Abstract
Growth-factor receptor bound protein 10 (Grb10) is a signal adapter protein encoded by an imprinted gene that has roles in growth control, cellular proliferation, and insulin signaling. Additionally, Grb10 is critical for the normal behavior of the adult mouse. These functions are paralleled by Grb10’s unique tissue-specific imprinted expression; the paternal copy of Grb10 is expressed in a subset of neurons whereas the maternal copy is expressed in most other adult tissues in the mouse. The mechanism that underlies this switch between maternal and paternal expression is still unclear, as is the role for paternally expressed Grb10 in neurons. Here, we review recent work and present complementary data that contribute to the understanding of Grb10 gene regulation and function, with specific emphasis on growth and neuronal development. Additionally, we show that in vitro differentiation of mouse embryonic stem cells into alpha motor neurons recapitulates the switch from maternal to paternal expression observed during neuronal development in vivo. We postulate that this switch in allele-specific expression is related to the functional role of Grb10 in motor neurons and other neuronal tissues.
Keywords: genomic imprinting, epigenetics, Grb10, motor neurons, neurodevelopment
Grb10 is an adapter protein that is a member of the Grb7/Gr10/Grb14 protein family. These proteins interact with numerous receptor tyrosine kinases, impacting a variety of signaling pathways (1). Specifically, Grb10 has been implicated in the regulation of cell proliferation, apoptosis, and metabolism, among other functions (2–4). The first characterized Grb10 knockout mouse exhibited overgrowth in numerous organs and tissues, suggesting that Grb10 acts as a potent growth suppressor in vivo (5). Other Grb10 knockout mouse models have additionally shown that Grb10 plays an important role in social dominance behavior in mice (6).
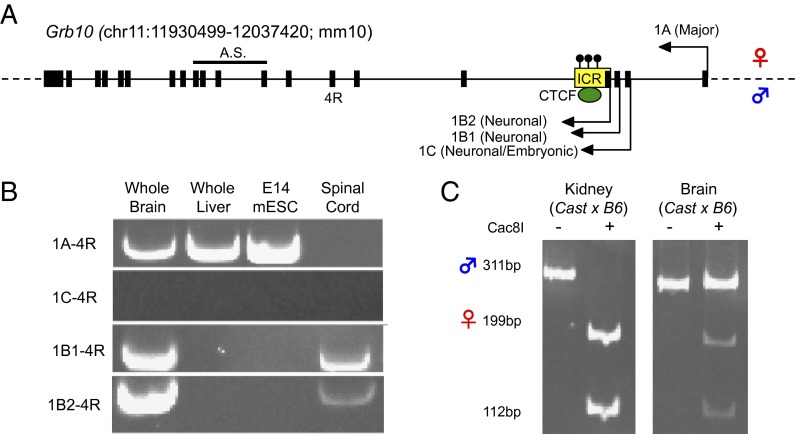
Interestingly, these phenotypes are dependent on the parental origin of the Grb10 mutation. The overgrowth phenotype is exhibited only when the mutation is transmitted through the maternal lineage whereas the behavior phenotype is revealed upon paternal transmission of the mutation. These surprising allele-specific phenotypes can be explained by the tissue-specific genomic imprinting of Grb10. Genomic imprinting refers to an infrequent regulatory phenomenon in which a gene is expressed in a parent-of-origin–specific manner. Genomic imprinting is dependent on cis-regulatory elements called imprinting control regions (ICRs) that are epigenetically modified on a single parental allele to induce allele-specific expression (7). Grb10 is expressed from the maternal chromosome in most tissues in the mouse but is expressed from the paternal allele in a subset of neurons. Maternal expression occurs from the Grb10 major promoter whereas Grb10 paternal expression comes from three downstream alternative promoters (Fig. 1). The region surrounding one of these alternative promoters has been identified as an ICR, which exhibits DNA methylation only on the maternal allele in all examined tissues (8). On the paternal allele, the unmethylated Grb10 ICR is bound by CCCTC-binding factor (CTCF), a multifunctional transcription factor, which is recruited in a DNA methylation-sensitive manner and has been implicated in the regulation of imprinted expression at other loci (Fig. 1A).
Fig. 1.
Grb10 is expressed maternally from most adult tissues and paternally in neuronal tissues. (A) A schematic of the Grb10 locus is shown, with the allelic-specific expression of alternative promoters indicated (maternal and paternal alleles, arrow above the line and below the line, respectively). The imprinting control region (ICR) is designated by a yellow box. Maternal allele-specific DNA methylation, evident in all characterized tissues, is indicated by black lollipops, above the line. The reverse primer location for expression analysis is shown by its name, 4R. The location of the amplicon used for allele-specific (A.S.) analysis is similarly indicated. (B) Expression of transcripts from alternative promoters of Grb10 is shown in adult brain, adult liver, mESCs, and adult spinal cord. (C) Allele-specific expression as measured via SNP-dependent Cac8I restriction enzyme digests is shown in brain and liver from Mus Castaneus/EiJ (Cast) × C57BL/6J (B6) hybrid adult mice. Expression of the maternal allele is indicated by the 199-bp and 112-bp bands after restriction digest whereas the 311-bp band indicates paternal expression.
Thus, Grb10 is an imprinted gene with multiple functions and a complex tissue-specific imprinted expression pattern. Here, we review work that has illuminated the functional roles of Grb10 in embryonic development, cellular growth, and behavior. We also review the various regulatory mechanisms that have been implicated in the control of Grb10 tissue-specific expression and imprinting. Additionally, we present data addressing the regulation of Grb10 expression during neuronal development in vitro. We show that in vitro differentiation of mouse embryonic stem cells (mESCs) into motor neurons results in the switch of Grb10 promoter use. Specifically, the maternally expressed major promoter is repressed whereas the paternally expressed neuron-specific promoter is activated, concordant with neuronal maturation. We postulate that this switch in expression is required for the appropriate function of Grb10 in the nervous system.
