Abstract
Iron acquisition is critical for the growth and pathogenesis of Legionella pneumophila, the causative agent of Legionnaires’ disease. L. pneumophila utilizes two main modes of iron assimilation, namely ferrous iron uptake via the FeoB system and ferric iron acquisition through the action of the siderophore legiobactin. This review highlights recent studies concerning the mechanism of legiobactin assimilation, the impact of c-type cytochromes on siderophore production, the importance of legiobactin in lung infection and a newfound role for a bacterial pyomelanin in iron acquisition. These data demonstrate that key aspects of L. pneumophila iron acquisition are significantly distinct from those of long-studied, ‘model’ organisms. Indeed, L. pneumophila may represent a new paradigm for a variety of other intracellular parasites, pathogens and under-studied bacteria.
Keywords: bacterial virulence, c-type cytochromes, FeoB, Gram-negative bacteria, iron acquisition, legiobactin siderophore, Legionella pneumophila, Legionnaires’ disease, pyomelanin pigment
Legionella pneumophila is a Gram-negative bacterium that is ubiquitous in both natural and man-made water systems [1,2]. One of 59 species within the Legionella genus [3,4], L. pneumophila is best known as the main etiologic agent of Legionnaires’ disease, a potentially fatal form of pneumonia [2]. Of note, the incidence of Legionnaires’ disease in the USA has increased approximately threefold since 2001, with similar increases occurring in Europe and Canada [5,6]. In aquatic environments, L. pneumophila flourishes as an intracellular parasite of amoebae and as a constituent of multiorganismal biofilms [1,7–8]. Humans are infected with the bacterium primarily by inhaling contaminated water droplets from aerosol-generating devices, including, most notoriously, cooling towers [9]. Once in the lung, L. pneumophila invades and grows in alveolar macrophages [10]. The ecology and pathogenesis of L. pneumophila is governed, to a great extent, by a remarkably large number of secreted proteins [8,11,12]. Iron acquisition is yet another key component of the organism’s physiology and virulence [13]. As one form of iron acquisition, L. pneumophila assimilates ferrous iron through the action of the inner-membrane protein FeoB [14]. This transport system is required for optimal intracellular infection of amoebae and macrophages as well as for full virulence in a murine model of pneumonia [14]. As a second form of iron uptake, L. pneumophila secretes legiobactin, a low-molecular weight, nonprotein, ferric iron chelator [15,16]. L. pneumophila secretes legiobactin, when it is grown in low-iron, chemically defined media [15]. The iron-chelating activity of the siderophore is readily detected by the chrome azurol S (CAS) assay. Legiobactin is also defined by its ability to stimulate the growth of iron-starved legionellae, including wild-type bacteria and a feoB mutant [16]. Early work demonstrated that legiobactin production is governed by the iron-regulated lbtA and lbtB genes [16]. LbtA has homology to known siderophore synthetases, and LbtB is related to inner membrane proteins that are involved in the export of other siderophores. Thus, cytoplasmic LbtA is likely involved in the synthesis of legiobactin, while LbtB mediates movement of legiobactin across the inner membrane (IM) prior to its final export. This review will discuss recent data concerning the mechanism of legiobactin utilization, the effect of c-type cytochromes on legiobactin production and the role of the siderophore in infection [17–20]. Additionally, it will describe a newly uncovered role for pyomelanin in iron acquisition [21,22]. As in the earlier studies, these recent data derive from the analysis of L. pneumophila strain 130b, a clinical isolate belonging to serogroup 1. The various genes that have been implicated in iron acquisition and will be discussed here are listed in Table 1, along with their open-reading-frame (ORF) designations in strain 130b as well as strains Paris and Philadelphia-1.
Table 1. Genes that promote Legionella pneumophila iron acquisition.
| Gene | 130b ORF† | Paris ORF† | Phil-1 ORF† | Role in iron acquisition |
|---|---|---|---|---|
| ccmC ‡ | lpw09401 | lpp0920 | lpg0858 | Legiobactin production |
| cyc4 | lpw01241 | lpp0138 | lpg0124 | Legiobactin production |
| feoB | lpw29101 | lpp2711 | lpg2657 | Ferrous iron uptake |
| frgA | lpw30551 | lpp2846 | lpg2800 | Putative siderophore synthetase |
| fur | lpw03201 | lpp0438 | lpg0232 | Transcriptional regulator |
| hbp | lpw00251 | lpp0024 | lpg0024 | Hemin-binding protein |
| hisC2 | lpw20551 | lpp1979 | lpg1998 | HGA production |
| hmgA | lpw13001 | lpp1248 | lpg1285 | HGA degradation |
| iraB | lpw08291 | lpp0812 | lpg0746 | Putative peptide transporter |
| iroT/mavN | lpw30711 | lpp2867 | lpg2815 | Ferrous iron uptake |
| lbtA | lpw13341 | lpp1280 | lpg1325 | Legiobactin biosynthesis |
| lbtB | lpw13331 | lpp1279 | lpg1324 | IM export of legiobactin |
| lbtC | lpw13321 | lpp1278 | lpg1323 | IM import of legiobactin |
| lbtU | lpw13361 | lpp1281 | lpg1326 | OM receptor for legiobactin |
| lly | lpw24691 | lpp2232 | lpg2278 | HGA production |
| mcoL | lpw03531 | lpp0339 | lpg0265 | Multicopper oxidase |
| phhA | lpw28991 | lpp2700 | lpg2647 | HGA production |
| proA/mspA | lpw05471 | lpp0532 | lpg0467 | Degrades transferrin |
| tatB | lpw31801 | lpp2974 | lpg2906 | Growth on low-iron media |
The ORF designations are derived from the genome sequencing data that have been recently reported for L. pneumophila strains 130b, Paris and Philadelphia-1 (Phil-1) [23–25]. Utilizing the ORF designations listed in this table, the corresponding ORFs in L. pneumophila strains Alcoy, Corby and Lens can been obtained by visiting the National Center for Biotechnology Information (NCBI) [26] or the Legionella genome website [27].
The entire ccm operon is implicated in iron acquisition, but for sake of brevity, only the ccmC gene is listed here.
HGA: Homogentisic acid; IM: Inner membrane; OM: Outer membrane; ORF: Open reading frame.
