Abstract
Background
MicroRNA-19a (miR-19a), an oncogenic microRNA, has been recently reported to target CD22 in B cell lymphoma cell lines, but its role in inflammatory response is unclear. CD22 is a negative regulator for BCR signaling, and we hypothesize that miR-19a regulates B cell response by targeting CD22 in sepsis.
Material/Methods
In order to determine whether miR-19a-CD22 pathway was involved in sepsis, and what role it played in the regulatory mechanisms, we detected the levels of miR-19a in B cells obtained from patients with sepsis, and measured the levels of miR-19a and CD22 expression in B cells activated by LPS in vitro. Additionally, we investigated the correlation between miR-19a and CD22, as well as the influence of this pathway on BCR signaling, in transfected B cells.
Results
We found that septic patients displayed up-regulated miR-19a in B cells. In vitro, miR-19a was increased in activated B cells, with CD22 expression initially enhanced but subsequently decreased. Moreover, overexpression of miR-19a resulted in an amplified BCR signaling, while overexpression of CD22 attenuated the effect of miR-19a and increased its expression.
Conclusions
Our study demonstrated that miR-19a and CD22 comprised a feedback loop for B cell response in sepsis, providing a potential therapeutic target to recover the immune homeostasis.
Keywords: B-Lymphocytes, MicroRNAs, Sepsis, Sialic Acid Binding Ig-like Lectin 2
Background
Sepsis refers to the systemic inflammatory response syndrome (SIRS) induced by invading pathogens. At present, sepsis is the most frequent cause of mortality among patients admitted to intensive care units [1–3] and is responsible for the death of patients with intra-abdominal infections [4–8]. B cells are important in immune responses induced by sepsis. Apart from the traditional understanding of their roles in antibody production and antigen presentation, recent findings suggest that B cells can enhance early innate-immune responses during bacterial sepsis [9–11]. Unfortunately, a combination of an enhanced harmful auto-reactive B cell response and an impaired protective antigen-specific antibody production has been observed in septic animals [12], which matches the inflammatory response in human patients with sepsis [13]. Therefore, a dilemma, in which either immune-enhancing or immune-suppressing drugs would aggravate the immune imbalance, obstructs the success of immunotherapy for sepsis. Indeed, the regulatory molecular mechanisms of the sepsis-induced B cell response are still unclear.
MicroRNAs (miRs) are endogenous short non-coding RNAs of about 19–23 nucleotides, which bind to the 3′ untranslated regions (3′ UTRs) of mRNAs, inhibiting translation or inducing mRNA degradation [14–16]. Significantly, miRs influence a variety of biological processes and disease states [17,18]. Many studies have been performed to characterize the expression of various miRs in sepsis [19,20], suggesting their potential roles in the mechanisms of sepsis. MiR-19a is a member of the miR-17-92 cluster, and is regarded as an oncogenic microRNA [21]. Recently, miR-19a was reported to directly target the transcripts of CD22 in human B cell lymphoma cell lines and to have a lymphomagenic effect [22], but its role in normal B cell response is still unclear. In normal B cells, CD22 acts as a negative regulator for B cell receptor (BCR) signaling [23–25]. Ligation of BCR by antigen leads to increased phosphorylation of the immunoreceptor tyrosine inhibitory motifs (ITIMs) of CD22, which results in the recruitment of the protein tyrosine phosphatase SHP-1 [26]. SHP-1 subsequently acts to dephosphorylate components of the BCR signaling cascade, such as phosphorylated B cell linker protein (p-BLNK) [27], to dampen BCR signaling. Therefore, we wondered whether the miR-19a-CD22 pathway is involved in the B cell response induced by sepsis, and what role it plays in regulatory mechanisms.
In the present study, we detected the levels of miR-19a and CD22 expression in B cells obtained from patients with sepsis or non-infected SIRS, and investigated the correlation between miR-19a and CD22, as well as the influence of this pathway on BCR signaling, in vitro. Our present study demonstrated that B cells obtained from patients with sepsis displayed up-regulated miR-19a. We also found that miR-19a was increased in LPS-activated B cells, and CD22 expression was initially enhanced but subsequently down-regulated. Additionally, artificial overexpression of miR-19a in activated B cells resulted in enhanced BCR signaling and suppressed CD22 expression, while overexpression of CD22 partially attenuated the effect of miR-19a and increased its expression. These results demonstrate that miR-19a and CD22 comprised a feedback loop for B cell response in sepsis.
Material and Methods
Ethics statement
The research protocol was approved by the Ethics Committee of Tongji Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, China (approval number: 20140402), and was registered with the Chinese Clinical Trial Registry (registration number: ChiCTR-RCC-14004598). The study was conducted according to the principles of the Declaration of Helsinki. All patients were required to provide written informed consent before inclusion in this study.
