Abstract
Post-translational histone modifications play critical roles in regulating transcription, the cell cycle, DNA replication and DNA damage repair1. The identification of new histone modifications critical for transcriptional regulation at initiation, elongation, or termination is of particular interest. Here, we report a new layer of regulation in transcriptional elongation that is conserved from yeast to mammals, based on a phosphorylation of a highly-conserved tyrosine residue, Y57, in histone H2A that is mediated by an unsuspected tyrosine kinase activity of casein kinase 2 (CK2). Mutation of H2A-Y57 in yeast or inhibition of CK2 activity impairs transcriptional elongation in yeast as well as in mammalian cells. Genome-wide binding analysis reveals that CK2α, the catalytic subunit of CK2, binds across RNA polymerase II-transcribed coding genes and active enhancers. Mutation of Y57 causes a loss of H2B mono-ubiquitylation as well as H3K4me3 and H3K79me3, histone marks associated with active transcription. Mechanistically, both CK2 inhibition and H2A-Y57F mutation enhance the H2B deubiquitylation activity of the SAGA complex, suggesting a critical role of this phosphorylation in coordinating the activity of the SAGA during transcription. Together, these results identify a new component of regulation in transcriptional elongation based on CK2-dependent tyrosine phosphorylation of the globular domain of H2A.
Keywords: tyrosine phosphorylation, histone H2A, transcriptional elongation, Casein Kinase 2 (CK2), yeast, SAGA complex
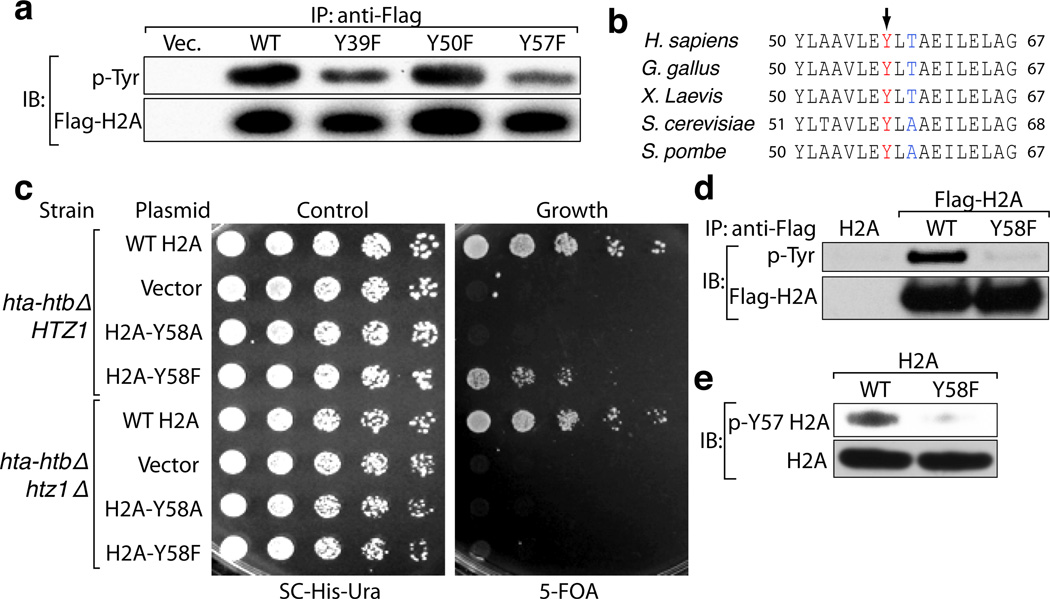
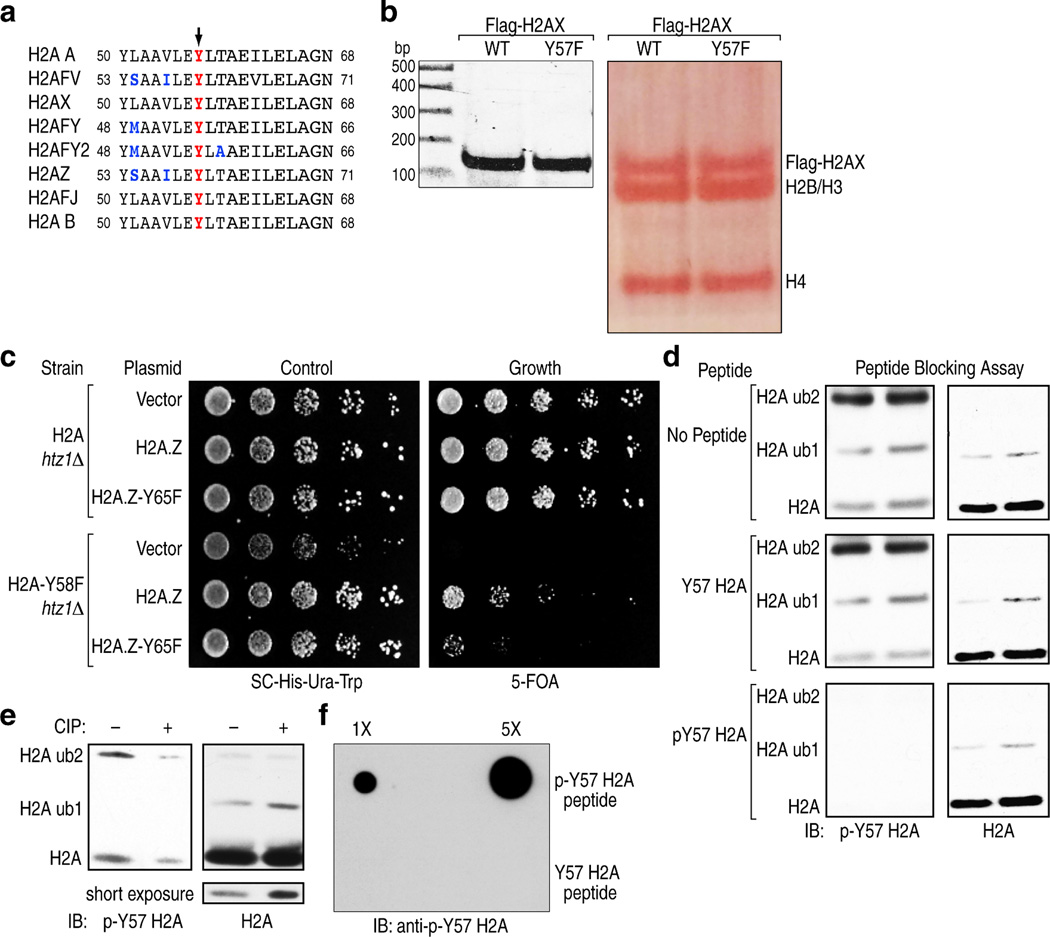
To assess potential tyrosine phosphorylation events in H2A, we mutated every tyrosine residue in H2A to phenylalanine and expressed the mutants in 293T cells. Mutation of Y39 and Y57 resulted in a decrease in tyrosine phosphorylation compared to the wild-type (WT) protein indicating that these residues might be phosphorylated (Fig. 1a). Mass spectrometry (MS) confirmed phosphorylation of these residues in histone extracts from 293T cells (Supplemental Table 1). The Y57 residue, along with neighboring residues, is conserved from yeast to mammals (Fig. 1b), and is present in all the variants of H2A (Extended Data Fig. 1a). In budding yeast, where genetic manipulation of histones is possible, mutation of the corresponding residue to alanine is lethal, suggesting a critical structural and/or functional contribution of this tyrosine residue2,3. Analysis of histones in mononucleosomes containing WT and H2A-Y57F mutant showed similar stoichiometry, suggesting that the Y57F mutation is unlikely to affect the structural integrity of nucleosomes (Extended Data Fig. 1b). Hence, we tested whether the more structurally conservative substitution of tyrosine with phenylalanine would produce a different phenotype in yeast. Y58 in yeast H2A corresponds to Y57 in mammalian H2A. H2A-Y58F mutant was viable and exhibited a slow growth phenotype (Fig. 1c). Interestingly, the same mutation proved to be lethal in the HTZ1 (the gene encoding H2AZ) null background, and double mutation of the tyrosine residue in both H2A and H2AZ resulted in an extremely slow growth phenotype (Fig. 1c, Extended Data Fig. 1c). Next, we tested if this site is phosphorylated in yeast. Immunoprecipitated Flag-tagged H2A-Y58F showed reduced tyrosine phosphorylation compared to the WT protein (Fig. 1d), suggesting that this residue is phosphorylated.
Figure 1. The conserved Y57 residue in H2A is phosphorylated.
