Significance
Social organisms benefit from group behaviors that endow favorable fitness consequences among kin. We describe such a behavior in the bacterium Myxococcus xanthus in which damaged members of a population are repaired by their kin by exchange of outer membrane material. This behavior rescues lethal cellular damage, restores antibiotic resistance to a compromised cell membrane, and increases the overall fitness of a heterogeneous population. To our knowledge, we provide the first evidence that a social bacterium can use cell-content sharing to repair damaged siblings, leading to beneficial fitness outcomes for both the donor and recipient.
Keywords: Myxococcus xanthus, outer membrane, lipopolysaccharide, lpxC, fusion
Abstract
Bacterial cells in their native environments must cope with factors that compromise the integrity of the cell. The mechanisms of coping with damage in a social or multicellular context are poorly understood. Here we investigated how a model social bacterium, Myxococcus xanthus, approaches this problem. We focused on the social behavior of outer membrane exchange (OME), in which cells transiently fuse and exchange their outer membrane (OM) contents. This behavior requires TraA, a homophilic cell surface receptor that identifies kin based on similarities in a polymorphic region, and the TraB cohort protein. As observed by electron microscopy, TraAB overexpression catalyzed a prefusion OM junction between cells. We then showed that damage sustained by the OM of one population was repaired by OME with a healthy population. Specifically, LPS mutants that were defective in motility and sporulation were rescued by OME with healthy donors. In addition, a mutant with a conditional lethal mutation in lpxC, an essential gene required for lipid A biosynthesis, was rescued by Tra-dependent interactions with a healthy population. Furthermore, lpxC cells with damaged OMs, which were more susceptible to antibiotics, had resistance conferred to them by OME with healthy donors. We also show that OME has beneficial fitness consequences to all cells. Here, in merged populations of damaged and healthy cells, OME catalyzed a dilution of OM damage, increasing developmental sporulation outcomes of the combined population by allowing it to reach a threshold density. We propose that OME is a mechanism that myxobacteria use to overcome cell damage and to transition to a multicellular organism.
A fundamental question in biology is how cells cope with damage. Microbes occupy diverse habitats fraught with physical, biological, and chemical insults (1, 2). UV radiation, desiccation, predation, extracellular enzymes, antimicrobial compounds, pH, temperature, and osmolarity changes are all stresses to the individual cell. In addition, when cells are in nutrient-poor environments, cell division can be rare, taking days to months to complete (3). In a slow-growing state, cell-surface components that may not be undergoing active repair can accumulate damage through natural aging processes such as oxidation (4, 5) and protein denaturation. Although internal cell stress response pathways are known (6), mechanisms to cope with cell surface damage are less well understood.
Although cell damage threatens the fitness of the individual, social organisms have strength in numbers. The strategy of kin selection allows evolutionarily viable cooperation between individuals in a closely related population (7). Communication between individuals and sharing of resources establishes the potential for assistance between individual population members. Social support can be beneficial when the fitness of individuals in a group depends on collaborative behaviors such as prey hunting or the development of complex structures such as biofilms or fruiting bodies. These behaviors require contributions from many individuals and thus are threatened by heterogeneity among members. Such heterogeneity might develop both from individual cells being exposed to different microenvironments present in the soil (8) and from diversity in cell ages (9). Thus, starving or damaged members of a population bear a damage burden for the community that, when prevalent, could be detrimental to group behaviors. The mechanisms that social organisms use to cope with this damage are largely unexplored.
Myxobacteria are highly social microbes that inhabit a diverse range of soil and water habitats (10) and are subject to cellular damage. Individual members function within communities that transcend the typical paradigm of a microbial biofilm (11). Their highly cooperative behaviors resemble a tissue, because they can undergo cooperative multicellular development to form fruiting bodies and can move and hunt prey in a coordinated manner (reviewed in ref. 12). How myxobacteria cope with damaged cells and population heterogeneity to retain a functional collective remains unknown.
Members of a myxobacteria swarm exchange bulk amounts of their outer membrane (OM), which are essentially “public good” commodities, upon cell–cell contact in a process known as OM exchange (OME) (13). This process is dependent on the cell-surface receptor TraA and the TraB cohort protein (14). Exchange between cells is highly selective, because TraA has evolved a polymorphic region that enables kin recognition (15, 16). Although TraA is conserved across myxobacteria, only strains within the same compatibility group, determined by the traA allele they express, exchange OM components. The requirement to selectively identify kin as exchange partners implies that OME confers an advantage to the group that is guarded from exploitation by similar, but nonidentical, groups. traA has been described as a social “greenbeard” gene, because it can identify and confer preferential treatment to others that display the same allele (15, 16).
The OM of Gram-negative bacteria defines the cell boundary and interacts directly with the extracellular environment. The outer leaflet of the OM is composed of the lipid-linked glycan structure lipopolysaccharide (LPS), which serves as the major permeability barrier. In addition to being a major structural component of the cell envelope, LPS also has species-specific functions. In Myxococcus xanthus, LPS O-antigen is required for development and social (S) gliding motility (17), i.e., the ability of cells to move as groups in a biofilm powered by type-4 pili extension and retraction (18). In contrast, individual cells can move by a separate system called “A-motility” (19). Although the function of LPS in S-motility is unknown, as a major constituent of the cell surface, it is likely to have a key role in how M. xanthus cells interact with each other and their environment. Here, we asked if TraAB-mediated cell interactions could transfer LPS and investigated the outcomes of this behavior.
The utility of OME in myxobacteria physiology is not fully understood. Because, to take place, OME requires two or more cells and because material transfer is bidirectional, it is a social behavior that may have evolved to improve the fitness of the population. Here, we hypothesized that OME could be used to dilute a damage load acquired in the environment and from cellular senescence to heal affected cells. Diluted damage may be easier to repair when spread among multiple cells. In turn, the damaged individuals would gain a direct benefit, and the now larger population could gain fitness through multicellular functions that require a quorum, such as fruiting body formation. By an analogous process, eukaryotic mitochondria are thought to repair damage by undergoing rounds of fusion and fission to dilute damaged lipids and proteins, allowing the population to maintain homeostasis more effectively (20). To test our hypothesis at the cellular level, we devised models that genetically mimic damaged membranes that lead to impaired fitness or death in M. xanthus. We then showed that OME can rescue these mutant phenotypes. Our results reveal a novel strategy used by a social microorganism to cope with damage.
Results
Overexpression of TraAB Triggers the Formation of Cell–Cell Prefusion Junctions.