Patients and blood samples
Sixty-four patients who were clinically diagnosed with SIRS were recruited from the Department of General Surgery, Wuhan General Hospital of Guangzhou Military Command, Wuhan, China, from May to October 2014. The criteria for SIRS included 2 or more of the following conditions [28]: (i) core temperature >38°C or <36°C; (ii) pCO2 <32 mmHg or >20 breaths/min; (iii) pulse rate >90/min; and (iv) white blood cell count >12 000/mm3, <4000/mm3, or >10% of band forms. The patients were divided into 2 groups: the sepsis group (n=38), referring to SIRS induced by infection or so-called infected SIRS; and the non-infected SIRS group (n=26). The diagnosis of infection was conducted via microbiological tests. Peripheral venous blood samples were obtained from each patient suspected of having sepsis or SIRS at the time of admission and before starting any treatment, and were excluded from the study cohort subsequently if they were not diagnosed with sepsis or SIRS. Additionally, 15 healthy volunteers with no history of autoimmune, inflammatory, or tumorous diseases were enrolled as controls.
Cells preparation
Blood samples were collected in heparinized tubes, and were diluted 1:1 with serum-free RPMI 1640 (Gibco, USA) medium. Peripheral blood mononuclear cells (PBMCs) were separated by Ficoll (GE Healthcare, USA) density gradient centrifugation. For RNA isolation and Western blotting, purified B cells (>95%) were isolated using MagCellect Human B Cell Isolation Kit (R&D systems, USA). B cell purity was assessed by flow cytometry using APC labeled CD19 antibody (Biolegend, USA). For transfection and cell culture, PBMCs were obtained from healthy volunteers.
Expression plasmid and transfection
The CD22 expression plasmid (provided by Sango Biotech, China) was generated by amplification of the entire coding region of CD22. The amplified fragment was inserted into pcDNA3.1 (Invitrogen, USA) using NotI and EcoRI (Takara, China) and was named as pcCD22. PBMCs were transfected with pcCD22, miR-19a mimic/inhibitor (RiboBio, China) or corresponding controls using DMRIE-C Transfection Reagent (Invitrogen, USA) and Opti-MEM I Reduced Serum Medium (Gibco, USA) according to the manufacturer’s instructions.
Cell culture
PBMCs, with or without transfection, were cultured at 1×106/ml in RPMI-1640 (Gibco, USA), supplemented with 10% heat-inactivated fetal bovine serum (Gibco, USA), 0.1mM nonessential amino acid solution (Sigma-Aldrich, USA), 1 mM sodium pyruvate (Gibco, USA), 50 μg/ml transferrin (Sigma-Aldrich, USA), 50 U/ml IL-2 (PeproTech, USA), 100 U/ml penicillin (Gibco, USA), 100 μg/ml streptomycin (Gibco, USA) and 100 μg/ml Normocin (InvivoGen, USA), and were stimulated with 50 μg/ml LPS (Sigma-Aldrich, USA). All cultures were conducted at 37°C in a humidified atmosphere containing 5% CO2.
RNA isolation and quantitative real-time PCR (qRT-PCR)
Purified B cells were isolated from PBMCs. Total RNA was extracted from purified B cells by Trizol reagent (Invitrogen, USA) according to the manufacturer’s instructions. For detection of miR-19a, reverse transcription was performed with the TaqMan MiRNA Reverse Transcription Kit (Applied Biosystems, USA), and real-time PCR was performed with the TaqMan MiRNA Assay Kit (Applied Biosystems, USA). To detect CD22 mRNA, RNA was reversely transcribed into cDNA with random hexamers using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems, USA), and real-time PCR was performed using SYBR Green PCR Master Mix (Applied Biosystems, USA). The relative expression level of miRNA and mRNA were normalized to U6 and GAPDH, respectively, using 2–ΔΔCt method [29].
The primers used were as follows: miR-19a- forward 5′-CCG GTG TGC AAA TCT ATG CAA-3′, miR-19a- reverse 5′-CAG TGC AGG GTC CGA GGT AT-3′, U6- forward 5′-CTC GCT TCG GCA GCA CAT A-3′, U6- reverse 5′-CGA ATT TGC GTG TCA TCC T-3′, CD22- forward 5′-CAT CTC CTC GGC CCC TGG CT-3′, CD22- reverse 5′-ATC CAG ACG CAG GCC CCC TC-3′, GAPDH- forward 5′-GGT GGT CTC CTC TGA CTT CAA CA-3′, GAPDH- reverse 5′-GTT GCT GTA GCC AAA TTC GTT GT-3′.
