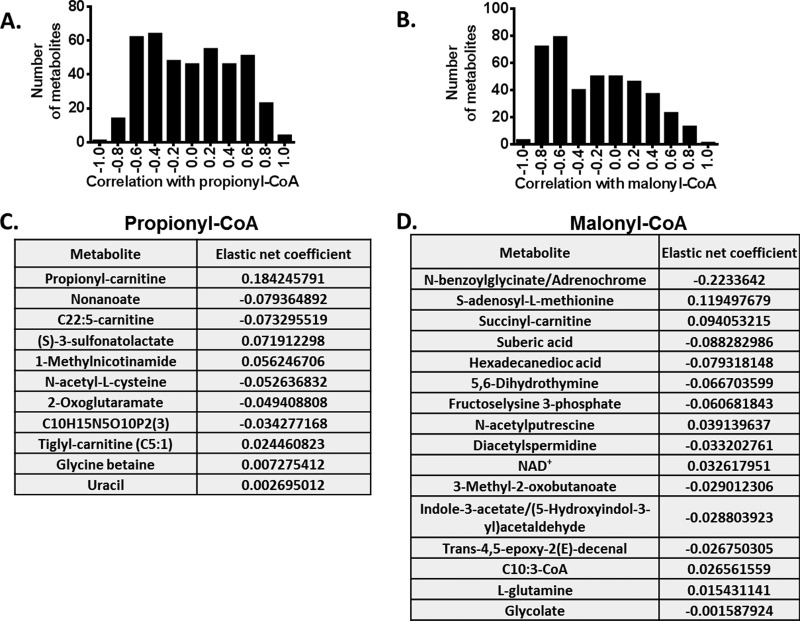
Metabolic Determinants of acyl-CoA Levels
Given the gross alterations of multiple pathways in response to changes in diet, we next investigated the relationships between metabolites and acyl-CoA compounds that vary across diet. We focused on propionyl-CoA and malonyl-CoA because they exhibited large changes in liver in response to diet. A histogram of spearman correlations between metabolites and propionyl-CoA and malonyl-CoA are plotted in Fig. 6A and 6B, respectively. From an analysis of these histograms, it was observed that over 40 metabolites had substantial (ρ > 0.8) correlations with propionyl-CoA and malonyl-CoA respectively. We next carried out a machine learning algorithm that aims to predict the features that are most indicative of propionyl-CoA and malonyl-CoA levels. With this elastic net regression calculation, metabolites with nonzero coefficients are plotted in Fig. 6C and 6D and can be interpreted as having the most direct influences on these acyl-CoA levels. Interestingly, the algorithm revealed that propionyl-carnitine has the strongest interaction with propionyl-CoA, whereas succinyl-carnitine has strongest interaction with malonyl-CoA. This could be caused by excess short chain acyl-CoA compounds being exported out of cells by conversion to their short chain carnitine conjugate. Other metabolites that interact most strongly with these acyl-CoA compounds are involved in several metabolic pathways including some far away in the network indicating several long range interactions in acyl-CoA metabolism. Notably metabolites in sulfur metabolism exhibited influences on both these acyl-CoA compounds providing a possible mechanistic link of the contribution of methionine metabolism to acyl-CoA metabolism through the effects on lipid head groups that are derived from sulfur metabolism, and the long chain fatty acid metabolism involved with acyl-CoA metabolites and the synthesis of acyl-CoA from cysteine.
Fig. 6.
Statistical modeling using elastic net regression. A, Histogram of Spearman correlation coefficient of metabolites with propionyl-CoA. B, Histogram of Spearman correlation coefficient of metabolites with malonyl-CoA. C, Elastic net regression coefficient of metabolites with propionyl-CoA levels. D, Elastic net regression coefficient of metabolites with malonyl-CoA levels.