Grb10 Binding Partners Define Its Role in Development
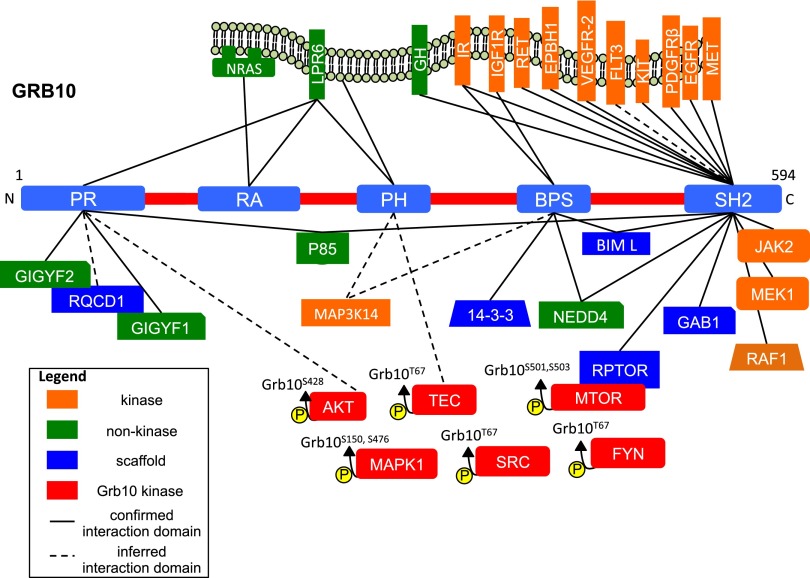
In mouse and human, Grb10 has five major protein domains: the N-terminal proline-rich region (PR), a centrally located pleckstrin homology domain (PH), a Ras-associating domain (RA), a C-terminal Src homology 2 domain (SH2), and the family-specific BPS domain, so named because it is between the PH and SH2 domains (4). Each domain is associated with protein binding partners that define the functions of Grb10 in development and growth (Fig. 2 and Table S1).
Fig. 2.
Grb10 protein partners. Shown is a schematic of the five major protein domains of Grb10: the proline-rich (PR) region, the ras-associating domain (RA), the pleckstrin homology domain (PH), the Src homology 2 domain (SH2), and the intervening domain (BPS). The known protein partners defined by in vivo interactions and immunoprecipitation are shown. Confirmed interactions with a specific Grb10 domain are indicated with a solid line whereas inferred interactions are indicated with a dotted line. The amino acids that are phosphorylated by Grb10 kinases are indicated above each protein.
The SH2 domain enables Grb10 to interact with phosphorylated tyrosine residues of other proteins and acts as the recruitment point for a variety of signaling molecules. Many of these proteins are receptors associated with growth, including the insulin receptor (IR) and the growth hormone receptor (GHR) (9, 10). The SH2 domain is critical for the role of Grb10 in the mitogen-activated protein kinase (MAPK) pathway, having direct interactions with numerous critical components including MEK1 and RAF1 (11). The SH2 domain also interacts with many receptor proteins implicated in cell proliferation, differentiation, and control of the cell cycle, including RET, KIT, MET, PDGFR, EGFR, FLT3, and VEGFR-2 (9, 12–16). Additionally, the SH2 domain has been implicated in Grb10 homodimerization, which is hypothesized to enable Grb10 to integrate signals from multiple pathways through the formation of larger protein complexes (17).
The BPS domain, specific to the Grb7/10/14 protein family, has also been shown to be important for Grb10’s role in insulin signaling through its binding to IR and additionally binds IGF1R (14). The BPS domain interacts with the 14-3-3 protein, another protein scaffold with numerous protein partners, and BCL2L11, a proapoptotic scaffold protein required for normal immune function (18, 19). The PH domain has the fewest interacting partners although it has been confirmed to bind phosphoinositides (PIs), critical components of the cell membrane that affect vesicle transport and cell signaling (20). This interaction is consistent with the requirement of the PH domain for Grb10 localization to the cell membrane. The RA domain of Grb7 and Grb14 extensively interacts with Ras-GTPases, important regulators of intercellular signaling related to cell growth and vesicle transport (21). Grb10 has been confirmed to bind at least one of these GTPases, NRAS (20). Finally, the PR domain binds to GIGYF1/2, proteins that are implicated in the modulation of insulin signaling and neuronal survival but have currently unclear molecular functions (22).
It is notable that, in human and mouse, the Grb10 mRNA is highly alternatively spliced, resulting in four to seven unique protein isoforms, most of which differ by truncations of the PR or PH domains (3). These splice variants are differentially expressed in various tissues although these expression patterns are not fully characterized. The differing levels of these isoforms and their respective protein partners may underlie tissue-specific functional roles for Grb10, which remain poorly understood.
Grb10 Is a Negative Regulator of Growth
Grb10 is involved in a host of signaling pathways, many of which affect cellular growth. Among these, Grb10’s role in insulin signaling is the most well understood, where it acts as an inhibitor. When IR binds insulin or another substrate, a cascade of phosphorylation events ultimately activates the PI3K/AKT and MAPK pathways (23). Activation of these two pathways leads to a variety of metabolic consequences affecting the storage and synthesis of glucose and fatty acids, in addition to promoting cellular growth through regulating proteins important for the cell cycle and other mitogenic factors. Additionally, both pathways can activate mTORC1, a protein complex that integrates growth signals and acts positively upon cell proliferation (24).
Grb10 potently inhibits cellular growth by acting at multiple levels in these pathways, among others. Grb10 inhibits insulin signaling by interacting with IR in response to insulin stimulation (14). Once bound, Grb10 inhibits the catalytic activity of IR in a noncompetitive manner in concert with Grb7 and Grb14 (25). Additionally, Grb10 has recently been described as a phosphorylation substrate of the mTORC1 complex (26). Phosphorylation and stabilization of Grb10 by mTORC1 leads to the feedback inhibition of PI3K/AKT and MAPK pathways, downstream of IR. Accordingly, overexpression of Grb10 inhibits the PI3K/AKT and MAPK pathways whereas Grb10 deficiency increases the insulin-dependent phosphorylation of proteins within these pathways, including AKT and MAPK1 (26–29). Conflicting studies, however, have reported that Grb10 has a positive effect on insulin signaling (9, 30). It has been proposed that these results are due to the overexpression of different Grb10 isoforms and/or the use of cell types that are not insulin-sensitive (25). Other pathways important for growth are similarly inhibited by Grb10. Insulin-like growth factor (IGF) signaling is inhibited by Grb10 directly binding and degrading IGF1R through its interaction with NEDD4, a ubiquitin protein ligase (31–33). Grb10 has also been implicated as a negative regulator of growth-hormone signaling through direct interaction with GRH (10).
