Abstract
Several retroviruses downmodulate the cell surface expression of envelope (Env) proteins through peptide sequences located in the cytoplasmic tail of the transmembrane (TM) subunit. We investigated whether cell surface expression of a chimeric protein containing the cytoplasmic domain of the TM protein (CTM) of bovine leukemia virus (BLV) was regulated by two membrane-proximal dileucine motifs or by tyrosine Y487 or Y498 in YXXL motifs. A chimeric protein composed of the extracellular and membrane-spanning portions of human CD8-α plus a wild-type (wt) BLV CTM was detectable on the surface of only 40% of the cells in which it was transiently expressed. Replacement of either dileucine pair with alanines increased the level of surface display of chimeric proteins. Nearly all cells became surface positive when both dileucine motifs were altered simultaneously and when either an N-terminal segment containing both dileucine motifs or a C-terminal segment containing all YXXL motifs was deleted. In contrast, replacement of Y487 or Y498 with alanine or phenylalanine enabled only small increases in surface display compared with wt levels. Chimeric proteins had similar stabilities but were downmodulated from the cell surface at three different rates. Point mutants segregated into each of the three groups of proteins categorized according to these different rates. Interestingly, Y487 mutants were downmodulated less efficiently than Y498 mutants, which behaved like wt. CD8-CTM chimeric proteins were phosphorylated on serine residues, but the native BLV Env protein was not phosphorylated either in transfected cells or in a lymphoid cell line constitutively producing BLV. Thus, both dileucine and YXXL motifs within the BLV CTM contribute to downmodulation of a protein containing this domain. Interactions with other proteins may influence surface exposure of Env protein complexes in virus-infected cells, assisting in viral evasion of adaptive immunity.
The cytoplasmic domains of several retroviral envelope (Env) proteins control the level of Env present on the surface of infected cells. A low level of surface expression is mediated by intracellular retention of Env proteins as well as their rapid endocytosis from the plasma membrane (22, 51, 54). The existence of mechanisms to ensure low-level cell surface display of Env proteins points to a feature of the retroviral life cycle that most probably evolved to reduce clearance of virus-producing cells by the immune system (21, 22, 35). The main target for antibody recognition of retroviral Env protein complexes is the surface (SU) protein subunit, which is anchored onto the membranes of infected cells and virions by the membrane-spanning domain of the transmembrane (TM) subunit of an Env monomer (26). A labile disulfide bond links the SU and TM subunits of bovine leukemia virus (BLV) (30), a deltaretrovirus. Upon binding of oligomers of retroviral SU to cellular receptors, the TM subunits mediate virus-cell fusion (26). In parallel with other retroviral TM proteins, the extracellular portion of BLV TM contains a putative oligomerization domain (19) and an N-terminal fusion peptide (60).
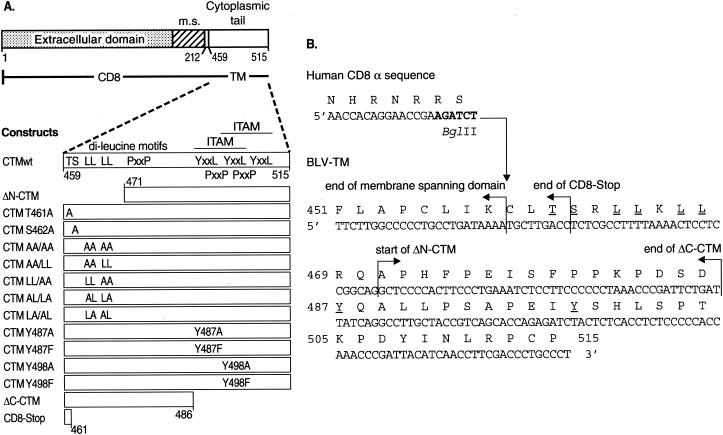
The 57-amino-acid cytoplasmic domain of the TM protein (CTM) of BLV is unusually long among oncogenic retroviruses (48, 52). It contains a number of amino acid sequences involved in protein-protein interactions (Fig. 1A), including two overlapping immunoreceptor tyrosine-based activation motifs (ITAMs) (47). Consensus ITAMs contain two YXXL sequences separated by seven amino acids and preceded by two precisely spaced acidic residues (D/EX7D/EX2YX2LX7YX2L/I) (9, 18, 47). Phosphorylation of the tyrosines in ITAMs creates binding sites for Src homology 2 (SH2) protein modules found in many cell-signaling proteins (42). The three YXXL sequences present in the BLV CTM could potentially form two overlapping ITAMs; the sequence of the first ITAM exactly fits the consensus, although the second lacks the most distal upstream acidic residue (1, 2).
FIG. 1.
(A) Schematic representation of CD8-CTM chimeric proteins. The top diagram represents the entire CD8-CTM fusion construct. The extracellular and membrane spanning (m.s.) domains plus four to seven cytoplasmic amino acids of CD8-α (aa 1 to 212) precede the BLV CTM (aa 459 to 515 of the BLV Env protein precursor). The expanded diagram of the wt CTM of T-15 BLV shows the locations of T461, S462, two dileucine motifs, three PXXP motifs, and the three YXXL motifs that form two overlapping ITAMs. Mutations are shown in their approximate locations. ΔN-CTM lacks 12 N-terminal amino acids of the BLV CTM, whereas ΔC-CTM lacks 29 C-terminal residues. CD8-Stop contains only the first three amino acids of the BLV CTM. (B) DNA and amino acid sequences of chimeric CD8-CTM constructs. The end of the CD8-α sequence encodes membrane-spanning and cytoplasmic amino acids just upstream of the junction with the BLV CTM; the BglII restriction site used to fuse the sequences is in bold type. The end of the predicted membrane-spanning domain (48, 52) of the BLV CTM is demarcated; the predicted cytoplasmic domain begins at C459, which is the first BLV-encoded amino acid in all CD8-CTM chimeras except ΔN-CTM. Arrows indicate the start of BLV-specific sequences of the ΔN-CTM chimera as well as the ends of the CD8-Stop and ΔC-CTM chimeras. Individual amino acids that were replaced are underlined.
The BLV CTM has the capacity to transmit activating signals elicited by the binding of an extracellular ligand to a membrane-spanning protein. Antibody cross-linking of a chimeric protein in which the BLV CTM replaces the cytoplasmic portion of murine CD8-α elicits rapid calcium release and increased tyrosine phosphorylation of proteins in murine B-lymphoma cells (1). Mutation of individual tyrosine and leucine residues within the YXXL motifs of the potential BLV ITAMs showed that the two YXXL motifs of the more membrane-proximal ITAM must be intact for cross-linking of CD8-CTM fusion proteins to induce early and late activation events in stably transfected mouse B- and T-lymphoma cells (2).
The YXXL motifs within the membrane-proximal ITAM could also be part of immunoreceptor tyrosine-based inhibition motifs (ITIMs) (23) having the sequences SXYXXL and IYSXL (57, 59). Upon phosphorylation of the tyrosine residues, ITIM-containing proteins are bound by SH2 domains, for example, in phosphatases and signaling is inhibited (reviewed in reference 23). SHP-1 phosphatase, an important negative regulator of B-cell signaling (38), associates with the BLV CTM in cultured peripheral blood mononuclear cells from BLV-infected cows when tyrosine phosphatases are inhibited prior to cell lysis (11). SHP-1, which contains two SH2 motifs, may bind to YXXLs in the BLV CTM when the tyrosines are phosphorylated.
In the context of viral replication, replacement of tyrosine within either of the first two YXXLs of the BLV CTM with aspartic acid, which introduces a constitutive negative charge, reduces or eliminates viral infectivity and proviral load in sheep injected with mutant proviruses (61). Moreover, replacement of the second tyrosine residue with alanine greatly reduces the propagation and infectivity of the virus in cultured fetal lamb kidney cells (27). Although propagation of signals by ITAMs and ITIMs depends on phosphorylation of their tyrosine residues, BLV TM has not been found to be phosphorylated at tyrosine when peripheral blood mononuclear cells from infected cows are cultured briefly to allow virus expression and synthesis of the Env protein (23).
Recognition and binding of PXXP motifs by proteins containing Src homology 3 (SH3) domains also brings together components of cell signaling pathways (42, 46). The BLV CTM contains three PXXP motifs (10). Replacement of the first three proline residues with alanine within the membrane-proximal PXXPX4PP motif decreases viral load in sheep injected with mutant proviruses, and the animals develop lower titers of serum antibodies specific for the viral capsid protein than do animals injected with wild-type (wt) proviruses (45). This suggests that the ability of BLV TM to associate with a protein containing an SH3 domain is important for normal levels of viral replication in vivo.
In contrast to that of BLV, the CTM domain of its close relative, human T-cell leukemia virus (HTLV) type 1, is only 23 amino acids long; it has no ITAM, ITIM, or proline-rich motifs but does contain a YXXI motif. When fused individually to the extracellular and membrane-spanning regions plus 13 cytoplasmic amino acids of the interleukin 2 receptor α (IL-2Rα) chain, the CTMs of HTLV and BLV each reduce the cell surface expression of the respective chimeric protein by mediating rapid internalization from the plasma membrane (22). Substitution of a serine for the tyrosine residue in the YSLI sequence of the HTLV CTM increases the residence of a CD8-CTM chimeric protein on the surface of transfected cells (3). The tyrosine of the YSLI sequence is essential for HTLV transmission from cell to cell (13), and it targets budding of a retrovirus pseudotyped with HTLV Env to the basolateral surface of polarized epithelial cells (35). The HTLV CTM interacts with isolated μ subunits of the clathrin-associated adaptor proteins (AP) 1 and 2 (3). Interactions of AP complexes with YXXΦ sequences, where Φ is a large hydrophobic amino acid, function in clathrin-mediated endocytosis of cell surface proteins (AP-2) and in trafficking of membrane-spanning proteins from the trans-Golgi network to endosomes and the cell surface (AP-1) (31).
