Abstract
Long noncoding RNAs (lncRNAs) have been implicated in numerous cellular processes including brain development. However, the in vivo expression dynamics and molecular pathways regulated by these loci are not well understood. Here, we leveraged a cohort of 13 lncRNA-null mutant mouse models to investigate the spatiotemporal expression of lncRNAs in the developing and adult brain and the transcriptome alterations resulting from the loss of these lncRNA loci. We show that several lncRNAs are differentially expressed both in time and space, with some presenting highly restricted expression in only selected brain regions. We further demonstrate altered regulation of genes for a large variety of cellular pathways and processes upon deletion of the lncRNA loci. Finally, we found that 4 of the 13 lncRNAs significantly affect the expression of several neighboring protein-coding genes in a cis-like manner. By providing insight into the endogenous expression patterns and the transcriptional perturbations caused by deletion of the lncRNA locus in the developing and postnatal mammalian brain, these data provide a resource to facilitate future examination of the specific functional relevance of these genes in neural development, brain function, and disease.
Keywords: long noncoding RNA, mouse, brain, development, in vivo
The exquisite complexity of the mammalian brain derives from its vast diversity of neuronal and glial cell types (1, 2). The specification and differentiation of such a variety of cell types during brain development is finely orchestrated spatiotemporally by the regulation of complex transcriptional programs. Increasing evidence points to a role for long noncoding RNAs (lncRNAs) as key regulatory elements of this process. Intriguingly, within the mammalian body, the largest repertoire and diversity of lncRNA genes outside the germ line occurs in the brain (3–10), where lncRNAs exhibit regional and cell-specific localization (6, 10). Although many unanswered questions remain regarding the functional activity and molecular mechanisms of lncRNA loci, the expression patterns of lncRNAs may serve as a proxy signal for important, context-specific biological activity.
A role for lncRNA genes in brain development and function is supported by the fact that ablation of two lncRNA loci, Evf2 and Pantr2 (linc-Brn1b), perturbs neuronal development (11, 12). Loss of Evf2, a developmentally regulated lncRNA that controls transcriptional activity through cooperation with the homeodomain protein DLX-2 (11), leads to abnormal development and synaptic function of hippocampal GABAergic interneurons (11). Similarly, ablation of the Pantr2 locus results in a decreased number of intermediate progenitors in the developing telencephalon, reduced neurons in L2/3 of the cerebral cortex, and disorganization of the barrel cortex (12). Furthermore, human genetic studies have pointed to lncRNAs as potential factors in brain disorders (10, 13–16).
To gain preliminary insights into the functional and physiological relevance of lncRNA loci in vivo, we previously generated knockout (KO) mouse models of 18 lncRNA loci by replacing each gene with a lacZ reporter cassette (12). Here we provide a map of the expression dynamics and regulatory effects of 13 lncRNA loci in the developing and mature brain (Table S1). These 13 strains were selected based on expression in embryonic stem (ES) cell-derived neural stem cells (17) and in brain RNA-seq datasets (12). Toward this goal, we first used the knocked-in lacZ reporter gene to determine the spatiotemporal expression profiles of these lncRNAs in the brain. We then performed massively parallel RNA sequencing of embryonic and adult whole brains from wildtype (WT) and KO strains to gain insights into the global transcriptional programs perturbed upon ablation of these lncRNA loci in vivo.
We find that lncRNAs exhibit a dynamic and wide range of expression profiles in the brain, with notable lncRNAs restricted to unique brain regions and cell types. The combined lncRNA expression profiles and gene KO transcriptional profiles generated through this work offer a resource to facilitate in-depth investigation of loci annotated as putative lncRNAs in brain development and physiology.
Results
Embryonic and Adult Expression of lncRNAs in the Brain.
A fundamental aspect of evaluating the functional relevance of a genomic locus is to determine when, and in what cellular contexts, it is active. We focused on 13 lncRNA mutant mouse models with evidence of expression in neural stem cells and in the brain (Table S1) (12). We set out to identify and describe the temporal expression of each lncRNA during development at embryonic day (E) 14.5 and in the adult mouse brain (age range 7.6–14.1 wk). E14.5 corresponds to a stage when key brain developmental events are taking place; neurogenesis already has reached its peak in both the nascent diencephalon and the telencephalon (2, 18, 19). Conversely, by 7–8 weeks of age, all major postnatal developmental milestones have been achieved, including myelination, and the mouse brain can be considered mature (20, 21).
The incorporation of a lacZ reporter in the targeting constructs of each lncRNA locus KO mouse strain allowed us to determine the precise location of expression of the lncRNA genes using heterozygote mice. We collected whole brains at E14.5 and adult time points for each of the 13 lncRNA mutant strains (Dataset S1). LncRNA expression patterns were assessed by selecting coronal sections collected every 80 µm (E14.5) or 240 µm (adult) and by staining to detect β-galactosidase (β-gal) activity. A series of rostro–caudal images spanning the entire length of the brains from these strains were also collected and made available as Dataset S2.
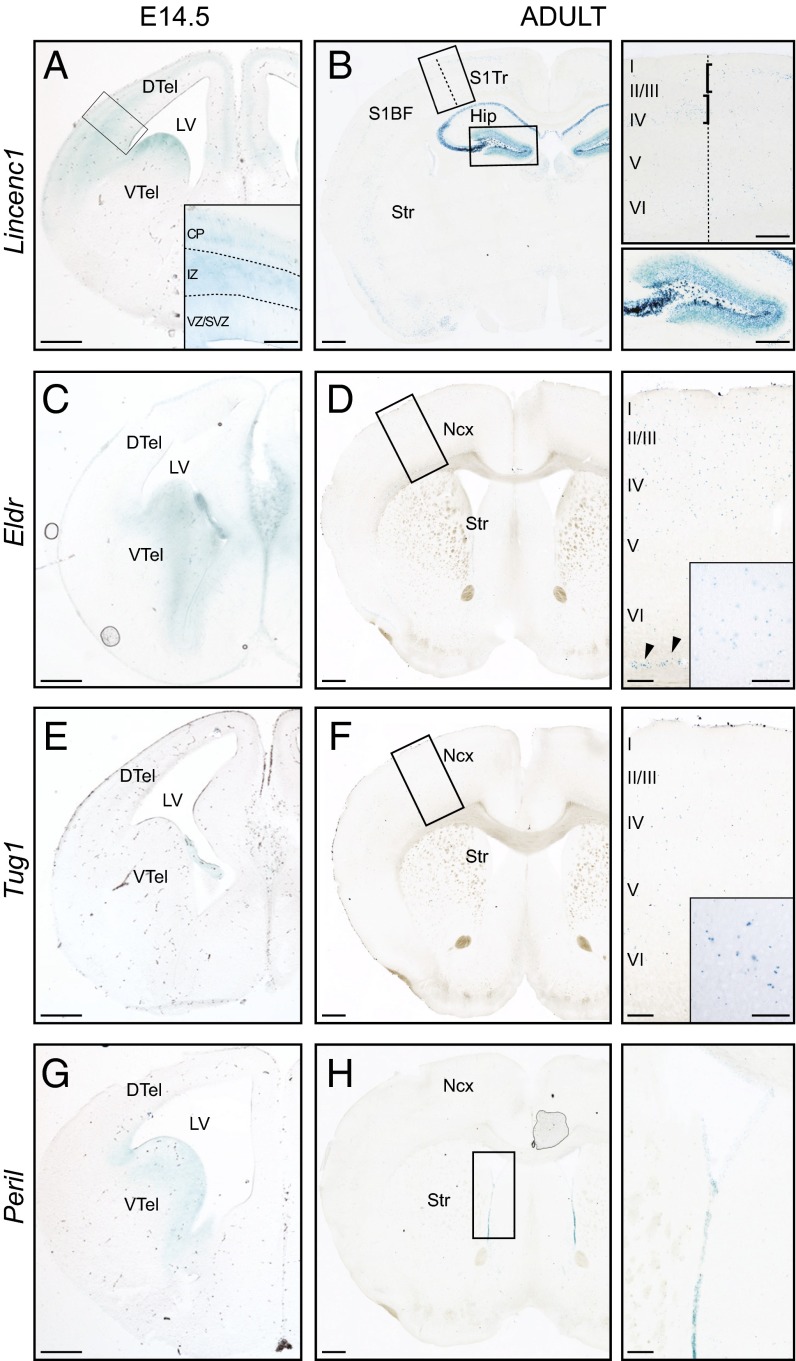
Of the 13 lncRNA strains analyzed by these methods, 10 showed clear β-gal signal in the adult brain. Consistent with our previous RNA-seq analysis of WT brains and ES cell-derived neuronal stem cells (12, 17), little or no β-gal expression was observed for Mannr, Halr1, and Trp53cor1 in the adult brain. Among those lncRNA loci with detectable β-gal signal in the adult brain, β-gal activity was also detected at E14.5 for the Lincenc1, Eldr, Pantr1, Pantr2 (12), and Peril loci. Several of these lncRNA genes demonstrated embryonic expression in regions known to give rise to the corresponding β-gal+ cell populations seen in the adult. At E14.5, Lincenc1 is expressed in the ventricular zone of the ventral telencephalon (VTel), but not of the dorsal telencephalon (DTel) (Fig. 1A). In the DTel, Lincenc1 is expressed in the intermediate zone (IZ) and lateral cortical plate (CP) (Fig. 1A, Inset). At this developmental stage, the lateral CP contains postmitotic neurons destined for the upper layers of the neocortex (22).