Consistent with a role for Grb10 as a negative regulator of insulin signaling and other growth-related pathways, overexpression of Grb10 in transgenic mice is associated with postnatal growth retardation, glucose intolerance, and insulin resistance (34, 35). Notably, Grb10 overexpression is linked to severe pancreatic dysfunction and dysmorphia in juvenile mice (36). Reduction in Grb10 expression is associated with widespread neonatal and postnatal overgrowth, along with increased sensitivity to insulin (6, 34, 37, 38). Pancreas-specific Grb10 deletion results in increased insulin production and improved glucose tolerance (39). Recent work has shown that, in human islets, shRNA-induced knockdown of Grb10 expression in pancreatic islets is associated with reduced insulin and glucagon secretion (40). In summary, Grb10’s role in the control of insulin is complex, impacting the recognition, production, and secretion of insulin.
The physiological underpinnings of the growth phenotypes observed in Grb10-deficient mice are also complex. Mice deficient for Grb10 have increased body size that is first evident during embryonic growth and persists through adulthood. Notably, embryonic growth is largely independent of insulin signaling, suggesting that Grb10’s role in other growth-related pathways is significant during embryonic development (41). Embryonic overgrowth also reflects the important role of Grb10 in placental development, with Grb10 deficiency resulting in increased placental size and efficiency (42). Postnatal overgrowth has been partially attributed to the increased lean mass observed in Grb10 deletion mice (37, 38, 43). Grb10 deletion mice have an increased number of muscle fibers and the molecular hallmarks of increased insulin signaling in muscle (44). Recent work has also implicated Grb10 as being critical for lipolysis and nonshivering thermogenesis in adipose tissues through down-regulation of mTORC1 signaling (45). Grb10 mutant mice fed a high-fat diet had greatly increased fat stores compared with their WT littermates. Strikingly, the proportion of lean to fat body mass in Grb10 mutant mice has also recently been shown to be dependent on Grb10 expression in both the mother and the pup (46). Grb10-deficient mothers had a larger effect on adiposity in the pup whereas the genotype of the pup was more strongly associated with influencing its own lean mass.
Paternal Grb10 Expression in Neurons Affects Behavior
In the first Grb10 knockout mouse, it was reported that the overgrowth associated with the mutation was not observed in the brain or spinal cord (5). Subsequent experiments in mice demonstrated that Grb10 is transcribed in neurons from a series of downstream alternative promoters, exclusively from the paternal allele (8, 47). This paternal expression is conserved in the human brain (48). In contrast, Grb10 expression in other somatic tissues is derived from the maternal allele in mouse but has been shown to be expressed from both alleles in a variety of human embryonic tissues (49). Paternally transcribed Grb10 is embryonically expressed throughout the ventral spinal cord, areas of the diencephalon, ventral midbrain, and medulla oblongata by embryonic day (E) 14.5 (6). These expression patterns are established by midembryonic development and are largely maintained in the adult. In the adult mouse brain, Grb10 paternal expression is evident within thalamic, hypothalamic, midbrain, hindbrain, and forebrain nuclei (6). Of note, nearly all monoaminergic cell populations within the midbrain and hindbrain show paternal Grb10 expression (6). Additionally, Grb10 paternal expression is excluded from the cortex both during development and in the adult (6). Grb10 paternal expression seems to be exclusive to neurons while being excluded from glia in adult tissues, and Grb10 expression in primary glial cultures and cell lines has been previously described as exclusively maternal (50). In situ hybridization for Grb10 in the adult mouse brain has shown that biallelic expression of Grb10 is limited to very few brain regions, suggesting that maternal glial expression in the adult brain is rare (6).
Heterozygous Grb10 mutant mice that inherited the deletion paternally exhibit no overgrowth in the central nervous system (CNS) and have no obvious anatomical changes (6). Despite no gross CNS phenotype, Grb10 paternal knockout mice display behaviors consistent with increased social dominance. In the tube test, two mice are placed on either side of a long narrow opening until one mouse forces the other to retreat (51). Grb10 heterozygous paternal knockout mice are less likely to back down in direct confrontation with an unfamiliar mouse in the tube test compared with WT littermates (6). Facial barbering is a dominance-related behavior that occurs during cogrooming, where a dominant mouse barbers other passive mice (52). Cagemates of paternal knockout mice had barbered whiskers, which regrew when caged with only WT littermates. Anxiety behavior, locomotor activity, olfactory function, and general aggression have also been assessed in paternal Grb10 knockout mice but were not significantly different from WT controls (6).
These behavior phenotypes, coupled with a lack of morphological changes in the central nervous system in paternal Grb10 knockout mice, suggest that Grb10 has a currently uncharacterized function unique to neurons. Grb10’s neuronal function is particularly difficult to parse because the brain regions involved in social dominance behavior have not been fully elucidated. The social hierarchy of mice has been shown to be predictably modified by changing the synaptic efficacy of neurons within the medial prefrontal cortex, but no Grb10 expression is evident within cortical tissues in the embryo or the adult (6, 53). Dominance-behavior paradigms often describe dominant and submissive like behavior being the result of an animal’s reaction toward acute and prolonged social stress (54). The limbic-hypothalamic-pituitary-adrenal (LHPA) axis is the biological system most associated with regulating stress response, among other functions (55). Paternal Grb10 expression is evident in LHPA-related brain regions, including the hypothalamus, making stress-related signaling in the LHPA a potential candidate for Grb10 neuronal function. Another imprinted gene with a role in novel exploration behavior, Nesp, has overlapping expression with Grb10 within brain regions involved in the LHPA (56). It has been suggested that these imprinted genes may be playing interconnected roles in controlling their respective behaviors although more directed experiments testing Grb10 function in the LHPA are necessary to connect these two proteins and to elucidate their role in the brain.
The Regulation of the Tissue-Specific and Imprinted Expression of Grb10 in Mice
As described in the Introduction, Grb10-imprinted expression is dependent on its ICR, which is DNA-methylated on the maternal chromosome and bound by CTCF on the paternal chromosome in all tissues (Fig. 1). Maternal expression, specific to nonneuronal tissues, is observed in mice from its major promoter whereas paternal expression, specific to neurons, initiates from at least three downstream alternative promoters. Paternal deletion of the Grb10 ICR in mice results in biallelic expression of the major isoform of Grb10 in all tissues, including neonatal brain (34). Paternal expression of neuron-specific Grb10 isoforms in these mice was ablated as at least one of the paternal-specific promoters is included in the deletion. This deletion results in global prenatal and postnatal undergrowth that persists through adulthood although the specific effects on brain size and morphology were not reported. Maternal deletion of the Grb10 ICR produced no observable adverse phenotype. These results suggest that the paternal ICR acts as a repressor of the major promoter of Grb10, likely through the paternal recruitment of CTCF, which has been characterized with repressor activity (57).
