Abstract
Increasing evidence has indicated that plenty of lncRNAs play important roles in many critical biological processes. Developing powerful computational models to construct lncRNA functional similarity network based on heterogeneous biological datasets is one of the most important and popular topics in the fields of both lncRNAs and complex diseases. Functional similarity network consturction could benefit the model development for both lncRNA function inference and lncRNA-disease association identification. However, little effort has been attempted to analysis and calculate lncRNA functional similarity on a large scale. In this study, based on the assumption that functionally similar lncRNAs tend to be associated with similar diseases, we developed two novel lncRNA functional similarity calculation models (LNCSIM). LNCSIM was evaluated by introducing similarity scores into the model of Laplacian Regularized Least Squares for LncRNA–Disease Association (LRLSLDA) for lncRNA-disease association prediction. As a result, new predictive models improved the performance of LRLSLDA in the leave-one-out cross validation of various known lncRNA-disease associations datasets. Furthermore, some of the predictive results for colorectal cancer and lung cancer were verified by independent biological experimental studies. It is anticipated that LNCSIM could be a useful and important biological tool for human disease diagnosis, treatment, and prevention.
There are estimated 20,000 protein-coding genes in the human genome, which account for only approximately 1.5% of the whole genome1,2,3,4,5,6,7,8,9. Therefore, more than 98% of the human genome does not encode protein sequences. Furthermore, plenty of evidences have demonstrated the critical regulative roles of noncoding RNAs (ncRNAs) in a broad range of fundamental and important biological processes10, which challenge the traditional view that RNA is just transcriptional noise and intermediary between gene and protein11,12. Especially, Taft et al. observed that the proportion of non-protein-coding sequence correspondingly increases with increased complexity of organisms13. Based on transcript lengths, ncRNAs can be divided into small ncRNAs and long ncRNAs. Long noncoding RNAs (lncRNAs) are defined as a class of important heterogeneous ncRNAs with the length more than 200 nucleotides6,14,15,16,17,18,19, which make up the largest fraction of the mammalian noncoding transcriptome10,14. Based on traditional gene mapping approaches, H19 and Xist were discovered in the early 1990 s20,21,22,23. However, these two lncRNAs were considered to be rare exceptions to the central dogma of molecular biology at that time. Guttman et al. used chromatin-state maps to develop a new genome wide approach for lncRNAs discovery and identified 1,600 large intervening non-coding RNAs (lincRNAs) across four mouse cell types24. Furthermore, a functional genomic approach has been developed to assign putative functions to each lincRNA, showing these lincRNAs has played various roles in fundamental and important biological processes24. Based on chromatin marks and RNA-sequencing (RNA-seq) data, Cabili et al. presented an integrative approach to generate the human lincRNA catalog, which included more than 8000 lincRNAs across 24 different human cell types and tissues25. These lincRNAs have also been characterized by a panorama of more than 30 properties, such as sequence, structural, transcriptional, and orthology features25. Nowadays, a lot of lncRNAs have been identified in eukaryotic organisms ranging from nematodes to humans, which benefits from the rapid development of both experimental technology and computational algorithms18,24,26,27,28,29. In comparison with protein-coding genes, lncRNAs tend to show a relatively lower expression level but much more tissue-specific pattern12,16,27,30,31,32,33,34,35,36. Furthermore, lncRNAs tend to be less conserved across species and have longer, but fewer, exons18,23,25.
Because of the low cross-species conservation, low expression levels and high tissue specificity of lncRNAs, people often argued against the functionality of lncRNAs in the past12,37. Increasing number of experimental studies in recent years have shown that plenty of lncRNAs are not transcriptional noise but play important roles in many critical biological processes, including transcriptional and post-transcriptional regulation, epigenetic regulation, organ or tissue development, cell differentiation, cell cycle control, cellular transport, metabolic processes, chromosome dynamics and so on7,8,9,11,14,15,24,29,38,39,40,41,42,43,44,45,46. Compared with the huge number of lncRNAs annotated by GENCODE16,18, only a few lncRNAs have been extensively studied, which have shed light on their possible functions and the underlying molecular mechanism of their functions23,46. Elucidating the functions of lncRNAs is a big challenge for both experimental studies and computational biology23. Considering the important roles of lncRNAs in various biological processes, it is no surprise that mutations and dysregulations of lncRNAs have been linked to the development and progression of a broad range of complex human diseases8,12,14,15,47,48,49,50, such as breast cancer51,52,53,54, hepatocellular cancer55,56,57,58,59,60, prostate cancer61,62,63,64,65, colon cancer66, bladder cancer67, thyroid cancer68, lung cancer69,70, ovarian cancer54, leukemia71,72, Alzheimer’s diseases73, diabetes74,75, and HIV76. lncRNA PCA3 has about 60 times expression levels in prostate tumors compared with normal tissues, therefore PCA3 has been treated as a well-known example of potential cancer diagnostic biomarker38,47,64. Another well-known example is HOTAIR, which is overexpressed from hundreds to nearly two-thousand-fold in breast cancer metastases based on quantitative PCR53. Furthermore, HOTAIR is also an independent prognostic marker of hepatocellular cancer recurrence for the patients after liver transplantation59. lncRNAs can be used as both potential biomarkers in disease diagnosis, treatment, prognosis and potential drug targets in drug discovery and clinical treatment47. So far, although plenty of biological datasets about lncRNA sequence and expression have been generated and stored in some publicly available databases, such as NRED77, lncRNAdb28, NONCODE29, the number of lncRNAs reported to be associated with diseases is still very limited.
Calculating lncRNA functional similarity could benefit the construction of computational model for lncRNA function inference and lncRNA-disease association identification based on the assumption that similar lncRNAs have similar functions and relevance with similar diseases78. In this way, potential lncRNA functions and lncRNA-disease associations could be verified based on further experimental validation. Therefore, the time and cost of biological experiments could be significantly reduced. Furthermore, it is well known that lncRNA function inference and disease-lncRNA association identification could benefit lncRNA functions understanding, biomarker identification and drug discovery for human disease diagnosis, treatment, prognosis and prevention. Computational methods have played important roles in ncRNA investigation in plenty of previous successful studies79,80,81,82,83,84,85,86,87,88. Therefore, developing powerful computational models based on heterogeneous biological datasets for lncRNA functional similarity calculation and functional network construction is one of the most important and popular topics in the fields of both lncRNAs and complex diseases.
In our previous work, we calculated the lncRNA functional similarity by integrating lncRNA expression similarity based on the Spearman correlation coefficient between the expression profiles of each lncRNA pair and lncRNA Gaussian interaction profile kernel similarity based on the assumption that similar lncRNAs tend to show a similar interaction and non-interaction pattern with the diseases78. Based on calculated lncRNA similarity, we further developed Laplacian Regularized Least Squares for LncRNA–Disease Association (LRLSLDA) in the semi-supervised learning framework78.