Fig. 1.
LncRNAs are temporally regulated in the developing and adult brain. β-Gal staining of lncRNA heterozygous mutant brain coronal sections at E14.5 (A, C, E, and G) and adult (B, D, F, and H) stages. (A) Lincenc1 is expressed in the VTel and in the IZ and lateral CP (Inset) of the DTel at E14.5. (B, Upper Right) In the adult brain, Lincenc1 is expressed in the primary somatosensory cortex, more specifically in layers II/III of the S1 S1Tr and layer IV in the S1BF. (Lower Right) β-Gal staining also shows expression in the hippocampus (Left) and the dentate gyrus. (C and D) At E14.5, Eldr is expressed in the VTel (C), but in the adult brain it is expressed in scattered cells in the upper layers of the neocortex as well as in layer VI cells lining the subcortical white matter (arrowheads) (D, Left, Right, and Inset). (E) No β-gal staining could be detected in Tug1+/− brains at E14.5, except in the choroid plexus. (F, Left, Right, and Inset) In adult brains, Tug1 expression was detected in scattered cells in the neocortex. (G and H) β-Gal staining indicates expression of Peril in the VTel at E14.5 (G) and in the ependymal zone of the ventricles in the adult brain (H, Right and Inset). Images are representative of staining performed on two animals. CP, cortical plate; DG, dentate gyrus; DTel, dorsal telencephalon; Hip, hippocampus; IZ, intermediate zone; LV, lateral ventricle; Ncx, neocortex; S1BF, primary somatosensory cortex barrel field; S1Tr, primary somatosensory of the trunk; Str, striatum; VTel, ventral telencephalon; VZ/SVZ, ventricular zone/subventricular zone. [Scale bars: 500 μm (A–H, lower magnification panels), 100 μm (B, D, F, H, higher magnification right panels and inset).]
The expression pattern of Lincenc1 at E14.5 is developmentally consistent with its expression in specific structures in the adult brain. Specifically, in the adult, Lincenc1 is expressed at high levels in the upper cortical layers II/III and IV (Fig. 1B), with highest number of labeled cells distributed in the primary somatosensory (S1) cortex. In this region, expression respects specific laminar and areal boundaries (Fig. 1 A and B). For example, Lincenc1 is restricted to layers II/III in the S1 region (representing sensory input from the trunk; S1Tr, Fig. 1B), whereas its distribution is restricted to layer IV within the S1 barrel field (S1BF, Fig. 1B). Outside this region, Lincenc1 is expressed in the CA1, CA2, and CA3 fields of the hippocampus and in the dentate gyrus, both in the granular and molecular cell layers and the hilus (Fig. 1B). Together these findings describe a lncRNA locus with an intricate expression pattern that is established during development and is maintained in corresponding structures of the adult brain.
Eldr is another lncRNA demonstrating correlated expression at E14.5 and in the adult brain. In the E14.5 embryonic brain, Eldr is expressed in the germinal and mantle zones of the VTel (Fig. 1C). In the adult brain, Eldr+ cells are found scattered throughout the neocortex in a pattern resembling that of cortical interneurons, which are developmentally generated in the VTel (Fig. 1D) (23, 24). In addition, Eldr is expressed in a specific subgroup of cortical layer VI cells lining the subcortical white matter (arrowheads in Fig. 1D, Inset). Lineage-fate mapping and combinatorial marker analysis will be required in the future to support a direct lineage relationship between Eldr+ cells in the embryonic and the adult brain.
Like Eldr, Tug1 expression was observed in scattered cells within the neocortex of the adult brain (Fig. 1F). However, unlike Eldr, in the embryonic E14.5 brain, Tug1 is expressed, albeit at low levels, only in the developing choroid plexus with no expression detected in the VTel.
The lncRNA Peril shows distinct expression in both the developing and adult brain (Fig. 1 G and H). At E14.5, Peril is expressed only in the VTel (Fig. 1G). In the adult brain, Peril is expressed primarily in the ependymal lining of the lateral ventricle, a region associated with adult neurogenesis (Fig. 1H) (25, 26). Expression of Peril was also detected in the olfactory bulb and dentate gyrus, albeit at lower intensity (Dataset S2). Interestingly, Peril is located ∼110 kb downstream of the protein-coding gene Sox2, a known neuronal stem cell regulator which is also expressed in similar regions of the brain (27), suggesting the possible involvement of Peril in the regulation of Sox2 transcription. These results highlight a few specific examples of lncRNAs whose expression maps to specific brain regions through development and into adulthood and that may be poised to exert functional roles for the establishment and function of these structures.
Regional Specificity of lncRNA Expression in the Adult Brain.
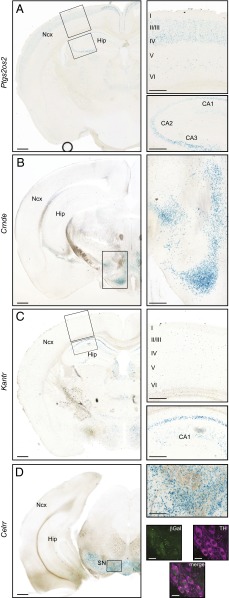
The majority of the lncRNAs in this study demonstrate some level of expression in the adult brain. Several of the lncRNAs analyzed showed restricted patterns of expression within multiple brain structures in the forebrain, midbrain, and hindbrain. Specific examples include the lncRNAs Ptgs2os2, Crnde, Kantr, and Celrr (Fig. 2).
Fig. 2.
LncRNAs are expressed in distinct and specific regions in the adult brain. (A) β-gal staining of a coronal section from Ptgs2os2+/− adult brain shows expression in layers II/III and IV of the neocortex (Ncx) and in CA1, CA2 and CA3 regions of the hippocampus. (B) β-gal staining shows Crnde is expressed in the hypothalamus and the thalamus. (C) Kantr is expressed mostly in scattered cells in the neocortex (Upper Right Inset) and in the CA1 of the hippocampus (Lower Right Inset). (D) Celrr is expressed at high levels in the substantia nigra, and β-gal (green) colocalizes with TH+ (magenta) dopaminergic neurons. Images are representative of staining performed on two animals. β-gal, β-galactosidase; CA, cornu ammonis; Hip, hippocampus; Ncx, neocortex; SN, substantia nigra; TH, tyrosine hydroxylase. [Scale bars: 500 μm (A–D, lower magnification left panels), 200 μm (A–D, higher magnification right panels), 20 μm (D, b-gal/TH-labeled confocal images).]