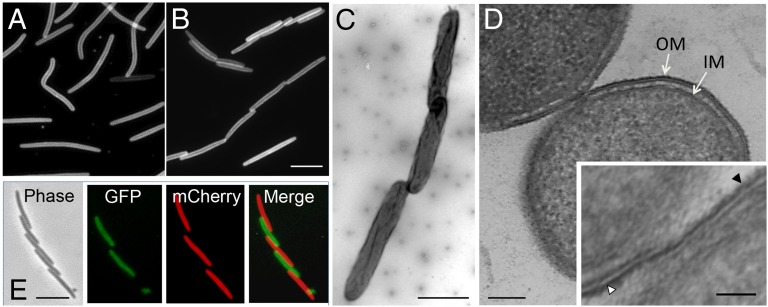
In a prior study (14) we found that TraAB functions as a cell–cell adhesin. Indeed, Fig. 1B shows that TraAB overexpression results in cells frequently adhering together side-by-side in liquid culture as well as forming end-to-end chains in which the cell poles are slightly offset relative to one another. In contrast, the parental strain grew in a dispersed manner and did not form cell–cell adhesions (Fig. 1A). Importantly, we note that both these strains contain a ΔpilA mutation that abolishes the cell–cell clumping or agglutination seen in WT cells (21). When viewed by transmission electron microscopy (TEM), offset cell chains were seen in detail (Fig. 1C). This cell-chaining pattern correlates with TraA cell-surface localization patterns (15), suggesting that TraA foci function as adhesion patches for cell–cell binding. The cellular junctions were investigated further by cryo-EM. A representative cross-section of two side-by-side cells demonstrates that the OM leaflets of the two cells are in direct contact and suggests that the membranes may be able to fuse (Fig. 1D). These tight cell–cell contacts were seen frequently by cryo-EM in the TraAB overexpression strain (Fig. S1) and were absent in the parent strain. Previously, we showed that OME did not occur in liquid culture (13, 22, 23); however, TraAB overexpression may have overcome the requirement that cells reside on a hard surface. To test this possibility, two differentially labeled strains that overexpress TraAB were cocultured in liquid CTT medium. As expected, these two strains adhered together in the liquid culture, although we did not detect transfer of the fluorescent reporters (Fig. 1E) (13). In contrast, under control conditions on a solid surface, these strains did carry out OME (Fig. S2). Thus, cell–cell binding between TraA receptors catalyzed tight cell–cell junctions that overcame steric and repulsion forces from bulky charged LPS molecules residing on the cell surface. The EM micrographs also suggested that TraAB overexpression led to the formation of a prefusion junction for the mixing of OM molecules that occurred only after the cells were placed on a hard surface.
Fig. 1.
TraAB overexpression triggers cell–cell prefusion junctions. (A) Parental strain DW1411, which was fluorescently labeled with a transferable OM reporter (lipoprotein SSOM-mCherry), exhibits dispersed growth because it contains a ΔpilA mutation. (B–D) The isogenic strain DW1463 (also ΔpilA) overexpresses TraAB, resulting in abundant cell–cell adhesion during liquid growth as seen by fluorescence microscopy (B), negative-stain TEM (C), and a cryo-EM thin section of two adhered cells (D). (Inset) At higher magnification the outer leaflets of the OM of the TraAB-overexpressing cells are in close proximity and appear to be fused. Following the outer leaflet of the OM of the upper cell (white triangle) the OM appears to be a continuous structure with the outer leaflet of the lower cell (black triangle). See Fig. S1 for additional cryo-EM micrographs. (E) A 1:1 coculture of two strains that overexpress TraAB and either the SSOM-mCherry (DW1463) or SSOM-sfGFP (DW2201) fluorescent markers. The reporters did not transfer in liquid culture but did transfer on a solid surface (see Fig. S2). OM, outer membrane; IM, inner membrane. [Scale bars: 2 µm in B (for both A and B), C, and E; 200 nm in D; 50 nm in Inset.]
OME Rescues the Motility and Development of O-Antigen Mutants.
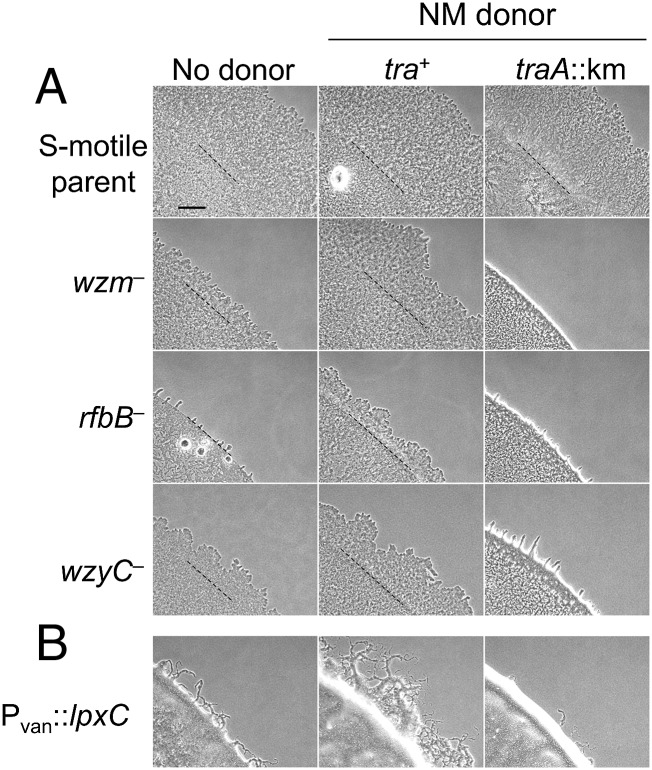
Because myxobacteria exchange large amounts of OM proteins and fluorescent lipid dyes and can transiently rescue phenotypic defects caused by missing proteins (13, 14, 24), we asked if OME could rescue motility defects of O-antigen LPS mutants. To this end, we introduced mutations to the previously identified wzm locus, which is required for O-antigen production and S-motility (17), and to two additional loci (rfbB and wzy) that were predicted from bioinformatic analysis to be required for LPS biosynthesis (Fig. S3). As expected, all three mutants showed an S-motility defect (Fig. 2A, Left). To test if OME from a donor strain with WT LPS could rescue the motility of the mutants, we examined the motility of each mutant in the presence of nonmotile donors. Motility was observed as swarm expansion from the edge of the inoculum. In the presence of the tra+ donor cells, the distance traveled from the edge of the inoculum increased relative to the controls in which no donor was present (in Fig. 2A, compare Left and Center). In contrast, when the donor contained a traA mutation and thus was OME deficient, motility was not stimulated (Fig. 2, Right). To eliminate the possibility that stimulation was caused by extracellular complementation of exopolysaccharide (EPS) molecules, we tested whether a dsp/dif mutant, which is defective in EPS production (25, 26), could be stimulated for motility. Using the same procedure, we observed that motility was not stimulated in this strain (Fig. S4). These results show that the motility defect of LPS mutants was rescued by OME and suggest that LPS molecules were transferred between cells in a TraA-dependent manner.
Fig. 2.
OME rescues motility defects of LPS mutants. (A, Left) LPS mutants constructed in an A−S+ background exhibit S-motility defects as seen by reduced swarm expansion compared with the parental strain (DK1217). (Center) LPS mutants mixed with nonmotile (NM) donors, which were Tra+ and expressed WT LPS (DK6204), were stimulated to swarm. (Right) In contrast, an isogenic traA::km donor (DW1409) did not rescue and in the case of the wzm mixture produced a nonmotile sharp colony edge phenotype. Dashed lines represent the edge of the liquid inoculum after it dried. (B) Cells depleted for lpxC were defective for S-motility and were rescued when mixed with a nonmotile Tra+ donor but not when mixed with a traA::km donor. Strains are listed in Table S1. (Scale bar in A: 300 µm in A and B.)
