Abstract
Malignant gliomas are the most common adult brain cancers. In spite of aggressive treatment, recurrence occurs in the great majority of patients and is invariably fatal. Polyunsaturated fatty acids are abundant in brain, particularly ω-6 arachidonic acid (AA) and ω-3 docosahexaenoic acid (DHA). Although the levels of ω-6 and ω-3 polyunsaturated fatty acids are tightly regulated in brain, the ω-6:ω-3 ratio is dramatically increased in malignant glioma, suggesting deregulation of fundamental lipid homeostasis in brain tumor tissue. The migratory properties of malignant glioma cells can be modified by altering the ratio of AA:DHA in growth medium, with increased migration observed in AA-rich medium. This fatty acid-dependent effect on cell migration is dependent on expression of the brain fatty acid binding protein (FABP7) previously shown to bind DHA and AA. Increased levels of enzymes involved in eicosanoid production in FABP7-positive malignant glioma cells suggest that FABP7 is an important modulator of AA metabolism. We provide evidence that increased production of eicosanoids in FABP7-positive malignant glioma growing in an AA-rich environment contributes to tumor infiltration in the brain. We discuss pathways and molecules that may underlie FABP7/AA-mediated promotion of cell migration and FABP7/DHA-mediated inhibition of cell migration in malignant glioma.
Keywords: astrocytoma, arachidonic acid, docosahexaenoic acid, fatty acid binding protein, eicosanoids, lipid metabolism, malignant glioma
1. Introduction
Gliomas are tumors that arise from glial or glial progenitor cells in the central nervous system. These tumors most commonly occur in the brain. A subtype of glioma, called astrocytoma, is characterized by expression of glial fibrillary acidic protein (GFAP), normally found in astrocytes. Anaplastic astrocytoma (grade III astrocytoma) and glioblastoma multiforme (grade IV astrocytoma) are collectively called high-grade astrocytomas or malignant gliomas. Malignant gliomas are the most common cancers of the central nervous system, accounting for ~70% of malignant primary brain tumors [1]. The prognosis of patients with malignant glioma is dismal with median survival times of 3 years and 15 months for patients with anaplastic astrocytoma and glioblastoma multiforme, respectively [2]. Although patients with grade II astrocytomas have a better prognosis, >35% of these tumors recur as high grade astrocytomas, further compounding the challenges associated with the treatment of astrocytoma tumors [3]. Surgical resection and adjuvant radiotherapy followed by chemotherapeutic agents such as temozolomide or nitrosourea is the standard treatment for malignant gliomas. In spite of this aggressive treatment, almost all (>90%) malignant gliomas recur, most commonly within 3 cm from the margin of the original tumor, suggesting that recurrence is due to infiltrative rather than invasive properties of the tumor cells [4–5]. Survival time is very short once recurrence has been diagnosed, usually 3–6 months [4].
Treatment options for recurring malignant glioma are limited because of toxicity and detrimental effects on brain function. For example, radical resection often cannot be considered for the treatment of recurrent tumors because of the associated decline in brain function, as measured by Karnofsky Performance Status, and/or surgery-related morbidity and infection [4, 6–7]. A combination of chemotherapy or stereotactic radiosurgery with repeated surgery was shown to improve survival of patients with recurrent glioblastoma compared to surgery alone, although none of the patients in this study survived beyond 44 weeks after treatment [8]. Furthermore, the use of radiotherapy is limited in recurrent tumors because of associated irreversible brain tissue damage and radiation-induced necrosis of normal brain [9]. The recommendation to wait at least 6 months before initiating a second round of radiation treatment further limits this treatment option [4]. Despite the above limitations, the standard treatment for recurrent malignant glioma is still a combination of radiotherapy and chemotherapy [10], highlighting the need of finding new therapeutic strategies that will limit or prevent tumor infiltration and recurrence.
There are reports indicating that lipid metabolism is deregulated in malignant glioma and that altered lipid metabolism is associated with a worse prognosis in these tumors [11–12]. Brain fatty acid-binding protein (B-FABP or FABP7), involved in the intracellular transport of polyunsaturated fatty acids (PUFA), is up-regulated in glioblastoma compared to normal brain tissue and low grade astrocytomas [13–14]. Furthermore, elevated levels of FABP7 in the nucleus are associated with a worse prognosis in glioblastoma [15–16]. We and others have shown that expression of FABP7 in malignant glioma cell lines increases cell motility and migration [13, 17]. Importantly, altering the DHA:AA ratio in the culture medium affects cell migration in a FABP7-specific manner, with an increased DHA:AA ratio associated with reduced cell migration [18]. In this review, we discuss how alterations in the lipid environment together with FABP7 expression may affect malignant glioma tumor growth. We propose that a better understanding of the consequences of lipid alterations in malignant glioma may shed light on the mechanisms driving tumor recurrence thereby revealing new approaches for the treatment of malignant glioma.
2. Normal brain lipid composition
Lipids constitute ~2% of the total cell mass in most organs. However, in the brain, lipids are major structural components with fatty acids making up about 50% of the total mass of neural membranes [19–20]. Long chain PUFA such as docosahexaenoic acid (DHA, C22:6, ω-3) and arachidonic acid (AA, C20:4, ω-6) are abundant in brain, constituting close to 20% of the dry weight of the brain, including 6% for AA and 8% for DHA [20]. The fatty acid composition of the three major types of brain cells (neurons, oligodendrocytes and astrocytes) has been reported in rats [21]. In 60-day old rats fed a soya oil diet, ω-3 and ω-6 fatty acids constitute ~30%, ~20% and ~29% of the total neuron, oligodendrocyte, and astrocyte lipid content, respectively, including 8%, 5% and 12% for DHA and 10%, 9% and 10% for AA [21].
Although there is no consensus on how fatty acids are taken up by brain, there is evidence that the unesterified fatty acid (albumin-bound) pool in plasma is a major contributor to the fatty acid pool in brain, at least in the case of AA and DHA [22]. The importance of low-density lipoproteins (LDL) and very low density lipoproteins (VLDL) in brain PUFA uptake was assessed using mice deficient for the LDL receptor (LDLr) or VLDL receptor (VLDLr); however, no differences in PUFA levels were detected between knockout mice and the wild type controls, suggesting that LDL and VLDL do not play a major role in PUFA uptake in the brain [23–24]. Fatty acids have also been postulated to enter the brain by passive diffusion and protein-mediated transport by membrane-associated proteins, such as fatty acid transport proteins and fatty acid translocases (CD36) [22]. Inside the cell, long chain fatty acids are transported by a group of intracellular lipid binding proteins called fatty acid binding proteins (FABPs) which are expressed in most tissues [25–34] (Table 1).
Table 1.
Names of different fatty acid binding proteins (FABPs) and the tissues from which they were first isolated.
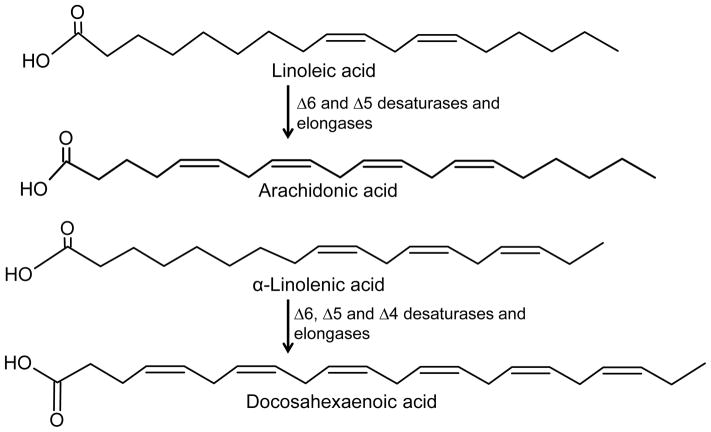
It is well established that the essential fatty acids, cis-linoleic acid (LA, 18:2, ω-6) and α-linolenic acid (ALA, 18:3, ω-3), the precursors of AA and DHA, respectively, have to be obtained from the diet because our bodies cannot synthesize them [20, 35]. In the liver, LA is converted to gamma-linolenic acid (GLA, 18:3, ω-6), dihomo-GLA (DGLA, 20:3, ω-6), and AA by different desaturases and elongases (Fig. 1) [35]. Similarly, ALA is converted to eicosapentaenoic acid (EPA, 20:5, ω-3) and DHA via the same combination of enzymes (Fig. 1). Both liver and diet are important determinants of brain DHA and AA composition. Studies done by Rapoport et al. (2007), who used positron emitting tomography to trace intravenously injected radiolabelled AA and DHA, showed that the rates of uptake of AA and DHA by human healthy brain were 17.8 mg/day/1.5 kg brain and 4.6 mg/day/1.5 kg, respectively [36]. Furthermore, incorporation rates of AA and DHA in the brain were approximated by their rates of loss from the brain [36]. The fact that the brain has little if any PUFA synthesis capacity, combined with the specific up-regulation of liver elongases and desaturases in animals fed a ω-3 deficient diet, led to the conclusion that the liver must play an important role in supplying the brain with AA and DHA [36–37].
Fig. 1.
Outline of the conversion of linoleic acid and α-linolenic acid into arachidonic acid and docosahexaenoic acid, respectively by a combination of desaturases and elongases. Note that conversions of linoleic acid to gamma-linolenic acid, dihomo-gammalinolenic acid and α-linolenic acid to eicosapentaenoic acid are not shown.