In the adult brain, Ptgs2os2, an immune response-activated lncRNA located proximal to the Prostaglandin-endoperoxide synthase 2 gene (Ptgs2) (17, 28), is expressed in the neocortex, hippocampus, and thalamus (Fig. 2A and Dataset S2). In the cortex, its expression profile resembles the distribution of excitatory pyramidal neurons in layers II/III and IV (Fig. 2A, Inset) (2). In the hippocampus, Ptgs2os2 is expressed in the CA1, CA2, and CA3 stratum pyramidale (Fig. 2A).
Similarly, multiple regions of the adult brain express Crnde. This lncRNA, which shares a bidirectional promoter with the Iroquois homeobox gene Irx5, is expressed in distinct nuclei of the hypothalamus (Fig. 2B, Inset). It is also weakly expressed in the adult ependymal layer of the lateral and third ventricles and in the habenula (Dataset S2).
We observed a distinct expression pattern in the adult brain for Kantr, a lncRNA located ∼20 kb downstream of the histone demethylase gene Kdm5c on the X chromosome. We find that Kantr is expressed in the adult neocortex, hippocampus, thalamus, and hypothalamus (Fig. 2C and Dataset S2). Within these regions, Kantr expression is spatially restricted to defined subregions. For example, in the hippocampus it is expressed largely in the CA1 stratum pyramidale (Fig. 2C, Inset), a region containing excitatory pyramidal neurons of different physiological and molecular identities (29–31).
Finally, we found that the lncRNA Celrr, located proximal to the Insulin-induced gene 2 (Insig2), is specifically expressed in the adult substantia nigra, where it colocalizes with tyrosine hydroxylase-positive (TH+) dopaminergic neurons (Fig. 2D).
Collectively, these findings indicate that lncRNAs can have very precise and restricted spatial expression in key structures of the adult brain. This broad catalog of brain images of lncRNA gene expression provides a unique resource to facilitate future investigation.
Gene-Expression Perturbations in Mutant Brains.
We next asked whether the deletion of these specific lncRNA genomic loci, while maintaining the act of transcription, contributed significantly to changes in gene expression. To address this question, whole brains from a minimum of three homozygous KO mice per strain were collected at both E14.5 and adult stages (age range 7.9–13.6 wk) (Dataset S3). Whole brains from WT E14.5 littermates and adults (age range 7.9–14 ± 2.0 wk) were also collected as control. To avoid potential sources of bias in sample collection and preparation, KO and WT brains were selected for each strain to maintain a balanced experimental design with respect to sex, litter (E14.5 only), and genetic background (Dataset S3). RNA was isolated from each sample as detailed in Materials and Methods and was used as input to generate Illumina TruSeq libraries.
Libraries were indexed and sequenced on the Illumina HiSeq 2500 platform across multiple flow cells, generating a total of 1.385 × 1010 50-bp paired-end reads at a median depth of 1.18 × 108 aligned reads per sample (Dataset S4). Reads were mapped and quantified as described in Materials and Methods and SI Materials and Methods. For quantification, a modified version of the GENCODE (vM2) transcriptome (Dataset S5) was used as a reference. For quality control, a preliminary expression analysis was conducted on all samples using Cuffnorm.
Through a combination of expression analysis and visual inspection of the RNA-seq read alignments to the presumptive deleted regions, we confirmed the genotype and sex of each lncRNA KO and WT sample (Dataset S6). Following this “digital genotyping” via lacZ expression, the 109 remaining samples passing our quality control were subjected to differential analysis using Cuffdiff2 (v2.2.1).
We first conducted a global analysis across all strains. Differential transcriptional regulatory events were aggregated from all pairwise lncRNA KO versus WT comparisons across all strains at E14.5 or adult time points. For both time points, we observed exceptionally low variability across all conditions with a moderate increase in cross-replicate variability at E14.5 (Fig. S1). To identify lncRNA KO strains with similar differential expression profiles, hierarchical clustering of either E14.5 or adult samples was performed using the Jensen–Shannon distance across the normalized expression profiles for the universe of differentially expressed genes in all pairwise comparisons. We observed a greater total number of differentially expressed genes at E14.5 (n = 2,389) than in the adult comparisons (n = 669) (Fig. S2A). Similar results were observed for differential isoforms and protein-coding sequences (CDS) (Fig. S2A). In adults, most lncRNA mutant strains demonstrated some significant transcriptional regulatory events, but the total number of differentially expressed genes, isoforms, and CDS was lower than observed at E14.5 for individual strains (Fig. S2A). Approximately 10% of the significant genes (n = 214), isoforms (n = 124), and CDS (n = 192) identified at E14.5 were also differentially expressed in the adult comparisons (Fig. S2B).
Next we globally assessed the gene pathways perturbed upon deletion of lncRNA loci in vivo. Gene ontology (GO) enrichment analysis was performed for the superset of differentially expressed genes at both E14.5 and adult stages. We observed a significant enrichment of gene sets associated with neuronal differentiation and cell fate commitment at E14.5 alongside gene sets associated with cell-cycle regulation (Fig. S2C). This finding is congruous with the observed expression of several lncRNAs in various germinal zones of the developing brain (Fig. 1 A, C, and G). In adult mice, the top significant GO gene sets included positive and negative regulators of cell differentiation and gene sets associated with PDGF and MAPK signaling pathways (Fig. S2D).
We further investigated the individual lncRNA mutant strains to determine which specific gene-expression pathways and programs are perturbed between WT and KO mice. We conducted differential expression analysis on individual KO versus WT comparisons for each strain at both E14.5 and adult time points. In these analyses, pathway and gene set enrichment studies were conducted using publicly available tools and annotated gene sets (detailed in SI Materials and Methods and Dataset S7).
One of the most abundant lncRNAs within this cohort is Kantr. Deletion of the Kantr locus results in a significant increase in gene sets involved in extracellular matrix organization, Ncam1 and L1cam interactions, NOTCH signaling, NGF and PDGF signaling, axon guidance, and neuronal development in the E14.5 brain (Fig. S3 A and B). We also observed significant enrichment for gene sets associated with neurodegeneration (oxidative phosphorylation, Alzheimer’s, Huntington’s, and Parkinson’s disease) (Fig. S3B). In the adult mouse brain, deletion of the Kantr locus resulted in a significant increase in gene sets associated with axon guidance and PDGF signaling.
In E14.5 Crnde−/− mouse brains, we observed significant down-regulation of genes associated with cell-cycle progression and regulation (Fig. S3 C and D). We also observed a significant down-regulation of genes with predicted binding sites in their upstream regulatory regions for the E2F family of transcription factors, known to influence cell-cycle progression (Dataset S6) (32). In adult Crnde−/− mouse brains, cell-cycle genes are no longer differentially expressed. However, an oncogenic signature gene set (MSigDB C6) that is up-regulated in cancers with overexpressed E2F3 remained significantly down-regulated relative to WT mouse brains.
Consistent with our observation that Peril is expressed in regions associated with neural stem and progenitor cells in both E14.5 and adult mouse brains (Fig. 1 G and H), deletion of the Peril locus (Fig. S2D) revealed specific effects on cell-cycle gene sets and key pathways involved in neural stem cell maintenance and differentiation. At E14.5, Peril−/− brains were enriched for genes associated with SHH and TGFβ/WNT signaling pathways and demonstrated a significant increase in the neural stem cell marker genes Notch1-3, Pax3, and Nestin (Fig. S2 E and F). Additionally, we detected a significant down-regulation of the VTel-restricted progenitor marker genes Dlx1, Dlx2, and Dlx5 (33, 34) relative to WT brains (Fig. S2E). Interestingly, and unlike E14.5 brains, adult brains of Peril−/− mice have a significant and specific reduction in the expression of the canonical neural progenitor marker Nestin and the cyclin-dependent kinase inhibitor Cdkn1a (p21) (Fig. S2G).