(a) Y57 in H2A is phosphorylated in 293T cells. Flag-tagged H2A mutants were expressed in 293T cells, immunoprecipitated under denaturing conditions, and immunoblotted as indicated. (b) Y57 in H2A is highly conserved. Comparison of H2A sequence surrounding the Y57 residue (arrow) in different organisms. (c) Y58 in H2A is functionally important in yeast. Five-fold serial dilutions of the indicated transformants were plated on SC-His-Ura for growth control and 5-FOA for the removal of pJH33. (d, e) Y58 in H2A is phosphorylated in S. cerevisiae. (d) Flag-tagged WT H2A and H2A-Y58F were immunoprecipitated under denaturing conditions and immunoblotted as indicated. (e) Whole cell extracts from yeast strains were prepared under denaturing conditions and immunoblotted as indicated. Data represent three independent experiments.
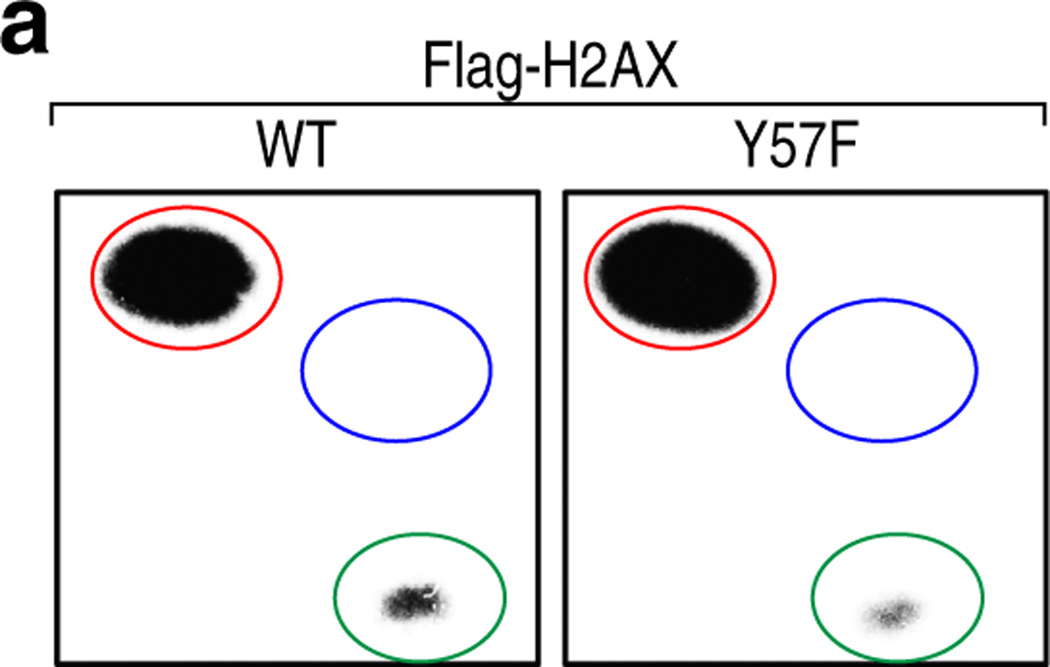
To confirm Y57 phosphorylation and investigate its function, an antibody specific for phospho-Y57 H2A was developed. This antibody detected proteins corresponding to the size of H2A and ubiquitylated H2A in 293T cells (Extended Data Fig. 1d). Peptide blocking assays and dot blot assays verified the specificity of the antibody and treatment with calf intestinal phosphatase further validated its phospho-specificity (Extended Data Fig. 1e, f). Use of this antibody confirmed H2A-Y58 phosphorylation in yeast (Fig. 1e). Collectively, these results demonstrate that Y57 in H2A is phosphorylated and that this phosphorylation is conserved from yeast to mammals.
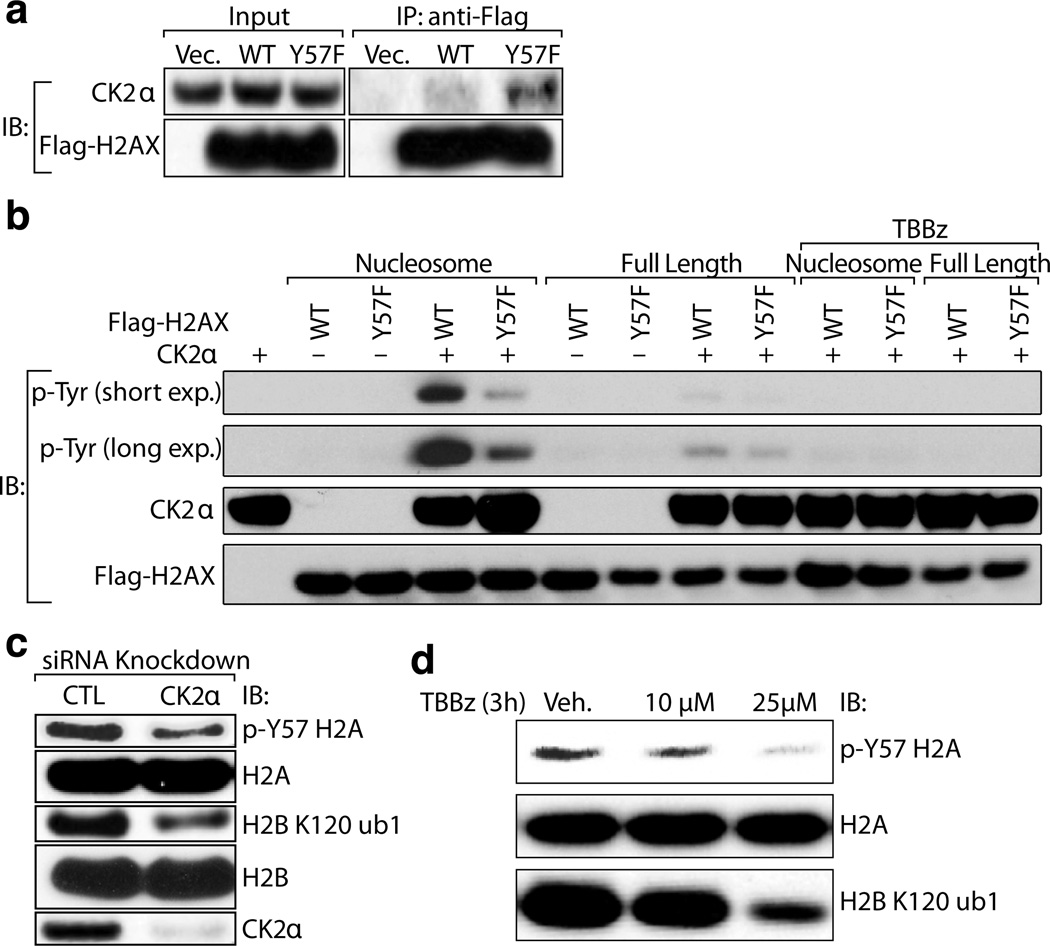
To identify the kinase(s) that mediate(s) phosphorylation of Y57 in H2A in mammals, we first performed MS analysis of proteins interacting with H2A in 293T cells. To be consistent with yeast, we used H2AX, a common H2A variant that has closer sequence homology to yeast H2A, for co-immunoprecipitation and in vitro kinase assays. The MS data revealed that the CK2α catalytic subunit of CK2, interacts preferentially with H2A-Y57F compared to WT H2A (Supplemental Table 2). This interaction was further verified by immunoblotting, which revealed a higher level of CK2α associated with H2A-Y57F (Fig. 2a). One implication of this interaction is that CK2 may phosphorylate Y57 in H2A, and Y57F mutation stabilizes the enzyme-substrate interaction, analogous to substrate trapping approaches that have been successfully used to identify substrates of tyrosine phosphatases4. Although CK2 is considered primarily to be a Ser/Thr kinase, two studies have reported its tyrosine phosphorylation activity, thus implicating it as a dual-specificity kinase5,6. To investigate potential roles of CK2 in H2A-Y57 phosphorylation, we tested whether CK2α phosphorylates the tyrosine residue (Y57) in H2A in an in vitro assay using full-length H2A or nucleosomes. The kinase assay revealed that CK2α phosphorylates Y57 in H2A, acting preferentially in the context of nucleosomes (Fig. 2b). This phosphorylation was inhibited by TBBz, a chemical inhibitor of CK27. To further establish the tyrosine kinase activity of CK2α, we performed phospho-amino acid analysis of H2A in nucleosomes using γ32P-labeled ATP and found that CK2α indeed phosphorylates Y57 in H2A as well as serine but not threonine residues (Extended Data Fig. 2a).
Figure 2. CK2 phosphorylates Y57 in H2A.
(a) CK2α interacts preferentially with the H2A-Y57F mutant. Flag-tagged WT and H2AX-Y57F were expressed in 293T cells, immunoprecipitated and immunoblotted. (b) CK2 phosphorylates Y57 in H2A in nucleosomes in vitro. In vitro kinase assays were performed using recombinant GST-CK2α, and full-length or nucleosomal Flag-tagged H2AX purified from 293T cells, and were immunoblotted to examine tyrosine phosphorylation. (c, d) CK2 phosphorylates Y57 in H2A in vivo. (c) CK2α was knocked down in 293T cells, and nuclear extracts were immunoblotted. CTL indicates scrambled siRNA. (d) Nuclear extract from 293T cells treated with TBBz were immunoblotted. Data represent three independent experiments.