The sequences in the BLV CTM that are responsible for rapid endocytosis have not been identified. In addition to its three YXXΦ sequences, the BLV CTM contains two membrane-proximal dileucine motifs (Fig. 1A). Dileucine motifs can participate in the downregulation of proteins from cell surface membranes by interacting with clathrin-associated AP complexes (33, 62). Phosphorylation of an upstream serine residue can stimulate dileucine-mediated endocytosis (15, 16, 20, 43).
To learn whether dileucine and YXXL motifs within the BLV CTM mediate low-level cell surface display of a chimeric protein, we fused the BLV CTM to the extracellular and membrane-spanning portions of the human CD8-α chain and then mutated the CTM. A deletion of the N terminus of the CTM removed the two dileucine pairs; a deletion of the C terminus removed all three YXXL motifs. Single amino acid substitutions changed individual tyrosines of the first two YXXL motifs to alanine or phenylalanine and the leucines of the dileucine motifs to alanines. A serine residue and a threonine residue preceding the dileucine motifs were individually replaced with alanine to assess whether a phosphoacceptor residue was necessary upstream of the dileucine motifs. We then assessed the stability of the CD8-CTM proteins in cells transiently expressing the chimeras, the extent of their display on the cell surface, and the rate of their internalization from the cell surface. Finally, given the regulatory role of phosphorylation in protein-protein interactions, we assessed the phosphorylation state of CD8-CTM chimeric proteins, of native BLV Env protein expressed by transiently transfected cells, and of native BLV Env protein present in cells from a BLV-infected lymphoid cell line.
MATERIALS AND METHODS
Construction of CD8-CTM expression vectors.
A DNA fragment encoding the extracytoplasmic and membrane-spanning portions of human CD8-α (28, 34) was cloned into a pcDNA3 expression plasmid (Invitrogen) in frame with the 57 amino acids of BLV proviral DNA that are predicted to encode the cytoplasmic tail of gp30-TM (48, 52). A HindIII-BglII DNA fragment encoding CD8-α was obtained from plasmid pRcCMV-CD8-NefHIV-1SF2 (55). DNA encoding the cytoplasmic portion of TM was obtained by PCR amplification of the T15-2 clone (14, 49) (GenBank accession number K02251). pcDNA3-CD8-CTM constructs (Fig. 1A) were created using two different approaches. In the first, single-step PCR was used to amplify DNA sequences that encode the wt CTM (CTMwt) sequence and amino- or carboxy-terminal deletions of CTM, as well as to mutagenize codons for T461, S462, L464, L465, L467, and L468. The 5′ primer contained a BamHI restriction endonuclease site to facilitate the in-frame fusion of the fragments encoding the cytoplasmic tail of TM and CD8-α, while the 3′ primer contained sequences complementary to those encoding the carboxy terminus of CTM plus an XbaI site to facilitate subsequent cloning. PCRs were carried out for 30 cycles consisting of 30-s incubations at 94, 55, and 72°C. Nuclease-digested PCR products were separated by agarose gel electrophoresis, purified on Quick Spin columns (QIAGEN, Inc.), and then ligated with pcDNA3. A HindIII-BglII fragment encoding CD8-α was inserted into pcDNA3-CTM intermediates that had been digested with HindIII and BamHI.
The second approach used recombinant PCR splice overlap extension (25) to mutagenize codons for Y487 and Y498. The first of two stages comprised two separate PCRs yielding DNA fragments with central overlapping regions. One half of the template DNA (pcDNA3-CD8-CTMwt) was amplified with a 5′ primer containing an EcoRV restriction site located in the CD8-encoding DNA sequence and a 3′ primer overlapping the CTM DNA sequence to be mutagenized. The remaining template DNA was amplified with a 5′ primer complementary to the 3′ primer of the previous reaction and the 3′ primer containing an XbaI site as described above. Both PCRs were carried out as described above.
In the second stage, 1 μl of DNA product from each of these two PCRs was denatured and strands were reannealed by their overlapping segments. Full-length fragments were amplified with the 5′ primer containing the EcoRV site and the 3′ primer with the XbaI site. The amplified PCR product was purified after agarose gel electrophoresis and subsequently cloned into pcDNA3-CD8. Details of mutagenesis and the sequences of primers used for PCR amplification (39) are available from the authors upon request. The CD8-Stop mutant contains three amino acids of the BLV CTM followed by a stop codon; it was isolated during preparation of Y487 mutants.
E. coli DH5α cells transformed with CD8-CTM constructs were grown for no longer than 13 h at 37°C in Luria-Bertani medium containing 100 μg of ampicillin per ml. Plasmids were purified using Endo-free Plasmid Maxi kits (QIAGEN). The identity of all plasmid constructs was confirmed by restriction endonuclease analysis and by semiautomated DNA sequencing (Applied Biosystems, Inc.) of both strands by using SP6 and T7 primers. Sequences were analyzed using Sequa software (PerkinElmerCetus Corp.).
Cell culture.
COS-7 monkey kidney cells were obtained from the American Type Culture Collection and were subcultured twice a week in high-glucose Dulbecco's modified Eagle medium (DMEM; Invitrogen-Life Technologies) supplemented with 10% fetal bovine serum (FBS), 2 mM l-glutamine, 100 U of penicillin per ml, and 100 μg of streptomycin per ml. BLV-negative BL3 cells (58) and BLV-positive BL3* cells (50) were cultured at 37°C with 5% CO2 in 50% minimum essential medium-50% Leibovitz L-15 medium (Invitrogen-Life Technologies) supplemented with 5% FBS, 0.1 mM nonessential amino acids, 2 mM l-glutamine, penicillin, and streptomycin.
Transfection.
COS-7 cells were seeded at 2.0 × 105 cells in 2 ml of DMEM-10% FBS per each 35-mm-diameter well of a 6-well plate. Twenty hours later, cells were transfected with 1 μg of DNA suspended in 100 μl of serum-free DMEM containing 3 μl of Fugene reagent (Roche, Indianapolis, Ind.) according to the manufacturer's protocol. When T-25 culture flasks were used, 5.0 × 105 cells were seeded in 6 ml of DMEM-10% FBS. Twenty-four hours later, cells were transfected with 2 μg of DNA suspended in 200 μl of serum-free DMEM containing 6 μl of Fugene reagent.
Metabolic radiolabeling and immunoprecipitation.
Metabolic radiolabeling and immunoprecipitation were performed according to previously described procedures (55). Steady-state metabolic labeling of COS-7 cells with [35S]Met/Cys was performed in 6-well plates at 24 to 48 h after transfection. Cells were washed twice in 1 ml of 37°C phosphate-buffered saline (PBS; 1.5 mM KH2PO4, 8 mM Na2HPO4, 2.7 mM KCl, 140 mM NaCl [pH 7.4]) and were incubated for 3 h at 37°C in 0.5 ml of Cys- and Met-free RPMI 1640 containing 5% dialyzed FBS and 25 μCi of [35S]Met/Cys (1,000 Ci/mmol; Trans35S-Label; ICN). Cells were washed twice in 37°C PBS and lysed in 1 ml of extraction buffer A (137 mM NaCl, 10% [vol/vol] glycerol, 50 mM Tris-HCl [pH 8.0], 0.5% [vol/vol] IGEPAL CA-630 [Sigma Chemical Co., St. Louis, Mo.; used as a substitute for NP-40], 2 mM EDTA) supplemented with 0.5 μg of leupeptin per ml, 2 μg of aprotinin per ml, and 10 μg of pepstatin A per ml. Lysates were rocked at 4°C for 30 min and then centrifuged at room temperature (RT) for 5 min at 15,000 × g to pellet insoluble material. The supernatant (1 ml) was transferred to a tube containing 4 μl of mouse monoclonal 3B5 anti-CD8 antibody (Caltag Laboratories) specific for the extracellular portion of the human CD8-α molecule and 100 μl of 20% (vol/vol) protein A-Sepharose suspended in wash buffer (extraction buffer without leupeptin, aprotinin, and pepstatin). After incubation at 4°C for 1 h, immune complexes were pelleted and washed four times with 1 ml of wash buffer. Immunoprecipitates were mixed with 28 μl of 2× gel-loading buffer (4% [wt/vol] sodium dodecyl sulfate [SDS], 4% [wt/vol] 2-mercaptoethanol, 10% [wt/vol] glycerol, 54 μg of bromophenol blue per ml). Samples were heated for 5 min at 100°C prior to SDS-12% polyacrylamide gel electrophoresis (PAGE). Dried gels were exposed to Biomax MR film (Eastman Kodak) at −80°C.
For metabolic labeling of BL3 or BL3* cells with [35S]Met/Cys, 107 cells were suspended in 2 ml of labeling medium. Cells were washed twice in ice-cold PBS containing 10 mM N-ethylmaleimide (NEM) (30) and lysed in 1 ml of extraction buffer B (150 mM NaCl, 25 mM Tris HCl [pH 8.0], 1% [wt/vol] IGEPAL CA-630, 1% [wt/vol] sodium deoxycholate, 2 mM EDTA, 10 mM NEM, 1 mg of ovalbumin per ml, 0.5 μg of leupeptin per ml, 2 μg of aprotinin per ml, 10 μg of pepstatin A per ml). The BLV Env protein was retrieved with 5 μl of BLV2 antibody specific for the D epitope of the SU subunit (Veterinary Medical Research and Development, Inc., Pullman, Wash.), and immune complexes were harvested with 100 μl of 50-mg/ml protein G.