LbtU, the unique receptor for legiobactin
Recent work has identified the receptor for legiobactin [18]. DNA sequence and RT-PCR analyses revealed the presence of an iron-repressed gene (lbtU) directly upstream of lbtA and lbtB. Based upon bioinformatic analysis, LbtU is an outer membrane (OM) protein with extracellular domains, a transmembrane β-barrel, and short periplasmic tails. Immunoblot analysis of cellular fractions confirmed this OM location. Mutants specifically lacking lbtU are impaired for growth on low-iron media. Although normal for legiobactin production, lbtU mutants are unable to utilize legiobactin for growth on iron-deplete media and display an impaired ability to uptake iron. Complemented lbtU mutants behave as the parental wild-type does, indicating that all mutant phenotypes are due specifically to the loss of LbtU. A cloned copy of lbtU can confer the ability to bind legiobactin upon a heterologous bacterium, Legionella longbeachae. Together, these data indicate that LbtU is involved in the uptake of legiobactin. Given its OM location, LbtU is most likely the receptor for legiobactin. Presumably, ferrilegiobactin binds to a surface domain(s) of LbtU and then passes through an OM-spanning pore created by the protein. Formally, an alternative hypothesis is that the ferric iron is released from the siderophore while still extracellular and LbtU provides transport for ‘free’ iron. However, since the lbtU mutant could, like wild-type bacteria, grow on low-iron media when provided with Fe3+ or Fe2+ salts, it seems unlikely that LbtU is a nonspecific transporter of ‘free’ iron.
LbtU appears to represent a new type of siderophore receptor [18]. LbtU is predicted to have eight external loops, a 16-stranded transmembrane β-barrel, and short N- and C-terminal periplasmic tails. This structure differs from those of previously characterized siderophore receptors, including FecA, FepA, FhuA, FpvA and FptA, all of which have a 22-stranded barrel and an extended N-terminus that binds the energy-transducing molecule TonB. This structural difference, coupled with the fact that L. pneumophila does not encode TonB or its interacting proteins ExbB and ExbD, implies that LbtU mediates iron uptake in a way that is mechanistically distinct from the existing paradigm. Compatible with this hypothesis, 3D-modeling by the I-TASSER and Phyre servers suggests that the 16-stranded barrel of LbtU provides a channel through the OM in a way that is different from the well-known 22-stranded β-barrel receptors. LbtU is not alone in being distinct from ‘traditional’ siderophore receptors. Indeed, a 14-stranded-β-barrel protein is a siderophore receptor for Francisella species [28]. However, LbtU and the Francisella proteins (e.g., FslE of F. tularensis) are different from each other in terms of the number of β-strands and there being an extended periplasmic tail in the Francisella protein. BLASTP results indicate that LbtU has similarity to hypothetical proteins that are predicted to be in the OM of Coxiella burnetii and Rickettsiella grylli, suggesting that LbtU may be the prototype of a new form of receptor.
In thinking about how LbtU might mediate iron acquisition, an important question is what fulfills the role of TonB-ExbBD in L. pneumophila [18]. One plausible answer is the Tol system (i.e., TolA, TolQ and TolR) which, in other bacteria, operates similarly to TonB-ExbBD, although with a different purpose, including the import of colicins [29]. In some cases, TolQ and TolR can function, albeit imperfectly, as replacements for ExbB and ExbD, even promoting iron uptake. A second possible answer to this question is that L. pneumophila uses a pathway that is completely distinct from TonB-ExbBD and TolAQR. Although more work is needed in order to distinguish between these two possibilities, it is clear that the energy generated by proton motive force is required for iron uptake by L. pneumophila [18]. Since LbtU lacks an extended periplasmic tail, another question is how the protein transitions between plugged/unplugged states in order to allow for the controlled import of siderophore and iron. One hypothesis would be that LbtU exists in a closed state, which can transition to an open state when siderophore is engaged. A second scenario is that another protein, perhaps the mimic of TonB, provides the plug, moving away from LbtU when iron is imported. Additional study of LbtU will have implications for other Gram-negative (like) bacteria that lack TonB, including species of Chlamydia, Chlamydophila, Coxiella, Ehrlichia, Francisella and Rickettsia [30].
LbtC, the IM transporter for legiobactin
After LbtU was defined, the LbtC protein was identified as an IM protein that is required for legiobactin utilization [20]. RT-PCR and DNA sequence analyses identified lbtC as an iron-repressed gene that is the last gene in the operon containing lbtA and lbtB. In silico analysis predicted that LbtC is a member of the major facilitator superfamily (MFS). More specifically, LbtC is within the DHA-12 subfamily in the MFS that is typically involved in the transport of small molecules across the IM. As was the case for lbtU mutants and lbtA mutants, lbtC mutants display impaired growth on low-iron media. Although elaborating wild-type levels of siderophore, lbtC mutants, such as lbtU mutants, cannot utilize legiobactin to stimulate their growth on low-iron media. The mutants also have an impaired capacity to assimilate radiolabeled iron. All mutant phenotypes can be complemented by reintroduction of lbtC. When both lbtC and lbtU are introduced into L. longbeachae, the bacterium acquires the ability to use legiobactin. Together, these data indicate that LbtC is required for the assimilation of legiobactin and based upon its location is likely the conduit for ferrilegiobactin transit across the IM.
The definition of LbtC provides new insight into bacterial siderophore transport [20]. In past, ATP-binding cassette (ABC)-type permeases have generally been defined as the conduit for ferrisiderophore import across the cytoplasmic membrane [31]. Indeed, only a few non-ABC-type systems have been identified as being important for siderophore transport across a bacterial membrane, including FptX of Pseudomonas aeruginosa and homologous RhtX of Sinorhizobium meliloti [32,33]. However, although RhtX and FptX are members of the MFS, they are placed into a subfamily that is distinct from that of LbtC [20]. Based upon BLASTP, LbtC has its greatest level of similarity with FslD/FigD, a protein encoded by the siderophore operon of Francisella [34]. Thus, the LbtC-like proteins and the RhtX-like proteins represent two types of MFS IM transporters involved in bacterial siderophore import. Since LbtC can confer upon L. longbeachae the ability to use legiobactin [20], it appears that a single MFS protein can mediate siderophore import across the IM. Compatible with this viewpoint is the fact that other MFS transporters act as single-protein carriers albeit for different sorts of molecules [35], and 3D-modeling predicts that LbtC has the capacity to form a pore in the IM [20].
A c-type cytochrome promotes siderophore production
Early studies determined that the ccm locus promotes L. pneumophila growth in low-iron conditions, suggesting that cytochrome c maturation has a role in intra- and extracellular iron acquisition [36,37]. The ccm locus is an eight-gene operon that encodes a protein complex which transports heme across the IM and then attaches it to apo-cytochromes in the periplasm as a final step in the maturation of c-type cytochromes [38,39]. Recently, it was reported that L. pneumophila Ccm is needed for expression of legiobactin [19]. Indeed, ccm mutants of L. pneumophila display a loss of siderophore, as measured by both the CAS assay and the Legionella-specific bioassay. Compatible with these data, ccm transcripts are expressed by legionellae when grown in deferrated medium. To discern the basis for this new role for Ccm, mutants lacking individual c-type cytochromes were made and examined. Whereas mutants lacking cytochrome c1 or cytochrome c5 have normal siderophore expression, cyc4 mutants defective for cytochrome c4 lack legiobactin [19]. These data, coupled with the expression pattern of cyc4 mRNA, affirm that cytochrome c4 promotes siderophore production. The data obtained from the study of L. pneumophila are the fourth case in which Ccm has been linked to siderophore expression, with past cases involving Paracoccus denitrificans, P. aeruginosa, Pseudomonas fluorescens and Rhizobium leguminosarum [19,40]. Since the four genera involved in these studies are quite distinct from each other as are the structures of their representative siderophores [17], the connection between Ccm and siderophore likely also exists in a variety of other bacteria.