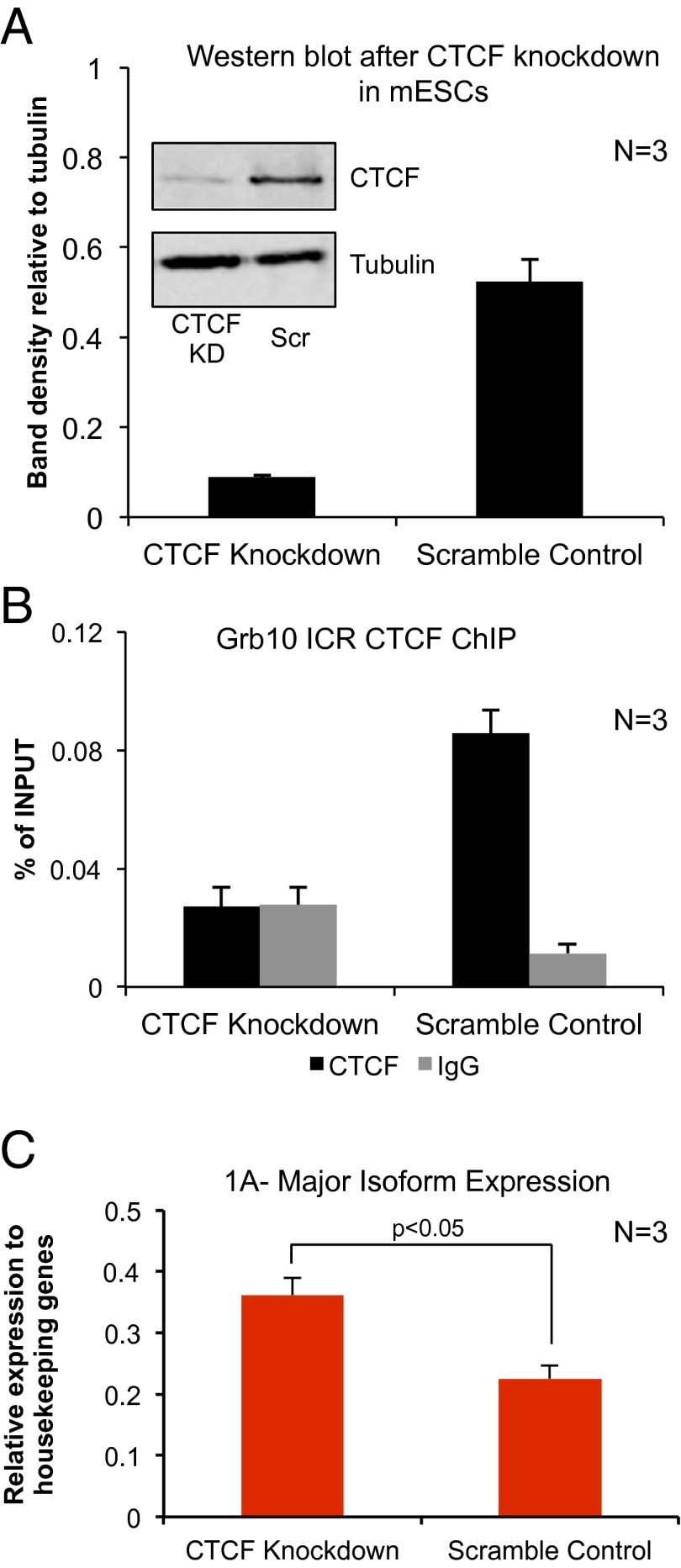
To test the role for CTCF in repression at the Grb10 locus, we depleted CTCF in mESCs using shRNA. CTCF depletion resulted in modest but significant up-regulation of the major Grb10 isoform (Fig. 3), but expression of the neuronal-specific isoform, which is repressed in mESCs, was unaffected and remained repressed. These results are consistent with CTCF acting as a repressor of the major promoter on the paternal allele although it is difficult to rule out indirect effects of CTCF knockdown. Other functions of CTCF could also explain this result. For example, CTCF acts as an enhancer blocker at the H19/Igf2-imprinted locus (58). This latter activity, however, would require the identification of an as yet uncharacterized Grb10 enhancer that drives expression of the major promoter. Moreover, it is unclear how CTCF’s role may differ in humans. Despite similar maternal methylation and paternally bound CTCF at the Grb10 ICR, the major promoter has been shown to be biallelically expressed in most human tissues and is repressed in neurons (49). It is notable, however, that Grb10 expression has been described to be maternally derived in human placenta and embryonic muscle, suggesting that Grb10 imprinting may be more nuanced and cell type-specific than in mouse.
Fig. 3.
Grb10 expression in mouse embryonic stem cells after CTCF knockdown results in the up-regulation of the major isoform. (A) Representative Western blot for CTCF and tubulin comparing cells infected with CTCF shRNA (KD) and scramble control (Scr). Average densitometry of the CTCF bands across three replicates is shown in the bar graph. (B) CTCF knockdown results in loss of CTCF enrichment over background at the Grb10 ICR compared with IgG. (C) CTCF knockdown causes a small but significant increase in the expression of the major Grb10 isoform (1A).
Because DNA methylation and CTCF binding are invariant across tissue types and species at the Grb10 ICR, they alone cannot explain tissue-specific promoter utilization or the difference in allele-specific expression between neuronal and nonneuronal tissues. Allele-specific histone modifications at the major promoter and the neuron-specific promoters have been implicated in Grb10-imprinted gene expression. In nonneuronal tissues in human and mouse, the DNA-methylated maternal ICR is enriched for repressive histone marks (H3K9me3 and H4K20me3) whereas the unmethylated paternal ICR is enriched for both active (H3K4me2) and repressive (H3K27me3) marks (47). In the brain, however, the ICR is enriched, for the active histone marks H3K9ac and H3K27ac on the expressed paternal allele (47). Thus, a bivalent chromatin domain (H3K4me2/H3K27me3) exists on the paternal Grb10 ICR in most tissues, repressing the neuronal-specific promoter, whereas, in neurons, active histone marks appear coincidentally with paternal transcription. Experiments using in vitro differentiation of neurospheres have also shown lowered enrichment of H3K27me3 and increased enrichment of H3K9ac on the paternal ICR, concordant with differentiation (47). Additionally, mESCs null for EED, a critical component of the polycomb repressive complex 2 (PRC2) that maintains repressed chromatin, express Grb10 from the neuronal-specific promoter, which is normally silent (47). Similar allele-specific histone modifications are also observed at the Grb10 major promoter. In cultured mouse fibroblasts, the major promoter is paternally enriched for repressive chromatin marks (H3K27me) and maternally enriched for activating chromatin marks (H3/4 pan-acetyl). In primary neuronal culture, however, the repressive marks are bound equally on both alleles, consistent with biallelic repression (50).
