Abstract
Essential viral proteins perform vital functions during morphogenesis via a complex interaction with other viral and cellular gene products. Here, we present a novel approach to comprehensive mutagenesis of essential cytomegalovirus genes and biological analysis in the 230-kbp-genome context. A random Tn7-based mutagenesis procedure at the single-gene level was combined with site-specific recombination via the FLP/FLP recognition target site system for viral genome reconstitution. We show the function of more than 100 mutants from a larger library of M50/p35, a protein involved in capsid egress from the nucleus. This protein recruits other viral proteins and cellular enzymes to the inner nuclear membrane. Our approach enabled us to rapidly discriminate between essential and nonessential regions within the coding sequence. Based on the prediction of the screen, we were able to map a site essential for viral protein-protein interaction at the amino acid level.
Cytomegaloviruses define the beta subgroup of the Herpesviridae, which are important animal and human pathogens. Human cytomegalovirus (HCMV) is detected with a high prevalence in the human population, and it causes severe and fatal disease in immunocompromised individuals. Murine cytomegalovirus (MCMV) infection serves as an animal model system. CMVs harbor large double-stranded DNA genomes of about 230 kbp (7, 32). The recent cloning of the HCMV and MCMV genomes as infectious bacterial artificial chromosomes (BACs) (4, 15, 24, 27, 45, 50) rendered these genomes accessible to advanced genetic manipulation (for a review, see reference 46).
Infectious herpesvirus particles are formed during lytic virus replication as a result of a complex maturation and egress process (for a review, see reference 28). This multistep process starts in the nucleus with the packaging of viral genomes into capsids. The final envelopment takes place in the trans-Golgi network. Each maturation step is controlled by particular multiprotein assemblies, which are formed by complex interactions between viral and cellular proteins. Efforts to reconstitute the first morphogenesis stage, the capsid assembly, have been successful for alphaherpesviruses (43). In the mature virion, some higher-order protein complexes were studied by crystallography and electron microscopy (for a review, see reference 9). Yet, the logistics of the dynamic transitory stages (e.g., capsid egress from the nucleus) are not accessible for high-resolution analysis using static physical methods. Artificial reconstitution of these intermediate stages would require regulated coexpression of a number of viral proteins in the context of an appropriate host cell environment. Thus, detailed structure-function analyses of protein machines, which are associated with nuclear egress of virus capsids, need the context of virus replication. Gene products involved in herpesvirus morphogenesis are often essential for viral growth. The genetic analysis of these genes is restricted to either the isolated expression of the protein in cell culture or transient transcomplementation. Recently, for retroviruses harboring genomes of about 10 kbp, a comprehensive mutational analysis of an essential viral gene or a selected region in the viral genome context was introduced (21, 37). A similar comprehensive analysis of essential herpesvirus genes in the genome context is not feasible, due to the ineffective generation of viral mutants and the lack of versatile complementing systems.
Here, we present an approach to increase the analytical resolution and efficiency of the mutagenesis of herpesvirus genes in the viral background. To this end, the MCMV M50 gene product was chosen for detailed structure-function analysis. The M50 gene codes for a 35-kDa type II transmembrane (TM) protein (M50/p35) that is associated with the nuclear egress machinery of MCMV (30). M50/p35 is located at the inner nuclear membrane. It recruits cellular protein kinase C to the inner nuclear membrane and interacts with another viral protein encoded by the M53 gene (M53/p38). The analysis of an M50/p35 deletion mutant proved that M50/p35 is essential for viral replication. By a combination of random Tn7-based mutagenesis (2) and site-specific reinsertion into the genome driven by the FLP/FLP recognition target site (FRT) system (25), a library of random M50 viral mutants was generated and tested for functionality during viral replication. The analysis of over 100 mutants in the viral context revealed that the N-terminal part of the protein and the TM domain harbor several essential functions, whereas the C-terminal part is not essential for viral replication. These data were confirmed by site-specific mutations that allowed the mapping of a domain that was essential for protein-protein interaction down to the single-amino-acid level.
MATERIALS AND METHODS
Cells and viruses.
NIH 3T3 fibroblasts (ATCC CRL 1658), M2-10B4 bone marrow stroma cells (ATCC CRL 1972), human 293 cells (ATCC CRL 1573), and mouse embryonic fibroblasts were propagated as described previously (23, 26). Wild-type (wt) MCMV and mutants were propagated on NIH 3T3 or M2-10B4 cells (26, 42). wt or mutant genomes were reconstituted to virus by transfection of BAC DNA into M2-10B4 cells with an MBS transfection kit (Stratagene, Cedar Creek, Tex.), following the instructions of the manufacturer. All virus stocks were prepared and titrated on NIH 3T3 cells as described previously (33).
Plasmids.
pOriR6K-zeo was constructed by combining the 0.4-kbp BamHI fragment of pGP704 containing the R6K origin of replication (29) and the phleomycin D1 (Zeocin) resistance gene of pZero1 (Invitrogen, Leek, The Netherlands), which was amplified by PCR with primers 3′-BglII-Zeo and 5′-BamHI-FRT-Zeo (primer sequences are given in Table 1). The M50 transcription unit (nucleotide positions 75276 to 76942 of MCMV strain Smith, according to Rawlinson et al. [32]) was amplified from pHindIII H (13) with primers 5′M50EKpnI and 3′M50EKpnI, thereby adding KpnI restriction sites to both ends of the 1.8-kbp fragment, and cloned into pOriR6K-zeo, resulting in plasmid pOriR6K-zeo-M50.
TABLE 1.
Primers used in this study
| Primer name | 5′ to 3′ sequencea |
|---|---|
| 3′-BglII-Zeocin | CCA AGA TCT TTA AAA ATG AAG TTT TAG CAC GTG TCA G |
| 5′-BamHI-FRT-Zeocin | ACT GGA TCC GAA GTT CCT ATT CTC TAG AAA GTA TAG GAA CTT CCG TTT ACA ATT TCG CCT GAT GCG G |
| 5′M50EKpnI | GTA GGT ACC GAA GAC GTG TCG CGT CAT GAC CT |
| 3′M50EKpnI | GTA GGT ACC GGA ACG AGT CGA TCG TCG TCA G |
| 5′M50KpnI | CGC GGT ACC ATG GAG ATC GAC A |
| 3′M50XhoI | CGC CTC GAG TCA CGG ATG ACC |
| Del5′-for | CGA AAG AAC TAT TTC CTT TTT GAC C |
| Del4-rev | GCC GGA AAT AGT TCC GCA GTC GGT GGG AAC G |
| Del3-rev | GCC GGA AAT AGT TCC CCA GCG CTG ACC CGG GGG AGG |
| Del2-rev | GCC GGA AAT AGT TCC CCC ACC TCC TGC GTA TCT TCG |
| Del1-rev | GCC GGA AAT AGT TCC GGC GGG GGG AGG CCG CTG GG |
| 5′M50YA | GTC GCT CCG GTC GAG GCT CTC CTC AGC TAT TGG |
| 3′M50YA | GAG AGC CTC GAC CGG AGC GAC GAT ATC GGT GGT |
| 5′M50Y1A | GTC GCT CCG GTC GAG TAT CTC CTC AGC TAT TGG |
| 3′M50Y1A | GAG ATA CTC GAC CGG AGC GAC GAT ATC GGT GGT |
| 5′M50Y2A | GTC TAT CCG GTC GAG GCT CTC CTC AGC TAT TGG |
| 3′M50Y2A | GAG AGC CTC GAC CGG ATA GAC GAT ATC GGT GGT |
| 5′M50EA | GTC TAT CCG GTC GCT TAT CTC CTC AGC TAT TGG |
| 3′M50EA | GAG ATA AGC GAC CGG ATA GAC GAT ATC GGT GGT |
| 5′-ΔM50 | GGC GAG GAT CAG GGC GCA GTG AAT AGC GTT GGC GTG CCT GGT CAA AAA GGA AAT AGT TCT CGA TTT ATT CAA CAA AGC CAC G |
| 3′-ΔM50 | GTC GGC TCG GGC GGC GCC ACT CGG ACG GCG GCG AGC TCA TCC GCG GCG GCG CCG GCG CGA GCC AGT GTT ACA ACC AAT TAA CC |
The sequences homologous to M50 for recombination are underlined.