These select examples highlight the breadth of information available in this RNA-seq dataset. Detailed reports for each KO versus WT brain comparison at E14.5 and adult time points are provided for each individual lncRNA locus in Dataset S6.
Cis Transcriptional Regulatory Dynamics of Specific lncRNA Loci.
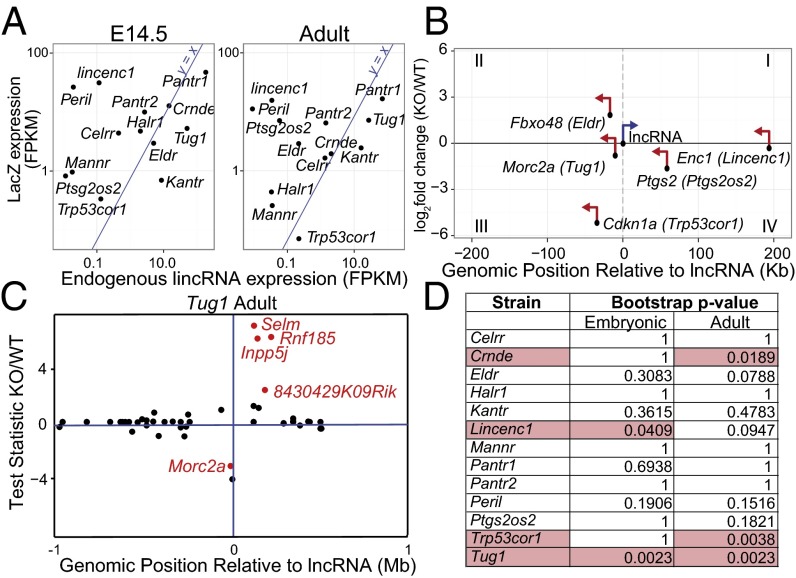
Because many lncRNA genes have been implicated in cis-regulatory functions (35–38), we asked whether we could observe any significant changes in expression from the local genomic region neighboring the selected lncRNA loci. We first asked whether we could observe a bias in the transcriptional output of these loci as a result of replacing the endogenous lncRNA with our lacZ reporter. To determine whether the expression level of the knocked-in lacZ reporter was similar to that of the endogenous lncRNA, we compared expression levels for lncRNAs across all WT samples with the lacZ reporter gene from each KO strain. We found that lacZ mRNA expression is positively but not strongly correlated with endogenous lncRNA expression (r2 = 0.4555; E14.5 = 0.47; adult = 0.29) (Fig. 3A). Although lacZ mRNA in several of the mutant strains such as Crnde−/−, Celrr−/−, and Pantr1−/− demonstrated expression levels consistent with that observed for the endogenous lncRNA (Fig. 3A), other strains exhibited more disparate expression levels. For example, lacZ was detected at higher levels than endogenous Lincenc1 and Peril when comparing KO with WT.
Fig. 3.
Cis effects of lncRNA deletion in the developing and adult brain. (A) Scatter plots of average lacZ expression (fragments per kilobase of exon per million fragments mapped, FPKM) in lncRNA KO versus endogenous lncRNA expression averaged across WT samples for all embryonic and adult samples. The line y = x is a reference for perfect correlation. (B) Summary of lncRNAs that significantly regulate their closest protein-coding neighbor. LncRNA mutant strains are shown in parenthesis. The x axis shows gene start distance (in kilobases) from the lncRNA transcriptional start site. The y axis shows log2-fold change of expression levels between WT and lncRNA KO. Genes in quadrants III and IV are down-regulated in the KO. The lncRNA is shown in blue, and the closest protein-coding neighbor is shown in red. The orientation of each gene is indicated with arrows. (C) Example of a cis-region plot. The x axis shows gene start distance (in megabases) from the lncRNA transcriptional start site. Here we show a truncated region spanning only a 2-Mb window rather than 4 Mb around the lncRNA of interest, Tug1. The y axis shows the test statistic (Cuffdiff2). Red color indicates significant differential expression between WT and KO. (D) Table of significance: P values for every cis-region plot. The five highlighted conditions have regional effects with significantly differentially regulated neighboring genes.
Next, we assessed the effect of each lncRNA locus on its local genomic environment by examining whether ablation of the lncRNA gene affected the expression of the closest protein-coding gene. For each strain and time point, we analyzed the KO vs. WT differential expression of nearest-neighbor protein-coding genes (Fig. 3B and Fig. S4). In 5 of the 13 mutant strains, deletion of the lncRNA genomic locus significantly affected the expression of the adjacent protein-coding gene at one or both time points (Fig. S5). We extended this analysis to the local 4-Mb flanking regions and identified several lncRNA loci with multiple differentially regulated genes in cis. We next asked whether this finding could represent a significant regional cis-regulatory effect as a result of the loss of the lncRNA locus. We detected five instances corresponding to four lncRNAs that demonstrated a significant (P < 0.05; bootstrap test) regional effect (Fig. 3D and Fig. S4). Interestingly, the majority of conditions, including those with a significant impact on the entire transcriptome, demonstrated no significant cis-regulatory effect. Collectively, these data point to variability among different lncRNAs regarding transcriptional regulation of neighboring loci. Further mechanistic work will be required to define cis regulation by each of the lncRNAs described here.
Differential Regulation and Spatial Distribution of lncRNAs Derived from the Pou3f3 Genomic Locus.
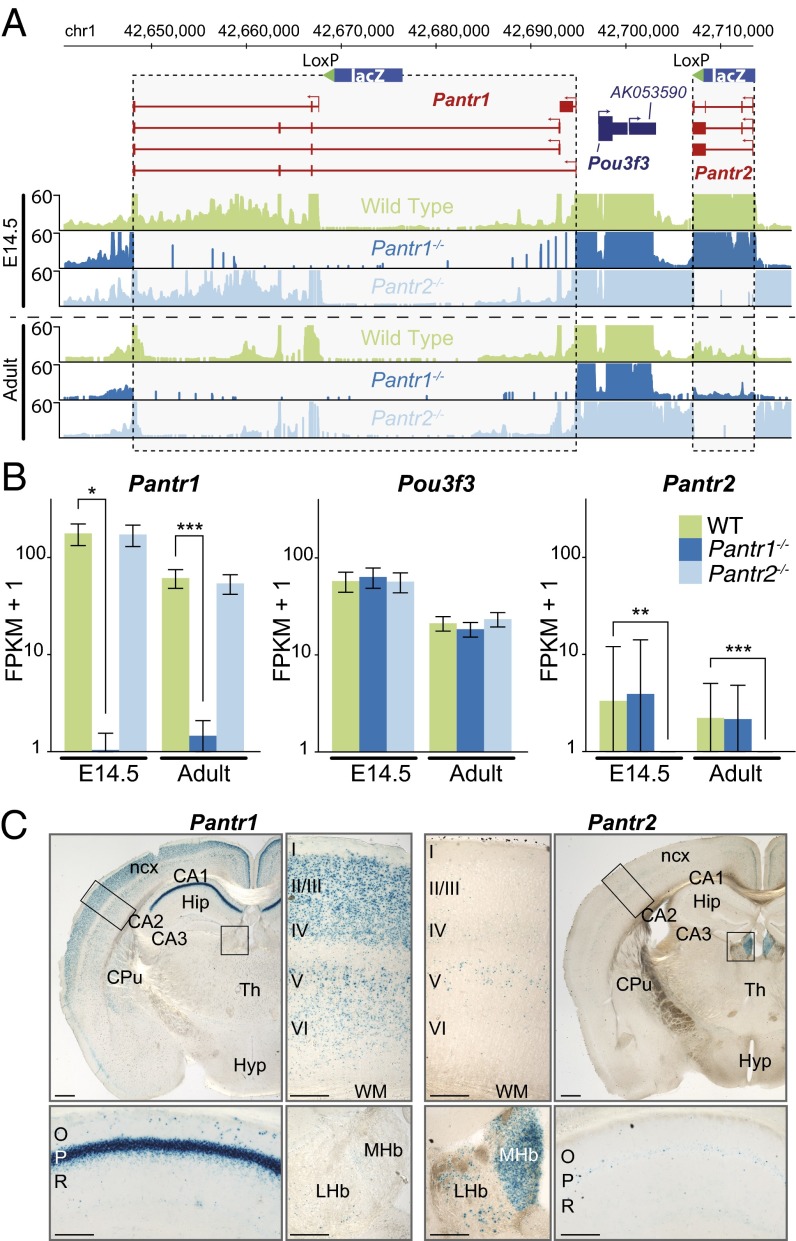
To gain specific insights into the distinct transcriptional programs that establish spatiotemporal segregation of lncRNAs within the brain, we focused on two lncRNAs, Pantr1 and Pantr2, that flank both sides of the POU-domain gene family member Pou3f3 (Brn1), a key transcription factor involved in cortical development (39, 40). Pantr1 shares a bidirectional promoter with Pou3f3, whereas Pantr2 is located ∼13.2 kb downstream of Pou3f3 in the opposite orientation (Fig. 4A).