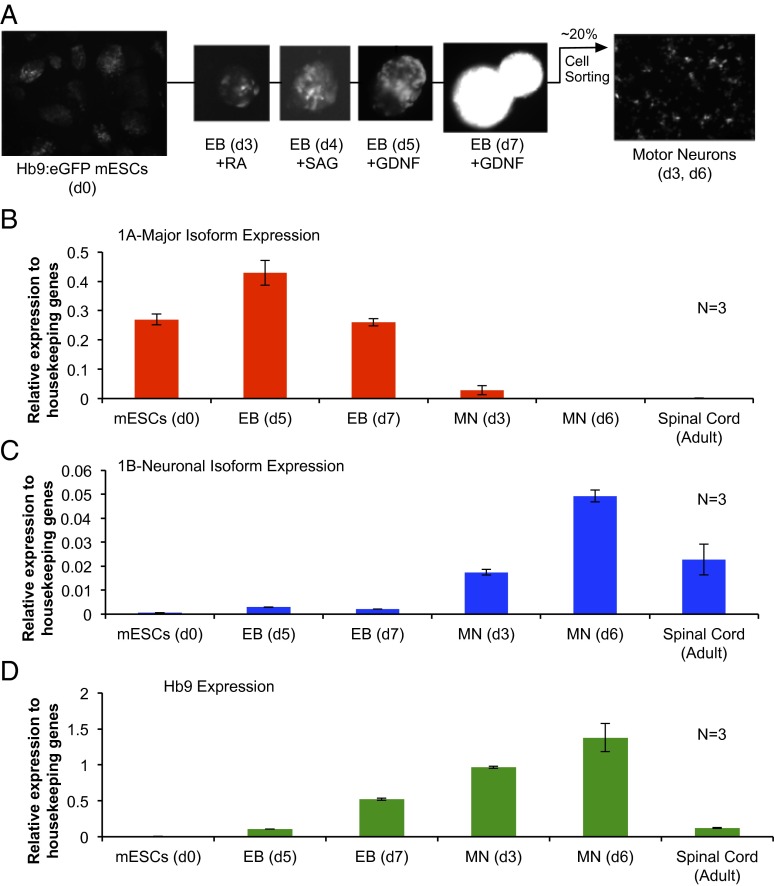
These and other studies of Grb10 expression in neurons have so far been conducted in a mixed population of neuronal and glial cells within tissues or primary culture. In particular, the adult brain is over 70% glia cells, meaning that the majority of cells used for such experiments do not express paternal Grb10. Thus, studying paternally expressed Grb10 during neuralization is particularly challenging, especially using in vivo systems. To address this issue, we have adapted an in vitro model for differentiating mESCs into a homogenous population of postmitotic alpha motor neurons (59). The maturation of these motor neurons corresponds with the repression of the major promoter and the activation of the neuron-specific promoter (Fig. 4 B–D). We observed even higher relative expression of this isoform than in adult spinal cord, likely because of the enrichment for a neuronal population over glia. These in vitro data suggest that Grb10 expression may be specific to neurons as opposed to glial populations during neurogenesis, as is evident in adult tissues. This differentiation model and others like it could be adapted for further study of Grb10 regulation and function specifically in neurons.
Fig. 4.
Promoter use switches from the maternal to paternal isoform of Grb10 during motor neuron differentiation in vitro. (A) Schematic of motor neuron differentiation. Mouse ESCs that express eGFP under the control of the promoter of an early motor neuron marker, Hb9 (d0), are used to form embryoid bodies (EBs). To induce neuralization, EBs are treated with retinoic acid (RA) on day 3 of differentiation. To induce ventralization of neural tissue and motor neurons, Smoothened agonist (SAG) is added to the EB culture on differentiation day 4. Glial cell-derived neurotrophic factor (GDNF) is added to EB culture post day 5 to promote neuronal survival. On day 7 of differentiation, EBs are dissociated and FACs-sorted based on eGFP expression, with an approximate 20% yield of motor neurons. (B–D) The maternally expressed major (1A) isoform of Grb10 is repressed during the differentiation process whereas the paternally expressed neuronal-specific (1B) isoform is up-regulated. These changes in expression correlate with the maturation of motor neurons as quantified by Hb9 expression, a marker for motor-neuron fate. For comparison, levels of expression of each of these genes in the adult spinal cord are shown.
Discussion and Future Directions
Numerous questions remain regarding the regulation and function of Grb10. How Grb10’s tissue-specific and allele-specific expression contributes to its disparate functional roles is currently conjectural. It is tempting to speculate that, in neurons, the paternally expressed Grb10 transcript encodes a unique Grb10 protein isoform. In mice, it is evident that both the major and the neuronal-specific promoters transcribe mRNAs that include all known coding exons. Public datasets show a number of truncated isoforms that originate from the neuronal-specific promoter, which could encode a truncated Grb10 protein. Theses transcripts, however, have yet to be validated in relevant tissue types. It is additionally possible that more-subtle alternative splicing differences between the paternal and maternal transcripts exist because alternative splicing of specific exons within Grb10 is still poorly understood. It is also likely that the function of Grb10 in neurons is distinct from its role in other somatic tissues because of differences in protein partners and relevant signaling molecules. A number of known Grb10 protein partners have critical roles in neuronal function and neurogenesis, including GIGYF2, NEDD4, and PDGFRβ (Table S1). Defining Grb10’s interactions with these proteins in neurons will elucidate possible neuronal-specific functions of the Grb10 paternal isoform. Additionally, as neuronal insulin signaling plays a critical role in the homeostatic regulation of the body’s metabolism, it is of interest to know whether the role of Grb10 in insulin signaling is maintained in neurons.
The regulation of Grb10 expression also requires further study. Specifically, the interplay between DNA methylation, CTCF, and histone-modifying enzymes has yet to be dissected at the locus. Again, tissue-specific differences in the transcriptional machinery must underlie the tissue-specific difference in expression. Specifically, it is possible that an uncharacterized tissue-specific transcription factor plays a role in the neuronal expression of Grb10. It is also likely that yet to be identified regulatory elements are involved in Grb10 expression. Recent work, for example, identified a downstream putative enhancer element that is critical for paternal expression in brain compared with the spinal cord (46). Further functional experiments that define regulatory regions in the locus are essential to elucidate the mechanisms that control Grb10 expression.