3. Malignant glioma: lipid metabolism imbalance
Similar to other types of cancers, fatty acid uptake and lipid metabolism is deregulated in malignant glioma [38–39]. Indeed, a recent report indicates marked metabolic differences between low and high grade gliomas with metabolomic differences serving as a reasonably accurate (71%) predictive tool for high versus low grade glioma [11]. Of note, accumulation of long chain fatty acid metabolites was consistently associated with the worst survival in malignant glioma patients [11]. These findings are in agreement with earlier studies showing that the lipid environment is altered in malignant glioma. Specifically, DHA levels were reduced by ~50% in malignant glioma samples compared to normal brain tissue [12, 40–41]. This reduction was evident whether total lipids or total phospholipids were analyzed. Upon testing the different phospholipid classes, DHA reduction was observed in phosphatidylethanolamine and phosphatidylserine fractions. The decrease in DHA observed in malignant glioma was accompanied by unchanged AA levels and an ~4-fold increase in LA, resulting in an overall increase in ω-6/ω-3 fatty acid ratio compared to normal brain (Fig. 2) [12]. Although fatty acid composition of phospholipids from different grades of astrocytomas has not been explored in depth, a study published by Albert et al. demonstrates that decreased DHA content in the phosphatidylethanolamine fraction is greater in grade IV astrocytomas (n=6) than in a grade II astrocytoma (n=1) [40].
Fig. 2.
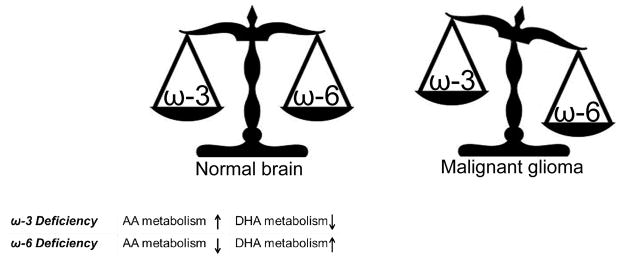
Diagrammatic representation of the balance between ω-3 and ω-6 fatty acids in normal brain and the deregulation of this balance in malignant glioma. A tightly controlled AA:DHA ratio is maintained in normal brain, with ω-3 deficiency and ω-6 deficiency resulting in enhanced AA/decreased DHA metabolism and enhanced DHA/decreased AA metabolism, respectively. This ratio is deregulated in malignant glioma tumors, with decreased DHA content and increased linoleic acid resulting in a significantly decreased ω-3:ω-6 ratio.
Using rat as a model, Igarashi et al. (2007) showed that in cases of DHA dietary deficiency, normal brain DHA content was maintained via liver conversion of ALA to DHA, provided that sufficient ALA was present in the diet [42]. In cases of ω-3 PUFA deficiency (greatly reduced ALA, no DHA) for 15 weeks, brain DHA content was reduced by 37% [37, 43]. To maintain brain DHA content, brain DHA metabolism, as measured by iPLA2 and COX-1, was reduced in ω-3 PUFA-deficient rats. In contrast, AA metabolism was up-regulated as the result of increased activity of AA-selective cytosolic phospholipase A2 (cPLA2), secretory PLA2 (sPLA2) and COX-2 [36]. These data suggest that there is a tight balance between brain AA and DHA turnover that serves to maintain the relative content of these two fatty acids at a favorable ratio (Fig. 2). In support of this idea, deprivation of ω-6 PUFAs was shown to increase brain DHA metabolism while maintaining AA levels by down-regulating enzymes involved in its metabolism [44]. Specifically, ω-6 PUFA deprivation resulted in up-regulation of the DHA selective intracellular phospholipase A2 (iPLA2 VIA) and 15-lipoxygenase (15-LOX). In a separate study, SREBP-1, a transcription factor involved in iPLA2 regulation, was also shown to be up-regulated upon deprivation of ω-6 PUFAs [45]. Predictably, expression of proteins responsible for AA-metabolism such as cPLA2 IVA and COX-2 were significantly reduced in ω-6-deprived rats, along with the transcription factors AP-2α and NF-κB p65 which are involved in the regulation of COX-2 and cPLA2 IVA [45]. Collectively, these data indicate that brain turnover of AA and DHA is closely linked and maintenance of a favorable or homeostatic DHA:AA ratio is critical for normal brain function.
An increase in the AA:DHA ratio has been consistently reported in malignant glioma (Fig. 2) [12, 40–41] suggesting that tumorigenicity is associated with enhanced DHA loss/metabolism, to the extent that brain DHA loss cannot be compensated for by liver or diet. The mechanisms driving DHA loss while maintaining AA levels and increasing LA content are not clear. However, recent studies have shed some light on the inhibitory effects of DHA on pro-inflammatory pathways such as COX-2, which is believed to be important for cancer proliferation and invasion [46–47]. Therefore, decreased DHA levels may be important for tumor survival and infiltration. As brain fatty acid-binding protein (FABP7) is negatively correlated with survival in glioblastoma patients and binds AA and DHA depending on their relative availability (although it has a higher affinity for DHA), it is reasonable to postulate that this lipid metabolic imbalance is linked to FABP7 overexpression.
4. FABPs, their fatty acid ligands and interacting partners
FABPs are intracellular fatty acid binding proteins involved in the binding and the intracellular trafficking of fatty acids and retinoids to different subcellular compartments: cytoplasmic, for metabolism and energy production, or nuclear, for regulation of gene transcription via activation of peroxisome proliferator-activated receptors (PPARs) [48]. Studies indicate that FABPs may serve as major determinants of the pharmacokinetics and metabolism of fatty acids and their metabolites. For example, FABP1 (L-FABP) (Table 1) expression strongly correlates with hepatic uptake of palmitate and the production of palmitic acid metabolites [49].
Different FABP genes are expressed at different stages of brain development, each with demonstrated binding preference for certain fatty acids [50]. For example, FABP3 (or H-FABP) is expressed after birth with its levels gradually increasing until adulthood [51]. FABP3 preferentially binds ω-6 PUFA [52]. FABP5 (E-FABP) is expressed in mid-term embryonic rat brain and reaches its peak at birth, then gradually decreases in postnatal life [51]. FABP5 preferentially binds saturated fatty acids [53]. FABP7 is expressed mainly in radial glial cells at early stages of brain development [51, 54–55]. Based on Isothermal Titration Calorimetry, FABP7 binds DHA and AA with dissociation constants of (Kd) of 53 nM and 207 nM, respectively [56], indicating that a preferred ligand for FABP7 is DHA. The expression of FABP7 decreases dramatically in neonatal and adult brain [51]. When mixed cultures of rodent glial cells and neurons were incubated with an anti-FABP7 antibody, inhibition of glial fiber formation and neuronal migration was observed, indicating a role for B-FABP in the establishment of the radial glial fiber system [55]. FABP7 has garnered attention because of its association with decreased survival in glioblastoma patients [13, 15–16, 57]. Another FABP, FABP4, has recently been shown to be preferentially expressed in grade IV astrocytomas compared to low grade astrocytomas and normal brain tissue [58]. FABP4 does not co-localize with FABP7 in astrocytoma tumors but rather is expressed in endothelial cells and other yet-to-be-identified cell types [58]. Like FABP7, FABP4 binds AA with high affinity (Kd of ~250 nM) [59–60]. The role of FABP4 in astrocytoma tumors remains to be elucidated.
PPARs, especially PPARα, have been shown to interact with and regulate FABPs. Specifically, FABP1, FABP2 and FABP3 were induced in liver, intestine and heart, respectively, in response to treatment with the PPARα agonist, Wy14643 [61]. The authors showed that interaction with PPAR response elements (PPRE) was required for transcriptional up-regulation of FABP1 by Wy14643 in the liver [61]. Using recombinant proteins and Fluorescence Resonance Energy Transfer (FRET), PPARα was shown to bind FABP1 with high affinity (Kd 56.5 nM) and at close proximity, with an average intermolecular distance of 52 Å [62]. Furthermore, double immunogold labeling electron microscopy revealed significant co-localization of FABP1 and PPARα in both the nuclei and cytoplasm of hepatocytes from wild-type FABP1+/+ mice [62]. These combined data suggest that PPARs and FABPs interact and that FABP expression is regulated through this interaction.
5. Brain fatty acid binding protein
5.1. FABP7 and its transcriptional regulators
PPAR ligands have been implicated in FABP7 induction during zebrafish development, with FABP7 levels significantly induced in the liver and intestine, but not the brain, of zebrafish treated with clofibrate, a specific ligand for PPARα and to a lesser extent for PPARγ [48, 63]. There is evidence suggesting that natural PPAR ligands such as fatty acids can also affect FABP7 expression. Nasrollahzadeh et al. used a rat C6 glioma model to demonstrate specific up-regulation of FABP7 gene expression in tumor cells when rats were fed a DHA oil-rich diet compared to linoleic acid-rich safflower oil diet [64]. In contrast to DHA, supplementation with the ω-6 fatty acid, γ-linolenic acid, had no effect on the expression of FABP7.
Several transcription factors other than PPARs have been implicated in FABP7 regulation. Among these are the Nuclear factor I (NFI) transcription factors, with NFIA/B/X up-regulating FABP7, and NFIC down-regulating FABP7 in malignant glioma cell lines [65]. PAX6, a transcription factor of the paired box and homeobox family, also up-regulates FABP7 in malignant glioma [66]. The importance of PAX6 in FABP7 regulation is still under investigation as most FABP7 positive malignant glioma cell lines and tumors express little or no PAX6 [66]. In fact, PAX6 is expressed at significantly lower levels in glioblastoma compared to anaplastic astrocytoma tumors, with the latter having PAX6 levels that are equivalent to those found in normal tissue [67].
Examination of 274 astrocytoma specimens has revealed a correlation between Notch1 expression, tumor grade and survival that is similar to that observed for FABP7 [13, 15, 68]. As Notch1 signaling has been implicated in the regulation of FABP7 in radial glial cells [69–70], it is conceivable that Notch1 may be involved in the up-regulation of FABP7 in astroctyoma tumors. Notch1 ligands, Delta-like-1 and Jagged-1, are also expressed in malignant glioma cell lines and tumor tissues, with levels of Jagged-1 being up-regulated in higher grade gliomas including glioblastoma [69]. Depletion of Notch1, Delta-like-1 and Jagged-1 in U251 malignant glioma cells by specific siRNA transfection resulted in ~90%, 50%, and 25% reduction in Notch1 transcriptional activity, respectively, as measured using the luciferase assay. These combined data demonstrate a potentially important link between Notch1, Notch ligands and FABP7 in malignant glioma. This is important as Jagged-1 has previously been shown to play a role in the activation of the transcription factor Activator Protein 1 (AP-1), which in turn regulates the expression of proteins involved in AA and DHA metabolism such as cyclooxygenase-2 (COX-2) and cytochrome P450 2J2 (CYP2J2) [71–74]. Thus, Notch and/or its ligands may contribute to the deregulation of lipid metabolism in malignant glioma.