Fig. 4.
LncRNAs at the Pou3f3 genomic locus are specifically and dynamically regulated in the developing and adult brain. (A) Pou3f3 genomic locus and targeting strategy of the adjacent lncRNAs Pantr1 and Pantr2. The RNA-seq representative read density profiles for E14.5 and adult brains collected from Pantr1−/− and Pantr2−/− mice confirm deletion of each respective lncRNA compared with WT samples. (B) RNA-seq expression estimates (average from triplicates) in E14.5 and adult brains collected from WT, Pantr1−/−, and Pantr2−/− mutant mice for Pantr1, Pou3f3, and Pantr2. (C) β-Gal staining of lncRNA Pantr1+/− (Left) and Pantr2+/− (Right) adult brain coronal sections shows strong expression of Pantr1 in the CA1 region of the hippocampus (Pantr1, Lower Left), in both upper (II/III, IV) and deep (V, VI) layers of the neocortex, and in the caudate putamen, whereas Pantr2 is expressed most strongly in the medial habenula and at lower levels in the lateral habenula, layer V of the neocortex, and the CA1 of the hippocampus (Pantr2, Lower Right). Images are representative of staining performed on two animals. CA, cornu ammonis; CPu, caudate putamen; Hip, hippocampus; Hyp, hypothalamus; LHb, lateral habenular nucleus; MHb, medial habenular nucleus; Ncx, neocortex; O, stratum oriens of the cornu ammonis; P, stratum pyramidale of the cornu ammonis; R, stratum radiatum of the cornu ammonis; Th, thalamus. [Scale bars: 500 μm (lower magnification whole brain panels), 200 μm (higher magnification middle and bottom panels).]
We first compared the gene-expression perturbations in Pantr1 and Pantr2 KO mice relative to WT mice. RNA-seq quantification indicated no significant effect on the expression level of Pou3f3 in either Pantr1−/− or Pantr2−/− E14.5 or adult mouse brains (Fig. 4B). However, we observed that loss of the Pantr1 genomic locus resulted in a significant, albeit modest, increase in the expression of Pou3f1, Pou3f2, and Pou3f4 in the E14.5 brain (Fig. S6). Conversely, deletion of the Pantr2 locus did not result in significant changes in expression of these paralogous regions.
Generally, we observed that, relative to all other lncRNA mutant mouse strains, the loss of the Pantr1 locus resulted in the most dramatic effects on the transcriptome of the whole brain at E14.5 (Fig. S2A). Notably, ablation of the Pantr1 locus resulted in significant up-regulation of the neuronal progenitor markers Ascl1, Nestin, Otx2, Notch1, Notch3, Msi1, Pax3, and Pax6, and significant down-regulation of mature neural cell-type markers including many canonical astrocyte markers such as Gfap, S100b, Aldh1l1, Fabp7, and Gap43 (Dataset S6). The observation that Pou3f3 and neighboring gene loci are not significantly affected in either Pantr1−/− or Pantr2−/− brains suggests that this effect may not be cis-mediated.
Interestingly, despite their genomic proximity to Pou3f3, we observed strikingly different expression patterns for these two lncRNAs in the adult brain. Although Pantr1 is strongly expressed in layers II/III–IV and at lower levels in layers V–VI (Fig. 4C, Upper), Pantr2+ cells are present only in low numbers in layer V (Fig. 4C, Upper). Similar differences are found in the hippocampus, where Pantr1 is strongly expressed in the pyramidal layer of CA1 and CA2, whereas Pantr2 is faintly present in a few scattered cells (Fig. 4C, Lower). Conversely, Pantr2 is very strongly expressed in the medial habenular nucleus, where Pantr1 is almost completely absent (Fig. 4C, Lower). These results, in conjunction with the previously demonstrated role for Pantr2 in the development and organization of the neocortex (12), suggest that loss of the Pantr1 and Pantr2 loci may impact neuronal development programs via distinct cellular and context-specific mechanisms. Collectively, these examples demonstrate the utility of tracking lncRNAs during development and monitoring perturbations in gene expression upon their deletion to facilitate the selection and development of additional experiments to disentangle the molecular mechanisms of these loci.
Discussion
We have explored the spatiotemporal expression patterns of 13 lncRNA genes and analyzed gene-expression changes resulting from the deletion of each locus at the whole-brain level. The publicly available resource of spatial lncRNA expression patterns and differential RNA-sequencing data generated in this study provides an initial map to facilitate the understanding of lncRNA loci in the development and function of the mammalian brain.
Although many strategies for genetic loss-of-function studies exist, the whole-gene ablation method used here is often a first approach to determine the functionality of a locus irrespective of its molecular nature. Although each of these loci contains a lncRNA, it is important to consider that any observation resulting from this strategy could reflect the loss of any regulatory element in the deleted region. Thus, we do not ascribe the expression perturbations identified in this work to a functional lncRNA molecule but rather to the genetic locus containing the targeted lncRNA. Despite this limitation, one advantage of this approach is the incorporation of the lacZ reporter, which can be used to determine the precise expression patterns of the lncRNA within the tissue. Moreover, because the reporter maintains transcriptional activity at the locus, it also serves to control, on some level, for phenotypes resulting from the loss of transcription.
Many questions remain as to the global mechanistic roles for lncRNA transcripts in regulating gene activity. The rate of lncRNA gene discovery has significantly outpaced our ability to evaluate both the physiological significance and function of these genes. It is difficult to predict whether the loss of any particular lncRNA locus will present a phenotype, but crucial information on the spatiotemporal dynamics of expression from each locus can provide significant direction and focus to downstream mechanistic studies by highlighting those loci most likely to have a physiological impact. Genetic approaches such as the gene-ablation method used in this study, and more broadly in large efforts such as the Knockout Mouse Project (KOMP) (41) and the International Knockout Mouse Consortium (IKMC) (42), have proven utility in determining if a genomic locus has any biological role even if broadly defined as the sum of multiple regulatory elements.