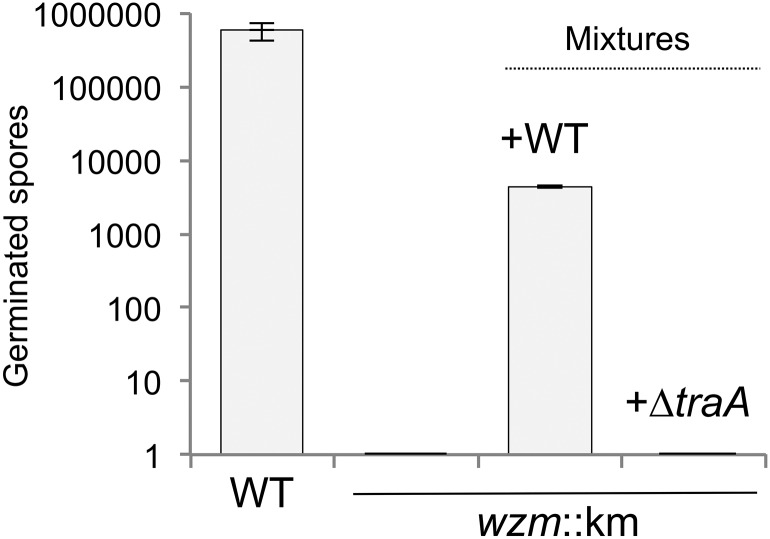
Under starvation conditions, myxobacteria undergo a complex, coordinated program of development culminating in fruiting body structures that harbor stress-resistant spores. LPS mutants are defective for this developmental program, because wzm mutants form abnormal fruiting structures, are delayed in sporulation, and produce relatively few spores compared with WT myxobacteria (17). After 72 h on starvation agar, 6 × 107 WT cells (DK1622) produced nearly 1 million spores, whereas a wzm mutant produced none (Fig. 3), underscoring the severe fitness consequences that result from LPS damage. We reasoned that in a population in which OME occurs, the fitness defect of a damaged OM may be relieved by exchange among neighboring cells. To test this notion, we mixed WT cells with wzm mutant cells in equal numbers on starvation agar and allowed the cells to develop for 72 h. The wzm mutant was able to sporulate at a significant level in the presence of WT cells but not in the presence of a ΔtraA mutant (Fig. 3), suggesting that OME can rescue development in cells that otherwise would be incapable of carrying out this process. As a control, under these conditions the ∆traA mutant sporulates at WT levels (14). These results suggest that OME confers a strong fitness advantage when OM damage impairs the developmental program.
Fig. 3.
OME rescues the sporulation defect of a wzm mutant. A DK1622 wzm mutant (DW2407) was unable to sporulate but was rescued when mixed with a WT strain. Rescue did not occur in the presence of an isogenic ∆traA strain. In mixture experiments, spore germination was assayed with kanamycin selection; thus only the wzm::km strain formed colonies. In this and subsequent figures, error bars represent SEM; n = 3 unless stated otherwise.
Depletion of LpxC Causes Morphological Defects and Cell Lysis.
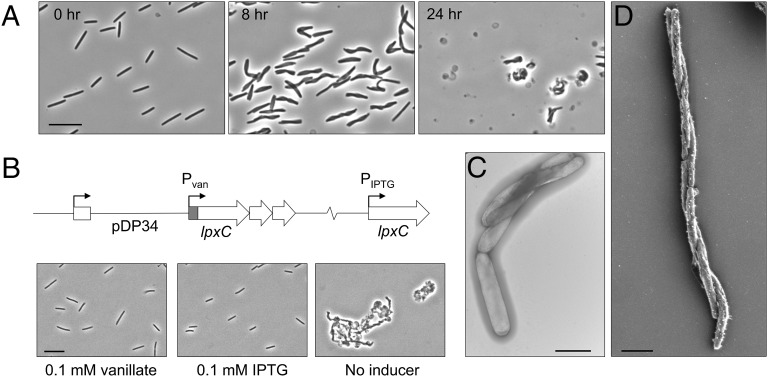
Based on the ability of OME to rescue motility and development in mutants with truncated LPS, we sought to create a model of membrane damage that interrupts the biosynthesis of the whole LPS molecule and thus affects cell viability. Lipid A is the acylated disaccharide that embeds LPS in the OM and is essential in most Gram-negative bacteria (27). The first committed step in the biosynthesis of lipid A is the deacetylation of UDP-3-O-acyl N-acetyl-glucosamine by cytoplasmically localized LpxC (27), a protein that is highly conserved and essential across Gram-negative bacterial species (28). We thus constructed a strain of M. xanthus in which the endogenous lpxC promoter was replaced with a vanillate-inducible promoter (29) (Pvan::lpxC) (Fig. S5). As predicted, in the presence of vanillate this strain grows like WT (Fig. 4A). However, when the CTT growth medium of a log-phase liquid culture was replaced with a vanillate-free medium, we observed defects in cell septation, which manifest as chains of cells. Interestingly, these cells twisted or wrapped around one another along their long axes (Fig. 4A). EM imaging revealed that the cells twisted in a right-handed helix (Fig. 4 C and D), although the significance of this directionality is unknown. After 8 h of vanillate starvation, nearly all cells exhibited the twisted phenotype (Fig. 4). On occasion, a twisted morphology has been reported in a WT population (30). When viewed by time-lapse microscopy, these twisted cells exhibited severe motility defects (Movies S1 and S2). Like O-antigen mutants, Pvan::lpxC cells deprived of vanillate were stimulated for motility by Tra-dependent interactions with cells that contained WT LPS (Fig. 2B). The twisted morphology also was observed in additional conditionally lethal LPS biosynthesis mutants (Fig. S6). Further inhibition of lpxC transcription resulted in cell lysis, which was most apparent after 24 h (Fig. 4A). Because lpxC is in an apparent operon (Fig. 4B), we wanted to confirm that the phenotype was specifically lpxC dependent. We thus complemented the mutation by ectopic lpxC expression from an isopropyl β-d-1-thiogalactopyranoside (IPTG)-inducible promoter (29). In the presence of either vanillate or IPTG, the strain behaved and morphologically appeared as WT, whereas in the absence of both inducers the cells lysed (Fig. 4B). We conclude that lpxC is essential in M. xanthus, and the viability of a Pvan::lpxC mutant is dependent on the presence of vanillate.
Fig. 4.
An lpxC-depleted strain twists and lyses. (A) Pvan::lpxC cells were morphologically WT when grown with 0.1 mM vanillate (0 h). Removal of vanillate resulted in a twisted cell morphology by 8 h and ultimately in lysis (24 h). (Scale bar: 10 µm.) (B) lpxC resides in an apparent three-gene operon, and the Pvan::lpxC mutation was complemented by ectopic expression from an IPTG-inducible promoter. (Scale bar: 10 µm.) (C and D) TEM (C) and SEM (D) micrographs show the twisted morphology of the lpxC-depleted strain. (Scale bars: 2 µm.)
Viability of an lpxC Mutant Can Be Rescued by OME.