To express genes from pOriR6K-zeo, the HCMV immediate-early promoter, a multiple cloning site and a poly(A) sequence derived from pCR3 (Invitrogen) were introduced into pOriR6K-zeo. The 1.3-kbp AflIII-DraIII fragment of pCR3 was cloned into the EcoRV and BamHI sites of pOriR6K-zeo, generating pOriR6K-zeo-ie.
For expression of M50/p35 and mutants of M50/p35 (M50mut) in eukaryotic cells, the M50 and the M50mut open reading frames (ORFs) were recloned from pOriR6K-zeo-M50 constructs into pCR3 and pOriR6K-zeo-ie, respectively. To this end, M50 and M50mut ORFs were amplified by PCR with primers 5′M50KpnI and 3′M50XhoI and cloned into pCR3 or pOriR6K-zeo-ie, generating plasmids pCR3-M50mut and pOriR6K-zeo-ie-M50mut.
To generate the M50-del1, -del2, -del3, and -del4 deletion constructs, pOriR6K-M50 was amplified by inverse PCR with various primers, with flanking XmnI sites that annealed at different positions within the M50 ORF. By recircularization of the PCR products, the following deletion constructs were created: M50-del4 with primers del5′-for and del4-rev, M50-del3 with primers del5′-for and del3-rev, M50-del2 with primers del5′-for and del2-rev, and M50-del1 with primers del5′-for and del1-rev. The M50 deletion constructs were recloned into the expression vector pOriR6K-zeo-ie, with primers 5′M50KpnI and 3′M50XhoI, generating pM50-del1, -del2, -del3, and -del4.
Single amino acid exchanges in the M50 ORF product were introduced with mutated overlapping primers, as follows. For M50Y53,57A, two M50 fragments were generated by PCR with primers 5′M50KpnI and 3′M50YA as well as 5′M50YA and 3′M50XhoI; for M50Y53A, primers 5′M50KpnI and 3′M50Y1A as well as 5′M50Y1A and 3′M50XhoI were used; for M50Y57A, primers 5′M50KpnI and 3′M50Y2A as well as 5′M50Y2A and 3′M50XhoI were used; and for M50E56A, primers 5′M50KpnI and 3′M50EA as well as 5′M50EA and 3′M50XhoI were used. To unify the two fragments, a PCR was run with the respective fragments as templates and with primers 5′M50KpnI and 3′M50XhoI, resulting in mutated M50 ORFs. These ORFs were subsequently cloned into pOriR6K-zeo-ie, resulting in pM50Y53,57A, pM50Y53A, pM50Y57A, and pM50E56A, respectively.
Mutagenesis and PCR screen for insertions in the M50 ORF.
The plasmid pOri6K-zeo-M50 was mutated by using the Tn7-based GPS linker scanning kit (NEB, Beverly, Mass.) following the manufacturer's protocol. The transposon was removed by PmeI digestion, and the plasmids were religated. Three micrograms of DNA from the library was introduced into Escherichia coli strain PIR1 (Invitrogen), and single colonies were picked for the PCR screen. Colonies were resuspended in 70 μl of H2O, incubated at 95°C for 15 min, and centrifuged for 5 min at 13,000 rpm (Eppendorf centrifuge, 5415 C). Two microliters of the supernatants served as a template for PCR. The M50 ORF was amplified with primers 5′M50KpnI and 3′M50XhoI. Clones with an insertion identified by PmeI digestion of the PCR products were sequenced.
Generation of recombinant viral BACs.
pSMfr3-ΔM50 and pΔM50-GFP, respectively, were constructed on the basis of the wt MCMV BAC pSM3fr (45) and pΔm152/GFP, which contains a green fluorescent protein (GFP) expression cassette at the position of gene m152 (26). A linear recombination fragment was generated by PCR with plasmid pACYC177 (NEB) as a template. To amplify the kanamycin cassette flanked by short sequences with homology to both ends of the M50 ORF, primers 5′-ΔM50 and 3′-ΔM50 were used. Most of the M50 ORF (nucleotide positions 75504 to 76449, according to Rawlinson et al. [32]) was deleted by insertion of the linear fragment into pΔm152-GFP and pSM3fr, with homologous recombination in E. coli as described previously (44).
The temperature-sensitive FLP expression plasmid pCP20 (8) was transformed into E. coli strain DH10B (Life Technologies, Karlsruhe, Germany) containing pΔM50-GFP to insert pOriR6K-zeo-M50 or pOriR6K-zeo-M50mut into pΔM50-GFP. In a second step, the rescue plasmids were introduced and the bacteria were incubated overnight at 30°C under selection conditions with 25 μg of chloramphenicol/ml and 33 μg of Zeocin/ml. Site-directed recombination yielded BACs pM50E and pM50Emut. Single clones were picked and incubated overnight at the nonpermissive temperature of 43°C for removal of pCP20. BAC DNA was isolated, and its presence was confirmed by restriction pattern analysis.
Metabolic labeling and coprecipitation.
NIH 3T3 cells and 293 cells were cotransfected with 5 μg of the construct pCR3-immunoglobulin M53 (pCR3-IgM53) (30) and pOriR6K-zeo-ie-M53 and 5 μg of plasmid pCR3-M50mut, pOriR6K-zeo-ie-M50, or pOriR6K-zeo-ie-M50mut by Ca2PO4 precipitation (38). Twenty-four hours posttransfection, a coprecipitation assay was performed as described previously (11, 30). Protein A-Sepharose (Amersham Biosciences, Freiburg, Germany) was used to pull down the Ig-tagged complexes. To precipitate M50/p35, a specific polyclonal rabbit antiserum was used (30). M53/p38 was precipitated on protein G-Sepharose (Amersham Biosciences) with a specific rat polyclonal antiserum raised against a synthetic peptide representing the 15 most N-terminal amino acids of the M53 ORF product (MFRSPEGEERDAADR).
Western blot analysis.
Samples were suspended in Laemmli sample buffer containing 5% β-mercaptoethanol and separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Proteins were transferred onto Hybond-P membranes (Amersham Biosciences) in the presence of blotting buffer (25 mM Tris, 192 mM glycine, 20% [vol/vol] methanol [pH 8.3]). Membranes were blocked in TBS-T (Tris-buffered saline, 0.05% Tween 20) containing 5% nonfat dry milk for 30 min at room temperature and then incubated with TBS-T containing the primary antibody. To detect M50/p35, a specific polyclonal rabbit antiserum was used (30). Membranes were washed with TBS-T and incubated with the appropriate horseradish peroxidase-conjugated secondary antibody (Dianova, Hamburg, Germany). The proteins were visualized with an ECL-Plus Western blot detection system (Amersham).
Confocal laser scanning microscopy.
Transfected NIH 3T3 cells were grown on glass coverslips and fixed as previously described (30). M50/p35 was visualized with a polyclonal rabbit antiserum as the primary antibody (30) and fluorescein- or Texas red-conjugated donkey anti-rabbit IgG (Dianova) as the secondary antibody. The Ig-tagged M53 was visualized directly by use of fluorescein- or Texas red-conjugated donkey anti-human IgG.
RESULTS
Analysis of a mutant library of a herpesvirus gene in the genome context.
