Abstract
Mouse and human genomes carry more than a dozen genes coding for closely related alpha interferon (IFN-α) subtypes. IFN-α, as well as IFN-β, IFN-κ, IFN-ɛ, and limitin, are thought to bind the same receptor, raising the question of whether different IFN subtypes possess specific functions. As some confusion existed in the identity and characteristics of mouse IFN-α subtypes, the availability of data from the mouse genome sequence prompted us to characterize the murine IFN-α family. A total of 14 IFN-α genes were detected in the mouse genome, in addition to three IFN-α pseudogenes. Four IFN-α genes (IFN-α1, IFN-α7/10, IFN-α8/6, and IFN-α11) exhibited surprising allelic divergence between 129/Sv and C57BL/6 mice. All IFN-α subtypes were found to be stable at pH 2 and to exhibit antiviral activity. Interestingly, some IFN subtypes (IFN-α4, IFN-α11, IFN-α12, IFN-β, and limitin) showed higher biological activity levels than others, whereas IFN-α7/10 exhibited lower activity. Most murine IFN-α turned out to be N-glycosylated. However, no correlation was found between N-glycosylation and activity. The various IFN-α subtypes displayed a good correlation between their antiviral and antiproliferative potencies, suggesting that IFN-α subtypes did not diverge primarily to acquire specific biological activities but probably evolved to acquire specific expression patterns. In L929 cells, IFN genes activated in response to poly(I•C) transfection or to viral infection were, however, similar.
Alpha/beta interferons (IFNs-α/β) were the first cytokines to be discovered. They were detected by their capacity to confer cell resistance to a viral challenge. IFNs play an important role in the host antiviral response but are also recognized for their antiproliferative and immunomodulatory activities. They are coded by an intronless multigene family clustered on murine chromosome 4 and in human chromosome 9. One IFN-β and multiple IFN-α genes were found in the mouse and human genomes (11, 13, 14, 15, 22, 25, 30, 35, 37, 39, 41, 44, 47, 50). Other IFN-α/β genes described include the murine limitin gene and the human IFN-ω gene, as well as the IFN-κ and IFN-ɛ/τ genes that were found in both the human and murine genomes (Table 1) (2, 9, 24, 32, 46). IFN subtypes coded by all these genes are thought to use a common cell surface receptor, raising the question of whether they play identical roles.
TABLE 1.
Human and murine IFN-α/β
| IFN subclass | No. of genes (pseudogenes) in human | No. of genes (pseudogenes) in mouse |
|---|---|---|
| IFN-αa | 13 (1) | 14 (3) |
| IFN-β | 1 (0) | 1 (0) |
| IFN-ω | 1 (6) | |
| Limitinb | ? | |
| IFN-κ | 1 (0) | 1 (0) |
| IFN-ɛ | 1 (0) | 1 (0) |
Two human IFN-α genes (IFN-α1 and IFN-α13) code for identical proteins.
The actual number of limitin genes in the mouse genome is unknown.
Both the human and the mouse genome code for more than a dozen closely related IFN-α subtypes. Phylogenetic analyses suggest that IFN-α subtypes have diverged by asymmetric crossover or gene conversion (14, 21) after the radiation of the major mammalian orders. Individual IFN-α genes could have evolved to acquire subtype-specific functions and/or subtype-specific expression patterns. Some experimental data support the hypothesis that the IFN-α genes might exert qualitatively distinct biological functions. For instance, Harle et al. recently showed that IFN-α/β subtypes differed in their antiviral potencies against two herpes simplex virus strains (19).
However, emerging experimental evidence supports the hypothesis of differential expression, regulated at the cellular level by the ratio between the different IFN regulatory factors (IRFs) (6, 5, 26). For example, murine IFN-α4 has been shown to be expressed early after viral infection, without a requirement for IRF-7 synthesis and activation, whereas the expression of other IFN-α subtypes depends on an autocrine or paracrine feedback loop mediated by this factor (27, 36). We recently showed that IFN-α13 was constitutively expressed at background levels in mouse cells (44). This IFN differs from other IFN-α subtypes by the fact that its expression is not influenced by viral infection. However, it is not known whether IFN-α13 plays a particular role in the organism.
Until recently, only the human IFN-α gene cluster had been extensively described (14). The number and characteristics of the murine IFN-α genes were still somewhat confused, as the sequences of certain IFN-α genes had not yet been deposited in the GenBank database, and others appeared to have been named twice. The availability of the whole mouse genome sequencing prompted us to make an inventory of the entire murine IFN-α gene family. We detected a total of 17 IFN-α subtype genes, including three pseudogenes. Allelic forms of certain IFN genes turned out to be surprisingly divergent.
We cloned the various murine IFN-α coding sequences as well as the IFN-β gene and one limitin gene for comparison. These IFNs were characterized and compared for their relative antiviral and antiproliferative activities in order to analyze whether the multiplicity of IFN-α subtypes was related to the acquisition of different biological activities.
MATERIALS AND METHODS
Viruses and cell culture.
Mengo virus was produced from the pMC24 cDNA clone kindly provided by Ann Palmenberg (University of Wisconsin, Madison) (16). Production and titration of this virus were done as described previously (45). B16 melanoma cells, kindly provided by Luc Pilotte and Benoît Van den Eynde (Ludwig Institute for Cancer Research, Brussels, Belgium) were cultured in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum, 100 IU of penicillin per ml, and 100 μg of streptomycin (Invitrogen, Life-technologies) per ml and supplemented with 116 μg of l-arginine per ml, 36 μg of l-asparagine per ml, and 216 μg of l-glutamine per ml. BHK-21, COS-7, BALB/3T3, and L929 cells were cultured as described previously (44).
Identification of murine gene sequences and multiple alignments.
Using the BLAST algorithm, we identified fragments from the whole mouse genome sequence database that aligned with more than 95% identity to the previously published IFN gene sequences (4). Raw fragments were aligned by using the MultiAlin program to reconstruct the sequence of the IFN-α genes (10). When necessary, data from the CELERA database were used to check for the presence of specific IFN genes in genomes of mouse strains other than C57BL/6.
Cloning of constructs expressing the various IFN-α subtypes.
The coding sequences of IFN-α1(129/Sv), IFN-α2, IFN-α8/6, IFN-α7/10, IFN-α9, IFN-α11, IFN-αA, IFN-αB, IFN-β, and limitin were amplified by PCR from 129/Sv mouse genomic DNA. IFN-α1, IFN-α12, IFN-α8/6(B6), IFN-α7/10(B6), and IFN-α11(B6) were amplified from C57BL/6 genomic DNA. The PCR products were cloned into the pcDNA3 vector (Invitrogen, Life Technologies) by using BamHI and XhoI restriction sites introduced in the PCR primers, as described previously for IFN-α4, IFN-α5, IFN-α6T, IFN-α13, and IFN-α14 (44). All the clones contain identical sequences upstream and downstream from the coding sequences, including a bona fide Kozak consensus sequence for translation.