Materials and Methods
Expression Analysis via PCR.
RNA extraction, reverse transcription, and PCR methodologies were used to assess transcript variants as previously described (60). Quantitative measurement of Grb10 and Hb9 expression via quantitative PCR (qPCR) was performed by comparing their expression levels with the geometric mean of the expression of the three housekeeping genes Nono, ArpO, and Gapdh as previously described (60). Expression primers and allele-specific expression-assay information are listed in Table S2.
CTCF Knockdown in Mouse Embryonic Stem Cells.
CTCF knockdown in mESCs was performed via lentiviral infection of shRNA as previously described (60, 61). ChIP and Western blot analysis were performed as previously described to confirm CTCF knockdown using antibodies against CTCF (07729; Millipore), normal rabbit IgG (SC2027; Santa Cruz), and α-tubulin (04-1117; Millipore). ChIP qPCR primers are listed in Table S2.
Culture of Mouse Embryonic Stem Cells.
Mouse embryonic cells stem cells were cultured with mitomycin C-treated mouse embryonic fibroblasts as previously described (60). CTCF knockdown experiments were conducted in E14 mESCs. The HBG3 mESC line was used for neuronal differentiation (59).
Neuron Differentiation and Isolation.
HBG3 mESCs, which express eGFP as a transgene under the Hb9 promoter, were differentiated toward a motor neuron lineage as previously described (59). In brief, 2E6 mESCs were seeded in a 10-cm dish in differentiation media [1:1 (vol/vol) DMEM/F12:neurobasal media, 10% (vol/vol) knockout serum, 0.1 μM beta-mercaptoethanol] to form embryoid bodies. On day 3 of differentiation, embryoid bodies were split 1:4 in differentiation media with 1 μM retinoic acid and 0.5 μM Smoothened agonist (SAG). Embryoid bodies were dissociated with 0.25% Trypsin on day 7 of differentiation and sorted based on eGFP expression in a BD biosciences FACS Aria III. Sorted cells were then suspended in motor neuron media (1:1 DMEM/F12:neurobasal media, 1× B27 supplement, 25 mM glutamate, 10 ng/mL GDNF) on plates treated with poly-D lysine and laminin. Motor neurons were collected for expression analysis 3 d and 6 d after plating (Fig. 4A).
Supplementary Material
Acknowledgments
We thank Hynek Wichterle for generously providing the Hb9::GFP mouse embryonic stem cell line and for continued advice and support. We also acknowledge Stewart Anderson, who aided in our collaboration with the Wichterle laboratory. We acknowledge the University of Pennsylvania Flow Cytometry and Cell Sorting Resource Laboratory for their assistance and expertise. This work was supported by National Institutes of Health Grants GM051279 (to M.S.B.) and T32GM008216 (to R.N.P.).
Footnotes
The authors declare no conflict of interest.
This paper results from the Arthur M. Sackler Colloquium of the National Academy of Sciences, “Epigenetic Changes in the Developing Brain: Effects on Behavior,” held March 28–29, 2014, at the National Academy of Sciences in Washington, DC. The complete program and video recordings of most presentations are available on the NAS website at www.nasonline.org/Epigenetic_changes.
This article is a PNAS Direct Submission. E.B.K. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1411254111/-/DCSupplemental.
References
- 1.Han DC, Shen TL, Guan JL. The Grb7 family proteins: Structure, interactions with other signaling molecules and potential cellular functions. Oncogene. 2001;20(44):6315–6321. doi: 10.1038/sj.onc.1204775. [DOI] [PubMed] [Google Scholar]
- 2.Holt LJ, Siddle K. Grb10 and Grb14: Enigmatic regulators of insulin action—and more? Biochem J. 2005;388(Pt 2):393–406. doi: 10.1042/BJ20050216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Riedel H. Grb10 exceeding the boundaries of a common signaling adapter. Front Biosci. 2004;9:603–618. doi: 10.2741/1227. [DOI] [PubMed] [Google Scholar]
- 4.Kabir NN, Kazi JU. Grb10 is a dual regulator of receptor tyrosine kinase signaling. Mol Biol Rep. 2014;41(4):1985–1992. doi: 10.1007/s11033-014-3046-4. [DOI] [PubMed] [Google Scholar]
- 5.Charalambous M, et al. Disruption of the imprinted Grb10 gene leads to disproportionate overgrowth by an Igf2-independent mechanism. Proc Natl Acad Sci USA. 2003;100(14):8292–8297. doi: 10.1073/pnas.1532175100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Garfield AS, et al. Distinct physiological and behavioural functions for parental alleles of imprinted Grb10. Nature. 2011;469(7331):534–538. doi: 10.1038/nature09651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee JT, Bartolomei MS. X-inactivation, imprinting, and long noncoding RNAs in health and disease. Cell. 2013;152(6):1308–1323. doi: 10.1016/j.cell.2013.02.016. [DOI] [PubMed] [Google Scholar]
- 8.Hikichi T, Kohda T, Kaneko-Ishino T, Ishino F. Imprinting regulation of the murine Meg1/Grb10 and human GRB10 genes: Roles of brain-specific promoters and mouse-specific CTCF-binding sites. Nucleic Acids Res. 2003;31(5):1398–1406. doi: 10.1093/nar/gkg232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang J, et al. Grb10, a positive, stimulatory signaling adapter in platelet-derived growth factor BB-, insulin-like growth factor I-, and insulin-mediated mitogenesis. Mol Cell Biol. 1999;19(9):6217–6228. doi: 10.1128/mcb.19.9.6217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moutoussamy S, Renaudie F, Lago F, Kelly PA, Finidori J. Grb10 identified as a potential regulator of growth hormone (GH) signaling by cloning of GH receptor target proteins. J Biol Chem. 1998;273(26):15906–15912. doi: 10.1074/jbc.273.26.15906. [DOI] [PubMed] [Google Scholar]
- 11.Nantel A, Mohammad-Ali K, Sherk J, Posner BI, Thomas DY. Interaction of the Grb10 adapter protein with the Raf1 and MEK1 kinases. J Biol Chem. 1998;273(17):10475–10484. doi: 10.1074/jbc.273.17.10475. [DOI] [PubMed] [Google Scholar]