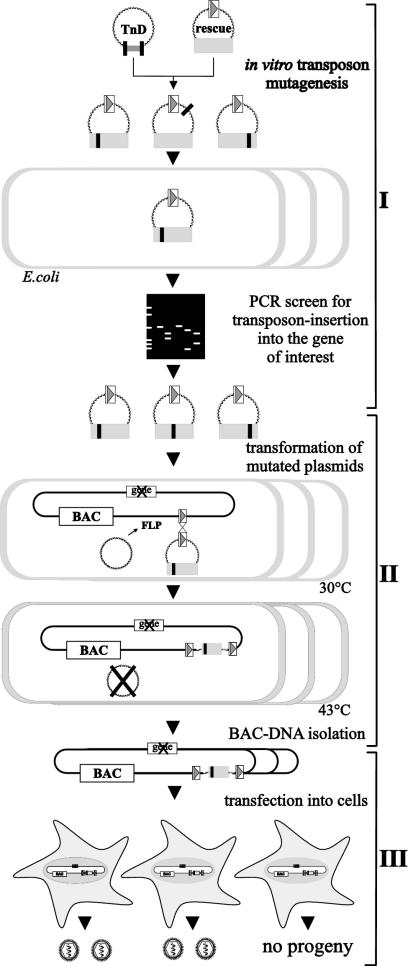
The objective of this study was to generate a large mutant pool of a subcloned viral gene and to establish procedures for the simple and efficient reinsertion of the mutants into the virus genome. For general applicability, the procedure should operate independently of the individual properties of a given gene and without a complementing cell line. This requires three steps: (i) the generation of a comprehensive mutant library of the subcloned sequence, (ii) the simple and efficient introduction of the gene mutants into the viral genome, and (iii) the analysis of gene function during virus replication. These requirements are fulfilled by the following strategy (Fig. 1). First, a gene of interest is subcloned into a rescue plasmid containing one FRT site and subsequently subjected to in vitro Tn7-based insertion mutagenesis (2), thereby creating a library of rescue plasmids containing insertions of 15 bp (Fig. 1, scheme I). The mutated plasmids are then screened by PCR for insertion events in the coding sequence of interest. Next, the plasmids are transformed into E. coli that contains a plasmid expressing the FLP recombinase and a viral BAC that has an FRT site but lacks the gene of interest (Fig. 1, scheme II). The FLP recombinase directs the site-specific recombination event between two FRT sites (25), located here on the BAC and the rescue plasmid. This recombination leads to the introduction of the mutated gene into the viral genome at an ectopic position. To test for functionality, the viral genomes are isolated and transfected into eukaryotic cells for virus reconstitution (Fig. 1, scheme III). If an essential function of the gene is affected by the 15-bp insertion, no viral progeny should be generated, thereby mapping important domains. M50/p35 was analyzed according to this strategy.
FIG. 1.
Strategy for random mutagenesis of an essential viral gene in the viral genome context. (I) The viral gene of interest (gray box) is subcloned into a rescue plasmid (rescue) containing one FRT site (open box with gray triangle). This plasmid is subjected to an in vitro Tn7-based random mutagenesis procedure (TnD), leading to a mutant library with 15-bp insertions (black box) into the target plasmid. This mutant library is transformed into E. coli (open boxes), and single clones are screened by PCR for insertions into the gene of interest. (II) To reinsert the gene mutants into the viral genome, the respective MCMV BAC and an FLP recombinase-expressing plasmid (FLP) are maintained in E. coli. Subsequently, the rescue plasmids are transformed into E. coli. FLP recombinase mediates site-specific recombination between the FRT sites and unifies the BAC and the rescue plasmid. Selection with chloramphenicol and Zeocin identifies BACs, with the inserted rescue plasmid carrying the mutated gene of interest. (III) Subsequently, BAC DNA is isolated and transfected into eukaryotic cells for virus reconstitution and cells are screened for viral plaques.
The M50 ORF is essential for viral growth and infectivity and can be reconstituted by ectopic reinsertion of the wt M50 gene into the MCMV genome.
M50/p35 is involved in the egress of the MCMV capsid from the nucleus (30). Inactivation of the homologous genes in certain alphaherpesviruses severely affects viral growth in cell culture (19, 31, 36). To test the essentiality of M50/p35 for virus replication, we deleted 93% of the M50 ORF. To this end, a kanamycin cassette was introduced by homologous recombination in E. coli (44) into either MCMV-BAC pSMfr3 (45) or pΔm152/GFP (26), generating pSMfr3-ΔM50 or pΔM50-GFP, respectively. These constructs were tested for infectivity by transfection into NIH 3T3 cells. We were unable to reconstitute virus from pSM3fr-ΔM50 or pΔM50-GFP; thus, the M50 ORF is essential for viral growth in cell culture (Table 2). To ensure that the null phenotype of the M50 deletion was due to the lack of the p35 protein, the M50 ORF was provided in trans. To this end, the expression plasmid pCR3-M50Flag (30) was introduced together with pΔM50-GFP into mouse embryonic fibroblasts. This introduction resulted in the formation of miniplaques, which did not spread during an observation time of 3 weeks, indicating that the gene complemented virus growth only in cotransfected cells that provided the protein in trans (Table 2).
TABLE 2.
Reconstitution of ΔM50 constructs
| BAC | Genea | Plaque formationb |
|---|---|---|
| pSM3fr | M50 | +++ |
| pΔm152/GFP | M50 | +++ |
| pSMfr-ΔM50 | ΔM50 | − |
| pΔM50-GFP | ΔM50 | − |
| pΔM50-GFP + pCR3-M50Flag | ΔM50 + M50 in trans | (+) |
| pM50E | ΔM50 + M50 ectopic | +++ |
M50 expression and M50/p35 in trans.
Formation of viral plaques 10 days postinfection. +++, wt plaque formation; −, no plaque formation; (+), formation of miniplaques.
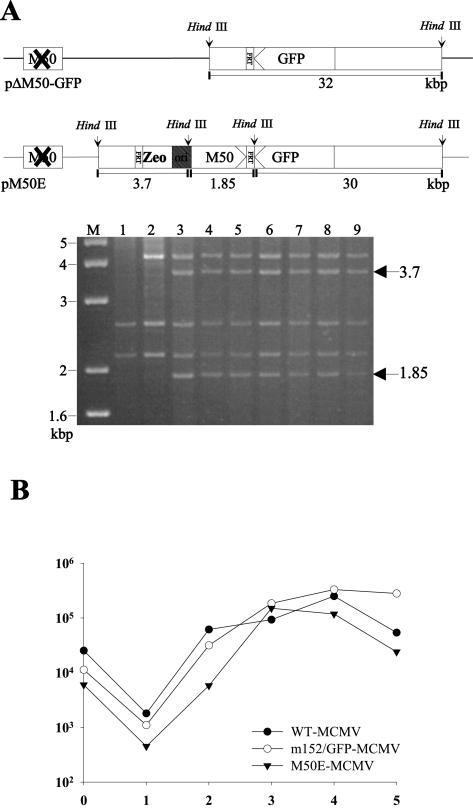
To revert to the M50 null phenotype, the M50 ORF was inserted at an ectopic position into pΔM50-GFP by the rescue plasmid pOriR6K-zeo-M50, generating BAC pM50E. The correct genome structure of pM50E was confirmed by restriction pattern analysis (Fig. 2A). Transfection of cells with pM50E DNA resulted in viral progeny, M50E-MCMV. Virus growth of M50E-MCMV was comparable to that of the parental BAC-derived viruses wt MCMV and Δm152/GFP-MCMV (Fig. 2B). Immunofluorescence studies with an M50-specific antiserum (30) confirmed early/late expression kinetics of the reinserted M50 in M50E-MCMV comparable to those of the wt (data not shown). Thus, it was possible to revert to the M50 null phenotype by the expression of the M50 ORF from an ectopic position.
FIG. 2.