It has been proposed that the role of cytochrome c4 in legiobactin production is due to there being an important electron-transfer step in the periplasm [19]. This could involve the shuttling of electrons to an enzyme that is needed for legiobactin maturation or secretion, since periplasmic enzymes are known to have roles in siderophore synthesis in some other bacteria [41]. On the other hand, cytochrome c4 might have a more indirect role, such as maintaining a redox state in the periplasm that is compatible with legiobactin processing. Because cytochromes c1 and c5 are not needed for legiobactin production, there is undoubtedly some specificity to the relationship between c-type cytochromes and the siderophore pathway.
The importance of legiobactin in infection
Another recent study documented the importance of legiobactin in lung infection by L. pneumophila [17]. Independently-derived lbtA mutants, but not a complemented derivative, exhibit a reduced ability to infect the lungs of A/J mice after intratracheal inoculation. The mutants display an in vivo defect that ranges from 3 to 13-fold over the 3-day course of infection. This defect, however, is not evident when the lbtA mutant and its parental strain are co-inoculated. These data indicate that siderophore released by the wild-type strain can enhance the growth of the mutant in trans. L. pneumophila lbtU mutants are also impaired for infection in a legiobactin-dependent manner [18]. Interestingly, lbtA mutants that are unable to produce legiobactin grow normally in murine lung macrophages and alveolar epithelial cells, suggesting that the siderophore is promoting something other than intracellular infection of resident cells [17]. In one scenario, legiobactin could be facilitating the growth or survival of a subset of bacteria that are residing in the extracellular spaces in the lung. That extracellular survival is a part of Legionella infection has been indicated before when other sorts of mutants were found to be more defective in lung infection than in intracellular infection assays [42]. In another scenario, legiobactin might be crucial for intracellular growth after the immune system has been engaged; for example, γ-interferon activated macrophages contain reduced levels of iron [43]. That legiobactin promotes infection is in keeping with our overall understanding of siderophores in infection [17]. Indeed, siderophores produced by Bordetella species, Burkholderia cenocepacia, Klebsiella pneumoniae and P. aeruginosa promote the extracellular growth and/or survival of the bacteria in the lungs. Moreover, siderophores are necessary for bacterial growth in macrophages, in the case of Bacillus anthracis, Brucella abortus, Mycobacterium tuberculosis and Salmonella enterica. Finally, in a situation most akin to that of L. pneumophila and legiobactin, siderophore mutants of Shigella flexneri are not defective for intracellular infection in vitro but are defective when examined in an animal model of disease.
The size of the defect displayed by the lbtA mutant is in agreement with our knowledge of iron acquisition in bacterial infection; in other words, since pathogens generally have various means for getting iron, the loss of one pathway often does not eliminate virulence and in some cases may have little to no effect on in vivo growth [44]. In the case of L. pneumophila, the other pathways that might compensate for the lack of legiobactin include the above-mentioned FeoB system, heme-binding capability, a putative iron-peptide transporter and a pyomelanin (see below) [14,21,37,45,46]. Based on its possession of an lbtA-like gene (frgA), L. pneumophila might even be capable of producing a second siderophore [16,47]. It is also worth considering that legiobactin could be promoting virulence in ways that are distinct from its role in iron assimilation. Along those lines, the pyoverdine siderophore made by P. aeruginosa acts as a signaling molecule, regulating the expression of other virulence factors [48,49], and another P. aeruginosa secreted factor, PQS, is both an iron chelator and quorum-sensing molecule [50]. Furthermore, P. aeruginosa pyochelin, by virtue of being a catalyst for generating hydroxyl radical, is a mediator of tissue damage [51].
L. pneumophila pyomelanin & its newfound role in iron acquisition
It has been known, for a long time, that L. pneumophila secretes a brown pigment [52]. As demonstrated by Steinert et al., this pigment is a polymerized form of homogentisic acid (HGA), a secondary metabolite that is secreted by the bacterium [53]. Depending upon the availability of L-tyrosine or L-phenylalanine, the initial synthesis of HGA in the bacterial cytoplasm occurs in either two or three steps [53,54]. If exogenous tyrosine is present in sufficient amount, the process begins with the conversion of L-tyrosine to 4-hydroxyphenylpyruvate through the action of the amino acid transferase encoded by the hisC2 gene. In the next and last step, 4-hydroxy-phenylpyruvate dioxygenase, encoded by the lly gene, converts 4-hydroxyphenylpyruvate to HGA. If exogenous tyrosine is absent or in low amount but L-phenylalanine is present, the process starts with the conversion of L-phenylalanine to L-tyrosine as catalyzed by phenylalanine hydroxylase encoded by the phhA gene. Once it is made, whether in two or three steps, HGA is secreted out of the bacterial cell by an as yet unknown mechanism, and then it can undergo oxidative polymerization resulting in HGA-melanin, a form of pyomelanin [21]. In addition to being subject to secretion, cytoplasmic HGA can also be converted to 4-maleylacetoacetate through the action of the homogentisate 1, 2-dioxygenase enzyme that is encoded by hmgA [21,54]. For years, the only role linked to L. pneumophila HGA-melanin was resistance to light [55]. During a screening of mutagenized L. pneumophila for strains that could not rescue the growth of the feoB mutant, an unusual mutant was obtained that had a strong inhibitory effect on the ferrous transport mutant [21]. The mutant proved to be an hmgA mutant that produced elevated levels of HGA-melanin. Thus, it was posited that secreted HGA-melanin is capable of conferring ferric reductase activity and that hyperpigmentation results in an excessive reduction of iron that can, in the case of the feoB mutant, slow growth. Supporting this hypothesis, culture supernatants of wild-type L. pneumophila contained ferric reductase activity [21]. Furthermore, a lly mutant defective for production of the pyomelanin lacked the reductase activity, whereas the hyperpigmented hmgA mutant had increased activity. In agreement with the nature of HGA-melanin, the secreted activity was enhanced by the presence of tyrosine in the growth media, resistant to protease treatment, acid-precipitable and heterogeneous in size [21]. Taken together, these data indicated that HGA-melanin, directly or indirectly, promotes the reduction of extracellular ferric iron, and therefore it might constitute an alternative pathway for iron acquisition.