It is well known that genes with similar functions tend to be associated with similar diseases and vice versa89. In the recent researches about non-coding RNAs, similar conclusions have been obtained11,78,82,90. Based on the logical extension of the basic assumption in the previous disease genes identification, Chen et al. and Lu et al. proposed and validated the following basic assumption for disease-related lncRNAs and miRNAs prediction: similar diseases tend to be associated with functionally similar lncRNAs and miRNAs and vice versa78,90, respectively. Therefore, the conclusion can be obtained that the functional similarity between two lncRNAs can be calculated by quantitatively measuring the similarity of diseases associated with these two lncRNAs. In this article, we developed two novel lncRNA functional similarity calculation models (LNCSIM) based on above conclusion. LNCSIM consists of the following two steps. Firstly, we developed two methods to calculate the semantic similarity between different diseases based on the structure of directed acyclic graph (DAG) which represents the relationships among different diseases. Secondly, the functional similarity of two lncRNAs was calculated by measuring the semantic similarity of their associated two groups of diseases. To validate the performance of LNCSIM, we introduced lncRNA functional similarity into the model of LRLSLDA for lncRNA-disease associations prediction developed in the previous work78. As a result, the reliable AUCs of 0.8130 and 0.8198 are obtained in the leave-one-out cross validation (LOOCV) of known experimentally confirmed lncRNA-disease association in the LncRNADisease for two versions of lncRNA similarity scores, increasing AUCs of 0.037 and 0.0438 than previous LRLSLDA, respectively. We also applied LRLSLDA with lncRNA functional similarity (LRLSLDAS) to Colorectal cancer and Lung cancer and further implemented global prediction for all the diseases simultaneously. Some of potential lncRNA-disease associations have been confirmed by recent biological experiments. Specially, 80% and 66.67% of top 15 potential associations based on global prediction have been confirmed, respectively, demonstrating the potential value of LNCSIM for disease-related lncRNA prediction and biomarker detection for human disease diagnosis, treatment, prognosis and prevention. Furthermore, when we applied LNCSIM to another lncRNA-disease association dataset in MNDR and integrated dataset consisting of lncRNA-disease associations obtained from LncRNADisease database and MNDR, significant performance improvement has also been demonstrated in the framework of LOOCV.
Results
lncRNA functional similarity
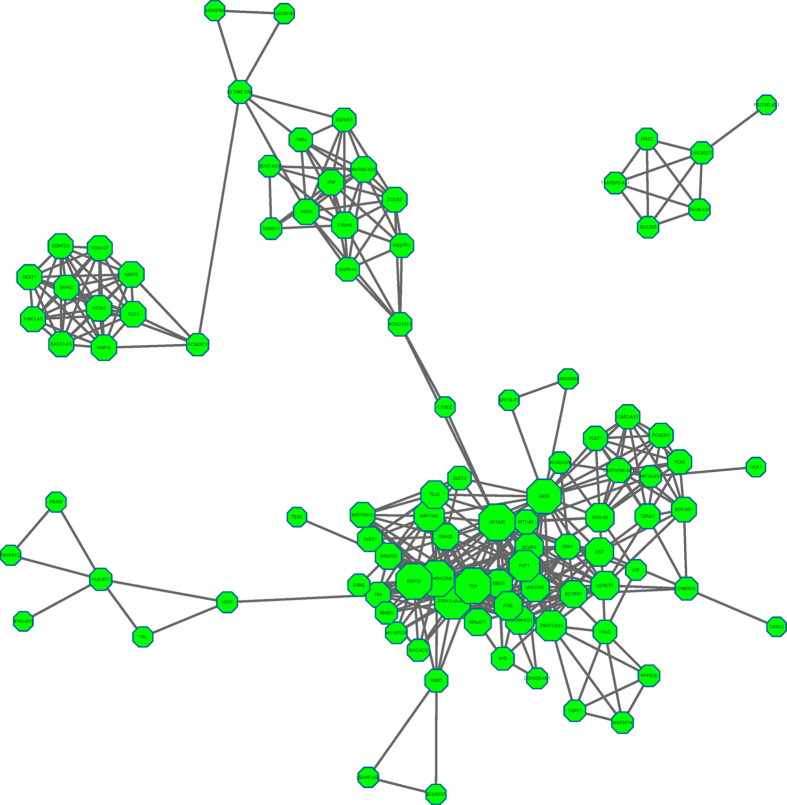
LNSCIM was applied to all the lncRNAs investigated in LncRNADisease database (See Fig. 1). Considering the fact that lncRNA functional similarity was calculated by measuring the semantic similarity of their associated disease groups in the current version of LNCSIM (See Fig. 1 and Methods section), LNCSIM can’t be applied to those lncRNAs without any known associated diseases. Therefore, we selected those lncRNAs with associated diseases in our dataset to implement LNCSIM. Therefore, we obtained the pairwise functional similarity among 104 lncRNAs (see Supplementary Table 1 and 2, respectively). Furthermore, the lncRNA functional network was constructed by setting up a functional similarity threshold and connecting lncRNA pairs with functional similarity greater than or equal to the threshold in the lncRNA functional network (see Fig. 2 and Supplementary Figure 1, respectively).
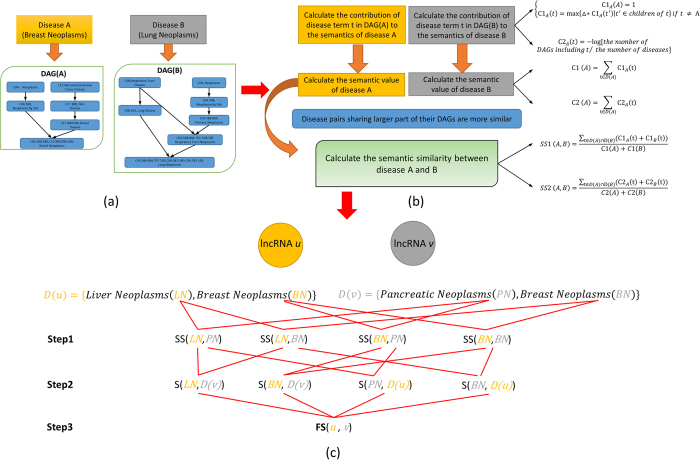
Figure 1. Flowchart of LNCSIM, demonstrating the basic ideas of calculating functional similarity between two lncRNAs.
The following three steps have been included: (a) identified diseases associated with lncRNA u and v and constructed their disease DAGs; (b) calculated semantic similarity between diseases in the disease groups associated with lncRNA u and v; (c) obtained functional similarity between lncRNA u and v by calculating the similarity between corresponding disease group associated with each lncRNA. DAG: directed acyclic graph; LN: Liver Neoplasms; BN: Breast Neoplasms; PN: Pancreatic Neoplasms; D(u) and D(v): the disease groups associated with lncRNA u and v; SS(LN,PN): disease semantic similarity matrix between disease LN and PN; S(LN, D(v)): the similarity between LN and the disease groups associated with lncRNA v: FS(u,v): the functional similarity between lncRNA u and v.
Figure 2.
lncRNA functional network was constructed by the model of LNCSIM based on disease semantic similarity model 1, where each node represents one lncRNA and the links was connected if lncRNA pair has a functional similarity equal to or greater than the similarity cutoff (here the cutoff is 0.3 considering the fact that known lncRNA-disease associations is seriously incomplete currently). The size of a node is proportional to the degree of the node. The network is visualized by cytoscape (http://cytoscape.github.io/).
Performance evaluation
The effectiveness of LNCSIM was validated by applying the functional similarity results into lncRNA-disease associations prediction based on the model of LRLSLDA developed in our previous work78. The aim is to confirm whether the performance of LRLSLDA can be further improved by introducing the information of functional similarity. In the previous version of LRLSLDA, disease similarity and lncRNA similarity scores were derived from Gaussian interaction profile kernel similarity and lncRNA expression similarity. Here, we combined new disease similarity from LNCSIM and disease Gaussian interaction profile kernel similarity into the integrated similarity by a simple mean operation. Furthermore, integrated lncRNA similarity is calculated based on the average of new lncRNA similarity from LNCSIM, lncRNA Gaussian interaction profile kernel similarity, and lncRNA expression similarity. New LRLSLDA models based on two different LNCSIM models were named LRLSLDA-LNCSIM1 and LRLSLDA-LNCSIM2, respectively.