We next investigated whether CK2 is necessary for H2A-Y57 phosphorylation in vivo. CK2α knockdown in 293T cells reduced the levels of H2A-Y57 phosphorylation (Fig. 2c), supporting an in vivo role of CK2α in regulating this phosphorylation. Moreover, a dose-dependent decrease in H2A-Y57 phosphorylation was observed upon treatment with TBBz (Fig. 2d), further strengthening the role of CK2 in H2A-Y57 phosphorylation. Together, these results provide strong evidence for a function of CK2 in H2A-Y57 phosphorylation.
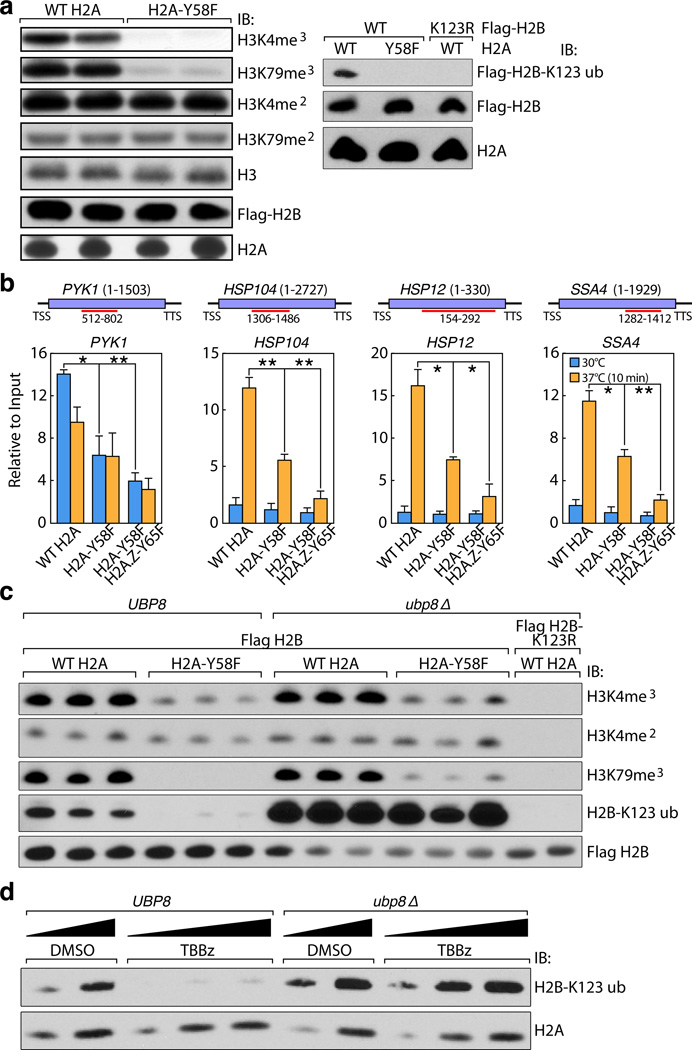
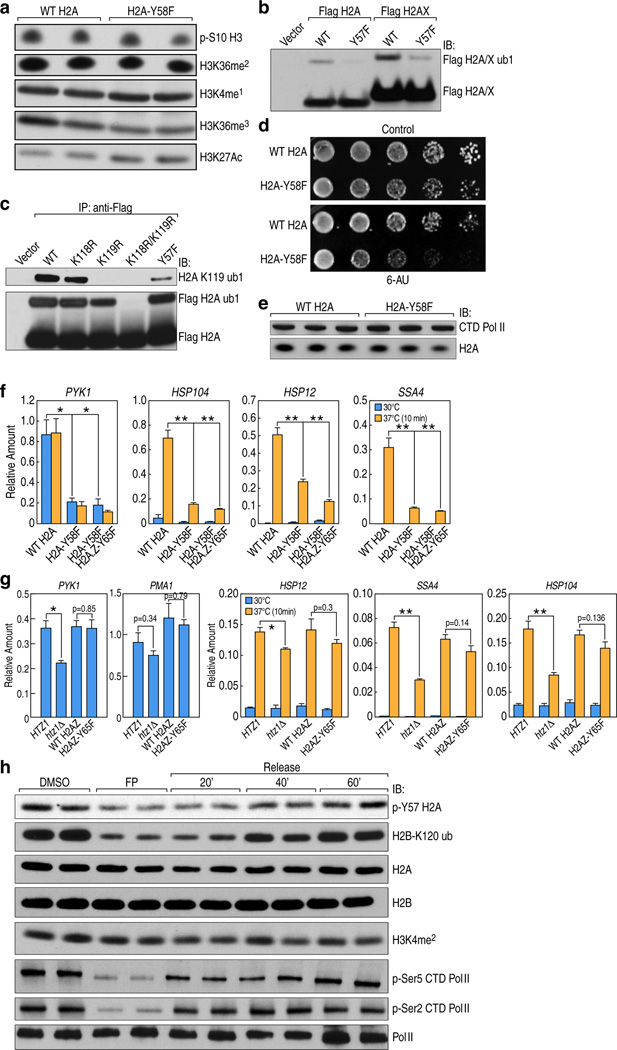
To investigate the physiological significance of H2A-Y57 phosphorylation, we first examined in yeast the impact of H2A-Y58F mutation on other important histone marks. We found that H2A-Y58F mutation resulted in a loss of H2B mono-ubiquitylation, as well as tri-methylation of H3K4 and H3K79 (Fig. 3a). H3K27 acetylation showed a modest increase, and all other histone modifications tested were unaffected (Extended Data Fig. 3a). Interestingly, Y57F mutation also lowered the level of H2A mono-ubiquitylation in 293T cells (Extended Data Fig. 3b, c). The role of H2A-Y58 phosphorylation as a potential regulator of H2B mono-ubiquitylation is of particular significance because this modification has an established role in transcriptional elongation8,9, thus potentially linking H2A-Y58 phosphorylation to transcriptional elongation. Further, yeast with H2A-Y58F mutation exhibited increased sensitivity to 6-azauracil (Extended Data Fig. 3d), indicating a defect in transcriptional elongation10.
Figure 3. H2A-Y58F mutation enhances H2B deubiquitylation, and impairs transcriptional elongation in yeast.
(a) The H2A-Y58F mutation affects several histone modifications. Whole cell extracts from indicated strains were immunoblotted. (b) H2A-Y58F mutation impairs transcriptional elongation. Pol II binding in the indicated genes was measured by ChIP-qPCR in yeast strains grown at 30°C or at 37°C for 10 min (n=3, mean ± SEM, * indicates p<0.05, ** indicates p<0.01). The ORF of the genes and the regions amplified by the primer pairs are shown. P-values were calculated by Student’s two-tailed t-test. (c) UBP8 deletion rescues the defect in H2B mono-ubiquitylation in the H2A-Y58F mutant yeast. Whole cell extracts from indicated yeast strains were immunoblotted. (d) CK2 prevents the deubiquitylation of H2B. WT or ubp8Δ cells were treated with vehicle (DMSO) or TBBz (25µM) for 3 hr and whole cell extracts were immunoblotted. Data represent three independent experiments.
To assess the role of Y58 phosphorylation in transcriptional elongation, binding of RNA polymerase II (Pol II) in actively transcribed genes was evaluated by chromatin immunoprecipitation (ChIP) followed by quantitative real-time polymerase chain reaction (qPCR). Pol II binding was reduced in the gene body of a housekeeping gene, PYK1, as well as a number of heat shock-induced genes11 upon heat shock in the H2A-Y58F mutant. Pol II binding was further reduced in H2A-Y58F and H2AZ-Y65F mutants (Fig. 3b). The decrease in Pol II binding in the H2A-Y58F mutant was not due to reduced Pol II expression (Extended Data Fig. 3e). Consistent with the defect in transcriptional elongation, a decreased level of messenger RNA (mRNA) of the corresponding genes was observed in H2A-Y58F mutants (Extended Data Fig. 3f), whereas H2AZ-Y65F mutants showed only a mild defect in transcription of heat shock-induced genes (Extended Data Fig. 3g). Furthermore, in 293T cells H2A-Y57 phosphorylation, like H2B-monoubiquitylation, was correlated with transcriptional elongation events, as demonstrated when transcriptional elongation was blocked with flavopiridol treatment12, and induced by washing out the drug (Extended Data Fig. 3h). H3K4me2, a control histone mark, did not change in this assay (Extended Data Fig. 3h). Collectively, these results indicate a conserved role of H2A-Y57/58 phosphorylation in regulating transcriptional elongation.