DISCUSSION
This high-resolution mass spectrometry based approach allowed for the simultaneous measurement of acyl-CoA compounds and a large set of polar metabolites. Meanwhile, it avoids difficult sample preparation steps and the use of stable isotope labeled internal standards. Unfortunately, our current method does not prevent the degradation of acyl-CoA compounds. However, the extract is relatively stable when it is stored at −80 °C as a dry pellet and we suspect that this may be the best strategy because acyl-CoA species show different degradation rates. Because of this issues, stable isotope dilution methods (using one or a few acyl-CoA compounds as internal standards to quantify a broad range of acyl-CoA) bring about substantial errors unless the isotopically labeled acyl-CoA standards for every acyl-CoA are synthesized or a calibration curve based on known concentrations of standards is made.
Two short chain acyl-CoA (malonyl-CoA and propionyl-CoA) and four medium to long chain acyl-CoA (C10:3, C16, C18:1, and C18:2-CoA) showed significant elevation in HF mouse liver, whereas many short to medium chain acyl-CoA compounds maintain constant levels (Fig. 3). Increasing malonyl-CoA levels lead to inhibition of carnitine-palmitoyl-transferase (CPT), and as a result less free long chain fatty acid will enter mitochondria for beta-oxidation. Meanwhile, malonyl-CoA is a precursor for fatty acid biosynthesis (28–30). Therefore, increases in malonyl-CoA levels could switch fatty acid metabolism from catabolic to anabolic reactions to avoid mitochondrial overloading. Propionyl-CoA is mainly generated from BCAAs and odd chain fatty acids, and converted to succinyl-CoA before entering the TCA cycle. Because odd chain fatty acids in diet are rare, the accumulation of propionyl-CoA is derived from BCAA degradation, which agrees with the previous study that enzymes in the BCAA pathway increase in response to HF diet(19). Propionyl-CoA can be further converted to succinyl-CoA to fuel the TCA cycle. However, succinyl-CoA did not increase suggesting additional mechanisms that preserve its concentration. Excess propionyl-CoA can also be converted to propionyl-carnitine, and eventually excreted to serum and urine. Indeed, a strong positive correlation of propionyl-carnitine with propionyl-CoA was observed in our study (Figs. 5 and 6), and an increase of propionyl-carnitine in serum of HF fed mouse has been reported (21). C10:3-CoA is an intermediate involved in unsaturated fatty acid metabolism, such as linoleic acid or conjugated linoleic acid (31), which indicates that unsaturated fatty acid metabolism is also altered even though they only occupy a very small portion of the HF diet. The increase of C16 and C18 acyl-CoA reflects the increase of fatty acid oxidation induced by high fat diet. Acetyl-CoA levels, however, stay constant, which may be explained by its multiple sources, including carbohydrate, amino acids and fatty acids, and exogenous acetate (32–35). Increased fatty acid-derived acetyl-CoA in HF diet could be balanced by decreased acetyl-CoA from carbohydrate.
The analysis of other metabolites, reveal widespread changes in several metabolic pathways. Not surprisingly, in the HF diet, where carbohydrate percentage drops from 67% to 20%, metabolites in amino sugar and nucleotide sugar metabolism all change (Fig. 4E and 4F, supplemental Fig. S3E). Very interestingly, one carbon donors derived from folate or from serine/glycine all show dramatic decreases, whereas choline phosphate, derived from glycerolphospolipids, shows a significant increase in HF samples (supplemental Fig. S3A and S3B). The depletion of the one carbon pool by folate and serine/glycine might explain the decrease of intermediates in pyrimidine and purine metabolism pathways (Fig. 4 and supplemental Fig. S3C and S3D), which are coupled to one carbon metabolism. Moreover, 1-methylnicotinamide also shows significant increases in HF fed mouse liver (supplemental Fig. S3F). It was reported that there is overexpression of nicotinamide N-methyltransferase in liver of obese or diabetic mice (36). HF diet does induce obese/diabetes in mice (23), so increases of 1-methylnicotinamide observed in HF fed mice may be metabolic readout of overexpression of nicotinamide N-methyltransferase.