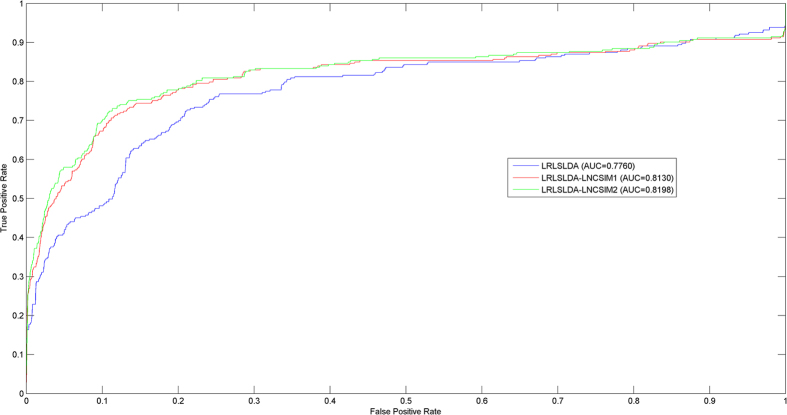
LOOCV was implemented on the known experimentally verified lncRNA-disease associations in the LncRNADisease database to compare the performance of LRLSLDA, LRLSLDA-LNCSIM1, and LRLSLDA-LNCSIM2. As a result, LRLSLDA, LRLSLDA-LNCSIM1, and LRLSLDA-LNCSIM2 achieved AUCs of 0.7760, 0.8130, and 0.8198, respectively (see Fig. 3). New predictive methods increased AUCs of 0.037 and 0.0438, respectively. Therefore, we can reach the conclusion that predictive accuracy has been improved by introducing new disease similarity and lncRNA functional similarity calculated from LNCSIM. In spite of less than two related lncRNAs for each disease on average in the known golden standard dataset, excellent predictive ability of LRLSLDA-LNCSIM1 and LRLSLDA-LNCSIM2 have been demonstrated.
Figure 3. Comparison between LRLSLDA, LRLSLDA-LNCSIM1, and LRLSLDA-LNCSIM2 in terms of ROC curve and AUC based on LOOCV.
As a result, new prediction method increase an AUC of 0.037 and 0.0438, respectively, demonstrating that predictive accuracy has been improved by the operation of introducing new disease similarity and lncRNA functional similarity calculated from LNCSIM.
According to Fig. 2, LRLSLDA-LNCSIM1 and LRLSLDA-LNCSIM2 showed similar predictive accuracy. Therefore, we wanted to know whether the similarity results based on LNCSIM1 and LNCSIM2 are complementary. Here, we used the mean, maximum, and minimum of the functional similarity calculated based on LNCSIM1 and LNCSIM2 as integrated functional similarity, respectively. Integrated functional similarity was introduced into the model of LRLSLDA to see whether the predictive performance could be further improved. New LRLSLDA models based on these three kinds of integrated similarity were named LRLSLDA-LNCSIM-mean, LRLSLDA-LNCSIM-max, and LRLSLDA-LNCSIM-min, respectively. We further implemented LOOCV on the known experimentally verified lncRNA-disease associations. As a result, LRLSLDA-LNCSIM-mean, LRLSLDA-LNCSIM-max, and LRLSLDA-LNCSIM-min achieved AUCs of 0.8168, 0.8199, and 0.8132, respectively (see Supplementary Figure 2). No significant performance differences from LRLSLDA-LNCSIM1 and LRLSLDA-LNCSIM2 could be observed, which indicated the similarity results based on LNCSIM1 and LNCSIM2 are not complementary.
Case studies
We regarded all the known experimentally confirmed lncRNA-disease associations in the LncRNADisease database as training samples and applied LRLSLDA-LNCSIM1 and LRLSLDA-LNCSIM2 to predict potential lncRNAs associated with several important diseases. Furthermore, we tried to search for recent experimental literatures to confirm the predictive results and evaluate the predictive ability of our models.
As the third most common cancer in males and the second in females, colorectal cancer accounts for approximately 8% of all cancer death91,92,93. Colorectal cancer most commonly occurs sporadically and only 25% of the patients have a family disease history, which indicates that lifestyle and environment risk factors could also promote the progression of colorectal cancer91,92. With the development of high-throughput sequencing technologies in the recent years, researchers have confirmed some critical mutations underlying the pathogenic mechanism of colorectal cancer, including some well-known frequently-mutated oncogenes or tumor suppressor genes (such as APC, KPRS, PIK3CA, and TP53) and a large number of mutated genes with a low frequency94,95,96. Nowadays, biological experiments have further linked mutations and dysregulations of some lncRNAs with the development and progression of colorectal cancer, such as HOTAIR, KCNQ1OT1, and MALAT1 in our training samples. For example, several independent experiments showed that HOTAIR could be considered as a negative prognostic marker in the blood of colorectal cancer patients23,97,98,99,100. We implemented LRLSLDA-LNCSIM1 and LRLSLDA-LNCSIM2 to prioritize candidate lncRNAs without the known relevance to colorectal cancer. As a result, four out of top 10 predicted colorectal cancer-related lncRNAs (CRNDE, H19, PVT1, and CASC2) have been confirmed to be associated with colorectal cancer based on recent experimental literatures101,102,103 (http://cpfd.cnki.com.cn/Article/CPFDTOTAL-KAXH201309001039.htm). For example, elevated expression of CRNDE in the tissue and plasma of almost all colorectal adenomas and adenocarcinomas has been detected based on microarray analysis, which showed CRNDE has the potential to be a biomarker for colorectal adenomas and cancers101. Furthermore, real time PCR demonstrated PVT1 may be a new oncogene and has the functional correlation with the proliferation and apoptosis of colorectal cancer cells102.
As the most common cause of cancer-related death worldwide in both men and women, there are estimated 1.4 million deaths resulting from lung cancer each year104,105,106,107. Lung cancer death is greater than the combination of following three most cancers: colon, breast, and prostate cancer104. Specially, five-year survival rate of lung cancer patients is only approximately 15% from the time of diagnosis, which is lower than other cancers types104,105,108,109. Furthermore, considering the important fact that lung cancer patients are not usually diagnosed until advanced stage and there are only few effective lung cancer risk biomarkers, it is necessary and urgent to investigate the mechanism of lung cancer and find new biomarkers for early diagnoses104,105,110,111,112. In the last decades, much attention has been paid to identify deregulation of protein-coding genes as diagnostic and therapeutic targets of lung cancer113. However, with the rapid development of lncRNA discovery and lncRNA function annotation, researchers have found that lncRNA plays a critical role in the development and progression of lung cancer49,114. Four known lung cancer related lncRNAs has been included in the golden standard dataset. For example, it has been observed that lncRNA BCYRN1 was expressed in the tissues of the breast, cervix, oesophagus, lung, ovary, parotid, and tongue cancer, respectively115. However, BCYRN1 was expressed not in corresponding normal tissues115. Another example is the association between lncRNA H19 and lung cancer. Based on a knockdown approach, experiments indicated that breast and lung cancer cell clonogenicity and anchorage-independent growth were decreased because of the down-regulation of H1951. We further prioritized candidate lncRNAs based on the scored calculated based on LRLSLDA-LNCSIM1 and LRLSLDA-LNCSIM2. Three out of top ten predicted lung cancer related lncRNAs (HOTAIR, UCA1, and GAS5) have been confirmed by independent experimental literatures102,116,117,118. A typical example is HOTAIR, which is ranked 2nd by both models. The expression of HOTAIR was upregulated in lung cancer cells based on a three-dimensional organotypic culture model116,118. One important fact must be pointed out that the known lncRNA-disease association dataset used in this paper for potential association prediction was generated before the publication of this paper. Therefore, this example could be considered as an independent validation of our model. Another biological experiment implemented in 72 NSCLC specimens by qRT-PCR revealed the expression of the tumor suppressor lncRNA GAS5 was significantly down-regulated in lung cancer tissues compared to adjacent noncancerous tissues117. Therefore, GAS5 is considered to be a potential diagnostic biomarker for lung cancer and a novel therapeutic target in patients with lung cancer117.