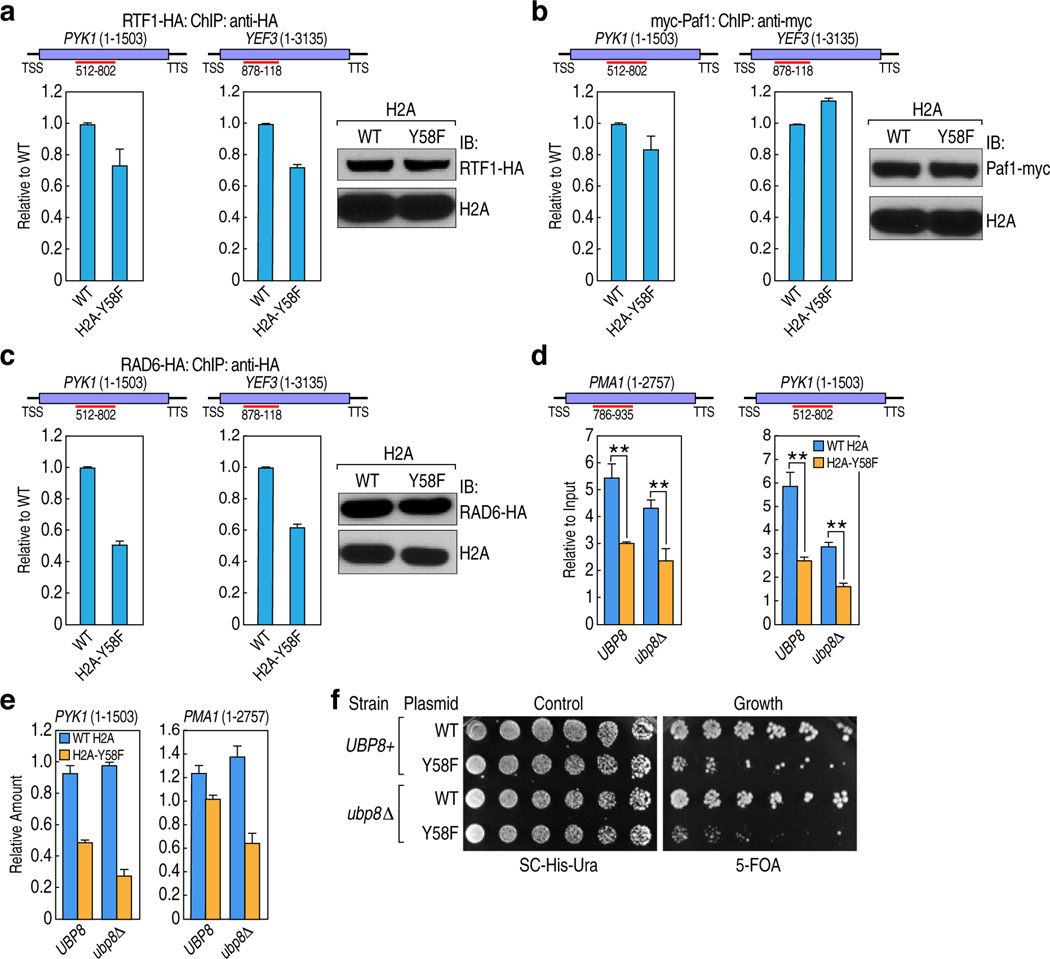
To address the mechanism through which H2A-Y57/Y58 phosphorylation regulates H2B ubiquitylation, we examined the recruitment of proteins known to be involved in establishing H2B mono-ubiquitylation, such as Paf1, Rtf1 and Rad613–15, by ChIP-qPCR. In yeast, binding of Paf1 and Rtf1 was comparable in WT and H2A-Y58, whereas Rad6 binding was slightly reduced in the genes tested in the mutant (Extended Data Fig. 4a–c). The effect of H2A-Y58F mutation was further evaluated in mutant UBP8, which encodes a major H2B deubiquitylase (DUB) that is a component of the SAGA complex. Deletion of UBP8 restored H2B mono-ubiquitylation in the H2A-Y58F mutant to the WT level (Fig. 3c), suggesting that the defect in H2B mono-ubiquitylation in the H2A-Y58F mutant occurs through Ubp8 DUB activity, and not through defective ubiquitylation machinery. Likewise, CK2 inhibition reduced H2B mono-ubiquitylation in WT yeast while having no effect in UBP8 mutants (Fig. 3d), further supporting the role of CK2-mediated H2A-Y58 phosphorylation in preventing H2B deubiquitylation. Surprisingly, despite the complete rescue of H2B mono-ubiquitylation, UBP8 deletion only partially rescued H3K79me3, and the level of H3K4me3 remained low in the H2A-Y58F mutant (Fig. 3c). Furthermore, deletion of UBP8 did not rescue defects in Pol II binding, the transcript levels, or the slow growth phenotype of the H2A-Y58F mutant (Extended Data Fig. 4d–f). These results suggest that the physiological effects of Y58 mutation in H2A are linked to, but extend beyond, the loss of H2B mono-ubiquitylation.
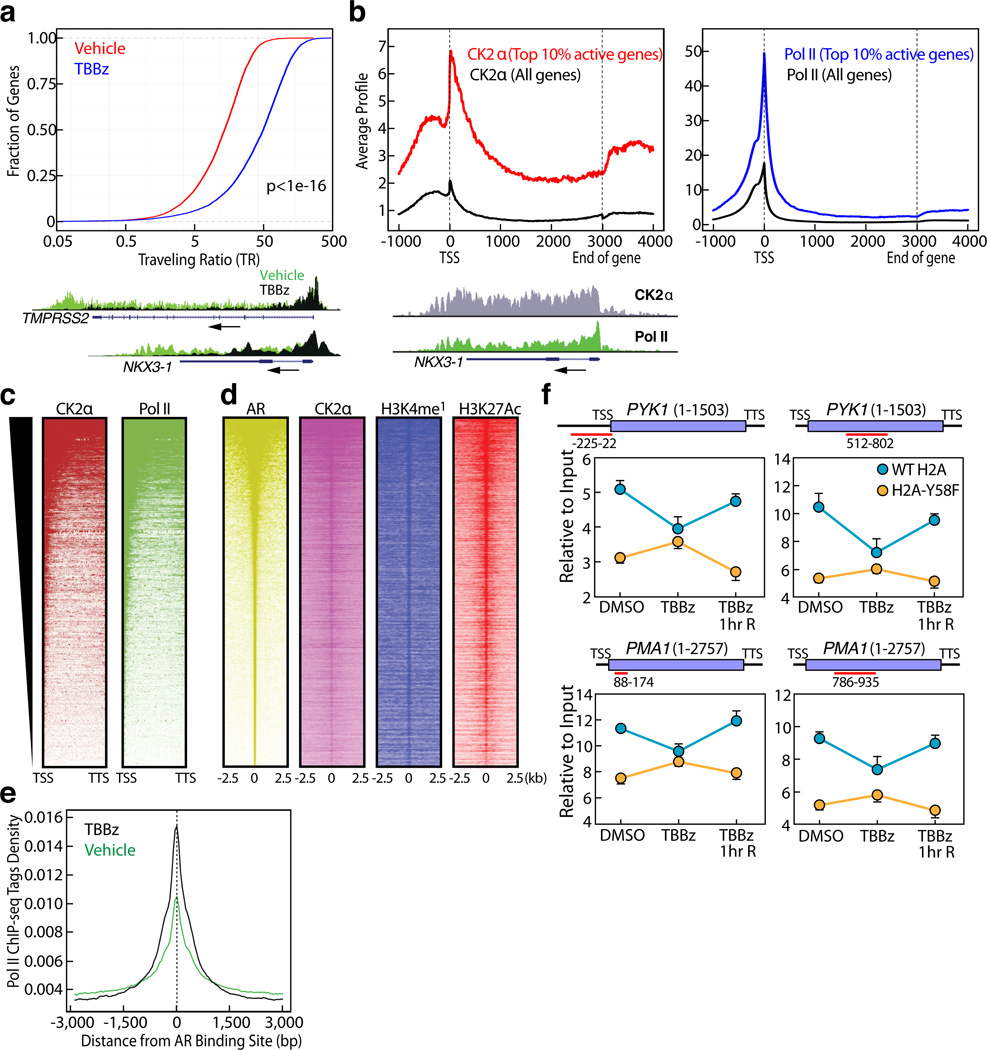
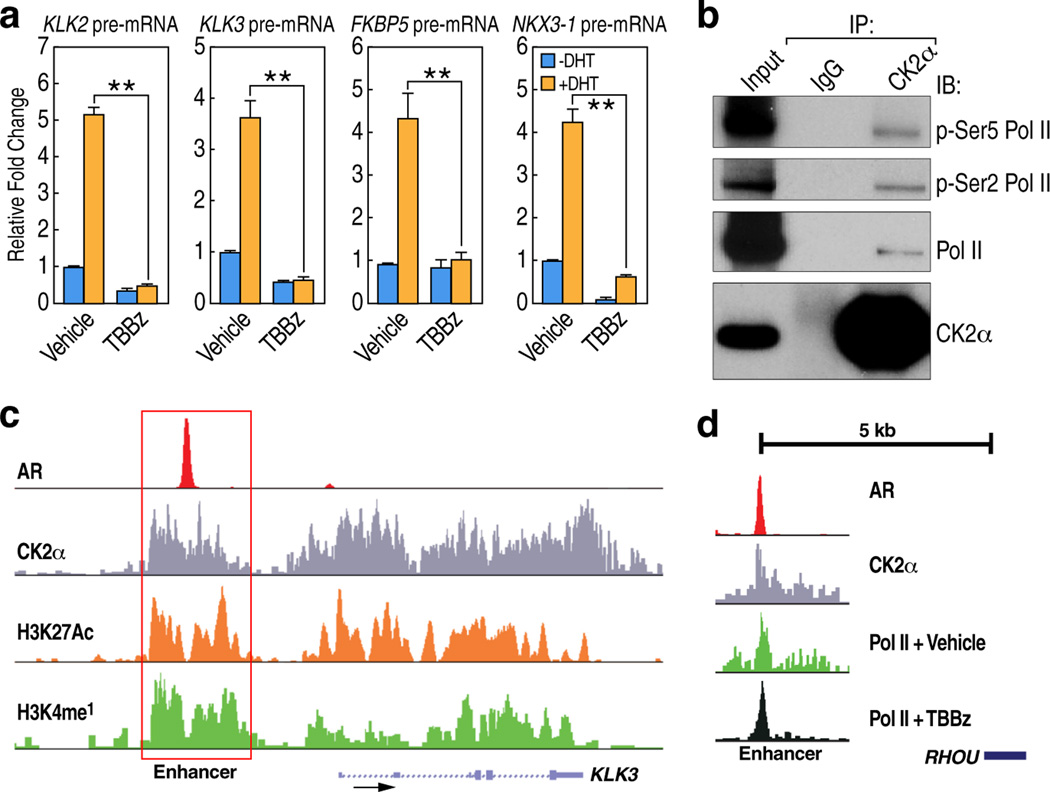
Next, we investigated whether knockdown or inhibition of CK2 phenocopies the effects of H2A-Y58F mutation in the regulation of transcriptional elongation. Consistent with the result in yeast, H2B mono-ubiquitylation was reduced upon CK2α knockdown or inhibition of CK2 kinase activity in 293T cells (Fig. 2c, d). Likewise, Pol II binding in active genes in LNCaP human prostate carcinoma cells was impaired in gene bodies but not in the promoter regions upon CK2 inhibition as determined by ChIP-sequencing (seq) (Fig. 4a). A traveling ratio (TR) plot of Pol II16,17 showed an obvious shift in CK2-inhibited cells (Fig. 4a), suggesting that CK2 kinase activity is required for transcriptional elongation in gene bodies. In agreement with this, dihydrotestosterone (DHT)-induced transcriptional activation of androgen receptor (AR)-regulated genes was impaired in LNCaP cells treated with TBBz (Extended Data Fig. 5a).
Figure 4. CK2 regulates transcriptional elongation.
(a) CK2 kinase activity is required for promoter-proximal pause release in mammalian cells. Pol II travelling ratio was plotted for LNCaP cells treated with vehicle or TBBz for 2 ½ hours (p< 1e-16). P-value was calculated by K-S test. Overlay of Pol II occupancy in representative genes (TMPRSS2 and NKX3-1) are shown. (b, c) CK2α binds across the actively transcribed genes globally. (b) CK2α and Pol II occupancy were determined in the top 10% active genes (n=3162) and all genes by CEAS24. The length of all gene bodies is normalized to 3kbp. Enrichment of CK2α and Pol II at a representative active gene (NKX3-1) is shown. (c) Heat map of Pol II and CK2α binding profile in top 10% active genes from Transcription Start Site (TSS) to Transcription Termination Site (TTS) is shown. (d) CK2α binds to active enhancers. Heat maps of CK2α, H3K4me118, and H3K27acetyl18 signals over androgen receptor (AR)18-enriched regions in LNCaP cells were determined. (e) CK2 regulates transcriptional elongation in enhancers. Shown are Pol II ChIP tag density plots centered at AR enriched enhancers. (f) H2A-Y58 phosphorylation is critical for CK2-mediated regulation of transcriptional elongation in yeast. WT and H2A-Y58F cells were treated with vehicle or TBBz (25µM) for 3 hr, or treated with TBBz for 3 hr followed by 1-hr inhibition release (TBBz 1hr R), and Pol II binding in the indicated genes was measured by ChIP-qPCR (n=3, mean ± SEM). The ORF of the genes and the regions amplified by the primer pairs are shown. Data represent two independent experiments for ChIP-seq (a–e) and three for ChIP-qPCR (f).