Metabolic labeling with H332PO4 was performed either with transfected COS-7 cells in confluent 6-well plates or with 107 BL3 or BL3* cells in suspension. Prior to labeling, cells were washed twice in 37°C Tris-buffered saline (150 mM NaCl-20 mM Tris-HCl [pH 7.4]) and then incubated at 37°C for 3 to 4 h in 2 ml of phosphate-free DMEM growth medium containing 125 μCi of H332PO4. After labeling, COS-7 cells were washed twice in RT PBS and lysed in 1 ml of extraction buffer A containing 50 mM NaF and 2 mM Na3VO4 to inhibit phosphatases. After 100 μl of 20% protein A-Sepharose (Sigma Chemical) was added, lysates were incubated at 4°C for 20 min. Samples were cleared by centrifugation for 5 min at 16,000 × g. The supernatant was transferred to a tube containing 5 μl of 3B5 anti-CD8 antibody and 100 μl of protein A-Sepharose (50 mg/ml) and was then treated as described for 35S-labeled immunoprecipitates.
32P-labeled lysates of COS-7 cells transfected with pcDNA3-BLV-Env (29) or of BL3/BL3* cells were immunoprecipitated as described for 35S-labeled lysates of BLV Env-containing cells, except that extraction buffer B was supplemented with 50 mM NaF and 2 mM Na3VO4.
Pulse-chase metabolic labeling and immunoprecipitation.
Twenty-four hours after 5.0 × 105 COS-7 cells had been transfected in T-25 flasks, the cells were washed twice in 2 ml of 37°C PBS and then rocked at 37°C for 15 min in 0.5 ml of prewarmed Cys- and Met-free RPMI 1640 containing 5% dialyzed FBS and 250 μCi of [35S]Met/Cys. Immediately thereafter, cells were washed three times with 1 ml of 37°C PBS and were incubated with 6 ml of prewarmed, nonradioactive DMEM growth medium. The zero time point sample was washed and immediately lysed in 1 ml of ice-cold extraction buffer A; the remaining flasks were incubated for 2, 4, 8, and 24 h prior to washing, extraction, and immunoprecipitation as described above.
Cell-surface and whole-cell expression of CD8-CTM proteins.
At 24 or 48 h after transfection, COS-7 cells were washed twice in 1 ml of PBS at RT and were removed from culture wells by repeated pipetting in 1 ml of PBS containing 2% (wt/vol) bovine serum albumin (PBS-B) and supplemented with 1 mM EDTA. Following two washes in 1 ml of PBS-B, cells to be stained for CD8-CTM proteins present throughout the cell were treated according to the manufacturer's protocol with the fixation and permeabilization reagents of a cell permeabilization kit (Caltag Laboratories). These cells and parallel samples to be stained only on the surface were resuspended in 100 μl of PBS-B containing 2 μg of phycoerythrin (PE)-conjugated mouse Leu 2A monoclonal anti-CD8 antibody (Becton Dickinson) and were incubated at RT in the dark for 30 min. Cells were then washed once in 1.5 ml of PBS-B, resuspended in 0.5 ml of PBS-B containing 1% (wt/vol) paraformaldehyde, and stored in the dark before analysis by flow cytometry.
Internalization of CD8-TM protein from the cell surface.
Following the procedure of Nakatsu et al. (37), COS-7 cells that had been transfected for 24 h were stained for 30 min on ice in 0.1 ml of PBS-B containing 2 μg of 3B5 monoclonal anti-CD8 antibody. After two washes in 4°C PBS-B, cells were resuspended in 1 ml of DMEM-10% FBS, divided into four equal portions, and incubated at 37°C for 0, 5, 15, and 30 min. After each interval, cells were chilled rapidly by adding 1 ml of ice-cold DMEM-10% FBS, centrifuged for 5 min at 325 × g, and washed once in cold PBS-B. The washed cells were incubated with 2 μg of secondary PE-conjugated goat anti-mouse IgG antibody (Caltag Laboratories) in 0.1 ml of PBS-B for 30 min on ice. Stained cells were then washed and fixed in 0.5 ml of PBS-B containing 1% paraformaldehyde and were kept in the dark at 4°C prior to analysis by flow cytometry.
Flow cytometry.
Immunostained cells were analyzed on a FACScan flow cytometer (Becton Dickinson). For each sample, 10,000 events were collected from single, live cells gated by forward and side scatter. Data were processed using CellQuest software (Becton Dickinson). For each experiment, the background fluorescence of antibody-stained, mock-transfected cells was used to establish a threshold, above which cells were scored positive.
Phosphoamino acid analysis.
One-dimensional separation of phosphoamino acids was performed according to the method of Hardin and Wolniak. (24) with minor modifications. Briefly, 32P-labeled, chimeric CD8-CTM proteins were transferred from SDS-polyacrylamide gels onto a polyvinylidene membrane by electrophoresis at 200 V for 1 h at 4°C in 120 mM glycine-15.6 mM Tris-HCl (pH 8.3). The membrane was rinsed in water for 1 min, air dried for 30 min, and exposed to X-ray film for 24 to 48 h at −80°C in the presence of an intensifying screen. Bands of appropriate mobility were excised and incubated in 200 μl of 100% methanol for 30 s. After membrane fragments were washed twice for 30 s in 200 μl of water, adhering protein was partially hydrolyzed by heating in 150 μl of 5.8 N HCl at 110°C for 90 min. The eluate was dried under vacuum, and the pellet was resuspended in 3 μl of phosphoamino acid standard solution (1 mg each of O-phospho-dl-serine, -threonine, and -tyrosine [Sigma Chemical] per ml of water).
The resulting samples, which contained 100 to 400 cpm by Cerenkov counting, were separated by low-voltage, thin-layer electrophoresis. Samples were spotted onto a 6.25- by 11.5-cm polyester-backed chromatography plate covered with a 250-μm layer of silica gel (Fisher Scientific). To visualize sample migration, 0.3 μl of tracking dyes (1% orange G and 1% phenol red) in electrophoresis buffer (pH 3.5; 0.5% [vol/vol] pyridine-5% [vol/vol] acetic acid in water) was applied over the spots. A Mini Sub-Cell electrophoresis unit (Bio-Rad) was set into an ice bath, and samples were separated at 250 V until the dye front reached the distal wick (approximately 1 h 45 min). After the plate was air dried for 2 h, phosphoamino acid standards were visualized by spraying the plate with 0.5% (wt/vol) ninhydrin in acetone and heating it for 15 min at 65°C. Locations of radioactive amino acids were determined by autoradiography using an intensifying screen.
RESULTS
To assess the role of key amino acid residues and motifs governing cell surface expression of BLV TM, we joined sequences encoding the C-terminal 57 amino acids that are predicted to constitute its cytoplasmic domain with sequences encoding the extracellular and membrane-spanning domains plus four to seven cytoplasmic amino acids of human CD8-α (Fig. 1B). Chimeric CD8-CTM constructs were prepared in vector pcDNA3 for transient expression in transfected COS-7 cells. By using this approach, extracellular epitopes of CD8-α could be detected with commercially available antibodies; no antibodies recognizing the extracellular domain of native BLV TM have been reported. Moreover, CD8-BLV CTM chimeric proteins have been shown to have signaling capabilities (1, 2, 23) that could affect cell survival during selection for stable expression. Transient expression reduces the chance of examining cells that survive selection because levels or compartmentalization of the expressed protein are altered.
We prepared 14 mutants of CD8-CTMwt (Fig. 1A). The ΔN-CTM construct contains the same portion of CTM used in a previous investigation of signaling by CD8-CTM chimeras (2). Among the 12 membrane-proximal amino acids missing in this deletion mutant, two dileucine pairs, a threonine, and a serine are potentially important in endocytosis. Leucines 464-465 and 467-468 were mutated to alanines in the following combinations: AA/AA, AA/LL, LL/AA, AL/LA, and LA/AL. The potential phosphoacceptor residues T461 and S462 were individually mutated to alanines (T461A and S462A).
The remainder of the cytoplasmic tail of BLV TM contains YXXL sequences that could interact with the clathrin-associated endocytic machinery (5). Tyrosines in the first and second YXXL motifs were individually replaced with alanine (Y487A and Y498A) or phenylalanine (Y487F and Y498F). Phenylalanine was tested because it cannot be phosphorylated and its space-filling properties are similar to those of tyrosine.
The C-terminal deletion mutant (ΔC-CTM) lacks all YXXL motifs but retains the membrane-proximal dileucine motifs and their upstream serine and threonine residues. Finally, the CD8-Stop construct, encoding only the first three amino acids of the BLV CTM, served as a control because it has a cytoplasmic domain lacking obvious regulatory sequences.
Stability of CD8-CTM chimeric proteins.
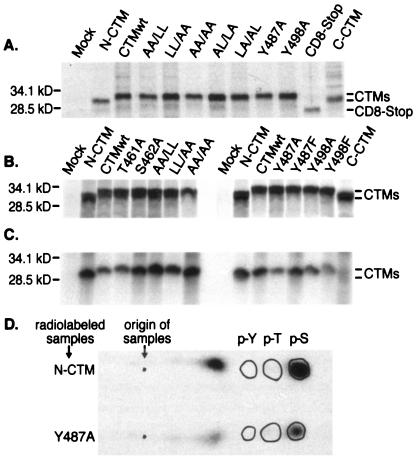
To determine whether the predicted chimeric proteins were synthesized and stable in transfected COS-7 cells, CD8-CTM proteins radiolabeled by metabolic incorporation of [35S]Met/Cys for 3 h were immunoprecipitated using CD8-specific monoclonal antibody. Chimeric proteins (Fig. 2A and B) of 28 kDa were encoded by all mutant constructs except ΔN- and ΔC-CTM, whose protein products each migrated as 25 kDa, and CD8-Stop, whose product migrated as 20 kDa. CD8-CTM chimeric proteins had similar levels of steady-state expression. The slight variability in the intensity of the bands shown in Fig. 2A and B was experiment specific and does not represent reproducible differences in levels of chimeric proteins. CD8-CTM chimeric proteins were quite sensitive to proteolysis; in some experiments, multiple degradation products were produced. No construct appeared to be more susceptible than the others. Thus, all transiently expressed chimeric proteins were present at similar steady-state levels, indicating that neither the introduced mutations nor the amino- and carboxy-terminal deletions greatly altered their abundance.