As a global ranking method, LRLSLDA-LNCSIM1 and LRLSLDA-LNCSIM2 can reconstruct the missing lncRNA-disease associations for all the diseases simultaneously. Therefore, these two models were applied to simultaneously rank all the candidate lncRNA-disease associations. Out of top 15 potential lncRNA-disease associations, 12 and 10 associations predicted by LRLSLDA-LNCSIM1 and LRLSLDA-LNCSIM2 have been confirmed by experimental literature, respectively (see Table 1 and Supplementary Table 3). Potential association between lncRNA MEG3 and heroin abuse was ranked 2nd and 4th out of 19413 candidate lncRNA-disease pairs by LRLSLDA-LNCSIM1 and LRLSLDA-LNCSIM2, respectively. Quantitative PCR confirmed our predictive result by demonstrating MEG3 was upregulated in human heroin abusers compared to matched drug-free control subjects119. Similar high-ranking evidences can also be found in our predictive list, which demonstrate the reliable performance by integrating LNCSIM and LRLSLDA.
Table 1. As a global ranking method, LRLSLDA-LNCSIM1 was applied to simultaneously rank all the candidate lncRNA-disease associations.
| Disease | lncRNA | Evidence (PMID) |
|---|---|---|
| Down’s syndrome | DGCR5 | Unconfirmed |
| heroin abuse | MEG3 | 21128942 |
| lung adenocarcinoma | H19 | 16707459 |
| colorectal cancer | CRNDE | 22393467 |
| velocardiofacial syndrome | NRON | Unconfirmed |
| colorectal neoplasia | HOTAIR | 24531795 |
| lung adenocarcinoma | MEG3 | Paper without PMID124 |
| lung adenocarcinoma | BCYRN1 | 9422992 |
| colorectal neoplasia | MALAT1 | 21503572 |
| colorectal neoplasia | KCNQ1OT1 | 23660942 |
| heroin addiction | MIAT | 21128942 |
| brain ischemia | B2 SINE RNA | 15016078 |
| liver injury | IFNG-AS1 | Unconfirmed |
| cervix cancer | H19 | 8570220 |
| breast cancer | MALAT1 | 24499465 |
The top 15 potential associations and the confirmation for their associations by experimental literature were listed here.
We have demonstrated reliable performance of LRLSLDA-LNCSIM1 and LRLSLDA-LNCSIM2 in the terms of LOOCV and the case studies of colorectal cancer and lung cancer. Therefore, we further implemented these two models to prioritize all the candidate lncRNAs for all the diseases in the LncRNADisease database by using all the known experimentally confirmed lncRNA-disease associations in the LncRNADisease database as training samples. Potential human disease-lncRNA association list for each disease were publicly released to benefit the biological experimental validation (see Supplementary Table 4 and 5). It is anticipated that potential disease-lncRNA associations predicted by our models could be confirmed by biological experiments and useful for complex disease research.
Further performance evaluation on another dataset
To further analysis and validate the results of LNCSIM, we applied LNCSIM to all the lncRNAs investigated in the manually curated diverse ncRNA-disease repository (MNDR)120. Pairwise functional similarity among 95 lncRNAs calculated based on two versions of LNCSIM was listed in Supplementary Table 6 and 7, respectively. Furthermore, we integrated the dataset in the LncRNADisease database and MNDR and implement LNCSIM to calculate lncRNA functional similarity among 169 lncRNAs investigated in the integrated dataset (see Supplementary Table 8 and 9, respectively).
Furthermore, LOOCV was implemented on the known experimentally verified lncRNA-disease associations in MNDR to compare the performance of LRLSLDA, LRLSLDA-LNCSIM1, and LRLSLDA-LNCSIM2 (see Supplementary Figure 3). Also for the integrated dataset, LOOCV was implemented based on LRLSLDA, LRLSLDA-LNCSIM1, and LRLSLDA-LNCSIM2 (see Supplementary Figure 4). It could be easily concluded that predictive accuracy has been improved by new disease similarity and lncRNA functional similarity calculated from LNCSIM.
Discussions
Quantitatively calculating lncRNA functional similarity is critical for lncRNA functions prediction and potential lncRNA-disease associations inference. Therefore, it has become an important goal and significant problem for computational biology research. In this article, the model of LNCSIM was developed to calculate lncRNA functional similarity on a large scale by integrating known lncRNA-disease associations and disease semantic similarity. LNCSIM was motivated based on the basic assumption that functionally similar lncRNAs tend to be associated with similar diseases and hence the lncRNA functional similarity can be calculated by measuring the similarity of diseases associated with them78,90. Furthermore, LNCSIM was introduced into lncRNA-disease association identification model LRLSLDA developed in our previous work to check whether the predictive performance of LRLSLDA can be further improved. The reliable performance improvement has been demonstrated in both cross validation and case studies about colorectal cancer and lung cancer. Potential lncRNA-disease associations for all the disease investigated in this article have been publicly released for further biological experiment confirmation. In our opinion, LNCSIM has potential value for lncRNA-related interactions prediction and lncRNA biomarker detection for human disease diagnosis, treatment, prognosis and prevention.
There are at least three limitations in the method design of LNCSIM. Firstly, considering the fact that lncRNA functional similarity was calculated by integrating lncRNA-disease association data and the disease DAG, LNCSIM may cause bias to lncRNAs with more associated diseases. Therefore, the performance of LNCSIM would be further improved when more known experimentally verified disease-lncRNA associations can be obtained. Secondly, semantic contribution decay factor appear in the current model and how to select this parameter is not still solved well. Finally, lncRNA functional similarity calculation could be improved greatly by integrating more reliable types of biological datasets, such as lncRNA-related various interactions, lncRNA sequence, disease phenotype information.
Methods
LncRNA-disease associations
Considering accumulating biological experiments have produced hundreds of lncRNA–disease associations, we manually collected experimentally reported disease-lncRNA associations and constructed the first publicly available lncRNA–disease association database, LncRNADisease (http://cmbi.bjmu.edu.cn/lncrnadisease) in the previous work11, which aims to provide a comprehensive resource of experimentally confirmed lncRNA–disease associations and lay the data fundament for lncRNA-related predictive research. The lncRNA-disease association dataset was downloaded from the LncRNADisease database in October, 2012. In LncRNADisease database, the same disease-lncRNA association based on the different experimental literature evidences has been considered to be different associations. Therefore, 486 associations have been recorded in this database. We got rid of those duplicate associations based on different evidences for the same lncRNA-disease pair. As a result, 293 distinct high-quality experimentally verified lncRNA–disease associations have been obtained, including 118 lncRNAs and 167 diseases (see Supplementary Table 10).
