Abstract
Branched five carbon (C5) alcohols are attractive targets for microbial production due to their desirable fuel properties and importance as platform chemicals. In this study, we engineered a heterologous isoprenoid pathway in E. coli for the high-yield production of 3-methyl-3-buten-1-ol, 3-methyl-2-buten-1-ol, and 3-methyl-1-butanol, three C5 alcohols that serve as potential biofuels. We first constructed a pathway for 3-methyl-3-buten-1-ol, where metabolite profiling identified NudB, a promiscuous phosphatase, as a likely pathway bottleneck. We achieved a 60% increase in the yield of 3-methyl-3-buten-1-ol by engineering the Shine-Dalgarno sequence of nudB, which increased protein levels by 9-fold and reduced isopentenyl diphosphate (IPP) accumulation by 4-fold. To further optimize the pathway, we adjusted mevalonate kinase (MK) expression and investigated MK enzymes from alternative microbes such as Methanosarcina mazei. Next, we expressed a fusion protein of IPP isomerase and the phosphatase (Idi1~NudB) along with a reductase (NemA) to diversify production to 3-methyl-2-buten-1-ol and 3-methyl-1-butanol. Finally, we used an oleyl alcohol overlay to improve alcohol recovery, achieving final titers of 2.23 g/L of 3-methyl-3-buten-1-ol (~70% of pathway-dependent theoretical yield), 150 mg/L of 3-methyl-2-buten-1-ol, and 300 mg/L of 3-methyl-1-butanol.
There has been considerable interest in the biosynthetic production of C2 – C5 alcohols as commodity chemicals and potential biofuels1,2. Butanol, for instance, serves as a platform chemical for thousands of compounds and can be used as a biogasoline or fuel additive3,4. Some of the most promising work in the microbial synthesis of these compounds has used refactored amino acid biosynthesis pathways to produce a variety of C3 – C5 alcohols at high titers5. Using this method, valuable alcohols such as isobutanol (C4) have been produced at impressively high yields and titers6.
Isoprenoid biosynthesis provides an additional route to energy-dense C5 alcohols, namely isopentenol (3-methyl-3- and 3-methyl-2-buten-1-ol, also known as isoprenol and prenol, respectively) and isopentanol (3-methyl-1-butanol)7. These alcohols have octane numbers and combustion properties that make them potential gasoline replacements8. Isopentanol, for example, has been evaluated for use in homogenous charge compression ignition (HCCI) engines and significantly outperforms ethanol9. In addition, these alcohols were shown to function as ideal anti-knock additives in spark ignition engines10. Although isoprenoids can be produced from both the methylerythritol phosphate (MEP) and mevalonate (MVA) pathways, initial engineering efforts towards isopentenol production have focused primarily on the MVA pathway11,12,13,14. Thus far, alcohol yields from these efforts compare favorably with analogous work using the MEP pathway15.
C5 alcohols can be produced from the MVA pathway following the dephosphorylation of isopentenyl diphosphate (IPP) and dimethylallyl diphosphate (DMAPP), the universal precursors of all isoprenoid compounds. A specific protein capable of catalyzing this dephosphorylation, NudF from B. subtilis, was first identified using a screening method based on prenyl diphosphate toxicity11. In a later study, an E. coli native enzyme (NudB) was also shown to effectively catalyze the dephosphorylation of IPP and DMAPP12. When paired with the IPP-overproducing mevalonate pathway, the expression of NudB produced 3-methyl-3-buten-1-ol at 8.3% theoretical yield. Using a fusion protein of IPP isomerase (Idi1) and NudB, IPP-derived 3-methyl-3-buten-1-ol and DMAPP-derived 3-methyl-2-buten-1-ol were produced concurrently. With the expression of an E. coli-native reductase (NemA), 3-methyl-2-buten-1-ol was successfully reduced to 3-methyl-1-butanol, albeit at low efficiency12.
Although optimization of the heterologous MVA pathway has improved the production of 3-methyl-3- and 3-methyl-2-buten-1-ol, yields are still considerably lower than those required for large-scale, economical production13,14. Furthermore, optimization towards the production of fully reduced 3-methyl-1-butanol has not been attempted. In the current work, we engineered the heterologous MVA pathway in E. coli to achieve the highest yields yet reported for 3-methyl-3-buten-1-ol. In addition, we improved the production of 3-methyl-1-butanol by more than 10-fold over previous results.
Results and Discussion
Pathway organization and engineering strategy
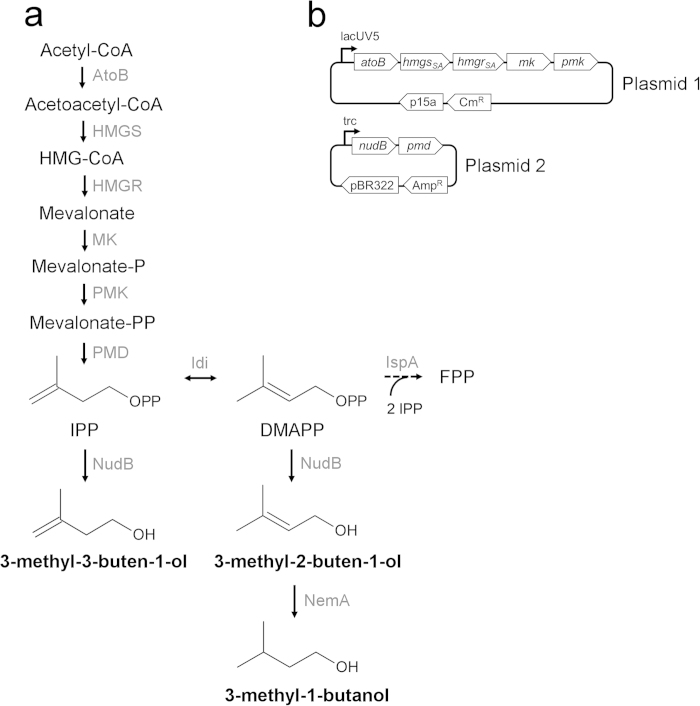
The heterologous MVA pathway in E. coli (Fig. 1a) allows for the biosynthesis of IPP and DMAPP, the universal precursors to all isoprenoid compounds. To produce 3-methyl-3-buten-1-ol from IPP, nudB, a gene encoding an E. coli native phosphatase, is overexpressed in addition to the mevalonate pathway. If an isomerase such as IDI1 from yeast is expressed, DMAPP and its corresponding alcohol 3-methyl-2-buten-1-ol will be formed. Expression of a reductase such as nemA will convert 3-methyl-2-buten-1-ol, but not 3-methyl-3-buten-1-ol, into fully reduced 3-methyl-1-butanol12 (Fig. 1a).
Figure 1. A heterologous MVA pathway for C5 alcohol production.