FIG. 2.
Steady-state levels of CD8-CTM chimeric proteins and their phosphorylation state. (A) Two days after transfection of COS-7 cells, CD8-CTM chimeric proteins were labeled by metabolic incorporation of [35S]Met/Cys for 3 h. Proteins were extracted, immunoprecipitated with anti-CD8 antibody, and separated by SDS-PAGE. The autoradiogram shows CD8-CTM proteins immunoprecipitated from 106 cells that were mock transfected or transfected with the listed constructs. Positions to which protein standards migrated are shown on the left of the autoradiogram (molecular mass is in kilodaltons) and CD8-CTM chimeric proteins are identified on the right. (B and C) Two sets of COS-7 cells were transfected in parallel, and after 24 h, they were radiolabeled for 3 h with [35S]Met/Cys to reveal the amounts of chimeric proteins synthesized (B) or with [32P]orthophosphate to reveal whether the proteins were phosphorylated (C). Autoradiograms of proteins immunoprecipitated with CD8-specific antibodies are shown. (D) Phosphoamino acid analysis of radiolabeled CD8-CTM chimeric proteins. Immunoprecipitated, 32P-labeled CD8-ΔN-CTM and CD8-Y487A proteins were excised from gels, hydrolyzed with acid, and separated by one-dimensional thin-layer electrophoresis. Positions to which phosphoamino acid standards (p-Y, phosphotyrosine; p-T, phosphothreonine; p-S, phosphoserine) migrated are encircled on the autoradiograms of ninhydrin-stained, thin-layer plates. The sample origin is marked.
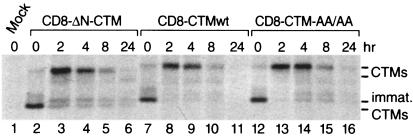
We measured the half-lives of selected chimeric proteins by pulsing transiently transfected cells with [35S]Met/Cys, replacing radioactive amino acids with unlabeled amino acids, and then immunoprecipitating radiolabeled proteins extracted 0, 2, 4, 8, and 24 h later. CD8-CTM bands of low molecular mass (approximately 20 kDa) were present at time zero (Fig. 3). By 2 h, the molecular mass of the predominant immunoprecipitated proteins had shifted to 30 to 33 kDa. This is consistent with posttranslational processing, as expected for the extracellular domain of CD8, which is glycosylated.
FIG. 3.
Stability of CD8-CTM chimeric proteins. Cells that had been transfected for 24 h were pulse-labeled for 15 min with 250 μCi of [35S]Met/Cys, washed, and then incubated in nonradioactive medium for 2, 4, 8, and 24 h. After extraction, chimeric proteins were immunoprecipitated with CD8-specific antibody and separated by SDS-PAGE. The autoradiogram (48-h exposure) shows CD8-CTM chimeric proteins retrieved from 2 × 106 cells that were mock transfected (lane 1) or transfected with ΔN-CTM (lanes 2 to 6), CTMwt (lanes 7 to 11), or AA/AA (lanes 12 to 16). Positions to which immature (immat.) and fully processed CD8-CTM fusion proteins migrated are indicated.
The CD8-CTMwt chimeric protein exhibited a 3- to 4-h half-life (Fig. 3, lanes 7 to 11), as did ΔN-CTM (lanes 2 to 6) and AA/AA (lanes 12 to 16) chimeric proteins. Since repeated measurements showed that steady-state levels of all the chimeric proteins were similar during the first 48 h after transient transfection, we infer that the other mutated proteins also have similar half-lives.
Some mutations increased cell surface expression of CD8-CTM chimeric proteins.
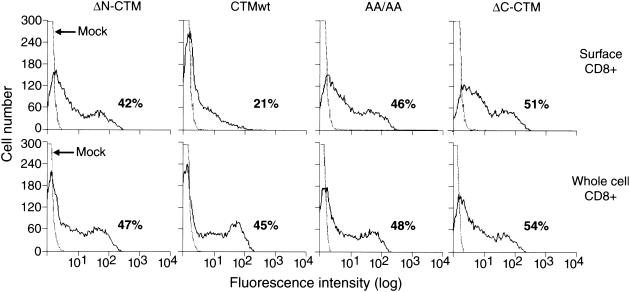
Since the wt cytoplasmic tail of BLV TM decreases display of an IL-2Rα chimeric protein on the cell surface (22), we used flow cytometry to determine how the altered protein motifs affected the residence of CD8-CTM proteins on the cell surface. At 48 h after transfection, COS-7 cells were stained directly on the surface with fluorochrome-conjugated anti-CD8 antibody. Chimeric proteins present throughout the cell were detected in parallel samples treated before staining with saponin-containing buffer to create openings in the cell membrane large enough for free passage of antibody. Percentages of CD8-positive cells did not differ reproducibly among permeabilized cells transfected with CTM constructs (representative samples are shown in Fig. 4, bottom panels). In contrast, surface display varied considerably (Fig. 4, top panels). Less than half of the cells containing the wt CTM chimeric generated a signal from surface protein that was above the background of fluorescence from mock-transfected cells. In contrast, equivalent percentages of transfected cells were positive for both surface and whole-cell expression of the ΔN-CTM, AA/AA, and ΔC-CTM chimeric proteins.
FIG. 4.
Display of CD8-CTM chimeric proteins on the cell surface. At 24 h after transfection, intact COS-7 cells were stained with PE-conjugated anti-CD8 antibody and analyzed by flow cytometry for surface display of CD8-CTM chimeric proteins (top panels). The total population of cells containing chimeric proteins was measured in parallel samples that were permeabilized and stained with the same antibody (bottom panels). Each panel shows a histogram of mock-transfected cells (dotted line) as well as a histogram generated by cells expressing one of the following chimeric proteins: ΔN-CTM, CTMwt, AA/AA, or ΔC-CTM (solid line). The percentage of cells staining for CD8 above the background staining of mock-transfected cells is shown on the lower right of each panel. Representative results from a single experiment are shown.
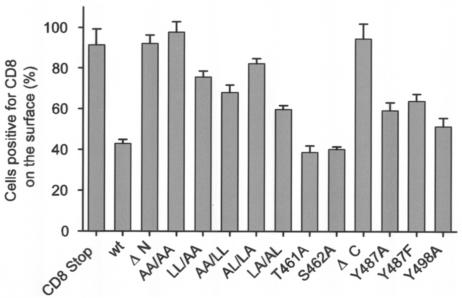
The proportions of CD8-CTM-positive cells that displayed detectable levels of the chimeric proteins on the cell surface are compared in Fig. 5; the percentages obtained in this steady-state measurement parallel mean fluorescence intensities (mfi) as exemplified by the data shown in Fig. 6B. The CD8-Stop protein lacking most cytoplasmic amino acids was displayed on the surface of nearly all the cells in which it was present (Fig. 5). In contrast, only 43% of the cells containing the CD8-CTMwt protein displayed detectable levels on the cell surface. Virtually all cells synthesizing the ΔN-CTM chimera displayed abundant amounts on the cell surface, and when both pairs of membrane-proximal leucines present in the N-terminal region of CTMwt were altered to alanines (AA/AA), the same surface-positive phenotype resulted in almost all cells. Thus, altering or removing both pairs of dileucines was sufficient to counteract the compartmentalization characteristic of the CD8-CTMwt chimeric protein. Differences between any of these constructs and wt were significant by the Tukey comparison of means (P = 0.01).
FIG. 5.
Steady-state cell surface display of CD8-CTM chimeric proteins by transiently transfected cells. Cells transfected 24 or 48 h earlier were stained with PE-conjugated anti-CD8 antibody and analyzed for surface and whole-cell CD8 as described in the legend to Fig. 4. Cells positive for CD8 on the surface are presented as a percentage of cells that contained the corresponding chimeric protein. The histogram displays averages from two to seven experiments per chimera (error bars show standard errors of the means).
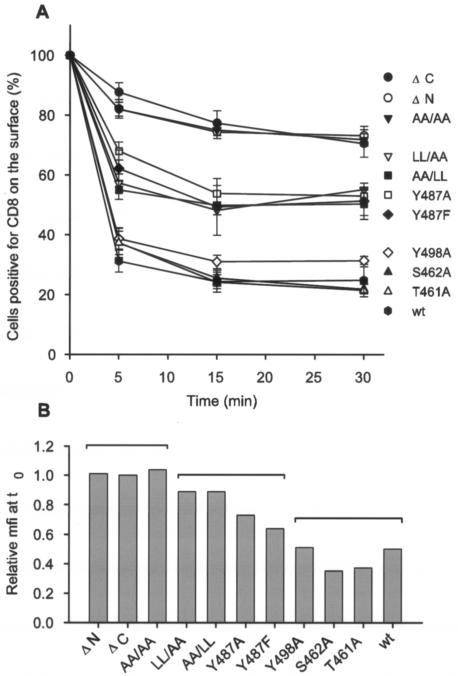
FIG. 6.
Internalization of antibody-tagged CD8-CTM chimeric proteins from the cell surface with time. Cells transfected 24 h earlier were incubated with unlabeled anti-CD8 antibody at 4°C. (A) After the time zero sample was taken, internalization of CD8-CTM antibody complexes was allowed to proceed at 37°C for 5, 15, and 30 min. Following these intervals, cells were washed, stained with a PE-conjugated secondary antibody, and analyzed by flow cytometry for the percentage displaying CD8 tagged with primary antibody on the cell surface. Those percentages are plotted as means ± standard errors from two to eight independent experiments relative to the surface-positive population measured at time zero (normalized to 100% for each chimeric protein). Differences among the means of the three groups were significant by the Tukey comparison of means (P = 0.001) at 5, 15, and 30 min. (B) The mfi at time zero of cells stained on the surface for each chimeric protein is plotted relative to the mfi (set at 1.0) of cells stained in parallel for surface ΔC-CTM protein. Brackets over the bars show the groups of chimeric proteins identified in panel A.