Next, it was determined that purified HGA and HGA-melanin can mediate the reduction of ferric nitrate, ferric chloride, ferric citrate and ferric pyrophosphate [22]. Importantly, HGA and HGA-melanin could also promote the uptake of radiolabeled iron by strains of L. pneumophila, L. anisa, L. jamestowniensis and L. micdadei [22]. In the case of a feoB ferrous iron transport mutant of L. pneumophila, this increase in iron acquisition was not evident. Together, these data indicate that HGA and its polymerized form directly promote the reduction of ferric iron and the ferrous iron that is generated is subject to assimilation by the bacteria. Interestingly, the HGA-melanin fraction that is found in bacterial supernatants contains ferric iron and ferrous iron and is capable of stimulating the growth of bacteria that had been depleted of iron [22]. Because material obtained from the culture supernatants of a nonpigmented mutant did not enhance growth, HGA-melanin is a potentiator of bacterial growth in low-iron conditions. In support of its role in iron assimilation, the amount of HGA-melanin in L. pneumophila supernatants is inversely related to siderophore activity; in other words, an lbtA mutant made fourfold more HGA-melanin than did the wild-type strain, and the hmgA mutant produced lower amounts of siderophore [22]. Compatible with a role in the biology of environmental L. pneumophila, HGA and HGA-melanin were able to reduce and release iron from insoluble ferric hydroxide. Suggestive of a role in pathogenesis, HGA and the pyomelanin were effective at reducing and releasing iron from ferritin and transferrin, two iron chelates that occur within the mammalian host [22].
These data from L. pneumophila are the first documentation of a role for HGA and a pyomelanin in bacterial iron acquisition. On the one hand, the ferrous iron that is generated by HGA and HGA-melanin might diffuse to the bacterial surface and be internalized by a yet-to-be-defined OM channel. On the other hand, since ferrous iron is typically unstable at neutral pH in aerobic conditions and because iron associates with HGA-melanin in the culture supernatants, the pyomelanin might function as a shuttle or trap ‘protecting’ and then bringing ferrous iron to the bacterial cell surface. In support of the latter scenario, HGA-melanin enhanced iron uptake to the same level as did several stronger reducing agents [22]. Thus, the extent of iron assimilation is likely influenced by the polymerized status of the reducing agent or the nature of the HGA-melanin complex. Since other bacteria, including B. cenocepacia, P. aeruginosa and Vibrio cholerae, elaborate HGA-melanin [21,56,57], the results obtained with L. pneumophila and its pigment have broad implications. It should also be noted that there are other types of secreted bacterial pigments, including the blue pigment pyocyanin of P. aeruginosa, which can mediate ferric reduction reactions [58].
Overall model for L. pneumophila iron acquisition
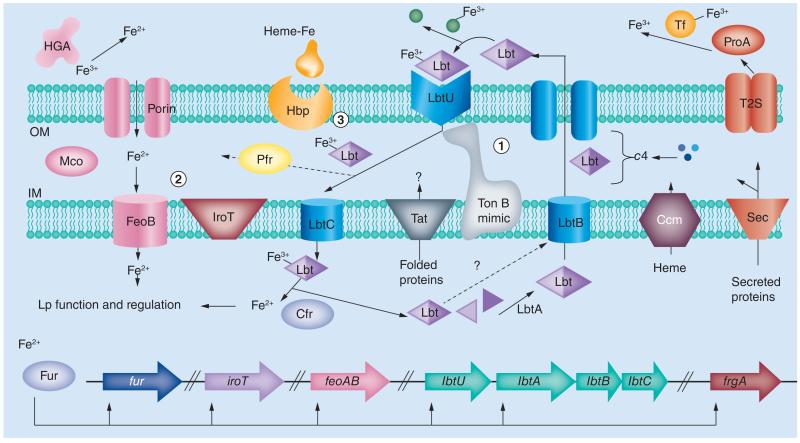
The current model for Legionella iron acquisition is depicted in Figure 1. Table 1 provides a listing of those genes that have been implicated in iron acquisition, as summarized below. L. pneumophila has two main modes of iron acquisition, namely siderophore-mediated Fe3+ uptake (mode 1) and FeoB-mediated Fe2+ uptake (mode 2) [13]. In support of this hypothesis, researchers have been unable to isolate a mutant lacking both FeoB and LbtA [16]. In mode 1, legiobactin (Lbt) is synthesized from precursors by the action of LbtA and then passes across the IM via LbtB, a member of the MFS [16,17]. The Ccm system is needed for full siderophore activity, with cytochrome c4 (secreted by Sec) facilitating siderophore maturation by donating electrons or maintaining the proper redox state in the periplasm [19,37]. Legiobactin likely exits the cell by passing through an OM channel, as occurs for other siderophores [59]. After scavenging Fe3+ from host and environmental chelators, Fe3+-Lbt is recognized at the cell surface by LbtU [18]. Because L. pneumophila does not have TonB-ExbBD, the organism undoubtedly has an alternative energy-transducing system (TonB-mimic) that conjoins with LbtU to import Fe3+-Lbt. After entry into the periplasm, Fe3+-Lbt passes through the IM via LbtC, another member of the MFS [20]. Upon delivery into the cytoplasm, Fe2+ is released by reductases that are known to exist there (Cfr) [60,61]. Alternately, reduction of Fe3+-Lbt might occur in the periplasm through the action of a periplasmic reductase (Pfr) [60,62], with the resultant Fe2+ then moving across the IM via FeoB. Although Cfr and Pfr have been identified by biochemical means, the genes encoding these reductases have not been defined. Once in the cytoplasm, Lbt may or may not be recycled, as both outcomes occur in other bacteria [63]. In mode 2, Fe2+is transported across the IM by FeoB [14]. As in other systems [64], a porin is likely the conduit for Fe2+ passage through the OM, and also as in others [65], a periplasmic multicopper oxidase (Mco) aids in growth in the presence of Fe2+ [66]. Recent work has identified the iron-regulated protein IroT/MavN as being important for ferrous iron uptake [67]. Predicted to be a membrane protein, IroT/MavN might be part of an IM transporter (as depicted in Figure 1), OM transporter, or it may aid in the formation of a transporter. L. pneumophila, secreted HGA-melanin can reduce Fe3+ and is an important source of Fe2+for import [21]. Twin-arginine translocation (Tat) is needed for L. pneumophila growth in low-iron conditions but not for siderophore activity or LbtU localization [21,68], suggesting that it might potentiate the trafficking of Fe3+-Lbt or Fe2+ in the periplasmic space. Another extracellular source of iron, which could be chelated by legiobactin or reduced, is the degradative release of Fe3+ from host transferrin (Tf) by the ProA/MspA protease that is secreted by L. pneumophila type II secretion (T2S) [11,61,69]. Underlying the FeoB and legiobactin pathways is transcriptional regulation by the iron-responsive Fur repressor, which controls fur, iroT/mavN, feoB, lbtU and lbtABC [14,16,67,70]. Another gene that is highly regulated by iron and Fur is frgA [47]. Based upon the strong sequence similarity between FrgA and other siderophore synthetases, including LbtA, it is possible that FrgA is involved in the production of a yet-to-be-defined siderophore [16,47]. In addition to the major FeoB and legiobactin pathways, L. pneumophila is able to bind and utilize heme (hemin) as yet another iron source (mode 3 in Figure 1) [46]. The molecular basis of heme utilization is minimally defined, although the Hbp protein is known to be required for optimal hemin-binding by L. pneumophila and is capable of conferring hemin-binding upon recombinant E. coli [46]. The cellular location of Hbp is likely to be either the OM (as depicted in Figure 1) or the periplasm, and sequence analysis indicates that the hbp gene is also subject to Fur regulation [46]. A possible fourth iron assimilation pathway that may be operative in L. pneumophila is the utilization of iron-loaded peptides [13]. This hypothesis is based upon the fact that IraB, which is homologous to di- and tripeptide transporters present in the IM of other bacteria, promotes L. pneumophila growth on iron-deplete media [45,71]. Additionally, recent experiments indicate that IraB is not required for legiobactin production or utilization [20]. Compatible with the role of iron acquisition in infection, many of the genes that have been implicated in iron acquisition, including ccmC, feoB, frgA, iraB, lbtABC, lly, phhA and proA (Table 1), are known to be expressed during intracellular infection of host cells [16,20,54,72–73]. Finally, several studies have begun to examine iron acquisition by the other species of Legionella. This work indicates that many but not all Legionella species secrete siderophore activity [3,74]. In a similar vein, pigment production is common but not universal among Legionella species [3]. These data may explain, in part, the varying degrees to which Legionella species grow under low-iron conditions [3].
