ABSTRACT
During development, the rate of cell proliferation must be constantly monitored so that an individual tissue achieves its correct size. Mutations in genes that normally promote tissue growth often result in undersized, disorganized and non-functional organs. However, mutations in genes that encode growth inhibitors can trigger the onset of tumorigenesis and cancer. The developing eye of the fruit fly, Drosophila melanogaster, has become a premier model system for studies that are focused on identifying the molecular mechanisms that underpin growth control. Here, we examine the mechanism by which the Notch pathway, a major contributor to growth, promotes cell proliferation in the developing eye. Current models propose that the Notch pathway directly influences cell proliferation by regulating growth-promoting genes such as four-jointed, cyclin D1 and E2f1. Here, we show that, in addition to these mechanisms, some Notch signaling is devoted to blocking the growth-suppressing activity of the bHLH DNA-binding protein Daughterless (Da). We demonstrate that Notch signaling activates the expression of extramacrochaetae (emc), which encodes a helix-loop-helix (HLH) transcription factor. Emc, in turn, then forms a biochemical complex with Da. As Emc lacks a basic DNA-binding domain, the Emc-Da heterodimer cannot bind to and regulate genomic targets. One effect of Da sequestration is to relieve the repression on growth. Here, we present data supporting our model that Notch-induced cell proliferation in the developing eye is mediated in part by the activity of Emc.
Keywords: Notch, Extramacrochaetae, Daughterless, Drosophila, Retina, Cell proliferation
Summary: The bHLH transcription factor Emc acts downstream of Notch signalling to promote proliferation in the Drosophila eye by sequestering the growth suppressor Da away from its target genes.
INTRODUCTION
An essential component of development is the regulation of cell proliferation. All tissues and organs originate from just a handful of early embryonic cells. These cells must divide rapidly in order to produce the requisite number of cells for the assigned tissue fate. As organs vary considerably in their final size, each precursor cell population must adjust its growth rate in order to develop synchronously with other tissues/organs. In addition, each organ must have a way to sense when its final size has been reached and then employ mechanisms to inhibit further cell proliferation. These processes are regulated by a multitude of signal transduction cascades and transcription factor networks, many of which directly impinge on the cell cycle and/or the apoptotic machinery.
Growth of the developing eye is controlled by two different phases of cell proliferation. The first phase can be broadly defined to include all cell divisions that take place during the first and second larval instars, as well as those ahead of the morphogenetic furrow during the third and final larval stage. This includes a coordinated wave of mitosis that occurs just ahead of the morphogenetic furrow called the ‘first mitotic wave’. This early growth phase is responsible for setting the overall number of ommatidia within the compound eye and is controlled by several signaling pathways, including JAK/STAT, Notch, Decapentaplegic (Dpp/TGFβ), Wingless (Wg) and Hippo (Panin et al., 1997; Cho and Choi, 1998; de Celis et al., 1998; Domínguez and de Celis, 1998; Go et al., 1998; Papayannopoulos et al., 1998; Kango-Singh et al., 2002; Tapon et al., 2002; Baonza and Freeman, 2005; Firth and Baker, 2005; Reynolds-Kenneally and Mlodzik, 2005; Fan and Bergmann, 2008; Gutierrez-Aviño et al., 2009; Koontz et al., 2013). The overlying peripodial membrane serves as an additional source of growth signals. Several studies have demonstrated that Dpp, Wg, Hedgehog (Hh) and Serrate (Ser), a Notch ligand, are all transported from the peripodial membrane and are required for the growth (and patterning) of the eye disc (Cho et al., 2000; Gibson and Schubiger, 2000; Gibson et al., 2002).
The second growth phase comprises the ‘second mitotic wave’, a synchronized band of mitoses that occurs behind the furrow and produce the following elements of each ommatidium: three of the eight photoreceptors (R1,6,7), all lens-secreting cone cells, all optical insulating pigment cells and all cells of the bristle complex (Ready et al., 1976; Wolff and Ready, 1991). These late cell divisions produce the vast majority of cells within each ommatidium and are therefore the major determinants of the overall number of cells within the adult retina. The Notch and EGF Receptor (Egfr) pathways regulate entry into and passage through the cell cycle of the second mitotic wave (Baker and Yu, 2001; Baonza et al., 2002; Yang and Baker, 2003; Baonza and Freeman, 2005; Firth and Baker, 2005, 2007; Sukhanova and Du, 2008). In addition to these growth signals, the Hh pathway controls compensatory proliferation as cells differentiate behind the furrow (Fan and Bergmann, 2008).
The establishment of the midline within the developing eye is essential for Notch activation and cell proliferation during early growth phases (Cho and Choi, 1998; Domínguez and de Celis, 1998; Papayannopoulos et al., 1998). The embryonic and first instar eye is composed of entirely ventral-fated tissue (Chern and Choi, 2002; Singh and Choi, 2003; Singh et al., 2006). These cells express fringe (fng), which encodes a glycosyltransferase (Irvine and Wieschaus, 1994). In the latter half of the first instar, pannier (pnr) expression is activated along the dorsal margin of the eye and in turn Pnr protein initiates a cascade of events that leads to the repression of fng with the dorsal half of the retina (Heitzler et al., 1996; Cho and Choi, 1998; Heberlein et al., 1998; Cavodeassi et al., 1999; Yang et al., 1999; Maurel-Zaffran and Treisman, 2000; Sato and Tomlinson, 2007; Oros et al., 2010). The juxtaposition of fng-positive and fng-negative cells leads to the activation of Notch signaling at the midline, which plays an essential role in promoting localized growth in the eye (Panin et al., 1997; Cho and Choi, 1998; Domínguez and de Celis, 1998; Papayannopoulos et al., 1998).
Here, we address the role that the helix-loop-helix (HLH) protein Extramacrochaetae (Emc) plays in mediating some of the Notch-induced growth. The emc locus was first identified by mutations that increased the number of macrochaetae class bristles (Craymer, 1980). emc, along with hairy (h), was subsequently shown to genetically modify the mutant phenotypes of members of the achaetae-scute complex (AS-C) (Botas et al., 1982; Moscoso del Prado and Garcia-Bellido, 1984). The diametrically opposing phenotypes of emc and AS-C loss-of-function mutants hinted at a biochemically antagonistic relationship. Members of the AS-C encode basic helix-loop-helix (bHLH) transcription factors, whereas Emc encodes a helix-loop-helix (HLH) protein (Villares and Cabrera, 1987; Murre et al., 1989a,b; Ellis et al., 1990; Garrell and Modolell, 1990). Emc binds to and forms heterodimers with several AS-C proteins and the class I bHLH factor Daughterless (Da) (Van Doren et al., 1991, 1992; Alifragis et al., 1997). However, as Emc lacks a basic domain, neither Emc itself nor Emc-bHLH heterodimers can interact with DNA (Van Doren et al., 1991, 1992). Therefore, Emc functions to sequester bHLH proteins away from downstream target genes. Mutations within emc have documented growth defects in both the developing wing and eye (García-Alonso and García-Bellido, 1988; de Celis et al., 1995; Baonza and García-Bellido, 1999; Baonza et al., 2000; Bhattacharya and Baker, 2009; Spratford and Kumar, 2013). However, the sequestration targets of Emc and the developmental mechanism by which it promotes cell proliferation are not well defined.
In several contexts, emc appears to function downstream of the Notch pathway (Baonza et al., 2000; Baonza and Freeman, 2001; Adam and Montell, 2004; Tapanes-Castillo and Baylies, 2004; Bhattacharya and Baker, 2009; Spratford and Kumar, 2015). Here, we show that the Notch pathway activates emc expression. Emc protein then promotes cell proliferation by binding to and sequestering Da away from endogenous DNA targets. We show that the sequestration of Da by Emc is sufficient to block the growth-inhibiting activity of Da. And finally, we provide evidence that entry into S phase may be slowed in emc mutant tissue. Our study complements another study that indicates that Emc-mediated regulation of Da also affects passage of dividing cells through the G2/M checkpoint (Andrade-Zapata and Baonza, 2014).
RESULTS
Emc is required for normal proliferation in the developing eye
Several studies have demonstrated that emc mutant clones proliferate poorly in comparison with wild-type cells (García-Alonso and García-Bellido, 1988; de Celis et al., 1995; Baonza and García-Bellido, 1999; Baonza et al., 2000; Bhattacharya and Baker, 2009). In this paper, we set out to determine the mechanism by which Emc regulates tissue growth in the developing eye disc. We began this study by measuring the growth rates of clones of wild-type and emcAP6 null mutant cells. Consistent with prior reports, we observe that the growth rate of emcAP6-null mutant clones is markedly less robust when compared with wild-type clones (Fig. 1A,B,E,F,I,J,M). However, if emcAP6-null clones are surrounded by tissue that is heterozygous for a Minute mutation, then the mutant tissue grows just as well as wild-type cells that are placed in the same environment (Fig. 1C,D,G,H,K,L,M). From these data we conclude that the defects in tissue growth are not due to a requirement for Emc in maintaining cell viability. We next considered a role for Emc in the suppression of apoptosis. To test this hypothesis, we used the MARCM method to overexpress p35 in both wild-type and emcAP6 mutant cells (Fig. 1N-R). We did not observe an increase in cleaved caspase 3 (CC3) levels in emcAP6 mutant clones (data not shown) nor did the expression of p35 restore growth to emcAP6 mutant cells (Fig. 1R); therefore, we conclude that the growth deficit in emc mutant tissue is also not due to an increase in apoptotic cell death. In light of this set of conclusions, we turned our attention to a possible role for Emc in promoting cell proliferation.
Fig. 1.