Replacement of one or the other pair of membrane-proximal leucines by alanines (AA/LL, LL/AA) partly counteracted wt regulation, increasing the proportion of cells displaying the mutant chimeric protein on the surface from wt levels (43%) to 68 to 76% of expressing cells (difference from wt is significant at P = 0.01). Interestingly, simultaneous alteration of the first and fourth leucines of the set (AL/LA) counteracted wt regulation (82% surface-positive cells; significantly different from wt at P = 0.01) much more strongly than did alteration of the second and third leucines (LA/AL; 60% surface-positive cells; not significantly different from wt at P = 0.01). Thus, the first and fourth leucines more strongly affect sequestration of the chimeric protein within cells.
Phosphorylation of T461 or S462 could affect the ability of the downstream dileucine motifs to mediate endocytosis. However, replacing either residue with alanine (T461A or S462A) to prohibit its phosphorylation did not alter wt distribution of the resulting chimeric proteins, which were displayed on the surfaces of only 39 to 40% of positive cells.
Deletion of the C-terminal 32 amino acids of CTM (ΔC-CTM) counteracted wt regulation and restored cell surface display of the chimeric proteins to virtually all (96%) CD8-positive cells. In contrast, replacement of either Y487 or Y498 within the first ITAM of the C terminus with alanine or phenylalanine slightly increased the proportion of cells displaying chimeric proteins on the surface from wt levels (43%) to 59% (Y487A), 64% (Y487F), and 51% (Y498A) (Y498F was also reduced [data not shown]). These values did not differ significantly from the level of wt (P = 0.01).
Thus, normal sequestration of the CD-CTMwt chimeric protein within cells was counteracted by removal of either an N- or a C-terminal segment of the CTM. Substitution of alanine for all four leucines within two dileucine motifs also conferred surface display on the altered chimeric proteins, and replacements of the first and fourth leucines were most influential. Replacement of either tyrosine within the first ITAM conferred small but reproducible increases in surface display of the chimeric proteins.
CD8-CTM chimeric proteins were internalized at different rates.
Differences in the rate of movement to and from the cell surface could account for the differential cell surface display levels of CD8-CTM proteins. To examine the role of internalization, we measured the percentage of cells retaining CD8 epitopes on the cell surface following initial binding of anti-CD8 antibody at 4°C. Antibody-tagged cells were incubated at 37°C to allow internalization of the antigen-antibody complex, and at 0-, 5-, 15-, and 30-min intervals, the cells were washed in cold growth medium. Staining of intact cells with a fluorochrome-conjugated secondary antibody revealed the percentage of cells still displaying antibody-tagged CD8 on the surface.
CTM chimeric proteins fell into three groups based on their rates of disappearance from the cell surface (Fig. 6A). Proteins in the first group, including CD8-CTMwt, were rapidly internalized. After only 5 min at 37°C, cells displaying tagged CTMwt, T461A, and S462A chimeric proteins at detectable levels on the surface dropped to 30 to 40% of the number present at the outset; the percentage of surface-positive cells then stabilized at 25%. Cells displaying the Y498A chimeric protein on the surface also dropped quickly to 38%, but 31% maintained surface display, a slightly higher proportion than the others in this group. Proteins in the second group had an intermediate rate of disappearance from the cell surface. Cells positive on the surface for the LL/AA, AA/LL, Y487A, and Y487F chimeric proteins dropped within 5 min to 55 to 68% of initially positive cells and to 48 to 55% by 15 and 30 min. Proteins in the third group disappeared most slowly from the cell surface. Over 80% of cells initially positive for the ΔC-CTM, ΔN-CTM, and AA/AA chimeric proteins displayed antibody-tagged protein on the surface at 5 min and 70 to 73% were surface positive at 30 min, nearly threefold more than those displaying the wt chimera at that time. Differences among means of the three groups were significant from each other at each time point by the Tukey comparison (P = 0.001). Importantly, these differences between the groups were not solely predicted by the relative mfi of the cells at time zero (Fig. 6B). The statistically significant segregation of Y487A and Y487F from Y498A into different internalization groups (Fig. 6A) could not have been predicted by comparison of the fluorescence intensities of these constructs at the outset (Fig. 6B).
Thus, the rapid disappearance of the CD8-CTMwt protein from the cell surface was markedly counteracted by deletion of either the N or C terminus of the CTM or by simultaneous mutation of both membrane-proximal dileucine pairs. Internalization was moderately counteracted by mutation of one of the two dileucine motifs or of the tyrosine in the most membrane-proximal YXXL motif. Mutation of the tyrosine in the middle YXXL motif did not prevent rapid internalization, nor did mutation of T461 or S462.
The BLV CTM was phosphorylated in COS-7 cells transiently expressing CD8-CTM chimeric proteins.
Phosphorylation of serine or threonine residues upstream of dileucine internalization motifs can affect their function (16, 20, 31), and phosphorylation of tyrosines in YXXL motifs within ITAMs is essential for their recognition by SH2 domains (42). Hence, we investigated the phosphorylation state of CD8-CTM chimeric proteins in transiently transfected COS-7 cells. All of the chimeric proteins were synthesized at similar levels, as shown by metabolic labeling with [35S]Met/Cys (Fig. 2A and B), and all but ΔC-CTM were phosphorylated in COS-7 cells labeled for 3 h with [32P]orthophosphate at 24 h posttransfection (Fig. 2C). Levels of phosphorylation of individual chimeric proteins differed somewhat among experiments, but no pattern was evident. The CD8-Stop chimera was never phosphorylated and the ΔC-CTM chimera was sometimes weakly phosphorylated when labeled at 48 h after transfection (39; data not shown). The CTMwt, ΔN-CTM, T461A, S462A, Y487A, and Y498A chimeric proteins contained phosphoserine (see examples in Fig. 2D). Neither phosphotyrosine nor phosphothreonine residues were detected. Thus, CD8-CTM chimeric proteins with intact C termini were phosphorylated at serine residues when transiently expressed in COS cells.
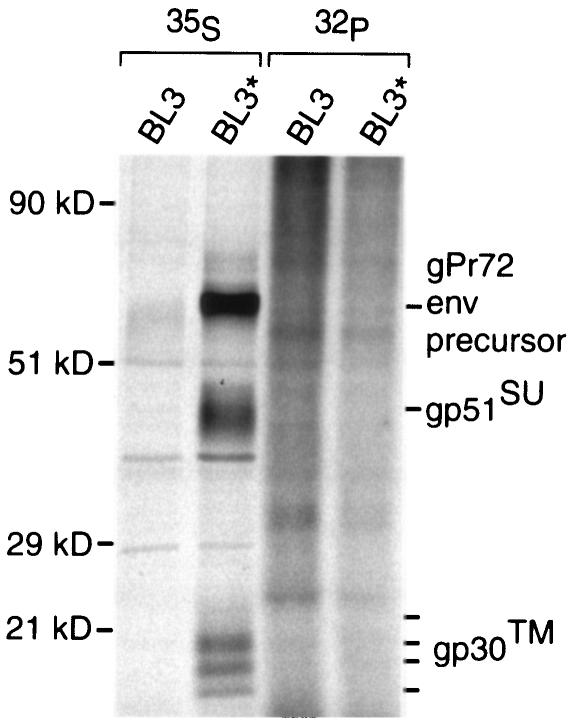
To determine whether the native BLV Env protein was phosphorylated in BLV-producing cells, we radiolabeled BLV-producing BL3* bovine lymphoid cells (50) and their parental uninfected BL3 cells (58) with [35S]Met/Cys or with [32P]orthophosphate. Lysates were prepared with NEM to stabilize disulfide binding between SU and TM, so that TM could be retrieved with a monoclonal antibody specific for SU (30). BLV Env proteins were not detectably phosphorylated in infected cells constitutively producing virus (Fig. 7), in agreement with results obtained by others using two different virus-producing, non-lymphoid cell lines (23). A difficulty with interpreting these results is that viral or cellular mutations could have occurred during the derivation of virus-producing cell lines and such a change might preclude phosphorylation of the CTM. As another approach, we investigated whether the BLV Env protein became phosphorylated when transiently expressed in cells in the absence of other viral proteins. TM was not phosphorylated when immunoprecipitated from radiolabeled COS-7 cells (39; data not shown) that had been transfected with a plasmid expressing the native BLV Env protein (29, 30).
FIG. 7.
Lack of phosphorylation of native BLV Env protein in B-lymphoid cells. BLV-producing BL3* cells and BLV-negative BL3 cells were incubated with [35S]Met/Cys or [32P]orthophosphate for 3 to 4 h. After lysates were prepared in the presence of NEM, Env proteins were immunoprecipitated using monoclonal antibodies specific for the D epitope of SU. Samples were treated with 2-mercaptoethanol and heated before proteins were separated by SDS-PAGE. Molecular mass standards are shown at left, and locations of the Env precursor protein (gPr72), SU (gp51), and TM (gp30) are shown at right.
