Summary
It is generally assumed that mRNAs undergoing translation are protected from decay. Here we show that mRNAs are in fact co-translationally degraded. This is a widespread and conserved process affecting most genes, where 5’-3’ transcript degradation follows the last translating ribosome, producing an in vivo ribosomal footprint. By sequencing the ends of 5’phosphorylated mRNA degradation intermediates, we obtain a genome-wide drug-free measurement of ribosome dynamics. We identify general translation termination pauses in both normal and stress conditions. In addition, we describe novel codon-specific ribosomal pausing sites in response to oxidative stress, which are dependent on the RNase Rny1. Our approach is simple and straightforward, and does not require the use of translational inhibitors or in vitro RNA footprinting that can alter ribosome protection patterns.
Introduction
For cells to change gene expression and adapt to new conditions, mRNAs need to be secluded from the translating pool or degraded through decay pathways. It is believed that mRNAs undergoing translation are protected from decay, recent evidence has however shown that the interplay between mRNA degradation and translation is likely more complex (Roy and Jacobson, 2013). Multiple lines of evidence suggest that elongating ribosomes limit mRNA decapping, a crucial first step in their degradation (Parker, 2012). In contrast, recent evidence from a handful of Saccharomyces cerevisiae mRNAs has shown that they can be decapped and undergo 5’-3’ exonucleolytic decay while associated with ribosomes (Hu et al., 2009). To date, it remains unclear whether this constitutes co-translational degradation and what its genome-wide implications would be.
The removal of mRNAs from the translating pool can either be transient, through sequestration in P-bodies or stress granules, or permanent, through degradation (Decker and Parker, 2012). Two general cytoplasmic mRNA degradation pathways exist: degradation of mRNA molecules from the 5’ end by 5’-3’ exonucleolytic decay (Hsu and Stevens, 1993; Muhlrad et al., 1994), and from the 3’ end by 3’-5’ exonucleolytic decay (Anderson and Parker, 1998). In addition, multiple specialized RNA degradation pathways exist for the elimination of faulty mRNAs, such as nonsense-mediated decay, no-go decay (endonucleolytic RNA cleavage) and non-stop decay (reviewed in (Parker, 2012; Roy and Jacobson, 2013)). Under normal conditions, shortening of the polyA tail, followed by decapping and subsequent 5’-3’ exonucleolytic decay by exonucleases like Xrn1 is the predominant mRNA degradation pathway (Parker, 2012). This sequence of steps gives rise to transient 5’ monophosphorylated (P) mRNA degradation intermediates with a shortened but present polyA tail. These RNAs, which we refer to here as 5’P mRNA degradation intermediates, can be readily isolated from cells and analysed to measure RNA decay activity in vivo. Here, we systematically analysed all polyadenylated RNAs each bearing different 5’ ends to obtain a characterization of RNA decay.
Most polyA RNAs in cells had been assumed to be capped and to function as a template for translation. However, there are other forms of RNA in cells that could be associated with ribosomes. Four populations of polyadenylated RNAs exist with different 5’ ends: capped, triphosphorylated, monophosphorylated (5’P), or hydroxylated (5’OH). Triphosphorylated RNAs have not yet been subjected to a capping reaction; 5’P mRNAs are products of exo- or endonucleolytic cleavage; while 5’OH mRNAs arise mainly from chemical cleavage. All of these classes have been assumed to be absent or of extremely low abundance in cells. However, genome-wide characterization of degradation intermediates has been largely lacking. Previous characterizations of RNA degradation focused on identifying of mRNA endonucleolytic cleavage sites using mutants with defective 5’-3’ RNA degradation in plants (Addo-Quaye et al., 2008; German et al., 2008; Gregory et al., 2008) and yeast (Harigaya and Parker, 2012). In addition, it had been assumed that 5’P mRNA degradation products that are present in wild-type samples arise from a random degradation process from which no biological information can be extracted.
In comparison to decay, our understanding of translation is more advanced through the study of ribosomal occupancy on mRNAs. Genome-wide approaches, such as polyribosome fractionation followed by analysis of ribosome-bound mRNAs by microarrays (Arava et al., 2003) or sequencing (Ingolia et al., 2009), have yielded immense insights. Ribosome profiling (Ingolia et al., 2009) a method in which polyribosomes are purified and subjected to in vitro RNA digestion and high-throughput sequencing, has revealed ribosomal footprints genome-wide. This method has been applied from bacteria to humans to study stress response (Gerashchenko et al., 2012), cellular differentiation (Brar et al., 2012; Ingolia et al., 2011), and details of the translation process itself such as ribosomal pausing (Li et al., 2012), and the rescue of stalled ribosomes (Guydosh and Green, 2014). Despite its potential to measure subcodon-resolution ribosome protection, in vitro sample-processing steps limit its ability to accurately reveal in vivo dynamics of ribosomes. Translation inhibitors, such a cycloheximide, are utilized to arrest ribosomes and to freeze them in their positions during the extensive in vitro sample processing steps (e.g. RNA extraction, sucrose fractionation and RNAse I footprinting) (Ingolia et al., 2012). In the case of yeast, ribosome profiling has been attempted without the use of an inhibitor, however this does not solve the problems caused by in vitro handling, and ribosomes can run off the mRNA if no inhibitor is present to freeze them in place (Lareau et al., 2014). For these reasons, rapid and complementary approaches to infer ribosome dynamics in vivo would be valuable.
Here to study mRNA turnover genome-wide, we developed 5PSeq, which identifies 5’P molecules that are a product of enzymatic decay in cells. This method turned out to also be effective for measuring ribosomal dynamics. By investigating the frequency distribution of 5’P positions of mRNA degradation intermediates in vivo, we discovered a characteristic three-nucleotide periodicity pattern in the coding region. By extensive experimental testing we show that this pattern is caused by general 5’-3’ mRNA co-translational decay. We infer that the exonuclease XRN1 follows the last translating ribosome, producing a progressive in vivo footprint of its 5’ position. By studying translation regulation in budding yeast upon oxidative stress, we reveal novel tRNA-specific ribosomal pause sites and delayed translational termination. Our 5PSeq approach thus provides a rapid and complementary method to measure ribosome dynamics in vivo, avoiding artefacts caused by translation inhibitors. Finally by applying it to Schizosaccharomyces pombe, we demonstrate that co-translational decay is conserved and our method can be generally applied to any RNA sample including those previously isolated.
Results
Quantifying mRNA degradation intermediates
We identified transcripts that belong to three populations of polyadenylated RNAs in S. cerevisiae cells with different 5’ ends: capped, monophosphorylated (5’P), or hydroxylated (5’OH) (Figure S1A). To characterize these RNAs, we selectively captured polyadenylated RNAs using oligo-dT reverse transcription coupled with modifications of oligo-capping (Pelechano et al., 2014; Pelechano et al., 2013). Specifically, 1) we identified 5’ capped molecules by first dephosphorylating all 5’P degradation intermediates using Calf Intestinal Phosphatase (CIP), rendering them unable to ligate in subsequent steps, followed by treatment with Tobacco Acid Pyrophosphatase (TAP) to remove the cap. In this approach, only previously capped molecules present a 5’P that can undergo single-stranded RNA ligation (Figure 1A). 2) Separately, 5’P molecules were identified by directly performing an RNA ligation step. 3) To identify the composite of both populations (5’cap and 5’P) the CIP treatment was omitted and after TAP treatment, both previously 5’capped and 5’P molecules were subjected to ligation (Figures 1A and S1B). We also attempted to profile 5’OH mRNA molecules, which are chemical cleavage products, by treatment with a 5’P specific 5’-3’ exonuclease, followed by the phosphorylation of the remaining 5’OH molecules (Figure S1B). Due to the low abundance of such products in cells, 5’OH mRNA molecules were not detectable in our samples.