Flow cytometry analysis
PBMCs were harvested and washed with PBS containing 1% serum, and were subsequently incubated with PE-Cy5 or FITC-labeled CD22-antibody (eBioscience, USA) and APC-labeled CD19-antibody (Biolegend, USA) for 30 min. After several washes, cells were measured on FACSCalibur flow cytometer (BD Biosciences, USA) and analyzed using the CellQuest or FlowJo software. At least 1×104 cells were analyzed per test.
Western blot analysis
Purified B cells were isolated from PBMCs. Protein lysates were separated by 10% SDS-PAGE gel electrophoresis and transferred to polyvinylidene difluoride (PVDF) membranes. Target proteins were detected with primary antibodies recognizing CD22 (1:1000, Santa Cruz Biotechnology, USA), BLNK (1:1000, Santa Cruz Biotechnology, USA), p-BLNK (1:1000, Santa Cruz Biotechnology, USA), and GAPDH (1:20000, Abcam, USA), respectively. After incubation with horseradish peroxidase-conjugated secondary antibody (1:2000, Abcam, USA), protein bands were visualized using the ECL system (Pierce, USA).
Statistical analysis
All in vitro experiments in this study were performed 3 times independently. Quantitative variables were analyzed using the t test or 1-way analysis of variance (ANOVA). Categorical variables were analyzed using the chi-square test. GraphPad Prism 5.0 was used for all statistical analyses. P values <0.05 were considered statistically significant.
Results
MiR-19a was up-regulated in B cells from patients with sepsis
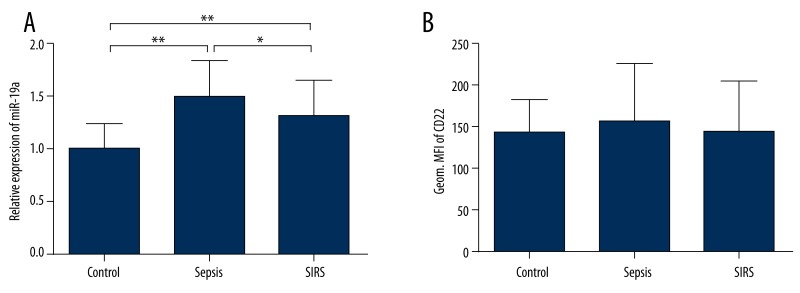
To determine whether miR-19a and CD22 were involved in the B cell response induced by sepsis, we measured the levels of miR-19a and CD22 expression in the B cells isolated from 38 septic patients, 26 non-infected SIRS patients, and 15 healthy controls using qRT-PCR and flow cytometry, respectively. The relative expression levels of miR-19a in different groups were calculated in comparison to the mean ΔCt values of healthy controls using 2−ΔΔCt method, and the levels of CD22 expression were displayed as geometrical mean fluorescence intensity (Geom. MFI). We found that miR-19a in B cells from septic patients and non-infected SIRS patients were significantly up-regulated compared with those from healthy volunteers (P<0.01), and the levels of miR-19a in B cells from septic patients were higher than those from non-infected SIRS patients (P<0.05) (Figure 1A). However, the levels of CD22 expression in B cells were variable and did not statistically differ among groups (P>0.05) (Figure 1B), suggesting a potential diversity in the changes of CD22 expression during inflammation.
Figure 1.
Expression of miR-19a and CD22 in B cells obtained from patients and controls. The relative expression levels of miR-19a in patients with sepsis (sepsis group) and in patients with non-infected SIRS (SIRS group) were significantly higher than that in healthy controls (control group), and the level in the sepsis group was elevated in comparison to that in the SIRS group (A). The levels of CD22 expression were displayed as geometrical mean fluorescence intensity (Geom. MFI) measured by flow cytometry, and were of no differences among different groups (B). Data are expressed as mean ±SD. * P<0.05, ** P<0.01.
For clinical characteristics, the Acute Physiology and Chronic Health Evaluation (APACHE) II scores were higher in septic patients than in non-infected SIRS patients (P<0.01), and subjects did not differ from each other with respect to age and sex (P>0.05) (Table 1). The types of bacteria in 36 septic patients were identified by microbiological tests. Another 2 septic patients with definite signs of infection (1 with suppurative appendicitis and 1 with suppurative cholangitis) had no microbial data and were regarded as having clinically diagnosed sepsis. Moreover, we did not find a correlation of miR-19a levels with the site of diseases or the type of bacteria (P>0.05) within the sepsis or SIRS group, indicating that the increased levels of miR-19a might not be regarded as pathogen- or organ-specific.
Table 1.