DISCUSSION
The BLV CTM is laden with peptide sequences that in other contexts mediate protein interactions involved in signaling and intracellular trafficking of membrane proteins. Alterations in YXXL and PXXPX4PP motifs reduce or eliminate BLV replication in vivo (45, 61) and in vitro (27), but the mechanisms and timing with which potential protein interactions could affect the viral life cycle remain unknown. To decipher these is difficult because there is no culture system in which single-cycle viral replication can be studied in B lymphocytes at appropriate stages of differentiation and activation. We chose to investigate the ability of the BLV CTM to affect the cell surface display of a heterologous protein and found that two membrane-proximal dileucine motifs and the membrane-proximal YXXL motif can modify wt downregulation. Among the amino acid substitutions tested, substitution of alanine for all four leucines in the membrane-proximal dileucine motifs most strongly countered protein internalization, and the effect of deleting the membrane-proximal 12 amino acids encompassing these residues was equivalent. Replacement of only one dileucine motif partially alleviated wt regulation. Individual tyrosine substitutions only weakly increased cell surface display of the proteins, but Y487 substitutions prolonged the residence of the chimeric protein on the cell surface. Removal of all YXXL motifs with a large C-terminal deletion increased surface display and slowed internalization as much as removal of the N-terminal 12 amino acids did, in spite of the fact that the residual cytoplasmic domain of CD8-ΔC-CTM retained the two membrane-proximal dileucine motifs, which otherwise exerted a powerful influence.
The strong surface display of both the ΔN- and ΔC-CTM proteins suggests that both the N- and C-terminal regions of the BLV CTM play roles in downmodulation. The very long CTM domains of simian immunodeficiency virus (SIV) and human immunodeficiency virus type 1 (HIV-1) contain membrane-proximal tyrosine residues (SIV Y721 and HIV-1 Y712) that mediate rapid internalization of Env (17, 54) and that interact with the clathrin adaptor complexes AP-2 and AP-1 (3, 4, 7, 32, 40). However, both viruses have C-terminal CTM sequences that also affect surface display (3, 7, 62). A C-terminal dileucine motif (LL855/856) in the HIV CTM interacts with AP-1; alanine substitution prevents the interaction with AP-1 and alters the intracellular location of Env (62). Alanine substitution does not by itself increase surface display, but when paired with the Y712A substitution, it augments surface display beyond that of Y712A alone. In contrast, replacing just one of the membrane-proximal dileucine motifs in the BLV CTM significantly increased surface display of the chimeric protein. We did not assess YXXL mutants in combination with dileucine mutants.
Although the sorting components interacting with YXXΦ and dileucine motifs can become saturated, the two motifs have not been found to compete for binding to adaptor molecules (36). We do not believe that saturation contributed significantly to differential levels of surface display of mutant chimeric proteins by the transfected COS cells. Overall levels of chimeric proteins were similar among cell populations, and equivalent numbers of cells synthesized the chimeric proteins. Even with the internalization machinery potentially challenged, 60% of cells expressing the CD8-CTMwt chimera (with all motifs intact) did not display detectable levels of the protein on the surface. Had the protein been expressed at lower levels, the range for detecting differences among mutant constructs might have expanded. Nonetheless, a number of differences from wt were significant.
The two membrane-proximal dileucines in the BLV CTM are embedded in a sequence, LTSRLLKLLR, that could form an amphipathic alpha helix having polybasic and hydrophobic faces. The positively charged side chains of the polybasic face could interact with negatively charged lipid head groups at the inner face of a membrane, orienting the bulky hydrophobic side chains toward the cytoplasm. In the BLV isolates sequenced to date, all leucines in this sequence are conserved except the second residue of the first dileucine motif, which is replaced by a serine in the proviral clone pBLV-A1 (GenBank accession no. D00647). The polar side chain of that particular residue would be at one edge of the hydrophobic patch formed by the other leucine side chains. Our results show that a large portion of this hydrophobic patch is necessary for wt sequestration of the CTM from the cell surface. However, even the whole patch is not sufficient for internalization when the C terminus of the CTM is missing. The dileucine residues forming the hydrophobic patch could potentially be recognized by trafficking components mediating endocytosis (6, 31). Dileucine motifs functioning in lysosomal and basolateral targeting usually have a negatively charged residue upstream (31), but in the BLV CTM, replacement of either S462 or T461 to prevent phosphorylation did not alter wt surface display and rapid internalization.
A CD8-CTM chimeric protein corresponding to our ΔN-CTM deletion mutant was used previously to show that the BLV CTM can transduce activating signals in murine lymphoma cells (2). All the mutations evaluated in that work were thus assessed in the context of a CTM lacking the putative N-terminal amphipathic helix. Stable transfectants expressing that parental chimeric protein exhibited low-level cell surface display relative to native murine CD8-α, whereas our transiently expressed CD8-ΔN-CTM protein was as highly displayed on the cell surface as our CD8-Stop construct. We speculate that selection during the derivation of stable transfectants (2) and perhaps the difference in host cell lineages account for the disparity. In contrast, a different chimeric protein composed of IL-2Rα and the BLV CTM was expressed at lower levels on the cell surface than those of authentic IL-2Rα (22), in agreement with our results, and the IL-2Rα-CTM and our CD8-CTM chimeric proteins had similar half-lives. Both the IL-2Rα-CTM and our CD8-CTMwt proteins were rapidly internalized. The partial colocalization of the IL-2Rα-CTM protein with transferrin (22), which is characteristically located in early and recycling endosomes, raises the possibility that our CD8-CTM chimeras are present in these compartments and could recycle to the plasma membrane.
Neither the Y487A/F nor the Y498A substitution increased the steady-state surface display of the respective chimeric proteins significantly above wt levels. However, a functional difference between Y487 and Y498 mutants became evident in the internalization experiment. In the 30 min after chimeric proteins were tagged with antibody on the cell surface, a significantly higher proportion of Y487A/F-expressing cells exhibited the tagged proteins on the surface than did cells expressing wt or Y498A chimeras. In addition to its placement in the most membrane-proximal YXXL of the first ITAM in the BLV CTM, we note that Y487 is embedded in a DXXXLL sequence. The peptide sequence D/EXXXLL/I is recognized by μ and/or β subunits of AP complexes (6, 31); its acidic residue is important for targeting proteins to late endosomes or lysosomes (44, 53). As part of this motif, Y487 could interact with a different trafficking component than Y498. Perhaps an interaction involving Y487 normally prevents recycling back onto the surface from early endosomes.
The fact that replacements of Y487 affect the extent of internalization as well as transmission of signals initiated by cross-linking of the extracellular domain of a CD8 chimera (2) suggests that this tyrosine residue plays multiple roles. Its phosphorylation could serve as a switch, since APs of the clathrin endocytic machinery cannot bind when the tyrosine of a YXXΦ motif is phosphorylated (5, 8, 41, 56). The YXXΦ sequence of the T-cell costimulatory receptor CTLA-4 exemplifies such a dual function: in resting T cells, it binds the μ subunit of AP-2 and the protein is rapidly internalized. Upon cellular activation, the tyrosine becomes phosphorylated, preventing interaction with μ2 and inhibiting internalization. The phosphorylated tyrosine can then become part of a ligand for SH2 domains of signaling molecules (12, 56, 63).
SHP-1 phosphatase, which contains two SH2 motifs, associates with the BLV CTM in cultured peripheral blood mononuclear cells from BLV-infected cows if tyrosine phosphatases are inhibited prior to cell lysis (11). Since only phosphoserine was present in CD8-CTM chimeric protein in transfected COS cells, our results suggest that an SHP-CTM interaction is unlikely to take place in those cells. The cytoplasmic tail of BLV TM may become phosphorylated at tyrosine and interact with SHP-1 only in virus-infected primary cells and only at certain stages of infection, because in some infected animals, no association of SHP-1 with TM could be detected (11). Disappointingly, systematic attempts to demonstrate phosphorylation of BLV TM in infected cells from infected animals have been unsuccessful (23). Success in such an endeavor may require replication of cell-cell signaling interactions that occur when BLV is being produced by B cells in vivo.
Other than for CD8-ΔC-CTM, we detected no reproducible differences in the levels of serine phosphorylation of mutant chimeric proteins. Of five serine residues in the BLV CTM, S478 and S485 reside in a region common to both N- and C-terminal deletion mutants. One or both of these serines may be modified, since both deletion chimeras were phosphorylated, albeit only weakly and late after transfection for ΔC-CTM. The relevance to the BLV life cycle of this interaction with a serine protein kinase is unclear. The CTM of Env may be phosphorylated at serine in vivo under certain conditions, but the protein has not yet been captured in that state. In addition, NEM treatment of Env, which was necessary to retrieve TM using anti-SU antibodies, may select a subpopulation of TM or may induce phosphatase activity.
The results presented here as well those published previously reveal functional distinctions between Y487 and Y498 that may reflect differential roles in cell signaling and protein trafficking. Here, the Y498A substitution did not by itself markedly alter wt downregulation of the CD8-CTM chimeric protein, although the residual level of tagged surface protein was subtly increased. This tyrosine, however, plays a critical role during BLV infection because a Y498D mutant provirus gives no signs of being infectious in sheep (61). Furthermore, a Y498A substitution completely prevents cell-to-cell propagation of the mutant virus in transfected fetal lamb kidney cells (27). The infectivity of viral particles produced after transfection of COS-1 cells with a provirus encoding the Y498A mutation is low, and the particles appear to lack Env protein (27). Cultured cells transfected with Y498D mutants shed particles containing the viral capsid protein, but the presence of Env protein, which is necessary for infectivity, was not assessed (61). Y498 may affect the assembly of viral cores with Env protein on some intracellular membrane. On the other hand, Y487 mutants do not have such severe effects on viral propagation. Sheep injected with Y487D mutant provirus have detectable viral loads and their blood cells can produce very small amounts of viral capsid protein in culture (61). Y487A mutants are not impaired in cell-to-cell transmission in culture (27). In our work described here, Y487A chimeric protein was present on the cell surface longer than Y498A protein. Together, these results suggest that altered trafficking could compensate in part for impairment of signaling function.