Sequences of IFN coding regions were determined either by direct sequencing of the PCR fragments or by sequencing two cloned fragments resulting from independent PCRs. Sequences diverging from previously sequenced alleles were deposited in GenBank (Table 2). The sequence of IFN-α12 had partially been published (35). Sequences of the pseudogenes, reconstructed from the fragments of C57BL/6 mice were deposited in the Third Party Annotation subset of the GenBank database (accession no. BK001229, BK001230, and BK001231). IFN-α pseudogene 1 [IFN-α(ψ1)] was previously identified by Le Roscouet et al. (25). Accession numbers of IFN-α/β gene sequences are given in Table 2.
TABLE 2.
Characteristics of murine alpha/beta IFNs
| Gene | Allelic forma | Strain | N-glycosylation sitesb | Activity level | Accession no. [strain(s)] |
|---|---|---|---|---|---|
| IFN-α1 | IFN-α1(129/Sv) | 129/Sv | 1 | Mean | AY226993 (129/Sv) |
| IFN-α1 | C57BL/6 | 1 | Mean | X01974 (BALB/c),c AY225950 (C57BL/6) | |
| IFN-α2 | 129/Sv | 1 | Mean | X01969 (BALB/c)d,g | |
| IFN-α4 | 129/Sv | 1 | High | X01973 (BALB/c), AY220463 (129/Sv)d | |
| IFN-α5 | 129/Sv | 1 | Mean | X01971 (BALB/c), AY220464 (129/Sv)h | |
| IFN-α6T | 129/Sv | 0 | Mean | AY220465 (129/Sv)h | |
| IFN-α7/10 | IFN-α7/10 | 129/Sv | 1 | Low | M13710 (Swiss)g |
| IFN-α7/10(B6) | C57BL/6 | 0 | Low | AY225952 (C57BL/6) | |
| IFN-α8/6 | IFN-α8/6 | 129/Sv | 1 | Mean | X01972 (BALB/c),g D00460 (Swiss), |
| IFN-α8/6(B6) | C57BL/6 | 1 | Mean | AY225953 (C57BL/6) | |
| IFN-α9 | 129/Sv | 1 | Mean | M13660 (BALB/c)g,h | |
| IFN-α11 | IFN-α11 | 129/Sv | 1 | High | M68944 (Swiss)g |
| IFN-α11 (B6) | C57BL/6 | 1 | High | AY225954 (C57BL/6) | |
| IFN-α12 | C57BL/6 | 1 | High | AY225951 (C57BL/6) | |
| IFN-α13 | 129/Sv | 2 | Mean | AY220461 (129/Sv)h | |
| IFN-α14 | 129/Sv | 0 | Mean | AY220462 (129/Sv)h | |
| IFN-αAi | 129/Sv | 0 | Mean | M28587 (BALB/c)g,h | |
| IFN-αB | 129/Sv | 1 | Mean | L38698 (BALB/c)e,g | |
| IFN-β | 129/Sv | 3 | High | X14029g,h | |
| Limitin | 129/Sv | 1 | High | AB024521 (C57BL/6*DBA/2), AY220466 (129/Sv)f | |
| IFN-τ/ɛ | C57BL/6 | 0 | NDj | AY190044 | |
| IFN-κ | 129/Sv | 0 | ND | AF547990 |
Alleles with less than 96% identity in the coding sequence.
Number of sites.
Five nucleotides diverge between the IFN-α1 alleles from BALB/c and C57BL/6 mice.
Two nucleotides diverge between the IFN-α2 and IFN-α4 alleles from 129/Sv and C57BL/6 mice.
Three nucleotides diverge between the IFN-αB alleles from 129/Sv and C57BL/6 mice.
Two nucleotides diverge between the limitin alleles from 129/Sv and C57BL/6 mice.
Sequence of the 129/Sv allele is identical.
Sequence of the C57BL/6 allele is identical.
Note that IFN-αA is also known as IFN-α3.
ND, not done.
Metabolic labeling.
COS-7 cells, seeded in 6-well plates at 150,000 cells per well, were transfected with the different IFN-α-expressing constructs by using Fugene-6 reagent (Roche). At 24 h after transfection, cells were washed two times and incubated for 24 h in 1 ml of methionine- and cysteine-deficient Dulbecco's modified Eagle's medium (ICN) containing 1/10 of the normal methionine concentration (3 mg/liter) and 70 or 80 μCi of 35S-labeled methionine-cysteine mixture (Redivue Pro-mix; Amersham-Pharmacia Biotech) per ml. After 24 h of incubation at 37°C, supernatants were collected, centrifuged at 15,000 × g to remove cell debris, and stored at −70°C.
Glycosylation analysis and quantification of the amount of secreted IFN.
35S-labeled supernatants were treated with N-glycosidase F (Calbiochem) and, when necessary, with a cocktail of enzymes required for O-glycosylation removal (Calbiochem deglycosylation kit). Crude or treated supernatants were diluted in sample buffer (62.5 mM Tris [pH 6.8], 2% β-mercaptoethanol, 3% sodium dodecyl sulfate [SDS], 10% glycerol, 0,1% bromophenol blue) and run on Tris-Tricine, SDS-11% polyacrylamide gel electrophoresis (PAGE). Gels were dried and exposed. In order to compare the relative amounts of the various IFNs produced, the bands of glycosylated and deglycosylated IFNs were quantified by PhosphorImager. Since certain highly glycosylated IFN subtypes (IFN-α13 and IFN-β) migrated in multiple bands, quantification by PhosphorImager might underestimate the amount of these proteins. The bands of deglycosylated IFNs were, therefore, also quantified. Quantification was corrected for the percentage of methionine and cysteine residues present in each IFN subtype. Since all the IFN subtypes were expressed from constructs with identical signals for transcription and translation, the mean variation in the yield of IFNs produced in parallel was less than twofold, except for the IFN-α8/6, IFN-α11, and IFN-α7/10(B6) subtypes, which were consistently found to be produced in lower amounts.
pH stability.
Supernatants issued from transfected COS-7 cells were treated at pH 2 for 24 h at 4°C, as described previously, and antiviral activities were then compared between treated and untreated samples (45).
Antiviral activity.
IFN antiviral activity was quantified, as described previously, by a standard cytopathic effect reduction assay performed in 96-well plates, on BALB/3T3 cells with Mengo virus (44). IFNs to be compared were always tested in parallel. For each experiment, the antiviral assay was performed twice and, for certain samples, three times if the relative antiviral activities diverged by more than fourfold between the first two measurements. Aberrant values were then excluded from the analysis. Relative IFN activities were calculated as the ratio between IFN activities and IFN protein amounts measured by PhosphorImager analysis. These activities were normalized to the activity calculated for IFN-α1 in the same experiment. A large series of IFN subtypes was produced and tested in parallel. For each IFN, at least three independent experiments (including production and activity testing) were performed.
Antiproliferative activity.
B16 melanoma cells were seeded in 96-well plates at a density of 500 cells per well. After 24 h, cells were incubated for 72 h with twofold serial dilutions of the IFN-containing supernatants to be tested. After incubation, the cell density was quantified by using the WST-1 cell proliferation reagent (Roche, Biochemicals). Briefly, 15 μl of reagent was added per well and incubated for 1 h at 37°. Absorbance was read at 450 nm. A standard curve was established by performing, in triplicate, twofold dilutions of cells, starting from 1,000 cells per well. The IFN dilution at which 50% inhibition of cell proliferation occurred was used to compare the relative antiproliferative activities between the IFN-α subtypes. For each independent experiment, the assay was performed twice and, for certain samples, three times if the activities diverged by more than fourfold between the first two measures. Aberrant values were excluded from the analysis. Activity is expressed relative to that of IFN-α1 and corrected for the quantification of the amount of IFN measured in the supernatants.