(a) Pathway overview. The heterologous mevalonate pathway in E. coli consists of 7 reactions to convert acetyl-CoA into IPP and DMAPP. Dephosphorylation of these compounds by NudB, a promiscuous E. coli phosphatase, produces 3-methyl-3-buten-1-ol and 3-methyl-2-buten-1-ol, respectively. NemA, an endogenous reductase, is capable of reducing 3-methyl-2-buten-1-ol, but not 3-methyl-3-buten-1-ol, into fully reduced 3-methyl-1-butanol. (b) Plasmid architecture. A two plasmid system for 3-methyl-3-buten-1-ol production served as the initial engineering platform (strain KG1). Plasmid 1 contained genes from atoB to PMK with a medium copy p15A ori and weak lacUV5 promoter16. Plasmid 2 contained nudB and PMD with a high copy pBR322 ori and strong trc promoter. To produce mixed alcohols, a fusion protein and reductase were expressed on plasmid two (see Fig. 4).
We initially assembled a two-plasmid system for 3-methyl-3-buten-1-ol production: plasmid 1 (pJBEI-6829), a medium copy vector with a lacUV5 promoter (pBbA5c16), contained mevalonate pathway genes from thiolase (atoB) to phosphomevalonate kinase (PMK) while plasmid 2 (pJBEI-6833), a high copy vector with a trc promoter (pTrc99A17), contained nudB and diphosphomevalonate decarboxylase (PMD) (Fig. 1b) (Table 1). In this pathway, atoB and nudB were derived from E. coli, mvaS and mvaA, which encode 3-hydroxy-3-methylglutaryl-CoA (HMG-CoA) synthase and reductase, respectively, were from Staphylococcus aureus18, and MK, PMK, and PMD were derived from yeast. These plasmids were transformed into E. coli DH1 to create strain KG1, which was used as an initial platform for subsequent engineering14.
Table 1. Plasmids and strains used in this study.
| Plasmids | Description | Reference |
|---|---|---|
| pJBEI-6829 | pBbA5c-MevTsa-MK-PMK | 14 |
| pJBEI-6833 | pTrc99A-nudBRBSo-PMD | 14 |
| JPUB-004498 | pTrc99A-nudBRBS1-PMD | This study |
| JPUB-004499 | pTrc99A-nudBRBS2-PMD | This study |
| JPUB-004500 | pTrc99A-nudBRBS3-PMD | This study |
| JPUB-004501 | pTrc99A-nudBRBS4-PMD | This study |
| JPUB-004502 | pTrc99A-nudBRBS5-PMD | This study |
| JPUB-004503 | pTrc99A-nudBRBS6-PMD | This study |
| JPUB-004504 | pTrc99A-nudBRBS7-PMD | This study |
| JPUB-004505 | pTrc99A-nudBRBS8-PMD | This study |
| JPUB-004506 | pTrc99A-nudBRBS9-PMD | This study |
| JPUB-004507 | pTrc99A-nudBRBS10-PMD | This study |
| pJBEI-6830 | pBbA5c-MevTsa-PMK-MK | 14 |
| pJBEI-6832 | pBbA5c-MevTsa-T1002-ptrc-PMK-MK | 14 |
| pJBEI-6823 | pBbA5c-MevTco-PMK-MK | 14 |
| JPUB-004508 | pBbA5c-MevTco-PMK-MKsa | This study |
| JPUB-004509 | pBbA5c-MevTco-PMK-MKmm | This study |
| JPUB-004510 | pBbA5c-MevTsa-PMK-MKmm | This study |
| JPUB-004511 | pTrc99A-idi1~nudB-nemA-PMD | This study |
| JPUB-004512 | pTrc99A-idi1rbs10~nudB-nemA-PMD | This study |
| pJBEI-6835 | pBbB8k-NudB | 14 |
| Strains | Description | Reference |
| KG1 | pJBEI-6829 + pJBEI-6833; base strain | 14 |
| KG1R1 | pJBEI-6829 + JPUB-004498; nudBRBS1 | This study |
| KG1R2 | pJBEI-6829 + JPUB-004499; nudBRBS2 | This study |
| KG1R3 | pJBEI-6829 + JPUB-004500; nudBRBS3 | This study |
| KG1R4 | pJBEI-6829 + JPUB-004501; nudBRBS4 | This study |
| KG1R5 | pJBEI-6829 + JPUB-004502; nudBRBS5 | This study |
| KG1R6 | pJBEI-6829 + JPUB-004503; nudBRBS6 | This study |
| KG1R7 | pJBEI-6829 + JPUB-004504; nudBRBS7 | This study |
| KG1R8 | pJBEI-6829 + JPUB-004505; nudBRBS8 | This study |
| KG1R9 | pJBEI-6829 + JPUB-004506; nudBRBS9 | This study |
| KG1R10 | pJBEI-6829 + JPUB-004507; nudBRBS10 | This study |
| KG2 | pJBEI-6832 + pJBEI-6833; increased MK expression | This study |
| KG2R3 | pJBEI-6832 + JPUB-004500; nudBRBS3 | This study |
| KG2R10 | pJBEI-6832 + JPUB-004507; nudBRBS10 | This study |
| KG3 | pJBEI-6830 + pJBEI-6833; MK control | This study |
| KG3R3 | pJBEI-6830 + JPUB-004500; nudBRBS3 | This study |
| KG3R10 | pJBEI-6830 + JPUB-004507; nudBRBS10 | This study |
| KG4 | pJBEI-6823 + pJBEI-6833; yeast MK | This study |
| KG4R3 | pJBEI-6823 + JPUB-004500; nudBRBS3 | This study |
| KG4R10 | pJBEI-6823 + JPUB-004507; nudBRBS10 | This study |
| KG5 | JPUB-004509 + pJBEI-6833; M. mazei MK | This study |
| KG5R3 | JPUB-004509 + JPUB-004500; nudBRBS3 | This study |
| KG5R10 | JPUB-004509 + JPUB-004507; nudBRBS10 | This study |
| KG6 | JPUB-004508 + pJBEI-6833-; S. aureusMK | This study |
| KG6R3 | JPUB-004508 + JPUB-004500; nudBRBS3 | This study |
| KG6R10 | JPUB-004508 + JPUB-004507; nudBRBS10 | This study |
| KG7 | JPUB-004510 + pJBEI-6833, M. mazeiMK | This study |
| KG7R3 | JPUB-004510 + JPUB-004500; nudBRBS3 | This study |
| KG7R10 | JPUB-004510 + JPUB-004507; nudBRBS10 | This study |
| KG8 | pJBEI-6829 + JPUB-004511; mixed alcohol production | This study |
| KG8R10 | pJBEI-6829 + JPUB-004512; Idi1RBS10 | This study |
| KG9 | pJBEI-6829 + JPUB-004511 + pJBEI-6835 | This study |
Our engineering strategy was to first optimize 3-methyl-3-buten-1-ol production before moving towards the production of mixed C5 alcohols. Rather than focusing on high-throughput strain generation or combinatorial pathway assembly, we applied robust metabolomics and proteomics methods to identify likely bottlenecks and rationally direct pathway engineering. Once we optimized the precursor pathway and improved 3-methyl-3-buten-1-ol production, we implemented additional engineering to produce 3-methyl-2-buten-1-ol and ultimately fully reduced 3-methyl-1-butanol.
