The lantibiotic-resistance protein NSR from S. agalactiae was crystallized in two different crystal forms, which diffracted X-rays to a maximum resolution of 2.2 Å.
Keywords: lantibiotic, nisin, resistance, immunity, lipoprotein, Lactococcus lactis
Abstract
Nisin is a 34-amino-acid antimicrobial peptide produced by Lactococcus lactis belonging to the class of lantibiotics. Nisin displays a high bactericidal activity against various Gram-positive bacteria, including some human-pathogenic strains. However, there are some nisin-non-producing strains that are naturally resistant owing to the presence of the nsr gene within their genome. The encoded protein, NSR, cleaves off the last six amino acids of nisin, thereby reducing its bactericidal efficacy. An expression and purification protocol has been established for the NSR protein from Streptococcus agalactiae COH1. The protein was successfully crystallized using the vapour-diffusion method in hanging and sitting drops, resulting in crystals that diffracted X-rays to 2.8 and 2.2 Å, respectively.
1. Introduction
The widespread use of antibiotics has led to the emergence of resistant pathogenic bacteria. Thus, there is an urgent need to develop new alternatives in order to fight infectious diseases. Promising candidates are antimicrobial peptides such as lantibiotics produced by some Gram-positive bacteria. Lantibiotics are ribosomally synthesized peptides that are characterized by the extensive modifications that they undergo and normally consist of dehydrated amino acids and (methyl)-lanthionine rings (Chatterjee et al., 2005 ▶; Lubelski et al., 2008 ▶). Lantibiotics target lipid II, an essential cell-membrane precursor, and form pores within the membrane, causing cell leakage and subsequently cell death (Breukink & de Kruijff, 2006 ▶). More than 50 lantibiotics have been discovered to date (Donaghy, 2010 ▶) and many of them have already entered the preclinical phase.
Nisin is the most studied and widely used lantibiotic. It is produced by Lactococcus lactis and has the characteristic one lanthionine and four methyl-lanthionine rings. Interestingly, nisin has been shown to be insensitive to proteolytic digestion owing to the presence of the five lanthionine rings. Nisin has a wide bactericidal activity spectrum against many Gram-positive bacteria, including L. lactis subsp. lactis and subsp. cremoris, L. bulgaricus, Staphylococcus aureus, Clostridium difficile, Streptococcus pneumoniae, MRSA, enterococci and Listeria monocytogenes. Nisin also prevents the outgrowth of many Clostridium and Bacillus species (Harris et al., 1992 ▶). The germicidal efficiency of nisin is owing to its capability to inhibit cell-wall biosynthesis and pore-forming ability, where it uses the cell-wall precursor lipid II as a docking molecule (Willey & van der Donk, 2007 ▶). Nisin has been found to be effective against S. pneumoniae, Mycobacterium tuberculosis and other multi-resistant Gram-positive pathogenic bacteria (Carroll et al., 2010 ▶; Cotter et al., 2005 ▶). Nisin also has potential for the treatment of various enterococcal infections, atopic dermatitis, peptic ulcers and bacterial mastitis (Cotter et al., 2005 ▶; de Arauz et al., 2009 ▶). However, there are some non-nisin-producing strains, including various pathogenic bacteria such as Streptococcus agalactiae and S. aureus, which are resistant against nisin (Harris et al., 1992 ▶). This resistance is owing to the presence of the nsr gene, which encodes the nisin resistance protein (referred to as NSR). It has been shown that NSR cleaves off the last six amino acids of nisin, thereby degrading nisin proteolytically. The cleaved nisin was found to have reduced affinity for the cell membrane and showed 100-fold less antibacterial efficiency (Sun et al., 2009 ▶). NSR is strongly hydrophobic at the N-terminus and was thus postulated to be membrane-associated (Froseth & McKay, 1991 ▶). Interestingly, there is a whole operon associated with the mechanism of nisin resistance, consisting of the nsr gene, a two-component regulatory system and an ABC transporter. This operon can be found in various species of Gram-positive human-pathogenic bacteria (Khosa et al., 2013 ▶).
Thus, the structure of NSR would lead to a deeper understanding of the overall mechanism of nisin resistance.
Here, we present the overexpression, purification and crystallization of the nisin resistance protein.