Ectopic rescue of the M50 deletion mutant. (A) Structure of the m152 region within pΔM50-GFP. The HindIII B fragment of BAC pΔM50-GFP contains one FRT site (pΔM50-GFP). In the HindIII H fragment of pΔM50-GFP, the M50 ORF was replaced by a kanamycin cassette (open box with X). pM50E shows this genomic region after the insertion of pOriR6Kzeo-M50 into pΔM50-GFP by site-specific recombination driven by the FLP recombinase. Arrowheads indicate the orientation of the respective ORFs. Arrows indicate HindIII restriction sites, and HindIII fragment sizes are indicated below the diagrams in kilobase pairs. DNAs of the MCMV BAC pΔm152/GFP (lane 1), pΔM50-GFP (lane2), and seven randomly picked clones of pM50E (lanes 3 to 9) were digested with HindIII and analyzed by gel electrophoresis. The sizes of the DNA bands characteristic for pM50E are indicated in kilobase pairs by arrows on the right. Marker bands are indicated on the left. (B) Growth kinetics of the viruses derived from the above-described BACs. NIH 3T3 cells were infected with viruses derived from pSMfr3 (WT-MCMV), pΔm152/GFP (m152/GFP-MCMV), or pM50E (M50E-MCMV) at a multiplicity of infection of 0.1. Supernatants of the infected cells were harvested on the indicated days, and virus titers were determined on NIH 3T3 cells by the 50% tissue culture infective dose method.
Generation of a library of random M50 mutants and test for functionality in the viral context.
For the identification of functionally important domains, the M50/p35 ORF was subjected to a Tn7-mediated random mutagenesis procedure. This procedure resulted in the random insertion of 5 amino acids (aa) or a stop codon into the protein coding sequence (2). From the initial library of mutated pOriR6K-zeo-M50 plasmids, 680 independent clones were screened by PCR. Plasmids with an insertion in the M50 ORF were sequenced to define the exact insertion site and to differentiate between the insertions leading to a stop or a read-through mutation. This screen resulted in 104 mutants, representing, on average, one insertion per 10 to 15 aa (Fig. 3A). Due to the nature of the Tn7 transposition, read-through and truncation mutants were found between the same amino acids (2). Mutants were individually reinserted into pΔM50-GFP. Cells were transfected with DNA of each mutant BAC to test the reconstitution of infectivity. The M50/p35 ORF codes for a type II TM protein, with a predicted TM domain from aa 289 to 305. The insertion of M50/p35 into the inner nuclear membrane is crucial for cellular function, since none of the truncation mutants (indicated in Fig. 3A) with the TM domain deleted gave rise to viral progeny. In contrast, the mutant with the truncation after aa 313 was functional. Localization studies revealed that mutants of M50 deprived of the TM domain are stalled in the cytoplasm and do not reach the nucleus (data not shown).
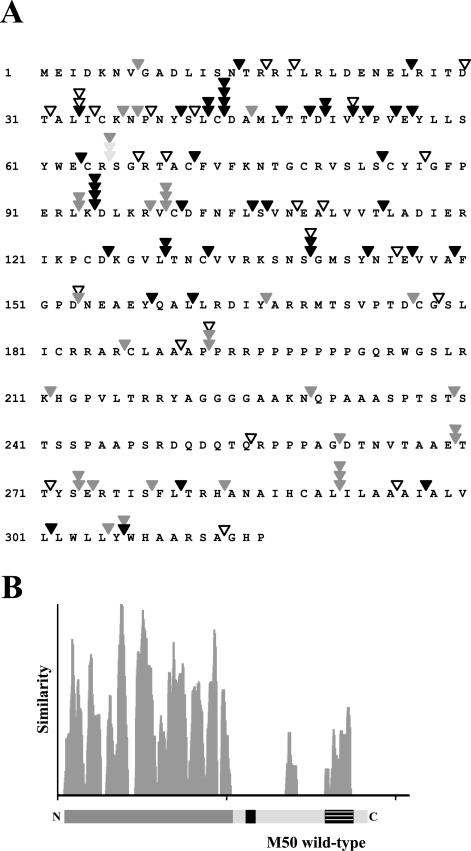
FIG. 3.
Analysis of 104 mutants with transposon insertions in the M50 ORF and screen for the functional relevance of these mutants in the viral background. (A) M50 mutants and their ability to rescue virus growth of the ΔM50 genome. Displayed is the amino acid sequence of M50. Arrowheads indicate transposon insertion sites. Open arrowheads indicate insertions leading to a stop codon; filled arrowheads represent insertions leading to the introduction of 5 aa into the M50 ORF. Dark gray arrowheads indicate mutants that rescued the ΔM50 phenotype. Light gray arrowheads indicate viruses with strongly attenuated growth. Black arrowheads indicate those mutants that were not able to rescue the ΔM50 phenotype. (B) Sequence comparison of M50/p35 homologues. The amino acid sequence of M50/p35 was aligned with those of UL50 (HCMV), R50 (rat cytomegalovirus), UL34 of HSV-1 and -2, and pseudorabies virus as well as ORF 67 of murine herpesvirus 68, herpesvirus saimiri, and human herpesvirus 8 by use of the Vector NTI AlignX program (InfoMax, Bethesda, Md.) via the BLOSUM62 similarity matrix. The depicted similarity plot was calculated using a 5-aa window size and with scores for identity and strong and weak similarities of 1.0, 0.5, and 0.2, respectively. The x axis represents the number of the amino acids of the consensus sequence. Below the diagram, a proportional representation of M50/p35 is displayed. The N-terminal region is marked in dark gray; the C-terminal region is marked in light gray. The striped box indicates the TM domain; the black box indicates the proline-rich region.
By studying in-frame mutants only (Fig. 3A), it became apparent that 40 of 59 mutants with insertions in the N-terminal part (aa 1 to 200) were lethal. In contrast, only 4 out of 20 mutants with insertions in the C-terminal part affected viral growth. The 16 viable C-terminal mutants showed viral plaques comparable to those of the wt. The insertion at aa 65 gave rise to severely attenuated viral progeny (Fig. 3A). Altogether, loss-of-function mutants accumulated in the N-terminal part of the protein, which is the most conserved region along the M50/p35 homologues (Fig. 3B).
Mapping the M53/p38 binding site in M50/p35.
wt M50/p35 recruits M53/p38 to the nuclear envelope. In the absence of M50/p35, M53/p38 is diffusely distributed in the nucleus. wt M50/p35 can be precipitated by using an Ig-tagged version of M53/p38 (30). We assumed that the binding of M50/p35 to M53/p38 might be essential for viral function. Therefore, we expected that some of the lethal M50 mutants would have lost the ability to bind to M53/p38. To exclude unspecific binding of M50/p35 to Ig-tagged M53/p38, we cotransfected 293 cells with M50/p35 and M53/p38 expression plasmids and the cell lysates were precipitated by specific antisera. Signals for both M50/p35 and M53/p38 were detected only in the cotransfected cells by use of an antiserum specific to either M50/p35 or M53/p38 (see Fig. S1 in the supplemental material). To map the interaction domain between M50/p35 and M53/p38, lethal in-frame mutants (Fig. 4A) were recloned into pCR3 for expression in cells. If more than one lethal insertion mutant was identified at the same site, only one of them was analyzed. 293 cells were cotransfected with the noncomplementing M50 constructs and an Ig-tagged M53/p38 (30). The protein complexes were precipitated by protein A-Sepharose and analyzed by SDS-PAGE. As an example, the coprecipitation of eight mutants is shown in Fig. 4B. For positive controls, we used either the wt construct of M50 (lane 2) or the M50 mutant with an insertion at aa 36, which rescued viral growth (lane 3). Three insertion mutants tested in this assay (Fig. 4B, lanes 4, 9, and 11), as well as the controls, coprecipitated with IgM53/p38, showing that the structural requirements to bind M53/p38 were maintained despite the 5-aa insertion. Five mutants lost the ability to bind to IgM53/p38, indicated by the lack of an M50/p35-specific band (Fig. 4B, lanes 5, 6, 7, 8, and 10). Four of those mutants clustered in the region from aa 53 to aa 56. In summary, 5 out of the 25 tested lethal M50 mutants lost the ability to interact with M53/p38, whereas 20 retained this property but the mutation rendered the protein nonfunctional for other reasons.