Figure 1. Current model of iron acquisition by Legionella pneumophila.
Two primary modes are depicted. Legiobactin-mediated ferric iron assimilation (mode 1) mainly involves LbtA- and LbtB-mediated siderophore production and LbtU- and LbtC-mediated siderophore uptake. Ferrous iron assimilation (mode 2) involves the HGA-melanin ferric reductase, the FeoB IM transporter and the accessory IroT/MavN protein. Also depicted is a third pathway involving Hbp-mediated heme-iron uptake (mode 3). See the text for further discussion and references.
IM: Inner membrane; OM: Outer membrane.
Conclusion & future perspective
Studies of L. pneumophila illustrate both the importance of iron acquisition in bacterial physiology and pathogenesis and the many ways in which iron can be assimilated. Importantly, recent reports also demonstrate that several key aspects of L. pneumophila iron acquisition are significantly different from those of other well-studied bacterial systems. Thus, many important questions remain to be answered, since the continued study of L. pneumophila has the potential to uncover new paradigms for iron uptake. For example, it will be interesting to discern how LbtU and LbtC conjoin to facilitate legiobactin transport across the bacterial cell envelope and how they do so in the absence of TonB-ExbBD. Additional mechanistic questions include determining how cytochrome c4 enhances siderophore production. Future efforts should also be directed toward understanding more precisely how legiobactin promotes pathogenesis as well as determining the role of HGA-melanin within the context of lung infection. Given that L. pneumophila culture supernatants contain a CAS-reactive substance(s) in addition to LbtA-dependent legiobactin as well as the fact that the LbtA-like protein FrgA is required for optimal intracellular infection [17,47], it will also be worthwhile to ascertain whether L. pneumophila in fact secretes a second siderophore. Lastly, there is merit to learning the importance and molecular mechanism of heme acquisition by L. pneumophila. Although certain aspects of Legionella iron acquisition are likely to be reflections of the bacterium’s unique environmental and intracellular niches, it is anticipated that many of the answers obtained from the work done with L. pneumophila will have relevance for understanding other environmentally and/or medically important microbes. Given its broad significance, iron acquisition systems can be considered as potential targets for industrial application as well as disease control and prevention. For example, increased understanding of siderophores may lead to the generation of siderophore inhibitors (analogues) or antireceptor vaccines that could control bacterial growth [63,75–81].
EXECUTIVE SUMMARY.
Iron acquisition is critical for the growth, intracellular infectivity and virulence of Legionella pneumophila. The key pathways for iron uptake by L. pneumophila are FeoB-mediated ferrous iron uptake and legiobactin-mediated ferric iron assimilation. Both of these pathways promote virulence in a murine model of pneumonia.
LbtU, the outer membrane receptor for legiobactin, and LbtC, the inner membrane importer of legiobactin, are rather distinct from their counterparts in well-studied bacterial systems. This, coupled with the absence of TonB-ExbB-ExbD in L. pneumophila, indicates that ferrilegiobactin assimilation is mechanistically unique. Yet, emerging data suggest that systems similar to that of legiobactin may be operative in other important bacteria that are, at present, relatively less characterized.
The HGA-melanin pigment that is secreted by L. pneumophila confers ferric reductase activity, and the ferrous iron that is generated is used to stimulate bacterial growth in low-iron conditions. This newfound role for a pyomelanin is likely to be relevant for a variety of other significant microbes.
Given the importance of iron for bacterial ecology and pathogenesis, the recently defined mediators of iron acquisition may represent potential target for industrial and biomedical applications.
Acknowledgments
Research in the Cianciotto laboratory is supported, in part, by NIH grants AI034937 and AI089712.