To further analysis and validate the results of LNCSIM, we downloaded human lncRNA-disease associations in the MNDR120 in March, 2015. Similar to the operations for the dataset from LncRNADisease database, we got rid of those duplicate records based on different experimental literature evidences for the same lncRNA-disease associations. As a result, we obtained 471 high-quality experimentally verified human disease-lncRNA associations, including 127 diseases and 241 lncRNAs (see Supplementary Table 11).
Disease MeSH descriptors and directed acyclic graph
MeSH descriptors of various diseases were downloaded from the National Library of Medicine (http://www.nlm.nih.gov/), which provided a strict system for disease classification for the research of the relationship among various diseases121. MeSH descriptors included 16 categories: Category A for anatomic terms, Category B for organisms, Category C for diseases, Category D for drugs and chemicals and so on. The MeSH descriptor of Category C for each disease was used in this paper. Furthermore, directed acyclic graph (DAG) ws constructed to demonstrate the relationship among various diseases, where the nodes represents disease MeSH descriptors and all the MeSH descriptors in the DAG are connected by a direct edge from a more general term (parent node) to a more specific term (child node) (See Fig. 1). Each MeSH descriptor has one or more tree numbers to numerically define its location in the DAG. The tree numbers of a child node are defined as the codes of its parent nodes appended by the child’s information. For the disease A, DAG is denoted as DAG(A) = (D(A),E(A)), where D(A) includes the nodes represent disease itself and its ancestor diseases and E(A) consisting of corresponding direct edges from a parent node to a child node represents the relationship between these two nodes (See Fig. 1).
Since the disease names in the LncRNADisease database and MNDR weren’t named based on MeSH descriptors, we mapped the diseases in these two disease-lncRNA association datasets into their MeSH descriptors. After getting rid of some diseases without any Mesh descriptors or tree numbers from these two disease-lncRNA association datasets and merging some diseases with the same Mesh descriptors, 254 and 260 distinct lncRNA-disease associations were obtained in LncRNADisease database and MNDR, respectively (see Supplementary Table 12 and 13).
Disease semantic similarity model 1
As mentioned, functional similarity between two lncRNAs is calculated based on the similarity of diseases associated with these two lncRNAs. Therefore, we developed two models to calculate disease semantic similarity based on disease DAGs (See Fig. 1).
Firstly, we calculated the disease similarity in the same way as described in the literature122. Disease can be described as a DAG. We defined the contribution of disease term t in DAG(A) to the semantic value of disease A as follows:
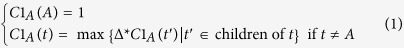 |
where 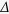 is the semantic contribution decay factor, which shows the contributions of other ancestor diseases to the semantic value of disease A decrease with the increase of the distance between this disease and disease A. In the DAG of disease A, disease A is located in the 0th layer, therefore it is the most specific disease term and its contribution to semantic value of disease A is defined as 1. Disease located in the 1st layer is considered to be a more general disease, so its contribution is multiplied by the semantic contribution decay factor. Based on above formula, the semantic contribution of diseases in different layers to semantic value of disease A are differentiated.
is the semantic contribution decay factor, which shows the contributions of other ancestor diseases to the semantic value of disease A decrease with the increase of the distance between this disease and disease A. In the DAG of disease A, disease A is located in the 0th layer, therefore it is the most specific disease term and its contribution to semantic value of disease A is defined as 1. Disease located in the 1st layer is considered to be a more general disease, so its contribution is multiplied by the semantic contribution decay factor. Based on above formula, the semantic contribution of diseases in different layers to semantic value of disease A are differentiated.
Therefore, summing all the contributions from ancestor diseases and disease A itself, the semantic value of disease A is defined as follows:
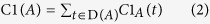 |
Furthermore, the semantic similarity between two diseases A and B can be defined based on the nodes shared by the two disease DAGs.
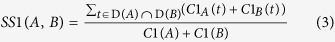 |
where SS1 is the disease semantic similarity matrix. The entity SS1(i,j) in row i column j is the disease semantic similarity between disease i and j based on disease semantic similarity model 1.
Disease semantic similarity model 2
Furthermore, the disease similarity was calculated in the same way as described in the literature123. According to disease semantic similarity model 1 defined above, the disease terms in the same layer of DAG(A) have the same contribution to the semantic value of disease A. However, different disease terms in the same layer of DAG(A) may appear in the different numbers of disease DAGs. For example, two diseases appear in the same layer of DAG(A) and the first disease appears in less disease DAGs than the second disease. Obviously, we can conclude that the first disease is more specific than the second disease. Therefore, it is less accurate to assign the same contribution value to these two diseases according to the above consideration. The contribution of the first disease to the semantic value of disease A should be higher than the second.
In conclusion, a more specific disease should have a greater contribution to the semantic value of disease A. Here, the contribution of disease term t in DAG(A) to the semantic value of disease A was defined as follows:
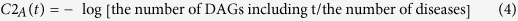 |
We defined the semantic similarity between disease A and B by summing all the contributions from ancestor diseases and disease A itself to define the semantic value of disease A in the similar way as model 1 and paying attention to the nodes shared by the two disease DAGs.
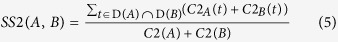 |
where SS2 is the disease semantic similarity matrix calculated based on model 2, C2(A) and C2(B) is the semantic value of disease A and B, respectively. The entity SS2(i,j) in row i column j is the disease semantic similarity between disease i and j based on disease semantic similarity model 2.
LNCSIM
Here, we developed the model of LNCSIM to quantitatively calculate lncRNA functional similarity by measuring the semantic similarity of their associated two groups of diseases (See Fig. 1). Taking the similarity calculation between lncRNA u and v as an example, we firstly defined D(u) and D(v) as the disease groups associated with lncRNA u and v, respectively. We calculated the similarity between D(u) and D(v) as the functional similarity between lncRNA u and v. To calculate the similarity between D(u) and D(v), the similarity between one of diseases associate with one lncRNA and the group of diseases associated with the other lncRNA should be defined. The similarity between one of diseases associated with lncRNA u, such as D1, and the group of diseases associated with lncRNA v was calculated as follows:
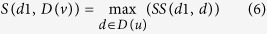 |
Finally, the functional similarity of lncRNA u and v was defined.
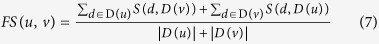 |
where FS is the lncRNA functional similarity matrix and the entity FS(i,j) in row i column j is the functional similarity between lncRNA i and j.
Performance evaluation
LOOCV was implemented to compare the performance of LRLSLDA, LRLSLDA-LNCSIM1, and LRLSLDA-LNCSIM2. Each known disease-lncRNA association was used as test sample in turn and how well this association was ranked relative to the candidate disease-lncRNA pair was observed. In this way, all other known experimentally confirmed disease-lncRNA associations and all the disease-lncRNA pairs without confirmed associations were considered as training samples and candidate disease-lncRNA pair, respectively. Receiver operating characteristics (ROC) curve and Area under the curve (AUC) was used to implement performance evaluation. ROC curve plots true-positive rate (TPR, sensitivity) versus false-positive rate (FPR, 1-specificity) at different rank cutoffs. When the rank cutoffs of successful prediction were varied, corresponding TPR and FPR can be obtained. Here, sensitivity represents the percentage of the test samples obtaining the ranking higher than a given rank cutoff; Specificity represents the percentage of samples obtaining the ranking lower than this given rank cutoff. In this way, ROC was drawn and Area under the curve (AUC) was calculated. AUC = 1 indicates perfect performance and AUC = 0.5 indicates random performance.
Additional Information
How to cite this article: Chen, X. et al. Constructing lncRNA functional similarity network based on lncRNA-disease associations and disease semantic similarity. Sci. Rep. 5, 11338; doi: 10.1038/srep11338 (2015).