To understand the molecular aspects of the role of CK2 in transcriptional elongation, genome-wide localization of CK2α in LNCaP cells was determined by ChIP-seq. Like Pol II, CK2α showed binding to actively transcribed genes across gene bodies, although its binding profile was distinct from that of Pol II (Fig. 4b). Meta analysis of the top 10% of active genes, based on global run-on sequencing (GRO-seq) results in LNCaP cells18, revealed that CK2α globally co-localizes with Pol II (Fig. 4c). Consistent with the genome-wide binding pattern, CK2α immunoprecipitated with the phosphorylated C-terminal domain (CTD) of Pol II, which is localized in the promoters (phospho-Ser 5) and gene bodies (phospho-Ser2) of active genes (Extended Data Fig. 5b). We also found CK2α binding in intergenic regions that co-localized with H3K4mono-methyl and H3K27acetyl marks, histone modifications that co-localize with active enhancers19, and LNCaP cell-type specific AR enhancers (Fig. 4d, Extended Data Fig. 5c), suggesting that the intergenic CK2α peaks are in enhancer regions. Inhibition of CK2 also caused stalling of Pol II in the AR-bound enhancers (Fig. 4e, Extended Data Fig. 5d), underpinning the function of CK2 in transcriptional elongation in both gene bodies and enhancers.
We asked if CK2 also regulates transcriptional elongation in yeast, and if so, whether H2A-Y58 phosphorylation is a key player, among the many other substrates of CK220. Inhibition of CK2 kinase activity resulted in a decrease in the recruitment of Pol II in both the promoter region as well as gene bodies of the tested genes in WT yeast, but did not have additive effects in the H2A-Y58F yeast (Fig. 4f). It is noteworthy that both CK2 inhibition and H2A-Y58F mutation did not result in promoter proximal pausing in the genes tested, consistent with the observation that regulation by promoter proximal pausing is rare in yeast21. Collectively, these results demonstrate that CK2 plays a deeply conserved role in transcriptional elongation both in gene bodies and enhancers, and that H2A-Y58 phosphorylation is critical in this regulation.
This study has identified a new H2A modification, phosphorylation of Y57/Y58, that provides new insight into how two important protein complexes, SAGA and Paf1, with opposite enzymatic activities on H2B-ubiquitylation, might be coordinated during transcriptional elongation. The data further emphasize the key significance of this delicate coordination as demonstrated by defects in transcriptional elongation upon mutation of the conserved phospho-site. A moderate increase in Gcn5-SAGA mediated H3K27 acetylation22 in the H2A-Y58F mutant yeast, suggests that the phosphorylation may antagonize multiple activities of SAGA. Such antagonism could explain the partial rescue of the defects in the H2A-Y58F mutant upon deletion of UBP8, as the other modules of SAGA remain functional in the absence of Ubp823. Although unlikely, the potential role in transcriptional elongation of the hydroxyl group of Y58, rather than phosphorylation itself, cannot yet be dismissed. Assays utilizing synthetic nucleosomes with constitutively phosphorylated H2A-Y57 may ultimately be used to further define the role of this site in transcriptional elongation. This study also emphasizes the functional significance of the tyrosine kinase activity of CK2, and invites search for other tyrosine substrates of CK2. Importantly, the identification of a highly conserved role of CK2 in regulating transcriptional elongation in both gene bodies and enhancer regions adds yet another layer to understanding transcription.