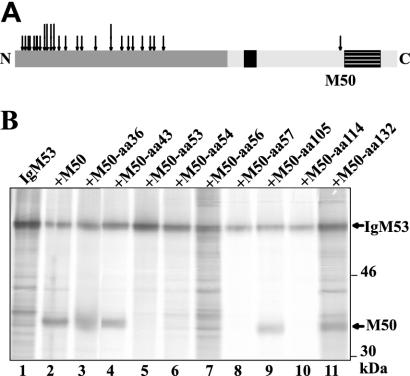
FIG. 4.
Coimmunoprecipitation of M50/p35 and M53/p38. (A) Mutants tested for the interaction between M50/p35 and M53/p38 proteins. Depicted is a schematic overview of the M50/p35 protein (see Fig. 2B). Arrows indicate the transposon insertions tested for interaction with M53/p38; long arrows represent those that lost the ability to interact with M53/p38. (B) Coimmunoprecipitation of M50/p35 and M53/p38. pCR3-M50 or pCR3-M50mut and pCR3-IgM53 were cotransfected into 293 cells. Cells were radioactively labeled and subsequently lysed. IgM53 was precipitated by protein A-Sepharose. Samples were then analyzed by SDS-PAGE. Individual mutants are characterized by their insertion sites. Arrows indicate M50/p35, M50/p35mut, and IgM53 protein bands. For a control, we used wt M50 (lane 2) and the functional M50 mutant with an insertion at aa 36 (M50-aa36).
Characterization of the interaction site of M50/p35 and M53/p38.
Most loss-of-function mutations (40 out of 59) concentrated in the N-terminal domain, whereas only 4 out of 20 insertions into the C-terminal domain affected viral growth. To test whether subdomains of the C terminus are nonessential, we constructed a series of targeted M50 deletion mutants. The N-terminal domain and the C-terminal TM region including the sequence that contained the lethal insertion at aa 280 were preserved, while the subdomains between (M50-del1, with aa 261 to 277 deleted [Δaa261-277], M50-del2 [Δaa225-277], M50-del3 [Δaa207-277], and M50-del4 [Δaa178-277]) were successively deleted. The deletion constructs were cotransfected with pCR3-IgM53 into 293 cells. In coprecipitation analyses, we could detect specific bands for all four deletion mutants, migrating at the expected molecular size. This result indicated that deletion of up to 99 aa in the C terminus of M50/p35 did not affect M53/p38 binding by the N-terminal domain (Fig. 5A).
FIG. 5.
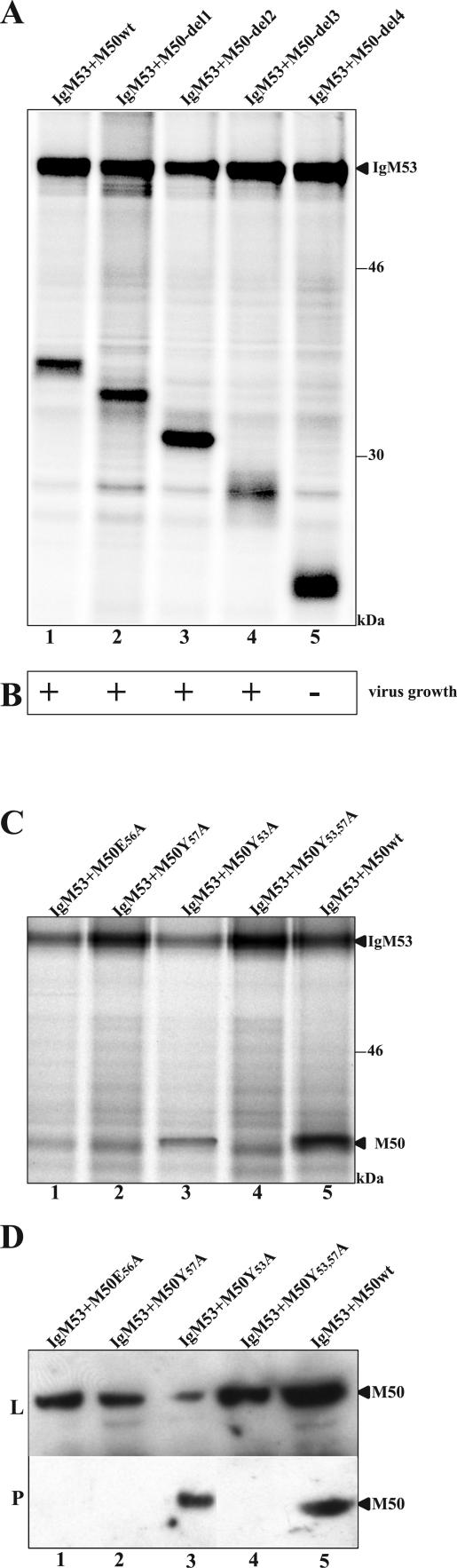
Functional analysis of M50 mutants. (A) Coprecipitation of IgM53 and M50/p35 deletion mutants. pCR3-IgM53 and M50-del1, M50-del2, M50-del3, M50-del4, and wt M50 were cotransfected into 293 cells. Cells were radioactively labeled, and protein complexes with IgM53 were precipitated with protein A-Sepharose. Samples were analyzed by SDS-PAGE. (B) Test for functionality of the M50 deletion mutants. For functional analysis, M50-del1, M50-del2, M50-del3, and M50-del4 were reinserted into pΔM50-GFP. BAC DNA was isolated and transfected into M2-10B4 cells. Ten days postinfection, cells were screened for plaque formation. The results are shown in the row below the depicted gel. (C) Coprecipitation of IgM53 and M50/p35 mutants affecting the potential binding motif. pCR3-IgM53 plus M50E56A, M50Y57A, M50Y53A, M50Y53,57A, or wt M50 was cotransfected into 293 cells. Cells were radioactively labeled, and protein complexes were precipitated with protein A-Sepharose. Samples were analyzed by SDS-PAGE. (D) Western blot analysis after pull-down of the M50-M53 complex. pCR3-IgM53 plus wt M50 M50Y53,57A, M50Y53A, M50E56A, or M50Y57A was cotransfected into 293 cells. Samples were analyzed by SDS-PAGE and plotted on membranes. M50/p35 was detected with an M50-specific antiserum in untreated cell lysate (L) and after pull-down of the M50/IgM53 complex with protein A-Sepharose (P). M50/p35-specific bands are indicated.
To test whether these deletion mutants rescue the growth phenotype of pΔM50-GFP, pM50-del1, pM50-del2, pM50-del3, and pM50-del4 were reintroduced into the MCMV BAC (Fig. 5B). The data showed that at least 70 aa from the C-terminal domain of M50/p35 are not required for the essential function of the protein, as predicted by the viability of individual insertion mutants from this region. Only the MCMV BAC with M50-del4 did not reconstitute viral progeny. This finding led to the conclusion that aa 178 to 207, a sequence with a prominent polyproline stretch, is probably not dispensable for virus growth.