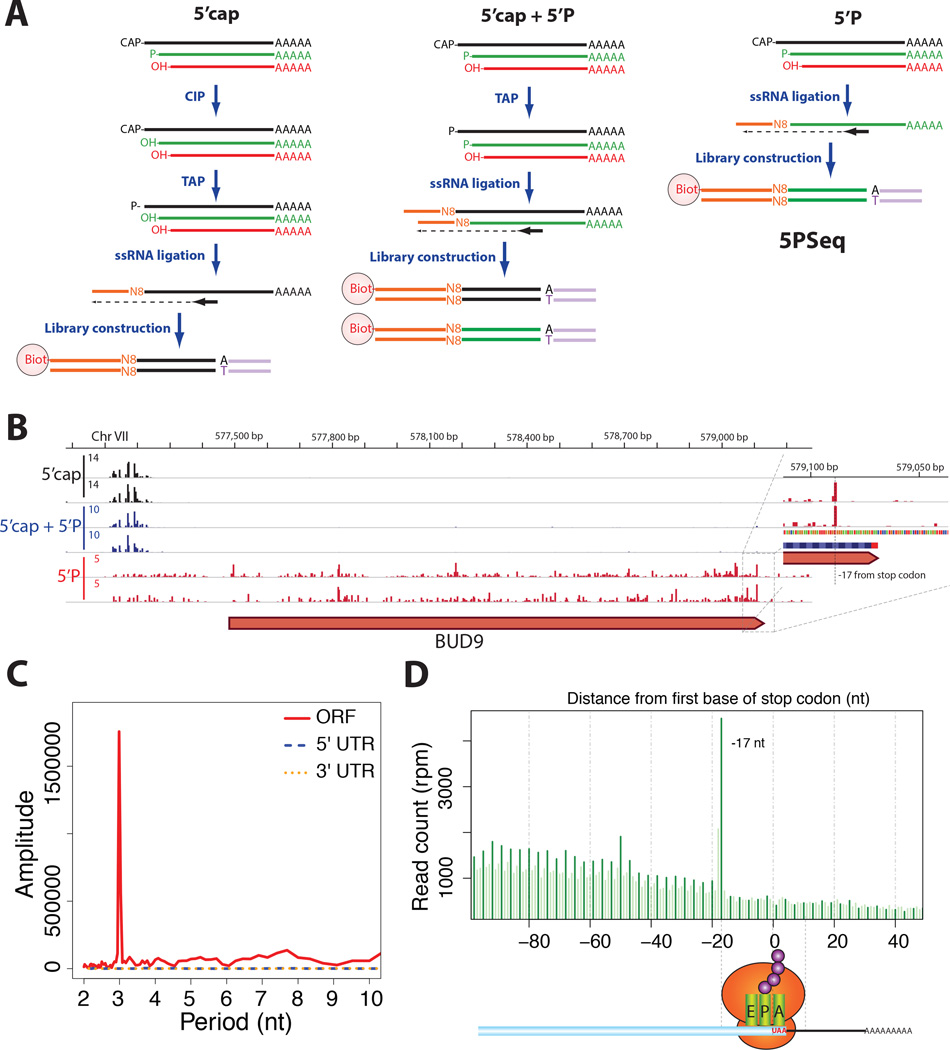
Figure 1. Genome-wide distribution of 5’P degradation intermediates.
A) Schematic diagram of 5’ specific sequencing methods that capture capped molecules, 5’P or the composite of both populations. In the case of 5PSeq, only 5’P RNA (in green) is able to ligate to a DNA/RNA oligo containing a molecular barcode (in orange).
B) Genome tracks of 5’ ends of capped (black), 5’P (red) or a composite of both capped and 5’P RNA molecules (blue). Coverage is expressed in reads per million (rpm), biological replicates are shown.
C) Discrete Fourier transformation of average 5PSeq reads across the ORF, 5’UTR or 3’ UTR.
D) Histogram of 5PSeq reads at each nucleotide position surrounding the stop codon. Protected frame is highlighted in dark green.
In comparison to the composite of the 5’P and 5’cap populations, we estimated that 5’P degradation intermediates represent ~12.4% of the cellular polyadenylated mRNA population. The ends of the 5’P mRNA molecules were heterogeneously and reproducibly distributed along gene bodies (Figure 1B and S1). To our surprise, although the distribution appeared random in the 5' and 3' untranslated regions (UTRs), it was periodic within coding regions, with a clear 3-nucleotide periodicity. (Figures 1C–D). This immediately suggested, that the translation process could be implicated.
The frequency distribution of 5’P mRNA ends implicates translation
To characterize this pattern further, we performed metagene analysis with respect to the open reading frames (ORFs). When aligning the 5’P reads based on the stop codons of all ORFs, we observed clear three-nucleotide periodicity with a large accumulation 17 nt upstream of the stop codon (Figures 1D and 2A). To rule out that this pattern was due to any underlying RNA sequence bias, we performed control experiments in which randomly fragmented RNA samples were re-phosphorylated and subjected to 5PSeq (Figure S1B). As expected no periodic, three-nucleotide pattern was observed in these controls (Figure 2A). Thus sequence context does not explain the observed pattern. It is known that a ribosome occupies a 28–29 nt footprint on the mRNA, of which the 5’ end position lies 15–16 nt upstream of the A-site (Ingolia et al., 2009; Lareau et al., 2014). Therefore, the 17 nt peak upstream of the stop codon corresponds roughly to the size of the footprint that an RNA ribosome occupies on the RNA when terminating translation at the stop codon (Ingolia et al., 2011). We therefore investigated if ribosomes paused at the stop could be protecting mRNAs undergoing degradation and lead to the accumulation of 5’P mRNA degradation intermediates.
Figure 2. Ribosome protection shapes the abundance of 5’P degradation intermediates.
A) Metagene analysis displaying the abundance of 5’P reads relative to ORF stop codons for cells in rich media (5PSeq YPD, in blue), in a mutant for the 5’-3’ RNA exonuclease (xrn1Δ, in pink), or after random fragmentation (5PSeq control, dotted light blue line). The blue peak at −17 nt corresponds to the protection of a putative termination-paused ribosome. Reads are represented in rpm (reads per million) and biological replicates are merged.
B) Three nucleotide periodicity pattern, displayed by histograms with the total number of reads overlapping each of the three frames along coding regions for 5PSeq in YPD (blue), random fragmentation (light blue), xrn1Δ strain (pink), and after cycloheximide treatment for isolated monosomes (yellow), polyribosome fractions (green) or total RNA (in purple).
C) Genome tracks of 5’ ends of 5’P molecules after cycloheximide treatment (in red), standard growth in YPD for a wild-type strain (in blue) and for xrn1Δ (in purple). For comparison a track displaying 5’ cap molecules (in grey) and random fragmented 5’ ends (in black) are shown. Coverage is expressed in reads per million (rpm). Horizontal bars represent regions significantly (FDR<0.1) enriched (in red) for 5PSeq coverage, when comparing CHX treatment with standard YPD conditions.
D) Metagene analysis displaying the abundance of 5’P reads relative to ORF start and stop codons as in panel A. 5’P reads after cycloheximide treatment is shown in purple (5PSeq CHX).
E) Proportion of 5PSeq reads in the ribosome-protected frame (Frame 1 in panel B), shown as smoothed lines (smooth splines) for genes without introns and longer than 600bp. 5PSeq samples from cells grown YPD (blue), after 5 min oxidative stress (brown), 3-AT treatment (orange), cycloheximide treatment (purple) or random fragmentation (light blue dashed line) are shown. The dotted black line shows the random expected frequency of 0.33.
F) Model for differential mRNA footprinting in 5PSeq compared to ribosome profiling.
One possibility is that the detected mRNA degradation intermediates originate from 5’-3’ mRNA decay (Parker, 2012; Stevens and Maupin, 1987) that occurs co-translationally. The prevalence of the three-nucleotide periodicity pattern suggests that co-translational degradation could be a general process affecting much of the transcriptome. In this model, the mRNA degradation machinery (putatively the 5’-3’ exonuclease, Xrn1) could closely follow the last ribosome producing a single nucleotide resolution in vivo footprint. We reasoned that this footprint could potentially provide information about ribosome dynamics and the translation status of the cell. To investigate the potential causative role of ribosomes in shaping 5’P degradation intermediates, we dissected the abundance and frequency distribution of 5’P molecules in multiple conditions, where the presence and position of both the ribosome and the 5’-3’ exonucleolytic machineries have been perturbed.