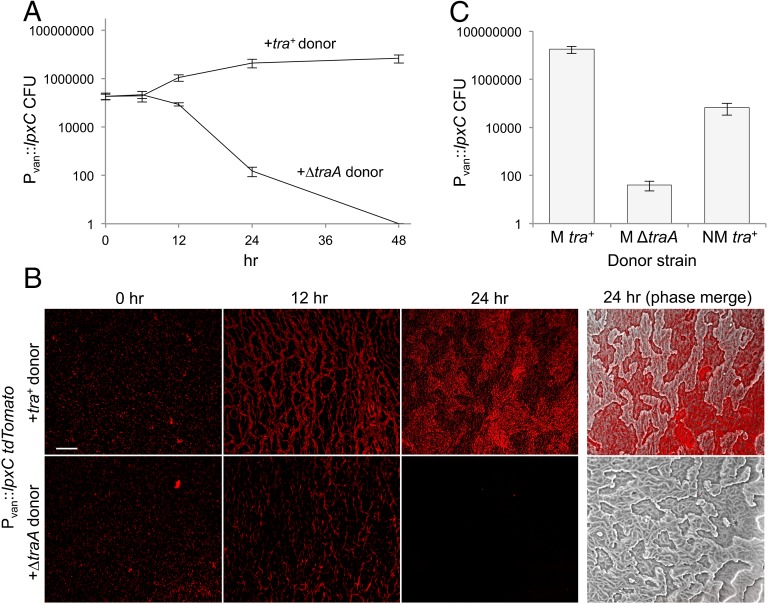
Because the Pvan::lpxC strain has a conditional lethal OM defect, we tested whether exchange from healthy donors could provide the LPS necessary to maintain viability of the damaged strain. We monitored the viability of Pvan::lpxC in mixed cultures with either OME-proficient (tra+) or OME-deficient (ΔtraA) isogenic strains. Vanillate was removed from the Pvan::lpxC culture medium, and these cells were mixed 1:1 with either of the two donor strains and placed on agar plates. At various times, mixed populations were harvested and plated on antibiotic selection plates containing vanillate to enumerate viable mutant cells. Based on the number of cfus at 12, 24, and 48 h after plating, Pvan::lpxC cells survived and grew in the presence of tra+ donors but died completely in the presence of ∆traA donors (Fig. 5A). To confirm this finding, we labeled Pvan::lpxC with tdTomato and conducted the same experiment by assessing the presence of fluorescent cells by microscopy. Again, we found that the LpxC-depleted cells thrived in the presence of tra+ donors but not with ∆traA cells (Fig. 5B).
Fig. 5.
OME rescues the viability of an lpxC-depleted strain. (A) The viability of the Pvan::lpxC kanamycin-resistant strain over time when mixed with donor strains (kanamycine sensitive) in the absence of vanillate on nutrient agar plates. Following incubation at indicated times, cells were harvested and viable Pvan::lpxC cfus were determined on kanamycin/vanillate plates. Tra+ cells (DK8615) rescued the viability of Pvan::lpxC cells, but ∆traA cells did not. (B) Pvan::lpxC cells (tdTomato) in the presence of unlabeled Tra+ (DK1622) or ∆traA donors on agar plates. Pvan::lpxC cells were visible in the presence of WT donors but died over time in the presence of ∆traA donors. (Scale bar: 100 µm.) (C) Pvan::lpxC cfus after 24 h on agar plates in the absence of vanillate when mixed with indicated donors. The nonmotile (NM) Tra+ donor (DK8601) exhibited reduced rescue compared with a motile (M) Tra+ donor (DK8615).
Motility is required for efficient OME (13, 22). We therefore hypothesized that the absence of motility in the Pvan::lpxC and donor strains would negatively impact OME and thus the rescue of viability. We repeated the above experiment with a nonmotile strain and found that nonmotile donors were able to rescue but did so 1,000-fold less effectively than motile donors (Fig. 5C). Therefore, under conditions in which OME was compromised, there was a corresponding defect in Pvan::lpxC rescue.
OME Restores the Permeability Barrier to LpxC-Deficient Cells.
Bacterial cells that have defective cell envelopes often are hypersensitive to certain antibiotics because of an increased ability for small molecules to permeate the OM barrier (31). Concordantly, we found that the Pvan::lpxC strain was hypersensitive to antibiotics, including polymyxin B, which binds LPS (32) and disrupts the OM (Fig. S7) (33). Next, we asked if OME with donor cells could restore the permeability barrier to cells with damaged membranes. To answer this question, we incubated the Pvan::lpxC strain in the absence of vanillate with tra+ or ΔtraA cells or with no donor on solid agar for 6 h to allow OME to occur. The cells were harvested and then exposed to polymyxin B. Under these conditions, LpxC-depleted cells that had exchanged membranes with donor cells survived better than those that had not exchanged membranes (Fig. 6). These results indicate that the transfer of WT membrane to damaged cells can restore OM function.
Fig. 6.
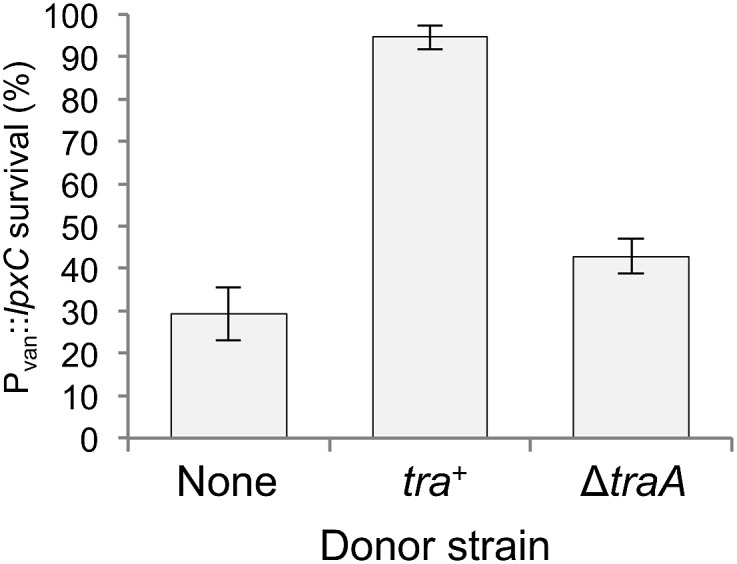
OME confers antibiotic resistance to the lpxC mutant. Percent survival of the Pvan::lpxC mutant when grown without vanillate and exposed to polymyxin B after 6-h incubation with the indicated donors. Percent survival was calculated by dividing the number of viable cells in the presence of polymyxin B by the number of viable cells when polymyxin B was not added to the medium. Viable cfus were determined with kanamycin selection; see Experimental Procedures for details.
Content Mixing Bestows a Fitness Advantage to Heterogeneous Sporulating Populations.