Nucleotide sequence accession numbers.
The following new sequences or sequences of alleles that differed from previously reported sequences have been deposited in the GenBank database: for C57Bl/6 mice, the sequences (accession no.) of IFN-α1 (AY225950), IFN-α7/10(B6) (AY225952), IFN-α8/6(B6) (AY225953), IFN-α11(B6) (AY225954), and IFN-α12 (AY225951); for 129/Sv mice, the sequences (accession no.) of IFN-α1(129/Sv) (AY226993) and limitin (AY220466).
RESULTS
Analysis of the mouse genome database and reconstitution of the IFN-α gene family.
The recent discovery of yet uncharacterized murine IFN-α subtypes and the availability of data from the mouse genome sequencing project prompted us to characterize the murine IFN-α gene family. Therefore, we screened the whole mouse genome sequence subset of the National Center for Biotechnology Information (NCBI) mouse genome database for fragments exhibiting more than 95% identity to known murine IFN-α genes. Raw sequence fragments from the database were aligned to reconstruct the various IFN-α gene sequences. A total of 17 IFN-α gene sequences were assembled from the genomic sequences of the C57BL/6 mouse: 3 corresponded to pseudogenes and 14 encoded distinct, potentially functional IFN-α subtypes (Table 2).
We found that four previously recognized IFN subtypes probably corresponded to only two distinct IFNs that had been named twice (IFN-α6 and IFN-α8 in one case and IFN-α7 and IFN-α10 in the other). For instance, the sequences of IFN-α6 and IFN-α8, identified in BALB/c and Swiss mice, respectively, shared more than 99% identity (22, 30). We found no evidence, either by PCR (data not shown) or by database mining, for the presence of these two genes in a single mouse strain. Thus, IFN-α6 and IFN-α8 likely represent a single subtype, hereafter referred to as IFN-α8/6. Similarly, the genes encoding IFN-α7 and IFN-α10 appeared to correspond to a single subtype, hereafter referred to as IFN-α7/10.
Divergence of allelic forms.
The coding sequences of the various IFN-α genes were cloned from 129/Sv genomic DNA and were sequenced (Table 2). In general, little divergence occurred between sequences of IFN genes cloned from 129/Sv mice and sequences of the corresponding genes found in the C57BL/6 genome database, with most genes exhibiting more than 99% identity between the two backgrounds. However, four genes (IFN-α1, IFN-α7/10, IFN-α8/6, and IFN-α11) presented substantial sequence divergence between alleles found in C57BL/6 and 129/Sv mice.
Screening the CELERA database confirmed the presence of divergent allelic forms for these genes in 129/Sv and DBA/2 mice.
Alleles of the IFN-α7/10, IFN-α8/6, and IFN-α11 genes, originally detected in mice of the Swiss and BALB/c backgrounds, were very similar to alleles found in 129/Sv and DBA/2 mice but differed strikingly from the corresponding alleles of the C57BL/6 mouse. These divergent alleles found in C57BL/6 mice were named IFN-α7/10(B6), IFN-α8/6(B6), and IFN-α11(B6), respectively.
In the case of IFN-α1, the allele originally found in BALB/c mice was similar to that of C57BL/6 mice but diverged from alleles found in 129/Sv and DBA/2 mice. As the 129/Sv allele diverged from the originally described allele, it was named IFN-α1(129/Sv).
Of the IFN-α genes identified, only the IFN-α12 gene failed to be cloned from 129/Sv genomic DNA, though one fragment from the CELERA database suggested the presence of this gene in mice of the 129/Sv background. This gene could, however, be cloned from C57BL/6 mouse DNA.
Characterization of IFN subtypes.
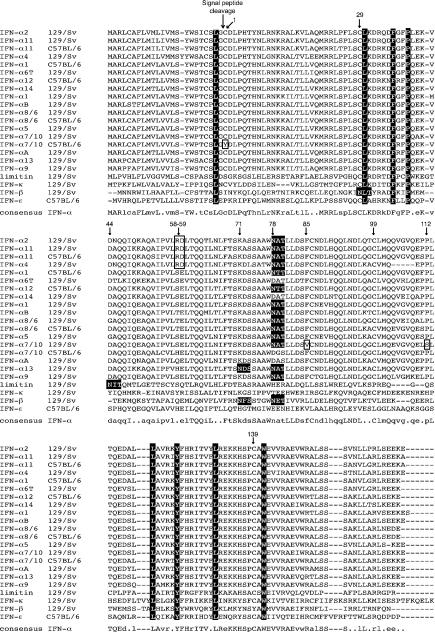
Alignment of the protein sequences shows a striking overall conservation of the various IFN-α subtypes (Fig. 1). In particular, the four cysteines that are involved in disulfide bond formation (Cys1-Cys99 and Cys 29-Cys139) are conserved in all the subtypes, with the noticeable exception of IFN-α7/10(B6), which lacks the first cysteine residue.
FIG. 1.
Multiple alignment of the murine IFN-α/β sequences. Sequences (Table 2) of IFNs-α/β from 129/Sv mice were aligned. The sequence of IFN-κ is from Vassileva et al. (46) (GenBank accession no. AF547990). Divergent alleles of IFN-α1, IFN-α7/10, IFN-α8/6, and IFN-α11, as well as the sequence of IFN-α12 and IFN-ɛ (9) from C57BL/6 mice were included in the analysis. Numbering refers to the mature sequences of IFN-α1 and IFN-α2. The predicted signal peptide cleavage site of IFN-α is indicated. Black columns show residues conserved in all the IFNs-α/β aligned. Predicted N-glycosylation sites [N-X-(S/T)] are outlined. Unique residues of the IFN-α2, IFN-α11, IFN-α11(B6), IFN-α4, and IFN-α7/10 proteins suspected to influence their activities are boxed. The cysteine residues involved in the formation of disulfide bridges are indicated by arrows and are numbered (residues 1 plus 99 and 29 plus 139). A consensus sequence for all murine IFNs-α is shown under the other sequences. Uppercase letters indicate conservation in all IFN-α subtypes. Lowercase letters were used when identical residues occurred in at least 14 of the 18 IFN-α sequences aligned.
To compare the properties of the different IFN subtypes, the coding sequences of all the identified IFN-α genes were cloned in the pcDNA3 expression vector, under the control of the cytomegalovirus immediate-early promoter. Coding sequences of limitin and IFN-β were also cloned for comparison.
The vectors expressing the various IFN subtypes were transiently transfected in COS-7 cells in parallel with the empty pcDNA3 vector. Supernatants from transfected 35S-labeled COS-7 cells were collected and analyzed.
Glycosylation of murine IFNs.