However, metabolites in central carbon metabolism, including glycolysis, pentose phosphate pathway, and TCA cycle intermediates showed no significant changes in response to dietary changes. The stress induced by limited availability of dietary glucose in HF diet might stimulate multiple gluconeogenic pathways to maintain the levels of glucose and metabolites in central carbon metabolism. In our targeted metabolomics data, glucose stays the same while other sugars decrease dramatically, such as fucose, which agrees with the decreasing carbohydrate uptake in HF diet. A previous study on mouse liver mitochondria shows high fat diet induced overexpression of gluconeogenesis enzymes, such as pyruvate carboxylase (19). Cellular energy state (ATP, adenosine diphosphate, and AMP level) and redox status (NAD+ and NADH level) are also observed in HF diet treated mouse liver through other compensatory pathways, such as fatty acid and amino acid oxidation.
The high correlation between acyl-CoA and acyl-carnitine levels confirms the utility of acyl-carnitines as suitable biomarkers of acyl-CoA levels in general (Fig. 5). However, some exceptions were apparent. Malonyl-CoA, surprisingly has poor correlation with malonyl-carnitine, which might indicate malonyl-CoA might be a poor substrate for malonyl-carnitine transferase, and instead, malonyl-CoA may tend to be used as a precursor of lipid biosynthesis or be converted back to acetyl-CoA through malonyl-CoA decarboxylase.
Because HF diet induced metabolic alterations included both acyl-CoA and nonacyl-CoA metabolites, we studied the interaction between propionyl-CoA/malonyl-CoA and other metabolites detected in both SC and HF samples. A spearman correlation calculation resulted in 15 metabolites strongly correlated (ρ > 0.8) with propionyl-CoA and 75 with malonyl-CoA. An elastic net regression simplified the interaction and produces 13 metabolites (with propionyl-CoA) and 16 metabolites (with malonyl-CoA) with nonzero correlation coefficient. Additional metabolites, such as nonanoate and S-adenosyl-l-methionine were identified in addition to propionyl-carnitine. Even though the interaction between these metabolites and propionyl-CoA and malonyl-CoA is not very clear, nevertheless the analysis gives us some guidance on the metabolic programs that regulate acyl-CoA levels.
Untargeted data analysis based on the full MS scan shows many more features with significant changes in HF samples compared with a targeted analysis, which is because of the limited number of metabolites in the targeted list and also the metabolite database is far from complete. To further investigate these features, additional methods are required, such as MS/MS spectrum, proposal of potential structures, synthesis of standards among other experiments are required.
CONCLUSION
We developed and validated a high throughput yet sensitive LC-QE-MS method with a broad coverage of acyl-CoA, hundreds of polar, a limited portion of nonpolar metabolites, along with thousands of unknown features. HF diet induced metabolic alterations in liver were widespread but affected mostly pathways outside of central carbon metabolism. Some of these metabolite level changes agree with previous enzyme expression level changes, and meanwhile, additional metabolites with large changes were enriched in pathways such as pyrimidine, glycine, serine and threonine metabolism, amino sugar and nucleotide sugar metabolism. A systematic study of acyl-CoA and acyl-carnitine shows that there is strong positive correlation between these two species, even though with some exceptions, which may give us some guidance on using acyl-carnitine to reflect acyl-CoA levels when the latter is below detection limit. Altogether, this study shows both feasibility of relatively straightforward sensitive acyl-CoA profiling along with the discovery of many features of metabolism that correspond to nutritional stress.
Supplementary Material
Acknowledgments
We thank Olga Malysheva, Jian Yan (Cornell University) and Locasale lab members for helpful discussions.