5.2. FABP7: link to prognosis, migration and lipid environment
In grade IV astrocytoma, FABP7 is highly expressed at sites of infiltration and surrounding blood vessels [15, 17]. Of note, a negative correlation was found between nuclear FABP7 and survival in glioblastoma patients [15–16]. Borderline (p=0.084) negative association of survival with cytoplasmic FABP7 has also been reported [16]. Furthermore, FABP7 is highly expressed in glioblastoma neurospheres [57, 66], with evidence of radiation-induced up-regulation of FABP7 [57]. Manipulating FABP7 levels by either expressing it in the FABP7-negative malignant glioma cell line U87 or depleting it from the FABP7-positive cell line U251 had a striking effect on malignant glioma growth properties, with FABP7 expression correlating with reduced proliferation and increased cell migration [13, 17–18]. Similar results were observed upon depleting FABP7 in glioblastoma neurospheres [57]. When combined with immunostaining data showing the presence of FABP7 at sites of tumor infiltration in grade IV astrocytomas, these data suggest a role for FABP7 in tumor infiltration and recurrence [17]. Intriguingly, the effect of FABP7 on malignant glioma cell migration was found to be dependent on the fatty acid composition of the culture medium, with a high DHA:AA ratio inhibiting cell migration, and a high AA:DHA ratio promoting cell migration [18]. Transfection of malignant glioma cells with FABP7 mutant expression constructs that either do not bind PUFAs or do not localize to the nucleus revealed that FABP7 bound to AA promotes cell migration even when FABP7 does not localize to the nucleus. In contrast, the inhibition in cell migration observed upon increasing the DHA:AA ratio was only observed when FABP7 localized to the nucleus [18]. FABP7/AA-mediated cell migration was accompanied by up-regulation of COX2 and increased levels of prostaglandin E2 (PGE2), whereas FABP7/DHA-mediated inhibition of cell migration appeared to be at least partially driven by PPARγ [18].
As mentioned earlier, DHA levels are decreased in malignant glioma tumor samples resulting in a higher AA:DHA in tumor compared to normal brain tissue [12, 41]. The up-regulation of FABP7 combined with the higher relative availability of AA may thus favor tumor infiltration. Therefore, it is conceivable that strategies aimed at correcting the DHA:AA ratio may result in the inhibition of tumor cell migration/infiltration and reduce tumor recurrence. The inhibitory effect of DHA on the migration of FABP7-positive malignant glioma cells in vitro lends further support to the idea that it may be possible to control malignant glioma infiltration through manipulation of the lipid environment. As glioblastoma tumor tissues also have elevated levels of the ω-6 LA, it would be worthwhile to investigate the effect of LA on the migration of FABP7-positive and FABP7-negative malignant glioma cells.
5.3. FABP7/fatty acid mechanisms controlling malignant glioma migration
To understand how AA and DHA affect malignant glioma growth, it is important to first understand the roles and utilization of these fatty acids in normal brain. Most AA and DHA are incorporated in the sn-2 position of phospholipids, with AA and DHA released from phospholipids by selective phospholipases (PLA2) upon activation [75–76]. Most of the released AA and DHA undergo reincorporation into available sn-2 positions of lysophospholipids via acyl-CoA synthetase and acyltransferase [36]. Normally this deacylation-reacylation occurs rapidly in association with neurotransmission involving receptors coupled to PLA2. A small fraction of the released AA and DHA is lost via either beta-oxidation or conversion into eicosanoids (for AA), or docosanoids (for DHA) [36]. It is worth mentioning that AA may also be cleaved from 1,2-diacylglycerol (which is cleaved from membrane phospholipids by the action of phospholipase C) through the action of diacylglycerol lipase [77].
Based on gene sequence comparisons, there are nine groups of PLA2 isoforms which are expressed in different species. PLA2 proteins are divided into three main groups based on their basic biochemical properties: (i) cytosolic Ca+2 dependent PLA2 or group IV PLA2 (cPLA2, 85 kDa), (ii) Ca+2-dependent extracellular secretory PLA2 or (groups IB, IIA, IID, IIE, V, and X) (sPLA2, 14 kDa), and (iii) Ca+2-independent intracellular PLA2 or group VI (iPLA2, 85–88 kDa) [78]. Unsaturated fatty acids are highly vulnerable to lipid peroxidation and it is thought that PLA2 preferentially releases peroxidized fatty acids from membranes. If these peroxidized fatty acids were to be retained in the membrane structure, this would lead to disruption of the membrane. PLA2s are therefore critical for the integrity of the membrane structure under normal physiological conditions [75]. Peroxidized fatty acids are then reduced by reaction with glutathione peroxidase and membrane repair is completed by reacylation with long-chain fatty acyl-CoA. Different PLA2 isoforms show different substrate selectivity with cPLA2 and iPLA2 being selective for AA and DHA-containing phospholipids, respectively. sPLA2 has no apparent selectivity requirement in vitro [75, 78–79]. Furthermore, iPLA2 appears to be involved in membrane phospholipid homeostasis and is up-regulated in response to increased phosphatidylcholine synthesis [80–81].
A fraction of the AA and DHA released from phospholipids is metabolized via three pathways, namely cyclooxygenases-2 (COX-2), lipoxygenases (LOX) and cytochrome P450 (CYP) epoxygenases and hydroxylases, thereby generating eicosanoids or docosanoids from AA or DHA, respectively. Eicosanoids include prostaglandins (PGs), leukotrienes, epoxyeicosatrienoic acids (EETs) and hydroxyeicosatrienoic acids (HETEs) [76]. Docosanoids include docosatrienes, protectins and resolvins [82–83].
6. Arachidonic acid-related mechanisms
6.1. Cytosolic phospholipase A2
cPLA2 is of special interest since cPLA2α knockout mice show defects in inflammation and generation of eicosanoids [84–85]. cPLA2α−/− mice have fertility defects and are partially protected in models of brain injury. When brain ischemia-reperfusion injury was induced in cPLA2α−/− mice, the infarct volume in the knockout mice was smaller (by 34%) than that of wild-type mice, with fewer functional neurological deficits observed in cPLA2α−/− compared to cPLA2α+/+ mice [86]. The activity of cPLA2 is regulated by Ca+2 levels and by phosphorylation. cPLA2 is mainly phosphorylated and activated by mitogen-activated protein kinase (MAPK or ERK1/2) which phosphorylates cPLA2 at Ser-505 [87]. Of note, MAPK has been reported to activate AP-1 in rat brain astrocytes through either direct phosphorylation of AP-1 proteins (Jun and Fos families) or phosphorylation of different factors involved in their transcription [88–89]. Importantly, MAPK is believed to be activated by Notch1 in some cancer types such as breast cancer and hematologic malignancies [90–91]. Activation of MAPK and cPLA2 through Notch in FABP7-expressing malignant glioma cells may therefore result in increased AA metabolism, resulting in increased cell migration and infiltration.
AA and its eicosanoid metabolites play important roles in actin remodeling, a process that underlies the shape and structural changes associated with cellular migration and response to inflammation and angiogenesis [92]. In NIH3T3 mouse fibroblasts, the initial transient burst in AA resulting from cPLA2 activation is responsible for cell adhesion, with the AA metabolites leukotrienes and PGs regulating cell spreading and cell migration, respectively [93–95]. Other reports have shown that cPLA2α and its Ser-505-phosphorylated form are rapidly and specifically recruited at sites of actin polymerization such as membrane ruffles and leading edges [92]. A role for cPLA2α in actin remodeling may be highly relevant in the context of FABP7/AA inducing malignant glioma cell migration [17–18], especially in light of the fact that: (i) the increase in FABP7-mediated malignant glioma cell migration is accompanied by increased expression of AA-metabolizing enzymes such as COX-2 and its metabolite PGE2 [18] and (ii) malignant glioma tumor tissues have a readily available supply of AA probably through an increased load of LA [12, 41]. However, studies of the detailed mechanisms underlying these observations and potential involvement of cPLA2 have yet to be carried out. We have used cDNA microarray analysis to compare gene expression in highly migratory U87 FABP7-positive versus non-migratory U87 FABP7-negative cells cultured in an AA-rich environment. Interestingly, cPLA2 IVA (group IV A) was up-regulated by 27-fold in the FABP7-positive cells. These results are in keeping with FABP7 promoting an AA cascade starting at the earliest step of AA release and ending with eicosanoid production.
6.2. Cyclooxygenase
AA is a major substrate for COX (prostaglandin-endoperoxide synthase) enzymes which catalyze its conversion to the key upstream prostanoid precursor prostaglandin H2 (PGH2). There are two main COX isoforms, namely COX-1, which is constitutively expressed and is responsible for production of physiological levels of PGs, and COX-2, which is inducible and up-regulated by different growth factors and cytokines [96]. Transcription factors NF-κB and AP-1 regulate COX-2 transcription in the brain [73, 97] and mitogen-activated protein kinase (MAPK) cascades (ERK1/2, JNK/SAPK, and p38) contribute to its induction [96].