The clinical characteristics of the subjects.
| Characteristics | Control (n=15) | SIRS (n=26) | Sepsis (n=38) | P |
|---|---|---|---|---|
| Age (years) | 46.5±14.5 | 45.1±18.6 | 58.4±19.3 | >0.05 |
| Gender (Male/Female) | 9/6 | 12/14 | 26/12 | >0.05 |
| APACHE II score | – | 8.4±2.4 | 12.3±3.1 | <0.01 |
| Disease | ||||
| Cholangitis | – | 0 | 10 | – |
| Cholecystitis | – | 0 | 6 | – |
| Peritonitis | – | 0 | 13 | – |
| Hepatapostema | – | 0 | 3 | – |
| Pancreatitis | – | 10 | 2 | – |
| Appendicitis | – | 0 | 4 | – |
| Gut hemorrhage | – | 4 | 0 | – |
| Trauma of spleen | – | 8 | 0 | – |
| Multiple trauma | – | 4 | 0 | – |
| Type of bacteria | ||||
| Escherichia coli | – | – | 14 | – |
| Pseudomonas aeruginosa | – | – | 6 | – |
| Klebsiella pneumoniae | – | – | 4 | – |
| Enterococcus faecalis | – | – | 6 | – |
| Proteus mirabilis | – | – | 3 | – |
| Other bacteria | – | – | 3 | – |
| No microbial data | – | – | 2 | – |
Continuous variables and categorical variables are present as mean ± SD and number respectively.
MiR-19a was increased in activated B cells, with CD22 expression initially up-regulated and subsequently decreased
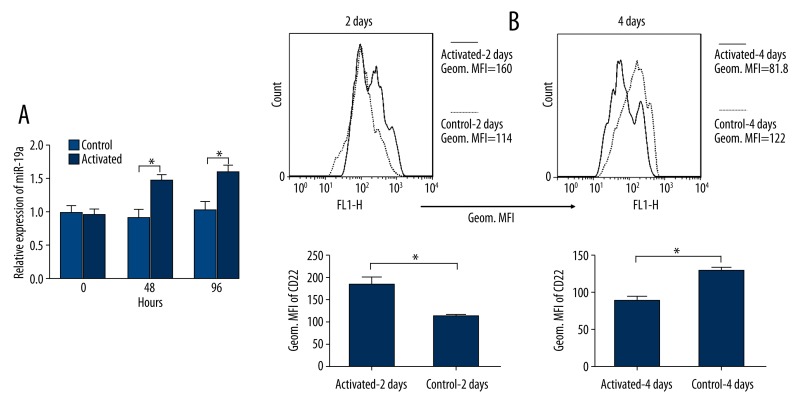
To confirm whether changes in miR-19a observed in septic patients resulted from B cell response, we stimulated PBMCs obtained from healthy volunteers in vitro with LPS, which is widely recognized as the prime stimulating factor in sepsis and is known to activate B cells [30]. We subsequently detected CD22 expression in stimulated PBMCs by flow cytometry and measured the levels of miR-19a in purified B cells isolated from PBMCs by qRT-PCR at 2 days and 4 days after stimulation, respectively. We found that the levels of miR-19a were increased by 2 days and 4 days in activated B cells compared to those in control B cells without stimulus (Figure 2A), suggesting that the increased levels of miR-19a could be attributed to B cell activation. For CD22 expression measured by flow cytometry, the Geom. MFI of CD22 in activated cells were increased by 2 days but decreased by 4 days (Figure 2B). The inverse correlation between miR-19a and CD22 after 2 days indicates a potential negative feedback in this pathway.
Figure 2.
Expressions of miR-19a and CD22 in activated B cells. PBMCs obtained from healthy volunteers were activated by LPS, and B cells were isolated after activation for the detection of miR-19a. The relative expression levels of miR-19a determined by qRT-PCR was increased by 2 days and 4 days in activated B cells compared to those in control B cells without stimulus (A). The mean fluorescence intensity (MFI) of CD22 on CD19 gated PBMCs determined by flow cytometry were increased by 2 days but decreased by 4 days (B). Data are expressed as mean ±SEM of 3 independent experiments. Histograms were obtained from 1 of 3 independent experiments that displayed similar results. * P<0.05.