More studies now have used whole-gene ablation to determine whether a lncRNA locus contributes to a particular phenotype or, as in the case of Visc-2, whether a lncRNA whose expression pattern in the developing brain is highly conserved across mammals results in no overt phenotype (43). The results from these whole-gene ablation studies inform the selection and focus of future studies to dissect a given locus further into its functional elements. It is important to note, however, that this approach cannot ascribe any observed biological activity to a functional lncRNA transcript, particularly in the absence of rescue experiments. Other in vivo loss-of-function approaches have also been applied to address the question of lncRNA functional roles (11, 44–48). However, it is important to stress that no single method exists that can account for all possible mechanisms of action of a noncoding locus (49). Within these limits, the phenotypes observed after ablation of specific lncRNA loci confirm that expression of this class of noncoding RNAs can serve as a proxy signal to identify functional genomic loci with physiological relevance to disease and development (12), independent of whether this activity is directly ascribed to a functional lncRNA molecule. For example, the perturbed pathways most commonly affected by knocking out the lncRNA loci analyzed here suggest that some of our mutant strains may have defects in neuronal differentiation during development.
The cellular heterogeneity of the mammalian brain can confound interpretation of RNA-seq studies of the whole brain for lncRNAs with discrete spatial expression. Examination of the whole brain allows the detection of very strong effects that result from the deletion of a genomic locus, including indirect effects in cell types that do not express the lncRNA in question. However, ablation of the lncRNA locus may have a direct impact on a relatively small population of cells, resulting in signal dilution over the entire brain. This limitation can be addressed only by transcriptional profiling of purified populations of cells. Despite the known cellular heterogeneity of the brain samples that we profiled, we observe large transcriptional impacts in several mutant strains (e.g., Kantr, Pantr1, and Peril), suggesting either a broad effect across multiple cell types or strong differential expression within a cell population. Although this lack of resolution can make direct interpretation of gene-expression perturbations difficult, it can be useful for identifying broad trends in differential transcriptional regulation and can begin to provide a picture of the impact of specific lncRNA loci within the developing and mature brain. In the future, sequencing of specific subpopulations of cells from these mutant strains should identify additional affected pathways. The availability of expression profiles for lncRNAs, as provided here, will aid in the selection of specific populations of cells for purification.
Our differential RNA-seq analysis provides biological contexts and interpretations that are, in most cases, borne out by the selective and specific positive β-gal stains. Thus, the combination of spatiotemporal in vivo expression and differential analysis can provide additive contextual clues and permit future hypothesis-driven analyses that can be used to determine whether a functional role exists in neural development and disorders for these lncRNAs.
The power of this combinatorial approach is apparent in several lncRNA mutant strains. For example, Peril RNA-seq and β-gal staining both suggest that this lncRNA is expressed in neural stem cell populations and may have a significant impact on their biology. Embryonically, Peril is expressed in regions containing neural progenitor cells. This distribution is maintained in Peril+/− adult mice, with β-gal+ staining observed in the ependymal lining of the ventricles (Fig. 2) and in the dentate gyrus of the hippocampus (Dataset S2), both regions that are associated with adult neurogenesis (50–52). Consistent with these expression patterns, RNA-seq analysis of the Peril−/− embryonic brain showed misregulation of cell-cycle genes critically important for proper maintenance and differentiation of neural progenitors (53). In adult whole brains, Peril−/− mice maintain a significant misregulation of Nestin (Fig. S2G). Interestingly, Peril−/− whole brains demonstrate a significant reduction in Cdkn1a (Fig. S2G) relative to WT, and the specific loss of this protein has been shown to induce premature terminal differentiation and quiescence in adult neural stem cells (54).
The Pou3f3-locus lncRNAs Pantr1 and Pantr2 provide other notable examples in which the in vivo expression pattern combined with RNA-seq analysis provides intriguing insights. We previously have demonstrated a functional role for the Pantr2 lncRNA locus in contributing to development of the neocortex in mice (12). However, the mechanism by which this locus mediates this effect remains unclear. Modulation of Pantr1 has been shown to affect Pou3f3 expression levels in cancer cells (55). However, in the context of whole-brain expression analysis in E14.5 and adult mutant mice, we observe no significant effect on the expression of the neighboring Pou3f3 gene for either Pantr1−/− or Pantr2−/− (Fig. 4B). Surprisingly, loss of the Pantr1 locus resulted in a significant, albeit modest, increase in the expression of the other POU3-family paralogs located on different chromosomes. These genes may act redundantly to coordinate neurogenesis cellular identity and migration in the developing cortex (39). Thus, although speculative at this stage, the Pantr1 locus might act to regulate coordinately the cellular processes governed by the POU3-family genes during brain development.
Our lncRNA KO strains are comprised of deletions of varying distance from the target promoter. However, we found no obvious trend related to gene-targeting design that could justify the discrepancy in some strains between lacZ expression levels in the brain and WT lncRNA levels. It is also important to note that the knocked-in lacZ reporter is driven off the endogenous lncRNA promoter and thus, in theory, experiences the same transcriptional regulatory dynamics as the lncRNA.
Here we have identified several lncRNA loci whose ablation in vivo results in significant differential expression of local (± 2 Mb) or neighboring (immediately adjacent) protein-coding genes. Different mechanisms could be proposed for such effects. A first possibility is that these specific lncRNAs may act in cis. For example, Tug1 is expressed in the brain, and Tug1−/− whole brains do show significant up-regulation of several proximal genes (Inpp5j, Selm, Rnf185, 8430429K09Rik), suggesting that this locus could exhibit cis-like regulatory effects.
A second possibility is that DNA regulatory elements are contained within lncRNA loci and that these elements are responsible for local gene regulation. A potential example for this modality is Trp53cor1 (linc-p21). We observed numerous differentially regulated protein-coding genes surrounding this locus in Trp53cor1−/− adult brains (Fig. S4). However, although this finding is consistent with a cis-regulatory effect for this locus (56), we did not detect expression of Trp53cor1 locus activity by β-gal staining or RNA-seq (Fig. 3A, Dataset S2, and Dataset S6). The absence of lncRNA expression combined with differential expression of surrounding genes points to a possible functional role for DNA elements within the locus. Of note, our KO strategy deletes a p53-binding motif a few base pairs upstream of the transcription start site of Trp53cor1. This observation suggests that this and potentially other DNA elements that reside within the Trp53cor1 locus may contribute to the local changes in expression levels.
Any investigation into the activity and impact of a lncRNA locus requires, as a foundational step, an examination of the in vivo expression patterns and resulting perturbations on gene expression in a loss-of-function context. In this study, we report the in vivo expression profiles for 13 lncRNAs during mammalian brain development and the consequent changes in gene-expression patterns after genomic ablation. However, the stable and reproducible production of an RNA transcript from a particular locus cannot be confounded with the assumption that this transcription results in a functional RNA molecule but rather serves as evidence of biological activity within the lncRNA locus. The precise molecular mechanisms then must be validated on a per-locus basis and require incorporating multiple genetic loss- and gain-of-function strategies. Here we provide a resource to facilitate further inquiry.
Materials and Methods
Mice were housed under controlled pathogen-free conditions in Harvard University’s Biological Research Infrastructure. All procedures were carried out in accordance with the guidelines of the National Institutes of Health's Guide for the Care and Use of Laboratory Animals (57) and were approved by the Harvard University Committee on the Use of Animals in Research and Teaching.
Mouse Tissue Collection and Processing.
LncRNA KO mice were generated in collaboration with Regeneron Pharmaceuticals by replacing the selected lncRNA gene with a lacZ reporter cassette as previously described (12). For β-gal expression, E14.5 whole brains were harvested from embryos fixed by immersion in 4% (vol/vol) paraformaldehyde (PFA) at 4 °C overnight. Adult brains were perfused with 4% (vol/vol) PFA and postfixed in 4% (vol/vol) PFA at 4 °C for 12 h. For RNA isolation, E14.5 whole brains were homogenized in TRIzol (Life Technology) (1 mL per brain). Adult brains were snap frozen in liquid nitrogen, and stored at −80 °C. Frozen brains were pulverized and homogenized in TRIzol (5 mL per brain).