Methods
Cell culture, siRNA, Primers, Plasmids, Transfection, Antibodies, Kinase inhibitors
LNCaP and 293T cells were cultured in F12 medium supplemented with 10% FBS and glutamine. For the DHT-treatment experiments, LNCaP cells were cultured in Deficient DME High Glucose medium with 5% FBS (charcoal dextran filtered) for 3–4 days. Short interfering RNA (siRNA) against CK2α was from Santa Cruz (sc-29918). Cells were transfected using lipofectamine 2000 (Invitrogen) using the manufacturer’s protocol. Mutagenesis was done using Quikchange Lightening Mutagenesis Kit following the manufacturer’s recommended protocol. The following antibodies were used in this study: p-Y57 H2A antibody was generated by Biomatik Company using phospho-Y57 H2A peptide as an antigen (LE(pY)LTAEILELAGNC), purified, and positively selected using p-Y57 H2A peptide column, and negatively selected using Y57 H2A peptide column; anti-H2A (Abcam Cat. No. ab18255), anti-CK2α (Cat. No. ab 70774), anti-H2B K120 ub (Cell Signaling Cat. No. 5546S), anti-Flag (Sigma M2), anti-RNA polymerase II (Santa Cruz N-20 (for mammalian cells), Abcam Cat. No. ab817 (for yeast cells)), anti-p-Ser2 Pol II (Abcam Cat. No. ab5095), anti-p-Ser5 Pol II (Abcam Cat. No. ab5131), anti-phosphotyrosine (Millipore Cat. No. 05-321 (4G10)), anti-H2A (yeast) (Active Motif Cat. No. 39236), anti-H4K4me3 (Active Motif Cat. No. 39159), anti-H3K4me2 (Millipore Cat. No. 07-030), anti-H3K4me1 (Millipore Cat. No. 07-436), anti-p-S10 H3 (Millipore Cat. No. 06-570), anti-H3K27 acetyl (Abcam Cat. No. ab4729), anti-H3K79me2 (Active Motif Cat. No. 39924), anti-H3K36me2 (Active Motif Cat. No. 39255) and anti-H2A-K119-ub (Cell Signaling Cat No. 8240S). TBBz and flavopiridol were from Sigma. Primers used in this study are listed in Supplemental Table 3.
Chromatin Immunoprecipitation
Cells were grown to 90-95% confluence, fixed with 1% formaldehyde for 15 min for Pol II ChIP, and with Di-Succinimidyl Glutarate (DSG) for 45 minutes followed by 10 min fixation with 1% formaldehyde for CK2α ChIP at room temperature. To terminate crosslinking, fixed cells were incubated with glycine (1.25 mM) for 10 min. Nuclei were prepared as described25, which were then sonicated in lysis buffer (150 mM NaCl, 1% Triton X-100, 20 mM Tris pH 8.0, 0.1% SDS) using a Bioruptor to fragment chromatin to less than 500bp. Chromatin was pre-cleared with Protein-G magnetic beads, and then immunoprecipitated using 5 µg of antibody per 10 cm plate for each samples. The chromatin antibody mix was incubated with 35 µl protein G-conjugated Dyna beads for 4 hr at 4°C, washed three times with wash buffer (1% Triton X-100, 50 mM Tris pH 8.0, 10% glycerol) with increasing concentrations of NaCl (150 mM, 300 mM and 400 mM), and two times with Tris-EDTA (TE) buffer. DNA was eluted in 1% SDS in TE buffer, crosslinking was reversed overnight at 65°C, and DNA was purified using Qiagen columns. Yeast ChIP was performed using 300–400 µg DNA and 2 µg antibody using the protocol as described26 with some modifications. Briefly, cells were fixed for 15 min, and then incubated with glycine at a final concentration of 2.5 mM for 5 min, and cells were lysed using glass beads (5 × 5 min beating with 2 min on ice intervals). Cells were then sonicated to obtain chromatin fragments of less than 1 kb, and the rest of the protocol was similar to the mammalian cells. All the buffers used included fresh complete protease inhibitors (Roche), 1 mM PMSF, 2 mM Na3VO4, 10 mM β-glycerol phosphate and 10 mM NaF.
Statistical Analysis
P-values for ChIP-qPCR and reverse transcription (RT)-PCR were calculated by Student's T-test, two tails, type two using Microsoft Excel. The statistical significance of the change of Travelling Ratio between control and CK2 inhibitor treated samples was determined using two-tailed Kolmogorov-Smirnov (K-S) test. In yeast experiments, independent transformants that were processed separately were considered biological replicates, and in mammalian experiments, cells cultured in different plates, and processed separately were considered biological replicates. Statistical analysis representing “n” means biological replicates. All independent experiments are biological replicates.
Identification of ChIP-seq peaks
ChIP-seq peaks were identified using HOMER (http://biowhat.ucsd.edu/homer). A 200bp sliding window was used for transcription factors and a 500bp sliding window was used for histone modifications with the requirement that two peaks are at least 500bp away for transcription factors, and 1250bp for histone modifications to avoid redundant peak identification. Tag density was calculated by using HOMER and average signal profiles surrounding AR enriched regions were generated with CEAS24 (cis-regulatory element annotation system) which were visualized with Java TreeView (http://jtreeview.sourceforge.net).
ChIP-seq Alignment
DNA was ligated to specific adaptors followed by HT sequencing on Illumina’s HiSeq 2000 system according to the manufacturer’s instructions. The first 48bp for each sequence tag returned was aligned to the hg18 (human) assembly using Bowtie2. The data were visualized by preparing custom tracks on the UCSC genome browser by using HOMER. The total number of mappable reads was normalized to 107 for each experiment presented.
ChIP-seq Data Deposition
ChIP-seq data has been deposited in GEO. The accession number is: GSE58607. Other published sequencing data used in the study are described18.
Traveling Ratio (TR) Calculation
Pol II TR was defined as the relative ratio of Pol II density in the promoter-proximal region and the gene body. The promoter proximal region refers to the window from −50bp to +300bp surrounding transcription start site (TSS), and the gene body refers to regions from 300bp downstream of TSS to 13kbp from TSS for genes longer than 13kbp, and to the transcription termination site (TTS) for the genes shorter than 13kbp. The significance of the change of TR between control and CK2 inhibitor-treated samples was displayed using box plot.
In vitro kinase assay and Phospho amino Acid Analysis (PAA)
In vitro kinase reactions were performed with 100 ng of recombinant GST-tagged CK2α (expressed in E. Coli with 0.2mM IPTG induction for 3 hr at 30°C) in 1× kinase buffer (20mM Tris-HCl, 50mM KCl, 10mM MgCl2pH 7.5) with the addition of 0.2 mM ATP for cold reaction (and 10 µM ATP mixed with 10 µCi of γ32P-labeled ATP for the radioactive reactions). The substrates (Flag-tagged WT H2AX and H2AX-Y57F) were purified from 293T cells expressing Flag-tagged H2AX constructs. For the full-length proteins, histone extracts were immunoprecipitated using Flag antibody, then washed several times with wash buffer (1% Triton X-100, 900 mM NaCl, 20mM Tris 8.0), treated with CIP for 30 min at 37°C, washed a few more times with wash buffer, and eluted with 3× Flag peptides. Mononucleosomes were prepared as described25 with minor changes in microccocal nuclease digestion. Briefly, nuclei were isolated from 15-cm fully confluent plates, and DNA was digested in 1.2 ml total volume with 2.5µl micrococcal nuclease (NEB Catalog # M0247S) for 10 min at 37°C, and the reaction was stopped by adding 5mM EGTA, and mononucleosomes were collected by centrifugation. Mononucleosomes were immunoprecipitated with Flag antibody, washed four times with Buffer A (340 mM Sucrose, 10 mM HEPES pH 7.5, 10% glycerol, 1.5mM MgCl2, 10 mM KCl) followed by three washes with kinase reaction buffer, then treatment with 500 µM FSBA (Sigma cat number F9128-25MG) for 25 min at 37°C to irreversibly inhibit any potential kinases interacting with the nucleosome. Following this were three washes with kinase buffer, treatment with CIP for 30 minutes at 37°C, and another three washes with buffer A. The bound nucleosomes were eluted with 3× Flag peptides in buffer A. The kinase reactions were carried out for 1 hr at 30°C. For PAA, the samples were separated by SDS-PAGE, transferred to PVDF membrane, and the membrane corresponding to the mobility of phosphorylated H2AX was excised, and PAA using two-dimensional electrophoresis on thin layer cellulose plates was performed as described27.
Whole cell extracts (WCE), Immunoprecipitation (IP) and Cell Fractionation
Yeast whole cell extracts were prepared either by breaking the cells with glass beads in PBS or boiling the cells in denaturing buffer (2% SDS with 30 mM DTT) for 10 min. To immunoprecipitate the Flag-tagged proteins under denaturing conditions, WCE were prepared as noted above in denaturing buffer, and the SDS concentration was adjusted to 0.1% by adding dilution buffer (150 mM NaCl, 1% Triton X-100, 20 mM Tris pH 8.0.), then immunoprecipitated overnight using anti-Flag (M2-Sigma) conjugated to agarose beads, and washed five times with dilution buffer. Bound proteins were eluted with 100 µg/mL 3× Flag peptides in TBS for 30 min at 8°C twice and the eluted proteins were precipitated using tricholoroacetic acid (TCA). In 293T cells, WCE for denaturing IP was prepared by boiling the cells in lysis buffer (1% SDS, 20 mM Tris pH 8.0, 10 mM DTT), and the SDS concentration was adjusted to 0.1% by adding dilution buffer before adding the Flag antibody for IP. Nuclear extracts were prepared using lysis buffer (10 mM HEPES pH 8.0, 1.5 mM MgCl2, 10 mM KCl, 1% NP40) to lyse cell membrane; the supernatant is the cytosolic fraction and the pellet is the nuclear fraction. For co-IP, the nuclear pellet was re-suspended in lysis buffer (0.1% NP40, 150 mM NaCl, 20 mM Tris pH 8.0 and 10% glycerol), and sonicated to disrupt the nuclei and chromatin. The antibody and nuclear extract were incubated overnight with 5µg of CK2α antibody or 20 µl of M2 Flag antibody conjugated to magnetic beads. The beads were washed three times with the same lysis buffer, and the proteins bound to the beads were analyzed by MS or by immunoblotting.