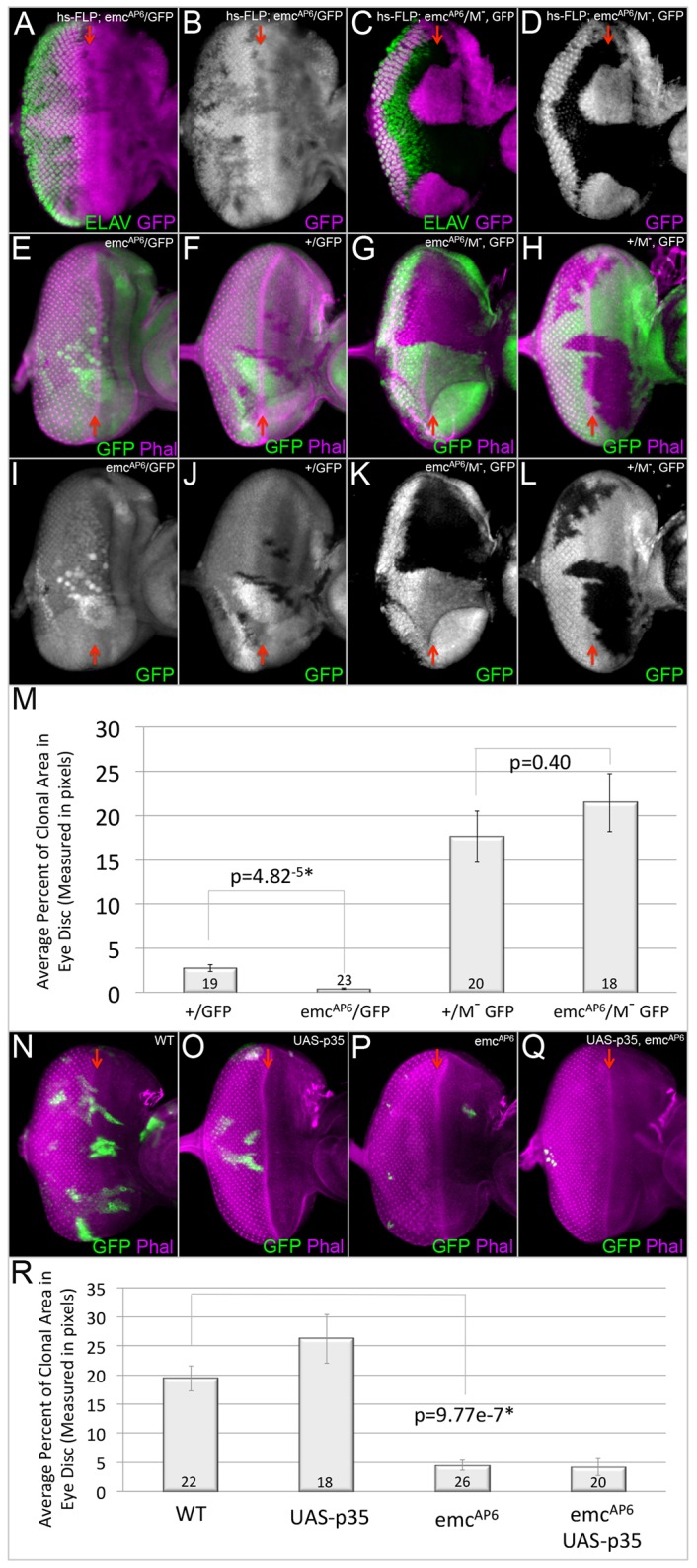
Emc is required for normal growth of the developing eye. (A-L,N-Q) Light microscope images of third instar eye discs containing loss-of-function or MARCM clones. Dorsal side is upwards and anterior is towards the right. The red arrows indicate the position of the morphogenetic furrow. All discs were photographed at 10× magnification. (A,B) emcAP6-null clones in a wild-type background proliferate poorly. (C,D) Growth is restored to emcAP6-null clones that are surrounded by Minute−/+ tissue. (E-L) Representative discs containing mitotic clones (lacking GFP) of differing genotypes. (E,I) emcAP6-null clones. (F,J) Wild-type clones. (G,K) emcAP6-null clones surrounded by Minute−/+ tissue. (H,L) Wild-type clones surrounded by Minute−/+ tissue. (M) The average percentage of disc area occupied by mitotic clones of the genotypes listed in E-L. Statistical significance was calculated using Student's t-test and equal or unequal variance was determined using a F-test. There is unequal variance when comparing +/GFP with emcAP6/M, whereas there is equal variance when comparing emcAP6/+ with +/M. The difference between wild-type and emcAP6 clones is statistically significant with a P-value of 4.82e−5. However, this statistical difference is eliminated when both types of clones are generated in a Minute background and compared with each other. This suggests that that emcAP6clones are outcompeted by surrounding wild-type cells. Error bars represent s.d. (N-Q) MARCM clones expressing GFP induced in a wild-type background. (N) Wild-type MARCM clones. (O) UAS-p35 MARCM clones. (P) emcAP6 MARCM clones. (Q) emcAP6, UAS-p35 MARCM clones. (R) The average percentage of the eye imaginal disc area occupied by MARCM clones of the genotypes listed in N-Q. The difference between wild-type and emcAP6 MARCM clones is statistically significant, with a P-value of 9.77e−7. The number of discs containing clones that were analyzed is listed in M and R. Statistical significance was calculated using Student's t-test, and equal or unequal variance was determined using a F-test. Error bars represent s.d.
Emc functions downstream of Notch-induced proliferation in the eye imaginal disc
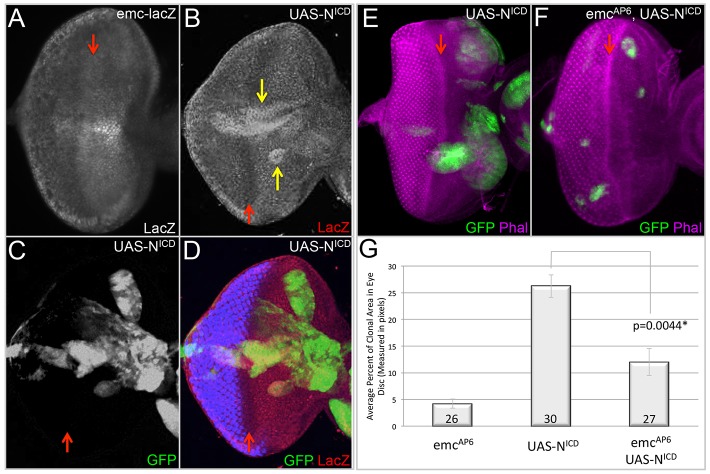
We focused on a potential connection between Notch signaling and Emc as several studies have demonstrated that Emc functions downstream of the Notch pathway in the wing and eye discs, embryonic mesodermal segments, and ovarian follicle cells (Baonza et al., 2000; Adam and Montell, 2004; Tapanes-Castillo and Baylies, 2004; Bhattacharya and Baker, 2009; Spratford and Kumar, 2015). In fact, emc expression appears to be dependent upon Notch pathway activity in several different contexts (Baonza et al., 2000; Baonza and Freeman, 2001; Bhattacharya and Baker, 2009; Spratford and Kumar, 2015). Activation of an emc transcriptional reporter (emc-lacZ) by overexpression of an activated Notch receptor (NICD) in the eye (Fig. 2A-D) is consistent with these findings. In an attempt to identify regulatory regions that may be responsive to Notch signaling, we analyzed the expression of eight genomic fragments surrounding the emc transcriptional start site (supplementary material Fig. S1A; Jory et al., 2012; Manning et al., 2012). Expression within the anterior compartment was of particular interest, as a significant level of Notch-dependent cell proliferation takes place within this region. Of the eight fragments that were tested, we found two putative enhancer regions, including E1 and GMR10H11, that have anterior expression (supplementary material Fig. S1B,F). Within both fragments are binding sites for Suppressor of Hairless [Su(H)], a transcription factor responsible for mediating Notch signaling (data not shown).
Fig. 2.
emc is activated by Notch and mediates its growth-promoting activity in the eye. (A-F) Light microscope images of third instar eye-antennal discs containing flp-out overexpression or MARCM clones. Dorsal side is upwards and anterior is towards the right. The red arrows indicate the position of the morphogenetic furrow. All discs were photographed at 10× magnification. (A) Expression pattern of emc-lacZ in a wild-type third instar eye imaginal disc. (B-D) UAS-NICD flp-out clones expressing GFP show increased levels of emc-lacZ in many compartments of the disc. The yellow arrows indicate clones in which emc-lacZ expression is activated in response to Notch signaling. (E,F) MARCM clones expressing GFP induced in a wild-type background. (E) UAS-NICD MARCM clones. (F) emcAP6, UAS-NICD MARCM clones. (G) The average percentage of the eye imaginal disc area occupied by MARCM clones of genotypes listed in E, F and Fig. 1P. Statistical significance was calculated using Student's t-test and equal or unequal variance was determined using a F-test. The difference between UAS-NICD and emcAP6, UAS-NICD clones is statistically significant with a P-value of 0.0044. The number of discs containing clones that were analyzed is listed within the panel. Error bars represent s.d.
We tested whether emc is required for the proliferative effects of Notch signaling in the eye disc by creating and comparing MARCM clones that overexpress NICD in wild-type and emcAP6 mutant clones. We find that the proliferative effects of Notch are partially mitigated by the loss of emc (Fig. 2E-G). From these results, we conclude that Emc mediates some of the Notch-induced proliferation in the eye disc. We did observe that the loss of emc was not sufficient to completely abrogate the effects of Notch signaling on growth (Fig. 2G). One plausible explanation for this result is that Notch and Emc may act on some common target genes. A similar phenomenon has been observed in the wing (Baonza et al., 2000; Campuzano, 2001).
Emc sequesters Da to promote proliferation in the eye
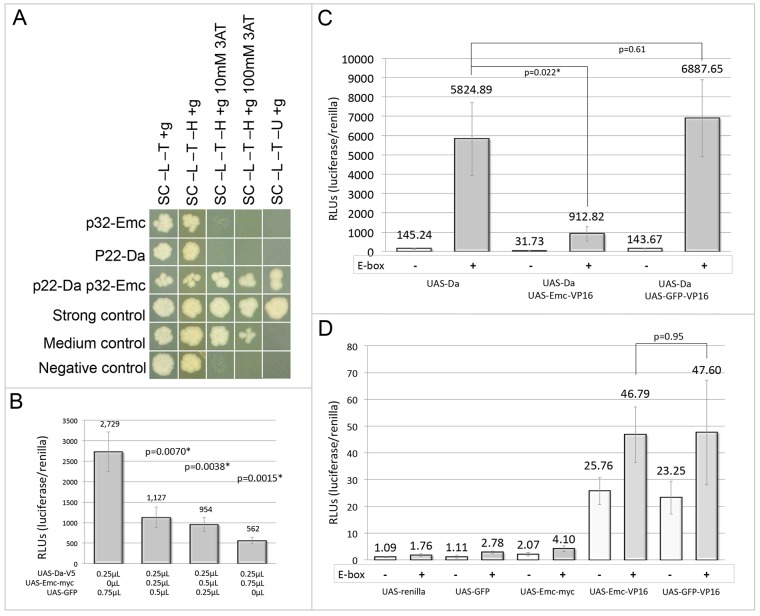
As Emc lacks a basic domain, neither Emc itself nor Emc-bHLH heterodimers should be capable of interacting with DNA (Ellis et al., 1990; Garrell and Modolell, 1990; Van Doren et al., 1991). Thus, Emc and its vertebrate homologs are thought to influence transcription by sequestering bHLH proteins away from target genes. In particular, genetic interaction studies and electromobility shift assays (EMSAs) have identified Da and AS-C members as targets of Emc sequestration (Van Doren et al., 1991, 1992; Martinez et al., 1993). Based on this mode of action, Emc is predicted to promote cell proliferation by sequestering one or more bHLH proteins that, on their own, are tasked with repressing tissue growth. One of the unresolved issues surrounding Emc is the identification of its sequestration targets in proliferating cells. We initiated a yeast two-hybrid screen for Emc-binding partners and identified several candidate bHLH proteins, including Da, Ac and Sage, a bHLH protein that is expressed exclusively in the salivary glands (supplementary material Fig. S2). We focused our attention on Da because flp-out overexpression clones display a severe undergrowth phenotype (Bhattacharya and Baker, 2011; Andrade-Zapata and Baonza, 2014), which is the expected phenotype if the sequestration of Da by Emc is required to promote tissue growth. We conducted a directed yeast two-hybrid assay to assess the strength of the interaction between Emc and Da, and find that it is equivalent to our medium/strong positive control colonies (Fig. 3A). We then confirmed that Emc could sequester Da away from a target sequence. To do this, we developed a luciferase transcriptional reporter assay that could be used in transfected Kc167 Drosophila cells. We multimerized an E-box sequence (Massari and Murre, 2000; Materials and Methods) and placed this synthetic regulatory element within a vector upstream of the firefly luciferase gene. This plasmid was transfected into Kc167 cells simultaneously with combinations of the following plasmids: (1) MT-GAL4, (2) UAS-Da, (3) UAS-Emc, (4) UAS-GFP and (5) UAS-renilla luciferase. Da, on its own, is capable of binding to and strongly activating the E-box-luciferase reporter (Fig. 3B). The addition of increasing amounts of Emc significantly inhibits reporter activation, thereby supporting the proposition that Emc antagonizes Da (and presumably other bHLH proteins) through sequestration (Fig. 3B).