A strong selective pressure must exist to maintain the BLV CTM as a whole because its membrane-proximal ITAM is bracketed by a direct repeat in the nucleotide sequence encoding the amino acids KPD (49). Loss of this segment by recombination would remove not only both ITAMs but also the DXXXLL and two PXXP motifs. The cytoplasmic domain would be shortened to 34 amino acids, leaving only one YXXL motif in a cytoplasmic domain more like that of HTLV TM. Other oncogenic retroviruses also contain a single YXXL motif, suggesting that it plays a common role in replication. Deletion mutants lacking the YXXI motif in the cytoplasmic tail of HTLV-1 TM fail in cell-to-cell transmission (13). Sequestration of Env inside the cell could circumvent certain mechanisms of immune-mediated destruction prior to, and perhaps during, assembly of new viral particles in an infected cell. The additional CTM sequences acquired and maintained by BLV must be intricately entwined in the signaling and protein trafficking pathways of its B-cell hosts.
Acknowledgments
Flow cytometry data were acquired in the UC Davis Optical Biology Shared Resource Facility, supported in part by the UC Davis Cancer Center. This research was supported by Public Health Service grants CA46374 from the National Cancer Institute (K.R.) and AI46145 from the National Institute of Allergy and Infectious Diseases (E.T.S.), by a University of California Davis Faculty Research Grant (K.R.), by funds from the UC Davis Cancer Center (K.R.), by funds from the Universitywide AIDS Research Program (E.T.S.), and by grants from the Prentice Foundation and the Educational Foundation of America. S.N. was supported in part through the University of California Systemwide Biotechnology Research and Education Program.
REFERENCES
- 1.Alber, G., K. M. Kim, P. Weiser, C. Riesterer, R. Sarsetti, and M. Reth. 1993. Molecular mimicry of the antigen receptor signalling motifs by transmembrane proteins of the Epstein-Barr virus and the bovine leukemia virus. Curr. Biol. 3:333-339. [DOI] [PubMed] [Google Scholar]
- 2.Beaufils, P., D. Choquet, R. Z. Mamoun, and B. Malissen. 1993. The (YXXL/I)2 signalling motif found in the cytoplasmic segments of the bovine leukaemia virus envelope protein and Epstein-Barr virus latent membrane protein 2A can elicit early and late lymphocyte activation events. EMBO J. 12:5105-5112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berlioz-Torrent, C., B. L. Shacklett, L. Erdtmann, L. Delamarre, I. Bouchaert, P. Sonigo, M. C. Dokhelar, and R. Benarous. 1999. Interactions of the cytoplasmic domains of human and simian retroviral transmembrane proteins with components of the clathrin adaptor complexes modulate intracellular and cell surface expression of envelope glycoproteins. J. Virol. 73:1350-1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boge, M., S. Wyss, J. S. Bonifacino, and M. Thali. 1998. A membrane-proximal tyrosine-based signal mediates internalization of the HIV-1 envelope glycoprotein via interaction with the AP-2 clathrin adaptor. J. Biol. Chem. 273:15773-15778. [DOI] [PubMed] [Google Scholar]
- 5.Boll, W., H. Ohno, Z. Songyang, I. Rapoport, L. C. Cantley, J. S. Bonifacino, and T. Kirchhausen. 1996. Sequence requirements for the recognition of tyrosine-based endocytic signals by clathrin AP-2 complexes. EMBO J. 15:5789-5795. [PMC free article] [PubMed] [Google Scholar]
- 6.Bonifacino, J. S., and L. M. Traub. 2003. Signals for sorting of transmembrane proteins to endosomes and lysosomes. Annu. Rev. Biochem. 72:395-447. [DOI] [PubMed] [Google Scholar]
- 7.Bowers, K., A. Pelchen-Matthews, S. Honing, P. J. Vance, L. Creary, B. S. Haggarty, J. Romano, W. Ballensiefen, J. A. Hoxie, and M. Marsh. 2000. The simian immunodeficiency virus envelope glycoprotein contains multiple signals that regulate its cell surface expression and endocytosis. Traffic 1:661-674. [DOI] [PubMed] [Google Scholar]
- 8.Bradshaw, J. D., P. Lu, G. Leytze, J. Rodgers, G. L. Schieven, K. L. Bennett, P. S. Linsley, and S. E. Kurtz. 1997. Interaction of the cytoplasmic tail of CTLA-4 (CD152) with a clathrin-associated protein is negatively regulated by tyrosine phosphorylation. Biochemistry 36:15975-15982. [DOI] [PubMed] [Google Scholar]
- 9.Cambier, J. C. 1995. Antigen and Fc receptor signaling. The awesome power of the immunoreceptor tyrosine-based activation motif (ITAM). J. Immunol. 155:3281-3285. [PubMed] [Google Scholar]
- 10.Cantor, G. H. 1996. A potential proline-rich motif upstream of the immunoreceptor tyrosine-based activation motif in bovine leukemia virus gp30, Epstein-Barr virus LMP2A, herpesvirus papio LMP2A, and African horsesickness virus VP7. Virology 220:265-266. [DOI] [PubMed] [Google Scholar]
- 11.Cantor, G. H., S. M. Pritchard, O. Orlik, G. A. Splitter, W. C. Davis, and R. Reeves. 1999. Bovine leukemia virus transmembrane protein gp30 physically associates with the down-regulatory phosphatase SHP-1. Cell Immunol. 193:117-124. [DOI] [PubMed] [Google Scholar]
- 12.Chuang, E., M. L. Alegre, C. S. Duckett, P. J. Noel, M. G. Vander Heiden, and C. B. Thompson. 1997. Interaction of CTLA-4 with the clathrin-associated protein AP50 results in ligand-independent endocytosis that limits cell surface expression. J. Immunol. 159:144-151. [PubMed] [Google Scholar]
- 13.Delamarre, L., C. Pique, A. R. Rosenberg, V. Blot, M. P. Grange, I. Le Blanc, and M. C. Dokhelar. 1999. The Y-S-L-I tyrosine-based motif in the cytoplasmic domain of the human T-cell leukemia virus type 1 envelope is essential for cell-to-cell transmission. J. Virol. 73:9659-9663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deschamps, J., R. Kettmann, and A. Burny. 1981. Experiments with cloned complete tumor-derived bovine leukemia virus information prove that the virus is totally exogenous to its target animal species. J. Virol. 40:605-609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dietrich, J., X. Hou, A. M. Wegener, and C. Geisler. 1994. CD3 gamma contains a phosphoserine-dependent di-leucine motif involved in down-regulation of the T cell receptor. EMBO J. 13:2156-2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dittrich, E., C. R. Haft, L. Muys, P. C. Heinrich, and L. Graeve. 1996. A di-leucine motif and an upstream serine in the interleukin-6 (IL-6) signal transducer gp130 mediate ligand-induced endocytosis and down-regulation of the IL-6 receptor. J. Biol. Chem. 271:5487-5494. [DOI] [PubMed] [Google Scholar]
- 17.Egan, M. A., L. M. Carruth, J. F. Rowell, X. Yu, and R. F. Siliciano. 1996. Human immunodeficiency virus type 1 envelope protein endocytosis mediated by a highly conserved intrinsic internalization signal in the cytoplasmic domain of gp41 is suppressed in the presence of the Pr55gag precursor protein. J. Virol. 70:6547-6556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Flaswinkel, H., M. Barner, and M. Reth. 1995. The tyrosine activation motif as a target of protein tyrosine kinases and SH2 domains. Semin. Immunol. 7:21-27. [DOI] [PubMed] [Google Scholar]
- 19.Gatot, J. S., I. Callebaut, J. P. Mornon, D. Portetelle, A. Burny, P. Kerkhofs, R. Kettmann, and L. Willems. 1998. Conservative mutations in the immunosuppressive region of the bovine leukemia virus transmembrane protein affect fusion but not infectivity in vivo. J. Biol. Chem. 273:12870-12880. [DOI] [PubMed] [Google Scholar]
- 20.Gibson, R. M., W. P. Schiemann, L. B. Prichard, J. M. Reno, L. H. Ericsson, and N. M. Nathanson. 2000. Phosphorylation of human gp130 at Ser-782 adjacent to the di-leucine internalization motif. Effects on expression and signaling. J. Biol. Chem. 275:22574-22582. [DOI] [PubMed] [Google Scholar]
- 21.Gould, S. J., A. M. Booth, and J. E. Hildreth. 2003. The Trojan exosome hypothesis. Proc. Natl. Acad. Sci. USA 100:10592-10597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grange, M. P., V. Blot, L. Delamarre, I. Bouchaert, A. Rocca, A. Dautry-Varsat, and M. C. Dokhelar. 2000. Identification of two intracellular mechanisms leading to reduced expression of oncoretrovirus envelope glycoproteins at the cell surface. J. Virol. 74:11734-11743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hamilton, V. T., D. M. Stone, S. M. Pritchard, and G. H. Cantor. 2002. Bovine leukemia virus gp30 transmembrane (TM) protein is not tyrosine phosphorylated: examining potential interactions with host tyrosine-mediated signaling. Virus Res. 90:155-169. [DOI] [PubMed] [Google Scholar]
- 24.Hardin, S. C., and S. M. Wolniak. 1998. Low-voltage separation of phosphoamino acids by silica gel thin-layer electrophoresis in a DNA electrophoresis cell. BioTechniques 24:344-346. [DOI] [PubMed] [Google Scholar]
- 25.Ho, S. N., H. D. Hunt, R. M. Horton, J. K. Pullen, and L. R. Pease. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77:51-59. [DOI] [PubMed] [Google Scholar]
- 26.Hunter, E. 1997. Viral entry and receptors, p. 71-119. In J. M. Coffin, S. H. Hughes, and H. E. Varmus (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. [PubMed]
- 27.Inabe, K., M. Nishizawa, S. Tajima, K. Ikuta, and Y. Aida. 1999. The YXXL sequences of a transmembrane protein of bovine leukemia virus are required for viral entry and incorporation of viral envelope protein into virions. J. Virol. 73:1293-1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Irving, B. A., and A. Weiss. 1991. The cytoplasmic domain of the T cell receptor zeta chain is sufficient to couple to receptor-associated signal transduction pathways. Cell 64:891-901. [DOI] [PubMed] [Google Scholar]
- 29.Johnston, E. R., L. M. Albritton, and K. Radke. 2002. Envelope proteins containing single amino acid substitutions support a structural model of the receptor-binding domain of bovine leukemia virus surface protein. J. Virol. 76:10861-10872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johnston, E. R., and K. Radke. 2000. The SU and TM envelope protein subunits of bovine leukemia virus are linked by disulfide bonds, both in cells and in virions. J. Virol. 74:2930-2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kirchhausen, T. 1999. Adaptors for clathrin-mediated traffic. Annu. Rev. Cell Dev. Biol. 15:705-732. [DOI] [PubMed] [Google Scholar]
- 32.LaBranche, C. C., M. M. Sauter, B. S. Haggarty, P. J. Vance, J. Romano, T. K. Hart, P. J. Bugelski, M. Marsh, and J. A. Hoxie. 1995. A single amino acid change in the cytoplasmic domain of the simian immunodeficiency virus transmembrane molecule increases envelope glycoprotein expression on infected cells. J. Virol. 69:5217-5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Letourneur, F., and R. D. Klausner. 1992. A novel di-leucine motif and a tyrosine-based motif independently mediate lysosomal targeting and endocytosis of CD3 chains. Cell 69:1143-1157. [DOI] [PubMed] [Google Scholar]
- 34.Littman, D. R., Y. Thomas, P. J. Maddon, L. Chess, and R. Axel. 1985. The isolation and sequence of the gene encoding T8: a molecule defining functional classes of T lymphocytes. Cell 40:237-246. [DOI] [PubMed] [Google Scholar]
- 35.Lodge, R., L. Delamarre, J. P. Lalonde, J. Alvarado, D. A. Sanders, M. C. Dokhelar, E. A. Cohen, and G. Lemay. 1997. Two distinct oncornaviruses harbor an intracytoplasmic tyrosine-based basolateral targeting signal in their viral envelope glycoprotein. J. Virol. 71:5696-5702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marks, M. S., L. Woodruff, H. Ohno, and J. S. Bonifacino. 1996. Protein targeting by tyrosine- and di-leucine-based signals: evidence for distinct saturable components. J. Cell Biol. 135:341-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nakatsu, F., M. Sakuma, Y. Matsuo, H. Arase, S. Yamasaki, N. Nakamura, T. Saito, and H. Ohno. 2000. A di-leucine signal in the ubiquitin moiety. Possible involvement in ubiquitination-mediated endocytosis. J. Biol. Chem. 275:26213-26219. [DOI] [PubMed] [Google Scholar]
- 38.Neel, B. G. 1997. Role of phosphatases in lymphocyte activation. Curr. Opin. Immunol. 9:405-420. [DOI] [PubMed] [Google Scholar]
- 39.Novakovic, S. 2002. Ph.D. thesis. University of California, Davis.