Metabolite profiling of KG1 identifies IPP accumulation as a likely bottleneck
Strain KG1 produced ~1.2 g/L of 3-methyl-3-buten-1-ol on 1% glucose after 48 hours, equivalent to 36% of pathway-dependent theoretical yield14. While an improvement over the original strain19, this titer was well below levels necessary for economical, large-scale production. We hypothesized that one or more pathway bottlenecks were limiting product yields in strain KG1. To accurately identify these bottlenecks, we performed a comprehensive analysis of pathway metabolites and proteins during the fermentation time-course.
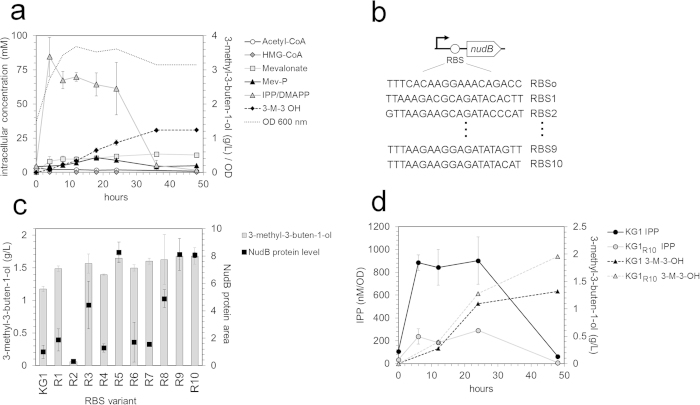
Quantification of MVA pathway intermediates in strain KG1 over a 48 hour time-course (Fig. 2a) revealed that IPP concentrations exceeded those of any other observed MVA pathway intermediate by more than 8-fold. The comparatively low concentrations of acetyl-CoA, HMG-CoA, mevalonate, and mevalonate phosphate suggested that flux through the upstream pathway was unlikely to be limiting. The accumulation of IPP alone implied that NudB, the protein responsible for the conversion of IPP into 3-methyl-3-buten-1-ol, was the primary bottleneck in strain KG1. IPP has previously been shown to be toxic, resulting in growth inhibition and a reduction in glucose uptake at high concentrations. Although no obvious growth defects were apparent in strain KG1, more subtle effects such as feedback inhibition20 could be deleterious to host and the pathway function. Even without exerting toxicity, the accumulation of a metabolite upstream of 3-methyl-3-buten-1-ol indicated suboptimal pathway performance. Consequently, reducing IPP accumulation was a priority in further engineering.
Figure 2. RBS engineering to improve 3-methyl-3-buten-1-ol titer and reduce IPP accumulation.
(a) Metabolite profiling of strain KG1. Mevalonate pathway intermediates were quantified in strain KG1 over a 48 hour time-course by LC-MS, showing IPP as the highest accumulating intermediate by a factor of 8. A growth curve for strain KG1 is shown as a dashed line. Error bars represent standard deviation (n = 3). 3-M-3-OH = 3-methyl-3-buten-1-ol. (b) RBS optimization of nudB. Ten RBS variants of nudB (see Table S1) were generated using RBS calculator and cloned into plasmid 2, forming JPUB-004498 to JPUB-004507. (c) 3-Methyl-3-buten-1-ol titer and NudB protein level in KG1R1 – KG1R10. Strains containing nudBRBS1 through nudBRBS10 (KG1R1 - KG1R10) were assayed for 3-methyl-3-buten-1-ol and NudB protein level. Bars represent 3-methyl-3-buten-1-ol titer after 36 hours. Squares show NudB protein area at 24 hours relative to that measured in strain KG1 (normalized to a value of 1). Error bars represent standard deviation (n = 3). (d) IPP accumulation and 3-methyl-3-buten-1-ol production in strains KG1 and KG1R10. IPP concentrations in KG1R10 were reduced by 4-fold relative to KG1, while 3-methyl-3-buten-1-ol titer increased by 60%. Error bars represent standard deviation (n = 3).
Quantification of pathway proteins in strain KG1 (Supplemental Figure S1) was conducted with targeted proteomics based on selected reaction monitoring (SRM)21,22 to assess protein stability and relative concentrations over a 48-hour time-course. In general, pathway proteins appeared stable throughout the fermentation other than the expected increase following pathway induction (0 to 6 h). Although this SRM method reports relative peak areas for each protein rather than absolute concentrations, low signal intensity often indicates poor protein expression and potential pathway bottlenecks14,22,23. Peak area was the lowest for HMG-CoA reductase (HMGR), implying weak expression of this enzyme, but the low steady-state levels of HMG-CoA (the substrate of HMGR) and rapid accumulation of IPP (a downstream product after the HMGR-catalyzed reaction) suggested that HMGR may not be limiting under current conditions. Protein areas of PMD and NudB, both of which were expressed from a high-copy plasmid, were an order of magnitude above the other pathway proteins. Although NudB is an E. coli native protein, the plasmid-borne copy was the primary source of quantifiable protein; background levels of NudB in DH1 wild type were more than 10-fold less than strain KG1 (data not shown).
Improved NudB protein expression reduces IPP accumulation and yields high 3-methyl-3-buten-1-ol titers
To prevent the accumulation of IPP, we could either slow down its formation through upstream pathway engineering or enhance its conversion to 3-methyl-3-buten-1-ol by improving NudB-catalyzed reaction efficiency. Since we did not want to decrease upstream pathway efficiency, we focused on improving flux to 3-methyl-3-buten-1-ol by increasing NudB expression. To increase NudB protein levels, we optimized the Shine-Dalgarno sequence of nudB using the RBS calculator24. Starting with the RBS sequence in strain KG1 (NudBRBSo) as an input, 10 sequences were generated that were predicted to improve NudB protein expression (Fig. 2b). We designed the 10 RBS sequences (RBS1 – RBS10) to achieve a range of expression to allow for the titration of NudB levels in subsequent engineering (Supplemental Table 1). After cloning these variants into plasmid 2, we co-transformed each of them into E. coli DH1 with pJBEI-6829 to generate strains KG1R1 (strain KG1 containing NudBRBS1) through KG1R10 (strain KG1 containing NudBRBS10) (Table 1). We performed a production test with these strains and collected protein samples to quantify levels of NudB (Fig. 2c).