Ribosome occupancy conditions 5’ ends of degradation intermediates
To assess if the observed pattern is due to in vivo ribosomal footprinting involving 5’-3’ degradation, we performed 5PSeq in a strain in which the cytoplasmic 5’-3’ RNA exonuclease Xrn1 is deleted. The three-nucleotide pattern is completely absent in an xrn1Δ strain and there is a clear accumulation of 5’P molecules mapping to the transcription start site at both the metagene and single-gene levels, resembling the distribution of 5’capped molecules (Figures 2A–C and S2A). Thus, in the absence of Xrn1, mRNAs can be decapped but not trimmed, and therefore they do not produce an in vivo ribosomal footprint.
We also inhibited ribosome elongation with cycloheximide (CHX). As expected a 5-minute treatment with CHX increased the observed three-nucleotide periodicity pattern in the ribosome protected frame (i.e., comparing 5’P reads in frame 1 with respect to frame 0 and 2; Figures 2B and 2D), as well as the amount of obtained 5PSeq-library products. These data are consistent with CHX arresting translation elongation, blocking access of the 5’-3’ RNA exonuclease and increasing the accumulation of 5’-3’ RNA co-translational degradation intermediates.
In addition, to confirm that our 5PSeq-measured degradation intermediates are bound by ribosomes, we carried out polyribosomal sucrose fractionation and extracted both polysome and monosome fractions. We extracted RNA from these two ribosome-bound fractions and applied 5PSeq to each. A clear three-nucleotide periodicity pattern was visible in these samples (Figures 2B and S2B). The observed peak profiles were very similar to the ones obtained from RNA extracted from whole cells after cycloheximide treatment, which is required for polyribosomal sucrose fractionation (Figure 2D). These data confirm that 5PSeq-measured degradation intermediates are bound by ribosomes.
Since our 5PSeq approach involved the enrichment of polyadenylated mRNA degradation intermediates, we assessed whether the subpopulation of degradation intermediates with at least partially remaining polyA tails after mRNA deadenylation was the only one preferentially co-translationally degraded. We thus repeated the same experiment substituting the polyA enrichment step by an rRNA removal step and obtained comparable results, reproducing a three-nucleotide periodicity pattern that was increased by CHX treatment (Figure S2C). Taking all this into account, the data suggest that a substantial fraction of the detected 5’P mRNA degradation intermediates, whether polyadenylated or not, originate from general co-translational 5’-3’ mRNA degradation.
Cycloheximide does not inhibit translation initiation or termination, and even in the case of translation elongation, it allows one complete translocation cycle before blocking the ribosome by binding to its E-site (Pestova and Hellen, 2003; Schneider-Poetsch et al., 2010). Thus, even if a ribosome pauses at the stop codon in vivo, treatment with CHX would not prevent the ribosome from running off the mRNA in vitro. Consequently, treatment with CHX reduces the 5PSeq peak 17 nt upstream of the stop codon both in our 5PSeq (Figure 2D and S2C), as well as in ribosome profiling experiments (Gerashchenko et al., 2012) (Figure S2D). Notably, when ribosome profiling was attempted in the absence of CHX in mouse cells, the peak upstream of the stop codon was observed, suggesting that CHX treatment causes its disappearance (Ingolia et al., 2011). However, in recent yeast, ribosome profiling data without CHX addition (Guydosh and Green, 2014; Lareau et al., 2014) this peak was not seen, possibly due to differences during the in vitro sample-processing steps.
We then analysed the patterns surrounding the start codon. We observed a clear three-nucleotide pattern near the start codon after CHX treatment. However, when analysing the protection pattern surrounding the start codon in a drug-free environment, we observed a very weak three-nucleotide periodicity pattern (Figure 2D and S2E). Instead, a distinct three-nucleotide pattern emerged only gradually over the course of ~300 nt downstream from the start codon (blue line, Figure 2E and S2F–G). This is likely because decapping is rate limiting, and thus makes it difficult for the exonuclease to catch up with the ribosome near the translation start site. Consistent with this interpretation, when translation elongation is limited after oxidative stress or treatment with the histidine biosynthesis inhibitor 3-amino-1,2,4-triazole (3-AT) (see below), the three-nucleotide pattern emerges closer to the start codon (Figure 2E). This effect is more pronounced after CHX treatment, which completely blocks translation elongation, producing a pattern around the translation start codon almost identical to the one observed during ribosome profiling (Figures 2D and S2D). This suggests that the typical ribosome accumulation pattern observed by ribosome profiling around the start codon, both at a metagene (Figure 2D) and individual gene level (Figure 2C), is mainly a secondary effect of the drug. These results are in agreement with the known ability of CHX to cause accumulation of ribosomes over the first few codons of an ORF. This is due to an inhibition of translation elongation by the drug, in the presence of continuing initiation of new ribosomes onto the mRNA (Gerashchenko and Gladyshev, 2014; Ingolia et al., 2011). Therefore, alternative ribosome profiling approaches omitting the addition of CHX prior to cell harvesting have been explored (Guydosh and Green, 2014; Lareau et al., 2014). These approaches greatly decrease the accumulation of ribosomes over the first few codons (Figure S2H), however a pattern can still be observed at the start codon, suggesting that translation initiation may still occur in vitro during the required sample handling steps.
5PSeq and ribosome profiling measure two related but distinct pools of RNA molecules. 5PSeq measures mRNAs undergoing 5’-3’ co-translational degradation in vivo and ribosome profiling measures the ribosome protected fragments from in vitro RNA digestion. Surprisingly, the addition of CHX produces a 5PSeq pattern remarkably similar to that obtained from ribosome profiling, however small differences exist (Figures 2D and S2D). 5’P degradation intermediates, produced by in vivo ribosome footprinting by Xrn1, are displaced by 2 nt with respect to the in vitro digestion performed by RNase I in ribosome profiling (Gerashchenko et al., 2012; Ingolia et al., 2009) (Figure 2F). This difference is likely explained by different accessibility of the two enzymes, as well as by the fact that 5PSeq measures mRNA degradation products naturally produced in cellular environments, while in ribosome profiling, digestion occurs in a purified polyribosomal fraction, from which most cellular components have been eliminated.
The clear three-nucleotide periodicity pattern surrounding the translation start site upon CHX treatment further confirms that the ribosome protected frame determines the abundance of specific 5’P mRNA degradation intermediates. Additionally, it suggests that a brief treatment with a translation inhibitor, followed by 5’P sequencing, is sufficient to experimentally identify translation start sites. Collectively, we conclude that general co-translational degradation of mRNAs determines the three-nucleotide periodicity pattern among 5PSeq products, and that these patterns enable inference of ribosome dynamics.
Changes in 5’P mRNA degradation intermediates mimic translational regulation
We investigated if particular groups of genes are preferentially targeted by co-translational 5’-3’ mRNA degradation. We computed a codon protection index as a self-controlled measure of the degree of co-translational degradation for each gene (Figure 3A). This codon protection index reflects the strength of the three-nucleotide periodicity pattern (i.e. difference between the ribosome protected frame and other frames) independent of read number. We identified groups of genes with significantly deviant codon protection indices for both polyA-enriched or rRNA-depleted samples (Figure S3A–B). Genes associated with translation (p-adjusted < 4·10−19), have lower codon protection indexes, suggesting that their degradation is less often associated with active translation (Figure S3A); while genes involved in transcription showed high degrees of co-translational degradation.
Figure 3. The abundance of co-translational mRNA degradation intermediates reflects translational regulation.
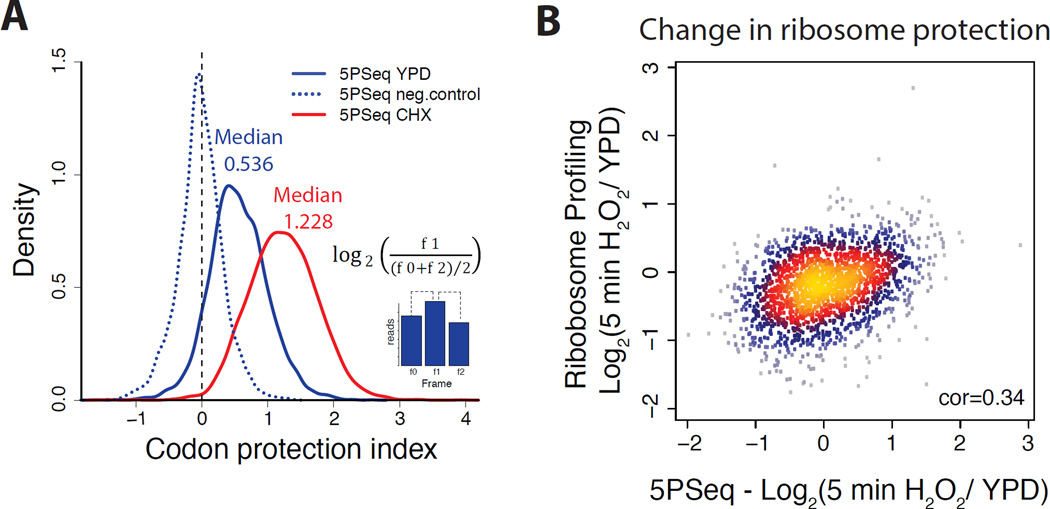
A) Histogram for codon protection index as a measure for co-translational degradation. Codon protection index was computed as the log2 ratio of the reads corresponding to the protected frame (f1) in respect to the average number of reads of the non-protected frames ((f0+f2)/2). This ratio was computed for cells in rich media (blue), after CHX treated (red) and randomly fragmented samples (dotted blue line). Only genes with at least 50 reads were considered for the analysis.