- 40.Ohno, H., R. C. Aguilar, M. C. Fournier, S. Hennecke, P. Cosson, and J. S. Bonifacino. 1997. Interaction of endocytic signals from the HIV-1 envelope glycoprotein complex with members of the adaptor medium chain family. Virology 238:305-315. [DOI] [PubMed] [Google Scholar]
- 41.Ohno, H., M. C. Fournier, G. Poy, and J. S. Bonifacino. 1996. Structural determinants of interaction of tyrosine-based sorting signals with the adaptor medium chains. J. Biol. Chem. 271:29009-29015. [DOI] [PubMed] [Google Scholar]
- 42.Pawson, T. 1995. Protein modules and signalling networks. Nature 373:573-580. [DOI] [PubMed] [Google Scholar]
- 43.Pitcher, C., S. Honing, A. Fingerhut, K. Bowers, and M. Marsh. 1999. Cluster of differentiation antigen 4 (CD4) endocytosis and adaptor complex binding require activation of the CD4 endocytosis signal by serine phosphorylation. Mol. Biol. Cell 10:677-691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pond, L., L. A. Kuhn, L. Teyton, M. P. Schutze, J. A. Tainer, M. R. Jackson, and P. A. Peterson. 1995. A role for acidic residues in di-leucine motif-based targeting to the endocytic pathway. J. Biol. Chem. 270:19989-19997. [DOI] [PubMed] [Google Scholar]
- 45.Reichert, M., A. Winnicka, L. Willems, R. Kettmann, and G. H. Cantor. 2001. Role of the proline-rich motif of bovine leukemia virus transmembrane protein gp30 in viral load and pathogenicity in sheep. J. Virol. 75:8082-8089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ren, R., B. J. Mayer, P. Cicchetti, and D. Baltimore. 1993. Identification of a ten-amino acid proline-rich SH3 binding site. Science 259:1157-1161. [DOI] [PubMed] [Google Scholar]
- 47.Reth, M. 1989. Antigen receptor tail clue. Nature 338:383-384. [PubMed] [Google Scholar]
- 48.Rice, N. R., R. M. Stephens, and R. V. Gilden. 1987. Sequence analysis of the bovine leukemia virus genome, p. 115-144. In A. Burny and M. Mammerickx (ed.), Enzootic bovine leukosis and bovine leukemia virus. Martinus Nijhoff, Leiden, The Netherlands.
- 49.Rice, N. R., R. M. Stephens, D. Couez, J. Deschamps, R. Kettmann, A. Burny, and R. V. Gilden. 1984. The nucleotide sequence of the env gene and post-env region of bovine leukemia virus. Virology 138:82-93. [DOI] [PubMed] [Google Scholar]
- 50.Romano, M. J., J. A. Stewart, and H. A. Lewin. 1989. Phenotypic characterization of bovine lymphoblastoid cell lines. Vet. Immunol. Immunopathol. 23:293-307. [DOI] [PubMed] [Google Scholar]
- 51.Rowell, J. F., P. E. Stanhope, and R. F. Siliciano. 1995. Endocytosis of endogenously synthesized HIV-1 envelope protein. Mechanism and role in processing for association with class II MHC. J. Immunol. 155:473-488. [PubMed] [Google Scholar]
- 52.Sagata, N., T. Yasunaga, J. Tsuzuku-Kawamura, K. Ohishi, Y. Ogawa, and Y. Ikawa. 1985. Complete nucleotide sequence of the genome of bovine leukemia virus: its evolutionary relationship to other retroviruses. Proc. Natl. Acad. Sci. USA 82:677-681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sandoval, I. V., S. Martinez-Arca, J. Valdueza, S. Palacios, and G. D. Holman. 2000. Distinct reading of different structural determinants modulates the dileucine-mediated transport steps of the lysosomal membrane protein LIMPII and the insulin-sensitive glucose transporter GLUT4. J. Biol. Chem. 275:39874-39885. [DOI] [PubMed] [Google Scholar]
- 54.Sauter, M. M., A. Pelchen-Matthews, R. Bron, M. Marsh, C. C. LaBranche, P. J. Vance, J. Romano, B. S. Haggarty, T. K. Hart, W. M. Lee, and J. A. Hoxie. 1996. An internalization signal in the simian immunodeficiency virus transmembrane protein cytoplasmic domain modulates expression of envelope glycoproteins on the cell surface. J. Cell Biol. 132:795-811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sawai, E. T., A. Baur, H. Struble, B. M. Peterlin, J. A. Levy, and C. Cheng-Mayer. 1994. Human immunodeficiency virus type 1 Nef associates with a cellular serine kinase in T lymphocytes. Proc. Natl. Acad. Sci. USA 91:1539-1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shiratori, T., S. Miyatake, H. Ohno, C. Nakaseko, K. Isono, J. S. Bonifacino, and T. Saito. 1997. Tyrosine phosphorylation controls internalization of CTLA-4 by regulating its interaction with clathrin-associated adaptor complex AP-2. Immunity 6:583-589. [DOI] [PubMed] [Google Scholar]
- 57.Sinclair, N. R. 2000. Immunoreceptor tyrosine-based inhibitory motifs on activating molecules. Crit. Rev. Immunol. 20:89-102. [PubMed] [Google Scholar]
- 58.Theilen, G. H., J. M. Miller, J. Higgins, R. N. Ruppanner, and W. Garrett. 1982. Vaccination against bovine leukaemia virus infection, p. 547-560. In O. C. Straub (ed.), Fourth international symposium on bovine leukosis. Martinius Nijhoff, The Hague, The Netherlands.
- 59.Vivier, E., and M. Daeron. 1997. Immunoreceptor tyrosine-based inhibition motifs. Immunol. Today 18:286-291. [DOI] [PubMed] [Google Scholar]
- 60.Voneche, V., D. Portetelle, R. Kettmann, L. Willems, K. Limbach, E. Paoletti, J. M. Ruysschaert, A. Burny, and R. Brasseur. 1992. Fusogenic segments of bovine leukemia virus and simian immunodeficiency virus are interchangeable and mediate fusion by means of oblique insertion in the lipid bilayer of their target cells. Proc. Natl. Acad. Sci. USA 89:3810-3814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Willems, L., J. S. Gatot, M. Mammerickx, D. Portetelle, A. Burny, P. Kerkhofs, and R. Kettmann. 1995. The YXXL signalling motifs of the bovine leukemia virus transmembrane protein are required for in vivo infection and maintenance of high viral loads. J. Virol. 69:4137-4141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wyss, S., C. Berlioz-Torrent, M. Boge, G. Blot, S. Honing, R. Benarous, and M. Thali. 2001. The highly conserved C-terminal dileucine motif in the cytosolic domain of the human immunodeficiency virus type 1 envelope glycoprotein is critical for its association with the AP-1 clathrin adapter. J. Virol. 75:2982-2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang, Y., and J. P. Allison. 1997. Interaction of CTLA-4 with AP50, a clathrin-coated pit adaptor protein. Proc. Natl. Acad. Sci. USA 94:9273-9278. [DOI] [PMC free article] [PubMed] [Google Scholar]