2. Materials and methods
2.1. Macromolecule production
2.1.1. Cloning and expression
The nsr gene from S. agalactiae COH1 was amplified using the genomic DNA into pET-28b in a similar manner as described previously (Khosa et al., 2013 ▶). Since the encoded protein has an N-terminal transmembrane region, the first 30 amino acids were removed (referred to as NSR30) and an 8×His tag was introduced at the N-terminus. The resulting plasmid (Table 1 ▶) was verified by sequencing and transformed into Escherichia coli BL21 (DE3) for expression. A single transformed colony was inoculated into 20 ml LB medium containing 30 µg ml−1 kanamycin. The culture was grown for 14 h at 310 K with shaking at 200 rev min−1. 2 l LB medium with 30 µg ml−1 kanamycin was inoculated with the overnight culture at an OD600 of 0.05 and grown at 310 K with shaking at 170 rev min−1 until an OD600 of 0.3 was reached. The temperature was lowered to 291 K and the cells were further grown to an OD600 of 0.8 before induction with 1 mM IPTG. The cells were grown for a further 15 h.
Table 1. Macromolecule-production information.
| Source organism | S. agalactiae |
| DNA source | Genomic DNA |
| Forward primer | TTAAATATTTATCTATTACCTCCTAGTAGCGAACGTTATGG |
| Reverse primer | ATGGCTGCCGCGCGGCACCAG |
| Cloning vector | pET-28b |
| Expression vector | pET-28b |
| Expression host | E. coli BL21 (DE3) |
| Complete amino-acid sequence of the construct produced | MGSSHHHHHHHHSSGLVPRGSHLNIYLLPPSSERYGRVILDRVEQRGLYSQGRQWQIIRQRSEKKLKTSKSYQESRNIVQEAVRYGGGKHSQILSKETVRRDTLDSRYPEYRRLNEDILLITIPSISKLDKRSISHYSGKLQNILMEKSYKGLILDLSNNTGGNMIPMIGGLASILPNDTLFHYTDKYGNKKTITMKNIPLEALKISRKTINTKHVPIAIITNHKTASSAEMTFLSFKGLPNVKSFGQATAGYTTVNETFMLYDGARLALTTGIVSDRQGYKYENTPILPDQVTSLPLQESQSWLKSRINQN |
The cells were harvested by centrifugation at 8000 rev min−1 for 20 min at 277 K. The harvested cell pellet was stored at 253 K until further use.
2.1.2. Purification
All steps were performed at 277 K. The stored cell pellet was thawed and resuspended in 10 ml buffer A (50 mM Tris pH 8.0, 50 mM NaCl, 10% glycerol), and 10 mg DNase (deoxyribonuclease I from bovine pancreas, Sigma–Aldrich) was added. The cells were lysed five times using a cell disruptor (Constant Cell Disruption Systems, United Kingdom) at 260 MPa. The lysate was centrifuged at 42 000 rev min−1 for 60 min using a Ti60 rotor to remove unlysed cells and debris.
Histidine was added to the cleared lysate to a final concentration of 1 mM. The lysate was then applied onto an Ni2+-loaded HiTrap HP Chelating column (GE Healthcare) pre-equilibrated with buffer B (20 mM Tris pH 8.0, 250 mM NaCl, 1 mM histidine) at a flow rate of 1 ml min−1. The column was washed with six column volumes of buffer B. The protein was then eluted with increasing concentrations of histidine from 1 to 120 mM in the form of a linear gradient spanning 60 min at a flow rate of 2 ml min−1. The fractions containing the protein of interest were pooled and concentrated to 12 mg ml−1 in an Amicon centrifugal filter concentrator with a 10 kDa cutoff membrane (Millipore). The concentrated protein was then further purified by size-exclusion chromatography using a Superose 12 GL 10/300 column (GE Healthcare) equilibrated with buffer C (25 mM MES pH 6.0, 150 mM NaCl). The protein eluted as a single homogeneous peak and the concerned fractions were pooled and concentrated to 8.6 mg ml−1 as described above. The purity of the protein was analyzed with SDS–PAGE and colloidal Coomassie stain (Dyballa & Metzger, 2009 ▶). The purified protein was directly used for crystallization.