COX-2’s importance in carcinogenesis, progression and survival of patients with different types of cancers is well established [98–100]. Clinical studies designed to assess the efficacy of combined COX-2 inhibitors and chemotherapy versus chemotherapy alone showed potential benefits only in those patients with elevated levels of COX-2 in their tumors [101–102]. A correlation between COX-2 expression and glioma grades has been described, with one report indicating that 71% of glioblastoma tumors had >50% COX-2-positive cells compared to 44% of anaplastic astrocytomas and 30% of low-grade astrocytomas [103–104]. Additionally, a significant correlation was observed between COX-2 expression and survival in glioblastoma patients [103]. We showed that COX-2 and PGE2 levels were increased in FABP7-positive U87 malignant glioma cells [18]. FABP7-positive U87 cells also had increased levels of PPARβ/δ compared to their FABP7-negative counterparts. Interestingly, PPARβ/δ activation was shown to induce COX-2 and PGE2 levels which in turn induced the phosphorylation of cPLA2α, in human cholangiocarcinoma cells [105]. Given the interaction of COX-2 with AA and PPARβ/δ, and the involvement of FABP7 with AA and PPARβ/δ in malignant glioma [18], it is possible that FABP7 is playing an important role in regulating COX-2 activity through PPARβ/δ activation.
6.3. Cytochrome P450
Recent studies indicate that some CYP-mediated metabolites of AA possess pro-tumorigenic, pro-angiogenic, pro-migratory and pro-invasive properties in different cancer types [106–109]. For example, the CYP epoxygenase 2J2 transforms AA into four regioisomeric epoxyeicosatrienoic acids (EETs). Overexpression of CYP2J2 or direct addition of EETs to four cancer cell lines derived from liver, lung, and tongue resulted in a 4.5 – 5.5X increase in migration based on the Transwell assay and a 3–3.5X increase in invasion based on the Matrigel assay [108]. Furthermore, MDA-MB-231 human breast carcinoma cells infected with adeno-associated viral vector containing CYP2J2 showed 60% more lung metastases compared with control mice [108]. It is noteworthy that the expression of some members of the CYP family is altered in U87 FABP7-positive versus U87 FABP7-negative malignant glioma cells. CYP2J2 in particular is up-regulated >100X at the RNA level based on our cDNA microarray data.
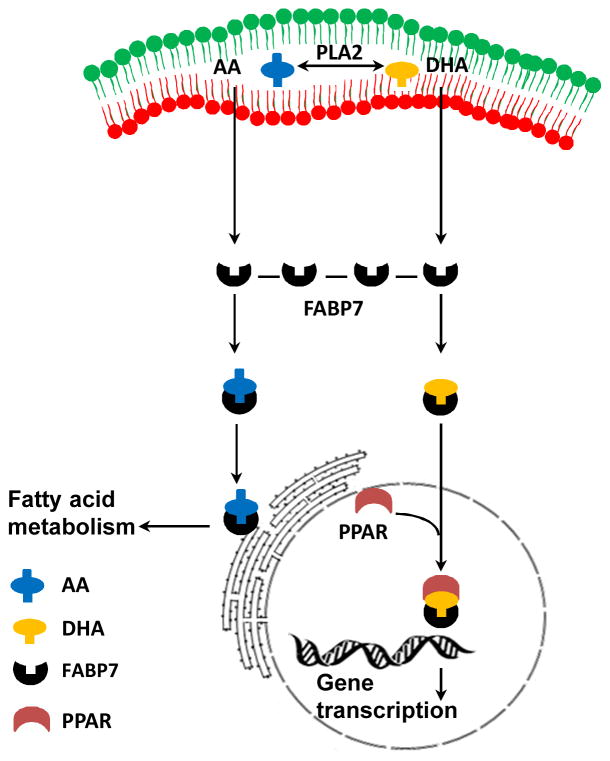
AA metabolism occurs mainly in the cytoplasm, at the luminal surface of the endoplasmic reticulum and on the inner and outer membranes of the nuclear envelope, where COX-2 and different CYP isoforms are expressed [110–111]. As we have shown that nuclear localization of FABP7 is not required, whereas fatty acid binding is essential, for induction of cell migration in U87 cells [18], our results suggest that cytoplasmic FABP7 bound to AA drives the increase in cell migration observed in FABP7-positive U87 cells. We propose that the effect of FABP7 on cell migration is driven by enhanced translocation of AA, mediated by FABP7, to the endoplasmic reticulum as well as the overexpression of AA metabolism enzymes discussed above (Fig. 3). It would be interesting to examine the effect of blocking FABP7 localization in the endoplasmic reticulum on AA-induced migration and eicosanoid production.
Fig. 3.
Diagram depicting the role of FABP7 in the intracellular trafficking of fatty acids. Fatty acids AA or DHA are cleaved from membrane phospholipids depending on which PLA2 isoform is activated. Upon release, AA or DHA binds to FABP7 which then translocates the fatty acid either to the nucleus, where it activates PPARs (for DHA) or to the endoplasmic reticulum for fatty acid metabolism (for AA).
7. Docosahexaenoic acid-related mechanisms
Inhibition of cell migration in U87 B-FABP-positive cells is observed upon increasing the DHA:AA ratio in the culture medium. Our laboratory has shown that the inhibitory effect of DHA on cell migration is partially mediated by PPARγ [18]. A documented natural ligand for PPARγ is DHA [112]. Furthermore, FABP7/DHA-dependent inhibition of migration is dependent on the nuclear localization of FABP7 [18]. These results suggest that DHA-bound-FABP7 is translocated to the nucleus where the complex interacts with and activates the PPARγ transcription factor (Fig. 3). In support of this hypothesis, Adida and Spener (2006) showed interaction between FABP7 and PPARγ in the monkey kidney (COS) cell line [113]. Target genes previously shown to be down-regulated by PPARγ include NF-κB and COX-2, involved in inflammation, and VCAM and ICAM, involved in cell adhesion [47]. Our results indicating that COX-2 is up-regulated and PPARγ is down-regulated, in FABP7-positive U87 cells cultured in an AA-rich environment, are in keeping with COX-2 being down-regulated by PPARγ in DHA-treated malignant glioma [18]. It is worth mentioning that some DHA metabolites such as 17-hydroxy DHA are more potent activators of PPARγ than DHA itself [112, 114] and therefore could contribute to the DHA effect.
Pathways other than PPARγ likely contribute to the inhibition of cell migration observed in FABP7-expressing U87 cells cultured in DHA-rich medium as efficient knock-down of PPARγ does not fully restore cell migration [50]. For example, production of anti-migratory DHA metabolites and inhibition of eicosanoid production, and hence migration, by direct competitive inhibition are possible mechanisms. A report from 30 years ago showed that DHA acts as a potent competitive inhibitor of AA metabolism by COX-2 [115]. More recent reports indicate that DHA also inhibits AA metabolism by CYP epoxygenases and hydroxylases [82]. In brain-metastatic melanoma and in a neuroblastoma cell line [116], DHA competition with AA resulted in decreased PGE2 levels, with COX-2-mediated DHA metabolite PGE3 significantly decreasing invasion in a dose-dependent manner [46]. DHA was also metabolized by recombinant COX-2 to 13-hydroxy DHA which has anti-inflammatory properties and down-regulates cytokine production and neutrophil recruitment to sites of inflammation [117]. Others have shown that metabolites derived from AA-COX and AA-LOX were significantly decreased in brain microvessels upon feeding marine fish oil to weanling rats [118]. In light of a recent report indicating that CYP2J2 is 4-fold more active in DHA metabolism than AA metabolism [82], it will be interesting to assess how DHA treatment of FABP7-positive malignant glioma cells affects CYP2J2 activity. These examples demonstrate the complexity of the interrelationship between the molecules and enzymes that utilize and process AA, DHA and their metabolites. We suggest that upstream regulators of genes encoding enzymes involved in AA and DHA metabolism such as Notch1 and PPARs may coordinate cellular responses depending on levels and availability of AA and DHA (Fig. 4).
Fig. 4.
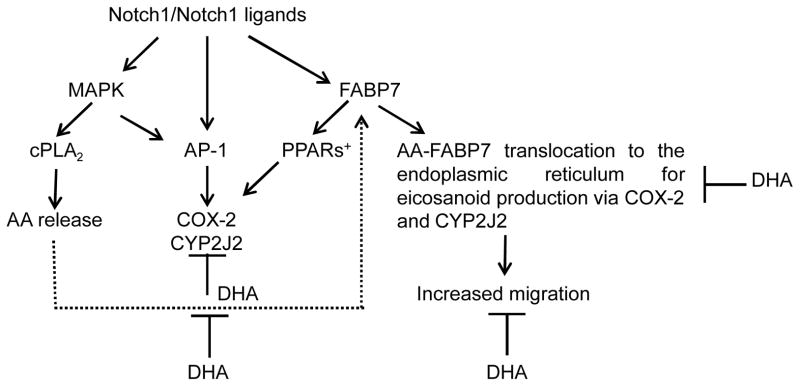
Diagrammatic representation of pathways that may be involved in FABP7-induced malignant glioma cell migration. Pro-migratory and anti-migratory pathways controlled through AA-FABP7 and DHA-FABP7, respectively, are indicated. + PPARβ/δ enhances COX-2 activity whereas PPARγ down-regulates COX-2.
8. Neuroimaging of AA and DHA: potential for astrocytoma detection and grading
The rates of uptake of some fatty acids in brain tumors are different from that of normal brain tissue. For example, Nariai et al. [39] used autoradiogrady to examine the uptake of intravenously-injected radiolabeled [9,10-3H] palmitate, [1-14C] AA and [1-14C] DHA into the lipids of Walker 256 carcinosarcoma cells intracerebrally implanted in rats. The authors showed that brain incorporation rates of these fatty acids was equal to their rates of metabolism by brain and that brain tumors had higher rates of uptake compared to normal brain tissue [39].