Protein levels of NudB were improved in all RBS sequence variants with the notable exception of NudBRBS2. The best-performing RBS sequences, RBS5, RBS9, and RBS10, yielded NudB protein levels 9-fold higher than the original sequence (Fig. 2c). As anticipated, strains with improved NudB expression also produced more 3-methyl-3-buten-1-ol (Fig. 2c). Surprisingly, the variability in 3-methyl-3-buten-1-ol titer was minor compared to the large changes in NudB protein level; even the slight increase in NudB protein in strain KG1R7 yielded a 3-methyl-3-buten-1-ol titer comparable to strain KG1R10, which produced 8-fold more NudB. This observation suggested that NudB expression was no longer limiting in the highest-performing strains, which produced ~1.6 g/L of isopentenol at 36 hours. Indeed, expression of additional NudB on a supplemental plasmid yielded no additional improvement in titer (Supplemental Figure S2). Still, we chose KG1R10 for further study, reasoning that this strain provided room for additional improvement in the precursor pathway and increased flux to IPP.
Metabolite analysis of KG1R10 confirmed that improved NudB expression relieved IPP accumulation. Over a 48-hour time-course, IPP levels in strain KG1R10 were reduced by 4-fold relative to KG1 (Fig. 2d). At 48 hours, the 3-methyl-3-buten-1-ol titer in strain KG1R10 reached 1.94 g/L, a 60% increase over strain KG1. Given the significant reduction in IPP level observed in strain KG1R10, we suspected that the pathway bottleneck might have shifted from NudB reaction to the upstream precursor pathway. If this were the case, further increases in IPP production could yield additional increases in 3-methyl-3-buten-1-ol titer.
Optimization of MK yields minor improvements in 3-methyl-3-buten-1-ol titer
Although the 3-methyl-3-buten-1-ol pathway consists of seven genes, MK is the primary determinant of flux to IPP14. Sufficient flux to IPP is required for high product yields, but excessive MK expression can lead to IPP accumulation and a reduction in growth and 3-methyl-3-buten-1-ol titer. Since we increased the “pull” on IPP using NudBRBS10 (Fig. 2d), we decided to increase the “push” to IPP through additional MK engineering. We employed two approaches: 1) pairing improved NudB expression with increased expression of MK, and 2) introducing MK homologs with desirable properties from other organisms. In each case, the MK gene was positioned downstream of PMK as this gene order previously increased MK protein levels14.
MK expression was enhanced by inserting a trc promoter upstream of MK (pJBEI-6832), which was shown to increase MK protein levels by ~4-fold14. We paired this plasmid with the previously characterized plasmids containing NudBRBSo (creating strain KG2), NudBRBS3 (strain KG2R3), and NudBRBS10 (strain KG2R10) to titrate low, medium, and high levels of NudB protein, respectively (Fig. 3). We compared strain KG2 with the control strain KG3, which contained an identical version of the pathway with the same gene order but no supplemental promoter (Table 1). 3-Methyl-3-buten-1-ol titer in KG2 was 268 mg/L, a 4-fold reduction compared to KG3. If this low titer was due to IPP accumulation, we anticipated that elevated NudB levels would dramatically increase 3-methyl-3-buten-1-ol titer. In accordance with our expectation, strain KG2R3, which expressed a higher level of NudB, produced 4-fold more 3-methyl-3-buten-1-ol than strain KG2. Even with this improvement, however, titers were about 20% less than the comparable control (strain KG3R3). With NudBRBS10 (i.e. strain KG2R10), we recorded an additional increase in titer to 1550 mg/L of 3-methyl-3-buten-1-ol, matching the performance of the control KG3R10 (Fig. 3). Attempts to further increase NudB levels with an additional plasmid (pBbB8k-NudB, pJBEI-6835) were unsuccessful (Supplemental Figure S2), suggesting that IPP accumulation was no longer problematic in strain KG2R10. Although elevated MK expression appeared to increase flux to IPP, the strong performance of the control strain suggested it was not necessary for peak pathway efficiency. Consequently, we focused on changing MK identity rather than expression level for further optimization.
Figure 3. Altering MK expression and identity to increase flux to IPP.
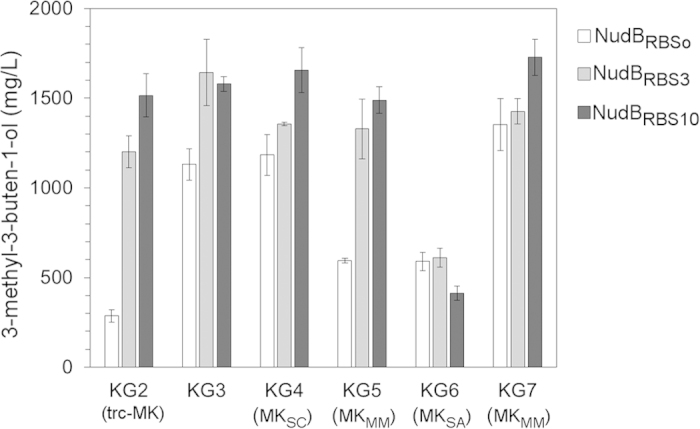
To increase pathway flux to IPP, MK expression and identity was altered and evaluated with different concentrations of NudB (see table 1 for a list of strains). 3-Methyl-3-buten-1-ol titers are shown at 36 hours post-induction. Error bars show standard deviation (n = 3).
Although most work on MVA pathway optimization has focused on S. cerevisiae-derived MK (MKSC), MK enzymes from alternative hosts such as Staphylococcus aureus (MKSA) and the archaea Methanosarcina mazei (MKMM) have promising characteristics. MKMM, for example, was shown to resist feedback inhibition from IPP, GPP, and FPP20. We tested these MK variations with a MVA pathway “top” portion that provided high flux to mevalonate (MevTco)14, reasoning that this context could better reveal differences in MK activity. Strains KG4, KG5, and KG6 contained this high flux pathway paired with MK enzymes derived from S. cerevisiae, M. mazei, and S. aureus, respectively (Table 1). As practiced above, we titrated each pathway with low (NudBRBSo), medium (NudBRBS3), and high (NudBRBS10) levels of NudB.