Figure 1 shows that 10 out of 14 IFN-α subtypes contain a putative N-glycosylation site (Asn-X-Ser/Thr). Only IFN-α6T, IFN-αA, IFN-α7/10(B6), and IFN-α14 lack such a site. All other IFN-α subtypes analyzed, as well as IFN-β, have a predicted N-glycosylation site in position 78 (76 for IFN-β). Note that the numbering of IFN-α residues in this section refers to the mature protein sequence of IFN-α1 and IFN-α2. Strikingly, IFN-α13, which was characterized in detail elsewhere, shows a second putative N-glycosylation site in position 71 (44). IFN-β contains three sites: two sites aligning with those of IFN-α13 (positions 69 and 76 for IFN-β) and a third site located at position 29. The putative N-glycosylation site of limitin, corresponding to position 44 of IFN-α, does not align with sites found in IFN-α or in IFN-β. Sequences of IFN-κ and IFN-τ/ɛ lack potential N-glycosylation sites.
To check whether the predicted N-glycosylation sites indeed carried N-linked sugars, 35S-labeled IFNs produced by transfected COS-7 cells were treated with N-glycosidase F or were left untreated. Treated and untreated IFNs were compared by SDS-PAGE analysis.
As shown in Fig. 2, all the IFN subtypes containing a predicted N-glycosylation site (Table 2) were, indeed, N-glycosylated. Upon treatment with N-glycosidase, all IFN subtypes migrated at around 18 kDa, confirming that slower migration on SDS-PAGE indeed resulted from the presence of N-linked sugars (Fig. 2B). No further shift in the migration was seen after O-glycosidase treatment, suggesting that murine IFNs lack O-glycosylation (data not shown).
FIG. 2.
N-glycosylation of murine IFNs-α/β. (A) Migration profile of IFNs from crude COS-7 cells supernatants. COS-7 cells were transfected with plasmids expressing the indicated IFN subtypes or the empty vector (−). IFN genes were from 129/Sv mice unless otherwise indicated. Supernatants from 35S-labeled cells were run on SDS-PAGE. Gels were dried and exposed. IFNs lacking N-glycosylation sites [IFN-α14, IFN-α7/10(B6), IFN-αA, and IFN-α6T] migrated at about 18 kDa, as expected from their calculated molecular mass. IFNs possessing one predicted N-glycosylation site [IFN-α1(B6), IFN-α2, IFN-α4, IFN-α5, IFN-α8/6, IFN-α7/10, IFN-α9, IFN-α11, IFN-α1(129/Sv), IFN-α8/6(B6), IFN-α11(B6), IFN-αB, and limitin] migrated at around 24 kDa. IFN-α13 and IFN-β migrated more slowly than IFN-α1, in agreement with the presence of two and three putative N-glycosylation sites in their sequences, respectively. Note that some small migration differences occurred, notably IFN-α12 reproducibly migrated more slowly than other IFN subtypes though its calculated molecular mass did not differ significantly. (B) Migration profile of IFN subtypes following N-glycosidase treatment. Following N-glycosidase treatment, all IFN subtypes migrated approximately at the same level as nonglycosylated IFNs. (C) Comparison between selected untreated (−) and treated samples. N-glyc, N-glycosidase.
pH 2 stability.
IFNs-α/β are remarkable in that they were found to resist pH 2 treatment. To know whether all the identified IFN-α subtypes shared this property, supernatants containing the various IFNs were treated at pH 2 for 24 h, and the activities of pH 2-treated and untreated supernatants against Mengo virus were compared. All the IFNs tested, except IFN-α7/10(B6), retained their antiviral activities after pH 2 treatment, the maximal loss of activity due to the treatment being between two- and fourfold. The effect of pH 2 treatment on IFN-α7/10(B6) supernatant was inconclusive, as this subtype repeatedly showed very low antiviral activity that was completely abolished by the treatment (data not shown).
Antiviral activities of the IFN subtypes.
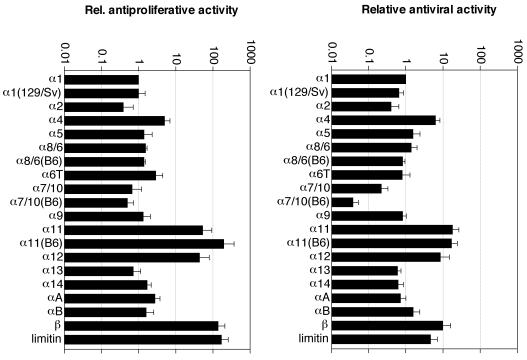
The relative amounts of the different 35S-labeled IFN subtypes present in the supernatants were quantified by PhosphorImager after SDS-PAGE. The antiviral activities of these supernatants were determined against Mengo virus. The measured antiviral activities were then corrected for the amount of IFN to calculate the relative antiviral activities of the various IFN subtypes. No measurable activity could be detected in the supernatants issued from cells transfected with the vector alone. Figure 3 shows that most of the IFN subtypes [IFN-α1(129/Sv), IFN-α5, IFN-α6T, IFN-α8/6, IFN-α8/6(B6), IFN-α9, IFN-α13, IFN-α14, IFN-αA, and IFN-αB] display antiviral activities in the range of the activity of IFN-α1. IFN-α2 and IFN-α7/10 show moderately reduced activity levels (three- and fivefold, respectively) compared to IFN-α1. The IFN-α7/10(B6) protein, in which the first cysteine residue is replaced by a tyrosine residue, shows 30-fold less activity than IFN-α1. Limitin and IFN-α4 display moderate (4- and 6-fold, respectively) increases in relative activities, whereas IFN-α11, IFN-α12, IFN-α11(B6) and IFN-β show the highest antiviral potencies, being between 9- and 18-fold more active than IFN-α1.
FIG. 3.
Relative (Rel.) antiviral and antiproliferative activities of IFN subtypes. Activities are expressed relative to the activity of IFN-α1. Note that the scale is logarithmic. A good correlation is observed between the antiviral and antiproliferative activities of the IFN subtypes.
Antiproliferative activity.
To determine whether some of the IFN-α subtypes evolved to acquire distinct biological functions, the relative antiproliferative activities of the different IFN subtypes were also determined by looking at the growth inhibition of B16 murine melanoma cells. Results presented in Fig. 3 show a striking correlation between the antiviral and antiproliferative potencies of the different subtypes. IFN-α11, IFN-α12, IFN-α11(B6), IFN-β, and limitin were again the most potent subtypes, showing 44- to 197-fold more activity than IFN-α1. IFN-α4 antiproliferative activity was moderately increased (fivefold) relative to that of IFN-α1. Most IFN-α subtypes [IFN-α2, IFN-α5, IFN-α8/6, IFN-α7/10, IFN-α9, IFN-α13, IFN-α14, IFN-α1(129/Sv), IFN-α8/6(B6), and IFN-αB] exhibited activity levels comparable to the activity of IFN-α1. The antiproliferative activity of IFN-α7/10(B6) was only slightly lower than that of IFN-α1, whereas its antiviral activity was significantly lower. Finally, there seems to be a small but reproducible threefold increase in the antiproliferative activities of IFN-αA and IFN-α6T relative to IFN-α1, whereas the antiviral activities of these subtypes were similar to, or lower than, the antiviral activity of IFN-α1.
IFN-α subtype gene expression profiling.