Footnotes
Financial & competing interests disclosure
The author has no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
- 1.Cianciotto NP, Hilbi H, Buchrieser C. Legionnnaires’ Disease. In: Rosenberg E, DeLong EF, Thompson F, Lory S, Stackebrandt E, editors. The Prokaryotes – Human Microbiology. 4th Edition Springer; Berlin, Heidelberg, Germany: 2013. pp. 147–217. [Google Scholar]
- 2.Mercante JW, Winchell JM. Current and emerging Legionella diagnostics for laboratory and outbreak investigations. Clin. Microbiol. Rev. 2015;28(1):95–133. doi: 10.1128/CMR.00029-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pearce MM, Theodoropoulos N, Mandel MJ, Brown E, Reed KD, Cianciotto NP. Legionella cardiaca sp. nov., isolated from a case of native valve endocarditis in a human heart. Int. J. Syst. Evol. Microbiol. 2012;62:2946–2954. doi: 10.1099/ijs.0.039248-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rizzardi K, Winiecka-Krusnell J, Ramliden M, Alm E, Andersson S, Byfors S. Legionella norrlandica sp. nov., isolated from the biopurification system of a wood processing plant in northern Sweden. Int. J. Syst. Evol. Microbiol. 2014;65(Pt 2):598–603. doi: 10.1099/ijs.0.068940-0. [DOI] [PubMed] [Google Scholar]
- 5.Parr A, Whitney EA, Berkelman RL. Legionellosis on the rise: a review of guidelines for prevention in the United States. J. Public Health Manag. Pract. 2014 doi: 10.1097/PHH.0000000000000123. doi:10.1097/PHH.0000000000000123. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Farnham A, Alleyne L, Cimini D, Balter S. Legionnaires’ disease incidence and risk factors, New York, USA, 2002–2011. Emerg. Infect. Dis. 2014;20:1795–1802. doi: 10.3201/eid2011.131872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buse HY, Schoen ME, Ashbolt NJ. Legionellae in engineered systems and use of quantitative microbial risk assessment to predict exposure. Water Res. 2012;46:921–933. doi: 10.1016/j.watres.2011.12.022. [DOI] [PubMed] [Google Scholar]
- 8.Tyson JY, Vargas P, Cianciotto NP. The novel Legionella pneumophila type II secretion substrate NttC contriubtes to infection of amoebae Hartmannella vermiformis and Willaertia magna. Microbiology. 2014;160(12):2732–2744. doi: 10.1099/mic.0.082750-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pagnier I, Merchat M, La Scola B. Potentially pathogenic amoeba-associated microorganisms in cooling towers and their control. Future Microbiol. 2009;4:615–629. doi: 10.2217/fmb.09.25. [DOI] [PubMed] [Google Scholar]
- 10.Newton HJ, Ang DK, Van Driel IR, Hartland EL. Molecular pathogenesis of infections caused by Legionella pneumophila. Clin. Microbiol. Rev. 2010;23(2):274–298. doi: 10.1128/CMR.00052-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cianciotto NP. Many substrates and functions of type II protein secretion: lessons learned from Legionella pneumophila. Future Microbiol. 2009;4:797–805. doi: 10.2217/FMB.09.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rolando M, Buchrieser C. Legionella pneumophila type IV effectors hijack the transcription and translation machinery of the host cell. Trends Cell Biol. 2014;24(12):771–778. doi: 10.1016/j.tcb.2014.06.002. [DOI] [PubMed] [Google Scholar]
- 13.Cianciotto NP. Iron acquisition by Legionella pneumophila. Biometals. 2007;20:323–331. doi: 10.1007/s10534-006-9057-4. [DOI] [PubMed] [Google Scholar]
- 14.Robey M, Cianciotto NP. Legionella pneumophila feoAB promotes ferrous iron uptake and intracellular infection. Infect. Immun. 2002;70:5659–5669. doi: 10.1128/IAI.70.10.5659-5669.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liles MR, Aber Scheel T, Cianciotto NP. Discovery of a nonclassical siderophore, legiobactin, produced by strains of Legionella pneumophila. J. Bacteriol. 2000;182:749–757. doi: 10.1128/jb.182.3.749-757.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Allard KA, Viswanathan VK, Cianciotto NP. lbtA and lbtB are required for production of the Legionella pneumophila siderophore legiobactin. J. Bacteriol. 2006;188(4):1351–1363. doi: 10.1128/JB.188.4.1351-1363.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Allard KA, Dao J, Sanjeevaiah P, et al. Purification of legiobactin and the importance of this siderophore in lung infection by Legionella pneumophila. Infect. Immun. 2009;77:2887–2895. doi: 10.1128/IAI.00087-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chatfield CH, Mulhern BJ, Burnside DM, Cianciotto NP. Legionella pneumophila LbtU acts as a novel, TonB-independent receptor for the legiobactin siderophore. J. Bacteriol. 2011;193(7):1563–1575. doi: 10.1128/JB.01111-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yip ES, Burnside DM, Cianciotto NP. Cytochrome c4 is required for siderophore expression by Legionella pneumophila, whereas cytochromes c1 and c5 promote intracellular infection. Microbiology. 2011;157(3):868–878. doi: 10.1099/mic.0.046490-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chatfield CH, Mulhern BJ, Viswanathan VK, Cianciotto NP. The major facilitator superfamily-type protein LbtC promotes the utilization of the legiobactin siderophore by Legionella pneumophila. Microbiology. 2012;158:721–735. doi: 10.1099/mic.0.055533-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chatfield CH, Cianciotto NP. The secreted pyomelanin pigment of Legionella pneumophila confers ferric reductase activity. Infect. Immun. 2007;75(8):4062–4070. doi: 10.1128/IAI.00489-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zheng H, Chatfield CH, Liles MR, Cianciotto NP. Secreted pyomelanin of Legionella pneumophila promotes bacterial iron uptake and growth under iron-limiting conditions. Infect. Immun. 2013;81:4182–4191. doi: 10.1128/IAI.00858-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schroeder GN, Petty NK, Mousnier A, et al. The genome of Legionella pneumophila strain 130b contains a unique combination of type IV secretion systems and encodes novel Dot/Icm secretion system effector proteins. J. Bacteriol. 2010;192:6001–6016. doi: 10.1128/JB.00778-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cazalet C, Rusniok C, Bruggemann H, et al. Evidence in the Legionella pneumophila genome for exploitation of host cell functions and high genome plasticity. Nat. Genet. 2004;36(11):1165–1173. doi: 10.1038/ng1447. [DOI] [PubMed] [Google Scholar]
- 25.Chien M, Morozova I, Shi S, et al. The genomic sequence of the accidental pathogen Legionella pneumophila. Science. 2004;305(5692):1966–1968. doi: 10.1126/science.1099776. [DOI] [PubMed] [Google Scholar]
- 26.National Center for Biotechnology Information (NCBI) www.ncbi.nlm.nih.gov.
- 27.LegioList http://genolist.pasteur.fr/LegioList.