With low levels of NudB (i.e. NudBRBSo), MKSC outperformed MKMM and MKSA: strain KG4 produced 2-fold more 3-methyl-3-buten-1-ol at 36 hours than KG5 and KG6 (Fig. 3). Surprisingly, growth inhibition consistent with IPP accumulation was clearly observed in strain KG5 (Supplemental Figure S3). This observation implied that the flux to IPP was actually increased when using MKMM. Consistent with this expectation, increased NudB expression by RBS engineering relieved the growth inhibition in strain KG5 (Supplemental Figure S3) and doubled 3-methyl-3-buten-1-ol production (Fig. 3). Despite this increase, maximum titers matched those of strain KG4, which produced 1650 mg/L of 3-methyl-3-buten-1-ol when paired with NudBRBS10. Surprisingly, 3-methyl-3-buten-1-ol production in strain KG6 (containing MKSA) did not increase with additional NudB protein. This observation suggested that S. aureus MK did not provide adequate pathway flux to IPP.
Since the preliminary results for MKMM were promising, we investigated this homolog further by pairing the enzyme with a different MVA pathway “top” portion, MevTSA, to create strain KG7 (Table 1). In this pathway context, a maximum 3-methyl-3-buten-1-ol titer of 1730 mg/L was recorded with NudBRBS10 (Fig. 3). Although MKMM did appear to improve flux to IPP relative to MKSC, similar maximum titers were reached with each enzyme. This apparent “plateau” in 3-methyl-3-buten-1-ol titer may suggest that further gains from MVA pathway engineering will be difficult to achieve. Although NudB protein expression is no longer a bottleneck, the poor kinetics of the enzyme12 may play a key role in limiting 3-methyl-3-buten-1-ol production. It is possible that an improvement in NudB kinetics is required to facilitate additional titer increases in the current pathway context.
Expression of Idi1~NudB fusion protein and NemA results in the production of 3-methyl-3-buten-1-ol, 3-methyl-2-buten-1-ol, and 3-methyl-1-butanol
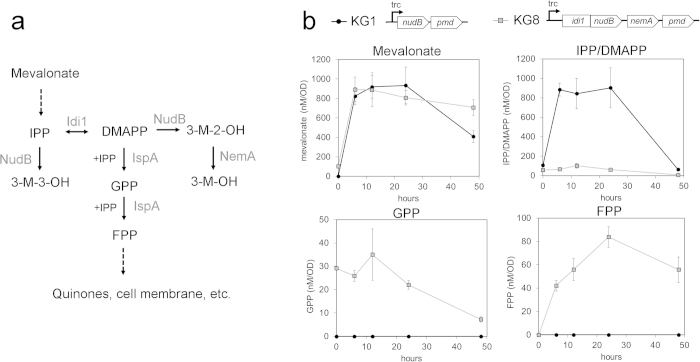
Since we successfully optimized a strain for 3-methyl-3-buten-1-ol production, we next sought to improve the production of 3-methyl-2-buten-1-ol and subsequently 3-methyl-1-butanol. To produce these alcohols, we over-expressed the yeast-derived IPP isomerase Idi1 and the E. coli-derived promiscuous reductase NemA to create strain KG8 (Table 1). Idi1 and NudB were expressed together as a fusion protein as described previously12 (Fig. 4a).
Figure 4. Engineering to produce mixed alcohols.
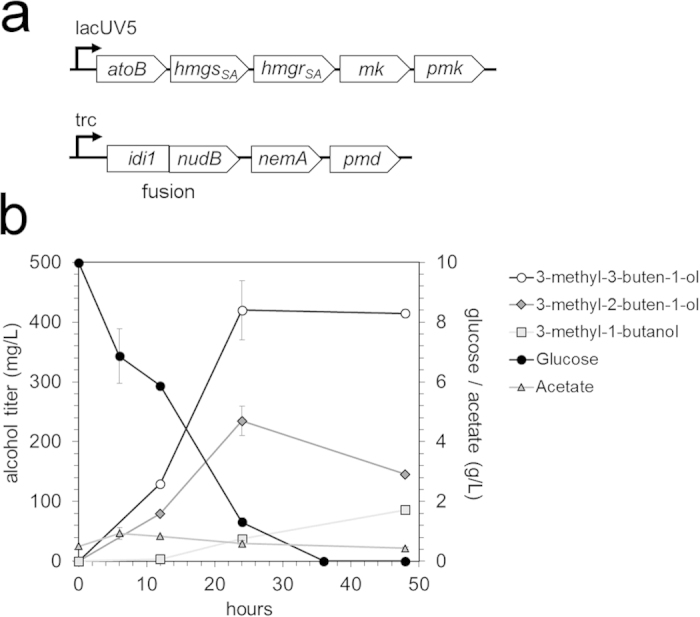
(a) Plasmid architecture. To produce 3-methyl-2-buten-1-ol and 3-methyl-1-butanol in addition to 3-methyl-3-buten-1-ol, plasmid 1 from strain KG1 (CmR, p15A ori) was paired with a mixed alcohol production plasmid (AmpR, pBR322 ori) encoding an Idi1~NudB fusion protein and a reductase (NemA)12 to create strain KG8. (b) Mixed alcohol production in strain KG8. A fermentation time-course revealed that strain KG8 produced three C5 alcohols. By 48 hours, strain KG8 produced over 400 mg/L of 3-methyl-1-buten-1-ol, 150 mg/L of 3-methyl-2-buten-1-ol, and 100 mg/L of 3-methyl-1-butanol. All glucose was consumed by 36 hours, and acetate secretion was minimal. Error bars represent standard deviation (n = 3).
Strain KG8 produced 3-methyl-3-buten-1-ol, 3-methyl-2-buten-1-ol, and fully reduced 3-methyl-1-butanol as anticipated (Fig. 4b). At 48 hours, total C5 alcohol content (i.e. the summed total 3-methyl-3-buten-1-ol, 3-methyl-2-buten-1-ol, and 3-methyl-1-butanol) reached 700 mg/L, a 4-fold improvement in titer over the previously reported result12. From 24 to 48 hours, the increase in 3-methyl-1-butanol production appeared to correlate with the reduction of 3-methyl-2-buten-1-ol as expected given its route of formation (Fig. 1a).
Compared to strain KG1, total C5 alcohol content in KG8 decreased by over 40%. We hypothesized that competition with endogenous pathways was responsible for this reduction in overall titer. We suspected that expression of Idi1—required for DMAPP and 3-methyl-2-buten-1-ol production—would divert carbon towards the formation of geranyl diphosphate (GPP) and farnesyl diphosphate (FPP), both of which are used in a variety of cellular processes (Fig. 5a). Consistent with this hypothesis, GPP and FPP were observed in strain KG8, but below detection in the 3-methyl-3-buten-1-ol production strain KG1 (Fig. 5b). A 10-fold decrease in the concentration of IPP/DMAPP was also observed in KG8, likely reflecting the multiple routes of IPP/DMAPP depletion in this strain. The detection of GPP and FPP in strain KG8 makes E. coli FPP synthase (ispA) an attractive engineering target; it is likely that reducing or eliminating IspA activity will yield increased production of 3-methyl-2-buten-1-ol and subsequently 3-methyl-1-butanol. Though levels of IPP, GPP, and FPP varied significantly between KG8 and KG1, mevalonate concentrations were similar in both strains. This suggested that carbon flow through the upstream mevalonate pathway was unaffected by the implementation of downstream engineering.