- 12.Pandey A, Duan H, Di Fiore PP, Dixit VM. The Ret receptor protein tyrosine kinase associates with the SH2-containing adapter protein Grb10. J Biol Chem. 1995;270(37):21461–21463. doi: 10.1074/jbc.270.37.21461. [DOI] [PubMed] [Google Scholar]
- 13.Jahn T, Seipel P, Urschel S, Peschel C, Duyster J. Role for the adaptor protein Grb10 in the activation of Akt. Mol Cell Biol. 2002;22(4):979–991. doi: 10.1128/MCB.22.4.979-991.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.He W, Rose DW, Olefsky JM, Gustafson TA. Grb10 interacts differentially with the insulin receptor, insulin-like growth factor I receptor, and epidermal growth factor receptor via the Grb10 Src homology 2 (SH2) domain and a second novel domain located between the pleckstrin homology and SH2 domains. J Biol Chem. 1998;273(12):6860–6867. doi: 10.1074/jbc.273.12.6860. [DOI] [PubMed] [Google Scholar]
- 15.Kazi JU, Rönnstrand L. FLT3 signals via the adapter protein Grb10 and overexpression of Grb10 leads to aberrant cell proliferation in acute myeloid leukemia. Mol Oncol. 2013;7(3):402–418. doi: 10.1016/j.molonc.2012.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Giorgetti-Peraldi S, Murdaca J, Mas JC, Van Obberghen E. The adapter protein, Grb10, is a positive regulator of vascular endothelial growth factor signaling. Oncogene. 2001;20(30):3959–3968. doi: 10.1038/sj.onc.1204520. [DOI] [PubMed] [Google Scholar]
- 17.Stein EG, Ghirlando R, Hubbard SR. Structural basis for dimerization of the Grb10 Src homology 2 domain: Implications for ligand specificity. J Biol Chem. 2003;278(15):13257–13264. doi: 10.1074/jbc.M212026200. [DOI] [PubMed] [Google Scholar]
- 18.Urschel S, et al. Phosphorylation of grb10 regulates its interaction with 14-3-3. J Biol Chem. 2005;280(17):16987–16993. doi: 10.1074/jbc.M501477200. [DOI] [PubMed] [Google Scholar]
- 19.Hu Z-Q, et al. Grb10 interacts with Bim L and inhibits apoptosis. Mol Biol Rep. 2010;37(7):3547–3552. doi: 10.1007/s11033-010-0002-9. [DOI] [PubMed] [Google Scholar]
- 20.Depetris RS, Wu J, Hubbard SR. Structural and functional studies of the Ras-associating and pleckstrin-homology domains of Grb10 and Grb14. Nat Struct Mol Biol. 2009;16(8):833–839. doi: 10.1038/nsmb.1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mizuno-Yamasaki E, Rivera-Molina F, Novick P. GTPase networks in membrane traffic. Annu Rev Biochem. 2012;81:637–659. doi: 10.1146/annurev-biochem-052810-093700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Giovannone B, et al. GIGYF2 gene disruption in mice results in neurodegeneration and altered insulin-like growth factor signaling. Hum Mol Genet. 2009;18(23):4629–4639. doi: 10.1093/hmg/ddp430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Siddle K. Signalling by insulin and IGF receptors: Supporting acts and new players. J Mol Endocrinol. 2011;47(1):R1–R10. doi: 10.1530/JME-11-0022. [DOI] [PubMed] [Google Scholar]
- 24.Laplante M, Sabatini DM. mTOR signaling at a glance. J Cell Sci. 2009;122(Pt 20):3589–3594. doi: 10.1242/jcs.051011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Desbuquois B, Carré N, Burnol A-F. Regulation of insulin and type 1 insulin-like growth factor signaling and action by the Grb10/14 and SH2B1/B2 adaptor proteins. FEBS J. 2013;280(3):794–816. doi: 10.1111/febs.12080. [DOI] [PubMed] [Google Scholar]
- 26.Yu Y, et al. Phosphoproteomic analysis identifies Grb10 as an mTORC1 substrate that negatively regulates insulin signaling. Science. 2011;332(6035):1322–1326. doi: 10.1126/science.1199484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu F, Roth RA. Grb-IR: A SH2-domain-containing protein that binds to the insulin receptor and inhibits its function. Proc Natl Acad Sci USA. 1995;92(22):10287–10291. doi: 10.1073/pnas.92.22.10287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Langlais P, et al. Negative regulation of insulin-stimulated mitogen-activated protein kinase signaling by Grb10. Mol Endocrinol. 2004;18(2):350–358. doi: 10.1210/me.2003-0117. [DOI] [PubMed] [Google Scholar]
- 29.Wick KR, et al. Grb10 inhibits insulin-stimulated insulin receptor substrate (IRS)-phosphatidylinositol 3-kinase/Akt signaling pathway by disrupting the association of IRS-1/IRS-2 with the insulin receptor. J Biol Chem. 2003;278(10):8460–8467. doi: 10.1074/jbc.M208518200. [DOI] [PubMed] [Google Scholar]
- 30.Deng Y, Zhang M, Riedel H. Mitogenic roles of Gab1 and Grb10 as direct cellular partners in the regulation of MAP kinase signaling. J Cell Biochem. 2008;105(5):1172–1182. doi: 10.1002/jcb.21829. [DOI] [PubMed] [Google Scholar]
- 31.Morrione A, et al. Grb10: A new substrate of the insulin-like growth factor I receptor. Cancer Res. 1996;56(14):3165–3167. [PubMed] [Google Scholar]
- 32.Vecchione A, Marchese A, Henry P, Rotin D, Morrione A. The Grb10/Nedd4 complex regulates ligand-induced ubiquitination and stability of the insulin-like growth factor I receptor. Mol Cell Biol. 2003;23(9):3363–3372. doi: 10.1128/MCB.23.9.3363-3372.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morrione A, et al. mGrb10 interacts with Nedd4. J Biol Chem. 1999;274(34):24094–24099. doi: 10.1074/jbc.274.34.24094. [DOI] [PubMed] [Google Scholar]
- 34.Shiura H, et al. Paternal deletion of Meg1/Grb10 DMR causes maternalization of the Meg1/Grb10 cluster in mouse proximal Chromosome 11 leading to severe pre- and postnatal growth retardation. Hum Mol Genet. 2009;18(8):1424–1438. doi: 10.1093/hmg/ddp049. [DOI] [PubMed] [Google Scholar]
- 35.Yamamoto Y, et al. Type 2 diabetes mellitus in a non-obese mouse model induced by Meg1/Grb10 overexpression. Exp Anim. 2008;57(4):385–395. doi: 10.1538/expanim.57.385. [DOI] [PubMed] [Google Scholar]