B) Scatter plot comparing the variation of abundance of 5PSeq reads in the body of the gene (xaxis) with the variation of ribosome profiling reads (y-axis) (Gerashchenko et al., 2012) after 5 minutes treatment with H2O2 0.2mM. Spearman correlation coefficient is shown.
To estimate the proportion of mRNA molecules that undergoes co-translational degradation (where Xrn1 speed is limited by ribosomes), we compared the codon protection index in the absence of ribosomes (assumed median 0, random fragmented sample) with the codon protection index resulting from CHX treatment (which blocks ribosomes and thus enforces a maximal rate of co-translational degradation, median 1.228 Figure 3A). Since the median codon protection index from 5PSeq is 0.536, this analysis suggests that at least 34% of degradation intermediates reflect co-translational degradation (Extended experimental procedures).
Altogether, on an individual gene basis, co-translational degradation is not correlated with translation rate as measured by ribosome profiling (Gerashchenko et al., 2012) (Figures S3C–D), or by polyribosome fractionation followed by microarray hybridization (Arava et al., 2005) (Figures S3E–G). However, the change in translation rate between two conditions is captured by all three data types. For this analysis, we measured the change in 5’P mRNA intermediates normalised by the mRNA abundance in response to oxidative stress (5 minutes in 0.2mM H2O2). We compared these data to those from the matching ribosome profiling experiment (Gerashchenko et al., 2012) and found a reasonable correlation between both methods (Spearman cor = 0.34, Figure 3B). This was striking, especially considering that 5PSeq studies the subpopulation of transcripts undergoing co-translational degradation, while ribosome profiling focuses on mature mRNAs. Although there are method-specific differences, shared gene categories can be identified as differentially regulated by both methods. Specifically, upon oxidative stress, genes related to ribosome biogenesis and translation increase their ribosome protection, and genes related to budding and cell cycle decrease their ribosome protection (Figures S3H–I).
At a first glance, this observed increase in ribosome protection of translation-related genes could appear contradictory to previous reports, where transcription has been reported to be down-regulated in response to stress (Gasch et al., 2000). However, it is important to remember that both ribosome profiling and 5PSeq measure ribosome protection and not necessarily protein production. Previous reports have shown that brief H2O2 stress promotes polyribosome association but does not necessary lead to increased protein production (Shenton et al., 2006). Specifically Shenton et al. showed that, in addition to the expected inhibition of translation initiation, short H2O2 stress causes a reduction in the rate of ribosomal run-off that is consistent with an inhibition of translation elongation or termination (Shenton et al., 2006). Our results not only support these conclusions, but also offer a new opportunity to identify where this putative translation elongation or termination regulation is occurring (see below). Additionally, our results indicate that translational regulation affecting mRNAs, rapidly impacts the subpopulation of mRNAs undergoing co-translational degradation (i.e., the ones measured by 5PSeq). Taken together, we conclude that changes in abundance of 5’P degradation intermediates inform us of upstream variation in ribosome occupancy, and could potentially be used as a proxy for the study of translational regulation.
Inferring ribosome dynamics from 5’P mRNA degradation intermediates
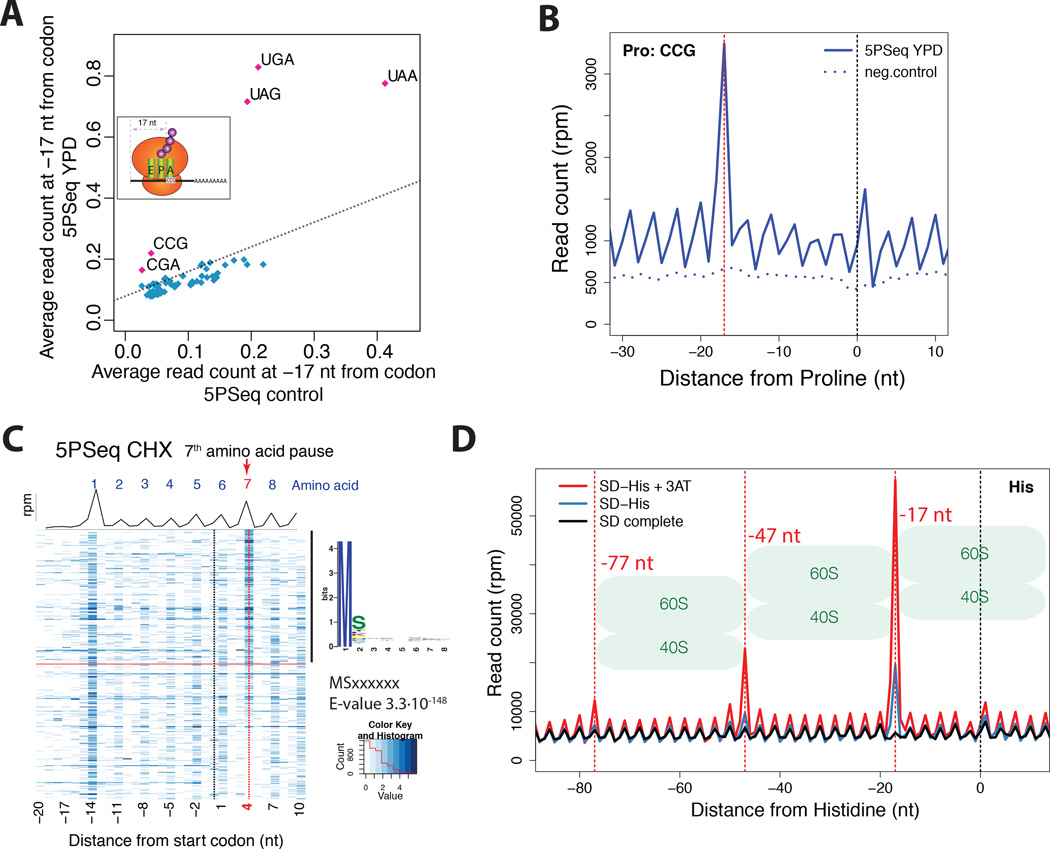
The main advantage of 5PSeq is that it allows the study of in vivo codon-specific ribosome protection patterns in a drug-free environment and without complex sample manipulation or in vitro RNA digestion. Our metagene analysis showed an accumulation of 5’P molecules corresponding to ribosomal pausing at the stop codon (i.e., UAA, UAG and UGA) (Figures 2A and 4A). In addition, we found a clear pause site corresponding to the rare proline codon CCG, and an arginine codon CGA that decreases translation efficiency due to wobble decoding (Letzring et al., 2010) (Figure 4A–B and S4C). Cycloheximide treatment, which allows one complete translocation cycle before blocking the ribosome (Pestova and Hellen, 2003; Schneider-Poetsch et al., 2010), diffuses this CCG codon protection pattern (Figure S4A). Thus CCG pausing cannot be detected with conventional ribosome profiling using CHX (Figures S4B). However, CCG pausing can be detected if CHX is omitted in ribosome profiling experiments (Lareau et al., 2014) (Figure S4D).
Figure 4. 5PSeq allows exploration of ribosome dynamics at codon resolution.
A) 5PSeq coverage 17 nt upstream of each codon (y-axis) compared with the random fragmented sample (x-axis). Stop codons (UGA, UAG and UAA), CGA (Arginine) and CCG (Proline) present an increased pausing. The dashed line was estimated by comparing the average percentage of reads at frame 1 in all codons between 5PSeq and random fragmented control.