We next analyzed if the same IFN-α subtypes were preferentially expressed in response to two different stimuli known to induce transcriptional activation of IFN genes: viral infection or double-stranded RNA. Therefore, L929 cells were either infected with the H53 strain of Newcastle disease virus (NDV) or with the attenuated clone 13 of Rift Valley fever virus (RVFV) or transfected with 10 μg of poly(I · C) per ml. The profile of the different IFN-α subtypes transcribed was then examined 9 and 12 h after stimulation, times corresponding to high transcriptional activation of IFN-α. Results shown in Table 3 demonstrate that the IFN-α genes expressed in response to the different stimuli did not differ substantially. Both in the case of viral infection and of poly(I · C) stimulation, very few IFN subtypes were expressed (Table 3). IFN-α4 was the predominant subtype detected [41 out of 47 clones in the case of NDV infection and 43 out of 57 clones in the case of poly(I · C) stimulation]. IFN-α5 and IFN-α2 were transcribed in lower proportions. The expression of IFN-α(ψ3) and IFN-α6T in poly(I · C)-stimulated cells (one clone each) probably reflects a background of constitutive transcription. We have previously shown that IFN-α(ψ3) was transcribed in uninfected L929 cells (44). No expression of other IFN-α subtypes was detected in the analyzed series.
TABLE 3.
Expression of IFN-α subtypes in L929 cells infected by NDV or RVFV or stimulated by poly(I · C)a
| Stimulus | Exposure time (h) | No. of clones obtained by IFN subtype
|
||||
|---|---|---|---|---|---|---|
| IFN-α2 | IFN-α4 | IFN-α5 | IFN-α6T | IFN-αψ3 | ||
| NDV | 9 | 22 | 4 | |||
| NDV | 12 | 1 | 19 | 1 | ||
| RVFV | 9 | 1 | 9 | 1 | ||
| Poly(I · C) | 9 | 1 | 25 | 1 | 1 | 1 |
| Poly(I · C) | 12 | 5 | 18 | 5 | ||
The proportions of the different IFN-α subtypes was analyzed in L929 cells. Total IFN-α was amplified by reverse transcription-PCR by using degenerated primers, as described previously (44). PCR products were then cloned and sequenced. Subtypes that are not shown in the table were not detected.
DISCUSSION
Map of the IFN gene cluster.
We found that 14 IFN-α genes and 3 IFN-α pseudogenes occur in the mouse genome. Figure 4 presents the clustering of IFN genes deduced from the provisional assembly of mouse chromosome 4 found at the NCBI database.
FIG. 4.
Map of the murine IFN-α/β locus on chromosome 4. The map of the IFN cluster was reconstructed from the partial supercontig assembly of chromosome 4 of the NCBI (accession no. NT_039271.2). Arrows indicate the direction of transcription. As breaks still occur in the assembled sequence, the order and orientation of presented segments (separated by //) might still be found to vary. A fragment encompassing three limitin genes, IFN-α7/10, IFN-α-11, IFN-α8/6, IFN-α5, and IFN-α4 occurs twice in the NCBI assembly. This duplication likely represents an assembly artifact since the duplicated segment is not connected to any other in the assembled sequence and since no experimental data suggest such a duplication in any mouse strain.
Interestingly, the IFN-α8/6, IFN-α7/10, and IFN-α11 genes, the coding sequences of which diverge between C57BL/6 and 129/5v mice, are clustered. They are, however, separated from the IFN-α1 gene, the other IFN gene showing higher divergence between strains.
The grouping of IFN-α13, IFN-α6T, and IFN-α(ψ3) may be of significance. Indeed, in contrast to other studied IFN genes, the IFN-α13 gene and the IFN-α(ψ3) pseudogene were shown to be transcribed constitutively at low levels, independently of viral infection (44). IFN-α6T was also found to be expressed in tissues of uninfected mice. It is not known whether this IFN gene is responsive to viral infection, but alignment of the promoter sequences of the IFN-α genes (Fig. 5) reveals that IFN-α6T contains mutations in the predicted IRF binding motifs.
FIG. 5.
Multiple alignment of the virus-responsive elements (VRE) of the IFN-α promoters. Four modules (A, B, C, and D) were reported to modulate IFN promoter activity in response to viral infection. Modules A and B correspond to the IRF-7 binding site, and module C corresponds to the IRF-3 binding site. Underlined nucleotides are those which do not match the reported VRE consensus (shown under the sequences). Note that the fourth nucleotide of the C module does not correspond to the consensus IRF binding sequence (GAAA repeat), although it was found to be functional (29). It was therefore not underlined. IFN-α4 is the only IFN subtype that shows a functional C module, in agreement with its role as an immediate-early IFN. In the B module, the last nucleotide of the consensus was not underlined as it diverged from the consensus, even for inducible IFN genes. The B module of the IFN-α13, IFN-α7, and IFN-α6T genes is predicted to be unresponsive to IRF-7.
The actual number of limitin genes is not yet defined. A tandem array of 16 consecutive, almost identical, limitin genes can be found in the NCBI contig assembly. However, assembly of this region is likely to contain artifacts, given the very high sequence identity between the tandem repeats (>98%). An additional IFN species, named IFN-ɛ, or IFN-τ, is present near the IFN-α/β cluster. The murine IFN-κ gene, also present on chromosome 4, is distant from the IFN-α/β cluster. The fact that it is the only IFN gene that contains an intron suggests that it could correspond to the ancestor of all the other IFNs-α/β.
Characterization of the murine IFNs-α/β.
Though many studies addressed the characteristics and the functions of particular IFN subtypes, this study is the first that systematically compares the properties of all IFN-α subtypes detected in the mouse genome.
We have observed that acid stability is a general feature of the IFN-α family and of limitin. In humans, although IFN-β is known to be N-glycosylated, only 2 out of 13 IFN-α subtypes have been found to carry glycosylations. IFN-α2 was shown to be O-glycosylated, and IFN-α14 was shown to be N-glycosylated (1, 31).
In the mouse, some IFN subtypes were previously reported to be N-glycosylated. These include IFN-β, IFN-α1, IFN-α2, IFN-α4, IFN-α5, and IFN-α13 (8, 43, 44). In contrast, IFN-α6T and IFN-α14 were known to lack N-glycosylation (43, 44). It was, therefore, of interest to analyze the glycosylation status of the other IFN subtypes.
Contrary to the situation encountered in humans, the majority of IFN-α subtypes turned out to be N-glycosylated in the mouse. The role of IFN glycosylation is not known. Globally, glycosylated IFN subtypes did not differ substantially from nonglycosylated subtypes with respect to their biological activities. Glycosylation could possibly participate in the stabilization of circulating IFN molecules in vivo, rather than influence their intrinsic activities. The purpose of the multiple glycosylations of IFN-α13 and IFN-β remains to be elucidated.
Relative biological activities of IFN subtypes.
Data concerning the relative activities of murine IFN subtypes are scarce. Our results show that some IFN-α species [IFN-α4, IFN-α11, IFN-α11(B6), and IFN-α12] and the more distantly related IFN-β and limitin display significantly higher activities than other IFN subtypes. As reported for certain human IFN-α subtypes, the observed differences in activities might be related to differences in receptor binding affinities (3, 28, 34, 48). The alignment of IFN protein sequences (Fig. 1) fails to highlight residues that would be conserved in IFN subtypes with high biological activities, suggesting that subtle modifications of the protein structure account for the variation in biological activities.