More recently, 1-11C-AA and 1-11C-DHA radiotracers have been used to image AA and DHA metabolism in the brain by positron emission tomography (PET) [36, 119–122]. Studies are underway to develop 18F-fluoro tracers, as 18F has a half life of 110 min compared to 20 min for 11C, thus allowing the capture of higher resolution images. While AA and DHA-based radiotracers have not yet been used to image brain tumors, PET imaging using 1-11C-acetate and 2-18F-fluoro-2-deoxy-D-glucose tracers have shown promise for the detection of primary astrocytomas in humans [123]. Furthermore, standardized uptake values that correlated with astrocytoma histological grade were observed with 2-18F-fluoro-2-deoxy-D-glucose [123]. We propose that the altered uptake/turnover of AA and DHA in malignant glioma compared to normal brain tissue could be exploited for the imaging and grading of astrocytomas. High resolution imaging of radiolabeled AA and DHA may also allow detection of small tumor lesions in brain as well as identify multiple sites of tumor growth in recurrent astrocytoma.
9. Conclusion
The PUFA-rich lipid environment of the brain is critical to our understanding of malignant glioma behaviour. Key proteins in AA and DHA metabolism have already been identified in brain, resulting in the production of AA and DHA metabolites with opposing roles in migration and invasion. We propose that FABP7 coordinates the utilization of AA and DHA in the brain and in malignant glioma depending on the relative availability of these two PUFAs. In turn, the amount of AA and DHA available for binding to FABP7 may be determined by specific PLA2‘s which release AA and DHA from phospholipids. Furthermore, PLA2’s and other enzymes involved in AA and DHA metabolism may themselves be regulated through AA-and DHA-dependent pathways (e.g. PPARs). A better understanding of the opposing effects of AA and DHA in malignant glioma may lead to the identification of new therapeutic approaches based on modifying the lipid environment of the tumour.
Acknowledgments
M.E. was supported by a fellowship from the Alberta Cancer Foundation. This work was supported by grants from the Alberta Cancer Foundation and Canadian Institutes of Health Research.
Abbreviations
- AA
arachidonic acid
- ALA
α-linolenic acid
- AP
Activator Protein
- COX
cyclooxygenases
- CYP
cytochrome P450
- DHA
docosahexaenoic acid
- DGLA
dihomo-gammalinolenic acid
- EPA
eicosapentaenoic acid
- FABP7 (B-FABP)
brain fatty acid binding protein
- GFAP
glial fibrillary acidic protein
- GLA
gamma-linolenic acid
- LA
cis-linoleic acid
- LDL
low density lipoprotein
- LOX
lipoxygenase
- MAPK
mitogen-activated protein kinase
- NFI
Nuclear Factor I
- PG
prostaglandin
- PLA2
phospholipase A2
- PPAR
peroxisome proliferator-activated receptor
- PPRE
PPAR response element
- PUFA
polyunsaturated fatty acid
- VLDL
very low density lipoprotein
References
- 1.Wen PY, Kesari S. Malignant gliomas in adults. N Engl J Med. 2008;359:492–507. doi: 10.1056/NEJMra0708126. [DOI] [PubMed] [Google Scholar]
- 2.Chamberlain MC. Emerging clinical principles on the use of bevacizumab for the treatment of malignant gliomas. Cancer. 2010;116:3988–99. doi: 10.1002/cncr.25256. [DOI] [PubMed] [Google Scholar]
- 3.Wessels PH, Weber WE, Raven G, Ramaekers FC, Hopman AH, Twijnstra A. Supratentorial grade II astrocytoma: biological features and clinical course. Lancet Neurol. 2003;2:395–403. doi: 10.1016/s1474-4422(03)00434-4. [DOI] [PubMed] [Google Scholar]
- 4.Niyazi M, Siefert A, Schwarz SB, Ganswindt U, Kreth FW, Tonn JC, et al. Therapeutic options for recurrent malignant glioma. Radiother Oncol. 2011;98:1–14. doi: 10.1016/j.radonc.2010.11.006. [DOI] [PubMed] [Google Scholar]
- 5.Hou LC, Veeravagu A, Hsu AR, Tse VC. Recurrent glioblastoma multiforme: a review of natural history and management options. Neurosurg Focus. 2006;20:E5. doi: 10.3171/foc.2006.20.4.2. [DOI] [PubMed] [Google Scholar]
- 6.Barker FG, 2nd, Chang SM, Gutin PH, Malec MK, McDermott MW, Prados MD, et al. Survival and functional status after resection of recurrent glioblastoma multiforme. Neurosurgery. 1998;42:709–20. doi: 10.1097/00006123-199804000-00013. discussion 20–3. [DOI] [PubMed] [Google Scholar]
- 7.Dirks P, Bernstein M, Muller PJ, Tucker WS. The value of reoperation for recurrent glioblastoma. Can J Surg. 1993;36:271–5. [PubMed] [Google Scholar]
- 8.Mandl ES, Dirven CM, Buis DR, Postma TJ, Vandertop WP. Repeated surgery for glioblastoma multiforme: only in combination with other salvage therapy. Surg Neurol. 2008;69:506–9. doi: 10.1016/j.surneu.2007.03.043. discussion 9. [DOI] [PubMed] [Google Scholar]
- 9.Mayer R, Sminia P. Reirradiation tolerance of the human brain. Int J Radiat Oncol Biol Phys. 2008;70:1350–60. doi: 10.1016/j.ijrobp.2007.08.015. [DOI] [PubMed] [Google Scholar]
- 10.VanderSpek L, Fisher B, Bauman G, Macdonald D. 3D conformal radiotherapy and cisplatin for recurrent malignant glioma. Can J Neurol Sci. 2008;35:57–64. doi: 10.1017/s0317167100007563. [DOI] [PubMed] [Google Scholar]
- 11.Chinnaiyan P, Kensicki E, Bloom G, Prabhu A, Sarcar B, Kahali S, et al. The metabolomic signature of malignant glioma reflects accelerated anabolic metabolism. Cancer Res. 2012;72:5878–88. doi: 10.1158/0008-5472.CAN-12-1572-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martin DD, Robbins ME, Spector AA, Wen BC, Hussey DH. The fatty acid composition of human gliomas differs from that found in nonmalignant brain tissue. Lipids. 1996;31:1283–8. doi: 10.1007/BF02587914. [DOI] [PubMed] [Google Scholar]
- 13.Liang Y, Diehn M, Watson N, Bollen AW, Aldape KD, Nicholas MK, et al. Gene expression profiling reveals molecularly and clinically distinct subtypes of glioblastoma multiforme. Proc Natl Acad Sci U S A. 2005;102:5814–9. doi: 10.1073/pnas.0402870102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tso CL, Shintaku P, Chen J, Liu Q, Liu J, Chen Z, et al. Primary glioblastomas express mesenchymal stem-like properties. Mol Cancer Res. 2006;4:607–19. doi: 10.1158/1541-7786.MCR-06-0005. [DOI] [PubMed] [Google Scholar]
- 15.Liang Y, Bollen AW, Aldape KD, Gupta N. Nuclear FABP7 immunoreactivity is preferentially expressed in infiltrative glioma and is associated with poor prognosis in EGFR-overexpressing glioblastoma. BMC Cancer. 2006;6:97. doi: 10.1186/1471-2407-6-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaloshi G, Mokhtari K, Carpentier C, Taillibert S, Lejeune J, Marie Y, et al. FABP7 expression in glioblastomas: relation to prognosis, invasion and EGFR status. J Neurooncol. 2007;84:245–8. doi: 10.1007/s11060-007-9377-4. [DOI] [PubMed] [Google Scholar]
- 17.Mita R, Coles JE, Glubrecht DD, Sung R, Sun X, Godbout R. B-FABP-expressing radial glial cells: the malignant glioma cell of origin? Neoplasia. 2007;9:734–44. doi: 10.1593/neo.07439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mita R, Beaulieu MJ, Field C, Godbout R. Brain fatty acid-binding protein and omega-3/omega-6 fatty acids: mechanistic insight into malignant glioma cell migration. J Biol Chem. 2010;285:37005–15. doi: 10.1074/jbc.M110.170076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guyton AC, Hall JE. Textbook of medical physiology. Elsevier Saunders; 2006. [Google Scholar]
- 20.Owada Y. Fatty acid binding protein: localization and functional significance in the brain. Tohoku J Exp Med. 2008;214:213–20. doi: 10.1620/tjem.214.213. [DOI] [PubMed] [Google Scholar]
- 21.Bourre JM, Pascal G, Durand G, Masson M, Dumont O, Piciotti M. Alterations in the fatty acid composition of rat brain cells (neurons, astrocytes, and oligodendrocytes) and of subcellular fractions (myelin and synaptosomes) induced by a diet devoid of n-3 fatty acids. J Neurochem. 1984;43:342–8. doi: 10.1111/j.1471-4159.1984.tb00906.x. [DOI] [PubMed] [Google Scholar]
- 22.Chen CT, Green JT, Orr SK, Bazinet RP. Regulation of brain polyunsaturated fatty acid uptake and turnover. Prostaglandins Leukot Essent Fatty Acids. 2008;79:85–91. doi: 10.1016/j.plefa.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 23.Chen CT, Ma DW, Kim JH, Mount HT, Bazinet RP. The low density lipoprotein receptor is not necessary for maintaining mouse brain polyunsaturated fatty acid concentrations. J Lipid Res. 2008;49:147–52. doi: 10.1194/jlr.M700386-JLR200. [DOI] [PubMed] [Google Scholar]
- 24.Rahman T, Taha AY, Song BJ, Orr SK, Liu Z, Chen CT, et al. The very low density lipoprotein receptor is not necessary for maintaining brain polyunsaturated fatty acid concentrations. Prostaglandins Leukot Essent Fatty Acids. 2010;82:141–5. doi: 10.1016/j.plefa.2009.11.003. [DOI] [PubMed] [Google Scholar]