Footnotes
Author contributions: X.L., and H.L. contributed new reagents or analytic tools and J.W.L. designed research; X.L. performed research; X.L., S. Sadhukhan, S. Sun, G.R.W., M.D.H., and L.Q. contributed new reagents or analytic tools; X.L., S. Sadhukhan, and J.W.L. analyzed data; X.L. and J.W.L. wrote the paper; H.L. commented on manuscript.
* This work was supported by the National Institutes of Health awards R00CA168997, R01AI110613 and R01GM098596.
 This article contains supplemental Figs. S1 to S4 and Table S1.
This article contains supplemental Figs. S1 to S4 and Table S1.
1 The abbreviations used are:
- acyl-CoA
- acyl-coenzyme A
- LC-MS
- liquid chromatography mass spectrometry
- HF
- high fat
- BCAA
- branched chain amino acid
- PCA
- principle component analysis
- HILIC
- hydrophilic interaction chromatography
- HCD
- high energy collision dissociation.
REFERENCES
- 1. Starai V. J., Celic I., Cole R. N., Boeke J. D., Escalante-Semerena J. C. (2002) Sir2-dependent activation of acetyl-CoA synthetase by deacetylation of active lysine. Science 298, 2390–2392 [DOI] [PubMed] [Google Scholar]
- 2. Wellen K. E., Hatzivassiliou G., Sachdeva U. M., Bui T. V., Cross J. R., Thompson C. B. (2009) ATP-citrate lyase links cellular metabolism to histone acetylation. Science 324, 1076–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Abu-Elheiga L., Matzuk M. M., Abo-Hashema K. A., Wakil S. J. (2001) Continuous fatty acid oxidation and reduced fat storage in mice lacking acetyl-CoA carboxylase 2. Science 291, 2613–2616 [DOI] [PubMed] [Google Scholar]
- 4. Beckers A., Organe S., Timmermans L., Scheys K., Peeters A., Brusselmans K., Verhoeven G., Swinnen J. V. (2007) Chemical inhibition of acetyl-CoA carboxylase induces growth arrest and cytotoxicity selectively in cancer cells. Cancer Res. 67, 8180–8187 [DOI] [PubMed] [Google Scholar]
- 5. Wagner G. R., Hirschey M. D. (2014) Nonenzymatic protein acylation as a carbon stress regulated by sirtuin deacylases. Mol. Cell 54, 5–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Du J., Zhou Y., Su X., Yu J. J., Khan S., Jiang H., Kim J., Woo J., Kim J. H., Choi B. H., He B., Chen W., Zhang S., Cerione R. A., Auwerx J., Hao Q., Lin H. (2011) Sirt5 is a NAD-dependent protein lysine demalonylase and desuccinylase. Science 334, 806–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tan M., Luo H., Lee S., Jin F., Yang J. S., Montellier E., Buchou T., Cheng Z., Rousseaux S., Rajagopal N., Lu Z., Ye Z., Zhu Q., Wysocka J., Ye Y., Khochbin S., Ren B., Zhao Y. (2011) Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell 146, 1016–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tan M., Peng C., Anderson K. A., Chhoy P., Xie Z., Dai L., Park J., Chen Y., Huang H., Zhang Y., Ro J., Wagner G. R., Green M. F., Madsen A. S., Schmiesing J., Peterson B. S., Xu G., Ilkayeva O. R., Muehlbauer M. J., Braulke T., Muhlhausen C., Backos D. S., Olsen C. A., McGuire P. J., Pletcher S. D., Lombard D. B., Hirschey M. D., Zhao Y. (2014) Lysine glutarylation is a protein posttranslational modification regulated by SIRT5. Cell Metab. 19, 605–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Magnes C., Suppan M., Pieber T. R., Moustafa T., Trauner M., Haemmerle G., Sinner F. M. (2008) Validated comprehensive analytical method for quantification of coenzyme A activated compounds in biological tissues by online solid-phase extraction LC/MS/MS. Anal. Chem. 80, 5736–5742 [DOI] [PubMed] [Google Scholar]