MiR-19a enhanced BCR signaling in activated B cells by suppressing CD22 expression
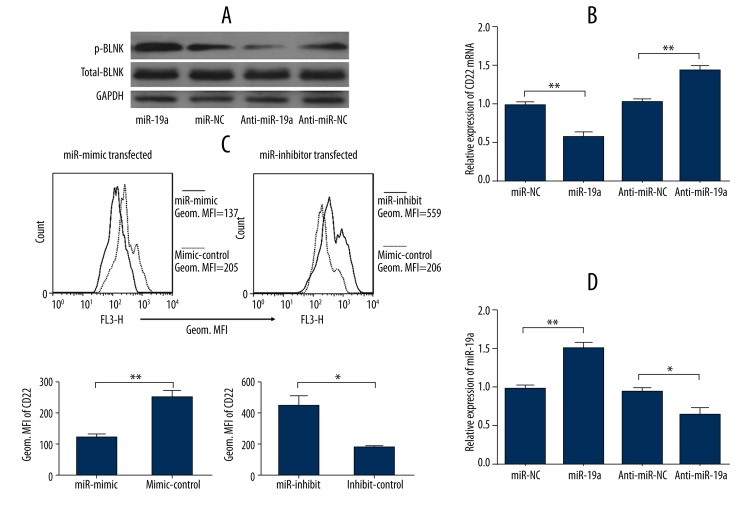
To investigate the function of miR-19a in B cell response, PBMCs transfected with miR-19a mimic or inhibitor were activated by LPS for 48 h. We performed Western blot analysis to detect the level of p-BLNK, which was used as a gauge for BCR signaling [31], in B cells isolated from the cultured PBMCs. B cells transfected with miR-19a mimic exhibited higher levels of p-BLNK when activated, and the results were reversed when miR-19a expression was repressed (Figure 3A), indicating the amplifying effect of miR-19a in B cell response. In addition, we detected CD22 mRNA levels in purified B cells and analyzed CD22 expression in PBMCs after activation, using qRT-PCR and flow cytometry respectively, to identify whether CD22 was the direct target of miR-19a. We found that overexpression of miR-19a induced decreases in CD22 mRNA and protein levels while silence of miR-19a resulted in an increased CD22 expression (Figure 3B, 3C), indicating that miR-19a directly targeted CD22 expression. The effect of miR-19a mimic/inhibitor transfection was confirmed by qRT-PCR (Figure 3D).
Figure 3.
Function of miR-19a in B cell activation. PBMCs transfected with miR-19a mimic or inhibitor (anti-miR19a), or corresponding controls (miR-NC, anti-miR-NC) were activated by LPS for 48 h. Activated B cells transfected with miR-19a mimic exhibited a higher level of p-BLNK, which was measured by Western blotting, while inhibition of miR-19a resulted in a suppressed level of p-BLNK (A). Overexpression of miR-19a induced decreases in CD22 mRNA while inhibition of miR-19a resulted in increased levels of CD22 mRNA (B). B cells transfected with miR-19a exhibited down-regulated CD22 expression, and inhibition of miR-19a enhanced CD22 expression (C). The effect of miR-19a mimic/inhibitor transfection was confirmed by the detection of miR-19a using qRT-PCR (D). Data are expressed as mean ±SEM of 3 independent experiments. Histograms and protein bands were obtained from 1 of 3 independent experiments that displayed similar results. * P<0.05, ** P<0.01.
CD22 overexpression partially attenuated the amplifying effect of miR-19a and increased its expression
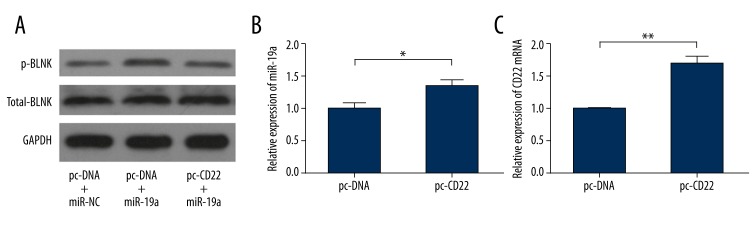
To explore how CD22 influenced the function of miR-19a, PBMCs were co-transfected with CD22 expression plasmid/control and miR-19a mimic/control, and were activated by LPS for 48 h. Western blotting was performed to determine the levels of p-BLNK. The results revealed that CD22 overexpression could partially attenuate the amplifying effect of miR-19a on BCR signaling (Figure 4A). Moreover, we transfected PBMCs with CD22 expression plasmid/control and measured the levels of miR-19a in isolated B cells during activation by LPS for 48 h to detect changes in miR-19a when CD22 was overexpressed. The qRT-PCR analysis demonstrated that overexpression of CD22 up-regulated the level of miR-19a (Figure 4B). The effect of CD22 expression plasmid was confirmed by qRT-PCR (Figure 4C).
Figure 4.
Influence of CD22 overexpression on miR-19a. PBMCs were co-transfected with CD22 expression plasmid (pc-CD22) or control (pc-DNA), and with miR-19a mimic or control (miR-NC), and were activated by LPS for 48 h. Western blotting revealed that CD22 overexpression partially attenuated the up-regulation of p-BLNK induced by miR-19a transfection (A). qRT-PCR analysis demonstrated that overexpression of CD22 resulted in an increased level of miR-19a in activated B cells (B). The effect of CD22 plasmid was confirmed by the detection of CD22 mRNA using qRT-PCR (C). Data are expressed as mean ±SEM of 3 independent experiments. Protein bands were obtained from 1 of 3 independent experiments that displayed similar results. * P<0.05, ** P<0.01.