β-Gal Staining and Immunostaining.
Brain-wide expression of the lacZ reporter gene was assessed in all mutant strains by histochemical detection of β-gal (X-gal staining) in biological replicates. Sequential sections, obtained at every 80 µm for E14.5 brains and at every 240 µm for adult brains, were imaged at 5× and 10× magnification using a Zeiss Axio Scan.Z1, a Nikon 90i microscope equipped with a Retiga Exi camera (QIMAGING), or a Zeiss LSM700 confocal microscope. Immunohistochemistry for β-gal and immunostaining for the interneuron marker TH was performed using standard methods. Primary antibodies and dilutions were chicken anti-βgal (1:500; CGAL-45; ICL) and rabbit anti-TH (1:5,000; AB152; Chemicon). Appropriate secondary antibodies were from the Molecular Probes Alexa series. Images were acquired and processed with the Zen (Zeiss) or Volocity analysis software v4.0.1 (Improvision).
mRNA-Seq Library Preparation and Sequencing.
For all samples, RNA-seq was performed as previously described (TruSeq RNA Sample Preparation Kit v2; Illumina) (12). Libraries were sequenced using a paired-end, 50-bp read-length sequencing protocol (NWL Bauer Core, Harvard University FAS Center for System Biology). Sequencing reads were aligned to mm10 using Tophat2 (58) with the following additional parameters: –no-coverage-search –max-multihits 10 -p 8. To standardize the analysis of each differential comparison, we created a report template that integrated components from several R/Bioconductor (59) packages as detailed in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Venus Lai, David Frendewey, and David Valenzuela from Regeneron Pharmaceuticals for generating the lncRNA KO mice; Manolis Kellis (Computer Science and Artificial Intelligence Laboratory, MIT) for mentorship and support of L.A.G.; all members of the J.L.R. and P.A. laboratories for advice and input on the manuscript, and David Kelley for helpful comments; the Harvard Center for Biological Imaging for microscopy support; and the Bauer Sequencing Core within Harvard University’s Faculty of Arts and Sciences Division of Science for sequencing support. This study was principally supported by National Institutes of Health (NIH)/National Institute of Mental Health Grant R01MH102416-0) (to J.L.R.) and in part by NIH/National Institute of Neurological Disorders and Stroke Grants R01NS062849 and R01MH101268 (to P.A.). L.A.G. is supported in part by National Science Foundation/Postdoctoral Research Fellowship in Biology DBI-0905973. A.F.G. is supported by National Science Foundation Graduate Research Fellowship DGE1144152. P.A. is a New York Stem Cell Foundation Robertson Investigator.
Footnotes
The authors declare no conflict of interest.
This paper results from the Arthur M. Sackler Colloquium of the National Academy of Sciences, “Epigenetic Changes in the Developing Brain: Effects on Behavior,” held March 28–29, 2014, at the National Academy of Sciences in Washington, DC. The complete program and video recordings of most presentations are available on the NAS website at www.nasonline.org/Epigenetic_changes.
This article is a PNAS Direct Submission.
Data deposition: The sequence reported in this paper has been deposited in the Gene Expression Omnibus databank (accession no. GSE61716).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1411263112/-/DCSupplemental.
References
- 1.Migliore M, Shepherd GM. Opinion: An integrated approach to classifying neuronal phenotypes. Nat Rev Neurosci. 2005;6(10):810–818. doi: 10.1038/nrn1769. [DOI] [PubMed] [Google Scholar]
- 2.Molyneaux BJ, Arlotta P, Menezes JRL, Macklis JD. Neuronal subtype specification in the cerebral cortex. Nat Rev Neurosci. 2007;8(6):427–437. doi: 10.1038/nrn2151. [DOI] [PubMed] [Google Scholar]
- 3.Qureshi IA, Mattick JS, Mehler MF. Long non-coding RNAs in nervous system function and disease. Brain Res. 2010;1338:20–35. doi: 10.1016/j.brainres.2010.03.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mehler MF, Mattick JS. Noncoding RNAs and RNA editing in brain development, functional diversification, and neurological disease. Physiol Rev. 2007;87(3):799–823. doi: 10.1152/physrev.00036.2006. [DOI] [PubMed] [Google Scholar]
- 5.St Laurent G, 3rd, Wahlestedt C. Noncoding RNAs: Couplers of analog and digital information in nervous system function? Trends Neurosci. 2007;30(12):612–621. doi: 10.1016/j.tins.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 6.Mercer TR, Dinger ME, Sunkin SM, Mehler MF, Mattick JS. Specific expression of long noncoding RNAs in the mouse brain. Proc Natl Acad Sci USA. 2008;105(2):716–721. doi: 10.1073/pnas.0706729105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chodroff RA, et al. Long noncoding RNA genes: Conservation of sequence and brain expression among diverse amniotes. Genome Biol. 2010;11(7):R72. doi: 10.1186/gb-2010-11-7-r72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ponjavic J, Oliver PL, Lunter G, Ponting CP. Genomic and transcriptional co-localization of protein-coding and long non-coding RNA pairs in the developing brain. PLoS Genet. 2009;5(8):e1000617. doi: 10.1371/journal.pgen.1000617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qureshi IA, Mehler MF. Emerging roles of non-coding RNAs in brain evolution, development, plasticity and disease. Nat Rev Neurosci. 2012;13(8):528–541. doi: 10.1038/nrn3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cabili MN, et al. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011;25(18):1915–1927. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bond AM, et al. Balanced gene regulation by an embryonic brain ncRNA is critical for adult hippocampal GABA circuitry. Nat Neurosci. 2009;12(8):1020–1027. doi: 10.1038/nn.2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sauvageau M, et al. Multiple knockout mouse models reveal lincRNAs are required for life and brain development. eLife. 2013;2:e01749. doi: 10.7554/eLife.01749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johnson R. Long non-coding RNAs in Huntington’s disease neurodegeneration. Neurobiol Dis. 2012;46(2):245–254. doi: 10.1016/j.nbd.2011.12.006. [DOI] [PubMed] [Google Scholar]
- 14.Qureshi IA, Mehler MF. Non-coding RNA networks underlying cognitive disorders across the lifespan. Trends Mol Med. 2011;17(6):337–346. doi: 10.1016/j.molmed.2011.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Salta E. 2012. Non-coding RNAs with essential roles in neurodegenerative disorders. The Lancet Neurology 11(2):189–200.