Fig. 3.
Emc binds and sequesters Da away from DNA. (A) In a directed yeast two-hybrid experiment, Emc and Da interact with a strength similar to that of the medium/strong control colonies. (B-D) Results from luciferase transcriptional activation assays. Experiments in which an E-box-containing vector is present are represented by gray bars; those lacking the E-box-containing vector are represented by white bars. In all cases, UAS-renilla and UAS-GFP were used as negative controls. Error bars represent s.d. Statistical significance was calculated using Student's t-test and equal or unequal variance was determined using a F-test. (B) Emc sequesters Da away from the E-box multimer. The addition of increasing amounts of Emc-myc to steady amounts of Da-V5 progressively reduces E-box reporter transcription. Columns 2 and 4 had unequal variance, whereas column 3 had equal variance. (C) A Da-Emc complex does not bind to the E-box multimer. The addition of Emc-VP16 to Da-V5 significantly reduces Da activation of the E-box reporter, while the activation of the reporter by Da-V5 is unaffected by the addition of GFP-VP16 to Da-V5 and does not significantly alter reporter levels. Unequal variance was found when comparing experiments in which Da was expressed in the absence or presence of the E-box target (columns 1 and 2). Equal variance was found for comparisons between experiments in which Da was expressed alone or in the presence of either Emc-VP16 or GFP-VP16 fusions (columns 2, 4 and 6). A statistically significant difference in luciferase activity was observed when comparing the expression of Da alone and Da+ Emc-VP16. This indicates that Emc sequesters Da away from the target E-box. (D) Emc is unlikely to bind the E-box multimer directly as there is no statistically significant difference between Emc-VP16 and the negative control, GFP-VP16. Variance was determined to be equal when comparing experiments in which Emc-VP16 was expressed in the absence or presence of an E-box (columns 7 and 8) as well as in experiments in which Emc-VP16 and GFP-VP16 were expressed in the presence of the E-box. Unequal variance was found when comparing experiments in which Emc or Emc-VP16 were expressed in the presence of an E-box. There is no statistical difference in luciferase activity when comparing Emc-VP16 and GFP-VP16, indicating that Emc does not bind to the E-box.
We also tested whether Emc could interfere with Da activity through additional mechanisms. In particular, we focused our efforts on determining whether the Emc-Da heterodimer could still bind to DNA as an inactive complex and/or whether Emc could directly interact with DNA, thus preventing bHLH proteins from binding to the same site. To test the first model, we fused the strong activation domain of VP16 to Emc (Emc-VP16) and compared the ability of Emc-VP16 to modulate the transcriptional activity of Da. If Emc and Da are capable of interacting with DNA as a heterodimer, then the Da-Emc-VP16 complex should activate the reporter at higher levels than Da alone. Our results show a significantly decreased level of activation instead, which indicates that the Da-Emc complex does not interact with DNA as a heterodimer (Fig. 3C). To test the ability of Emc to directly compete for DNA-binding sites with Da we compared the ability of wild type Emc, Emc-VP16 and GFP-VP16 (a negative control) to activate the E-box luciferase reporter. Wild-type Emc failed to activate the reporter above basal levels and we observed no difference between Emc-VP16 and GFP-VP16 (Fig. 3D). Last, an Emc-GAL4DBD chimeric protein is unable to activate transcription of several reporters that were placed under UAS control in yeast cells (data not shown). Together, these results suggest that Emc itself completely lacks transcriptional activation potential and does not compete with Da (or other bHLH proteins) for E-box sites in the genome. The sequestration of bHLH proteins appears to be the only method by which Emc antagonizes Da.
To test whether the sequestration of Da by Emc is required to promote growth in the developing eye, we generated and measured the size of clones that overexpress emc, da or both genes together. Although our heat-shock regime (see Materials and methods), is sufficient to induce healthy wild-type clones (Fig. 4A,E), we failed to recover any da overexpressing clones (Fig. 4B,E). This suggests that Da normally inhibits tissue growth and it is consistent with the undergrowth phenotype that has been previously observed with its overexpression (Bhattacharya and Baker, 2011). Growth is restored to da overexpression clones by the co-expression of emc (Fig. 4D,E), indicating that Emc can sequester Da in vivo to promote cell proliferation. Last, overexpression of emc, on its own, is also sufficient to induce clones that are significantly larger than da-expressing clones (Fig. 4C,E). In this case, it is likely that the exogenous Emc is promoting growth by sequestering endogenous Da. Finally, we attempted to generate MARCM clones in which da is overexpressed within emcAP6-null mutant clones but these clones fail to survive. This is consistent with the undergrowth phenotype that is observed when da is overexpressed by itself. Taken together, our data suggest that the sequestration of Da by Emc is required to promote cell proliferation in the eye.
Fig. 4.
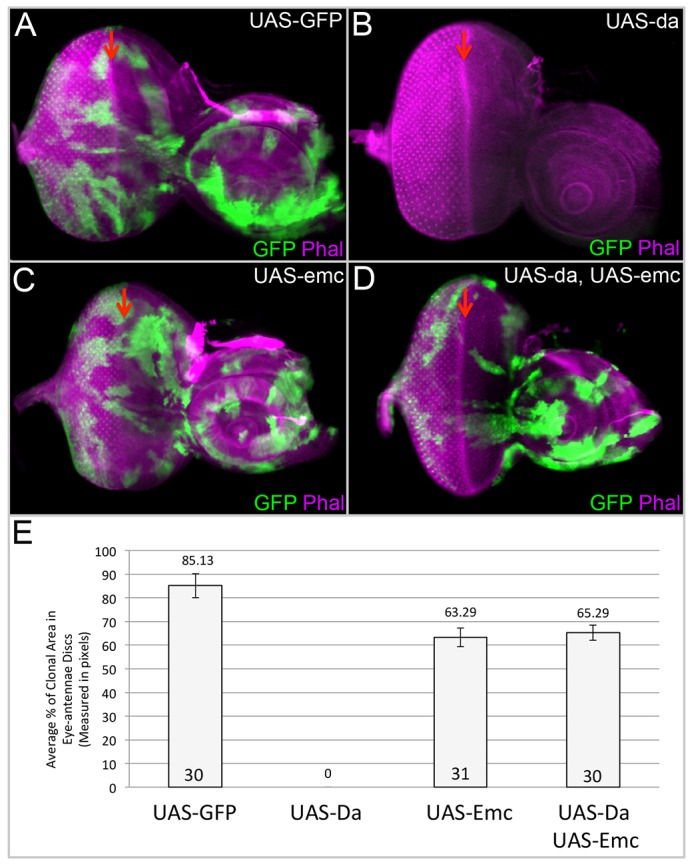
Emc inhibits the growth-suppressing activity of da over-expression. (A-D) Light microscope images of third instar eye-antennal discs containing flp-out clones of varying genotypes marked positively with GFP. Dorsal side is upwards and anterior is towards the right. The red arrow indicates the position of the morphogenetic furrow. All discs were photographed at 10× magnification. (A) Wild-type flp-out clones grow well in a wild-type background. (B) UAS-Da clones do not survive in the eye-antennal disc. (C) UAS-Emc flp-out clones are able to grow similar to wild-type clones. (D) Clones that overexpress both Da and Emc proliferate. (E) Chart depicting the average percentage of eye-antenna imaginal disc area occupied by overexpression clones of genotypes described in A-D. Statistical significance was calculated using Student's t-test and equal or unequal variance was determined using a F-test. The number of discs containing clones that were analyzed can be found in E. We screened 250 discs for Da-expressing clones and failed to recover a single such clone. Error bars represent s.d.
It has been reported that the removal of Notch signaling (using a temperature-sensitive allele) abolishes the high level of da expression that exists between neighboring proneural clusters (Lim et al., 2008). Consistent with this result, the hyper-activation of Notch signaling in MARCM clones leads to an increase in da expression (supplementary material Fig. S3A-C). Notch activation of da appears to be dependent upon emc, as the activation of Notch signaling in emcAP6-null clones failed to activate da expression (supplementary material Fig. S3D-F). The apparent requirement for Emc in Notch-mediated activation of da is at odds with another report in which Emc was shown to repress da expression (Bhattacharya and Baker, 2011). The authors demonstrated that, in emc mutant clones, da expression was elevated compared with surrounding wild-type tissue. Although the reasons underlying these conflicting results are not clear, our results are consistent with a model in which Notch-mediated growth requires Emc to sequester Da.
Emc genetically interacts with Mnt
In addition to Da, three other bHLH proteins (Myc, Max and Mnt) are also important for regulating cell proliferation and differentiation (Grandori et al., 2000; Orian et al., 2003) and thus could be potential sequestration targets of Emc. Similar to da, clones that overexpress Mnt are rarely recovered and are small in size (Loo et al., 2005). We overexpressed Mnt throughout the entire eye using an ey-GAL4 driver and observe a range of rough eye phenotypes (Fig. 5A-D, 136 females and 117 males were analyzed). The small rough eye (Fig. 5C) is consistent with earlier efforts to overexpress Mnt in the eye (Loo et al., 2005). We were able to partially suppress the eye defects by co-expressing emc (Fig. 5D, 142 females and 112 males were analyzed). Expression of emc alone does not alter the structure or size of the compound eye (Fig. 5D, 156 females and 147 males analyzed) and is indistinguishable from controls (Fig. 5D, 180 females and 202 males analyzed). Based on this genetic interaction, we used directed yeast two-hybrid assays and immunoprecipitation in rabbit reticulocyte lysates to test for physical interactions (and thus potential sequestration) between the two proteins but we failed to detect Emc-Mnt complexes in either assay (Fig. 5E, data not shown).
Fig. 5.