- 28.Ramakrishnan G, Sen B, Johnson R. Paralogous outer membrane proteins mediate uptake of different forms of iron and synergistically govern virulence in Francisella tularensis tularensis. J. Biol. Chem. 2012;287(30):25191–25202. doi: 10.1074/jbc.M112.371856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lloubes R, Goemaere E, Zhang X, Cascales E, Duche D. Energetics of colicin import revealed by genetic cross-complementation between the Tol and Ton systems. Biochem. Soc. Trans. 2012;40(6):1480–1485. doi: 10.1042/BST20120181. [DOI] [PubMed] [Google Scholar]
- 30.Chu BC, Peacock RS, Vogel HJ. Bioinformatic analysis of the TonB protein family. Biometals. 2007;20(3–4):467–483. doi: 10.1007/s10534-006-9049-4. [DOI] [PubMed] [Google Scholar]
- 31.Rees DC, Johnson E, Lewinson O. ABC transporters: the power to change. Nat. Rev. Mol. Cell. Biol. 2009;10(3):218–227. doi: 10.1038/nrm2646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cuiv PO, Clarke P, Lynch D, O’Connell M. Identification of rhtX and fptX, novel genes encoding proteins that show homology and function in the utilization of the siderophores rhizobactin 1021 by Sinorhizobium meliloti and pyochelin by Pseudomonas aeruginosa, respectively. J. Bacteriol. 2004;186(10):2996–3005. doi: 10.1128/JB.186.10.2996-3005.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Michel L, Bachelard A, Reimmann C. Ferripyochelin uptake genes are involved in pyochelin-mediated signalling in Pseudomonas aeruginosa. Microbiology. 2007;153(5):1508–1518. doi: 10.1099/mic.0.2006/002915-0. [DOI] [PubMed] [Google Scholar]
- 34.Kiss K, Liu W, Huntley JF, Norgard MV, Hansen EJ. Characterization of fig operon mutants of Francisella novicida U112. FEMS Microbiol. Lett. 2008;285(2):270–277. doi: 10.1111/j.1574-6968.2008.01237.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Law CJ, Maloney PC, Wang DN. Ins and outs of major facilitator superfamily antiporters. Annu. Rev. Microbiol. 2008;62:289–305. doi: 10.1146/annurev.micro.61.080706.093329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Viswanathan VK, Kurtz S, Pedersen LL, et al. The cytochrome c maturation locus of Legionella pneumophila promotes iron assimilation and intracellular infection and contains a strain-specific insertion sequence element. Infect. Immun. 2002;70(4):1842–1852. doi: 10.1128/IAI.70.4.1842-1852.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Naylor J, Cianciotto NP. Cytochrome c maturation proteins are critical for in vivo growth of Legionella pneumophila. FEMS Microbiol. Lett. 2004;241(2):249–256. doi: 10.1016/j.femsle.2004.10.028. [DOI] [PubMed] [Google Scholar]
- 38.Cianciotto NP, Cornelis P, Baysse C. Impact of the bacterial type I cytochrome c maturation system on different biological processes. Mol. Microbiol. 2005;56(6):1408–1415. doi: 10.1111/j.1365-2958.2005.04650.x. [DOI] [PubMed] [Google Scholar]
- 39.Sanders C, Turkarslan S, Lee DW, Daldal F. Cytochrome c biogenesis: the Ccm system. Trends Microbiol. 2010;18(6):266–274. doi: 10.1016/j.tim.2010.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Baert B, Baysse C, Matthijs S, Cornelis P. Multiple phenotypic alterations caused by a c-type cytochrome maturation ccmC gene mutation in Pseudomonas aeruginosa. Microbiology. 2008;154(1):127–138. doi: 10.1099/mic.0.2007/008268-0. [DOI] [PubMed] [Google Scholar]
- 41.Yeterian E, Martin LW, Guillon L, Journet L, Lamont IL, Schalk IJ. Synthesis of the siderophore pyoverdine in Pseudomonas aeruginosa involves a periplasmic maturation. Amino Acids. 2010;38(5):1447–1459. doi: 10.1007/s00726-009-0358-0. [DOI] [PubMed] [Google Scholar]
- 42.Debroy S, Dao J, Soderberg M, Rossier O, Cianciotto NP. Legionella pneumophila type II secretome reveals unique exoproteins and a chitinase that promotes bacterial persistence in the lung. Proc. Natl acad. Sci. USA. 2006;103(50):19146–19151. doi: 10.1073/pnas.0608279103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Klein TW, Yamamoto Y, Brown HK, Friedman H. Interferon-gamma induced resistance to Legionella pneumophila in susceptible A/J mouse macrophages. J. Leuk. Biol. 1991;49(1):98–103. doi: 10.1002/jlb.49.1.98. [DOI] [PubMed] [Google Scholar]
- 44.Fischbach MA, Lin H, Liu DR, Walsh CT. How pathogenic bacteria evade mammalian sabotage in the battle for iron. Nat. Chem. Biol. 2006;2(3):132–138. doi: 10.1038/nchembio771. [DOI] [PubMed] [Google Scholar]
- 45.Viswanathan VK, Edelstein PH, Pope CD, Cianciotto NP. The Legionella pneumophila iraAB locus is required for iron assimilation, intracellular infection, and virulence. Infect. Immun. 2000;68:1069–1079. doi: 10.1128/iai.68.3.1069-1079.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.O’Connell WA, Hickey EK, Cianciotto NP. A Legionella pneumophila gene that promotes hemin binding. Infect. Immun. 1996;64:842–848. doi: 10.1128/iai.64.3.842-848.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hickey EK, Cianciotto NP. An iron- and fur-repressed Legionella pneumophila gene that promotes intracellular infection and encodes a protein with similarity to the Escherichia coli aerobactin synthetases. Infect. Immun. 1997;65:133–143. doi: 10.1128/iai.65.1.133-143.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Beare PA, For RJ, Martin LW, Lamont IL. Siderophore-mediated cell signalling in Pseudomonas aeruginosa: divergent pathways regulate virulence factor production and siderophore receptor synthesis. Mol. Microbiol. 2003;47(1):195–207. doi: 10.1046/j.1365-2958.2003.03288.x. [DOI] [PubMed] [Google Scholar]
- 49.Lamont IL, Beare PA, Ochsner U, Vasil AI, Vasil ML. Siderophore-mediated signaling regulates virulence factor production in Pseudomonas aeruginosa. Proc. Natl Acad. Sci. USA. 2002;99(10):7072–7077. doi: 10.1073/pnas.092016999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bredenbruch F, Geffers R, Nimtz M, Buer J, Haussler S. The Pseudomonas aeruginosa quinolone signal (PQS) has an iron-chelating activity. Environ. Microbiol. 2006;8(8):1318–1329. doi: 10.1111/j.1462-2920.2006.01025.x. [DOI] [PubMed] [Google Scholar]
- 51.Britigan BE, Rasmussen GT, Cox CD. Augmentation of oxidant injury to human pulmonary epithelial cells by the Pseudomonas aeruginosa siderophore pyochelin. Infect. Immun. 1997;65(3):1071–1076. doi: 10.1128/iai.65.3.1071-1076.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Baine WB, Rasheed JK. Aromatic substrate specificity of browning by cultures of the Legionnaires’ disease bacterium. Ann. Int. Med. 1979;90(4):619–620. doi: 10.7326/0003-4819-90-4-619. [DOI] [PubMed] [Google Scholar]
- 53.Steinert M, Flugel M, Schuppler M, et al. The Lly protein is essential for p-hydroxyphenylpyruvate dioxygenase activity in Legionella pneumophila. FEMS Microbiol. Lett. 2001;203(1):41–47. doi: 10.1111/j.1574-6968.2001.tb10818.x. [DOI] [PubMed] [Google Scholar]