Several authors have reported, in concordance with our results, that IFN-α4 is 5 to 10 times more active than IFN-α1 (42, 43). The higher activity of IFN-α4 was attributed in part to the region comprised between amino acids 55 and 67 (49). Interestingly, in this region residues Arg58 and Asp59 are conserved in IFN-α11 and IFN-α11(B6), two IFNs with high activity levels, as well as in IFN-α2, a subtype with low to moderate activity (Fig. 1). If residues Arg58 and Asp59 are involved in the high activities of IFN-α4, IFN-α11, and IFN-α11(B6), one must suppose that other residues in the C terminus of the closely related IFN-α2 protein could negatively influence the activity of the latter subtype.
The low activity of IFN-α7/10(B6) could be due to the lack of the Cys1 residue which, in other subtypes, is involved in disulfide bond formation between cysteines 1 and 99 (Fig. 1). Beilharz et al. have shown that this bond was essential for the activity of human IFN-α1 (7). On the other hand, the IFN-α7/10 allele of 129/Sv and BALB/c mice was also reported to have lower activity, though this allele conserved the Cys1 residue (41). Trapman et al. mapped the low activity of this subtype to the region comprised between residues 68 and 123. Two residues, Val85 and Ser112, which occur in this region are unique to IFN-α7/10, suggesting that these amino acids might be responsible for the low activity of this subtype.
Correlation between the antiviral and antiproliferative activities of IFN subtypes.
Our results show a good correlation between the antiviral and antiproliferative potencies of the IFN subtypes. Indeed, IFNs that displayed high antiviral activities also displayed high antiproliferative potencies. This suggests that the signaling pathways activated by the IFNs, leading to an antiviral state or to inhibition of cell proliferation, largely overlap. Differences between subtypes were more pronounced in the antiproliferative assay. The reason for this discrepancy is not known. One could hypothesize that this reflects differences in IFN stability or in receptor binding kinetics, as the antiviral and antiproliferative assays involved different cell types and different kinetics.
Comparing the antiviral activities of a limited number of IFN-α subtypes against Mengo virus, vesicular stomatitis virus, and Theiler's murine encephalomyelitis virus also failed to reveal any functional specialization of specific IFN subtypes (data not shown).
Up to now, there has been little experimental evidence showing a specialization in the function of the IFN-α subtypes. Human IFN-α17 is the only subtype identified for which there is a clear dissociation between its antiviral activity and its ability to activate NK cells (33). More recently, Cull et al. have shown that murine IFN-α subtypes differentially activated STAT3 and STAT5 signaling in J2E erythroleukemic cells (12). Harle et al. also reported that IFN-α subtypes differed in their antiviral potencies against two strains of herpes virus (19).
Our results do not completely rule out the possibility for such qualitative differences between the biological activities of IFN subtypes, since small differences between samples (lower than fourfold) could not be detected in our study, and effects other than antiviral or antiproliferative effects were not investigated. However, our data suggest that the main reason for the diversity of IFN-α subtypes does not derive from the specificity of their biological activities but, rather, relates to their expression specificity.
Differential induction of IFN genes.
To test whether the nature of the inducer influences IFN subtype gene expression, we compared the profiles of IFN subtypes expressed by L929 cells in response to NDV or RVFV infection and to poly(I · C) transfection. In all these cases, IFN-α4 was the predominant subtype detected. IFN-α2 and IFN-α5 were expressed at lower levels than IFN-α4.
The expression of IFN genes was reported to occur as a two-step mechanism (27, 36). Transcription of IFN-α4 and IFN-β genes termed immediate-early IFNs depends on IRF-3, a transcription factor constitutively expressed in the cell and activated early after viral infection by phosphorylation cascades (18, 38). The transcription of the other IFN-α genes further depends on IRF-7. Expression of IRF-7 itself depends on the priming of cells with exogenous IFN-α/β. Thus, viral infection is thought to activate the release of IFN-α4 and IFN-β, which prime the cells and trigger the production of the other IFN-α subtypes.
Accordingly, sequence alignments (Fig. 5) show that the IFN-α4 promoter is the sole IFN-α gene promoter to contain an intact IRF-3 binding sequence. The predominance of IFN-α4 in L929 cells infected with NDV, RVFV, or transfected with poly(I · C) is thus not surprising and had been reported previously in the case of NDV infection of L929 cells (Kelley et al. [22] and Hoss-Homfeld et al. [20]). However, it is not clear why IFN-α2 and IFN-α5 promoters are preferentially induced compared to other delayed-type IFN-α genes. Comparison of the putative promoter sequences of the IFN-α genes (Fig. 5) indicates that most of the IFN-α genes (with the exception of IFN-α13, IFN-α6T, and IFN-α7/10) contain a functional IRF-7 binding site. One would have expected, in terms of IFN response efficacy, that the IFNs exhibiting the highest antiviral activities (IFN-α11 and IFN-α12) would be the most prominently expressed in case of viral infection.
Induction of specific human IFN-α subtypes was reported to be regulated by the balance between IRF-3, IRF-7, and IRF-5 (5, 6, 26). As the concentration of these factors seems to vary according to the cell type, it will be of interest to investigate further the pattern of expression of IFN subtypes in different cell types.
Recent work reported the identification of a new cytokine family (called interleukin-28/interleukin-29 or IFNs-λ) and of their receptor (17, 23, 40). Though they bind to a receptor unrelated to the IFN-α/β receptor, these cytokines exert antiviral (23, 40) and antiproliferative (17a) activities strikingly similar to those of IFNs-α/β. Again, this functional redundancy suggests that the difference between IFN-λ and IFN-α/β could lie in their expression and further adds interest to characterizing the expression of all these cytokines and of their receptors at the cellular level.
Acknowledgments
We thank Pierre Rensonnet for expert technical assistance. We greatly appreciated the help from Peter Staeheli with the NDV and RVFV infections and poly(I · C) activation experiments.
T.M. is a research associate, and V.V.P is a research fellow with the FNRS (Belgian Fund for Scientific Research). This work was supported by the national fund for Medical Scientific Research (FRSM convention 3.4549.02), by FNRS (crédit aux chercheurs), by the French Association pour la recherche sur la Sclérose en Plaques (ARSEP), and by the Fonds Spécial de Recherche (FSR) of the University of Louvain.