- 10. Lu W., Clasquin M. F., Melamud E., Amador-Noguez D., Caudy A. A., Rabinowitz J. D. (2010) Metabolomic analysis via reversed-phase ion-pairing liquid chromatography coupled to a stand alone orbitrap mass spectrometer. Anal. Chem. 82, 3212–3221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Basu S. S., Mesaros C., Gelhaus S. L., Blair I. A. (2011) Stable isotope labeling by essential nutrients in cell culture for preparation of labeled coenzyme A and its thioesters. Anal. Chem. 83, 1363–1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Magnes C., Sinner F. M., Regittnig W., Pieber T. R. (2005) LC/MS/MS method for quantitative determination of long-chain fatty acyl-CoAs. Anal. Chem. 77, 2889–2894 [DOI] [PubMed] [Google Scholar]
- 13. Zimmermann M., Thormann V., Sauer U., Zamboni N. (2013) Nontargeted profiling of coenzyme A thioesters in biological samples by tandem mass spectrometry. Anal. Chem. 85, 8284–8290 [DOI] [PubMed] [Google Scholar]
- 14. Li Q., Zhang S., Berthiaume J. M., Simons B., Zhang G. F. (2014) Novel approach in LC-MS/MS using MRM to generate a full profile of acyl-CoAs: discovery of acyl-dephospho-CoAs. J. Lipid Res. 55, 592–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Xie Z., Dai J., Dai L., Tan M., Cheng Z., Wu Y., Boeke J. D., Zhao Y. (2012) Lysine succinylation and lysine malonylation in histones. Mol. Cell. Proteomics 11, 100–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhao S., Xu W., Jiang W., Yu W., Lin Y., Zhang T., Yao J., Zhou L., Zeng Y., Li H., Li Y., Shi J., An W., Hancock S. M., He F., Qin L., Chin J., Yang P., Chen X., Lei Q., Xiong Y., Guan K. L. (2010) Regulation of cellular metabolism by protein lysine acetylation. Science 327, 1000–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhang J., Sprung R., Pei J., Tan X., Kim S., Zhu H., Liu C. F., Grishin N. V., Zhao Y. (2009) Lysine acetylation is a highly abundant and evolutionarily conserved modification in Escherichia coli. Mol. Cell. Proteomics 8, 215–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Liu X., Ser Z., Locasale J. W. (2014) Development and quantitative evaluation of a high-resolution metabolomics technology. Anal. Chem. 86, 2175–2184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Guo Y., Darshi M., Ma Y., Perkins G. A., Shen Z., Haushalter K. J., Saito R., Chen A., Lee Y. S., Patel H. H., Briggs S. P., Ellisman M. H., Olefsky J. M., Taylor S. S. (2013) Quantitative proteomic and functional analysis of liver mitochondria from high fat diet (HFD) diabetic mice. Mol. Cell. Proteomics 12, 3744–3758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wendel A. A., Li L. O., Li Y., Cline G. W., Shulman G. I., Coleman R. A. (2010) Glycerol-3-phosphate acyltransferase 1 deficiency in ob/ob mice diminishes hepatic steatosis but does not protect against insulin resistance or obesity. Diabetes 59, 1321–1329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Newgard C. B., An J., Bain J. R., Muehlbauer M. J., Stevens R. D., Lien L. F., Haqq A. M., Shah S. H., Arlotto M., Slentz C. A., Rochon J., Gallup D., Ilkayeva O., Wenner B. R., Yancy W. S., Jr., Eisenson H., Musante G., Surwit R. S., Millington D. S., Butler M. D., Svetkey L. P. (2009) A branched-chain amino acid-related metabolic signature that differentiates obese and lean humans and contributes to insulin resistance. Cell Metab. 