- 36.Shiura H, et al. Meg1/Grb10 overexpression causes postnatal growth retardation and insulin resistance via negative modulation of the IGF1R and IR cascades. Biochem Biophys Res Commun. 2005;329(3):909–916. doi: 10.1016/j.bbrc.2005.02.047. [DOI] [PubMed] [Google Scholar]
- 37.Smith FM, et al. Mice with a disruption of the imprinted Grb10 gene exhibit altered body composition, glucose homeostasis, and insulin signaling during postnatal life. Mol Cell Biol. 2007;27(16):5871–5886. doi: 10.1128/MCB.02087-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang L, et al. Peripheral disruption of the Grb10 gene enhances insulin signaling and sensitivity in vivo. Mol Cell Biol. 2007;27(18):6497–6505. doi: 10.1128/MCB.00679-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang J, et al. Disruption of growth factor receptor-binding protein 10 in the pancreas enhances β-cell proliferation and protects mice from streptozotocin-induced β-cell apoptosis. Diabetes. 2012;61(12):3189–3198. doi: 10.2337/db12-0249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Prokopenko I, et al. A central role for GRB10 in regulation of islet function in man. PLoS Genet. 2014;10(4):e1004235. doi: 10.1371/journal.pgen.1004235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kitamura T, Kahn CR, Accili D. Insulin receptor knockout mice. Annu Rev Physiol. 2003;65:313–332. doi: 10.1146/annurev.physiol.65.092101.142540. [DOI] [PubMed] [Google Scholar]
- 42.Charalambous M, et al. Maternally-inherited Grb10 reduces placental size and efficiency. Dev Biol. 2010;337(1):1–8. doi: 10.1016/j.ydbio.2009.10.011. [DOI] [PubMed] [Google Scholar]
- 43.Holt LJ, et al. Dual ablation of Grb10 and Grb14 in mice reveals their combined role in regulation of insulin signaling and glucose homeostasis. Mol Endocrinol. 2009;23(9):1406–1414. doi: 10.1210/me.2008-0386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Holt LJ, et al. Grb10 regulates the development of fiber number in skeletal muscle. FASEB J. 2012;26(9):3658–3669. doi: 10.1096/fj.11-199349. [DOI] [PubMed] [Google Scholar]
- 45.Liu M, et al. Grb10 promotes lipolysis and thermogenesis by phosphorylation-dependent feedback inhibition of mTORC1. Cell Metab. 2014;19(6):967–980. doi: 10.1016/j.cmet.2014.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cowley M, et al. Developmental programming mediated by complementary roles of imprinted Grb10 in mother and pup. PLoS Biol. 2014;12(2):e1001799. doi: 10.1371/journal.pbio.1001799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sanz LA, et al. A mono-allelic bivalent chromatin domain controls tissue-specific imprinting at Grb10. EMBO J. 2008;27(19):2523–2532. doi: 10.1038/emboj.2008.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Arnaud P, et al. Conserved methylation imprints in the human and mouse GRB10 genes with divergent allelic expression suggests differential reading of the same mark. Hum Mol Genet. 2003;12(9):1005–1019. doi: 10.1093/hmg/ddg110. [DOI] [PubMed] [Google Scholar]
- 49.Monk D, et al. Reciprocal imprinting of human GRB10 in placental trophoblast and brain: Evolutionary conservation of reversed allelic expression. Hum Mol Genet. 2009;18(16):3066–3074. doi: 10.1093/hmg/ddp248. [DOI] [PubMed] [Google Scholar]
- 50.Yamasaki-Ishizaki Y, et al. Role of DNA methylation and histone H3 lysine 27 methylation in tissue-specific imprinting of mouse Grb10. Mol Cell Biol. 2007;27(2):732–742. doi: 10.1128/MCB.01329-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lindzey G, Winston H, Manosevitz M. Social dominance in inbred mouse strains. Nature. 1961;191:474–476. doi: 10.1038/191474a0. [DOI] [PubMed] [Google Scholar]
- 52.Sarna JR, Dyck RH, Whishaw IQ. The Dalila effect: C57BL6 mice barber whiskers by plucking. Behav Brain Res. 2000;108(1):39–45. doi: 10.1016/s0166-4328(99)00137-0. [DOI] [PubMed] [Google Scholar]
- 53.Wang F, et al. Bidirectional control of social hierarchy by synaptic efficacy in medial prefrontal cortex. Science. 2011;334(6056):693–697. doi: 10.1126/science.1209951. [DOI] [PubMed] [Google Scholar]
- 54.Tamashiro KLK, Nguyen MMN, Sakai RR. Social stress: From rodents to primates. Front Neuroendocrinol. 2005;26(1):27–40. doi: 10.1016/j.yfrne.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 55.Ulrich-Lai YM, Herman JP. Neural regulation of endocrine and autonomic stress responses. Nat Rev Neurosci. 2009;10(6):397–409. doi: 10.1038/nrn2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dent CL, Isles AR. Brain-expressed imprinted genes and adult behaviour: the example of Nesp and Grb10. Mamm Genome. 2014;25(1-2):87–93. doi: 10.1007/s00335-013-9472-0. [DOI] [PubMed] [Google Scholar]
- 57.Ohlsson R, Renkawitz R, Lobanenkov V. CTCF is a uniquely versatile transcription regulator linked to epigenetics and disease. Trends Genet. 2001;17(9):520–527. doi: 10.1016/s0168-9525(01)02366-6. [DOI] [PubMed] [Google Scholar]
- 58.Hark AT, et al. CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature. 2000;405(6785):486–489. doi: 10.1038/35013106. [DOI] [PubMed] [Google Scholar]
- 59.Wichterle H, Lieberam I, Porter JA, Jessell TM. Directed differentiation of embryonic stem cells into motor neurons. Cell. 2002;110(3):385–397. doi: 10.1016/s0092-8674(02)00835-8. [DOI] [PubMed] [Google Scholar]
- 60.Plasschaert RN, et al. CTCF binding site sequence differences are associated with unique regulatory and functional trends during embryonic stem cell differentiation. Nucleic Acids Res. 2014;42(2):774–789. doi: 10.1093/nar/gkt910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lin S, Ferguson-Smith AC, Schultz RM, Bartolomei MS. Nonallelic transcriptional roles of CTCF and cohesins at imprinted loci. Mol Cell Biol. 2011;31(15):3094–3104. doi: 10.1128/MCB.01449-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.