2.2. Crystallization
Crystallization screening was performed at 285 K using an NT8 robot (Formulatrix) and the sitting-drop vapour-diffusion method in Corning 3553 sitting-drop plates. For initial screening, various commercial crystallization screens were used (The JCSG Core Suite I, Classics Suite, PEGs Suite and MPD Suite from Qiagen, Germany and MIDAS from Molecular Dimensions, England). Nanodrops consisting of 0.1 µl each of protein and reservoir solution were mixed and equilibrated against 50 µl reservoir solution. The screening yielded some initial rod-shaped crystals after 3 d in the condition 0.5 M lithium sulfate, 15%(w/v) PEG 8000 (The Classics I Suite condition F5). The initial crystals were optimized by varying the concentration of PEG [5, 10, 15, 20, 25 and 30%(w/v)] and lithium sulfate (0.4, 0.5, 0.6 and 0.7 M) using hanging and sitting-drop vapour-diffusion methods at 297 and 285 K, respectively. Each drop consisted of 1 µl protein solution (concentration of 9 mg ml−1) mixed with 1 µl reservoir solution and was equilibrated against a reservoir volume of 500 µl. Crystals were obtained after 1 d and grew to their maximum dimensions within 5 d. Crystallization information is summarized in Table 2 ▶.
Table 2. Crystallization.
| Method | Vapour diffusion |
| Plate type | Hanging/sitting drop |
| Temperature (K) | 297/285 |
| Protein concentration (mgml1) | 9 |
| Buffer composition of protein solution | 25mM MES pH 6.0, 150mM NaCl |
| Composition of reservoir solution | 0.5M lithium sulfate, 15%(w/v) PEG 8000 |
| Ratio of drop | 1:1 |
| Volume of reservoir (l) | 500 |
2.3. Data collection and processing
Drops containing the optimized crystals were overlaid with 2 µl mineral oil before the crystals were harvested and flash-cooled in liquid nitrogen. X-ray diffraction data were collected on the ID23-EH2 beamline at the European Synchrotron Radiation Facility (ESRF; Flot et al., 2010 ▶), Grenoble using a Pilatus detector. After the initial diffraction tests, a data-collection strategy was calculated using the EDNA software available at the beamline (Incardona et al., 2009 ▶) and the data subsequently collected were processed and scaled using XDS and XSCALE (Kabsch, 2010 ▶).
3. Results and discussion
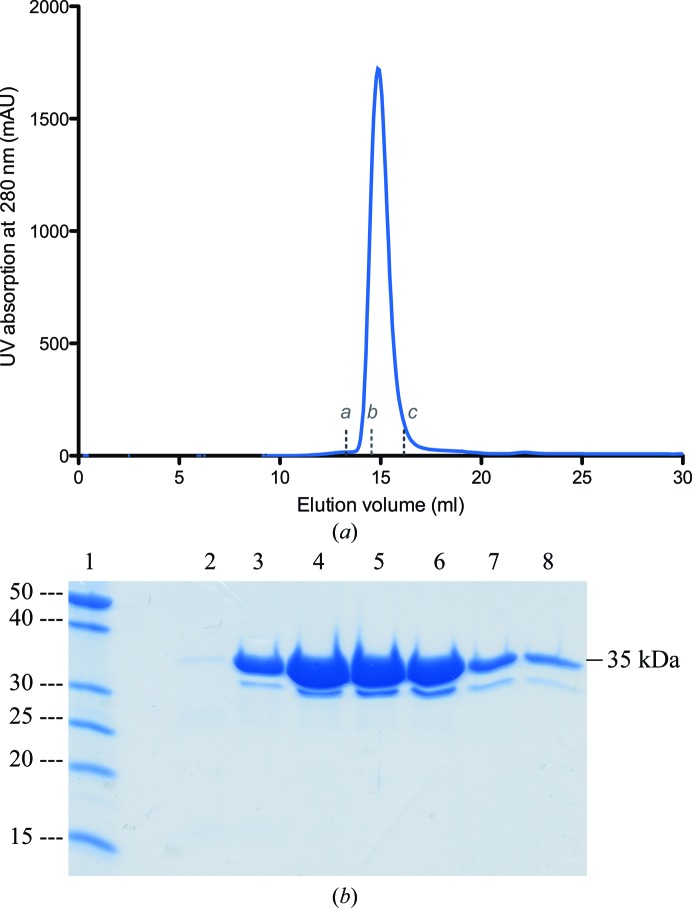
NSR30 was successfully cloned and overexpressed in a soluble form in E. coli BL21 (DE3) cells. The protein was purified via a two-step purification protocol. Nickel-affinity chromatography was performed first, followed by size-exclusion chromatography (Fig. 1 ▶ a). NSR30 eluted from the size-exclusion chromatogram at 14.8 ml, which corresponds to a molecular mass of 30–40 kDa, indicating that NSR30 is a monomer in solution. The yield of the protein was around 5 mg per litre of cell culture. Protein homogeneity and purity was assessed by SDS–PAGE (Fig. 1 ▶ b). The molecular mass of the purified protein was comparable to the theoretically calculated molecular weight of 35.3 kDa (Gasteiger et al., 2005 ▶).