B) Metagene displaying the abundance of 5’P reads in respect to the rare proline codon (CCG) for cells in rich media (blue) and a randomly fragmented samples (dotted blue line). The expected 5’ endpoint protection (−17 nt) is displayed by a dotted red line
C) Metagene representing both the average coverage (black line) and gene-specific coverage (blue heatmap) for cells grown after cycloheximide treatment. Only genes with at least 10 reads in the displayed regions were considered. Genes were sorted by the ratio of reads at position 4 nt (red dotted line) to the reads corresponding to the surrounding +/−2 codons. To identify specific peptide sequences, the first 8 amino acids of the top 50% of genes were compared to the bottom half using MEME (Bailey et al., 2009).
D) Metagene displaying the abundance of 5’P reads with respect to histidine codons for 5PSeq of cells in synthetic defined media (SD, in black), SD media without Histidine (in blue) and SD media without histidine poisoned with 3-AT (in red). At −17, −47 and −77 nt, dotted red lines corresponding to the expected 5' endpoints of protection by a ribosome, disome or trisomes halted at the histidine codon are displayed.
We also detected a sharp accumulation of 5’P molecules 4 nt downstream of the start codon in untreated wild type and CHX treated samples, but not in negative controls (Figures 2D and S2E). Considering the ribosomal footprint, this position corresponds to the 7th amino acid. The 5’P accumulation signal thus suggests a generalized pause associated with the addition of the amino acid at this position. It is known, that certain genes encode peptides containing arrest sequences that interact with the ribosome exit tunnel and arrest their own translation, often in an environmentally regulated manner (Ito and Chiba, 2013; Wilson and Beckmann, 2011). However, here we identify a subtle but generalized pause specifically associated with peptides containing serine at the second position (pattern as MSxxxxx, E-value 3.3·10−148 analysed by MEME (Bailey et al., 2009); Figures 4C and S4E).
To further confirm that 5PSeq can detect translational pause sites in vivo, we induced widespread accumulation of ribosomes at histidine codons by treating yeast with 3-AT (Guydosh and Green, 2014; Klopotowski and Wiater, 1965). We compared cells grown in minimal defined media (SD), minimal media without histidine (SD -His), and minimal media without histidine and treated with 3-AT (SD -His +3AT). We saw increased pausing at all histidine codons after histidine depletion (Figure 4D). Additionally, the pausing is so extreme that especially with 3-AT treatment, peaks corresponding to 2 or even 3 ribosomes in close proximity to one another appear upstream of histidine codons. These chains of sterically blocked ribosomes are in agreement with the disome particles recently detected by modified ribosome profiling, using the dom34Δ mutant involved in the dissociation of stalled ribosomes (Guydosh and Green, 2014), and have also been seen in response to amino acid starvation (Subramaniam et al., 2014). These findings also suggest that for strongly translated genes in vivo pausing by steric interference among ribosomes translating the same mRNAs could be common.
Oxidative stress causes tRNA-specific translation pauses dependent on the RNase Rny1p
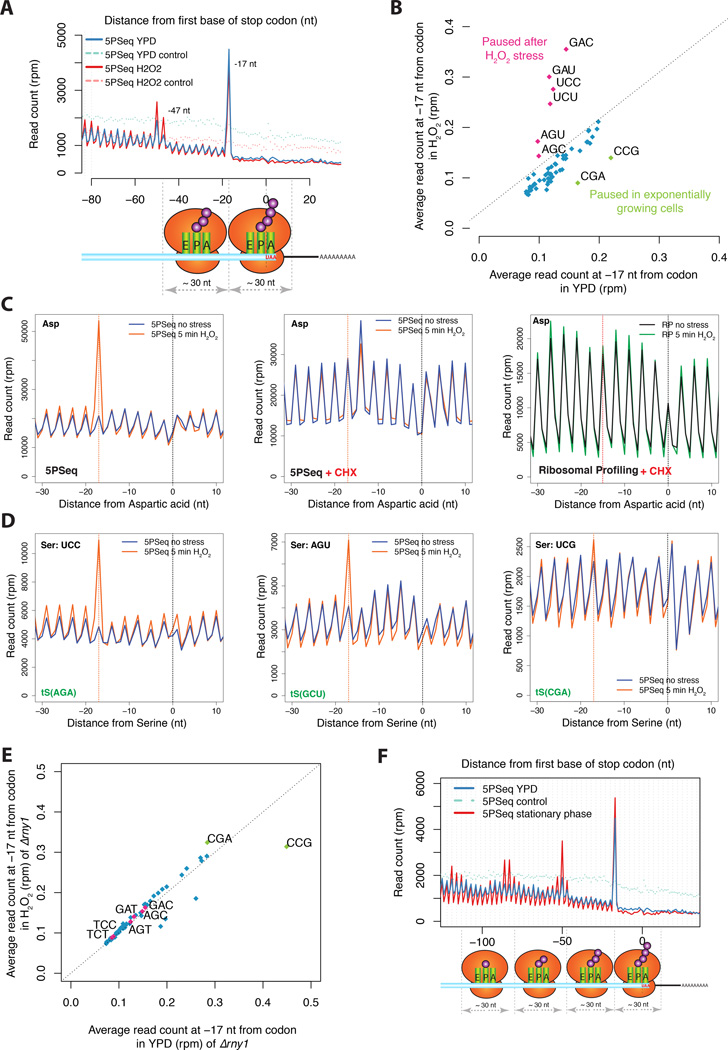
We then applied 5PSeq to the question of how translational dynamics are altered by oxidative stress. Previous studies have found a slower rate of ribosomal run-off after oxidative stress, which is consistent with the inhibition of either translational elongation or termination (Shenton et al., 2006). How such regulation occurs is not known. After a 5-minute H2O2 treatment, an accumulation of 5’P degradation intermediates appears ~30 nt upstream of ribosomes paused at the stop codon (−50 and −47 nt relative to the stop codon) (Figure 5A). This accumulation suggests that in response to oxidative stress, there is a generalized translational pause at termination, generating disomes. In addition, to the pause at the stop codon, we identify clear ribosome elongation pausing sites at all aspartic acid codons (GAU and GAC) and at most serine codons (especially UCU, UCC, AGU and AGC) (Figures 5B–D and S5). These pauses are lost upon the addition of CHX, both for 5PSeq and ribosome profiling (Figures 5C and S5A), suggesting that the generalized translational inhibition caused by CHX is sufficient to mask naturally occurring ribosome pausing sites. This is consistent with ribosomal pausing having been difficult to detect by ribosome profiling (Tarrant and von der Haar, 2014). Additionally, the pausing observed for CGA and CCG codons in normal conditions (Figure 4A) is lost upon oxidative stress (Figure 5B). As the identified pauses do not affect all codons encoding each amino acid (e.g., UCG and UCA encoding for serine, Figures 5D and S5C), this process is likely regulated at the level of availability of charged tRNAs, rather than at the level of amino acid abundance.
Figure 5. Drug free approach allows the exploration of new ribosome dynamics.
A) Metagene displaying the abundance of 5’P reads with respect to stop codons for 5PSeq of cells in rich media (in blue) or after 5 minutes H2O2 0.2mM treatment (in pink). Randomly fragmened controls are shown in dashed lines.
B) 5PSeq coverage 17 nt upstream of each codon after H2O2 treatment (y-axis) compared to exponentially growing cells (x-axis) as in Figure 4A. Codons paused after stress are highlighted in pink and codons paused in exponential growth are highlighted in green. Only amino acid coding codons are shown.
C) Metagene displaying the Asp specific ribosome pausing after 5 minutes H2O2 0.2mM stress. The clear protection pattern is lost both by the treatment with cycloheximide or if analysed by ribosome profiling (Gerashchenko et al., 2012). The expected 5’ endpoint protection (−17 nt, or −15 for ribosome profiling) is displayed by a dotted red line
D) Upon oxidative stress (orange line) a clear pause can be observed at the codons UCC and AGU, but not at UCG. As shown in panel C.
E) Relative codon pausing after oxidative stress in an rny1Δ strain as shown in Figure 5B.