Mass Spectrometry
Protein samples were prepared as described28. Briefly, the protein samples were diluted in TNE (50 mM Tris pH 8.0, 100 mM NaCl, 1 mM EDTA) buffer. RapiGest SF reagent (Waters Corp.) was added to the mix to a final concentration of 0.1% and samples were boiled for 5 min. TCEP (Tris (2-carboxyethyl) phosphine) was added to 1 mM (final concentration) and the samples were incubated at 37°C for 30 min. Subsequently, the samples were carboxymethylated with 0.5 mg/ml of iodoacetamide for 30 min at 37°C followed by neutralization with 2 mM TCEP (final concentration). Proteins samples prepared as above were digested with trypsin (trypsin:protein ratio - 1:50) overnight at 37°C. RapiGest was degraded and removed by treating the samples with 250 mM HCl at 37°C for 1 h followed by centrifugation at 14000 rpm for 30 min at 4°C. The soluble fraction was then added to a new tube and the peptides were extracted and desalted using C18 desalting columns (Thermo Scientific).
Trypsin-digested peptides were analyzed by ultra high pressure liquid chromatography (UPLC) coupled with tandem mass spectroscopy (LC-MS/MS) using nano-spray ionization as described29. The nano-spray ionization experiments were performed using a TripleTof 5600 hybrid mass spectrometer (ABSCIEX) interfaced with nano-scale reversed-phase UPLC (Waters corporation nano ACQUITY) using a 20 cm-75 micron ID glass capillary packed with 2.5-µm C18 (130) CSH™ beads (Waters corporation). Peptides were eluted from the C18 column into the mass spectrometer using a linear gradient (5–80%) of ACN (acetonitrile) at a flow rate of 250 µl/min for 1hr. The buffers used to create the ACN gradient were: Buffer A (98% H2O, 2% ACN, 0.1% formic acid, and 0.005% TFA) and Buffer B (100% ACN, 0.1% formic acid, and 0.005% TFA). MS/MS data were acquired in a data-dependent manner in which the MS1 data was acquired for 250 ms at m/z of 400 to 1250 Da and the MS/MS data was acquired from m/z of 50 to 2,000 Da. The Independent data acquisition (IDA) parameters were as follow; MS1-TOF acquisition time of 250 ms, followed by 50 MS2 events of 48 ms acquisition time for each event. The threshold to trigger MS2 event was set to 150 counts when the ion had the charge state +2, +3 and +4. The ion exclusion time was set to 4 seconds. Finally, the collected data were analyzed using Protein Pilot 4.5 (ABSCIEX) for peptide identifications.
Yeast strains and yeast plasmids used in the study
Yeast strains and yeast plasmids used in the study are described in Supplemental Table 4.
Extended Data
Extended Data Figure 1. The conserved Y57 residue in H2A is phosphorylated.
(a) The Y57 residue is conserved in all variants of H2A. Sequence of H2A variants in mammals surrounding the Y57 residue (arrow) is shown. (b) The Y57F mutation in H2A does not affect the structural integrity of nucleosomes. Mononucleosomes containing Flag-tagged WT or H2AX-Y57F were immunoprecipitated, and histones and DNA were visualized by Ponceau staining (Right) and by Ultra Violet light (Left) respectively. (c) The H2A-Y58 residue has overlapping functions with the H2AZ-Y65 residue in yeast. Five-fold serial dilutions of the indicated transformants were plated on SC-His-Ura-Trp for growth and 5-FOA for the loss of pJH33. (d, e) H2A-Y57 is phosphorylated in 293T cells. (d) Nuclear extracts from 293T cells were immunoblotted with anti-p-Y57 H2A pre-incubated with indicated peptides, and re-probed with anti-H2A. (e) Histone extracts from 293T cells were treated with calf intestinal phosphatase (CIP) for 1 hr at 37°C, and immunoblotted. (f) The anti-phospho-Y57-H2A antibody specifically recognizes H2A peptide phosphorylated at Y57 but not the non-phosphorylated peptide. Indicated peptides were spotted on nitrocellulose, and probed with anti-phospho-Y57 H2A. Data represent three independent experiments.
Extended Data Figure 2. CK2α phosphorylates Y57 in H2A.
(a) An in vitro kinase reaction was performed using recombinant GST-CK2α, 10 µCi of γ32P labeled ATP supplemented with 10 µM cold ATP, and nucleosomes containing Flag-tagged WT or H2AX-Y57F from 293T cells followed by phospho amino acid analysis (PAA) of the Flag-tagged H2AX. The red circle indicates phospho-Ser, the blue circle indicates phospho-Thr and the green circle indicates phospho-Tyr. Data represent two independent experiments.
Extended Data Figure 3. H2A Y57 phosphorylation regulates transcriptional elongation.
(a) H2A-Y58F mutation does not affect several other histone marks. Whole cell extracts from WT or H2A-Y58F yeast cells were immunoblotted. (b, c) H2A-Y57F mutation affects H2A ubiquitylation in 293T cells. (b) Flag-tagged WT H2A/H2AX and Y57F mutants were expressed in 293T cells, and mono-ubiquitylation was assessed by immunoblotting with Flag antibody. (c) The Flag-tagged H2A mutants were expressed in 293T cells, immunoprecipitated under denaturing conditions, and immunoblotted. (d) H2A-Y58F mutant cells are defective in transcriptional elongation. Five-fold serial dilutions of WT and H2A-Y58F cells were plated on SC supplemented with NH4OH (solvent) or 100µg/mL 6-azauracil (6-AU). (e) Pol II protein level is comparable in WT and H2A-Y58F yeast. Whole cell extracts from WT or H2A-Y58F yeast were immunoblotted. (f, g) H2A-Y58F mutation affects transcription. (f) WT H2A, H2A-Y58F, and H2A-Y58F H2AZ-Y65F yeast, and (g) WT, and htz1Δ strains transformed with vector, HTZ1 or htz1-Y65F were grown at 30°C or shifted to 37°C for 10min. RNA was extracted, and transcript levels of the indicated genes were measured by RT-qPCR and normalized to SCR1, a Pol III transcript (n=3, mean ± SEM, * indicates p<0.05, **indicates p<0.01). P-values were calculated by Student’s two-tailed t-test. (h) Y57 in H2A is phosphorylated during transcriptional elongation. 293T cells were treated with vehicle (DMSO) or flavopiridol (FP) (1 µM) for 4 ½ hr, or treated with FP for 4 ½ hr, then FP was washed out (Release) and cells were harvested at the indicated min (‘), and the nuclear extracts were immunoblotted. Data represent two (a, d, h) or three independent experiments (b, c, e-g).
Extended Data Figure 4. H2A-Y58F mutation enhances H2B deubiquitylation.
(a–c) The H2A-Y58 mutation has moderate to no effect on the recruitment of the H2B ubiquitylation machinery. Binding of (a) Rtf1-HA, (b) Paf1-myc, and (c) Rad6-HA was measured by ChIP-qPCR in the indicated genes in WT and H2A-Y58F yeasts. Whole cell extracts from the yeast strains were immunoblotted to compare the protein levels. ORF of the genes, and the region amplified by the primer pairs are shown (n=2, mean ± SEM). (d) UBP8 deletion does not rescue Pol II binding in the H2A-Y58F mutant. Pol II binding in the indicated strains was measured by ChIP-qPCR. (n=3, mean ± SEM, * indicates p<0.05, ** indicates p<0.01). P-values were calculated by Student’s two-tailed t-test. The ORF of the genes and the regions amplified by the primer pairs are shown. (e) UBP8 deletion does not rescue the defect in transcriptional output in the H2A-Y58F yeast. The mRNA levels of the indicated genes were determined by RT-qPCR and normalized to SCR1 transcript. (n=2, mean ± SEM). (f) UBP8 deletion does not rescue the growth defect in the H2A-Y58F yeast. UBP8 and ubp8Δ strains transformed with WT H2A or H2A-Y58F were plated at 2.5-fold serial dilutions on SC-His-Ura for growth and 5-FOA for the removal of pJH33. Data represent two (a-c, e, f) or three independent experiments (d).
Extended Data Figure 5. CK2 regulates transcriptional elongation.
(a) CK2 kinase activity is necessary for normal gene expression. LNCaP cells were treated with vehicle (DMSO) or TBBz (25 µM) for 60 minutes, and then treated with vehicle (EtOH) or 100 nM DHT for 90 min, and induction of the indicated AR target genes was measured by RT-qPCR (n=3, mean ± SEM, * indicates p<0.05, ** indicates p<0.01). P-values were calculated by Student’s two-tailed t-test. (b) Nuclear extracts from 293T cells were immunoprecipitated using CK2α antibody and immunoblotted. (c) Enrichment of CK2α, H3K4me118, H3K27acetyl18 and AR18 at a representative AR enhancer (KLK3) is shown. (d) Pol II tag density in cells treated with vehicle or TBBz at a representative RHOU enhancer is shown. Data represent two (b–d) or three independent experiments (a).