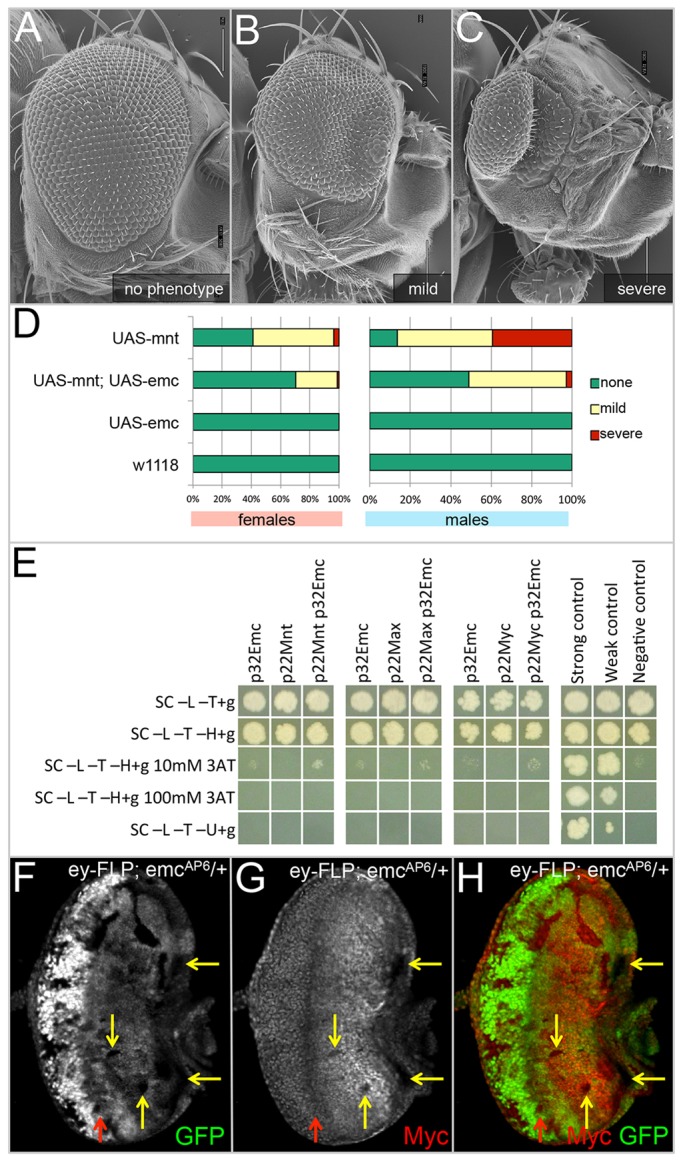
Emc interacts genetically with Mnt and regulates Myc expression. (A-C) SEM images displaying the range of phenotypic severity that is observed in ey-GAL4, UAS-Mnt animals. Adult heads were photographed at 180× magnification. (D) Charts displaying the percentage of adult animals categorized according to the phenotypes imaged in A-C. (E) Directed yeast two-hybrid results showing no interaction between Emc and Mnt, Max or Myc. (F-H) Light microscope images of third instar eye-antennal discs containing emcAP6 loss-of-function clones. Dorsal side is upwards and anterior is towards the right. The red arrow indicates the position of the morphogenetic furrow. All discs were photographed at 10× magnification. Levels of Myc antibody staining (yellow arrow) are decreased in emcAP6-null clones.
Emc influences Myc expression in the developing eye
Myc, which is encoded by the diminutive (dm) gene, is an important player in cell proliferation and cell competition in several Drosophila tissues, including the wing and eye (Johnston et al., 1999; de la Cova et al., 2004). The overexpression of Myc throughout the eye, unlike Mnt, does not alter the overall size and structure of the adult retina (Bellosta et al., 2005); therefore, a simple genetic interaction assay could not be performed. However, we did examine emcAP6-null mutant clones and observed that the level of Myc protein is decreased (Fig. 5F-H). The reduction in Myc levels suggests that the undergrowth phenotype of emc mutant tissue may be due in part to reductions in Myc levels. A complete knockdown of Myc is seen only in a subset of emc mutant clones. The partial knockdown seen in other clones may suggest that additional inputs (independent of Emc) regulate Myc expression. A transcriptional link between Myc and Emc/Id also appears to exist in vertebrates, but interestingly in this case Myc appears to lie upstream and activates the expression of Id2, a homolog of Emc (Lasorella et al., 2000). Despite these differences, the data from both systems suggest that there is a transcriptional chain linking Myc and Emc/Id. To test whether Emc also binds and sequesters Myc, we used directed yeast two-hybrid and immunoprecipitation assays to probe for the formation of an Emc-Myc protein complex. We failed to detect such a complex (Fig. 5E, data not shown); thus, it is likely that Emc and Myc are connected mainly via transcriptional regulation and not through protein sequestration.
In mammalian systems, Id2 binds to Retinoblastoma (Rb) protein (Lasorella et al., 2000). This is consistent with Emc being capable of binding to non-bHLH proteins in Drosophila (Giot et al., 2003). Sequestration of vertebrate Rb or its two fly homologs, Rbf1 and Rbf2, (Du et al., 1996; Stevaux et al., 2002) is predicted to increase the levels of E2F that would be available to promote proliferation through the transition into S phase (Nevins, 1992a,b; Weinberg, 1995; Sherr, 1996). Both Rbf1 and Rbf2 were tested for interactions with Emc via a directed yeast two-hybrid assay and co-immunoprecipitation in rabbit reticulocyte lysates but were not found to interact (supplementary material Fig. S4). We also used a PCNA::GFP reporter, which contains several E2F-binding sites as readout for E2F levels. The PCNA::GFP reporter remains unchanged in clones that overexpress emc (Fig. 6Q-T). If Emc bound to either Rbf1 or Rbf2 in vivo then we would expect to see increased levels of PCNA::GFP as increased levels of E2F would be available to bind and activate the reporter. However, the data from our biochemical studies and the PCNA::GFP reporter support the overall conclusion that Emc does not regulate Rb/E2F, but instead promotes proliferation by sequestering/inhibiting Da and by genetically interacting with Myc and Mnt via mechanisms that are unlikely to involve their direct protein sequestration.
Fig. 6.
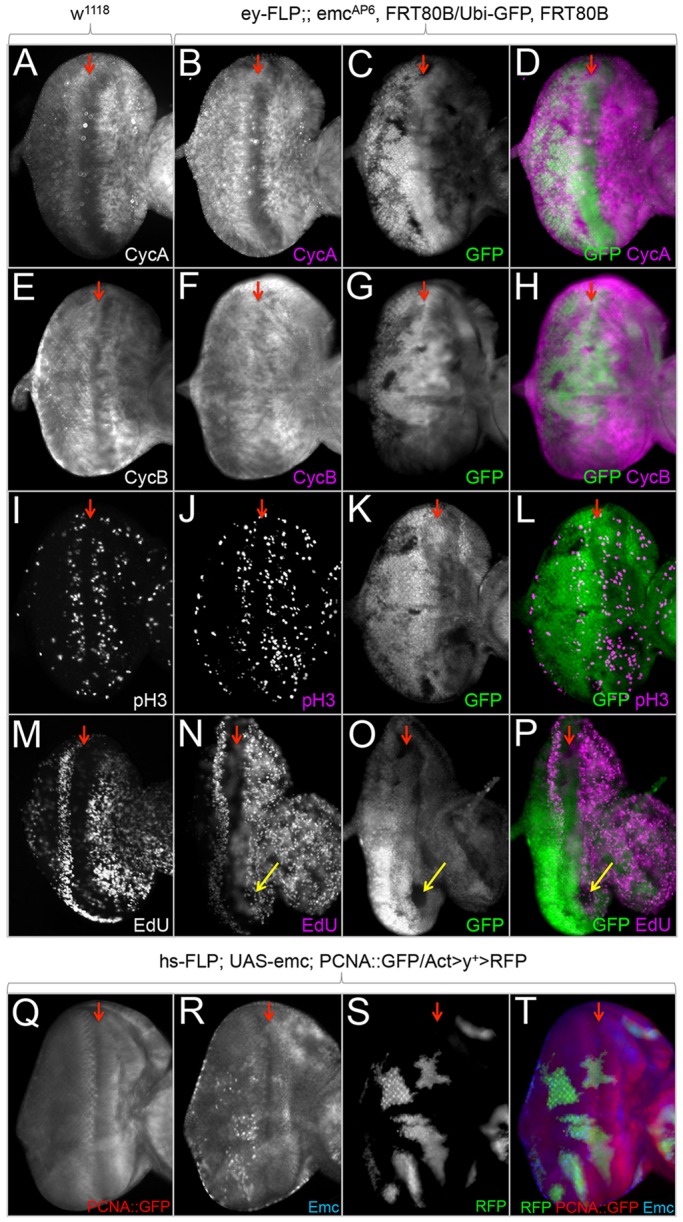
Emc delays entry into S phase in the developing eye. (A-T) Light microscope images of third instar eye-antennal discs containing loss-of-function and flp-out overexpression clones. Dorsal side is upwards and anterior is towards the right. The red arrows indicate the position of the morphogenetic furrow. All discs were photographed at 10× magnification. (A,E,I,M,Q) Normal localization of CycA, CycB and pH3, EdU incorporation, and a readout for E2F in wild-type eye discs. (B-D,F-H,J-L,N-P) Mitotic emcAP6 clones induced continuously throughout eye development using ey-FLP. (B-D,F-H,J-L) emcAP6-null clones (lacking GFP) do not show dramatic alterations in levels of CycA, CycB or pH3 staining. (N-P) emcAP6 clones show decreased number of cells incorporating EdU (yellow arrows) compared with surrounding wild-type tissue. (R-T) Overexpression clones of emc induced with hs-FLP do not show an increase in the PCNA::GFP reporter, indicating that E2F binding to the reporter is not elevated.
Inhibition of cell cycle progression in emc mutants
As emc mutant tissue grows poorly, we set out to determine whether entry into or passage through the cell cycle is negatively impacted. To do this, we analyzed the expression profiles of several cell cycle markers in emcAP6 loss-of-function clones. Most of the cell cycle regulators that we tested appear normal (Fig. 6A-L). However, we did notice that EdU incorporation is reduced in emcAP6 clones (Fig. 6M-P; supplementary material Fig. S5A-F). An analysis of 48 clones anterior to the furrow indicates that in 22.9% of emcAP6 clones (11/48) there is an observable reduction in EdU incorporation when compared with surrounding wild-type tissue. EdU incorporation in the remaining emcAP6 clones is comparable with wild-type cells. These results suggests that entry into S phase may be either delayed or partially blocked, and that, in normal cells, Emc may participate in the entry into or influence the length of time that is spent in S phase. Such a role for Emc at this point in the cell cycle would be consistent with the known role that Notch signaling plays in cell cycle progression through the second mitotic wave (Baonza and Freeman, 2005; Firth and Baker, 2005; Sukhanova and Du, 2008). A delay or partial block in the entry into S phase is also consistent with the undergrowth phenotype that is associated with emc loss-of-function mutant tissue and da overexpression clones.