REFERENCES
- 1.Adolf, G. R., I. Kalsner, H. Ahorn, I. Maurer-Fogy, and K. Cantell. 1991. Natural human interferon-alpha 2 is O-glycosylated. Biochem. J. 276:511-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adolf, G. R., I. Maurer-Fogy, I. Kalsner, and K. Cantell. 1990. Purification and characterization of natural human interferon omega 1. Two alternative cleavage sites for the signal peptidase. J. Biol. Chem. 265:9290-9295. [PubMed] [Google Scholar]
- 3.Aguet, M., M. Grobke, and P. Dreiding. 1984. Various human interferon alpha subclasses cross-react with common receptors: their binding affinities correlate with their specific biological activities. Virology 132:211-216. [DOI] [PubMed] [Google Scholar]
- 4.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barnes, B. J., A. E. Field, and P. M. Pitha-Rowe. 2003. Virus-induced heterodimer formation between IRF-5 and IRF-7 modulates assembly of the IFNA enhanceosome in vivo and transcriptional activity of IFNA genes. J. Biol. Chem. 278:16630-16641. [DOI] [PubMed] [Google Scholar]
- 6.Barnes, B. J., P. A. Moore, and P. M. Pitha. 2001. Virus-specific activation of a novel interferon regulatory factor, IRF-5, results in the induction of distinct interferon alpha genes. J. Biol. Chem. 276:23382-23390. [DOI] [PubMed] [Google Scholar]
- 7.Beilharz, M. W., I. T. Nisbet, M. J. Tymms, P. J. Hertzog, and A. W. Linnane. 1986. Antiviral and antiproliferative activities of interferon-alpha 1: the role of cysteine residues. J. Interferon Res. 6:677-685. [DOI] [PubMed] [Google Scholar]
- 8.Civas, A., B. Fournet, C. Coulombel, D. Le Roscouet, A. Honvault, F. Petek, J. Montreuil, and J. Doly. 1988. Purification and carbohydrate structure of natural murine interferon-beta. Eur. J. Biochem. 173:311-316. [DOI] [PubMed] [Google Scholar]
- 9.Conklin, D., F. J. Grant, M. Rixon, and W. Kindsvogel. December 2001. Interferon-epsilon. U.S. patent 6,329,175.
- 10.Corpet, F. 1988. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 16:10881-10890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coulombel, C., G. Vodjdani, and J. Doly. 1991. Isolation and characterization of a novel interferon-alpha-encoding gene, IFN-alpha 11, within a murine IFN cluster. Gene 104:187-195. [DOI] [PubMed] [Google Scholar]
- 12.Cull, V. S., P. A. Tilbrook, E. J. Bartlett, N. L. Brekalo, and C. M. James. 2003. Type I interferon differential therapy for erythroleukemia: specificity of STAT activation. Blood 101:2727-2735. [DOI] [PubMed] [Google Scholar]
- 13.Daugherty, B., D. Martin-Zanca, B. Kelder, K. Collier, T. C. Seamans, K. Hotta, and S. Pestka. 1984. Isolation and bacterial expression of a murine alpha leukocyte interferon gene. J. Interferon Res. 4:635-643. [DOI] [PubMed] [Google Scholar]
- 14.Diaz, M. O., H. M. Pomykala, S. K. Bohlander, E. Maltepe, K. Malik, B. Brownstein, and O. I. Olopade. 1994. Structure of the human type-I interferon gene cluster determined from a YAC clone contig. Genomics 22:540-552. [DOI] [PubMed] [Google Scholar]
- 15.Dion, M., G. Vodjdani, and J. Doly. 1986. Sequence and expression of a novel murine interferon alpha gene-homology with enhancer elements in the regulatory region of the gene. Biochem. Biophys. Res. Commun. 138:826-834. [DOI] [PubMed] [Google Scholar]
- 16.Duke, G. M., and A. C. Palmenberg. 1989. Cloning and synthesis of infectious cardiovirus RNAs containing short, discrete poly(C) tracts. J. Virol. 63:1822-1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dumoutier, L., D. Lejeune, S. Hor, H. Fickenscher, and J. C. Renauld. 2003. Cloning of a new type II cytokine receptor activating signal transducer and activator of transcription (STAT)1, STAT2 and STAT3. Biochem. J. 370:391-396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17a.Dumoutier, L., A. Tounsi, T. Michiels, C. Sommereyns, S. V. Kotenko, and J.-C. Renauld. 27 May 2004. Role of the interleukin-28 receptor tyrosine residues for antiviral and antiproliferative activity of IL-29/IFN-λ1: similarities with type 1 interferon signalling. J. Biol. Chem. PMID: 15166220. [DOI] [PubMed]
- 18.Fitzgerald, K. A., S. M. McWhirter, K. L. Faia, D. C. Rowe, E. Latz, D. T. Golenbock, A. J. Coyle, S. M. Liao, and T. Maniatis. 2003. IKKepsilon and TBK1are essential components of the IRF-3 signaling pathway. Nat Immunol. 4:491-496. [DOI] [PubMed] [Google Scholar]
- 19.Harle, P., V. Cull, L. Guo, J. Papin, C. Lawson, and D. J. Carr. 2002. Transient transfection of mouse fibroblasts with type I interferon transgenes provides various degrees of protection against herpes simplex virus infection. Antiviral Res. 56:39-49. [DOI] [PubMed] [Google Scholar]
- 20.Hoss-Homfeld, A., E. C. Zwarthoff, and R. Zawatzky. 1989. Cell type specific expression and regulation of murine interferon alpha and beta genes. Virology 173:539-550. [DOI] [PubMed] [Google Scholar]
- 21.Hughes, A. L. 1995. The evolution of the type I interferon gene family in mammals. J. Mol. Evol. 41:539-548. [DOI] [PubMed] [Google Scholar]
- 22.Kelley, K. A., and P. M. Pitha. 1985. Characterization of a mouse interferon gene locus I. Isolation of a cluster of four alpha interferon genes. Nucleic Acids Res. 13:805-823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kotenko, S. V., G. Gallagher, V. V. Baurin, A. Lewis-Antes, M. Shen, N. K. Shah, J. A. Langer, F. Sheikh, H. Dickensheets, and R. P. Donnelly. 2003. IFN-lambdas mediate antiviral protection through a distinct class II cytokine receptor complex. Nat. Immunol. 4:69-77. [DOI] [PubMed] [Google Scholar]
- 24.LaFleur, D. W., B. Nardelli, T. Tsareva, D. Mather, P. Feng, M. Semenuk, K. Taylor, M. Buergin, D. Chinchilla, V. Roshke, G. Chen, S. M. Ruben, P. M. Pitha, T. A. Coleman, and P. A. Moore. 2001. Interferon-kappa, a novel type I interferon expressed in human keratinocytes. J. Biol. Chem. 276:39765-39771. [DOI] [PubMed] [Google Scholar]
- 25.Le Roscouet, D., G. Vodjdani, Y. Lemaigre-Dubreuil, M. G. Tovey, M. Latta, and J. Doly. 1985. Structure of a murine alpha interferon pseudogene with a repetitive R-type sequence in the 3′ flanking region. Mol. Cell. Biol. 5:1343-1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Levy, D. E., I. Marie, and A. Prakash. 2003. Ringing the interferon alarm: differential regulation of gene expression at the interface between innate and adaptive immunity. Curr. Opin. Immunol. 15:52-58. [DOI] [PubMed] [Google Scholar]