Figure 5. Metabolite profiling of strain KG8 reveals competition with endogenous pathways.
(a) Competition with endogenous pathways. Expression of yeast Idi1 increases flux to DMAPP and subsequently the longer chain terpenes GPP and FPP. Competition between 3-methyl-2-buten-1-ol and these terpenes likely limits alcohol production. 3-M-3-OH = 3-methyl-3-buten-1-ol. 3-M-2-OH = 3-methyl-2-buten-1-ol. 3-M-OH = 3-methyl-1-butanol. (b)Detection of GPP and FPP in strain KG8. GPP and FPP were detected in mixed alcohol strain KG8, but not in strain KG1, the 3-methyl-3-buten-1-ol production strain. Although downstream metabolite concentrations varied substantially, steady state levels of mevalonate were nearly identical, suggesting that flux through the upstream pathway was not affected. Error bars show standard deviation (n = 3).
We increased the level of NudB protein to optimize the pathway given its positive effect in strain KG1 (Fig. 2). Since NudB in strain KG8 was part of a protein fusion with Idi1, we increased NudB levels by using a third plasmid, pBbB8k-NudB (pJBEI-6835)14. We designated this 3-plasmid strain as KG9 (Table 1). Compared to strain KG8, total mixed alcohol content at 48 hours increased more than 20% to 880 mg/L (Fig. 6). However, 3-methyl-3-buten-1-ol accounted for the entirety of this increase—levels of 3-methyl-2-buten-1-ol remained constant while those of 3-methyl-1-butanol slightly decreased. More engineering is clearly required to direct carbon flow from DMAPP towards 3-methyl-2-buten-1-ol and 3-methyl-1-butanol and away from GPP and FPP. One promising approach would be the development or discovery of a phosphatase specific for DMAPP. Although NudB is effective in producing high titers of C5 alcohol, the enzyme is promiscuous and does not appear to discriminate between IPP or DMAPP, both of which are non-native substrates25. More broadly, carbon loss to GPP and FPP should be mitigated through a selective knockdown of IspA activity. This may prove challenging, however, since the formation of FPP and thus ispA is essential for E. coli growth26.
Figure 6. Increased NudB expression improves total C5 alcohol content.
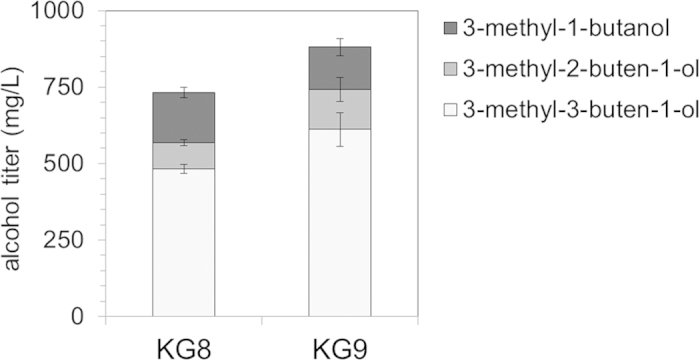
The introduction of a 3rd plasmid containing an additional copy of nudB (pBbB8k-NudB, see Table 1) in strain KG9 yielded a 20% increase in total alcohol content at 48 hours post induction. Error bars show standard deviation (n = 3).
An oleyl alcohol overlay improves yields of each C5 alcohol
Since short-chain alcohols and terpenes are often volatile, calculated yields are commonly underestimated during fermentation. In the production of 3-methyl-3-buten-1-ol, an apparent decrease in recoverable alcohol observed from 48 to 72 hours (Supplemental Figure S4) suggested that evaporation was occurring. To quantify the amount of 3-methyl-3-buten-1-ol, 3-methyl-2-buten-1-ol, and 3-methyl-1-butanol lost during production assays, we spiked each alcohol into culture tubes containing growth medium at various concentrations and monitored recovery over time at 30°C (Supplemental Figure S5). After 48 hours, losses of 20%, 10%, and 40% were observed for 3-methyl-3-buten-1-ol, 3-methyl-2-buten-1-ol, and 3-methyl-1-butanol, respectively. When the same tubes were incubated at 4°C, 100% recovery of each alcohol was observed after 48 hours (data not shown).
Hydrophobic overlays such as decane or dodecane are often used to prevent evaporation during microbial fermentations, particularly in the case of fatty acids or longer chain isoprenoids27,28. For the recovery of alcohols such as 1-butanol or 3-methyl-1-butanol, however, oleyl alcohol has proven a better choice in both Clostridia29 and E. coli30. We assessed the efficacy of an oleyl alcohol overlay with KG1R10 and KG9, high producers of 3-methyl-3-buten-1-ol and mixed alcohols, respectively. Using a 20% oleyl alcohol overlay (10 mL overlay added to 50 mL of culture) with strain KG1R10, a 3-methyl-3-buten-1-ol titer of 2.23 g/L (70% theoretical) was recorded after 48 hours, a 20% increase over the same strain without an overlay (Fig. 7). With strain KG9, an overlay led to a 26% increase in total C5 alcohol content (Fig. 7). The largest increase was in 3-methyl-1-butanol, where titers more than doubled to ~300 mg/L. The increased volatility of 3-methyl-1-butanol relative to 3-methyl-3- and 3-methyl-2-buten-1-ol (Supplemental Figure S5) and differential partitioning of 3-methyl-1-butanol into oleyl alcohol (Supplemental Figure S6) provide potential explanations for the improved recovery of this alcohol. We suspected this was due to the increased volatility (Supplemental Figure S5) and partitioning (Supplemental Figure S6) of this alcohol relative to 3-methyl-3- and 3-methyl-2-buten-1-ol.
Figure 7. An oleyl alcohol overlay improves C5 alcohol recovery.
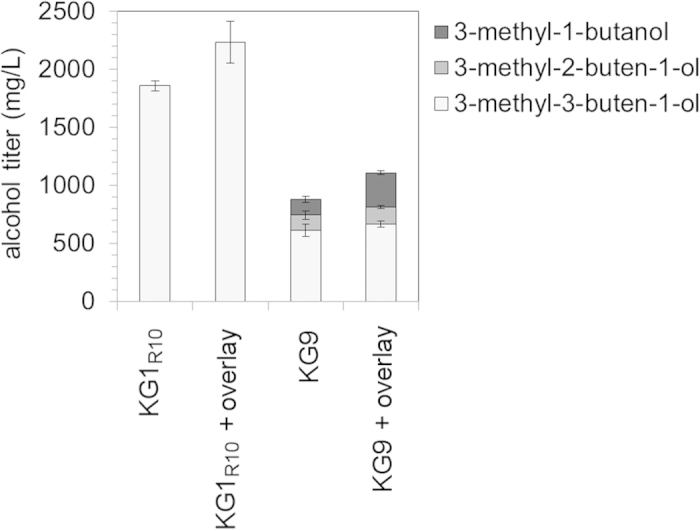
The addition of a 20% overlay of oleyl alcohol increased the titers of strains KG1R10 and KG9 by 20% and 26%, respectively. Concentrations at 48 hours post induction are shown. Error bars represent standard deviation (n = 3).