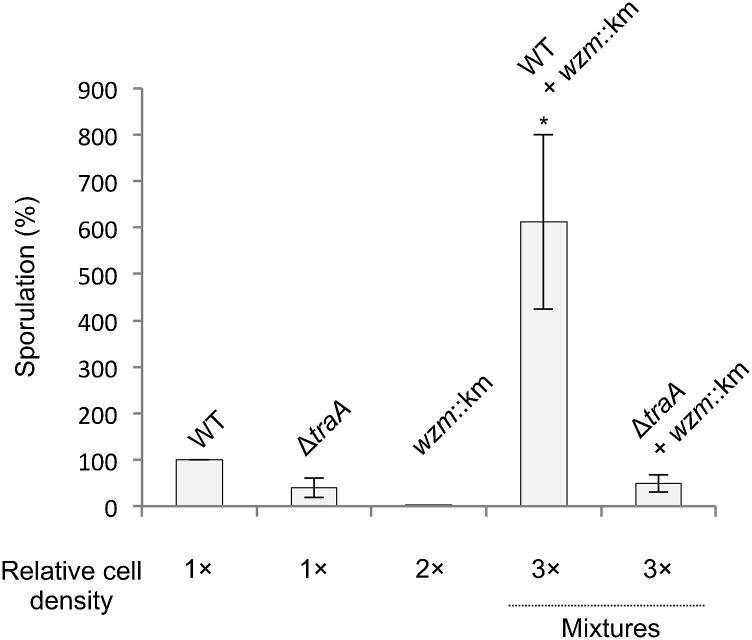
Healthy cells that exchange membranes with damaged cells likely do so at a cost, because they acquire defective membranes from their damaged kin. From an evolutionary perspective, this behavior must have a benefit that outweighs the cost. Although a direct fitness benefit for the recipient cells was clear, we hypothesized that when damaged and healthy populations merge and exchange OMs the subsequent increase in population density could help offset the cost to the donor. In this regard, myxobacteria require a threshold cell density to develop (34, 35). During starvation, M. xanthus monitors population density and will continue development if the density exceeds a threshold of ∼3 × 108 cells/mL (36). In addition, the expression of developmental genes increases exponentially from the threshold density until reaching a maximum around 109 cells/mL (37). From these observations, we assayed the sporulation ability of WT, ΔtraA, and wzm strains at a lower-than-optimal cell density for sporulation (3 × 108, 3 × 108, and 6 × 108 cells/mL, respectively). As described above, the wzm mutant again was unable to sporulate, while the other strains sporulated at low efficiencies. To simulate the joining of damaged and healthy populations, we mixed the same total numbers of WT and wzm cells from the above experiment in an equal volume of buffer. This mixing increased the total cell density three times compared with the WT condition alone, but the cocultures contained the same total number of cells as the WT and wzm populations combined. The mixed population sporulated roughly sixfold more efficiently than the WT alone population (Fig. 7), indicating a synergistic effect. In contrast, ∆traA cells mixed with wzm cells sporulated at the same level as ∆traA alone (Fig. 7). Importantly, the increase in population density conferred by the mixing of damaged and healthy populations was beneficial to sporulation only when OME could take place (Fig. 7). This result indicates that increasing the cell population so that it reached a threshold density conferred a sporulation advantage despite the introduction of damage and sporulation-defective cells into the population.
Fig. 7.
OME increases the sporulation fitness of heterogeneous populations. WT, ΔtraA, and wzm::km strains were assayed for sporulation at lower-than-optimal cell densities (3 × 108, 3 × 108, and 6 × 108 cfu/mL, respectively). The percentage of sporulation is shown relative to WT. Development proceeded on starvation agar for 72 h. When the wzm::km mutant and WT were mixed (2:1 ratio) in an equal volume, consequently increasing the cell density, sporulation efficiency increased (t test, *P = 0.035 relative to WT alone; n = 4). Conversely, when the wzm::km and ΔtraA mutants were similarly mixed, the increase in cell density did not increase sporulation efficiency.
Taken together these results indicate that, because tra+ cells rescued sporulation-deficient cells (Fig. 3), the resulting increase in the density of sporulation-proficient cells was advantageous to the total population. Because ∆traA cells were unable to rescue sporulation of wzm cells (Fig. 3), the increase in cell density did not increase the number of sporulation-competent cells and therefore did not change sporulation efficiency compared with ∆traA cells incubated alone (Fig. 7). This experiment suggests that OME can be critical for the survival of heterogeneous populations. That is, OME can mediate the repair of damaged individuals, thereby increasing the density of functional members to achieve the density necessary to meet a population threshold or quorum.
Discussion
Microbes in their natural habitats must adapt to complex and changing microenvironments (38) that often are unfavorable, resulting in growth delay, stress, and cell damage. As a result, solitary cells either will survive or perish. In contrast, social species have the potential to aid distressed siblings to create a more fit population. In line with this lifestyle, myxobacteria coordinate single cells to form communities with abilities that exceed those of the individual cells. These factors led us to investigate interactions between compromised and healthy populations. We found that myxobacteria can exploit the social behavior of OME to repair one another to build a more homogeneous and fit community. From these findings, we propose a model (Fig. 8) in which two heterogeneous groups merge; if these populations express identical traA alleles (15), they will recognize one another physically and catalyze OME. Hence, damage from one population will be diluted among a healthy population that is better suited to repair envelope damage. In addition, OME promotes the ability of a population to establish envelope homeostasis and to increase in size. Although damage is introduced into a healthy population, we have found that even a 2:1 ratio of damaged to healthy cells in merging populations can be beneficial to sporulation efficiency of the total population (Fig. 7). We propose that the ability to recognize and receive kin into a group while tolerating the addition of a damage load is a beneficial trait for social microbes in which population size and density correlate directly with fitness (Fig. 7) (34).
Fig. 8.
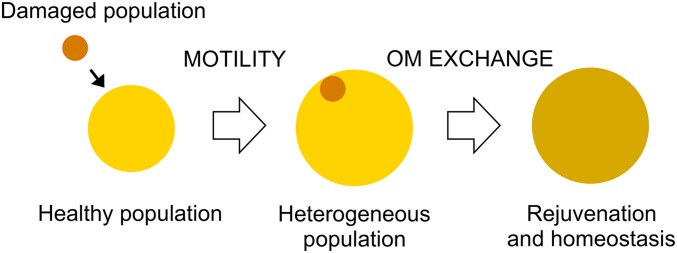
Model of OME repair of a damaged population and social consequences. In this model, a mixture of damaged and healthy cells creates a heterogeneous population. OME facilitates the dilution of OM damage among cells as the population moves toward homeostasis. The more homogenous population has increased in population size/critical mass, thus creating a more fit population for multicellular tasks.
In other biological systems, content mixing can lead to beneficial outcomes. We envision that our model (Fig. 8) is functionally analogous to the way mitochondrial fusion and fission cycling plays a critical role for the physiological health of the organelle. In this example, content mixing is important to phenotypically complement mitochondria that have sustained damage or deleterious mutations (20, 39). In fact, when mitochondria contain an error-prone DNA polymerase, abrogation of the machinery required for fusion and fission is lethal (40). This example highlights the benefit of sharing a collective membrane between individuals.
OME may provide other advantages in addition to the benefit of content mixing between populations. Gram-negative bacteria modify the chemical composition of their LPS armor in response to environmental insults (41). For example, the PmrA–PmrB systems of Escherichia coli and Pseudomonas aeruginosa enzymatically modify LPS composition to confer resistance to antimicrobials (42, 43). This resistance mechanism includes reducing the overall negative charge on LPS by covalently modifying phosphate atoms to endow resistance to certain cationic antimicrobials. Within soil microenvironments bacteria experience different stimuli that can trigger cellular responses, including adaptations to LPS composition. For instance, LPS in M. xanthus is modified during development by methylation of a galactosamine moiety (44). By extension of our findings, the ability of cells to distribute modified membrane lipids and proteins directly provides a rapid and efficient means for myxobacteria to adapt to their environment. In such a scenario “scout cells,” which have explored the surrounding terrain beyond the central collective, can relay information about how they adapted to their environment by exchanging OM molecules. Recipient cells thus would become preadapted to environmental stresses before the stresses are encountered. Although it is not well known how myxobacteria alter their OMs in response to stress, we did show through a surrogate experiment with LPS mutants that TraAB-mediated transfer can confer resistance to cells that otherwise would be susceptible to antibiotic stress (Fig. 6). We also note that antibiotic exposure is likely a common stress for myxobacteria, because they and other soil microbes produce a wide variety of such compounds (45).