- 16.Abe Y, et al. Xq26.1-26.2 gain identified on array comparative genomic hybridization in bilateral periventricular nodular heterotopia with overlying polymicrogyria. Dev Med Child Neurol. 2014;56(12):1221–1224. doi: 10.1111/dmcn.12553. [DOI] [PubMed] [Google Scholar]
- 17.Guttman M, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458(7235):223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Altman J, Bayer SA. Development of the diencephalon in the rat. V. Thymidine-radiographic observations on internuclear and intranuclear gradients in the thalamus. J Comp Neurol. 1979;188(3):473–499. doi: 10.1002/cne.901880309. [DOI] [PubMed] [Google Scholar]
- 19.Grove EA, Tole S. Patterning events and specification signals in the developing hippocampus. Cereb Cortex. 1999;9(6):551–561. doi: 10.1093/cercor/9.6.551. [DOI] [PubMed] [Google Scholar]
- 20.Baumann N, Pham-Dinh D. Biology of oligodendrocyte and myelin in the mammalian central nervous system. Physiol Rev. 2001;81(2):871–927. doi: 10.1152/physrev.2001.81.2.871. [DOI] [PubMed] [Google Scholar]
- 21.Tomassy GS, et al. Distinct profiles of myelin distribution along single axons of pyramidal neurons in the neocortex. Science. 2014;344(6181):319–324. doi: 10.1126/science.1249766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bayer SA, Altman J. 1991. Neocortical Development (Raven, New York)
- 23.Marín O, Rubenstein JL. A long, remarkable journey: Tangential migration in the telencephalon. Nat Rev Neurosci. 2001;2(11):780–790. doi: 10.1038/35097509. [DOI] [PubMed] [Google Scholar]
- 24.Marín O, Rubenstein JLR. Cell migration in the forebrain. Annu Rev Neurosci. 2003;26:441–483. doi: 10.1146/annurev.neuro.26.041002.131058. [DOI] [PubMed] [Google Scholar]
- 25.Temple S. The development of neural stem cells. Nature. 2001;414(6859):112–117. doi: 10.1038/35102174. [DOI] [PubMed] [Google Scholar]
- 26.Alvarez-Buylla A, Seri B, Doetsch F. Identification of neural stem cells in the adult vertebrate brain. Brain Res Bull. 2002;57(6):751–758. doi: 10.1016/s0361-9230(01)00770-5. [DOI] [PubMed] [Google Scholar]
- 27.Ferri ALM, et al. Sox2 deficiency causes neurodegeneration and impaired neurogenesis in the adult mouse brain. Development. 2004;131(15):3805–3819. doi: 10.1242/dev.01204. [DOI] [PubMed] [Google Scholar]
- 28.Carpenter S, et al. A long noncoding RNA mediates both activation and repression of immune response genes. Science. 2013;341(6147):789–792. doi: 10.1126/science.1240925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lein ES, Zhao X, Gage FH. Defining a molecular atlas of the hippocampus using DNA microarrays and high-throughput in situ hybridization. J Neurosci. 2004;24(15):3879–3889. doi: 10.1523/JNEUROSCI.4710-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lorente De Nó R. Studies on the structure of the cerebral cortex. II. Continuation of the study of the ammonic system. Journal für Psychologie und Neurologie. 1934;46:113–177. [Google Scholar]
- 31.Corsellis JA, Bruton CJ. Neuropathology of status epilepticus in humans. Adv Neurol. 1983;34:129–139. [PubMed] [Google Scholar]
- 32.Chen H-Z, Tsai S-Y, Leone G. Emerging roles of E2Fs in cancer: An exit from cell cycle control. Nat Rev Cancer. 2009;9(11):785–797. doi: 10.1038/nrc2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bulfone A, et al. Spatially restricted expression of Dlx-1, Dlx-2 (Tes-1), Gbx-2, and Wnt-3 in the embryonic day 12.5 mouse forebrain defines potential transverse and longitudinal segmental boundaries. J Neurosci. 1993;13(7):3155–3172. doi: 10.1523/JNEUROSCI.13-07-03155.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gulacsi AA, Anderson SA. Beta-catenin-mediated Wnt signaling regulates neurogenesis in the ventral telencephalon. Nat Neurosci. 2008;11(12):1383–1391. doi: 10.1038/nn.2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ulitsky I, Bartel DP. lincRNAs: Genomics, evolution, and mechanisms. Cell. 2013;154(1):26–46. doi: 10.1016/j.cell.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ørom UA, et al. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143(1):46–58. doi: 10.1016/j.cell.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang KC, et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472(7341):120–124. doi: 10.1038/nature09819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dominguez MH, Ayoub AE, Rakic P. POU-III transcription factors (Brn1, Brn2, and Oct6) influence neurogenesis, molecular identity, and migratory destination of upper-layer cells of the cerebral cortex. Cereb Cortex. 2013;23(11):2632–2643. doi: 10.1093/cercor/bhs252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McEvilly RJ, de Diaz MO, Schonemann MD, Hooshmand F, Rosenfeld MG. Transcriptional regulation of cortical neuron migration by POU domain factors. Science. 2002;295(5559):1528–1532. doi: 10.1126/science.1067132. [DOI] [PubMed] [Google Scholar]
- 41.Austin CP, et al. The knockout mouse project. Nat Genet. 2004;36(9):921–924. doi: 10.1038/ng0904-921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Collins FS, Rossant J, Wurst W. International Mouse Knockout Consortium A mouse for all reasons. Cell. 2007;128(1):9–13. doi: 10.1016/j.cell.2006.12.018. [DOI] [PubMed] [Google Scholar]
- 43.Oliver PL, et al. Disruption of Visc-2, a brain-expressed conserved long noncoding RNA, does not elicit an overt anatomical or behavioral phenotype. Cereb Cortex. 2014 doi: 10.1093/cercor/bhu196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Grote P, et al. The tissue-specific lncRNA Fendrr is an essential regulator of heart and body wall development in the mouse. Dev Cell. 2013;24(2):206–214. doi: 10.1016/j.devcel.2012.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fitzpatrick GV, Soloway PD, Higgins MJ. Regional loss of imprinting and growth deficiency in mice with a targeted deletion of KvDMR1. Nat Genet. 2002;32(3):426–431. doi: 10.1038/ng988. [DOI] [PubMed] [Google Scholar]
- 46.Sleutels F, Zwart R, Barlow DP. The non-coding Air RNA is required for silencing autosomal imprinted genes. Nature. 2002;415(6873):810–813. doi: 10.1038/415810a. [DOI] [PubMed] [Google Scholar]
- 47.Nakagawa S, et al. Malat1 is not an essential component of nuclear speckles in mice. RNA. 2012;18(8):1487–1499. doi: 10.1261/rna.033217.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nakagawa S, Naganuma T, Shioi G, Hirose T. Paraspeckles are subpopulation-specific nuclear bodies that are not essential in mice. J Cell Biol. 2011;193(1):31–39. doi: 10.1083/jcb.201011110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bassett AR, et al. Considerations when investigating lncRNA function in vivo. eLife. 2014;3:e03058. doi: 10.7554/eLife.03058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Eriksson PS, et al. Neurogenesis in the adult human hippocampus. Nat Med. 1998;4(11):1313–1317. doi: 10.1038/3305. [DOI] [PubMed] [Google Scholar]
- 51.Chiasson BJ, Tropepe V, Morshead CM, van der Kooy D. Adult mammalian forebrain ependymal and subependymal cells demonstrate proliferative potential, but only subependymal cells have neural stem cell characteristics. J Neurosci. 1999;19(11):4462–4471. doi: 10.1523/JNEUROSCI.19-11-04462.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kempermann G, Kuhn HG, Gage FH. More hippocampal neurons in adult mice living in an enriched environment. Nature. 1997;386(6624):493–495. doi: 10.1038/386493a0. [DOI] [PubMed] [Google Scholar]
- 53.Götz M, Huttner WB. The cell biology of neurogenesis. Nat Rev Mol Cell Biol. 2005;6(10):777–788. doi: 10.1038/nrm1739. [DOI] [PubMed] [Google Scholar]
- 54.Porlan E, et al. Transcriptional repression of Bmp2 by p21(Waf1/Cip1) links quiescence to neural stem cell maintenance. Nat Neurosci. 2013;16(11):1567–1575. doi: 10.1038/nn.3545. [DOI] [PubMed] [Google Scholar]
- 55.Li L, Chang HY. Physiological roles of long noncoding RNAs: Insight from knockout mice. Trends Cell Biol. 2014;24(10):594–602. doi: 10.1016/j.tcb.2014.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dimitrova N, et al. LincRNA-p21 activates p21 in cis to promote Polycomb target gene expression and to enforce the G1/S checkpoint. Mol Cell. 2014;54(5):777–790. doi: 10.1016/j.molcel.2014.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Committee on Care and Use of Laboratory Animals (1996) Guide for the Care and Use of Laboratory Animals (Natl Inst Health, Bethesda), DHHS Publ No (NIH) 85-23.
- 58.Kim D, et al. TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14(4):R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gentleman RC, et al. Bioconductor: Open software development for computational biology and bioinformatics. Genome Biol. 2004;5(10):R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.