Conclusions
In this work we report the successful metabolic engineering of E. coli for the production of three isoprenoid-derived C5 alcohols: 3-methyl-3-buten-1-ol, 3-methyl-2-buten-1-ol, and 3-methyl-1-butanol. Using targeted metabolomics and proteomics, we rapidly identified pathway bottlenecks and improved titer. This approach was particularly successful for 3-methyl-3-buten-1-ol, where final yields approached 70% of pathway-dependent theoretical maximum. To achieve high C5 alcohol yields from high flux mevalonate pathway strains and prevent IPP accumulation, efficient phosphatase activity was critical. We significantly reduced IPP accumulation through RBS engineering of NudB, achieving a 9-fold improvement in protein level, a 4-fold reduction in IPP levels, and a 60% increase in 3-methyl-3-buten-1-ol production. Although increased NudB expression resulted in high yields in our most productive strains, significant improvements in pathway productivity will be required for fermentation scale-up. Future work to develop a more catalytically active phosphatase should be undertaken to achieve this goal.
Production titers of 3-methyl-2-buten-1-ol and 3-methyl-1-butanol were improved by more than 10-fold compared to previous work, where titers on 0.2% glucose were <10 mg/L12. Furthermore, we demonstrated that an oleyl alcohol overlay minimizes product loss due to evaporation and is effective for C5 alcohol fermentations. Still, additional engineering is required to improve total C5 alcohol content and optimize for the production of 3-methyl-2-buten-1-ol and 3-methyl-1-butanol. The detection of GPP and FPP, metabolites of endogenous isoprenoid metabolism, makes E. coli FPP synthase (ispA) an attractive engineering target. A reduction in IspA expression or activity should reduce carbon loss to FPP and improve flux to 3-methyl-2-buten-1-ol. Although NemA successfully catalyzes the conversion of 3-methyl-2-buten-1-ol to fully reduced 3-methyl-1-butanol, it does so at a low efficiency12. Protein engineering to improve the kinetics of this reaction should result in the complete conversion to 3-methyl-1-butanol. As titers of C5 alcohols increase, product toxicity is likely to become an issue31. Although use of oleyl alcohol may reduce toxicity, host engineering for increased tolerance32 may eventually be required to reach production goals.
Methods
Chemicals, solvents and media components were purchased and used without modification from Sigma-Aldrich (St. Louis, MO), Fisher Scientific (Pittsburgh, PA), or VWR (West Chester, PA) unless otherwise noted. E. coli strains DH10B (Invitrogen, Carlsbad, CA) and DH1 (ATCC) were used for plasmid construction and production experiments, respectively. For targeted proteomics experiments, mass spectrometric-grade trypsin was obtained from Sigma-Aldrich and prepared according to manufacturer's instructions.
Plasmid and strain construction
E. coli DH10B was used as the host for all cloning and plasmid manipulations. The BglBrick standard33 was used to assemble all plasmids as previously described. With the exception of pTrc99A17, plasmids were derived from the BglBrick plasmid library16. E. coli DH1 was used as the host for all production assays. Plasmids used in this study are available on the JBEI public registry and listed in Table 1 along with a brief description of production strains.
Production assays
Starter cultures of E. coli DH1 harboring production plasmids were grown overnight in LB medium containing appropriate antibiotics at 37 °C and shaken at 200 rpm in rotary shakers. Chloramphenicol, ampicillin, and kanamycin were provided at final concentrations of 25 mg/L, 100 mg/L, and 25 mg/L, respectively. Production assays were performed in triplicate in EZ-Rich defined medium (Teknova) containing 1% glucose. Briefly, starter cultures were used to inoculate 5 mL of production media in a culture tube or 50 mL of production media in a 250 mL Erlenmeyer flask to an OD600 of 0.1. Production cultures were grown in rotary shakers (200 rpm) at 37 °C to an OD600 of 0.4-0.6 and induced with 500 μM isopropyl β-D-1-thiogalactopyranoside (IPTG). Strains harboring pJBEI-6835 were also induced with 20 mM arabinose. Following induction, cultures were moved to 30°C for the duration of the assay. At set times, samples were taken for C5 alcohol quantification analysis by GC-FID as described previously12. For samples containing oleyl alcohol, the entire culture volume was extracted with ethyl acetate after 48 hours. Samples were analyzed by GC-FID as previously described30.
Metabolite quantification
Glucose and organic acids were quantified in filter-sterilized supernatant by high performance liquid chromatography (HPLC) at set time points using an Agilent 1200 Series HPLC system. Intracellular concentrations of mevalonate and IPP were measured by liquid chromatography mass spectrometry (LC-MS). Please see references 14 and 34 for complete protocols.
Volatility assays
To assess the evaporation of each C5 alcohol, 3-methyl-3-buten-1-ol, 3-methyl-2-buten-1-ol, and 3-methyl-1-butanol were spiked into 5 mL of EZ-Rich media in culture tubes (triplicate) at various concentrations and placed at 30 °C while shaking (200 rpm) for 48 hours. One set of tubes was placed at 4 °C to act as a control. Samples were collected for alcohol quantification by GC-FID at times 0, 24, and 48 hours.
Targeted proteomics analysis
At 24 hours, 1.5 mL of production culture was collected and pelleted by centrifugation at 8000 × g (4 °C). After the supernatant was decanted, cell pellets were frozen in liquid nitrogen and stored at −80 °C. Sample preparation and protein extraction was performed as described previously14,22.
Additional Information
How to cite this article: George, K. W. et al. Metabolic engineering for the high-yield production of isoprenoid-based C5 alcohols in E. coli. Sci. Rep. 5, 11128; 10.1038/srep11128 (2015).
Supplementary Material
Acknowledgments
This work was part of the DOE Joint BioEnergy Institute (http://www.jbei.org) supported by the U.S. Department of Energy, Office of Science, Office of Biological and Environmental Research, through contract DE-AC02-05CH11231 between Lawrence Berkeley National Laboratory and the U.S. Department of Energy
Footnotes
Author Contributions K.W.G., J.D.K. and T.S.L. designed the experiments. K.W.G., M.G.T., A.K., E.B., G.W., L.J.G.C., C.J.P. performed the experiments. K.W.G., M.G.T., P.D.A., C.J.P. and T.S.L. analyzed the data. K.W.G., M.G.T., J.D.K. and T.S.L. wrote the manuscript.