- 27.Marie, I., J. E. Durbin, and D. E. Levy. 1998. Differential viral induction of distinct interferon-alpha genes by positive feedback through interferon regulatory factor-7. EMBO J. 17:6660-6669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meister, A., G. Uze, K. E. Mogensen, I. Gresser, M. G. Tovey, M. Grutter, and F. Meyer. 1986. Biological activities and receptor binding of two human recombinant interferons and their hybrids. J. Gen. Virol. 67:1633-1643. [DOI] [PubMed] [Google Scholar]
- 29.Morin, P., P. Genin, J. Doly, and A. Civas. 2002. The virus-induced factor VIF differentially recognizes the virus-responsive modules of the mouse IFNA4 gene promoter. J. Interferon Cytokine Res. 22:77-86. [DOI] [PubMed] [Google Scholar]
- 30.Navarro, S., M. Dion, G. Vodjdani, F. Berlot-Picard, and J. Doly. 1989. Isolation and characterization of a functional murine interferon alpha gene which is not expressed in fibroblasts upon virus induction. J. Gen. Virol. 70:1381-1389. [DOI] [PubMed] [Google Scholar]
- 31.Nyman, T. A., N. Kalkkinen, H. Tolo, and J. Helin. 1998. Structural characterisation of N-linked and O-linked oligosaccharides derived from interferon-alpha2b and interferon-alpha14c produced by Sendai-virus-induced human peripheral blood leukocytes. Eur. J. Biochem. 253:485-493. [DOI] [PubMed] [Google Scholar]
- 32.Oritani, K., K. L. Medina, Y. Tomiyama, J. Ishikawa, Y. Okajima, M. Ogawa, T. Yokota, K. Aoyama, I. Takahashi, P. W. Kincade, and Y. Matsuzawa. 2000. Limitin: an interferon-like cytokine that preferentially influences B-lymphocyte precursors. Nat. Med. 6:659-666. [DOI] [PubMed] [Google Scholar]
- 33.Ortaldo, J. R., R. B. Herberman, C. Harvey, P. Osheroff, Y. C. Pan, B. Kelder, and S. Pestka. 1984. A species of human alpha interferon that lacks the ability to boost human natural killer activity. Proc. Natl. Acad. Sci. USA 81:4926-4929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Piehler, J., and G. Schreiber. 1999. Mutational and structural analysis of the binding interface between type I interferons and their receptor Ifnar2. J. Mol. Biol. 294:223-237. [DOI] [PubMed] [Google Scholar]
- 35.Riego, E., A. Perez, R. Martinez, F. O. Castro, R. Lleonart, and J. de la Fuente. 1995. Differential constitutive expression of interferon genes in early mouse embryos. Mol. Reprod. Dev. 41:157-166. [DOI] [PubMed] [Google Scholar]
- 36.Sato, M., H. Suemori, N. Hata, M. Asagiri, K. Ogasawara, K. Nakao, T. Nakaya, M. Katsuki, S. Noguchi, N. Tanaka, and T. Taniguchi. 2000. Distinct and essential roles of transcription factors IRF-3 and IRF-7 in response to viruses for IFN-alpha/beta gene induction. Immunity 13:539-548. [DOI] [PubMed] [Google Scholar]
- 37.Seif, I., and J. De Maeyer-Guignard. 1986. Structure and expression of a new murine interferon-alpha gene: MuIFN-alpha I9. Gene 43:111-121. [DOI] [PubMed] [Google Scholar]
- 38.Sharma, S., B. tenOever, N. Grandvaux, G. P. Zhou, R. Lin, and J. Hiscott. 2003. Triggering the interferon antiviral response through an IKK-related pathway. Science 300:1148-1151. [DOI] [PubMed] [Google Scholar]
- 39.Shaw, G. D., W. Boll, H. Taira, N. Mantei, P. Lengyel, and C. Weissmann. 1983. Structure and expression of cloned murine IFN-alpha genes. Nucleic Acids Res. 11:555-573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sheppard, P., W. Kindsvogel, W. Xu, K. Henderson, S. Schlutsmeyer, T. E. Whitmore, R. Kuestner, U. Garrigues, C. Birks, J. Roraback, C. Ostrander, D. Dong, J. Shin, S. Presnell, B. Fox, B. Haldeman, E. Cooper, D. Taft, T. Gilbert, F. J. Grant, M. Tackett, W. Krivan, G. McKnight, C. Clegg, D. Foster, and K. M. Klucher. 2003. IL-28, IL-29 and their class II cytokine receptor IL-28R. Nat. Immunol. 4:63-68. [DOI] [PubMed] [Google Scholar]
- 41.Trapman, J., M. van Heuvel, P. de Jonge, I. J. Bosveld, P. Klaassen, and E. C. Zwarthoff. 1988. Structure-function analysis of mouse interferon alpha species: MuIFN-alpha 10, a subspecies with low antiviral activity. J. Gen. Virol. 69:67-75. [DOI] [PubMed] [Google Scholar]
- 42.Van Heuvel, M., I. J. Bosveld, P. Klaassen, E. C. Zwarthoff, and J. Trapman. 1988. Structure-function analysis of murine interferon-alpha: antiviral properties of novel hybrid interferons. J. Interferon Res. 8:5-14. [DOI] [PubMed] [Google Scholar]
- 43.Van Heuvel, M., I. J. Bosveld, A. A. Mooren, J. Trapman, and E. C. Zwarthoff. 1986. Properties of natural and hybrid murine alpha interferons. J. Gen. Virol. 67:2215-2222. [DOI] [PubMed] [Google Scholar]
- 44.van Pesch, V., and T. Michiels. 2003. Characterization of IFN-α13, a novel constitutive murine interferon-alpha subtype. J. Biol. Chem. 278:46321-46328. [DOI] [PubMed] [Google Scholar]
- 45.van Pesch, V., O. van Eyll, and T. Michiels. 2001. The leader protein of Theiler's virus inhibits immediate-early alpha/beta interferon production. J. Virol. 75:7811-7817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vassileva, G., S. C. Chen, M. Zeng, S. Abbondanzo, K. Jensen, D. Gorman, B. M. Baroudy, Y. Jiang, N. Murgolo, and S. A. Lira. 2003. Expression of a novel murine type I IFN in the pancreatic islets induces diabetes in mice. J. Immunol. 170:5748-5755. [DOI] [PubMed] [Google Scholar]
- 47.Vodjdani, G., C. Coulombel, and J. Doly. 1988. Structure and characterization of a murine chromosomal fragment containing the interferon beta gene. J. Mol. Biol. 204:221-231. [DOI] [PubMed] [Google Scholar]
- 48.Yamaoka, T., S. Kojima, S. Ichi, Y. Kashiwazaki, T. Koide, and Y. Sokawa. 1999. Biologic and binding activities of IFN-alpha subtypes in ACHN human renal cell carcinoma cells and Daudi Burkitt's lymphoma cells. J. Interferon Cytokine Res. 19:1343-1349. [DOI] [PubMed] [Google Scholar]
- 49.Zwarthoff, E. C., A. Gennissen, I. J. Bosveld, J. Trapman, and M. van Heuvel. 1987. Two domains in alpha interferons influence the efficacy of the antiviral response. Biochem. Biophys. Res. Commun. 147:47-55. [DOI] [PubMed] [Google Scholar]
- 50.Zwarthoff, E. C., A. T. Mooren, and J. Trapman. 1985. Organization, structure and expression of murine interferon alpha genes. Nucleic Acids Res. 13:791-804. [DOI] [PMC free article] [PubMed] [Google Scholar]