DISCUSSION
Cells lacking emc have been reported to grow poorly in several developing tissues, including the ovary, wing and eye (de Celis et al., 1995; Baonza and García-Bellido, 1999; Baonza et al., 2000; Adam and Montell, 2004; Bhattacharya and Baker, 2009; Spratford and Kumar, 2013; Andrade-Zapata and Baonza, 2014). In this manuscript, we have attempted to bring several observations together and demonstrate that Emc mediates some of the Notch-mediated growth in the developing eye by sequestering the type I bHLH protein Da. Several previous findings are crucial for this study: first, in several developmental contexts, emc appears to lie downstream of the Notch signaling pathway (de Celis et al., 1995; Baonza and García-Bellido, 1999; Baonza et al., 2000; Adam and Montell, 2004; Bhattacharya and Baker, 2009; Spratford and Kumar, 2015). This is an important observation as Notch signaling is a key promoter of growth in the eye (Cho and Choi, 1998; de Celis et al., 1998; Domínguez and de Celis, 1998; Go et al., 1998; Papayannopoulos et al., 1998; Kenyon et al., 2003; Chao et al., 2004; Dominguez et al., 2004; Baonza and Freeman, 2005; Firth and Baker, 2005; Reynolds-Kenneally and Mlodzik, 2005). Second, Emc antagonizes the activity of several bHLH proteins, including Da, by binding to and sequestering them away from target DNA sequences (Van Doren et al., 1991, 1992; Martinez et al., 1993; Alifragis et al., 1997). Finally, the overexpression of Da, leads to an undergrowth phenotype similar to that seen with emc loss-of-function mutants (Bhattacharya and Baker, 2011).
We started this study by analyzing the proliferative state of cells that lack emc. Although cells lacking emc grow very poorly, the mutant cells are not subviable. When placed in competition with cells that have been given a growth disadvantage, emc mutant cells grow as well as wild-type cells (Fig. 1A-M). This undergrowth phenotype does not appear to be due to an increase in apoptotic levels, as the expression of p35, which blocks all caspase-dependent cell death, cannot restore normal growth rates to emc mutant cells (Fig. 1N-R). Thus, emc mutant cells are neither intrinsically compromised in their ability to survive nor do they undergo apoptotic cell death. Instead, it appears that cells lacking emc are deficient in the ability to proliferate. As emc has been implicated in Notch-mediated growth in the developing wing, we sought to determine whether a similar relationship exists within the eye. To accomplish this goal, we analyzed the growth of three different types of cell populations: (1) emc-null mutant cells, (2) wild-type cells overexpressing activated Notch and (3) emc-null mutant cells overexpressing activated Notch. The proliferative effects of Notch are greatly diminished in emc mutant cells (Fig. 2E-G) – this indicates that, as in the wing, Emc mediates some of the Notch-dependent cell proliferation in the eye. Interestingly, the loss of emc does not completely abrogate Notch-mediated growth (Fig. 2E-G), suggesting that the Notch pathway may have targets that are not regulated by Emc.
We set out to determine the mechanism by which Emc modulates Notch-dependent growth in the eye. Current models suggest that several growth-promoting genes such as four-jointed, cyclin A, cyclin B and E2f1 are directly activated by the Notch pathway via Su(H)/Mam and/or the E(spl) complex (Knoblich and Lehner, 1993; Baonza and Freeman, 2005; Gutierrez-Aviño et al., 2009). Our data hint at an additional mechanism in which the Notch pathway activates emc expression and, in turn, Emc protein promotes cell proliferation by binding to and sequestering one or more growth-inhibiting bHLH proteins. We have demonstrated here that Notch signaling can activate emc expression in proliferating cells ahead of the morphogenetic furrow (Fig. 2A-C; Spratford and Kumar, 2015). These results are similar to those obtained by others in the wing and in developing photoreceptor neurons. We did not test the possibility that Su(H)/Mam directly binds to the emc locus and activates its expression, but it is an attractive model as two Su(H)-binding sites exist close to the transcriptional start site and emc expression is lost in Su(H)- and mam-null mutant clones (Spratford and Kumar, 2015).
Once emc expression is activated by Notch signaling, we propose that Emc protein goes on to promote growth by sequestering Da. Clones that overexpress da grow very poorly, suggesting that Da functions to inhibit cell proliferation (Bhattacharya and Baker, 2011; Fig. 4A,B,E). This is the expected growth phenotype for a target of Emc and therefore makes Da an ideal candidate for sequestration by Emc in proliferating cells. Consistent with this model, Emc can bind to Da in yeast two-hybrid assays and can interfere with the ability of Da to interact with DNA in electromobility shift assays and in transcriptional activation assays conducted in Drosophila Kc167 cells (Van Doren et al., 1991, 1992; Alifragis et al., 1997; Fig. 3; supplementary material Fig. S2). Here, we further demonstrate that the Emc-Da interaction is an important step in tissue growth, as the expression of emc within the eye disc is sufficient to block the growth inhibiting properties of da overexpression (Fig. 4A-E). Consistent with earlier in vitro protein-DNA binding assays, our transcriptional activation assays in yeast and Drosophila Kc167 cells indicate that protein sequestration appears to be the major, if not only, mechanism by which Emc antagonizes Da (Fig. 3).
Last, we attempted to determine how Emc regulates the cell cycle in the eye disc. Multiple studies have demonstrated that Notch signaling and Da exert opposing activities on the G1/S transition. In particular, Notch signaling promotes entry into S phase, while Da counteracts this activity by activating dacapo (dap) (Fig. 7; Baonza and Freeman, 2005; Firth and Baker, 2005; Sukhanova et al., 2007; Sukhanova and Du, 2008). In addition, da inhibits the transcription of string (stg) (Andrade-Zapata and Baonza, 2014). String, the Drosophila homolog of yeast cdc25, plays an important role in transitioning cells through the G2/M checkpoint in Drosophila (Edgar and O'Farrell, 1989). These observations, coupled with our genetic studies placing Emc between the Notch pathway and Da, suggest that both the G1/S and G2/M transitions may be the points in the cell cycle that are controlled by Emc. Consistent with this model, we find that EdU incorporation is lower in emc mutant cells when compared with wild-type cells (Fig. 6M-P; supplementary material Fig. S5). As emc mutant cells can proliferate when surrounded by growth-deficient cells, it is unlikely that these cells are completely blocked from entering S phase. Rather, it is more likely that emc mutant cells are delayed in S-phase entry. In further support of a role for Emc in regulating G1/S, we find that levels of Myc, which is required for entry and progression through S phase, are severely reduced in emc mutant clones (Fig. 5F-H). A role for Emc in regulating the G2/M transition is proposed in a recent study in which the authors found that emc mutant cells remain in G2 (Andrade-Zapata and Baonza, 2014). In our study, we analyzed cell cycle profile of emc-null mutant cells using a variety of markers. In the other study, the authors used RNAi lines targeting emc and da overexpression constructs to manipulate protein levels for FACS analysis. It is likely that the use of different methods in the two studies have allowed for the identification of roles for Emc at the two different cell cycle checkpoints.
Fig. 7.
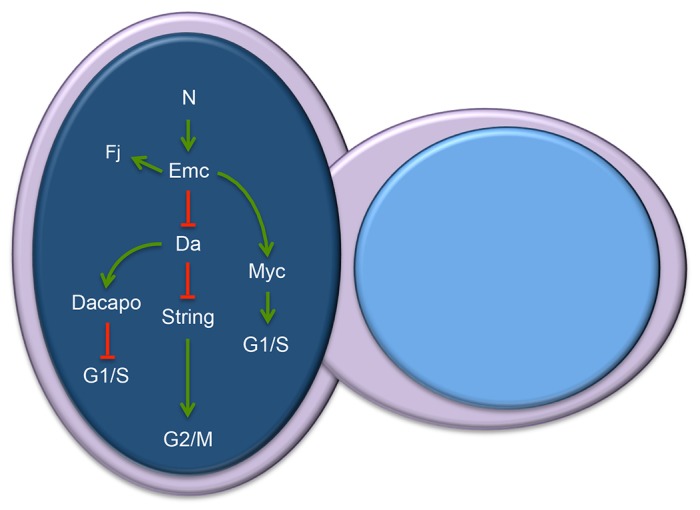
Potential mechanisms by which Emc promotes Notch-induced growth. Da has been shown to regulate G1/S via modulation of dacapo and string expression levels. Our data indicate that Emc-mediated sequestration of Da is required to promote growth in the eye. We also have evidence that Emc regulates expression levels of Myc, another regulator of the G1/S checkpoint. The model summarizes current data and proposes that Notch activation of emc leads to the sequestration of Da by Emc and the activation of Myc. These events ultimately result in the passage of the cell through the G1/S checkpoint.
Vertebrate genomes harbor four homologs of emc and are referred to as inhibitors of differentiation 1-4 (Id1-Id4). The expression of the Id genes is mediated by several oncoproteins and is upregulated in numerous cancers (reviewed by Perk et al., 2005). Multiple Id proteins themselves are also known to regulate cell proliferation and tissue growth (Norton, 2000; Zebedee and Hara, 2001; Sikder et al., 2003; Wong et al., 2004; Iavarone and Lasorella, 2006; Ling et al., 2006). We have demonstrated that Emc promotes cell proliferation via the sequestration of Da. Interestingly, all four Id proteins form biochemical complexes with E12 and E47, the vertebrate homologs of Da (Cochrane et al., 2009; Sun et al., 2009; Teachenor et al., 2012). A significant advance will be made if it emerges that the sequestration of Da/E2A by Emc/Id is a universal mechanism by which cell proliferation and tissue growth is promoted in both flies and mammals. It will also be interesting to determine the extent to which sequestration of bHLH proteins by Emc/Id is used during development. In addition to the known sequestration targets, our Y2H screen also identified Sage, a bHLH protein that is specifically expressed in the salivary gland. A comprehensive analysis of potential bHLH-Emc/Id interactions could shed considerable light on the extent to which Emc/Id regulates bHLH-dependent processes in both flies and vertebrate systems.
MATERIALS AND METHODS
Fly stocks
Fly stocks used were: (1) hs-FLP22; (2) y1 w* hs-FLP22;; FRT80B emcAP6/TM6B; (3) y1 w* hs-FLP22;; FRT80B Ubi-GFP M(3)i55/TM6B; (4) w1118; FRT80B Ubi-GFP(S65T)nls/TM3 Sb1; (5) w*; FRT80B ry506; (6) y1 w* hs-FLP22 UAS-mCD8::GFP.L Ptp4ELL4; Pin1/CyO; (7) y1 w*; Act5C-GAL4/CyO; (8) y1 w*; FRT80B tub-GAL80; (9) UAS-p35; (10) UAS-NICD; FRT80B tub-GAL80; (11) y1 w*; AyGAL4 UAS-GFP(S65T) Myo31DF; (12) w1118; (13) UAS-da; (14) UAS-emc4M; (15) UAS-da; UAS-emc4M; (16) ey-GAL4; (17) UAS-MntT233; (18) UAS-emc; UAS-MntT233; (19) PCNA. ΔNhe::GFP; and (20) w1118; Act5C-GAL4 UAS-RFP.W/TM3 Sb1. A list of full genotypes to accompany the figures can be found in supplementary material Table S1.