Discussion
In the present study, we show that miR-19a was up-regulated in the B cells obtained from patients with sepsis or non-infected SIRS compared to those from healthy controls. We demonstrated that miR-19a was potentially involved in sepsis-induced B cell response, and the up-regulation of miR-19a in B cells was attributed to inflammatory response rather than specific bacterial infection. Additionally, the levels of miR-19a were higher in septic patients than in non-infected SIRS patients, suggesting that miR-19a level was associated with the severity of inflammatory response. MiR-19a has been recently reported to amplify B cell signaling via inhibition of CD22, an inhibitory coreceptor of BCR. Therefore, one of the possible regulatory mechanisms in sepsis is that up-regulation of miR-19a enhances B cell response by suppressing CD22 expression. However, the levels of CD22 expression in B cells obtained from patients were variable and we did not find any difference between septic patients and non-infected SIRS patients. To investigate the correlation between miR-19a and CD22, we stimulated PBMCs in vitro with LPS. LPS is an outer-membrane component of Gram-negative bacteria, and has been widely recognized as the prime stimulating factor in sepsis [32,33]. In our study, most pathogens identified in the septic patients were Gram-negative bacteria; thus, LPS approximatively recapitulated the stimulus in vivo. We found that miR-19a was increased in LPS-activated B cells, which matches the observation in patients. In contrast to the suppressing effect of increased miR-19a, however, the level of CD22 expression was initially up-regulated in activated B cells. Subsequently, the expression of CD22 decreased and the level of miR-19a kept increasing. The subsequent inverse correlation between miR-19a and CD22 expression accorded with the inhibitory function of miR-19a. These in vitro data indicated that up-regulation of miR-19a might not be a promoter for enhanced B cell response, but rather is a feedback reaction to overexpression of CD22 resulting from B cell activation. Additionally, the initial up-regulation and subsequent decrease in CD22 expression during B cell activation could explain the variability of CD22 levels in patients, because the time courses of disease in individuals were different.
Our study also showed that overexpression of miR-19a significantly decreased the level of CD22 mRNA, as well as its expression level on the B cell surface. CD22 has been previously reported to be the direct target of miR-19a in human B cell lymphoma cell lines, and our study showed that miR-19a likewise suppressed the transcript of CD22 in normal human B cells. CD22 was reported to decrease BCR signaling [34] and mediate inhibition of BCR-induced B cell proliferation [35]. Expectedly, the positively regulatory effect of miR-19a on B cell response by suppressing CD22 expression was observed in our present study. The BCR signaling was enhanced in miR-19a-overexpressed B cells, corresponding to the suppressed CD22 expression. These data show that miR-19a positively regulated B cell response by suppressing CD22 expression. Moreover, we found CD22 overexpression partially attenuated the amplifying effect of miR-19a on BCR signaling, confirming that miR-19a played its roles via suppression of CD22. Additionally, artificial overexpression of CD22 in activated B cells could up-regulated the level of miR-19a, indicating that increased miR-19a was the feedback consequence of up-regulation of CD22 expression.
CD22 acts as an inhibitory coreceptor for B cells. It has been reported that B cells that are deficient in CD22 undergo accelerated cell division after stimulation, resulting in rapid generation of plasma cells and antibody production [36]. In the present study, we observed the positively regulatory effect of miR-19a on B cell response by suppressing CD22 expression, indicating miR-19a as a pro-inflammatory factor that could accelerate B cell response and antibody production. MiR-19a has been reported to mediate several immune-associated pathways, such as TNF-α [37] and TGF-β [38] signaling, indicating its multiple functions in regulating immune response. During sepsis, the recognition of bacterial antigen leads to universal triggers of immune system and subsequently results in SIRS, involving many types of immune cells and signaling pathways. Our present study showed the role of the miR-19a-CD22 pathway in B cell response. Indeed, the crosstalk between B cells and other activated immune cells, as well as other pathways involved in sepsis, might influence the changes of miR-19a, and further studies are required to achieve full understanding of the role of miR-19a in sepsis.
Conclusions
The present study demonstrates that CD22 and miR-19a comprise a feedback regulatory loop for B cell response in sepsis. The initiation of BCR signaling induces an up-regulation of CD22 expression, which suppresses the BCR signaling. The overexpression of CD22 subsequently results in an increased level of miR-19a, which amplifies the BCR signaling by suppressing CD22. This pathway has so-called double-negative feedback loops or bi-stability, which potentially help preserve the balance between pro- and anti-inflammation in B cell response. Immune drugs targeting the components in this feed-back loop might invert the enhanced auto-reactive B cell response and the impaired antigen-specific antibody production, and recover immune homeostasis in septic patients. Our work provides a new regulatory mechanism for sepsis-induced B cell response, and might potentially provide a therapeutic strategy for the treatment of sepsis.