OME provides a platform for myxobacteria to transfer cell OM content bidirectionally and to communicate. In this work we showed that these exchanges could lead to beneficial outcomes. However, as documented in other bacterial cell–cell transport systems, the transfer of cell cargo also can lead to detrimental outcomes to recipient cells, such as in toxin delivery (46–48). In a separate study, we indeed have found that OME can function to police or kill certain recipient cells. The finding of opposing outcomes for OME is not surprising, because hundreds of different proteins, lipids, glycans, and likely other classes of molecules are exchanged (13). Thus, depending on the genotype and physiology of the partnering cells, OME can be either beneficial or detrimental to cells. In eukaryotic tissue, the interplay between beneficial and antagonistic interactions allows multicellular organisms to improve fitness (49). Similarly, we hypothesize that OME facilitates myxobacteria to transition from solitary to multicellular life. Such a transition requires cooperative and policing functions.
Our findings extend the knowledge of OME. First, we have shown that lipids, proteins, lipoproteins, and now glycolipids (i.e., LPS) are exchangeable commodities between cells. Therefore, the TraAB pathway transfers all major OM constituents. Second, our microscopy studies have shown that TraAB overexpression leads to tight cell–cell adhesion junctions (Fig. 1D and Fig. S1), which overcome steric and repulsion forces created by bulky charged LPS molecules. Although myxobacteria have a peculiar ability to produce long filamentous OM tubes, which some have argued are conduits for exchange, we find no evidence that these structures are required for OM exchange (23). Instead, we suggest that TraAB, the only known determinants for OME (50), catalyze the formation of prefusion junctions that lead to OM fusion and exchange.
Experimental Procedures
Strains, Plasmids, and Media.
The bacterial strains, plasmids, and primers used in this study are listed in Table S1. M. xanthus was routinely grown in CTT medium [1% casitone; 10 mM Tris⋅HCl (pH 7.6); 8 mM MgSO4; 1 mM KPO4] in the dark at 33 °C. E. coli DH5α was grown in LB at 37 °C. As needed for selection or induction, medium was supplemented with kanamycin (50 µg/mL), oxytetracycline (10 µg/mL), vanillate (0.1 mM), or IPTG (0.1 mM). TPM buffer (CTT without casitone) was used to wash cells or for starvation-induced development experiments. CTT agar was used as a solid growth medium. For microscopy, 1% agarose pads were made on glass slides.
Insertion mutations were created by PCR amplification of ∼500 bp of the gene of interest, and ligation of the amplicon into the pCR2.1 TOPO TA vector (Life Technologies), followed by electroporation into DH5α and antibiotic selection. Constructs were confirmed by PCR and subsequently used to transform M. xanthus by electroporation. To create promoter-replacement strains in which the endogenous promoter was replaced with a heterologous inducible promoter, primers with engineered restriction sites were used to PCR amplify ∼500 bp of the 5′ end of the gene of interest, with the forward primer beginning at the start codon. Amplicons were ligated into pMR3690 containing a deletion of the MXAN_18-19 homologous region (pDP30) to direct homologous recombination to the chromosomal locus of interest. Constructs were electroporated into M. xanthus, and strains were confirmed by a vanillate-dependent growth phenotype. To create pPC1, the sfGFP DNA sequence, including an OM signal sequence, was synthesized (GenScript) with codon use optimized for expression in GC-rich organisms. This DNA then was subcloned into pXW6 (13), so that sfGFP expression was driven by PpilA.
Motility and Stimulation Assays.
Insertion mutations in LPS biosynthesis genes were created in an A-motility–defective, S-motile background (DK1217). Strains were plated on starvation agar (TPM, 1% agar, 2 mM CaCl2) to assess motility, because this condition was best for resolving S-motility phenotypes of LPS mutants while still supporting OME. DK1217 and mutants were grown in CTT medium to log phase, harvested, washed with TPM buffer, and adjusted to ∼3 × 108 cells/mL. A 5-μL aliquot of each suspension was plated on starvation agar. After a 72-h incubation at 33 °C, motility was assessed microscopically. In mixed-culture stimulation experiments, strains were grown as described and mixed at a 1:1 ratio with nonmotile (ΔmglBA) donors that contained either a WT traA allele or a traA insertion mutation.
Developmental Rescue.
DK1622, DK1622 wzm::km, and DK1622 ΔtraA were grown in CTT (plus antibiotic as needed) and harvested at log phase. Cells were washed and adjusted to 3 × 109 cells/mL. DK1622 wzm::km cells were mixed at a 1:1 ratio with either DK1622 or DK1622 ΔtraA cells, and 20 μL of each mixture, in addition to 20-µL aliquots of WT cells and wzm::km cells alone, were plated on TPM agar. After 72 h, fruiting bodies were harvested, and viable spores were assayed as described (51). Briefly, fruiting bodies were harvested and incubated at 50 °C for 2 h followed by sonication. A serial dilution of each spore preparation in TPM was plated on CTT agar or CTT agar with 50 µg/mL kanamycin and assessed microscopically for cfus after 3-d incubation. These and subsequent experiments were performed at least in triplicate, unless otherwise stated.
Viability Rescue Assays.
DW2404 was grown to log phase in CTT with 50 µg/mL kanamycin and with 0.1 mM vanillate (CTT kanamycin/vanillate). Cells were washed twice and adjusted to 3 × 108 cells/mL. The suspension was mixed at a 1:1 ratio with a suspension consisting of 3 × 108 cells/mL of isogenic donors (DK8615, DK8615 ΔtraA, or DK8601). Mixed cultures were plated as 20-μL droplets on 1/2 CTT agar (4 mM MgSO4). At the time points described, cells were harvested, and serial dilutions were performed in a suspension of 6 × 108 cells/mL of a companion strain (DK8615) to improve plating efficiency. The dilutions were plated on CTT kanamycin/vanillate to kill the donor and companion strains and support growth of the Pvan::lpxC strain. Colonies were counted under a dissecting microscope. Identity of the Pvan::lpxC strain was confirmed by their nonmotile colony morphology. The fluorescence rescue assays were performed similarly. Cells were observed microscopically through a phase-contrast 20× lens and Texas Red filter set when needed, as described (13).
Rescue of Polymyxin B Hypersensitivity.
DK8601 Pvan::lpxC was grown to log phase in CTT kanamycin/vanillate. Cells were washed twice and adjusted to 3 × 109 cells/mL. The suspension (100 μL) was mixed with 100 μL of the same concentration of donor cells, and the mixture was spread on CTT agar. After a 6-h incubation at 33 °C, cells were harvested, centrifuged, and resuspended in 1 mL TPM buffer. The suspension was split into two equal volumes, centrifuged, and resuspended in 1 mL CTT or CTT with 50 μg/mL polymyxin B (Calbiochem). Cells were incubated at 33 °C with shaking for 1 h. Each sample was centrifuged and washed three times, serial dilutions were plated, and cfus were counted as described above. Data are presented as the number of cfus after culturing with CTT containing 50 μg/mL polymyxin B divided by the number of cfus when cultured with plain CTT.
Density-Dependent Sporulation Fitness Assay.
DK1622, DW1480, and DW2407 were grown to log phase, harvested, and adjusted to 3 × 108, 3 × 108, and 6 × 108 cells/mL, respectively. Mixtures of 2:1 DW2407/DK1622 or DW2407/DW1480 were prepared at a final concentration of 9 × 108 cells/mL. A 20-µL aliquot of each of the five preparations (including the individual strains) was plated onto TPM agar. Development and spore quantification was carried out in quadruplicate as described above.
TEM.