- 25.Ockner RK, Manning JA. Fatty acid-binding protein in small intestine. Identification, isolation, and evidence for its role in cellular fatty acid transport. J Clin Invest. 1974;54:326–38. doi: 10.1172/JCI107768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rustow B, Kunze D, Hodi J, Egger E. A fatty acid binding peptide of rat liver cytosol: characterization and origin. FEBS Lett. 1979;108:469–72. doi: 10.1016/0014-5793(79)80590-6. [DOI] [PubMed] [Google Scholar]
- 27.Unterberg C, Heidl G, von Bassewitz DB, Spener F. Isolation and characterization of the fatty acid binding protein from human heart. J Lipid Res. 1986;27:1287–93. [PubMed] [Google Scholar]
- 28.Haq RU, Christodoulides L, Ketterer B, Shrago E. Characterization and purification of fatty acid-binding protein in rat and human adipose tissue. Biochim Biophys Acta. 1982;713:193–8. doi: 10.1016/0005-2760(82)90236-3. [DOI] [PubMed] [Google Scholar]
- 29.Siegenthaler G, Hotz R, Chatellard-Gruaz D, Didierjean L, Hellman U, Saurat JH. Purification and characterization of the human epidermal fatty acid-binding protein: localization during epidermal cell differentiation in vivo and in vitro. Biochem J. 1994;302(Pt 2):363–71. doi: 10.1042/bj3020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stengelin S, Apel S, Becker W, Maier M, Rosenberger J, Bewersdorf U, et al. The rabbit ileal lipid-binding protein. Gene cloning and functional expression of the recombinant protein. Eur J Biochem. 1996;239:887–96. doi: 10.1111/j.1432-1033.1996.0887u.x. [DOI] [PubMed] [Google Scholar]
- 31.Shimizu F, Watanabe TK, Shinomiya H, Nakamura Y, Fujiwara T. Isolation and expression of a cDNA for human brain fatty acid-binding protein (B-FABP) Biochim Biophys Acta. 1997;1354:24–8. doi: 10.1016/s0167-4781(97)00115-2. [DOI] [PubMed] [Google Scholar]
- 32.Majava V, Polverini E, Mazzini A, Nanekar R, Knoll W, Peters J, et al. Structural and functional characterization of human peripheral nervous system myelin protein P2. PLoS One. 5:e10300. doi: 10.1371/journal.pone.0010300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu RZ, Li X, Godbout R. A novel fatty acid-binding protein (FABP) gene resulting from tandem gene duplication in mammals: transcription in rat retina and testis. Genomics. 2008;92:436–45. doi: 10.1016/j.ygeno.2008.08.003. [DOI] [PubMed] [Google Scholar]
- 34.Kido T, Namiki H. Expression of testicular fatty acid-binding protein PERF 15 during germ cell apoptosis. Dev Growth Differ. 2000;42:359–66. doi: 10.1046/j.1440-169x.2000.00520.x. [DOI] [PubMed] [Google Scholar]
- 35.Das UN. Essential fatty acids and their metabolites as modulators of stem cell biology with reference to inflammation, cancer, and metastasis. Cancer Metastasis Rev. 2011;30:311–24. doi: 10.1007/s10555-011-9316-x. [DOI] [PubMed] [Google Scholar]
- 36.Rapoport SI, Rao JS, Igarashi M. Brain metabolism of nutritionally essential polyunsaturated fatty acids depends on both the diet and the liver. Prostaglandins Leukot Essent Fatty Acids. 2007;77:251–61. doi: 10.1016/j.plefa.2007.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rapoport SI, Igarashi M. Can the rat liver maintain normal brain DHA metabolism in the absence of dietary DHA? Prostaglandins Leukot Essent Fatty Acids. 2009;81:119–23. doi: 10.1016/j.plefa.2009.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Santos CR, Schulze A. Lipid metabolism in cancer. FEBS J. 2012;279:2610–23. doi: 10.1111/j.1742-4658.2012.08644.x. [DOI] [PubMed] [Google Scholar]
- 39.Nariai T, DeGeorge JJ, Greig NH, Genka S, Rapoport SI, Purdon AD. Differences in rates of incorporation of intravenously injected radiolabeled fatty acids into phospholipids of intracerebrally implanted tumor and brain in awake rats. Clin Exp Metastasis. 1994;12:213–25. doi: 10.1007/BF01753889. [DOI] [PubMed] [Google Scholar]
- 40.Albert DH, Anderson CE. Fatty acid composition at the 2-position of ether-linked and diacyl ethanolamine and choline phosphoglycerides of human brain tumors. Lipids. 1977;12:722–8. doi: 10.1007/BF02570902. [DOI] [PubMed] [Google Scholar]
- 41.Marszalek R, Pisklak M, Jankowski W, Lukaszkiewicz J, Horsztynski D, Wawer I. NMR and gas chromatography studies of lyophilized human brain tumors. Acta Pol Pharm. 2010;67:129–36. [PubMed] [Google Scholar]
- 42.Igarashi M, DeMar JC, Jr, Ma K, Chang L, Bell JM, Rapoport SI. Docosahexaenoic acid synthesis from alpha-linolenic acid by rat brain is unaffected by dietary n-3 PUFA deprivation. J Lipid Res. 2007;48:1150–8. doi: 10.1194/jlr.M600549-JLR200. [DOI] [PubMed] [Google Scholar]
- 43.Igarashi M, Ma K, Chang L, Bell JM, Rapoport SI. Dietary n-3 PUFA deprivation for 15 weeks upregulates elongase and desaturase expression in rat liver but not brain. J Lipid Res. 2007;48:2463–70. doi: 10.1194/jlr.M700315-JLR200. [DOI] [PubMed] [Google Scholar]
- 44.Igarashi M, Kim HW, Chang L, Ma K, Rapoport SI. Dietary n-6 polyunsaturated fatty acid deprivation increases docosahexaenoic acid metabolism in rat brain. J Neurochem. 2012;120:985–97. doi: 10.1111/j.1471-4159.2011.07597.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim HW, Rao JS, Rapoport SI, Igarashi M. Dietary n-6 PUFA deprivation downregulates arachidonate but upregulates docosahexaenoate metabolizing enzymes in rat brain. Biochim Biophys Acta. 2011;1811:111–7. doi: 10.1016/j.bbalip.2010.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Denkins Y, Kempf D, Ferniz M, Nileshwar S, Marchetti D. Role of omega-3 polyunsaturated fatty acids on cyclooxygenase-2 metabolism in brain-metastatic melanoma. J Lipid Res. 2005;46:1278–84. doi: 10.1194/jlr.M400474-JLR200. [DOI] [PubMed] [Google Scholar]
- 47.Deckelbaum RJ, Worgall TS, Seo T. n-3 fatty acids and gene expression. Am J Clin Nutr. 2006;83:1520S–5S. doi: 10.1093/ajcn/83.6.1520S. [DOI] [PubMed] [Google Scholar]
- 48.Venkatachalam AB, Lall SP, Denovan-Wright EM, Wright JM. Tissue-specific differential induction of duplicated fatty acid-binding protein genes by the peroxisome proliferator, clofibrate, in zebrafish (Danio rerio) BMC Evol Biol. 2012;12:112. doi: 10.1186/1471-2148-12-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hung DY, Burczynski FJ, Chang P, Lewis A, Masci PP, Siebert GA, et al. Fatty acid binding protein is a major determinant of hepatic pharmacokinetics of palmitate and its metabolites. Am J Physiol Gastrointest Liver Physiol. 2003;284:G423–33. doi: 10.1152/ajpgi.00328.2002. [DOI] [PubMed] [Google Scholar]
- 50.Liu RZ, Mita R, Beaulieu M, Gao Z, Godbout R. Fatty acid binding proteins in brain development and disease. Int J Dev Biol. 2010;54:1229–39. doi: 10.1387/ijdb.092976rl. [DOI] [PubMed] [Google Scholar]
- 51.Owada Y, Yoshimoto T, Kondo H. Spatio-temporally differential expression of genes for three members of fatty acid binding proteins in developing and mature rat brains. J Chem Neuroanat. 1996;12:113–22. doi: 10.1016/s0891-0618(96)00192-5. [DOI] [PubMed] [Google Scholar]
- 52.Murphy EJ, Owada Y, Kitanaka N, Kondo H, Glatz JF. Brain arachidonic acid incorporation is decreased in heart fatty acid binding protein gene-ablated mice. Biochemistry. 2005;44:6350–60. doi: 10.1021/bi047292r. [DOI] [PubMed] [Google Scholar]
- 53.Hanhoff T, Lucke C, Spener F. Insights into binding of fatty acids by fatty acid binding proteins. Mol Cell Biochem. 2002;239:45–54. [PubMed] [Google Scholar]
- 54.Kurtz A, Zimmer A, Schnutgen F, Bruning G, Spener F, Muller T. The expression pattern of a novel gene encoding brain-fatty acid binding protein correlates with neuronal and glial cell development. Development. 1994;120:2637–49. doi: 10.1242/dev.120.9.2637. [DOI] [PubMed] [Google Scholar]
- 55.Feng L, Hatten ME, Heintz N. Brain lipid-binding protein (BLBP): a novel signaling system in the developing mammalian CNS. Neuron. 1994;12:895–908. doi: 10.1016/0896-6273(94)90341-7. [DOI] [PubMed] [Google Scholar]
- 56.Balendiran GK, Schnutgen F, Scapin G, Borchers T, Xhong N, Lim K, et al. Crystal structure and thermodynamic analysis of human brain fatty acid-binding protein. J Biol Chem. 2000;275:27045–54. doi: 10.1074/jbc.M003001200. [DOI] [PubMed] [Google Scholar]