F) Metagene displaying the abundance of 5’P reads with respect to stop codons in cells grown in rich media (in blue and 5PSeq control in light blue dashed line) or in a saturated culture (5PSeq stationary phase in red).
Previous reports have shown that in response to oxidative stress RNY1, a vacuolar RNase of the T(2) family, is released into the cytoplasm and tRNAs are cleaved (Thompson et al., 2008; Thompson and Parker, 2009). Therefore, we repeated our 5PSeq experiment in an rny1Δ strain. In this mutant, the tRNA-specific pauses were lost (Figure 5E). This suggests that RNY1, or the RNA fragments derived from its action (Thompson and Parker, 2009), are required for the observed novel ribosome pausing during stress. In addition, the fact that the rny1Δ strain presents a decreased physiological resistance to oxidative stress (Thompson et al., 2008), suggests that these codon-specific translation pauses are important for cellular adaptation to oxidative stress. To further investigate if tRNA-specific pauses are common in response to stress we also analysed 5PSeq data of cells in stationary phase (i.e. saturated culture after 3 days of growth). In this condition, we do not observe codon-specific translational pauses (Figure S5B), but rather a massive accumulation of ribosomes paused at the stop codon (Figure 5F). This suggests that the mechanism used to limit translation in the stationary phase (i.e. long term repression) follows a different strategy than the one used for the rapid translational repression exerted after oxidative stress.
Co-translational mRNA degradation is evolutionarily conserved
To determine whether co-translational degradation is evolutionarily conserved, we performed 5PSeq in evolutionarily distant fission yeast (S. pombe). We observed the same three-nucleotide 5’P pattern and it was enhanced by the addition of CHX (Figure 6). The observed pattern is subtler than in the case of S. cerevisiae. However, the fact that CHX treatment enhances the protection pattern and random fragmentation completely destroys it, suggests that co-translational degradation is an evolutionarily conserved process. Thus, our method could be widely used to investigate ribosome dynamics in other species.
Figure 6. Co-translational degradation is an evolutionarily conserved process.
A) Metagene displaying the abundance of 5’P reads with respect to stop codons for 5PSeq of cells in rich media (in blue), after 5 or 30 minutes treatment with cycloheximide (in pink and magenta respectively) or a randomly fragmented sample (dotted light blue line) for S. pombe. Shown as in Figure 2.
B) Histograms showing the total number of reads in each reading frame within coding regions for 5PSeq YES (in blue), 5PSeq after cycloheximide treatment (in pink and magenta), and randomly fragmented sample (in light blue) for S. pombe.
To determine whether the preferential co-translational degradation of specific gene categories is also conserved, we analyzed codon protection indexes for S. pombe and compared them to our data for S. cerevisiae. We found that translation-related genes have a low codon protection index in both species and thus their degradation is less often associated with active translation. Other categories related to cell division and vesicle transport tended to have a higher codon protection index in both species implying that their RNA is more frequently co-translationally degraded (Table S1). These data suggest that the preferential use of co-translational degradation for specific groups of genes is conserved throughout evolution.
Discussion
Here we show that 5’P mRNA degradation intermediates are widespread and that co-translational 5’-3’ decay is a general and conserved process that produces an in vivo ribosomal footprint. By measuring the relative abundance of 5’P mRNA degradation intermediates, we obtain a genome-wide drug-free measurement of ribosome dynamics, overcoming previous limitations caused by the use of translation inhibitors.
The distribution of 5'P ends (Figure 2) indicates that there is a pronounced general pausing of ribosomes at stop codons, and that this pattern is lost upon addition of cycloheximide (Figure 2). The accumulation of 5’-3’ mRNA degradation intermediates corresponding to the products produced by stalling at the stop codon is consistent with previously unexplained accumulation of Xrn1, detected by RNA crosslinking (CRAC), at the same position (Figure S2I) (Tuck and Tollervey, 2013).
It has been proposed that translation rates are lower at the 5’ end than in the body of coding regions, leading to the relative accumulation of ribosomes toward the 5' end (Tuller et al., 2010; Tuller et al., 2011). This hypothesis has been supported by results of ribosome profiling, which indicated a greater concentration of ribosomes in the 5' half of ORFs (Ingolia et al., 2009). Other studies suggested, however, that such accumulation is caused by other factors such as RNA secondary structure (Bentele et al., 2013; Yang et al., 2014) or confounding factors like the rapid initiation of short genes (Shah et al., 2013). Our results support neither hypothesis, and rather suggest that the accumulation of paused ribosomes in the 5’ region of genes is due to a secondary effect of the translation inhibitor used or due to continued ribosome initiation during sample handling (Figures 2C and S2). We show that by decreasing translation elongation, as occurs with the use of translation inhibitors, the accumulation of 5’ located ribosomes increases. Additionally, we detected less ribosomes in the 5’ region, which is consistent with a faster, rather than a slower, elongation at the beginning of genes (Figure 2E). As 5PSeq reveals the ribosome footprint of the last elongating ribosome, the relative depletion of ribosomes in the 5’ region is also compatible with the requirement for a minimum distance between the cap to the last trailing ribosome for efficient decapping and Xrn1-dependent mRNA trimming.
Our results show that the use of absolute values of experimentally measured ribosome protection data should be treated with caution. This is especially true when deriving kinetic models and when using translation inhibitors (Gerashchenko and Gladyshev, 2014; Ingolia et al., 2009). Even when ribosome profiling is performed in the absence of drugs, ribosomes can elongate for 50 codons during sample processing (Lareau et al., 2014). Nonetheless, CHX-treated samples are useful for identifying translation start codons and translation rates (Ingolia et al., 2011; Lee et al., 2012).
We have shown that co-translational mRNA degradation is a general and conserved process affecting all genes. Only highly translated genes, and especially genes associated with translation, are less often co-translationaly degraded (Figure S3). Changes in the abundance of 5’P degradation intermediates undergoing co-translational degradation between two conditions parallels the changes observed for the translation 5’ capped mRNA molecules (Figure 3B). Thus 5PSeq can be used as a complementary approach to study changes in ribosome association, especially in cases where it is not possible to apply methods such as ribosome profiling (Ingolia et al., 2009), or polyribosome fractionation (Arava et al., 2003).
The main advantage of 5PSeq is that it allows the determination of the position of the last translating ribosomes on multiple mRNAs from the same gene, and thereby provides drug-free measurements of their movements. We have detected ribosome accumulation at rare codons that could not be detected by previous methods (Figures 4A–B and S4). We have also shown the existence of a generalized translational elongation pause at the 7th amino acid (Figures 4C and S4E), which is consistent with an interaction of the nascent peptide with the ribosome exit tunnel (Ito and Chiba, 2013; Wilson and Beckmann, 2011). Recently, the existence of a sequence-independent post-initiation pause at the 5th amino acid has been reported (Han et al., 2014). The authors suggested that this pause is caused by the interaction of the nascent peptide with the ribosome exit tunnel (i.e. RPL4). They also suggested that this constitutes a general pause required to ensure elongation commitment. When analysing ribosome profiling data, we detect a similar pause at the 4th and 5th codon positions (Figure S2D), however with 5PSeq the pause is even clearer and is displaced downstream by two codons (Figures 2D and S2E). Analysing the 7th amino acid pause, we discovered that peptides containing a serine at the second position are more likely to induce this transient translational halt. Further research will be necessary to understand the structural cause of this pause and its biological significance.
We have identified codon- and terminator-specific ribosome pauses, which likely explain the transient accumulation of translationally inactive polyribosome fractions (Shenton et al., 2006). Since we observed that alternative codons for the same amino acid are differentially regulated, this process is likely controlled at the level of charged tRNAs availability rather than at the level of amino acid abundance (Figures 5 and S5). The fast response that we detected (i.e., 5 minutes) is too rapid to be explained by changes in tRNA synthesis, as tRNAs have a half-life in the range of hours (Rudra and Warner, 2004). Our data shows that Rny1, a vacuolar RNase of the T(2) family, is required for the tRNA-specific ribosomal pausing that we found upon oxidative stress. Further research will be necessary to determine if Rny1, or the RNA fragments derived from its action, cause these translational pauses. The existence of tRNA-specific pauses shows that codons encoding for the same amino acid can be independently regulated. Synonymous codons therefor offer different evolutionary advantages, especially for adaptation to changing environments. Independently of that, the existence of a fast translational response highlights the need for methods such as 5PSeq, that allow rapid kinetic experiments without the need for in vitro incubation steps. Finally, we have demonstrated the applicability of 5PSeq to study ribosome dynamics in other organisms, such as S. pombe (Figure 6). This highlights evolutionary conservation of co-translational degradation. Further research is needed to study to what degree this process is conserved in higher eukaryotes.