The N terminus of the protein sufficed for M50-M53 interaction. In order to characterize the potential binding motif predicted by the mutants with lethal mutations between aa 53 and 56, amino acids Y53, E56, and Y57, which are conserved among herpesviral homologues, were replaced by alanine. 293 cells were cotransfected with plasmids pM50Y53,57A, pM50Y53A, pM50Y57A, and pM50E56A together with pIgM53, and the interaction was tested by coprecipitation of the respective mutant with IgM53. wt M50 (Fig. 5C, lane 5) and M50Y53A (Fig. 5C, lane 3) were able to coprecipitate with IgM53, whereas M50E56A, M50Y57A, and M50Y53,57A (Fig. 5C, lanes 1, 2 and 4, respectively) lost the ability to interact with IgM53, as indicated by the lack of an M50/p35-specific band. Due to the direct pull-down with protein A-Sepharose, a weaker, unspecific background signal is present. Immunoprecipitation of the same lysates with a specific antibody directed against M50/p35 showed all constructs and showed that the lack of coprecipitation was indeed caused by the loss of interaction between M50/p35 and M53/p38 (see Fig. S2 in the supplemental material).
For further analysis, pM50Y53,57A, pM50Y53A, pM50Y57A, and pM50E56A were cotransfected with pIgM53 into 293 cells and tested by Western blot analysis (Fig. 5D). The M50-specific antiserum detected the protein in all lysates (Fig. 5D, upper panel). However, when the M50/IgM53 complex was pulled down with protein A-Sepharose and bound M50/p35 was subsequently detected using the antibody, only wt M50/p35 and M50Y53A gave a specific signal (Fig. 5D, lower panel).
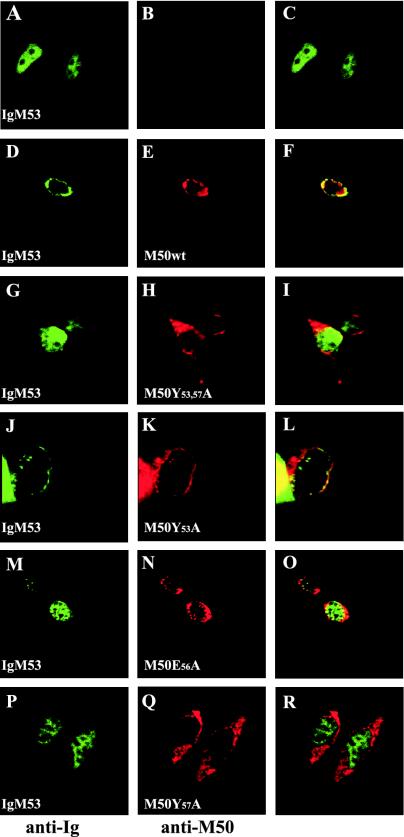
Mutated M50/p35 might still interact with M53/p38 but resist coprecipitation. We therefore studied the localization of wt and different mutants of M50/p35 with M53/p38 by immunofluorescence. To this end, plasmids pCR3-M50mut and pCR3-IgM53 were cotransfected into NIH 3T3 cells and detected after staining of the different proteins with specific antibodies. wt M50/p35 and IgM53/p38 colocalized at the inner nuclear membrane (Fig. 6D to F), whereas IgM53 alone showed a diffuse nuclear staining (Fig. 6A to C). Coexpression of IgM53 and M50Y53,57A, M50E56A, or M50Y57A resulted in a diffuse nuclear staining of M53/p38 (Fig. 6G to I, M to O, and P to R, respectively), comparable to isolated expression of IgM53. M50Y53,57A was localized in the nuclear membrane as well as in the adjacent endoplasmic reticulum compartment. In contrast, M50Y53A was located in the inner nuclear membrane and was still able to recruit IgM53. These data confirm that the interaction between M50/p35 and M53/p38 is necessary to provide a correct localization of both proteins at the inner nuclear membrane and to exert their function in the egress process of viral capsids. In summary, the immunofluorescence studies confirmed the observations made by coimmunoprecipitation. Thus, neighboring aa 56 and 57 are crucial for the interaction with M53, whereas the M50Y53A mutation did not influence the interaction of the two viral proteins.
FIG. 6.
Localization of M53/p38 in the presence of wtM50/p35 or mutants. NIH 3T3 cells were transiently transfected with pCR3-IgM53 alone or together with wt M50, M50Y53,57A, M50Y53A, M50E56A, or M50Y57A. Cells were stained with a fluorescein-conjugated antibody against the Ig tag to detect IgM53/p38 (green) and costained with an M50/p35-specific rabbit serum, which was detected by a Texas red-coupled secondary antibody (red). The discrepancy between the appearance of M50/p35-specific cytoplasmic staining shown here and that in reference 30 is due to the different cell lines used.
To test for functionality, M50Y53,57A was reintroduced into pΔM50-GFP and tested for the ability of the mutant to rescue the M50 deletion null phenotype. In repeated independent experiments, we were not able to reconstitute virus from MCMV BAC containing M50Y53,57A, indicating that the amino acid exchange abolished the function of M50/p35 (data not shown).
DISCUSSION
The comprehension of viral functions that play an important role in morphogenesis requires a detailed genetic analysis of the respective genes in the context of viral replication, where the relevant genes are expressed in operational conditions. This has been demonstrated recently by elegant screens for genetic footprinting of viral genes, subgenome fragments, and complete viral genomes of up to 10 kbp in size (18, 21, 37). These screens discriminate between virus mutants which preserve the ability to replicate and are retrieved after selection and mutants in which essential functions are affected and which cannot be retrieved (40).
Due to the fact that transfection of large viral DNA is not efficient enough for pool reconstitution, mutational analysis can be performed only by using individual mutants. A high DNA load, which is required for the reconstitution of large viruses, would inevitably lead to a high frequency of unwanted complementation events. For the small 10-kbp potato virus A genome, it was already found that the number of individual mutants able to propagate alone was lower than the number detected in the pool analysis, indicating transcomplementation during selection (18). Random transposon mutagenesis procedures described so far for herpesviruses create null mutants of the respective genes and serve to analyze nonessential genes or to define essentiality (6, 16, 17, 26, 41, 47). These genome-based insertion libraries cannot be used for the functional characterization of a coding sequence. First, this type of transposon cannot be removed from the genome; thus, most of the insertions destroy the target gene completely. Second, the number of insertions within a given gene in these libraries is far below the requirements of a comprehensive analysis.
Comprehensive mutant pools for cloned viral genes can be obtained through random mutagenesis procedures, which result in the efficient introduction of small insertions or deletions (2, 18, 40). However, a comprehensive set of the resulting mutants has to be reintroduced into the genome individually to allow functional analysis in the context of virus replication. As a consequence of the genome size, simple approaches using traditional molecular cloning techniques are not feasible in the case of large-DNA viruses.
To overcome these constraints, we developed a strategy combining a comprehensive mutagenesis procedure for the subcloned gene with fast reinsertion of the mutant at an ectopic position into the CMV genome. In principle, homologous recombination would be useful for the introduction of the mutants at their native sites (44). However, site-specific recombination is more efficient and technically much easier than homologous recombination. Therefore, to reintroduce the mutated gene at an ectopic position, we used FLP/FRT-mediated site-specific recombination (5, 26). In contrast to the commonly used approaches, in which the FRT sites are located on the same DNA molecule and mediate deletions or inversions, we used the FRT sites to induce intermolecular recombination, resulting in the unification of separate DNA molecules. The gene of interest can be subjected to random mutagenesis and can then be reintroduced into the viral genome in a single step. A functionally irrelevant insertion site was used to place mutated genes into the viral genome (26). This process allows gene insertion without influencing the expression of adjacent genes.