Figure 1.
Purification of NSR30. (a) Chromatogram representing the purification of NSR30 by size-exclusion chromatography. (b) 15% SDS–PAGE showing the purified NSR30 fractions. Lane 1 contains PageRuler Unstained Protein Ladder (labelled in kDa) and lanes 2–8 contain the purified NSR30 fractions at 35 kDa. The lower 30 kDa band also arises from NSR30, as verified by mass spectrometry, and is likely to be a degradation product. a, b and c refer to the protein standards BSA (molecular weight 67 kDa), β-lactoalbumin (35 kDa) and cytochrome c (12.7 kDa), respectively.
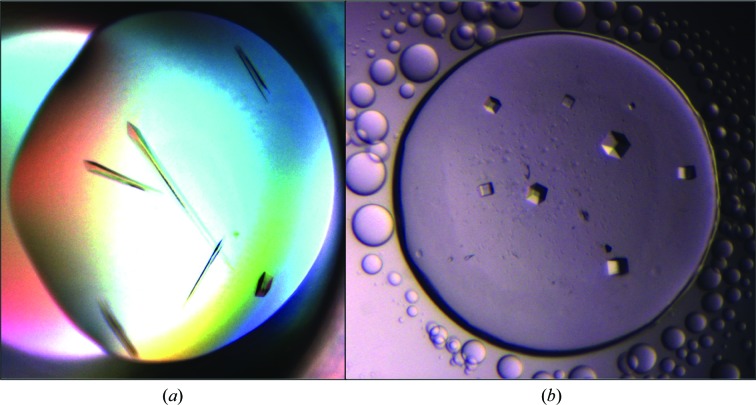
Initial crystals of NSR30 appeared after 3 d in The Classics Screen I condition F5 [0.5 M lithium sulfate, 15%(w/v) PEG 8000] with a sitting-drop setup. Optimizations were performed by varying the concentrations of lithium sulfate and PEG. Hanging-drop and sitting-drop vapour-diffusion methods were also tried. Crystals were obtained with 5–30%(w/v) PEG 8000 and 0.4–0.7 M lithium sulfate after 1 d (Fig. 2 ▶). Sitting-drop vapour diffusion at 285 K yielded rod-shaped crystals (Fig. 2 ▶ a) with maximum dimensions of 560 × 45 × 30 µm, whereas cubic-shaped crystals with dimensions of 125 × 125 × 40 µm were obtained using the hanging-drop vapour-diffusion method at 297 K (Fig. 2 ▶ b). The crystals obtained were dissolved and analysed by mass spectrometry, revealing the presence of the NSR30 protein (Supporting Information).
Figure 2.
Crystals of NSR30. (a) Rod-shaped crystals obtained by the sitting-drop vapour-diffusion method at 285 K. (b) Cubic crystals obtained by the hanging-drop vapour-diffusion method at 297 K.