Footnotes
Source of support: This study was supported by the National Natural Science Foundation of China (No. 81471587)
Conflict of interest
The authors declare no conflict of interest.
References
- 1.Martin GS, Mannino DM, Eaton S, Moss M. The epidemiology of sepsis in the United States from 1979 through 2000. N Engl J Med. 2003;348:1546–54. doi: 10.1056/NEJMoa022139. [DOI] [PubMed] [Google Scholar]
- 2.Lever A, Mackenzie I. Sepsis: definition, epidemiology, and diagnosis. BMJ. 2007;335:879–83. doi: 10.1136/bmj.39346.495880.AE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vincent JL, Opal SM, Marshall JC, Tracey KJ. Sepsis definitions: time for change. Lancet. 2013;381:774–75. doi: 10.1016/S0140-6736(12)61815-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blot S, De Waele JJ. Critical issues in the clinical management of complicated intra-abdominal infections. Drugs. 2005;65:1611–20. doi: 10.2165/00003495-200565120-00002. [DOI] [PubMed] [Google Scholar]
- 5.Solomkin JS, Mazuski J. Intra-abdominal sepsis: newer interventional and antimicrobial thera-pies. Infect Dis Clin North Am. 2009;23:593–608. doi: 10.1016/j.idc.2009.04.007. [DOI] [PubMed] [Google Scholar]
- 6.Lichtenstern C, Schmidt J, Knaebel HP, et al. Postoperative bacterial/fungal infections: a challenging problem in critically ill patients after abdominal surgery. Dig Surg. 2007;24:1–11. doi: 10.1159/000099009. [DOI] [PubMed] [Google Scholar]
- 7.Zhang H, Yang Q, Xiao M, et al. Antimicrobial susceptibility of Gram-negative bacteria causing intra-abdominal infections in China: SMART China 2011. Chin Med J. 2014;127:2429–33. [PubMed] [Google Scholar]
- 8.Hawser SP, Badal RE, Bouchillon SK, et al. Susceptibility of gram-negative aerobic bacilli from intra-abdominal pathogens to antimicrobial agents collected in the United States during 2011. J Infect. 2014;68:71–76. doi: 10.1016/j.jinf.2013.09.001. [DOI] [PubMed] [Google Scholar]
- 9.Kelly-Scumpia KM, Scumpia PO, Weinstein JS, et al. B cells enhance early innate immune responses during bacterial sepsis. J Exp Med. 2011;208:1673–82. doi: 10.1084/jem.20101715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rauch PJ, Chudnovskiy A, Robbins CS, et al. Innate response activator B cells protect against microbial sepsis. Science. 2012;335:597–601. doi: 10.1126/science.1215173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weber GF, Chousterman BG, He S, et al. Interleukin-3 amplifies acute inflammation and is a potential therapeutic target in sepsis. Science. 2015;347:1260–65. doi: 10.1126/science.aaa4268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mohr A, Polz J, Martin EM, et al. Sepsis leads to a reduced antigen-specific primary antibody response. Eur J Immunol. 2012;42:341–52. doi: 10.1002/eji.201141692. [DOI] [PubMed] [Google Scholar]
- 13.Volk HD, Reinke P, Döcke WD. Clinical aspects: from systemic inflammation to ‘immunoparalysis’. Chem Immunol. 2000;74:162–77. doi: 10.1159/000058753. [DOI] [PubMed] [Google Scholar]
- 14.Lim LP, Glasner ME, Yekta S, et al. Vertebrate microRNA genes. Science. 2003;299:1540. doi: 10.1126/science.1080372. [DOI] [PubMed] [Google Scholar]
- 15.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 16.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–33. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ardekani AM, Naeini MM. The Role of MicroRNAs in Human Diseases. Avicenna J Med Biotechnol. 2010;2:161–79. [PMC free article] [PubMed] [Google Scholar]
- 18.Schickel R, Boyerinas B, Park SM, Peter ME. MicroRNAs: key players in the immune system, differentiation, tumorigenesis and cell death. Oncogene. 2008;27:5959–74. doi: 10.1038/onc.2008.274. [DOI] [PubMed] [Google Scholar]
- 19.Essandoh K, Fan GC. Role of extracellular and intracellular microRNAs in sepsis. Biochim Biophys Acta. 2014;1842:2155–62. doi: 10.1016/j.bbadis.2014.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.How CK, Hou SK, Shih HC, et al. Expression Profile of MicroRNAs in Gram-Negative Bacte-rial Sepsis. Shock. 2015;43:121–27. doi: 10.1097/SHK.0000000000000282. [DOI] [PubMed] [Google Scholar]