Supplementary Material
Acknowledgments
The financial support from the National Natural Science of Foundation of China under Grant No. 11301517, 61472203, 61327902 and National Center for Mathematics and Interdisciplinary Sciences, CAS is highly appreciated.
Footnotes
Author Contributions X.C. conceived the project, developed the prediction method, designed and implemented the experiments, analyzed the result, and wrote the paper. C.G.Y. implemented the experiments and analyzed the result. C.L., W.J., Y.D.Z. and Q.H.D. analyzed the result. All authors read and approved the final manuscript.
References
- Bertone P. et al. Global identification of human transcribed sequences with genome tiling arrays. Science 306, 2242–2246 (2004). [DOI] [PubMed] [Google Scholar]
- Birney E. et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 447, 799–816 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carninci P. et al. Genome-wide analysis of mammalian promoter architecture and evolution. Nat Genet 38, 626–635 (2006). [DOI] [PubMed] [Google Scholar]
- Claverie J. M. Fewer genes, more noncoding RNA. Science 309, 1529–1530 (2005). [DOI] [PubMed] [Google Scholar]
- Core L. J., Waterfall J. J. & Lis J. T. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science 322, 1845–1848 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapranov P. et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science 316, 1484–1488 (2007a). [DOI] [PubMed] [Google Scholar]
- Lander E. S. et al. Initial sequencing and analysis of the human genome. Nature 409, 860–921 (2001). [DOI] [PubMed] [Google Scholar]
- Taft R. J., Pang K. C., Mercer T. R., Dinger M. & Mattick J. S. Non‐coding RNAs: regulators of disease. J Pathol 220, 126–139 (2010). [DOI] [PubMed] [Google Scholar]
- Wilusz J. E., Sunwoo H. & Spector D. L. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev 23, 1494–1504 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteller M. Non-coding RNAs in human disease. Nat Rev Genet 12, 861–874 (2011). [DOI] [PubMed] [Google Scholar]
- Chen G. et al. LncRNADisease: a database for long-non-coding RNA-associated diseases. Nucleic Acids Res 41, D983–D986 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponting C. P., Oliver P. L. & Reik W. Evolution and functions of long noncoding RNAs. Cell 136, 629–641 (2009). [DOI] [PubMed] [Google Scholar]
- Taft R. J., Pheasant M. & Mattick J. S. The relationship between non‐protein‐coding DNA and eukaryotic complexity. Bioessays 29, 288–299 (2007). [DOI] [PubMed] [Google Scholar]
- Mercer T. R., Dinger M. E. & Mattick J. S. Long non-coding RNAs: insights into functions. Nat Rev Genet 10, 155–159 (2009). [DOI] [PubMed] [Google Scholar]
- Wapinski O. & Chang H. Y. Long noncoding RNAs and human disease. Trends Cell Biol 21, 354–361 (2011). [DOI] [PubMed] [Google Scholar]
- Derrien T. et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res 22, 1775–1789 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M., Russell P., Ingolia N. T., Weissman J. S. & Lander E. S. Ribosome profiling provides evidence that large noncoding RNAs do not encode proteins. Cell 154, 240–251 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrow J. et al. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res 22, 1760–1774 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K. C. & Chang H. Y. Molecular mechanisms of long noncoding RNAs. Mol. Cell 43, 904–914 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borsani G. et al. Characterization of a murine gene expressed from the inactive X chromosome. Nature 351, 325–329 (1991). [DOI] [PubMed] [Google Scholar]
- Brannan C. I., Dees E. C., Ingram R. S. & Tilghman S. M. The product of the H19 gene may function as an RNA. Mol. Cell. Biol. 10, 28–36 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockdorff N. et al. The product of the mouse Xist gene is a 15 kb inactive X-specific transcript containing no conserved ORF and located in the nucleus. Cell 71, 515–526 (1992). [DOI] [PubMed] [Google Scholar]
- Hrdlickova B., de Almeida R. C., Borek Z. & Withoff S. Genetic variation in the non-coding genome: Involvement of micro-RNAs and long non-coding RNAs in disease. Biochim. Biophys. Acta. 1842, 1910–1922 (2014). [DOI] [PubMed] [Google Scholar]
- Guttman M. et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 458, 223–227 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabili M. N. et al. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev 25, 1915–1927 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalil A. M. et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci U S A 106, 11667–11672 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M. et al. Ab initio reconstruction of cell type-specific transcriptomes in mouse reveals the conserved multi-exonic structure of lincRNAs. Nat Biotechnol 28, 503–510 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaral P. P., Clark M. B., Gascoigne D. K., Dinger M. E. & Mattick J. S. lncRNAdb: a reference database for long noncoding RNAs. Nucleic Acids Res 39, D146–D151 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bu D. et al. NONCODE v3. 0: integrative annotation of long noncoding RNAs. Nucleic Acids Res 40, D210–D215 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babak T., Blencowe B. J. & Hughes T. R. A systematic search for new mammalian noncoding RNAs indicates little conserved intergenic transcription. BMC Genomics 6, 104 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bono H. et al. Systematic expression profiling of the mouse transcriptome using RIKEN cDNA microarrays. Genome Res 13, 1318–1323 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibb E. A., Brown C. J. & Lam W. L. The functional role of long non-coding RNA in human carcinomas. Mol Cancer 10, 38 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer T. R., Dinger M. E., Sunkin S. M., Mehler M. F. & Mattick J. S. Specific expression of long noncoding RNAs in the mouse brain. Proc Natl Acad Sci U S A 105, 716–721 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauli A. et al. Systematic identification of long noncoding RNAs expressed during zebrafish embryogenesis. Genome Res 22, 577–591 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsköld D., Wang E. T., Burge C. B. & Sandberg R. An abundance of ubiquitously expressed genes revealed by tissue transcriptome sequence data. PLoS Comput Biol 5, e1000598 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang G., Lu X. & Yuan L. LncRNA: A link between RNA and cancer. Biochim. Biophys. Acta. 839, 1097–1109 (2014). [DOI] [PubMed] [Google Scholar]
- Li J. et al. A bioinformatics method for predicting long noncoding RNAs associated with vascular disease. Sci China Life Sci 57, 852–857 (2014). [DOI] [PubMed] [Google Scholar]
- Bussemakers M. J. et al. DD3:: A New Prostate-specific Gene, Highly Overexpressed in Prostate Cancer. Cancer Res 59, 5975–5979 (1999). [PubMed] [Google Scholar]
- Managadze D., Rogozin I. B., Chernikova D., Shabalina S. A. & Koonin E. V. Negative correlation between expression level and evolutionary rate of long intergenic noncoding RNAs. Genome Biol Evol 3, 1390–1404 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattick J. S. The genetic signatures of noncoding RNAs. PLoS Genet 5, e1000459 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattick J. S. & Makunin I. V. Non-coding RNA. Hum Mol Genet 15, R17–R29 (2006). [DOI] [PubMed] [Google Scholar]