- 57.De Rosa A, Pellegatta S, Rossi M, Tunici P, Magnoni L, Speranza MC, et al. A Radial Glia Gene Marker, Fatty Acid Binding Protein 7 (FABP7), Is Involved in Proliferation and Invasion of Glioblastoma Cells. PLoS One. 2012;7:e52113. doi: 10.1371/journal.pone.0052113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cataltepe O, Arikan MC, Ghelfi E, Karaaslan C, Ozsurekci Y, Dresser K, et al. Fatty acid binding protein 4 is expressed in distinct endothelial and non-endothelial cell populations in glioblastoma. Neuropathol Appl Neurobiol. 2012;38:400–10. doi: 10.1111/j.1365-2990.2011.01237.x. [DOI] [PubMed] [Google Scholar]
- 59.Bernlohr DA, Coe NR, Simpson MA, Hertzel AV. Regulation of gene expression in adipose cells by polyunsaturated fatty acids. Adv Exp Med Biol. 1997;422:145–56. doi: 10.1007/978-1-4757-2670-1_12. [DOI] [PubMed] [Google Scholar]
- 60.Kane CD, Bernlohr DA. A simple assay for intracellular lipid-binding proteins using displacement of 1-anilinonaphthalene 8-sulfonic acid. Anal Biochem. 1996;233:197–204. doi: 10.1006/abio.1996.0028. [DOI] [PubMed] [Google Scholar]
- 61.Fujishiro K, Fukui Y, Sato O, Kawabe K, Seto K, Motojima K. Analysis of tissue-specific and PPARalpha-dependent induction of FABP gene expression in the mouse liver by an in vivo DNA electroporation method. Mol Cell Biochem. 2002;239:165–72. [PubMed] [Google Scholar]
- 62.Hostetler HA, McIntosh AL, Atshaves BP, Storey SM, Payne HR, Kier AB, et al. L-FABP directly interacts with PPARalpha in cultured primary hepatocytes. J Lipid Res. 2009;50:1663–75. doi: 10.1194/jlr.M900058-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ibabe A, Herrero A, Cajaraville MP. Modulation of peroxisome proliferator-activated receptors (PPARs) by PPAR(alpha)- and PPAR(gamma)-specific ligands and by 17beta-estradiol in isolated zebrafish hepatocytes. Toxicol In Vitro. 2005;19:725–35. doi: 10.1016/j.tiv.2005.03.019. [DOI] [PubMed] [Google Scholar]
- 64.Nasrollahzadeh J, Siassi F, Doosti M, Eshraghian MR, Shokri F, Modarressi MH, et al. The influence of feeding linoleic, gamma-linolenic and docosahexaenoic acid rich oils on rat brain tumor fatty acids composition and fatty acid binding protein 7 mRNA expression. Lipids Health Dis. 2008;7:45. doi: 10.1186/1476-511X-7-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Brun M, Coles JE, Monckton EA, Glubrecht DD, Bisgrove D, Godbout R. Nuclear factor I regulates brain fatty acid-binding protein and glial fibrillary acidic protein gene expression in malignant glioma cell lines. J Mol Biol. 2009;391:282–300. doi: 10.1016/j.jmb.2009.06.041. [DOI] [PubMed] [Google Scholar]
- 66.Liu RZ, Monckton EA, Godbout R. Regulation of the FABP7 gene by PAX6 in malignant glioma cells. Biochem Biophys Res Commun. 2012;422:482–7. doi: 10.1016/j.bbrc.2012.05.019. [DOI] [PubMed] [Google Scholar]
- 67.Zhou YH, Tan F, Hess KR, Yung WK. The expression of PAX6, PTEN, vascular endothelial growth factor, and epidermal growth factor receptor in gliomas: relationship to tumor grade and survival. Clin Cancer Res. 2003;9:3369–75. [PubMed] [Google Scholar]
- 68.Li J, Cui Y, Gao G, Zhao Z, Zhang H, Wang X. Notch1 is an independent prognostic factor for patients with glioma. J Surg Oncol. 2011;103:813–7. doi: 10.1002/jso.21851. [DOI] [PubMed] [Google Scholar]
- 69.Purow BW, Haque RM, Noel MW, Su Q, Burdick MJ, Lee J, et al. Expression of Notch-1 and its ligands, Delta-like-1 and Jagged-1, is critical for glioma cell survival and proliferation. Cancer Res. 2005;65:2353–63. doi: 10.1158/0008-5472.CAN-04-1890. [DOI] [PubMed] [Google Scholar]
- 70.Anthony TE, Mason HA, Gridley T, Fishell G, Heintz N. Brain lipid-binding protein is a direct target of Notch signaling in radial glial cells. Genes Dev. 2005;19:1028–33. doi: 10.1101/gad.1302105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Han S, Inoue H, Flowers LC, Sidell N. Control of COX-2 gene expression through peroxisome proliferator-activated receptor gamma in human cervical cancer cells. Clin Cancer Res. 2003;9:4627–35. [PubMed] [Google Scholar]
- 72.Marden NY, Murray M. Characterization of a c-Jun-responsive module in the 5′-flank of the human CYP2J2 gene that regulates transactivation. Biochem J. 2005;391:631–40. doi: 10.1042/BJ20050798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Allport VC, Slater DM, Newton R, Bennett PR. NF-kappaB and AP-1 are required for cyclo-oxygenase 2 gene expression in amnion epithelial cell line (WISH) Mol Hum Reprod. 2000;6:561–5. doi: 10.1093/molehr/6.6.561. [DOI] [PubMed] [Google Scholar]
- 74.LaVoie MJ, Selkoe DJ. The Notch ligands, Jagged and Delta, are sequentially processed by alpha-secretase and presenilin/gamma-secretase and release signaling fragments. J Biol Chem. 2003;278:34427–37. doi: 10.1074/jbc.M302659200. [DOI] [PubMed] [Google Scholar]
- 75.Chakraborti S. Phospholipase A(2) isoforms: a perspective. Cell Signal. 2003;15:637–65. doi: 10.1016/s0898-6568(02)00144-4. [DOI] [PubMed] [Google Scholar]
- 76.Phillis JW, Horrocks LA, Farooqui AA. Cyclooxygenases, lipoxygenases, and epoxygenases in CNS: their role and involvement in neurological disorders. Brain Res Rev. 2006;52:201–43. doi: 10.1016/j.brainresrev.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 77.Dennis EA, Rhee SG, Billah MM, Hannun YA. Role of phospholipase in generating lipid second messengers in signal transduction. FASEB J. 1991;5:2068–77. doi: 10.1096/fasebj.5.7.1901288. [DOI] [PubMed] [Google Scholar]
- 78.Vance DE, Vance JE. Biochemistry of Lipids, Lipoproteins and Membranes. 5. Elsevier; Amsterdam: 2008. Phospholipid biosynthesis in eukaryotes. [Google Scholar]
- 79.Strokin M, Sergeeva M, Reiser G. Docosahexaenoic acid and arachidonic acid release in rat brain astrocytes is mediated by two separate isoforms of phospholipase A2 and is differently regulated by cyclic AMP and Ca2+ Br J Pharmacol. 2003;139:1014–22. doi: 10.1038/sj.bjp.0705326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Winstead MV, Balsinde J, Dennis EA. Calcium-independent phospholipase A(2): structure and function. Biochim Biophys Acta. 2000;1488:28–39. doi: 10.1016/s1388-1981(00)00107-4. [DOI] [PubMed] [Google Scholar]
- 81.Barbour SE, Kapur A, Deal CL. Regulation of phosphatidylcholine homeostasis by calcium-independent phospholipase A2. Biochim Biophys Acta. 1999;1439:77–88. doi: 10.1016/s1388-1981(99)00078-5. [DOI] [PubMed] [Google Scholar]
- 82.Arnold C, Markovic M, Blossey K, Wallukat G, Fischer R, Dechend R, et al. Arachidonic acid-metabolizing cytochrome P450 enzymes are targets of {omega}-3 fatty acids. J Biol Chem. 2010;285:32720–33. doi: 10.1074/jbc.M110.118406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Serhan CN. Novel eicosanoid and docosanoid mediators: resolvins, docosatrienes, and neuroprotectins. Curr Opin Clin Nutr Metab Care. 2005;8:115–21. doi: 10.1097/00075197-200503000-00003. [DOI] [PubMed] [Google Scholar]
- 84.Kita Y, Ohto T, Uozumi N, Shimizu T. Biochemical properties and pathophysiological roles of cytosolic phospholipase A2s. Biochim Biophys Acta. 2006;1761:1317–22. doi: 10.1016/j.bbalip.2006.08.001. [DOI] [PubMed] [Google Scholar]
- 85.Sapirstein A, Bonventre JV. Specific physiological roles of cytosolic phospholipase A(2) as defined by gene knockouts. Biochim Biophys Acta. 2000;1488:139–48. doi: 10.1016/s1388-1981(00)00116-5. [DOI] [PubMed] [Google Scholar]
- 86.Bonventre JV, Huang Z, Taheri MR, O’Leary E, Li E, Moskowitz MA, et al. Reduced fertility and postischaemic brain injury in mice deficient in cytosolic phospholipase A2. Nature. 1997;390:622–5. doi: 10.1038/37635. [DOI] [PubMed] [Google Scholar]
- 87.Ghosh M, Tucker DE, Burchett SA, Leslie CC. Properties of the Group IV phospholipase A2 family. Prog Lipid Res. 2006;45:487–510. doi: 10.1016/j.plipres.2006.05.003. [DOI] [PubMed] [Google Scholar]
- 88.Tung WH, Lee IT, Hsieh HL, Yang CM. EV71 induces COX-2 expression via c-Src/PDGFR/PI3K/Akt/p42/p44 MAPK/AP-1 and NF-kappaB in rat brain astrocytes. J Cell Physiol. 2010;224:376–86. doi: 10.1002/jcp.22133. [DOI] [PubMed] [Google Scholar]
- 89.Karin M. The regulation of AP-1 activity by mitogen-activated protein kinases. J Biol Chem. 1995;270:16483–6. doi: 10.1074/jbc.270.28.16483. [DOI] [PubMed] [Google Scholar]
- 90.Leong KG, Karsan A. Recent insights into the role of Notch signaling in tumorigenesis. Blood. 2006;107:2223–33. doi: 10.1182/blood-2005-08-3329. [DOI] [PubMed] [Google Scholar]
- 91.Rogowski AS. [Master’s Theses] Chicago: Loyola University; 2011. Notch-1 Specifically Activates Erk1/2 in Multiple Breast Cancer Subtypes. [Google Scholar]