- 21.Olive V, Jiang I, He L. mir-17-92, a cluster of miRNAs in the midst of the cancer network. Int J Biochem Cell Biol. 2010;42:1348–54. doi: 10.1016/j.biocel.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Psathas JN, Doonan PJ, Raman P, et al. The Myc-miR-17-92 axis amplifies B-cell receptor signaling via inhibition of ITIM proteins: a novel lymphomagenic feed-forward loop. Blood. 2013;122:4220–29. doi: 10.1182/blood-2012-12-473090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Crocker PR, Paulson JC, Varki A. Siglecs and their roles in the immune system. Nat Rev Immunol. 2007;7:255–66. doi: 10.1038/nri2056. [DOI] [PubMed] [Google Scholar]
- 24.Walker JA, Smith KG. CD22: an inhibitory enigma. Immunology. 2008;123:314–25. doi: 10.1111/j.1365-2567.2007.02752.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nitschke L. The role of CD22 and other inhibitory co-receptors in B-cell activation. Curr Opin Immunol. 2005;17:290–97. doi: 10.1016/j.coi.2005.03.005. [DOI] [PubMed] [Google Scholar]
- 26.Doody GM, Justement LB, Delibrias CC, et al. A role in B cell activation for CD22 and the protein tyrosine phosphatase SHP. Science. 1995;269:242–44. doi: 10.1126/science.7618087. [DOI] [PubMed] [Google Scholar]
- 27.Gerlach J, Ghosh S, Jumaa H, et al. B cell defects in SLP65/BLNK-deficient mice can be par-tially corrected by the absence of CD22, an inhibitory coreceptor for BCR signaling. Eur J Immunol. 2003;33:3418–26. doi: 10.1002/eji.200324290. [DOI] [PubMed] [Google Scholar]
- 28.Bone RC, Balk RA, Cerra FB, et al. Definitions for sepsis and organ failure and guidelines for the use of innovative therapies in sepsis. The ACCP/SCCM Consensus Conference Committee. American College of Chest Physicians/Society of Critical Care Medicine. Chest. 1992;101:1644–55. doi: 10.1378/chest.101.6.1644. [DOI] [PubMed] [Google Scholar]
- 29.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantita-tive PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 30.Andersson J, Coutinho A, Lernhardt W, Melchers F. Clonal growth and maturation to immu-noglobulin secretion in vitro of every growth-inducible B lymphocyte. Cell. 1977;10:27–34. doi: 10.1016/0092-8674(77)90136-2. [DOI] [PubMed] [Google Scholar]
- 31.Fu C, Turck CW, Kurosaki T, Chan AC. BLNK: a central linker protein in B cell activation. Immunity. 1998;9:93–103. doi: 10.1016/s1074-7613(00)80591-9. [DOI] [PubMed] [Google Scholar]
- 32.Opal SM, Scannon PJ, Vincent JL, et al. Relationship between plasma levels of lipopolysac-charide (LPS) and LPS-binding protein in patients with severe sepsis and septic shock. J Infect Dis. 1999;180:1584–89. doi: 10.1086/315093. [DOI] [PubMed] [Google Scholar]
- 33.Morrison DC, Ryan JL. Endotoxins and disease mechanisms. Annu Rev Med. 1987;38:417–32. doi: 10.1146/annurev.me.38.020187.002221. [DOI] [PubMed] [Google Scholar]
- 34.Grimaldi CM, Cleary J, Dagtas AS, et al. Estrogen alters thresholds for B cell apoptosis and activation. J Clin Invest. 2002;109:1625–33. doi: 10.1172/JCI14873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Santos L, Draves KE, Boton M, et al. Dendritic cell-dependent inhibition of B cell proliferation requires CD22. J Immunol. 2008;180:4561–69. doi: 10.4049/jimmunol.180.7.4561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Onodera T, Poe JC, Tedder TF, Tsubata T. CD22 regulates time course of both B cell division and antibody response. J Immunol. 2008;180:907–13. doi: 10.4049/jimmunol.180.2.907. [DOI] [PubMed] [Google Scholar]
- 37.Liu M, Wang Z, Yang S, et al. TNF-α is a novel target of miR-19a. Int J Oncol. 2011;38:1013–22. doi: 10.3892/ijo.2011.924. [DOI] [PubMed] [Google Scholar]
- 38.Haj-Salem I, Fakhfakh R, Bérubé JC, et al. MicroRNA-19a enhances proliferation of bronchial epithelial cells by targeting TGFβR2 gene in severe asthma. Allergy. 2015;70:212–19. doi: 10.1111/all.12551. [DOI] [PubMed] [Google Scholar]