Antibodies and microscopy
Primary antibodies used were: (1) rat anti-Elav (1:100, DSHB); (2) mouse anti-Myc (1:100, DSHB); (3) mouse anti-cyclin A (1:4, DSHB); (4) mouse anti-cyclin B (1:4, DSHB); (5) mouse anti-cyclin E (1:4, DSHB); (6) rabbit anti-pH3 (1:20,000, Abcam, ab32107); and (7) mouse anti-β-gal (1:250, Promega, Z3781). Fluorophore-conjugated secondary antibodies and phalloidin-fluorophore conjugates were obtained from Jackson Laboratories and Molecular Probes. Imaginal discs were prepared as described previously (Anderson et al., 2012). All eye-antennal discs were photographed on a Zeiss Axioplan II compound microscope at 10× magnification, except for the discs in supplementary material Fig. S5, which were photographed at 40×. For scanning electron microscopy, adult flies were serially incubated in 25% ethanol, 50% ethanol, 75% ethanol, 100% ethanol, 50% ethanol: 50% hexamethyldisilazane (HMDS) and 100% HMDS, coated with gold-palladium, and viewed with a JEOL 5800LV SEM. All adult heads were photographed at 180× magnification.
EdU incorporation
This assay was performed using the Life Technologies Click-iT EdU Alexa Flour 594 Imaging Kit. Larval heads were dissected in phosphate buffer. Heads were individually incubated in 50 µM solution of EdU in phosphate buffer for 15 min. Heads were washed with phosphate buffer for 15 min then fixed individually with a 4% paraformaldehyde solution for 30 min. Fixed heads underwent three 5 min washes in 0.3% Triton, one 20 min wash in 0.6% Triton, followed by two 5 min washes in 3% BSA. The Click-iT reaction step was performed for 30 min followed by one 5 min wash in 3% BSA, then two 5 min washes in 0.1% Triton. Eye imaginal discs were then dissected from the head complex and treated as described in the antibodies and microscopy section
Clone size analysis
Adobe Photoshop CS6 was used to outline and measure the area of the eye imaginal disc (in pixels). Next, within this area, a color range selection was made to select all clonal tissue (GFP positive for flp-out and MARCM clones, and GFP negative for loss-of-function clones). The area of the clones was also measured in pixels. The clonal area was divided by the total area of the eye disc and then multiplied by 100 in order to calculate the percentage of the eye field that is occupied by clonal tissue. The individual percentages for each genotype were then averaged. Statistical significance was calculated using Student's t-test and equal or unequal variance was determined using a F-test. Error bars within the figures represent s.d.
Temperature shift regimes
All heat-shock clones were induced at 37°C 48 h after egg lay (AEL). emcAP6 loss-of-function clones in wild-type or Minute− backgrounds were induced by a 20 min heat pulse followed by a 60 min rest period (room temperature) and a second 20 min heat pulse. Clones in Fig. 1E-L were induced with a single 20 min heat pulse. Overexpression and MARCM clones were induced with 20 and 60 min heat pulses, respectively.
Cloning and analysis of putative Emc enhancers
The three putative enhancer regions shown in supplementary material Fig. S1A were amplified from w1118 genomic DNA using PCR and cloned into the placZ.attB plasmid (a gift from Konrad Basler, University of Zurich, Switzerland). PCR conditions and primer sequences, cloning strategies and sequence files for each construct are all available upon request. The p.enhancer.lacZ.attB constructs were transformed into flies using the phiC31 integrase system. Stable stocks were created and analyzed for lacZ expression in imaginal and brain tissue.
Yeast two-hybrid screen
The ProQuest Yeast 2-Hybrid System (Invitrogen) was used to identify proteins that physically associate with Emc. A library of cDNAs from Drosophila third instar larvae was cloned into the pDEST-22 vector, which contains the GAL4 activation domain (prey plasmid). Yeast MaV203 cells were transformed with the prey library and the bait plasmid pDEST-32 in which Emc is fused to the GAL4 DNA-binding domain. The IU Yeast Two-Hybrid Facility conducted this unbiased Y2H screen. Interactions between prey proteins and Emc were identified by the activation of three reporters (UAS-lacZ, UAS-HIS3 and UAS-URA3). Plasmids from positive clones were isolated and sequenced.
Directed yeast two-hybrid
Da, Myc, Mnt, Max, Rbf1 and Rbf2 cDNAs were individually cloned into the pDEST22 vector from Invitrogen to create corresponding p22 prey vectors. These were individually tested in pairwise transformations with the p32-Emc bait vector. The strong, weak and negative controls used are listed within the Invitrogen manual. Cultures were grown at 30°C in 500 ml SC-L-T+g media shaking at 200 rpm. After 48 h, 100 ml of each culture was diluted in 2 ml sterile water and replica stamped onto the following plate types: (1) SC-L-T+g, (2) SC-L-T-H+g, (3) SC-L-T-U+g, (4) SC-L-T-H+g 10 mM 3AT and (5) SC-L-T-H+g 100 mM 3AT. Plates were incubated at 30°C. After 1 week, colony growth was recorded and photographed. A da cDNA was also cloned into the p32 bait vector and tested against the Myc-, Mnt- and Max-containing p22 prey vectors.
Luciferase transcriptional reporter assay
An oligomer consisting of five repeats of the E-box CANNTG with spacers of ATATAT was constructed by Integrated DNA Technologies (IDT) and cloned into a plasmid containing the hsp70 promoter and firefly luciferase gene. da and emc cDNAs were cloned into vectors containing UAS regulatory sites. The Qiagen Effectene Transfection Reagent was used to transfect KC167 cells with the E-box reporter plasmid, the pMT-GAL4 driver plasmid, the UAS-Renilla control plasmid and differing combinations of UAS-GFP, UAS-da or UAS-emc plasmids. Transfected cells were incubated at 37°C for 24 h, at which time protein production was induced by adding CuSO4. Twenty-four hours later, cells were treated according to the Promega Dual-Luciferase Reporter Assay System protocol, and firefly luciferase and renilla luciferase levels were measured using a Glomax 20/20 1996 Luminometer. For each experiment, three independent biological samples were taken. In each case, the firefly luciferase reading was divided by the renilla luciferase control reading to obtain a ratio. The ratios from the three samples were then averaged. Statistical significance was calculated using Student's t-test and equal or unequal variance was determined using a F-test. Error bars within the figures represent s.d.
Supplementary Material
Acknowledgements
We thank Brandon and Bonnie Weasner for technical support with molecular cloning and biochemical assays, Bonnie Weasner and Laurel Bender for technical help with directed yeast two-hybrid assays, Scott Michaels and Laurel Bender for performing the unbiased Emc Y2H screen, and Rudi Turner for the SEM images. We also thank Spyros Artavanis-Tsakonas, Antonio Baonza, Nick Baker, Brian Calvi, Robert Holmgren, Yuh-Nung Jan, Gerald Rubin, Adi Salzberg, the Bloomington Drosophila Stock Center (BDSC) and the Drosophila Genomics Resource Center (DGRC) for fly stocks, antibodies and DNA plasmids. Finally, we thank Utpal Banerjee for generously allowing C.M.S. to complete some of the work for this manuscript in his lab.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Funding
This research was supported by a stipend from the National Institutes of Health GCMS Training Grant [T32-GM007757], by the Frank W. Putnam Research Fellowship and by a Robert Briggs Research Fellowship to C.M.S.; and by a grant from the National Eye Institute [R01 EY014863] to J.P.K. Deposited in PMC for release after 12 months.
Author contributions
C.M.S. and J.P.K. designed experiments, analyzed data and wrote the manuscript. C.M.S. carried out the experiments and collected the data.