- 54.Flydal MI, Chatfield CH, Zheng H, et al. Phenylalanine hydroxylase from Legionella pneumophila is a thermostable enzyme with a major functional role in pyomelanin synthesis. PLoS ONE. 2012;7(9):e46209. doi: 10.1371/journal.pone.0046209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Steinert M, Engelhard H, Flugel M, Wintermeyer E, Hacker J. The Lly protein protects Legionella pneumophila from light but does not directly influence its intracellular survival in Hartmannella vermiformis. Appl. Environ. Microbiol. 1995;61(6):2428–2430. doi: 10.1128/aem.61.6.2428-2430.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hunter RC, Newman DK. A putative ABC transporter, HatABCDE, is among molecular determinants of pyomelanin production in Pseudomonas aeruginosa. J. Bacteriol. 2010;192(22):5962–5971. doi: 10.1128/JB.01021-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Valeru SP, Rompikuntal PK, Ishikawa T, et al. A role of melanin pigment in expression of Vibrio cholerae virulence factors. Infect. Immun. 2009;77(3):935–942. doi: 10.1128/IAI.00929-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang Y, Wilks JC, Danhorn T, Ramos I, Croal L, Newman DK. Phenazine-1-carboxylic acid promotes bacterial biofilm development via ferrous iron acquisition. J. Bacteriol. 2011;193(14):3606–3617. doi: 10.1128/JB.00396-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Grass G. Iron transport in Escherichia coli: all has not been said and done. Biometals. 2006;19(2):159–172. doi: 10.1007/s10534-005-4341-2. [DOI] [PubMed] [Google Scholar]
- 60.Poch MT, Johnson W. Ferric reductases of Legionella pneumophila. Biometals. 1993;6:107–114. doi: 10.1007/BF00140111. [DOI] [PubMed] [Google Scholar]
- 61.James BW, Mauchline WS, Dennis PJ, Keevil CW. A study of iron acquisition mechanisms of Legionella pneumophila grown in chemostat culture. Curr. Microbiol. 1997;34:238–243. doi: 10.1007/s002849900176. [DOI] [PubMed] [Google Scholar]
- 62.Greenwald J, Hoegy F, Nader M, et al. Real time fluorescent resonance energy transfer visualization of ferric pyoverdine uptake in Pseudomonas aeruginosa. A role for ferrous iron. J. Biol. Chem. 2007;282(5):2987–2995. doi: 10.1074/jbc.M609238200. [DOI] [PubMed] [Google Scholar]
- 63.Miethke M, Marahiel MA. Siderophore-based iron acquisition and pathogen control. Microbiol. Mol. Biol. Rev. 2007;71(3):413–451. doi: 10.1128/MMBR.00012-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cartron ML, Maddocks S, Gillingham P, Craven CJ, Andrews SC. Feo--transport of ferrous iron into bacteria. Biometals. 2006;19(2):143–157. doi: 10.1007/s10534-006-0003-2. [DOI] [PubMed] [Google Scholar]
- 65.Huston WM, Jennings MP, Mcewan AG. The multicopper oxidase of Pseudomonas aeruginosa is a ferroxidase with a central role in iron acquisition. Mol. Microbiol. 2002;45(6):1741–1750. doi: 10.1046/j.1365-2958.2002.03132.x. [DOI] [PubMed] [Google Scholar]
- 66.Huston WM, Naylor J, Cianciotto NP, Jennings MP, Mcewan AG. Functional analysis of the multi-copper oxidase from Legionella pneumophila. Microbes Infect. 2008;10(5):497–503. doi: 10.1016/j.micinf.2008.01.011. [DOI] [PubMed] [Google Scholar]
- 67.Portier E, Zheng H, Sahr T, et al. IroT/mavN, a new iron-regulated gene involved in Legionella pneumophila virulence against amoebae and macrophages. Environ. Microbiol. 2014;17(4):1338–1350. doi: 10.1111/1462-2920.12604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rossier O, Cianciotto NP. The Legionella pneumophila tatB gene facilitates secretion of phospholipase C, growth under iron-limiting conditions, and intracellular infection. Infect. Immun. 2005;73(4):2020–2032. doi: 10.1128/IAI.73.4.2020-2032.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cianciotto NP. Type II secretion and Legionella virulence. Curr. Top. Microbiol. Immunol. 2013;376:81–102. doi: 10.1007/82_2013_339. [DOI] [PubMed] [Google Scholar]
- 70.Hickey EK, Cianciotto NP. Cloning and sequencing of the Legionella pneumophila fur gene. Gene. 1994;143:117–121. doi: 10.1016/0378-1119(94)90615-7. [DOI] [PubMed] [Google Scholar]
- 71.Pope CD, O’Connell W, Cianciotto NP. Legionella pneumophila mutants that are defective for iron acquisition and assimilation and intracellular infection. Infect. Immun. 1996;64:629–636. doi: 10.1128/iai.64.2.629-636.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bruggemann H, Hagman A, Jules M, et al. Virulence strategies for infecting phagocytes deduced from the in vivo transcriptional program of Legionella pneumophila. Cell. Microbiol. 2006;8(8):1228–1240. doi: 10.1111/j.1462-5822.2006.00703.x. [DOI] [PubMed] [Google Scholar]
- 73.Faucher SP, Mueller CA, Shuman HA. Legionella pneumophila transcriptome during intracellular multiplication in human macrophages. Front. Microbiol. 2011;2:215158. doi: 10.3389/fmicb.2011.00060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Starkenburg SR, Casey JM, Cianciotto NP. Siderophore activity among members of the Legionella genus. Curr. Microbiol. 2004;49:203–207. doi: 10.1007/s00284-004-4342-3. [DOI] [PubMed] [Google Scholar]
- 75.Braun V, Pramanik A, Gwinner T, Koberle M, Bohn E. Sideromycins: tools and antibiotics. Biometals. 2009;22(1):3–13. doi: 10.1007/s10534-008-9199-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Stirrett KL, Ferreras JA, Jayaprakash V, Sinha BN, Ren T, Quadri LE. Small molecules with structural similarities to siderophores as novel antimicrobials against Mycobacterium tuberculosis and Yersinia pestis. Bioorg. Med. Chem. Lett. 2008;18(8):2662–2668. doi: 10.1016/j.bmcl.2008.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Quadri LE. Strategic paradigm shifts in the antimicrobial drug discovery process of the 21st century. Infect. Disord. Drug Targets. 2007;7(3):230–237. doi: 10.2174/187152607782110040. [DOI] [PubMed] [Google Scholar]
- 78.Alteri CJ, Hagan EC, Sivick KE, Smith SN, Mobley HLT. Mucosal immunization with iron receptor antigens protects against urinary tract infection. PLoS Pathog. 2009;5:e1000586. doi: 10.1371/journal.ppat.1000586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Balado M, Osorio CR, Lemos ML. FvtA is the receptor for the siderophore vanchrobactin in Vibrio anguillarum: utility as a route of entry for vanchrobactin analogues. Appl. Environ. Microbiol. 2009;75(9):2775–2783. doi: 10.1128/AEM.02897-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Frederick RE, Mayfield JA, Dubois JL. Iron trafficking as an antimicrobial target. Biometals. 2009;22(4):583–593. doi: 10.1007/s10534-009-9236-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mollmann U, Heinisch L, Bauernfeind A, Kohler T, Ankel-Fuchs D. Siderophores as drug delivery agents: application of the “Trojan Horse” strategy. Biometals. 2009;22(4):615–624. doi: 10.1007/s10534-009-9219-2. [DOI] [PubMed] [Google Scholar]