Notably, the prevalence of 5’P co-translational degradation intermediates we observed here could have implications even for previous studies. For example, cellularly abundant 5’P molecules are likely contaminants in most genome-wide measurements of RNA secondary structure that include a single-stranded RNA ligation step (Ding et al., 2014; Kertesz et al., 2010). Studies using PARS (Kertesz et al., 2010) or structure-Seq (Ding et al., 2014) have described analogous three-nucleotide periodicity patterns (reviewed in (Mortimer et al., 2014)), but interpreted them to reflect RNA secondary structure. However, in these studies, the authors did not consider the possibility that 5'P molecules with a defined biological origin (not random) could be present in the samples. This would be especially relevant in regions with less prominent structure and where potential 5’P contamination could lead to erroneous attribution of structural readouts, such as the three-nucleotide pattern of 5'P ends caused by ribosome protection. To disentangle the effects of RNA structure from those of ribosome protection, these approaches (reviewed in (Mortimer et al., 2014)) might benefit from a pre-treatment with a 5’P specific exonuclease prior to sample treatment or library construction.
Here, we presented a rapid and straightforward approach to study ribosome dynamics in vivo by measuring natural mRNA degradation intermediates. This approach can be applied to different organisms and to any previously isolated RNA samples. By examining ribosome dynamics we can uncover new layers of translational regulation, distinguish those acting at the level of translation termination or elongation through specific codons, and improve the understanding of how complex biological systems respond to external stimuli.
Experimental Procedures
Growth conditions and sample preparation
Saccharomyces cerevisiae strains BY4741 (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0), xrn1Δ (xrn1Δ::kanMX4, BY4741 background) or rny1Δ (rny1Δ::kanMX4, BY4741 background) were grown using YPD or Complete Synthetic Media (CSM). Schizosaccharomyces pombe (h-) was grown using YES media. All samples were collected by centrifugation and frozen in liquid N2. Total RNA was isolated by the standard phenol extraction and DNA was removed by DNase I treatment. For cycloheximide (CHX) treatment, CHX was added to 0.1mg/mL final and incubated either 5 or 30 min. For 3-amino-1,2,4-triazole (3-AT) treatment we used S96 strain (MATa lys5Δ gal2Δ, S288c background) grown in CSM-His. For polyribosome fractionation, CHX treated samples were separated using a 10–50% sucrose gradient.
Preparation of sequencing libraries
For the construction of 5’-specific sequencing libraries we used 10 µg of DNA-free total RNA for the 5’cap libraries and 50–100 µg for the 5’P libraries (5PSeq). To select the 5’ modification of interest different treatments were performed on the initial (schematized in Figure S1A). After the initial library-specific treatment, all protocols converged in the same RNA ligation step and downstream library construction. For 5’P sequencing (5PSeq), samples were directly subjected to RNA ligation. PolyA-enriched (or rRNA depleted) RNA was fragmented and reverse transcribed using random hexamers. cDNA molecules containing the 5’ ligated oligo was captured with Streptavidin beads and used to generate Illumina compatible sequencing libraries. Detailed information is provided in the extended experimental procedures.
Sequence analysis
Sequencing read analysis was performed using R and Bioconductor (http://www.bioconductor.org/). 5' ends of reads passing quality filtering were collapsed to unique molecules (based on random barcodes) for further analysis. Detailed information is provided in the extended experimental procedures, Supplementary tables S1–S5, and at http://steinmetzlab.embl.de/5PSeq/
Supplementary Material
Highlights.
Co-translational RNA degradation produces an in vivo ribosomal footprint.
Profiling mRNA degradation provides a measurement of ribosome dynamics.
Oxidative stress causes tRNA-specific translation pausing sites dependent on RNY1.
Acknowledgments
We thank the members of the Steinmetz laboratory for helpful discussions and critical comments on the manuscript. We thank P. Jacob for technical assistance, and M. Petitt, A. Jones and R. Aiyar for help editing the manuscript. This study was technically supported by the EMBL Genomics Core Facility. This study was financially supported by the National Institutes of Health (NIH R01 GM068717 and NIH P01 HG000205), Deutsche Forschungsgemeinschaft (1422/3-1) and a European Research Council Advanced Investigator Grant (AdG-294542) to L.M.S.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession Numbers
Sequencing data is deposited at GEO under the accession GSE63120.
Author Contributions
V.P., W.W. and L.M.S. conceived the project. V.P. developed the methods and performed the experiments. W.W. and V.P. performed the analysis. V.P., W.W. and L.M.S wrote the manuscript. V.P. and W.W. contributed equally to the study.
The authors declare no conflict of interest.
References
- Addo-Quaye C, Eshoo TW, Bartel DP, Axtell MJ. Endogenous siRNA and miRNA targets identified by sequencing of the Arabidopsis degradome. Current biology : CB. 2008;18:758–762. doi: 10.1016/j.cub.2008.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson JS, Parker RP. The 3' to 5' degradation of yeast mRNAs is a general mechanism for mRNA turnover that requires the SKI2 DEVH box protein and 3' to 5' exonucleases of the exosome complex. The EMBO journal. 1998;17:1497–1506. doi: 10.1093/emboj/17.5.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arava Y, Boas FE, Brown PO, Herschlag D. Dissecting eukaryotic translation and its control by ribosome density mapping. Nucleic acids research. 2005;33:2421–2432. doi: 10.1093/nar/gki331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arava Y, Wang Y, Storey JD, Liu CL, Brown PO, Herschlag D. Genome-wide analysis of mRNA translation profiles in Saccharomyces cerevisiae. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:3889–3894. doi: 10.1073/pnas.0635171100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. MEME SUITE: tools for motif discovery and searching. Nucleic acids research. 2009;37:W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentele K, Saffert P, Rauscher R, Ignatova Z, Bluthgen N. Efficient translation initiation dictates codon usage at gene start. Molecular systems biology. 2013;9:675. doi: 10.1038/msb.2013.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brar GA, Yassour M, Friedman N, Regev A, Ingolia NT, Weissman JS. High-resolution view of the yeast meiotic program revealed by ribosome profiling. Science. 2012;335:552–557. doi: 10.1126/science.1215110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker CJ, Parker R. P-bodies and stress granules: possible roles in the control of translation and mRNA degradation. Cold Spring Harbor perspectives in biology. 2012;4:a012286. doi: 10.1101/cshperspect.a012286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y, Tang Y, Kwok CK, Zhang Y, Bevilacqua PC, Assmann SM. In vivo genome-wide profiling of RNA secondary structure reveals novel regulatory features. Nature. 2014;505:696–700. doi: 10.1038/nature12756. [DOI] [PubMed] [Google Scholar]
- Gasch AP, Spellman PT, Kao CM, Carmel-Harel O, Eisen MB, Storz G, Botstein D, Brown PO. Genomic expression programs in the response of yeast cells to environmental changes. Molecular biology of the cell. 2000;11:4241–4257. doi: 10.1091/mbc.11.12.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerashchenko MV, Gladyshev VN. Translation inhibitors cause abnormalities in ribosome profiling experiments. Nucleic acids research. 2014;42:e134. doi: 10.1093/nar/gku671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerashchenko MV, Lobanov AV, Gladyshev VN. Genome-wide ribosome profiling reveals complex translational regulation in response to oxidative stress. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:17394–17399. doi: 10.1073/pnas.1120799109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- German MA, Pillay M, Jeong DH, Hetawal A, Luo S, Janardhanan P, Kannan V, Rymarquis LA, Nobuta K, German R, et al. Global identification of microRNA-target RNA pairs by parallel analysis of RNA ends. Nature biotechnology. 2008;26:941–946. doi: 10.1038/nbt1417. [DOI] [PubMed] [Google Scholar]