To image whole cells, cultures were grown to midlog phase in CTT medium. Glow-discharged, carbon-coated gold grids were floated on a drop of culture medium for 2 min. The grids then were rinsed twice with water and stained for 1 min with 2% uranyl acetate. For embedding and thin sectioning, a liquid culture at midlog phase was plated on modified CTT agar (buffered with 10 mM Hepes, pH 8.0, to replace Tris) and grown overnight at 32 °C. The cells were fixed by gently covering the plate with 0.5% glutaraldehyde and incubating at room temperature for 45 min. Cells were collected by centrifugation, and the cell pellet was washed three times with 20 mM Hepes buffer (pH 7.5), cryo-fixated by high-pressure freezing using a Leica EM-PACT2 system, and freeze-substituted according to standard protocols (52). Briefly, the frozen cells were kept for 80 h in pure acetone containing 2% osmium tetroxide at −87 °C, warmed stepwise (10 °C/h) to room temperature, and washed with acetone to remove excess fixative. Samples were infiltrated with Epon before sectioning, and thin sections were mounted on 200-mesh copper grids and stained with uranyl acetate and lead citrate (53). For all samples, images were recorded using an XR-80 CCD camera from Advanced Microscopy Techniques and the AMTV602 Image Capture Engine software at nominal magnifications ranging from 11,000× to 135,000×.
SEM.
Cultures were grown to midlog phase in liquid culture and collected by centrifugation. Cells were fixed for 90 min at room temperature with 2.5% glutaraldehyde in PM buffer (20 mM NaPO4, 1 mM MgSO4, pH 7.4), washed with PM buffer, and adhered to a glass coverslip. The sample then was fixed with 1% osmium tetroxide in PM buffer, washed, and stained with 2% uranyl acetate. Cells were dehydrated with ethanol, sputter coated with a 20-nm layer of Au/Pd, and imaged using a Leo Gemini 1530 field-emission SEM at 1 kV as described (54).
Supplementary Material
Acknowledgments
We thank Montse Elías-Arnanz and Larry Shimkets for plasmids. This work was supported by NIH Grants GM101449 (to D.W.), GM85024 (to E.H.), and T32AI007417 (in support of D.M.Z.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1503553112/-/DCSupplemental.
References
- 1.Ramos JL, et al. Responses of Gram-negative bacteria to certain environmental stressors. Curr Opin Microbiol. 2001;4(2):166–171. doi: 10.1016/s1369-5274(00)00183-1. [DOI] [PubMed] [Google Scholar]
- 2.Russell AD. Bacterial outer membrane and cell wall penetration and cell destruction by polluting chemical agents and physical conditions. Sci Prog. 2003;86(Pt 4):283–311. doi: 10.3184/003685003783238608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Matin A. The molecular basis of carbon-starvation-induced general resistance in Escherichia coli. Mol Microbiol. 1991;5(1):3–10. doi: 10.1111/j.1365-2958.1991.tb01819.x. [DOI] [PubMed] [Google Scholar]
- 4.Desnues B, et al. Differential oxidative damage and expression of stress defence regulons in culturable and non-culturable Escherichia coli cells. EMBO Rep. 2003;4(4):400–404. doi: 10.1038/sj.embor.embor799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nyström T. Role of oxidative carbonylation in protein quality control and senescence. EMBO J. 2005;24(7):1311–1317. doi: 10.1038/sj.emboj.7600599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Visick JE, Clarke S. Repair, refold, recycle: How bacteria can deal with spontaneous and environmental damage to proteins. Mol Microbiol. 1995;16(5):835–845. doi: 10.1111/j.1365-2958.1995.tb02311.x. [DOI] [PubMed] [Google Scholar]
- 7.Hamilton WD. The genetical evolution of social behaviour. I. J Theor Biol. 1964;7(1):1–16. doi: 10.1016/0022-5193(64)90038-4. [DOI] [PubMed] [Google Scholar]
- 8.Ranjard L, Richaume A. Quantitative and qualitative microscale distribution of bacteria in soil. Res Microbiol. 2001;152(8):707–716. doi: 10.1016/s0923-2508(01)01251-7. [DOI] [PubMed] [Google Scholar]
- 9.Saint-Ruf C, et al. Massive diversification in aging colonies of Escherichia coli. J Bacteriol. 2014;196(17):3059–3073. doi: 10.1128/JB.01421-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reichenbach H. The ecology of the myxobacteria. Environ Microbiol. 1999;1(1):15–21. doi: 10.1046/j.1462-2920.1999.00016.x. [DOI] [PubMed] [Google Scholar]
- 11.Pathak DT, Wei X, Wall D. Myxobacterial tools for social interactions. Res Microbiol. 2012;163(9-10):579–591. doi: 10.1016/j.resmic.2012.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Velicer GJ, Vos M. Sociobiology of the myxobacteria. Annu Rev Microbiol. 2009;63:599–623. doi: 10.1146/annurev.micro.091208.073158. [DOI] [PubMed] [Google Scholar]
- 13.Wei X, Pathak DT, Wall D. Heterologous protein transfer within structured myxobacteria biofilms. Mol Microbiol. 2011;81(2):315–326. doi: 10.1111/j.1365-2958.2011.07710.x. [DOI] [PubMed] [Google Scholar]
- 14.Pathak DT, et al. Cell contact-dependent outer membrane exchange in myxobacteria: Genetic determinants and mechanism. PLoS Genet. 2012;8(4):e1002626. doi: 10.1371/journal.pgen.1002626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pathak DT, Wei X, Dey A, Wall D. Molecular recognition by a polymorphic cell surface receptor governs cooperative behaviors in bacteria. PLoS Genet. 2013;9(11):e1003891. doi: 10.1371/journal.pgen.1003891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wall D. Molecular recognition in myxobacterial outer membrane exchange: Functional, social and evolutionary implications. Mol Microbiol. 2014;91(2):209–220. doi: 10.1111/mmi.12450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bowden MG, Kaplan HB. The Myxococcus xanthus lipopolysaccharide O-antigen is required for social motility and multicellular development. Mol Microbiol. 1998;30(2):275–284. doi: 10.1046/j.1365-2958.1998.01060.x. [DOI] [PubMed] [Google Scholar]
- 18.Wall D, Kaiser D. Type IV pili and cell motility. Mol Microbiol. 1999;32(1):1–10. doi: 10.1046/j.1365-2958.1999.01339.x. [DOI] [PubMed] [Google Scholar]
- 19.Nan B, McBride MJ, Chen J, Zusman DR, Oster G. Bacteria that glide with helical tracks. Curr Biol. 2014;24(4):R169–R173. doi: 10.1016/j.cub.2013.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chan DC. Fusion and fission: Interlinked processes critical for mitochondrial health. Annu Rev Genet. 2012;46:265–287. doi: 10.1146/annurev-genet-110410-132529. [DOI] [PubMed] [Google Scholar]
- 21.Wu SS, Wu J, Kaiser D. The Myxococcus xanthus pilT locus is required for social gliding motility although pili are still produced. Mol Microbiol. 1997;23(1):109–121. doi: 10.1046/j.1365-2958.1997.1791550.x. [DOI] [PubMed] [Google Scholar]
- 22.Wall D, Kaiser D. Alignment enhances the cell-to-cell transfer of pilus phenotype. Proc Natl Acad Sci USA. 1998;95(6):3054–3058. doi: 10.1073/pnas.95.6.3054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wei X, Vassallo CN, Pathak DT, Wall D. Myxobacteria produce outer membrane-enclosed tubes in unstructured environments. J Bacteriol. 2014;196(10):1807–1814. doi: 10.1128/JB.00850-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nudleman E, Wall D, Kaiser D. Cell-to-cell transfer of bacterial outer membrane lipoproteins. Science. 2005;309(5731):125–127. doi: 10.1126/science.1112440. [DOI] [PubMed] [Google Scholar]