Supplementary Material
Acknowledgements
We acknowledge M. Ghassemian (UCSD Mass Spectrometry Facility) for assistance in mass spectrometry analysis. We also acknowledge K. Arndt (University of Pittsburgh) for kindly providing the UBP8 null yeast strain. CK2 constructs were kindly provided by D. Litchfield (University of Western Ontario). We acknowledge J. Hightower for assistance in figure preparation. We acknowledge K. Tumaneng and I. Bassets for critical reading of the manuscript and DJ. Forbes for discussion. This work was supported by grants from NIH to MGR, UC-CRCC to LP, NIH-GM033279 support for XBS and NCI CA82683 grant to TH. MGR is an Investigator with HHMI. TH is a Frank and Else Schilling American Cancer Society Professor and holds the Renato Dulbecco Chair in Cancer Research.
Footnotes
Author Contributions
MGR and HB conceived the idea, and wrote the manuscript with contributions from LP, XBS and TH. MGR and HB designed the experiments with mammalian cells, and HB performed the experiments. LP, XBS and HB designed the yeast experiments, and XBS and HB performed the experiments. YT did the bioinformatics analysis. DM aligned the ChIP-seq data. KAO prepared the ChIP-seq library, and conducted the high-throughput sequencing. TH designed the PAA experiments, and JM performed the experiments. All authors read the manuscript, and approve the content.
The authors declare no competing financial interests.
References
- 1.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 2.Nakanishi S, et al. A comprehensive library of histone mutants identifies nucleosomal residues required for H3K4 methylation. Nat. Struct. Mol. Biol. 2008;15:881–888. doi: 10.1038/nsmb.1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dai J, et al. Probing nucleosome function: a highly versatile library of synthetic histone H3 and H4 mutants. Cell. 2008;134:1066–1078. doi: 10.1016/j.cell.2008.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Flint AJ, Tiganis T, Barford D, Tonks NK. Development of ‘substrate-trapping’ mutants to identify physiological substrates of protein tyrosine phosphatases. Proc. Natl. Acad. Sci. U. S. A. 1997;94:1680–1685. doi: 10.1073/pnas.94.5.1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wilson LK, Dhillon N, Thorner J, Martin GS. Casein kinase II catalyzes tyrosine phosphorylation of the yeast nucleolar immunophilin Fpr3. J. Biol. Chem. 1997;272:12961–12967. doi: 10.1074/jbc.272.20.12961. [DOI] [PubMed] [Google Scholar]
- 6.Vilk G, et al. Protein kinase CK2 catalyzes tyrosine phosphorylation in mammalian cells. Cell. Signal. 2008;20:1942–1951. doi: 10.1016/j.cellsig.2008.07.002. [DOI] [PubMed] [Google Scholar]
- 7.Pagano MA, et al. 2-Dimethylamino-4,5,6,7-tetrabromo-1H-benzimidazole: a novel powerful and selective inhibitor of protein kinase CK2. Biochem. Biophys. Res. Commun. 2004;321:1040–1044. doi: 10.1016/j.bbrc.2004.07.067. [DOI] [PubMed] [Google Scholar]
- 8.Xiao T, et al. Histone H2B ubiquitylation is associated with elongating RNA polymerase II. Mol. Cell. Biol. 2005;25:637–651. doi: 10.1128/MCB.25.2.637-651.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tanny JC, Erdjument-Bromage H, Tempst P, Allis CD. Ubiquitylation of histone H2B controls RNA polymerase II transcription elongation independently of histone H3 methylation. Genes Dev. 2007;21:835–847. doi: 10.1101/gad.1516207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Uptain SM, Kane CM, Chamberlin MJ. Basic mechanisms of transcript elongation and its regulation. Annu. Rev. Biochem. 1997;66:117–172. doi: 10.1146/annurev.biochem.66.1.117. [DOI] [PubMed] [Google Scholar]
- 11.Ferguson SB, et al. Protein kinase A regulates constitutive expression of small heat-shock genes in an Msn2/4p-independent and Hsf1p-dependent manner in Saccharomyces cerevisiae. Genetics. 2005;169:1203–1214. doi: 10.1534/genetics.104.034256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Robzyk K, Recht J, Osley MA. Rad6-dependent ubiquitination of histone H2B in yeast. Science. 2000;287:501–504. doi: 10.1126/science.287.5452.501. [DOI] [PubMed] [Google Scholar]
- 13.Wood A, Schneider J, Dover J, Johnston M, Shilatifard A. The Paf1 complex is essential for histone monoubiquitination by the Rad6-Bre1 complex, which signals for histone methylation by COMPASS and Dot1p. J. Biol. Chem. 2003;278:34739–34742. doi: 10.1074/jbc.C300269200. [DOI] [PubMed] [Google Scholar]
- 14.Ng HH, Dole S, Struhl K. The Rtf1 component of the Paf1 transcriptional elongation complex is required for ubiquitination of histone H2B. J. Biol. Chem. 2003;278:33625–33628. doi: 10.1074/jbc.C300270200. [DOI] [PubMed] [Google Scholar]
- 15.Rahl PB, et al. c-Myc regulates transcriptional pause release. Cell. 2010;141:432–445. doi: 10.1016/j.cell.2010.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reppas NB, Wade JT, Church GM, Struhl K. The transition between transcriptional initiation and elongation in E. coli is highly variable and often rate limiting. Mol. Cell. 2006;24:747–757. doi: 10.1016/j.molcel.2006.10.030. [DOI] [PubMed] [Google Scholar]
- 17.Wang D, et al. Reprogramming transcription by distinct classes of enhancers functionally defined by eRNA. Nature. 2011;474:390–394. doi: 10.1038/nature10006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heintzman ND, et al. Histone modifications at human enhancers reflect global cell type-specific gene expression. Nature. 2009;459:108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meggio F, Pinna LA. One-thousand-and-one substrates of protein kinase CK2? FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2003;17:349–368. doi: 10.1096/fj.02-0473rev. [DOI] [PubMed] [Google Scholar]
- 20.Wade JT, Struhl K. The transition from transcriptional initiation to elongation. Curr. Opin. Genet. Dev. 2008;18:130–136. doi: 10.1016/j.gde.2007.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Suka N, Suka Y, Carmen AA, Wu J, Grunstein M. Highly specific antibodies determine histone acetylation site usage in yeast heterochromatin and euchromatin. Mol. Cell. 2001;8:473–479. doi: 10.1016/s1097-2765(01)00301-x. [DOI] [PubMed] [Google Scholar]
- 22.Henry KW, et al. Transcriptional activation via sequential histone H2B ubiquitylation and deubiquitylation, mediated by SAGA-associated Ubp8. Genes Dev. 2003;17:2648–2663. doi: 10.1101/gad.1144003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shin H, Liu T, Manrai AK, Liu XS. CEAS: cis-regulatory element annotation system. Bioinforma. Oxf. Engl. 2009;25:2605–2606. doi: 10.1093/bioinformatics/btp479. [DOI] [PubMed] [Google Scholar]
- 24.O’Neill LP, Turner BM. Immunoprecipitation of native chromatin: NChIP. Methods San Diego Calif. 2003;31:76–82. doi: 10.1016/s1046-2023(03)00090-2. [DOI] [PubMed] [Google Scholar]
- 25.Strahl-Bolsinger S, Hecht A, Luo K, Grunstein M. SIR2 and SIR4 interactions differ in core and extended telomeric heterochromatin in yeast. Genes Dev. 1997;11:83–93. doi: 10.1101/gad.11.1.83. [DOI] [PubMed] [Google Scholar]
- 26.Kamps MP, Sefton BM. Acid and base hydrolysis of phosphoproteins bound to immobilon facilitates analysis of phosphoamino acids in gel-fractionated proteins. Anal. Biochem. 1989;176:22–27. doi: 10.1016/0003-2697(89)90266-2. [DOI] [PubMed] [Google Scholar]
- 27.Chao S-H, Price DH. Flavopiridol inactivates P-TEFb and blocks most RNA Polymerase II transcription in vivo. J. Biol. Chem. 2001;276:31793–31799. doi: 10.1074/jbc.M102306200. [DOI] [PubMed] [Google Scholar]
- 28.Guttman M, et al. Interactions of the NPXY microdomains of the low density lipoprotein receptor-related protein 1. Proteomics. 2009;9:5016–5028. doi: 10.1002/pmic.200900457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McCormack AL, et al. Direct analysis and identification of proteins in mixtures by LC/MS/MS and database searching at the low-femtomole level. Anal. Chem. 1997;69:767–776. doi: 10.1021/ac960799q. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.