- Gregory BD, O'Malley RC, Lister R, Urich MA, Tonti-Filippini J, Chen H, Millar AH, Ecker JR. A link between RNA metabolism and silencing affecting Arabidopsis development. Developmental cell. 2008;14:854–866. doi: 10.1016/j.devcel.2008.04.005. [DOI] [PubMed] [Google Scholar]
- Guydosh NR, Green R. Dom34 rescues ribosomes in 3' untranslated regions. Cell. 2014;156:950–962. doi: 10.1016/j.cell.2014.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y, Gao X, Liu B, Wan J, Zhang X, Qian SB. Ribosome profiling reveals sequence-independent post-initiation pausing as a signature of translation. Cell research. 2014;24:842–851. doi: 10.1038/cr.2014.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harigaya Y, Parker R. Global analysis of mRNA decay intermediates in Saccharomyces cerevisiae. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:11764–11769. doi: 10.1073/pnas.1119741109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu CL, Stevens A. Yeast cells lacking 5'-->3' exoribonuclease 1 contain mRNA species that are poly(A) deficient and partially lack the 5' cap structure. Molecular and cellular biology. 1993;13:4826–4835. doi: 10.1128/mcb.13.8.4826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W, Sweet TJ, Chamnongpol S, Baker KE, Coller J. Co-translational mRNA decay in Saccharomyces cerevisiae. Nature. 2009;461:225–229. doi: 10.1038/nature08265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Brar GA, Rouskin S, McGeachy AM, Weissman JS. The ribosome profiling strategy for monitoring translation in vivo by deep sequencing of ribosome-protected mRNA fragments. Nature protocols. 2012;7:1534–1550. doi: 10.1038/nprot.2012.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324:218–223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Lareau LF, Weissman JS. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell. 2011;147:789–802. doi: 10.1016/j.cell.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito K, Chiba S. Arrest peptides: cis-acting modulators of translation. Annual review of biochemistry. 2013;82:171–202. doi: 10.1146/annurev-biochem-080211-105026. [DOI] [PubMed] [Google Scholar]
- Kertesz M, Wan Y, Mazor E, Rinn JL, Nutter RC, Chang HY, Segal E. Genome-wide measurement of RNA secondary structure in yeast. Nature. 2010;467:103–107. doi: 10.1038/nature09322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klopotowski T, Wiater A. Synergism of aminotriazole and phosphate on the inhibition of yeast imidazole glycerol phosphate dehydratase. Archives of biochemistry and biophysics. 1965;112:562–566. doi: 10.1016/0003-9861(65)90096-2. [DOI] [PubMed] [Google Scholar]
- Lareau LF, Hite DH, Hogan GJ, Brown PO. Distinct stages of the translation elongation cycle revealed by sequencing ribosome-protected mRNA fragments. eLife. 2014;3:e01257. doi: 10.7554/eLife.01257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Liu B, Lee S, Huang SX, Shen B, Qian SB. Global mapping of translation initiation sites in mammalian cells at single-nucleotide resolution. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:E2424–E2432. doi: 10.1073/pnas.1207846109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letzring DP, Dean KM, Grayhack EJ. Control of translation efficiency in yeast by codon-anticodon interactions. Rna. 2010;16:2516–2528. doi: 10.1261/rna.2411710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li GW, Oh E, Weissman JS. The anti-Shine-Dalgarno sequence drives translational pausing and codon choice in bacteria. Nature. 2012;484:538–541. doi: 10.1038/nature10965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortimer SA, Kidwell MA, Doudna JA. Insights into RNA structure and function from genome-wide studies. Nature reviews Genetics. 2014;15:469–479. doi: 10.1038/nrg3681. [DOI] [PubMed] [Google Scholar]
- Muhlrad D, Decker CJ, Parker R. Deadenylation of the unstable mRNA encoded by the yeast MFA2 gene leads to decapping followed by 5'-->3' digestion of the transcript. Genes & development. 1994;8:855–866. doi: 10.1101/gad.8.7.855. [DOI] [PubMed] [Google Scholar]
- Parker R. RNA degradation in Saccharomyces cerevisae. Genetics. 2012;191:671–702. doi: 10.1534/genetics.111.137265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelechano V, Wei W, Jakob P, Steinmetz LM. Genome-wide identification of transcript start and end sites by transcript isoform sequencing. Nature protocols. 2014;9:1740–1759. doi: 10.1038/nprot.2014.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelechano V, Wei W, Steinmetz LM. Extensive transcriptional heterogeneity revealed by isoform profiling. Nature. 2013;497:127–131. doi: 10.1038/nature12121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova TV, Hellen CU. Translation elongation after assembly of ribosomes on the Cricket paralysis virus internal ribosomal entry site without initiation factors or initiator tRNA. Genes & development. 2003;17:181–186. doi: 10.1101/gad.1040803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy B, Jacobson A. The intimate relationships of mRNA decay and translation. Trends in genetics : TIG. 2013;29:691–699. doi: 10.1016/j.tig.2013.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudra D, Warner JR. What better measure than ribosome synthesis? Genes & development. 2004;18:2431–2436. doi: 10.1101/gad.1256704. [DOI] [PubMed] [Google Scholar]
- Schneider-Poetsch T, Ju J, Eyler DE, Dang Y, Bhat S, Merrick WC, Green R, Shen B, Liu JO. Inhibition of eukaryotic translation elongation by cycloheximide and lactimidomycin. Nature chemical biology. 2010;6:209–217. doi: 10.1038/nchembio.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah P, Ding Y, Niemczyk M, Kudla G, Plotkin JB. Rate-limiting steps in yeast protein translation. Cell. 2013;153:1589–1601. doi: 10.1016/j.cell.2013.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shenton D, Smirnova JB, Selley JN, Carroll K, Hubbard SJ, Pavitt GD, Ashe MP, Grant CM. Global translational responses to oxidative stress impact upon multiple levels of protein synthesis. The Journal of biological chemistry. 2006;281:29011–29021. doi: 10.1074/jbc.M601545200. [DOI] [PubMed] [Google Scholar]
- Stevens A, Maupin MK. A 5'----3' exoribonuclease of Saccharomyces cerevisiae: size and novel substrate specificity. Archives of biochemistry and biophysics. 1987;252:339–347. doi: 10.1016/0003-9861(87)90040-3. [DOI] [PubMed] [Google Scholar]
- Subramaniam AR, Zid BM, O'Shea EK. An integrated approach reveals regulatory controls on bacterial translation elongation. Cell. 2014;159:1200–1211. doi: 10.1016/j.cell.2014.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarrant D, von der Haar T. Synonymous codons, ribosome speed, and eukaryotic gene expression regulation. Cellular and molecular life sciences : CMLS. 2014;71:4195–4206. doi: 10.1007/s00018-014-1684-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson DM, Lu C, Green PJ, Parker R. tRNA cleavage is a conserved response to oxidative stress in eukaryotes. Rna. 2008;14:2095–2103. doi: 10.1261/rna.1232808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson DM, Parker R. The RNase Rny1p cleaves tRNAs and promotes cell death during oxidative stress in Saccharomyces cerevisiae. The Journal of cell biology. 2009;185:43–50. doi: 10.1083/jcb.200811119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuck AC, Tollervey D. A transcriptome-wide atlas of RNP composition reveals diverse classes of mRNAs and lncRNAs. Cell. 2013;154:996–1009. doi: 10.1016/j.cell.2013.07.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuller T, Carmi A, Vestsigian K, Navon S, Dorfan Y, Zaborske J, Pan T, Dahan O, Furman I, Pilpel Y. An evolutionarily conserved mechanism for controlling the efficiency of protein translation. Cell. 2010;141:344–354. doi: 10.1016/j.cell.2010.03.031. [DOI] [PubMed] [Google Scholar]
- Tuller T, Veksler-Lublinsky I, Gazit N, Kupiec M, Ruppin E, Ziv-Ukelson M. Composite effects of gene determinants on the translation speed and density of ribosomes. Genome biology. 2011;12:R110. doi: 10.1186/gb-2011-12-11-r110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson DN, Beckmann R. The ribosomal tunnel as a functional environment for nascent polypeptide folding and translational stalling. Current opinion in structural biology. 2011;21:274–282. doi: 10.1016/j.sbi.2011.01.007. [DOI] [PubMed] [Google Scholar]
- Yang JR, Chen X, Zhang J. Codon-by-codon modulation of translational speed and accuracy via mRNA folding. PLoS biology. 2014;12:e1001910. doi: 10.1371/journal.pbio.1001910. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.