- 25.Lancero H, et al. Mapping of Myxococcus xanthus social motility dsp mutations to the dif genes. J Bacteriol. 2002;184(5):1462–1465. doi: 10.1128/JB.184.5.1462-1465.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang Z, et al. Myxococcus xanthus dif genes are required for biogenesis of cell surface fibrils essential for social gliding motility. J Bacteriol. 2000;182(20):5793–5798. doi: 10.1128/jb.182.20.5793-5798.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Young K, et al. The envA permeability/cell division gene of Escherichia coli encodes the second enzyme of lipid A biosynthesis. UDP-3-O-(R-3-hydroxymyristoyl)-N-acetylglucosamine deacetylase. J Biol Chem. 1995;270(51):30384–30391. doi: 10.1074/jbc.270.51.30384. [DOI] [PubMed] [Google Scholar]
- 28.Beall B, Lutkenhaus J. Sequence analysis, transcriptional organization, and insertional mutagenesis of the envA gene of Escherichia coli. J Bacteriol. 1987;169(12):5408–5415. doi: 10.1128/jb.169.12.5408-5415.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Iniesta AA, García-Heras F, Abellón-Ruiz J, Gallego-García A, Elías-Arnanz M. Two systems for conditional gene expression in Myxococcus xanthus inducible by isopropyl-β-D-thiogalactopyranoside or vanillate. J Bacteriol. 2012;194(21):5875–5885. doi: 10.1128/JB.01110-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pelling AE, Li Y, Shi W, Gimzewski JK. Nanoscale visualization and characterization of Myxococcus xanthus cells with atomic force microscopy. Proc Natl Acad Sci USA. 2005;102(18):6484–6489. doi: 10.1073/pnas.0501207102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nikaido H. Molecular basis of bacterial outer membrane permeability revisited. Microbiol Mol Biol Rev. 2003;67(4):593–656. doi: 10.1128/MMBR.67.4.593-656.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morrison DC, Jacobs DM. Binding of polymyxin B to the lipid A portion of bacterial lipopolysaccharides. Immunochemistry. 1976;13(10):813–818. doi: 10.1016/0019-2791(76)90181-6. [DOI] [PubMed] [Google Scholar]
- 33.Vaara M, Vaara T. Polycations as outer membrane-disorganizing agents. Antimicrob Agents Chemother. 1983;24(1):114–122. doi: 10.1128/aac.24.1.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kadam SV, Velicer GJ. Variable patterns of density-dependent survival in social bacteria. Behav Ecol. 2006;17(5):833–838. [Google Scholar]
- 35.Wireman JW, Dworkin M. Morphogenesis and developmental interactions in myxobacteria. Science. 1975;189(4202):516–523. doi: 10.1126/science.806967. [DOI] [PubMed] [Google Scholar]
- 36.Kaplan HB, Plamann L. A Myxococcus xanthus cell density-sensing system required for multicellular development. FEMS Microbiol Lett. 1996;139(2-3):89–95. doi: 10.1111/j.1574-6968.1996.tb08185.x. [DOI] [PubMed] [Google Scholar]
- 37.Kuspa A, Plamann L, Kaiser D. A-signalling and the cell density requirement for Myxococcus xanthus development. J Bacteriol. 1992;174(22):7360–7369. doi: 10.1128/jb.174.22.7360-7369.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Blackburn N, Fenchel T. Influence of bacteria, diffusion and sheer on micro-scale nutrient patches, and implications for bacterial chemotaxis. Mar Ecol Prog Ser. 1999;189:1–7. [Google Scholar]
- 39.Youle RJ, van der Bliek AM. Mitochondrial fission, fusion, and stress. Science. 2012;337(6098):1062–1065. doi: 10.1126/science.1219855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen H, et al. Mitochondrial fusion is required for mtDNA stability in skeletal muscle and tolerance of mtDNA mutations. Cell. 2010;141(2):280–289. doi: 10.1016/j.cell.2010.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Raetz CR, Reynolds CM, Trent MS, Bishop RE. Lipid A modification systems in gram-negative bacteria. Annu Rev Biochem. 2007;76:295–329. doi: 10.1146/annurev.biochem.76.010307.145803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McPhee JB, Lewenza S, Hancock RE. Cationic antimicrobial peptides activate a two-component regulatory system, PmrA-PmrB, that regulates resistance to polymyxin B and cationic antimicrobial peptides in Pseudomonas aeruginosa. Mol Microbiol. 2003;50(1):205–217. doi: 10.1046/j.1365-2958.2003.03673.x. [DOI] [PubMed] [Google Scholar]
- 43.Froelich JM, Tran K, Wall D. A pmrA constitutive mutant sensitizes Escherichia coli to deoxycholic acid. J Bacteriol. 2006;188(3):1180–1183. doi: 10.1128/JB.188.3.1180-1183.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Panasenko SM, Jann B, Jann K. Novel change in the carbohydrate portion of Myxococcus xanthus lipopolysaccharide during development. J Bacteriol. 1989;171(4):1835–1840. doi: 10.1128/jb.171.4.1835-1840.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wenzel SC, Müller R. Myxobacteria—‘microbial factories’ for the production of bioactive secondary metabolites. Mol Biosyst. 2009;5(6):567–574. doi: 10.1039/b901287g. [DOI] [PubMed] [Google Scholar]
- 46.Cascales E, Christie PJ. The versatile bacterial type IV secretion systems. Nat Rev Microbiol. 2003;1(2):137–149. doi: 10.1038/nrmicro753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Konovalova A, Petters T, Søgaard-Andersen L. Extracellular biology of Myxococcus xanthus. FEMS Microbiol Rev. 2010;34(2):89–106. doi: 10.1111/j.1574-6976.2009.00194.x. [DOI] [PubMed] [Google Scholar]
- 48.Hayes CS, et al. 2014. Mechanisms and biological roles of contact-dependent growth inhibition systems. Cold Spring Harb Perspect Med 4(2):1–12.
- 49.Morata G, Ballesteros-Arias L. Developmental biology. Death to the losers. Science. 2014;346(6214):1181–1182. doi: 10.1126/science.aaa2345. [DOI] [PubMed] [Google Scholar]
- 50.Dey A, Wall D. A genetic screen in Myxococcus xanthus identifies mutants that uncouple outer membrane exchange from a downstream cellular response. J Bacteriol. 2014;196(24):4324–4332. doi: 10.1128/JB.02217-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kuspa A, Kaiser D. Genes required for developmental signalling in Myxococcus xanthus: Three asg loci. J Bacteriol. 1989;171(5):2762–2772. doi: 10.1128/jb.171.5.2762-2772.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hoiczyk E, Baumeister W. Envelope structure of four gliding filamentous cyanobacteria. J Bacteriol. 1995;177(9):2387–2395. doi: 10.1128/jb.177.9.2387-2395.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Reynolds ES. The use of lead citrate at high pH as an electron-opaque stain in electron microscopy. J Cell Biol. 1963;17:208–212. doi: 10.1083/jcb.17.1.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Koch MK, McHugh CA, Hoiczyk E. BacM, an N-terminally processed bactofilin of Myxococcus xanthus, is crucial for proper cell shape. Mol Microbiol. 2011;80(4):1031–1051. doi: 10.1111/j.1365-2958.2011.07629.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.