References
- Atsumi S. & Liao J. C. Metabolic engineering for advanced biofuels production from Escherichia coli. Curr. Opin. Biotechnol. 19, 414–419 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peralta-Yahya P. P., Zhang F., del Cardayre S. B. & Keasling J. D. Microbial engineering for the production of advanced biofuels. Nature 488, 320–328 (2012). [DOI] [PubMed] [Google Scholar]
- Atsumi S. et al. Metabolic engineering of Escherichia coli for 1-butanol production. Metab. Eng. 10, 305–311 (2008). [DOI] [PubMed] [Google Scholar]
- Jang Y.-S. et al. Bio-based production of C2-C6 platform chemicals. Biotechnol. Bioeng. 109, 2437–2459 (2012). [DOI] [PubMed] [Google Scholar]
- Atsumi S., Hanai T. & Liao J. C. Non-fermentative pathways for synthesis of branched-chain higher alcohols as biofuels. Nature 451, 86–89 (2008). [DOI] [PubMed] [Google Scholar]
- Baez A., Cho K.-M. & Liao J. C. High-flux isobutanol production using engineered Escherichia coli: a bioreactor study with in situ product removal. Appl. Microbiol. Biotechnol. 90, 1681–1690 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- George K. W., Alonso-Gutierrez J., Keasling J. D. & Lee T. S. Isoprenoid drugs, biofuels and chemicals - artemisinin, farnesene and beyond. Adv. Biochem. Eng. Biotechnol. 1–35 (2015). 10.1007/10_2014_288 [DOI] [PubMed] [Google Scholar]
- Hull A., Golubkov I., Kronberg B., Marandzheva T. & Stam J. van. An Alternative Fuel for Spark Ignition Engines. Int. J. Engine Res. 7, 203–214 (2006). [Google Scholar]
- Yang Y., Dec J., Dronniou N. & Simmons B. Characteristics of Isopentanol as a Fuel for HCCI Engines. SAE Int. J. Fuels Lubr. 3, 725–741 (2010). [Google Scholar]
- Mack J. H., Rapp V. H., Broeckelmann M., Lee T. S. & Dibble R. W. Investigation of biofuels from microorganism metabolism for use as anti-knock additives. Fuel 117, Part B, 939–943 (2014). [Google Scholar]
- Withers S. T., Gottlieb S. S., Lieu B., Newman J. D. & Keasling J. D. Identification of isopentenol biosynthetic genes from Bacillus subtilis by a screening method based on isoprenoid precursor toxicity. Appl. Environ. Microbiol. 73, 6277–6283 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou H. H. & Keasling J. D. Synthetic pathway for production of five-carbon alcohols from isopentenyl diphosphate. Appl. Environ. Microbiol. 78, 7849–7855 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y. et al. Metabolic engineering of Escherichia coli for high-specificity production of isoprenol and prenol as next generation of biofuels. Biotechnol. Biofuels 6, 57 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- George K. W. et al. Correlation analysis of targeted proteins and metabolites to assess and engineer microbial isopentenol production. Biotechnol. Bioeng. 111, 1648–1658 (2014). [DOI] [PubMed] [Google Scholar]
- Liu H. et al. MEP Pathway-mediated isopentenol production in metabolically engineered Escherichia coli. Microb. Cell Fact. 13, 135 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T. S. et al. BglBrick vectors and datasheets: A synthetic biology platform for gene expression . J. Biol. Eng. 5, 12 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amann E., Ochs B. & Abel K. J. Tightly Regulated Tac Promoter Vectors Useful for the Expression of Unfused and Fused Proteins in Escherichia-Coli. Gene 69, 301–315 (1988). [DOI] [PubMed] [Google Scholar]
- Tsuruta H. et al. High-level production of amorpha-4,11-diene, a precursor of the antimalarial agent artemisinin, in Escherichia coli. PLoS One 4, e4489 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou H. H. & Keasling J. D. Synthetic pathway for production of five-carbon alcohols from isopentenyl diphosphate. Appl. Env. Microbiol. 78, 7849–7855 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Primak Y. A. et al. Characterization of a feedback-resistant mevalonate kinase from the archaeon Methanosarcina mazei. Appl. Env. Microbiol. 77, 7772–7778 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange V., Picotti P., Domon B. & Aebersold R. Selected reaction monitoring for quantitative proteomics: a tutorial. Mol. Syst. Biol. 4, 222 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redding-Johanson A. M. et al. Targeted proteomics for metabolic pathway optimization: application to terpene production. Metab. Eng. 13, 194–203 (2011). [DOI] [PubMed] [Google Scholar]
- Dahl R. H. et al. Engineering dynamic pathway regulation using stress-response promoters. Nat. Biotechnol. 31, 1039–1046 (2013). [DOI] [PubMed] [Google Scholar]
- Salis H. M., Mirsky E. A. & Voigt C. A. Automated design of synthetic ribosome binding sites to control protein expression. Nat. Biotechnol. 27, 946–950 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLennan A. G. The Nudix hydrolase superfamily. Cell. Mol. Life Sci. 63, 123–143 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba T. et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2, 2006.0008 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh E.-B., Baidoo E. E. K., Keasling J. D. & Beller H. R. Engineering of bacterial methyl ketone synthesis for biofuels. Appl. Environ. Microbiol. 78, 70–80 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman J. D. et al. High-level production of amorpha-4,11-diene in a two-phase partitioning bioreactor of metabolically engineered Escherichia coli. Biotechnol. Bioeng. 95, 684–691 (2006). [DOI] [PubMed] [Google Scholar]
- Lee S. Y. et al. Fermentative butanol production by Clostridia. Biotechnol. Bioeng. 101, 209–228 (2008). [DOI] [PubMed] [Google Scholar]
- Connor M. R., Cann A. F. & Liao J. C. 3-Methyl-1-butanol production in Escherichia coli: random mutagenesis and two-phase fermentation. Appl. Microbiol. Biotechnol. 86, 1155–1164 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunlop M. J. et al. Engineering microbial biofuel tolerance and export using efflux pumps. Mol. Syst. Biol. 7, 487 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foo J. L. et al. Improving microbial biogasoline production in Escherichia coli using tolerance engineering. mBio 5, e01932 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson J. C. et al. BglBricks: A flexible standard for biological part assembly. J. Biol. Eng. 4, 1 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver L. J. et al. A kinetic-based approach to understanding heterologous mevalonate pathway function in E. coli. Biotechnol. Bioeng. 112, 111–119 (2014). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.