Using this strategy, we tested 104 mutants of M50/p35 for functionality in the MCMV genome context and classified the M50 ORF regions into functional and nonfunctional domains. M50/p35 is conserved among the Herpesviridae (7, 32). For the alphaherpesvirus homologue UL34, a crucial role in egress of the viral capsid from the nucleus was demonstrated (19, 30, 31, 35, 36, 39). Recently, a mutational approach to the investigation of the protein function of UL34 of HSV-1 has been carried out (3). Based on the assumption that charged cluster mutants will interrupt exposed areas of the protein, 19 targeted mutants were used to complement a UL34-defective virus in trans. Remarkably, no correlation could be found between conserved parts of the protein and protein functionality. The higher resolution of our random approach, however, shows that in MCMV, essential functions of M50/p35 indeed accumulate in the conserved N-terminal part, whereas large parts of the C-terminal domain can be deleted without a loss of function. Results with M50-del3 also showed that the potential LEM domain between aa 190 and 243 (30) is not essential. However, M50-del4 is able to bind to M53/p38 but does not rescue virus growth, indicating a potential essential sequence between aa 178 and 207, including a polyproline stretch that is conserved among betaherpesviruses. In the literature, data regarding the essentiality of the TM domain of UL34 in HSV-1 are contradictory (3, 49). For the betaherpesviruses, we demonstrate that the TM domain is crucial for the function of M50/p35. All stop mutants that lost the TM domain were nonfunctional, and even two mutants with independent insertions in the TM domain were lethal. A conserved feature in the alpha- and betaherpesvirus subgroups is the interaction between M50/p35 homologues and M53/p38 homologues (UL31) (14, 30, 34, 35, 48, 49). Therefore, all 25 in-frame loss-of-function mutants identified in our screen were tested in coprecipitation and colocalization analysis for interaction with M53/p38. The region around aa 52 to 56 is conserved among homologues of M50/p35. Single amino acid exchanges at positions 56 and 57, E to A and Y to A, respectively, rendered M50/p35 nonfunctional and defective for M53/p38 interaction. The change of Y to A at aa 53 did not influence the M50/p35 function or the interaction with M53/p38, confirming that the mutations at aa 56 and 57 affected the binding of M50/p35 to M53/p38. Thus, we defined a novel region essential for protein-protein interaction and virus replication.
Altogether, our random approach allows three conclusions. (i) The conserved N-terminal domain of M50/p35 comprises the majority of essential functions, including the interaction site with M53/p38. We hypothesize that this site has the same function in homologous proteins of other herpesviruses. (ii) The TM domain is essential for the function of at least the betaherpesvirus protein. (iii) A large domain of the nonconserved C-terminal region is not essential for viral replication and varies in length among herpesviruses. This region may be used to insert tags to monitor virus morphogenesis.
Only recently has the cloning of large-DNA viruses as infectious clones in bacteria been successful (1, 12, 22). Here, we present a comprehensive mutagenesis procedure that is generally applicable to any gene of a large cloned viral genome. This procedure will help to define sites of protein-protein interactions, either between viral proteins or between viral and cellular proteins, which play a key role in the virus life cycle. Protein-protein interaction sites may provide templates for the rational design of peptides or peptidomimetics with antiviral activity (10, 20). Based on the results of the comprehensive analysis, null phenotypes can be associated with a particular biochemical or cell biological feature.
Supplementary Material
Acknowledgments
We thank W. Muranyi for providing pCR3-IgM53 and antisera specific for M50/p35 and M53/p38. We also thank S. Boos for excellent technical assistance and B. Rupp for critical reading of the manuscript.
This work was supported by the Deutsche Forschungsgemeinschaft through SFB 455, “Viral functions and immune modulation.”
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org.
REFERENCES
- 1.Almazan, F., J. M. Gonzalez, Z. Penzes, A. Izeta, E. Calvo, J. Plana-Duran, and L. Enjuanes. 2000. Engineering the largest RNA virus genome as an infectious bacterial artificial chromosome. Proc. Natl. Acad. Sci. USA 97:5516-5521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Biery, M. C., F. J. Stewart, A. E. Stellwagen, E. A. Raleigh, and N. L. Craig. 2000. A simple in vitro Tn7-based transposition system with low target site selectivity for genome and gene analysis. Nucleic Acids Res. 28:1067-1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bjerke, S. L., J. M. Cowan, J. K. Kerr, A. E. Reynolds, J. D. Baines, and R. J. Roller. 2003. Effects of charged cluster mutations on the function of herpes simplex virus type 1 UL34 protein. J. Virol. 77:7601-7610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borst, E.-M., G. Hahn, U. H. Koszinowski, and M. Messerle. 1999. Cloning of the human cytomegalovirus (HCMV) genome as an infectious bacterial artificial chromosome in Escherichia coli: a new approach for construction of HCMV mutants. J. Virol. 73:8320-8329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Borst, E.-M., S. Mathys, M. Wagner, W. Muranyi, and M. Messerle. 2001. Genetic evidence of an essential role for cytomegalovirus small capsid protein in viral growth. J. Virol. 75:1450-1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brune, W., C. Menard, J. Heesemann, and U. H. Koszinowski. 2001. A ribonucleotide reductase homolog of cytomegalovirus and endothelial cell tropism. Science 291:303-305. [DOI] [PubMed] [Google Scholar]
- 7.Chee, M. S., A. T. Bankier, S. Beck, R. Bohni, C. M. Brown, R. Cerny, T. Horsnell, C. A. Hutchison III, T. Kouzarides, J. A. Martignetti, et al. 1990. Analysis of the protein-coding content of the sequence of human cytomegalovirus strain AD169. Curr. Top. Microbiol. Immunol. 154:125-169. [DOI] [PubMed] [Google Scholar]
- 8.Cherepanov, P. P., and W. Wackernagel. 1995. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158:9-14. [DOI] [PubMed] [Google Scholar]
- 9.Chiu, W., and F. J. Rixon. 2002. High resolution structural studies of complex icosahedral viruses: a brief overview. Virus Res. 82:9-17. [DOI] [PubMed] [Google Scholar]
- 10.Coen, D. M., and P. A. Schaffer. 2003. Antiherpesvirus drugs: a promising spectrum of new drugs and drug targets. Nat. Rev. Drug Discov. 2:278-288. [DOI] [PubMed] [Google Scholar]
- 11.del Val, M., H. Hengel, H. Hacker, U. Hartlaub, T. Ruppert, P. Lucin, and U. H. Koszinowski. 1992. Cytomegalovirus prevents antigen presentation by blocking the transport of peptide-loaded major histocompatibility complex class I molecules into the medial-Golgi compartment. J. Exp. Med. 176:729-738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Domi, A., and B. Moss. 2002. Cloning the vaccinia virus genome as a bacterial artificial chromosome in Escherichia coli and recovery of infectious virus in mammalian cells. Proc. Natl. Acad. Sci. USA 99:12415-12420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ebeling, A., G. M. Keil, E. Knust, and U. H. Koszinowski. 1983. Molecular cloning and physical mapping of murine cytomegalovirus DNA. J. Virol. 47:421-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fuchs, W., B. G. Klupp, H. Granzow, N. Osterrieder, and T. C. Mettenleiter. 2002. The interacting UL31 and UL34 gene products of pseudorabies virus are involved in egress from the host-cell nucleus and represent components of primary enveloped but not mature virions. J. Virol. 76:364-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hahn, G., H. Khan, F. Baldanti, U. H. Koszinowski, M. G. Revello, and G. Gerna. 2002. The human cytomegalovirus ribonucleotide reductase homolog UL45 is dispensable for growth in endothelial cells, as determined by a BAC-cloned clinical isolate of human cytomegalovirus with preserved wild-type characteristics. J. Virol. 76:9551-9555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hobom, U., W. Brune, M. Messerle, G. Hahn, and U. H. Koszinowski. 2000. Fast screening procedures for random transposon libraries of cloned herpesvirus genomes: mutational analysis of human cytomegalovirus envelope glycoprotein genes. J. Virol. 