Supplementary material
Supplementary material available online at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.121855/-/DC1
References
- Adam J. C. and Montell D. J. (2004). A role for extra macrochaetae downstream of Notch in follicle cell differentiation. Development 131, 5971-5980. 10.1242/dev.01442 [DOI] [PubMed] [Google Scholar]
- Alifragis P., Poortinga G., Parkhurst S. M. and Delidakis C. (1997). A network of interacting transcriptional regulators involved in Drosophila neural fate specification revealed by the yeast two-hybrid system. Proc. Natl. Acad. Sci. USA 94, 13099-13104. 10.1073/pnas.94.24.13099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson A. M., Weasner B. M., Weasner B. P. and Kumar J. P. (2012). Dual transcriptional activities of SIX proteins define their roles in normal and ectopic eye development. Development 139, 991-1000. 10.1242/dev.077255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrade-Zapata I. and Baonza A. (2014). The bHLH factors Extramacrochaetae and Daughterless control cell cycle in Drosophila imaginal discs through the transcriptional regulation of the cdc25 phosphatase string. PLoS Genet. 10, e1004233 10.1371/journal.pgen.1004233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker N. E. and Yu S.-Y. (2001). The EGF receptor defines domains of cell cycle progression and survival to regulate cell number in the developing Drosophila eye. Cell 104, 699-708. 10.1016/S0092-8674(01)00266-5 [DOI] [PubMed] [Google Scholar]
- Baonza A. and Freeman M. (2001). Notch signalling and the initiation of neural development in the Drosophila eye. Development 128, 3889-3898. [DOI] [PubMed] [Google Scholar]
- Baonza A. and Freeman M. (2005). Control of cell proliferation in the Drosophila eye by Notch signaling. Dev. Cell 8, 529-539. 10.1016/j.devcel.2005.01.019 [DOI] [PubMed] [Google Scholar]
- Baonza A. and Garcı´a-Bellido A. (1999). Dual role of extramacrochaetae in cell proliferation and cell differentiation during wing morphogenesis in Drosophila. Mech. Dev. 80, 133-146. 10.1016/S0925-4773(98)00198-1 [DOI] [PubMed] [Google Scholar]
- Baonza A., de Celis J. F. and Garcia-Bellido A. (2000). Relationships between extramacrochaetae and Notch signalling in Drosophila wing development. Development 127, 2383-2393. [DOI] [PubMed] [Google Scholar]
- Baonza A., Murawsky C. M., Travers A. A. and Freeman M. (2002). Pointed and Tramtrack69 establish an EGFR-dependent transcriptional switch to regulate mitosis. Nat. Cell Biol. 4, 976-980. 10.1038/ncb887 [DOI] [PubMed] [Google Scholar]
- Bellosta P., Hulf T., Balla Diop S., Usseglio F., Pradel J., Aragnol D. and Gallant P. (2005). Myc interacts genetically with Tip48/Reptin and Tip49/Pontin to control growth and proliferation during Drosophila development. Proc. Natl. Acad. Sci. USA 102, 11799-11804. 10.1073/pnas.0408945102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya A. and Baker N. E. (2009). The HLH protein Extramacrochaetae is required for R7 cell and cone cell fates in the Drosophila eye. Dev. Biol. 327, 288-300. 10.1016/j.ydbio.2008.11.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya A. and Baker N. E. (2011). A network of broadly expressed HLH genes regulates tissue-specific cell fates. Cell 147, 881-892. 10.1016/j.cell.2011.08.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botas J., Moscoso del Prado J. and Garcia-Bellido A. (1982). Gene-dose titration analysis in the search of trans-regulatory genes in Drosophila. EMBO J. 1, 307-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campuzano S. (2001). Emc, a negative HLH regulator with multiple functions in Drosophila development. Oncogene 20, 8299-8307. 10.1038/sj.onc.1205162 [DOI] [PubMed] [Google Scholar]
- Cavodeassi F., Diez del Corral R., Campuzano S. and Dominguez M. (1999). Compartments and organising boundaries in the Drosophila eye: the role of the homeodomain Iroquois proteins. Development 126, 4933-4942. [DOI] [PubMed] [Google Scholar]
- Chao J.-L., Tsai Y.-C., Chiu S.-J. and Sun Y. H. (2004). Localized Notch signal acts through eyg and upd to promote global growth in Drosophila eye. Development 131, 3839-3847. 10.1242/dev.01258 [DOI] [PubMed] [Google Scholar]
- Chern J. J. and Choi K. W. (2002). Lobe mediates Notch signaling to control domain-specific growth in the Drosophila eye disc. Development 129, 4005-4013. [DOI] [PubMed] [Google Scholar]
- Cho K.-O. and Choi K.-W. (1998). Fringe is essential for mirror symmetry and morphogenesis in the Drosophila eye. Nature 396, 272-276. 10.1038/24394 [DOI] [PubMed] [Google Scholar]
- Cho K.-O., Chern J., Izaddoost S. and Choi K.-W. (2000). Novel signaling from the peripodial membrane is essential for eye disc patterning in Drosophila. Cell 103, 331-342. 10.1016/S0092-8674(00)00124-0 [DOI] [PubMed] [Google Scholar]
- Cochrane S. W., Zhao Y., Welner R. S. and Sun X.-H. (2009). Balance between Id and E proteins regulates myeloid-versus-lymphoid lineage decisions. Blood 113, 1016-1026. 10.1182/blood-2008-06-164996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craymer L. (1980). New mutants report. D.I.S. 55, 197-200. [Google Scholar]
- de Celis J. F., Baonza A. and García-Bellido A. (1995). Behavior of extramacrochaetae mutant cells in the morphogenesis of the Drosophila wing. Mech. Dev. 53, 209-221. 10.1016/0925-4773(95)00436-5 [DOI] [PubMed] [Google Scholar]
- de Celis J. F., Tyler D. M., de Celis J. and Bray S. J. (1998). Notch signalling mediates segmentation of the Drosophila leg. Development 125, 4617-4626. [DOI] [PubMed] [Google Scholar]
- de la Cova C., Abril M., Bellosta P., Gallant P. and Johnston L. A. (2004). Drosophila myc regulates organ size by inducing cell competition. Cell 117, 107-116. 10.1016/S0092-8674(04)00214-4 [DOI] [PubMed] [Google Scholar]
- Domínguez M. and de Celis J. F. (1998). A dorsal/ventral boundary established by Notch controls growth and polarity in the Drosophila eye. Nature 396, 276-278. 10.1038/24402 [DOI] [PubMed] [Google Scholar]
- Dominguez M., Ferres-Marco D., Gutierrez-Aviño F. J., Speicher S. A. and Beneyto M. (2004). Growth and specification of the eye are controlled independently by Eyegone and Eyeless in Drosophila melanogaster. Nat. Genet. 36, 31-39. 10.1038/ng1281 [DOI] [PubMed] [Google Scholar]
- Du W., Vidal M., Xie J. E. and Dyson N. (1996). RBF, a novel RB-related gene that regulates E2F activity and interacts with cyclin E in Drosophila. Genes Dev. 10, 1206-1218. 10.1101/gad.10.10.1206 [DOI] [PubMed] [Google Scholar]
- Edgar B. A. and O'Farrell P. H. (1989). Genetic control of cell division patterns in the Drosophila embryo. Cell 57, 177-187. 10.1016/0092-8674(89)90183-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis H. M., Spann D. R. and Posakony J. W. (1990). extramacrochaetae, a negative regulator of sensory organ development in Drosophila, defines a new class of helix-loop-helix proteins. Cell 61, 27-38. 10.1016/0092-8674(90)90212-W [DOI] [PubMed] [Google Scholar]
- Fan Y. and Bergmann A. (2008). Distinct mechanisms of apoptosis-induced compensatory proliferation in proliferating and differentiating tissues in the Drosophila eye. Dev. Cell 14, 399-410. 10.1016/j.devcel.2008.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firth L. C. and Baker N. E. (2005). Extracellular signals responsible for spatially regulated proliferation in the differentiating Drosophila eye. Dev. Cell 8, 541-551. 10.1016/j.devcel.2005.01.017 [DOI] [PubMed] [Google Scholar]
- Firth L. C. and Baker N. E. (2007). Spitz from the retina regulates genes transcribed in the second mitotic wave, peripodial epithelium, glia and plasmatocytes of the Drosophila eye imaginal disc. Dev. Biol. 307, 521-538. 10.1016/j.ydbio.2007.04.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Alonso L. A. and García-Bellido A. (1988). Extramacrochaetae, a trans-acting gene of the achaete-scute complex of Drosophila involved in cell communication. Rouxs Arch. Dev. Biol. 197, 328-338. 10.1007/BF00375952 [DOI] [PubMed] [Google Scholar]
- Garrell J. and Modolell J. (1990). The Drosophila extramacrochaetae locus, an antagonist of proneural genes that, like these genes, encodes a helix-loop-helix protein. Cell 61, 39-48. 10.1016/0092-8674(90)90213-X [DOI] [PubMed] [Google Scholar]
- Gibson M. C. and Schubiger G. (2000). Peripodial cells regulate proliferation and patterning of Drosophila imaginal discs. Cell 103, 343-350. 10.1016/S0092-8674(00)00125-2 [DOI] [PubMed] [Google Scholar]
- Gibson M. C., Lehman D. A. and Schubiger G. (2002). Lumenal transmission of Decapentaplegic in Drosophila imaginal discs. Dev. Cell 3, 451-460. 10.1016/S1534-5807(02)00264-2 [DOI] [PubMed] [Google Scholar]
- Giot L., Bader J. S., Brouwer C., Chaudhuri A., Kuang B., Li Y., Hao Y. L., Ooi C. E., Godwin B., Vitols E. et al. (2003). A protein interaction map of Drosophila melanogaster. Science 302, 1727-1736. 10.1126/science.1090289 [DOI] [PubMed] [Google Scholar]
- Go M. J., Eastman D. S. and Artavanis-Tsakonas S. (1998). Cell proliferation control by Notch signaling in Drosophila development. Development 125, 2031-2040. [DOI] [PubMed] [Google Scholar]
- Grandori C., Cowley S. M., James L. P. and Eisenman R. N. (2000). The Myc/Max/Mad network and the transcriptional control of cell behavior. Annu. Rev. Cell Dev. Biol. 16, 653-699. 10.1146/annurev.cellbio.16.1.653 [DOI] [PubMed] [Google Scholar]
- Gutierrez-Aviño F. J., Ferres-Marco D. and Dominguez M. (2009). The position and function of the Notch-mediated eye growth organizer: the roles of JAK/STAT and four-jointed. EMBO Rep. 10, 1051-1058. 10.1038/embor.2009.140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heberlein U., Borod E. R. and Chanut F. A. (1998). Dorsoventral patterning in the Drosophila retina by wingless. Development 125, 567-577. [DOI] [PubMed] [Google Scholar]
- Heitzler P., Haenlin M., Ramain P., Calleja M. and Simpson P. (1996). A genetic analysis of pannier, a gene necessary for viability of dorsal tissues and bristle positioning in Drosophila. Genetics 143, 1271-1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iavarone A. and Lasorella A. (2006). ID proteins as targets in cancer and tools in neurobiology. Trends Mol. Med. 12, 588-594. 10.1016/j.molmed.2006.10.007 [DOI] [PubMed] [Google Scholar]
- Irvine K. D. and Wieschaus E. (1994). fringe, a Boundary-specific signaling molecule, mediates interactions between dorsal and ventral cells during Drosophila wing development. Cell 79, 595-606. 10.1016/0092-8674(94)90545-2 [DOI] [PubMed] [Google Scholar]
- Johnston L. A., Prober D. A., Edgar B. A., Eisenman R. N. and Gallant P. (1999). Drosophila myc regulates cellular growth during development. Cell 98, 779-790. 10.1016/S0092-8674(00)81512-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jory A., Estella C., Giorgianni M. W., Slattery M., Laverty T. R., Rubin G. M. and Mann R. S (2012). A survey of 6,300 genomic fragments for cis-regulatory activity in the imaginal discs of Drosophila melanogaster. Cell Rep. 2, 1014-1024. 10.1016/j.celrep.2012.09.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kango-Singh M., Nolo R., Tao C., Verstreken P., Hiesinger P. R., Bellen H. J., and Halder G (2002). Shar-pei mediates cell proliferation arrest during imaginal disc growth in Drosophila. Development 129, 5719-5730. 10.1242/dev.00168 [DOI] [PubMed] [Google Scholar]