- Qureshi I. A., Mattick J. S. & Mehler M. F. Long non-coding RNAs in nervous system function and disease. Brain Res 1338, 20–35 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibb E. A. et al. Human cancer long non-coding RNA transcriptomes. PLoS One 6, e25915 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Q. et al. Large-scale prediction of long non-coding RNA functions in a coding–non-coding gene co-expression network. Nucleic Acids Res 39, 3864–3878 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran V. A., Perera R. J. & Khalil A. M. Emerging functional and mechanistic paradigms of mammalian long non-coding RNAs. Nucleic Acids Res 40, 6391–6400 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu M.-T., Hu J.-W., Yin R. & Xu L. Long noncoding RNA: an emerging paradigm of cancer research. Tumour Biol 34, 613–620 (2013). [DOI] [PubMed] [Google Scholar]
- Spizzo R., Almeida M., Colombatti A. & Calin G. Long non-coding RNAs and cancer: a new frontier of translational research&quest? Oncogene 31, 4577–4587 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheetham S., Gruhl F., Mattick J. & Dinger M. Long noncoding RNAs and the genetics of cancer. Br J Cancer 108, 2419–2425 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutschner T. & Diederichs S. The hallmarks of cancer: a long non-coding RNA point of view. RNA Biol 9, 703–719 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Xuan Z. & Liu C. Long non-coding RNAs and complex human diseases. Int J Mol Sci 14, 18790–18808 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barsyte-Lovejoy D. et al. The c-Myc oncogene directly induces the H19 noncoding RNA by allele-specific binding to potentiate tumorigenesis. Cancer Res 66, 5330–5337 (2006). [DOI] [PubMed] [Google Scholar]
- Guffanti A. et al. A transcriptional sketch of a primary human breast cancer by 454 deep sequencing. BMC Genomics 10, 163 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta R. A. et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 464, 1071–1076 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Y. et al. Amplification of PVT1 contributes to the pathophysiology of ovarian and breast cancer. Clin Cancer Res 13, 5745–5755 (2007). [DOI] [PubMed] [Google Scholar]
- Calin G. A. et al. Ultraconserved regions encoding ncRNAs are altered in human leukemias and carcinomas. Cancer Cell 12, 215–229 (2007). [DOI] [PubMed] [Google Scholar]
- Lin R., Maeda S., Liu C., Karin M. & Edgington T. A large noncoding RNA is a marker for murine hepatocellular carcinomas and a spectrum of human carcinomas. Oncogene 26, 851–858 (2006). [DOI] [PubMed] [Google Scholar]
- Panzitt K. et al. Characterization of HULC, a novel gene with striking up-regulation in hepatocellular carcinoma, as noncoding RNA. Gastroenterology 132, 330–342 (2007). [DOI] [PubMed] [Google Scholar]
- Wang J. et al. CREB up-regulates long non-coding RNA, HULC expression through interaction with microRNA-372 in liver cancer. Nucleic Acids Res 38, 5366–5383 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. et al. Overexpression of long non-coding RNA HOTAIR predicts tumor recurrence in hepatocellular carcinoma patients following liver transplantation. Ann Surg Oncol 18, 1243–1250 (2011). [DOI] [PubMed] [Google Scholar]
- Liu Y. et al. A genetic variant in long non-coding RNA HULC contributes to risk of HBV-related hepatocellular carcinoma in a Chinese population. PLoS One 7, e35145 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung S. et al. Association of a novel long non‐coding RNA in 8q24 with prostate cancer susceptibility. Cancer Sci 102, 245–252 (2011). [DOI] [PubMed] [Google Scholar]
- de Kok J. B. et al. DD3PCA3, a very sensitive and specific marker to detect prostate tumors. Cancer Res 62, 2695–2698 (2002). [PubMed] [Google Scholar]
- Széll M., Bata-Csörgő Z. & Kemény L. The enigmatic world of mRNA-like ncRNAs: their role in human evolution and in human diseases. Semin Cancer Biol 18, 141–148 (2008). [DOI] [PubMed] [Google Scholar]
- van Poppel H. et al. The relationship between Prostate CAncer gene 3 (PCA3) and prostate cancer significance. BJU Int 109, 360–366 (2011). [DOI] [PubMed] [Google Scholar]
- Cui Z. et al. The prostate cancer-up-regulated long noncoding RNA PlncRNA-1 modulates apoptosis and proliferation through reciprocal regulation of androgen receptor. Urol Oncol 31, 1117–1123 (2013). [DOI] [PubMed] [Google Scholar]
- Pibouin L. et al. Cloning of the mRNA of overexpression in colon carcinoma-1: a sequence overexpressed in a subset of colon carcinomas. Cancer Genet Cytogenet 133, 55–60 (2002). [DOI] [PubMed] [Google Scholar]
- Zhang Z. et al. Evaluation of novel gene UCA1 as a tumor biomarker for the detection of bladder cancer. Zhonghua Yi Xue Za Zhi 92, 384–387 (2012). [PubMed] [Google Scholar]
- Jendrzejewski J. et al. The polymorphism rs944289 predisposes to papillary thyroid carcinoma through a large intergenic noncoding RNA gene of tumor suppressor type. Proc Natl Acad Sci U S A 109, 8646–8651 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X. et al. A pituitary-derived MEG3 isoform functions as a growth suppressor in tumor cells. J Clin Endocrinol Metab 88, 5119–5126 (2003). [DOI] [PubMed] [Google Scholar]
- Ji P. et al. MALAT-1, a novel noncoding RNA, and thymosin β4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene 22, 8031–8041 (2003). [DOI] [PubMed] [Google Scholar]
- Calin G. A. et al. Ultraconserved regions encoding ncRNAs are altered in human leukemias and carcinomas. Cancer Cell 12, 215–229 (2007). [DOI] [PubMed] [Google Scholar]
- Zhang X. et al. A myelopoiesis-associated regulatory intergenic noncoding RNA transcript within the human HOXA cluster. Blood 113, 2526–2534 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faghihi M. A. et al. Expression of a noncoding RNA is elevated in Alzheimer’s disease and drives rapid feed-forward regulation of β-secretase. Nat Med 14, 723–730 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez M. L. & DiStefano J. K. Functional characterization of the plasmacytoma variant translocation 1 gene (PVT1) in diabetic nephropathy. PLoS One 6, e18671 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasmant E., Sabbagh A., Vidaud M. & Bièche I. ANRIL, a long, noncoding RNA, is an unexpected major hotspot in GWAS. FASEB J 25, 444–448 (2011). [DOI] [PubMed] [Google Scholar]
- Zhang Q., Chen C.-Y., Yedavalli V. S. & Jeang K.-T. NEAT1 Long Noncoding RNA and Paraspeckle Bodies Modulate HIV-1 Posttranscriptional Expression. MBio 4, e00596–00512 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinger M. E. et al. NRED: a database of long noncoding RNA expression. Nucleic Acids Res 37, D122–D126 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X. & Yan G.-Y. Novel human lncRNA–disease association inference based on lncRNA expression profiles. Bioinformatics 29, 2617–2624 (2013). [DOI] [PubMed] [Google Scholar]
- Liu B., Fang L., Liu F., Wang X. & Chou K.-C. iMiRNA-PseDPC: microRNA precursor identification with a pseudo distance-pair composition approach. J Biomol Struct Dyn, 10.1080/07391102.2015.1014422 (2015). [DOI] [PubMed] [Google Scholar]
- Liu B., Fang L., Chen J., Liu F. & Wang X. miRNA-dis: microRNA precursor identification based on distance structure status pairs. Mol Biosyst 11, 1194–1204 (2015). [DOI] [PubMed] [Google Scholar]