74:7720-7729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jenkins, F. J., M. J. Casadaban, and B. Roizman. 1985. Application of the mini-Mu-phage for target-sequence-specific insertional mutagenesis of the herpes simplex virus genome. Proc. Natl. Acad. Sci. USA 82:4773-4777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kekarainen, T., H. Savilahti, and J. P. Valkonen. 2002. Functional genomics on potato virus A: virus genome-wide map of sites essential for virus propagation. Genome Res. 12:584-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klupp, B. G., H. Granzow, and T. C. Mettenleiter. 2000. Primary envelopment of pseudorabies virus at the nuclear membrane requires the UL34 gene product. J. Virol. 74:10063-10073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lai, L., and W. J. Britt. 2003. The interaction between the major capsid protein and the smallest capsid protein of human cytomegalovirus is dependent on two linear sequences in the smallest capsid protein. J. Virol. 77:2730-2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Laurent, L. C., M. N. Olsen, R. A. Crowley, H. Savilahti, and P. O. Brown. 2000. Functional characterization of the human immunodeficiency virus type 1 genome by genetic footprinting. J. Virol. 74:2760-2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luckow, V. A., S. C. Lee, G. F. Barry, and P. O. Olins. 1993. Efficient generation of infectious recombinant baculoviruses by site-specific transposon-mediated insertion of foreign genes into a baculovirus genome propagated in Escherichia coli. J. Virol. 67:4566-4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lutarewych, M. A., M. R. Quirk, B. A. Kringstad, W. Li, C. M. Verfaillie, and M. C. Jordan. 1997. Propagation and titration of murine cytomegalovirus in a continuous bone marrow-derived stromal cell line (M2-10B4). J. Virol. Methods 68:193-198. [DOI] [PubMed] [Google Scholar]
- 24.Marchini, A., H. Liu, and H. Zhu. 2001. Human cytomegalovirus with IE-2 (UL122) deleted fails to express early lytic genes. J. Virol. 75:1870-1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McLeod, M., S. Craft, and J. R. Broach. 1986. Identification of the crossover site during FLP-mediated recombination in the Saccharomyces cerevisiae plasmid 2μm circle. Mol. Cell. Biol. 6:3357-3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Menard, C., M. Wagner, Z. Ruzsics, K. Holak, W. Brune, A. E. Campbell, and U. H. Koszinowski. 2003. Role of murine cytomegalovirus US22 gene family members in replication in macrophages. J. Virol. 77:5557-5570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Messerle, M., I. Crnkovic, W. Hammerschmidt, H. Ziegler, and U. H. Koszinowski. 1997. Cloning and mutagenesis of a herpesvirus genome as an infectious bacterial artificial chromosome. Proc. Natl. Acad. Sci. USA 94:14759-14763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mettenleiter, T. C. 2002. Herpesvirus assembly and egress. J. Virol. 76:1537-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miller, V. L., and J. J. Mekalanos. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. Bacteriol. 170:2575-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Muranyi, W., J. Haas, M. Wagner, G. Krohne, and U. H. Koszinowski. 2002. Cytomegalovirus recruitment of cellular kinases to dissolve the nuclear lamina. Science 297:854-857. [DOI] [PubMed] [Google Scholar]
- 31.Neubauer, A., J. Rudolph, C. Brandmuller, F. T. Just, and N. Osterrieder. 2002. The equine herpesvirus 1 UL34 gene product is involved in an early step in virus egress and can be efficiently replaced by a UL34-GFP fusion protein. Virology 300:189-204. [DOI] [PubMed] [Google Scholar]
- 32.Rawlinson, W. D., H. E. Farrell, and B. G. Barrell. 1996. Analysis of the complete DNA sequence of murine cytomegalovirus. J. Virol. 70:8833-8849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reddehase, M. J., F. Weiland, K. Münch, S. Jonjic, A. Lüske, and U. H. Koszinowski. 1985. Interstitial murine cytomegalovirus pneumonia after irradiation: characterization of cells that limit viral replication during established infection of the lungs. J. Virol. 55:264-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reynolds, A. E., B. J. Ryckman, J. D. Baines, Y. Zhou, L. Liang, and R. J. Roller. 2001. UL31 and UL34 proteins of herpes simplex virus type 1 form a complex that accumulates at the nuclear rim and is required for envelopment of nucleocapsids. J. Virol. 75:8803-8817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reynolds, A. E., E. G. Wills, R. J. Roller, B. J. Ryckman, and J. D. Baines. 2002. Ultrastructural localization of the herpes simplex virus type 1 UL31, UL34, and US3 proteins suggests specific roles in primary envelopment and egress of nucleocapsids. J. Virol. 76:8939-8952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roller, R. J., Y. Zhou, R. Schnetzer, J. Ferguson, and D. DeSalvo. 2000. Herpes simplex virus type 1 UL34 gene product is required for viral envelopment. J. Virol. 74:117-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rothenberg, S. M., M. N. Olsen, L. C. Laurent, R. A. Crowley, and P. O. Brown. 2001. Comprehensive mutational analysis of the Moloney murine leukemia virus envelope protein. J. Virol. 75:11851-11862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 39.Shiba, C., T. Daikoku, F. Goshima, H. Takakuwa, Y. Yamauchi, O. Koiwai, and Y. Nishiyama. 2000. The UL34 gene product of herpes simplex virus type 2 is a tail-anchored type II membrane protein that is significant for virus envelopment. J. Gen. Virol. 81:2397-2405. [DOI] [PubMed] [Google Scholar]
- 40.Singh, I. R., R. A. Crowley, and P. O. Brown. 1997. High-resolution functional mapping of a cloned gene by genetic footprinting. Proc. Natl. Acad. Sci. USA 94:1304-1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smith, G. A., and L. W. Enquist. 1999. Construction and transposon mutagenesis in Escherichia coli of a full-length infectious clone of pseudorabies virus, an alphaherpesvirus. J. Virol. 73:6405-6414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thäle, R., U. Szepan, H. Hengel, G. Geginat, P. Lucin, and U. H. Koszinowski. 1995. Identification of the mouse cytomegalovirus genomic region affecting major histocompatibility complex class I molecule transport. J. Virol. 69:6098-6105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thomsen, D. R., L. L. Roof, and F. L. Homa. 1994. Assembly of herpes simplex virus (HSV) intermediate capsids in insect cells infected with recombinant baculoviruses expressing HSV capsid proteins. J. Virol. 68:2442-2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wagner, M., A. Gutermann, J. Podlech, M. J. Reddehase, and U. H. Koszinowski. 2002. Major histocompatibility complex class I allele-specific cooperative and competitive interactions between immune evasion proteins of cytomegalovirus. J. Exp. Med. 196:805-816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wagner, M., S. Jonjić, U. H. Koszinowski, and M. Messerle. 1999. Systematic excision of vector sequences from the BAC-cloned herpesvirus genome during virus reconstitution. J. Virol. 73:7056-7060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wagner, M., Z. Ruzsics, and U. H. Koszinowski. 2002. Herpesvirus genetics has come of age. Trends Microbiol. 10:318-324. [DOI] [PubMed] [Google Scholar]
- 47.Weber, P. C., M. Levine, and J. C. Glorioso. 1987. Rapid identification of nonessential genes of herpes simplex virus type 1 by Tn5 mutagenesis. Science 236:576-579. [DOI] [PubMed] [Google Scholar]
- 48.Yamauchi, Y., C. Shiba, F. Goshima, A. Nawa, T. Murata, and Y. Nishiyama. 2001. Herpes simplex virus type 2 UL34 protein requires UL31 protein for its relocation to the internal nuclear membrane in transfected cells. J. Gen. Virol. 82:1423-1428. [DOI] [PubMed] [Google Scholar]
- 49.Ye, G. J., and B. Roizman. 2000. The essential protein encoded by the UL31 gene of herpes simplex virus 1 depends for its stability on the presence of UL34 protein. Proc. Natl. Acad. Sci. USA 97:11002-11007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yu, D., G. A. Smith, L. W. Enquist, and T. Shenk. 2002. Construction of a self-excisable bacterial artificial chromosome containing the human cytomegalovirus genome and mutagenesis of the diploid TRL/IRL13 gene. J. Virol. 76:2316-2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.