- Kenyon K. L., Ranade S. S., Curtiss J., Mlodzik M. and Pignoni F. (2003). Coordinating proliferation and tissue specification to promote regional identity in the Drosophila head. Dev. Cell 5, 403-414. 10.1016/S1534-5807(03)00243-0 [DOI] [PubMed] [Google Scholar]
- Knoblich J. A. and Lehner C. F. (1993). Synergistic action of Drosophila cyclins A and B during the G2-M transition. EMBO J. 12, 65-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koontz L. M., Liu-Chittenden Y., Yin F., Zheng Y., Yu J., Huang B., Chen Q., Wu S. and Pan D. (2013). The Hippo effector Yorkie controls normal tissue growth by antagonizing scalloped-mediated default repression. Dev. Cell 25, 388-401. 10.1016/j.devcel.2013.04.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasorella A., Noseda M., Beyna M., and Iavarone A. (2000). Id2 is a retinoblastoma protein target and mediates signalling by Myc oncoproteins. Nature 407, 592-598. 10.1038/35036504 [DOI] [PubMed] [Google Scholar]
- Lim J., Jafar-Nehad H., Hsu Y.-C., and Choi K.-W (2008). Novel function of the class I bHLH protein Daughterless in the negative regulation of proneural gene expression in the Drosophila eye. EMBO Rep. 9, 1128-1133. 10.1038/embor.2008.166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling M.-T., Wang X., Zhang X. and Wong Y.-C. (2006). The multiple roles of Id-1 in cancer progression. Differentiation 74, 481-487. 10.1111/j.1432-0436.2006.00083.x [DOI] [PubMed] [Google Scholar]
- Loo L. W. M., Secombe J., Little J. T., Carlos L.-S., Yost C., Cheng P.-F., Flynn E. M., Edgar B. A. and Eisenman R. N. (2005). The transcriptional repressor dMnt is a regulator of growth in Drosophila melanogaster. Mol. Cell. Biol. 25, 7078-7091. 10.1128/MCB.25.16.7078-7091.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning L., Heckscher E. S., Purice M. D., Roberts J., Bennett A. L., Kroll J. R., Pollard J. L., Strader M. E., Lupton J. R., Dyukareva A. V. et al. (2012). A resource for manipulating gene expression and analyzing cis-regulatory modules in the Drosophila CNS. Cell Rep. 2, 1002-1013. 10.1016/j.celrep.2012.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez C., Modolell J. and Garrell J. (1993). Regulation of the proneural gene achaete by helix-loop-helix proteins. Mol. Cell. Biol. 13, 3514-3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massari M. E. and Murre C. (2000). Helix-loop-helix proteins: regulators of transcription in eucaryotic organisms. Mol. Cell. Biol. 20, 429-440. 10.1128/MCB.20.2.429-440.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurel-Zaffran C. and Treisman J. E. (2000). pannier acts upstream of wingless to direct dorsal eye disc development in Drosophila. Development 127, 1007-1016. [DOI] [PubMed] [Google Scholar]
- Moscoso del Prado J. M. and Garcia-Bellido A. (1984). Genetic regulation of the achaete-scute complex of Drosophila melanogaster. Wilhelm Rouxs Arch. Dev. Biol. 193, 242-245. 10.1007/BF01260345 [DOI] [PubMed] [Google Scholar]
- Murre C., McCaw P. S. and Baltimore D. (1989a). A new DNA binding and dimerization motif in immunoglobulin enhancer binding, daughterless, MyoD, and myc proteins. Cell 56, 777-783. 10.1016/0092-8674(89)90682-X [DOI] [PubMed] [Google Scholar]
- Murre C., McCaw P. S., Vaessin H., Caudy M., Jan L. Y., Jan Y. N., Cabrera C. V., Buskin J. N., Hauschka S. D., Lassar A. B. et al. (1989b). Interactions between heterologous helix-loop-helix proteins generate complexes that bind specifically to a common DNA sequence. Cell 58, 537-544. 10.1016/0092-8674(89)90434-0 [DOI] [PubMed] [Google Scholar]
- Nevins J. R. (1992a). E2F: a link between the Rb tumor suppressor protein and viral oncoproteins. Science 258, 424-429. 10.1126/science.1411535 [DOI] [PubMed] [Google Scholar]
- Nevins J. R. (1992b). A closer look at E2F. Nature 358, 375-376. 10.1038/358375a0 [DOI] [PubMed] [Google Scholar]
- Norton J. D. (2000). ID helix-loop-helix proteins in cell growth, differentiation and tumorigenesis. J. Cell Sci. 113, 3897-3905. [DOI] [PubMed] [Google Scholar]
- Orian A., van Steensel B., Delrow J., Bussemaker H. J., Li L., Sawado T., Williams E., Loo L. W. M., Cowley S. M., Yost C. et al. (2003). Genomic binding by the Drosophila Myc, Max, Mad/Mnt transcription factor network. Genes Dev. 17, 1101-1114. 10.1101/gad.1066903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oros S. M., Tare M., Kango-Singh M. and Singh A. (2010). Dorsal eye selector pannier (pnr) suppresses the eye fate to define dorsal margin of the Drosophila eye. Dev. Biol. 346, 258-271. 10.1016/j.ydbio.2010.07.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panin V. M., Papayannopoulos V., Wilson R. and Irvine K. D. (1997). Fringe modulates Notch-ligand interactions. Nature 387, 908-912. 10.1038/43191 [DOI] [PubMed] [Google Scholar]
- Papayannopoulos V., Tomlinson A., Panin V. M., Rauskolb C. and Irvine K. D. (1998). Dorsal-ventral signaling in the Drosophila eye. Science 281, 2031-2034. 10.1126/science.281.5385.2031 [DOI] [PubMed] [Google Scholar]
- Perk J., Iavarone A. and Benezra R. (2005). Id family of helix-loop-helix proteins in cancer. Nat. Rev. Cancer 5, 603-614. 10.1038/nrc1673 [DOI] [PubMed] [Google Scholar]
- Ready D. F., Hanson T. E. and Benzer S. (1976). Development of the Drosophila retina, a neurocrystalline lattice. Dev. Biol. 53, 217-240. 10.1016/0012-1606(76)90225-6 [DOI] [PubMed] [Google Scholar]
- Reynolds-Kenneally J. and Mlodzik M. (2005). Notch signaling controls proliferation through cell-autonomous and non-autonomous mechanisms in the Drosophila eye. Dev. Biol. 285, 38-48. 10.1016/j.ydbio.2005.05.038 [DOI] [PubMed] [Google Scholar]
- Sato A. and Tomlinson A. (2007). Dorsal-ventral midline signaling in the developing Drosophila eye. Development 134, 659-667. 10.1242/dev.02786 [DOI] [PubMed] [Google Scholar]
- Sherr C. J. (1996). Cancer cell cycles. Science 274, 1672-1677. 10.1126/science.274.5293.1672 [DOI] [PubMed] [Google Scholar]
- Sikder H. A., Devlin M. K., Dunlap S., Ryu B. and Alani R. M. (2003). Id proteins in cell growth and tumorigenesis. Cancer Cell 3, 525-530. 10.1016/S1535-6108(03)00141-7 [DOI] [PubMed] [Google Scholar]
- Singh A. and Choi K. W. (2003). Initial state of the Drosophila eye before dorsoventral specification is equivalent to ventral. Development 130, 6351-6360. 10.1242/dev.00864 [DOI] [PubMed] [Google Scholar]
- Singh A., Shi X. and Choi K.-W. (2006). Lobe and Serrate are required for cell survival during early eye development in Drosophila. Development 133, 4771-4781. 10.1242/dev.02686 [DOI] [PubMed] [Google Scholar]
- Spratford C. M. and Kumar J. P. (2013). Extramacrochaetae imposes order on the Drosophila eye by refining the activity of the Hedgehog signaling gradient. Development 140, 1994-2004. 10.1242/dev.088963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spratford C. M. and Kumar J. P. (2015). Extramacrochaetae functions in dorsal-ventral patterning of Drosophila imaginal discs. Development 142, 1006-1015. 10.1242/dev.120618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevaux O., Dimova D., Frolov M. V., Taylor-Harding B., Morris E. and Dyson N. (2002). Distinct mechanisms of E2F regulation by Drosophila RBF1 and RBF2. EMBO J. 21, 4927-4937. 10.1093/emboj/cdf501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukhanova M. J. and Du W. (2008). Control of cell cycle entry and exiting from the second mitotic wave in the Drosophila developing eye. BMC Dev. Biol. 8, 7 10.1186/1471-213X-8-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukhanova M. J., Deb D. K., Gordon G. M., Matakatsu M. T. and Du W. (2007). Proneural basic helix-loop-helix proteins and epidermal growth factor receptor signaling coordinately regulate cell type specification and cdk inhibitor expression during development. Mol. Cell. Biol. 27, 2987-2996. 10.1128/MCB.01685-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun R., Su Y., Zhao X., Qi J., Luo X., Yang Z., Yao Y., Luo X. and Xia Z. (2009). Human calcium/calmodulin-dependent serine protein kinase regulates the expression of p21 via the E2A transcription factor. Biochem J. 419, 457-466. 10.1042/BJ20080515 [DOI] [PubMed] [Google Scholar]
- Tapanes-Castillo A. and Baylies M. K. (2004). Notch signaling patterns Drosophila mesodermal segments by regulating the bHLH transcription factor twist. Development 131, 2359-2372. 10.1242/dev.01113 [DOI] [PubMed] [Google Scholar]
- Tapon N., Harvey K. F., Bell D. W., Wahrer D. C., Schiripo T. A., Haber D. and Hariharan I. K (2002). Salvador promotes both cell cycle exit and apoptosis in Drosophila and is mutated in human cancer cell lines. Cell 110, 467-478. 10.1016/S0092-8674(02)00824-3 [DOI] [PubMed] [Google Scholar]
- Teachenor R., Beck K., Wright L. Y. T., Shen Z., Briggs S. P. and Murre C. (2012). Biochemical and phosphoproteomic analysis of the helix-loop-helix protein E47. Mol. Cell. Biol. 32, 1671-1682. 10.1128/MCB.06452-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Doren M., Ellis H. M. and Posakony J. W. (1991). The Drosophila extramacrochaetae protein antagonizes sequence-specific DNA binding by daughterless/achaete-scute protein complexes. Development 113, 245-255. [DOI] [PubMed] [Google Scholar]
- Van Doren M., Powell P. A., Pasternak D., Singson A. and Posakony J. W. (1992). Spatial regulation of proneural gene activity: auto- and cross-activation of achaete is antagonized by extramacrochaetae. Genes Dev. 6, 2592-2605. 10.1101/gad.6.12b.2592 [DOI] [PubMed] [Google Scholar]
- Villares R. and Cabrera C. V. (1987). The achaete-scute gene complex of Drosophila melanogaster: conserved domains in a subset of genes required for neurogenesis and their homology to myc. Cell 50, 415-424. 10.1016/0092-8674(87)90495-8 [DOI] [PubMed] [Google Scholar]
- Weinberg R. A. (1995). The retinoblastoma protein and cell cycle control. Cell 81, 323-330. 10.1016/0092-8674(95)90385-2 [DOI] [PubMed] [Google Scholar]
- Wolff T. and Ready D. F. (1991). The beginning of pattern formation in the Drosophila compound eye: the morphogenetic furrow and the second mitotic wave. Development 113, 841-850. [DOI] [PubMed] [Google Scholar]
- Wong Y.-C., Wang X. and Ling M.-T. (2004). Id-1 expression and cell survival. Apoptosis 9, 279-289. 10.1023/B:APPT.0000025804.25396.79 [DOI] [PubMed] [Google Scholar]
- Yang L. and Baker N. E. (2003). Cell cycle withdrawal, progression, and cell survival regulation by EGFR and its effectors in the differentiating Drosophila eye. Dev. Cell 4, 359-369. 10.1016/S1534-5807(03)00059-5 [DOI] [PubMed] [Google Scholar]
- Yang C. H., Simon M. A. and McNeill H. (1999). mirror controls planar polarity and equator formation through repression of fringe expression and through control of cell affinities. Development 126, 5857-5866. [DOI] [PubMed] [Google Scholar]
- Zebedee Z. and Hara E. (2001). Id proteins in cell cycle control and cellular senescence. Oncogene 20, 8317-8325. 10.1038/sj.onc.1205092 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.