- Chen X., Liu M. X., Cui Q. H. & Yan G. Y. Prediction of Disease-Related Interactions between MicroRNAs and Environmental Factors Based on a Semi-Supervised Classifier. PLoS One 7, e43425 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Liu M. X. & Yan G. RWRMDA: predicting novel human microRNA-disease associations. Mol Biosyst 8, 2792–2798 (2012). [DOI] [PubMed] [Google Scholar]
- Chen X. & Yan G.-Y. Semi-supervised learning for potential human microRNA-disease associations inference. Sci Rep 4, 5501 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M.-X., Chen X., Chen G., Cui Q.-H. & Yan G.-Y. A computational framework to infer human disease-associated long noncoding RNAs. PloS one 9, e84408 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y. et al. Connect the dots: a systems level approach for analyzing the miRNA-mediated cell death network. Autophagy 9, 436–439 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H. et al. Walking the interactome to identify human miRNA-disease associations through the functional link between miRNA targets and disease genes. BMC Syst Biol 7, 101 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J. et al. Prioritizing Candidate Disease miRNAs by Topological Features in the miRNA Target–Dysregulated Network: Case Study of Prostate Cancer. Mol Cancer Ther 10, 1857–1866 (2011). [DOI] [PubMed] [Google Scholar]
- Liu B. et al. Identification of Real MicroRNA Precursors with a Pseudo Structure Status Composition Approach. PloS one 10, e0121501 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh K.-I. et al. The human disease network. Proc Natl Acad Sci U S A 104, 8685–8690 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu M. et al. An analysis of human microRNA and disease associations. PLoS One 3, e3420 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweiger M. R., Hussong M., Röhr C. & Lehrach H. Genomics and epigenomics of colorectal cancer. Wiley Interdiscip Rev Syst Biol Med 5, 205–219 (2013). [DOI] [PubMed] [Google Scholar]
- Migheli F. & Migliore L. Epigenetics of colorectal cancer. Clin Genet 81, 312–318 (2012). [DOI] [PubMed] [Google Scholar]
- Jemal A. et al. Global cancer statistics. CA Cancer J Clin 61, 69–90 (2011). [DOI] [PubMed] [Google Scholar]
- Zhu J. et al. Deciphering genomic alterations in colorectal cancer through transcriptional subtype-based network analysis. PLoS One 8, e79282 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fearon E. R. Molecular genetics of colorectal cancer. Annu Rev Pathol 6, 479–507 (2011). [DOI] [PubMed] [Google Scholar]
- Wood L. D. et al. The genomic landscapes of human breast and colorectal cancers. Science 318, 1108–1113 (2007). [DOI] [PubMed] [Google Scholar]
- Maass P. G., Luft F. C. & Bähring S. Long non-coding RNA in health and disease. J Mol Med 92, 337–346 (2014). [DOI] [PubMed] [Google Scholar]
- Kogo R. et al. Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res 71, 6320–6326 (2011). [DOI] [PubMed] [Google Scholar]
- Harries L. Long non-coding RNAs and human disease. Biochem. Soc. Trans. 40, 902–906 (2012). [DOI] [PubMed] [Google Scholar]
- Svoboda M. et al. HOTAIR long non-coding RNA is a negative prognostic factor not only in primary tumors, but also in the blood of colorectal cancer patients. Carcinogenesis 3, 1510–1515 (2014). [DOI] [PubMed] [Google Scholar]
- Graham L. D. et al. Colorectal neoplasia differentially expressed (CRNDE), a novel gene with elevated expression in colorectal adenomas and adenocarcinomas. Genes Cancer 2, 829–840 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X., Sun M., Liu H., Yao Y. & Song Y. Long non-coding RNAs: a new frontier in the study of human diseases. Cancer Lett 339, 159–166 (2013). [DOI] [PubMed] [Google Scholar]
- Baldinu P. et al. Identification of a novel candidate gene, CASC2, in a region of common allelic loss at chromosome 10q26 in human endometrial cancer. Hum Mutat 23, 318–326 (2004). [DOI] [PubMed] [Google Scholar]
- Xue Z., Wen J., Chu X. & Xue X. A microRNA gene signature for identification of lung cancer. Surg Oncol 23, 126–131 (2014). [DOI] [PubMed] [Google Scholar]
- Wang J., Zhao Y., Lu Y. & Ma C. Integrated bioinformatics analyses identify dysregulated miRNAs in lung cancer. Eur Rev Med Pharmacol Sci 18, 2270–2274 (2014). [PubMed] [Google Scholar]
- Jemal A., Siegel R., Xu J. & Ward E. Cancer statistics, 2010. CA Cancer J Clin 60, 277–300 (2010). [DOI] [PubMed] [Google Scholar]
- Brambilla E., Travis W. D., Colby T., Corrin B. & Shimosato Y. The new World Health Organization classification of lung tumours. Eur Respir J 18, 1059–1068 (2001). [DOI] [PubMed] [Google Scholar]
- Scott W. J., Howington J., Feigenberg S., Movsas B. & Pisters K. Treatment of non-small cell lung cancer stage I and stage II: ACCP evidence-based clinical practice guidelines. Chest 132, 234S–242S (2007). [DOI] [PubMed] [Google Scholar]
- van Zandwijk N. Neoadjuvant strategies for non-small cell lung cancer. Lung Cancer 34, S145–S150 (2001). [DOI] [PubMed] [Google Scholar]
- Swensen S. J. CT screening for lung cancer. AJR Am J Roentgenol 179, 833–836 (2002). [DOI] [PubMed] [Google Scholar]
- Cagle P. T. & Allen T. C. Lung cancer genotype-based therapy and predictive biomarkers: present and future. Arch Pathol Lab Med 136, 1482–1491 (2012). [DOI] [PubMed] [Google Scholar]
- Lam W. K. & Watkins D. N. Lung cancer: future directions. Respirology 12, 471–477 (2007). [DOI] [PubMed] [Google Scholar]
- White N. M. et al. Transcriptome sequencing reveals altered long intergenic non-coding RNAs in lung cancer. Genome. Biol 15, 429 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prensner J. R. & Chinnaiyan A. M. The emergence of lncRNAs in cancer biology. Cancer Discov 1, 391–407 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W., Böcker W., Brosius J. & Tiedge H. Expression of neural BC200 RNA in human tumours. J Pathol 183, 345–351 (1997). [DOI] [PubMed] [Google Scholar]
- Li G. et al. Long Noncoding RNA Plays a Key Role in Metastasis and Prognosis of Hepatocellular Carcinoma. Biomed Res Int 2014, 780521 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X. et al. A critical role for the long non‐coding RNA GAS5 in proliferation and apoptosis in non‐small‐cell lung cancer. Mol Carcinog, In press (2013). [DOI] [PubMed] [Google Scholar]
- Zhuang Y. et al. Induction of long intergenic non-coding RNA HOTAIR in lung cancer cells by type I collagen. J Hematol Oncol 6, 35 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelhaugh S. K. et al. Mining Affymetrix microarray data for long non‐coding RNAs: altered expression in the nucleus accumbens of heroin abusers. J Neurochem 116, 459–466 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y. et al. Mammalian ncRNA-disease repository: a global view of ncRNA-mediated disease network. Cell Death Dis 4, e765 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipscomb C. E. Medical subject headings (MeSH). Bull Med Libr Assoc 88, 265–266 (2000). [PMC free article] [PubMed] [Google Scholar]
- Wang D., Wang J., Lu M., Song F. & Cui Q. Inferring the human microRNA functional similarity and functional network based on microRNA-associated diseases. Bioinformatics 26, 1644–1650 (2010). [DOI] [PubMed] [Google Scholar]
- Xuan P. et al. Prediction of microRNAs Associated with Human Diseases Based on Weighted k Most Similar Neighbors. PLoS One 8, e70204 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P. et al. Expression of long non-coding RNA MEG3 ST8SIA3 and TUG1 in lung cancer. J Environ Health 30, 988–990 (2013). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.