9, 311–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Newgard C. B. (2012) Interplay between lipids and branched-chain amino acids in development of insulin resistance. Cell Metab. 15, 606–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sun S., Xia S., Ji Y., Kersten S., Qi L. (2012) The ATP-P2X7 signaling axis is dispensable for obesity-associated inflammasome activation in adipose tissue. Diabetes 61, 1471–1478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lehotay D. C., Hall P., Lepage J., Eichhorst J. C., Etter M. L., Greenberg C. R. (2011) LC-MS/MS progress in newborn screening. Clin. Biochem. 44, 21–31 [DOI] [PubMed] [Google Scholar]
- 25. Bennett B. D., Yuan J., Kimball E. H., Rabinowitz J. D. (2008) Absolute quantitation of intracellular metabolite concentrations by an isotope ratio-based approach. Nat. Protoc. 3, 1299–1311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yanes O., Tautenhahn R., Patti G. J., Siuzdak G. (2011) Expanding coverage of the metabolome for global metabolite profiling. Anal. Chem. 83, 2152–2161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhang G. F., Kombu R. S., Kasumov T., Han Y., Sadhukhan S., Zhang J., Sayre L. M., Ray D., Gibson K. M., Anderson V. A., Tochtrop G. P., Brunengraber H. (2009) Catabolism of 4-hydroxyacids and 4-hydroxynonenal via 4-hydroxy-4-phosphoacyl-CoAs. J. Biol. Chem. 284, 33521–33534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ruderman N. B., Saha A. K., Vavvas D., Witters L. A. (1999) Malonyl-CoA, fuel sensing, and insulin resistance. Am. J. Physiol. 276, E1–E18 [DOI] [PubMed] [Google Scholar]
- 29. Iglesias M. A., Ye J. M., Frangioudakis G., Saha A. K., Tomas E., Ruderman N. B., Cooney G. J., Kraegen E. W. (2002) AICAR administration causes an apparent enhancement of muscle and liver insulin action in insulin-resistant high-fat-fed rats. Diabetes 51, 2886–2894 [DOI] [PubMed] [Google Scholar]
- 30. McGarry J. D., Mannaerts G. P., Foster D. W. (1977) A possible role for malonyl-CoA in the regulation of hepatic fatty acid oxidation and ketogenesis. J. Clin. Invest. 60, 265–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Liang X., Zhu D., Schulz H. (1999) Delta3,5,7,Delta2,4,6-trienoyl-CoA isomerase, a novel enzyme that functions in the beta-oxidation of polyunsaturated fatty acids with conjugated double bonds. J. Biol. Chem. 274, 13830–13835 [DOI] [PubMed] [Google Scholar]
- 32. Hardwick D. C. (1966) The fate of acetyl groups derived from glucose in the isolated perfused goat udder. Biochem. J. 99, 228–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Brown T. D., Jones-Mortimer M. C., Kornberg H. L. (1977) The enzymic interconversion of acetate and acetyl-coenzyme A in Escherichia coli. J. Gen. Microbiol. 102, 327–336 [DOI] [PubMed] [Google Scholar]
- 34. Kowit J. D., Goldberg A. L. (1977) Intermediate steps in the degradation of a specific abnormal protein in Escherichia coli. J. Biol. Chem. 252, 8350–8357 [PubMed] [Google Scholar]
- 35. Lazarow P. B. (1978) Rat liver peroxisomes catalyze the beta oxidation of fatty acids. J. Biol. Chem. 253, 1522–1528 [PubMed] [Google Scholar]
- 36. Kraus D., Yang Q., Kong D., Banks A. S., Zhang L., Rodgers J. T., Pirinen E., Pulinilkunnil T. C., Gong F., Wang Y. C., Cen Y., Sauve A. A., Asara J. M., Peroni O. D., Monia B. P., Bhanot S., Alhonen L., Puigserver P., Kahn B. B. (2014) Nicotinamide N-methyltransferase knockdown protects against diet-induced obesity. Nature 508, 258–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.