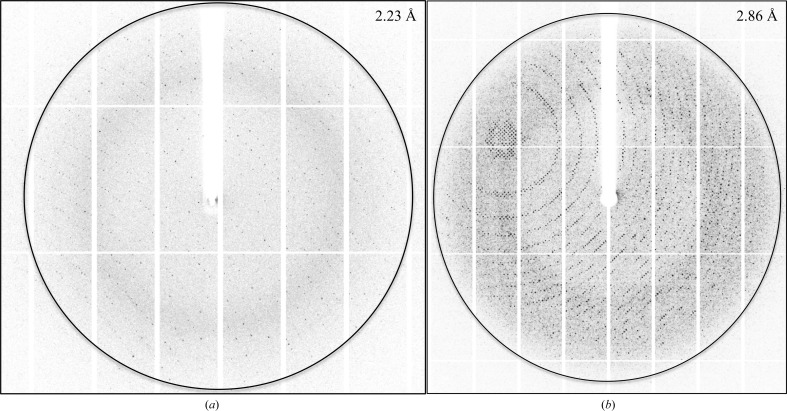
The initial rod-shaped crystals diffracted to 2.2 Å resolution using synchrotron X-rays (Fig. 3 ▶ a). The crystals belonged to space group P212121, with unit-cell parameters a = 58.8, b = 137.2, c = 164.0 Å, α = β = γ = 90° (Table 3 ▶). Matthews coefficient calculations resulted in a V M of 2.34 Å3 Da−1 and a solvent content of 47.34%, corresponding to four molecules in the asymmetric unit (Matthews, 1968 ▶; Kantardjieff & Rupp, 2003 ▶). However, the cubic crystals diffracted to 2.8 Å resolution using synchrotron X-rays (Fig. 3 ▶ b). The crystals belonged to space group P432, with unit-cell parameters a = b = c = 186.4 Å, α = β = γ = 90° (Table 3 ▶). Interestingly, Matthews coefficient calculations resulted in a V M of 3.81 or 2.54 Å3 Da−1 and a solvent content of 68 or 52%, corresponding to two or three molecules in the asymmetric unit (Matthews, 1968 ▶; Kantardjieff & Rupp, 2003 ▶). The exact number of molecules in the asymmetric unit will be determined when the structure has been solved. The self-rotation function revealed a noncrystallographic twofold axis for the P432 crystal form and a noncrystallographic fourfold axis for the crystal form displaying P212121 symmetry. Furthermore, no large peaks were seen in the native Patterson function for either of the crystal forms. Since there are no template structures which can be used for molecular replacement in the PDB that display a sequence identity of higher than 22%, the structure will be solved by using experimental phase determination, i.e. selenomethionine incorporation or heavy-atom derivatization, which is currently in progress.
Figure 3.
X-ray diffraction pattern of NSR30. Diffraction images of rod-shaped (a) and cubic (b) NSR30 crystals with an oscillation width of 1.0°. The images were used to calculate a data-collection strategy using EDNA (Incardona et al., 2009 ▶).
Table 3. Data collection and processing.
Values in parentheses are for the outer shell.
| Rod-shaped crystal | Cubic crystal | |
|---|---|---|
| Diffraction source | ID23-EH2, ESRF | ID23-EH2, ESRF |
| Wavelength () | 0.87260 | 0.87260 |
| Temperature (K) | 100 | 100 |
| Detector | Pilatus3 2M | Pilatus3 2M |
| Crystal-to-detector distance (mm) | 273.31 | 359.15 |
| Rotation range per image () | 0.05 | 0.05 |
| Total rotation range () | 120 | 40 |
| Exposure time per image (s) | 0.02 | 0.04 |
| Space group | P212121 | P432 |
| a, b, c () | 58.8, 137.2, 164.0 | 186.4, 186.4, 186.4 |
| , , () | 90, 90, 90 | 90, 90, 90 |
| Mosaicity () | 0.05 | 0.05 |
| Resolution range () | 100.02.21 (2.292.21) | 100.02.87 (2.972.87) |
| Total No. of reflections | 303210 (28341) | 208348 (19523) |
| No. of unique reflections | 67177 (6695) | 25913 (2471) |
| Completeness (%) | 99.6 (99.3) | 99.8 (100.0) |
| Multiplicity | 4.5 (4.2) | 8.03 (7.9) |
| I/(I) | 8.67 (1.79) | 9.69 (1.71) |
| R p.i.m. (%) | 6.0 (33.5) | 7.4 (41.1) |
| Overall B factor from Wilson plot (2) | 28.4 | 33.4 |
The nisin resistance protein could be a model for other lantibiotic resistance proteins observed in the genome of a wide variety of human pathogens. Initial experiments show that NSR cleaves the nisin peptide in vitro as observed in vivo, where it has been shown that NSR confers a 20-fold resistance towards nisin (Khosa et al., 2013 ▶). The structure of the nisin resistance protein would thus lead to a deeper understanding of its function and also pave the way for the development of new therapeutic agents with the potential of substituting for antibiotics.