- 92.Moes M, Boonstra J, Regan-Klapisz E. Novel role of cPLA(2)alpha in membrane and actin dynamics. Cell Mol Life Sci. 2010;67:1547–57. doi: 10.1007/s00018-010-0267-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Stockton RA, Jacobson BS. Modulation of cell-substrate adhesion by arachidonic acid: lipoxygenase regulates cell spreading and ERK1/2-inducible cyclooxygenase regulates cell migration in NIH-3T3 fibroblasts. Mol Biol Cell. 2001;12:1937–56. doi: 10.1091/mbc.12.7.1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Navarro-Tito N, Robledo T, Salazar EP. Arachidonic acid promotes FAK activation and migration in MDA-MB-231 breast cancer cells. Exp Cell Res. 2008;314:3340–55. doi: 10.1016/j.yexcr.2008.08.018. [DOI] [PubMed] [Google Scholar]
- 95.van Helden SF, Krooshoop DJ, Broers KC, Raymakers RA, Figdor CG, van Leeuwen FN. A critical role for prostaglandin E2 in podosome dissolution and induction of high-speed migration during dendritic cell maturation. J Immunol. 2006;177:1567–74. doi: 10.4049/jimmunol.177.3.1567. [DOI] [PubMed] [Google Scholar]
- 96.Tanabe T, Tohnai N. Cyclooxygenase isozymes and their gene structures and expression. Prostaglandins Other Lipid Mediat. 2002;68–69:95–114. doi: 10.1016/s0090-6980(02)00024-2. [DOI] [PubMed] [Google Scholar]
- 97.Nadjar A, Tridon V, May MJ, Ghosh S, Dantzer R, Amedee T, et al. NFkappaB activates in vivo the synthesis of inducible Cox-2 in the brain. J Cereb Blood Flow Metab. 2005;25:1047–59. doi: 10.1038/sj.jcbfm.9600106. [DOI] [PubMed] [Google Scholar]
- 98.Min BS, Choi YJ, Pyo HR, Kim H, Seong J, Chung HC, et al. Cyclooxygenase-2 expression in pretreatment biopsy as a predictor of tumor responses after preoperative chemoradiation in rectal cancer. Arch Surg. 2008;143:1091–7. doi: 10.1001/archsurg.143.11.1091. discussion 7. [DOI] [PubMed] [Google Scholar]
- 99.Muller-Decker K, Furstenberger G. The cyclooxygenase-2-mediated prostaglandin signaling is causally related to epithelial carcinogenesis. Mol Carcinog. 2007;46:705–10. doi: 10.1002/mc.20326. [DOI] [PubMed] [Google Scholar]
- 100.Singh AK, Parshad R, Pasi S, Madhavan T, Das SN, Mishra B, et al. Prognostic significance of cyclooxygenase-2 and response to chemotherapy in invasive ductal breast carcinoma patients by real time surface plasmon resonance analysis. DNA Cell Biol. 2011;30:801–7. doi: 10.1089/dna.2011.1215. [DOI] [PubMed] [Google Scholar]
- 101.Fidler MJ, Argiris A, Patel JD, Johnson DH, Sandler A, Villaflor VM, et al. The potential predictive value of cyclooxygenase-2 expression and increased risk of gastrointestinal hemorrhage in advanced non-small cell lung cancer patients treated with erlotinib and celecoxib. Clin Cancer Res. 2008;14:2088–94. doi: 10.1158/1078-0432.CCR-07-4013. [DOI] [PubMed] [Google Scholar]
- 102.Almhanna K, El-Rayes B, Sethi S, Dyson G, Heilbrun L, Philip PA, et al. Association between COX-2 expression and effectiveness of COX-2 inhibitors in a phase II trial in patients with metastatic colorectal adenocarcinoma. Anticancer Res. 2012;32:3559–63. [PMC free article] [PubMed] [Google Scholar]
- 103.Shono T, Tofilon PJ, Bruner JM, Owolabi O, Lang FF. Cyclooxygenase-2 expression in human gliomas: prognostic significance and molecular correlations. Cancer Res. 2001;61:4375–81. [PubMed] [Google Scholar]
- 104.Perdiki M, Korkolopoulou P, Thymara I, Agrogiannis G, Piperi C, Boviatsis E, et al. Cyclooxygenase-2 expression in astrocytomas. Relationship with microvascular parameters, angiogenic factors expression and survival. Mol Cell Biochem. 2007;295:75–83. doi: 10.1007/s11010-006-9275-7. [DOI] [PubMed] [Google Scholar]
- 105.Xu L, Han C, Wu T. A novel positive feedback loop between peroxisome proliferator-activated receptor-delta and prostaglandin E2 signaling pathways for human cholangiocarcinoma cell growth. J Biol Chem. 2006;281:33982–96. doi: 10.1074/jbc.M600135200. [DOI] [PubMed] [Google Scholar]
- 106.Panigrahy D, Kaipainen A, Greene ER, Huang S. Cytochrome P450-derived eicosanoids: the neglected pathway in cancer. Cancer Metastasis Rev. 2010;29:723–35. doi: 10.1007/s10555-010-9264-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Chen C, Li G, Liao W, Wu J, Liu L, Ma D, et al. Selective inhibitors of CYP2J2 related to terfenadine exhibit strong activity against human cancers in vitro and in vivo. J Pharmacol Exp Ther. 2009;329:908–18. doi: 10.1124/jpet.109.152017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Jiang JG, Ning YG, Chen C, Ma D, Liu ZJ, Yang S, et al. Cytochrome p450 epoxygenase promotes human cancer metastasis. Cancer Res. 2007;67:6665–74. doi: 10.1158/0008-5472.CAN-06-3643. [DOI] [PubMed] [Google Scholar]
- 109.Jiang JG, Chen CL, Card JW, Yang S, Chen JX, Fu XN, et al. Cytochrome P450 2J2 promotes the neoplastic phenotype of carcinoma cells and is up-regulated in human tumors. Cancer Res. 2005;65:4707–15. doi: 10.1158/0008-5472.CAN-04-4173. [DOI] [PubMed] [Google Scholar]
- 110.Spencer AG, Woods JW, Arakawa T, Singer II, Smith WL. Subcellular localization of prostaglandin endoperoxide H synthases-1 and -2 by immunoelectron microscopy. J Biol Chem. 1998;273:9886–93. doi: 10.1074/jbc.273.16.9886. [DOI] [PubMed] [Google Scholar]
- 111.Werck-Reichhart D, Feyereisen R. Cytochromes P450: a success story. Genome Biol. 2000;1:REVIEWS3003. doi: 10.1186/gb-2000-1-6-reviews3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Itoh T, Fairall L, Amin K, Inaba Y, Szanto A, Balint BL, et al. Structural basis for the activation of PPARgamma by oxidized fatty acids. Nat Struct Mol Biol. 2008;15:924–31. doi: 10.1038/nsmb.1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Adida A, Spener F. Adipocyte-type fatty acid-binding protein as inter-compartmental shuttle for peroxisome proliferator activated receptor gamma agonists in cultured cell. Biochim Biophys Acta. 2006;1761:172–81. doi: 10.1016/j.bbalip.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 114.Edwards IJ, O’Flaherty JT. Omega-3 Fatty Acids and PPARgamma in Cancer. PPAR Res. 2008;2008:358052. doi: 10.1155/2008/358052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Rao GH, Radha E, White JG. Effect of docosahexaenoic acid (DHA) on arachidonic acid metabolism and platelet function. Biochem Biophys Res Commun. 1983;117:549–55. doi: 10.1016/0006-291x(83)91235-4. [DOI] [PubMed] [Google Scholar]
- 116.Gleissman H, Yang R, Martinod K, Lindskog M, Serhan CN, Johnsen JI, et al. Docosahexaenoic acid metabolome in neural tumors: identification of cytotoxic intermediates. FASEB J. 2010;24:906–15. doi: 10.1096/fj.09-137919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Serhan CN, Hong S, Gronert K, Colgan SP, Devchand PR, Mirick G, et al. Resolvins: a family of bioactive products of omega-3 fatty acid transformation circuits initiated by aspirin treatment that counter proinflammation signals. J Exp Med. 2002;196:1025–37. doi: 10.1084/jem.20020760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kalman J, Gecse A, Farkas T, Joo F, Telegdy G, Lajtha A. Dietary manipulation with high marine fish oil intake of fatty acid composition and arachidonic acid metabolism in rat cerebral microvessels. Neurochem Res. 1992;17:167–72. doi: 10.1007/BF00966795. [DOI] [PubMed] [Google Scholar]
- 119.Esposito G, Giovacchini G, Der M, Liow JS, Bhattacharjee AK, Ma K, et al. Imaging signal transduction via arachidonic acid in the human brain during visual stimulation, by means of positron emission tomography. Neuroimage. 2007;34:1342–51. doi: 10.1016/j.neuroimage.2006.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Esposito G, Giovacchini G, Liow JS, Bhattacharjee AK, Greenstein D, Schapiro M, et al. Imaging neuroinflammation in Alzheimer’s disease with radiolabeled arachidonic acid and PET. J Nucl Med. 2008;49:1414–21. doi: 10.2967/jnumed.107.049619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Pichika R, Taha AY, Gao F, Kotta K, Cheon Y, Chang L, et al. The synthesis and in vivo pharmacokinetics of fluorinated arachidonic acid: implications for imaging neuroinflammation. J Nucl Med. 2012;53:1383–91. doi: 10.2967/jnumed.112.105734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Basselin M, Ramadan E, Rapoport SI. Imaging brain signal transduction and metabolism via arachidonic and docosahexaenoic acid in animals and humans. Brain Res Bull. 2011;87:154–71. doi: 10.1016/j.brainresbull.2011.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Liu RS, Chang CP, Chu LS, Chu YK, Hsieh HJ, Chang CW, et al. PET imaging of brain astrocytoma with 1-11C-acetate. Eur J Nucl Med Mol Imaging. 2006;33:420–7. doi: 10.1007/s00259-005-0023-0. [DOI] [PubMed] [Google Scholar]