Supplementary Material
Mass-spectrometric identification of NSR30.. DOI: 10.1107/S2053230X15006226/rl5098sup1.pdf
Acknowledgments
The authors are indebted to Lutz Schmitt for his kind support, advice and invaluable suggestions. This work was initiated by Philipp Ellinger. We also thank André Abts for fruitful scientific discussions and advice. We are grateful to the staff of ESRF ID23-2, especially Christoph Mueller-Dieckmann, for assistance and support during crystal screening and data collection. We are thankful to Heinrich Heine International Graduate School of Protein Science and Technology (iGRASPseed) for providing a scholarship to SK.
References
- Arauz, L. J. de, Jozala, A. F., Mazzola, P. G. & Vessoni Penna, T. C. (2009). Trends Food Sci. Technol. 20, 146–154.
- Breukink, E. & de Kruijff, B. (2006). Nature Rev. Drug Discov. 5, 321–323. [DOI] [PubMed]
- Carroll, J., Draper, L. A., O’Connor, P. M., Coffey, A., Hill, C., Ross, R. P., Cotter, P. D. & O’Mahony, J. (2010). Int. J. Antimicrob. Agents, 36, 132–136. [DOI] [PubMed]
- Chatterjee, C., Paul, M., Xie, L. & van der Donk, W. A. (2005). Chem. Rev. 105, 633–684. [DOI] [PubMed]
- Cotter, P. D., Hill, C. & Ross, R. P. (2005). Curr. Protein Pept. Sci. 6, 61–75. [DOI] [PubMed]
- Donaghy, J. (2010). Bioeng. Bugs, 1, 437–439. [DOI] [PMC free article] [PubMed]
- Dyballa, N. & Metzger, S. (2009). J. Vis. Exp., 2009, 1431. [DOI] [PMC free article] [PubMed]
- Flot, D., Mairs, T., Giraud, T., Guijarro, M., Lesourd, M., Rey, V., van Brussel, D., Morawe, C., Borel, C., Hignette, O., Chavanne, J., Nurizzo, D., McSweeney, S. & Mitchell, E. (2010). J. Synchrotron Rad. 17, 107–118. [DOI] [PMC free article] [PubMed]
- Froseth, B. R. & McKay, L. L. (1991). Appl. Environ. Microbiol. 57, 804–811. [DOI] [PMC free article] [PubMed]
- Gasteiger, E., Hoogland, C., Gattiker, A., Wilkins, M. R., Appel, R. D. & Bairoch, A. (2005). The Proteomics Protocols Handbook, edited by J. M. Walker, pp. 571–607. Totowa: Humana Press.
- Harris, L. J., Fleming, H. P. & Klaenhammer, T. R. (1992). Food. Res. Int. 25, 57–66.
- Incardona, M.-F., Bourenkov, G. P., Levik, K., Pieritz, R. A., Popov, A. N. & Svensson, O. (2009). J. Synchrotron Rad. 16, 872–879. [DOI] [PubMed]
- Kabsch, W. (2010). Acta Cryst. D66, 125–132. [DOI] [PMC free article] [PubMed]
- Kantardjieff, K. A. & Rupp, B. (2003). Protein Sci. 12, 1865–1871. [DOI] [PMC free article] [PubMed]
- Khosa, S., AlKhatib, Z. & Smits, S. H. (2013). Biol. Chem. 394, 1543–1549. [DOI] [PubMed]
- Lubelski, J., Rink, R., Khusainov, R., Moll, G. & Kuipers, O. (2008). Cell. Mol. Life Sci. 65, 455–476. [DOI] [PMC free article] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol. 33, 491–497. [DOI] [PubMed]
- Sun, Z., Zhong, J., Liang, X., Liu, J., Chen, X. & Huan, L. (2009). Antimicrob. Agents Chemother. 53, 1964–1973. [DOI] [PMC free article] [PubMed]
- Willey, J. M. & van der Donk, W. A. (2007). Annu. Rev. Microbiol. 61, 477–501. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Mass-spectrometric identification of NSR30.. DOI: 10.1107/S2053230X15006226/rl5098sup1.pdf