Absence of colonic keratin 8 causes intestinal inflammation and decreased levels of the ketogenic enzyme HMGCS2. Upstream, the butyrate transporter MCT1 is decreased, leading to increased luminal butyrate. Ketogenic conditions fail to induce HMGCS2 in the keratin 8–knockout colon, suggesting a role for keratins in colonocyte energy homeostasis.
Abstract
Simple-type epithelial keratins are intermediate filament proteins important for mechanical stability and stress protection. Keratin mutations predispose to human liver disorders, whereas their roles in intestinal diseases are unclear. Absence of keratin 8 (K8) in mice leads to colitis, decreased Na/Cl uptake, protein mistargeting, and longer crypts, suggesting that keratins contribute to intestinal homeostasis. We describe the rate-limiting enzyme of the ketogenic energy metabolism pathway, mitochondrial 3-hydroxy-3-methylglutaryl-CoA synthase 2 (HMGCS2), as a major down-regulated protein in the K8-knockout (K8−/−) colon. K8 absence leads to decreased quantity and activity of HMGCS2, and the down-regulation is not dependent on the inflammatory state, since HMGCS2 is not decreased in dextran sulfate sodium-induced colitis. Peroxisome proliferator–activated receptor α, a transcriptional activator of HMGCS2, is similarly down-regulated. Ketogenic conditions—starvation or ketogenic diet—increase K8+/+ HMGCS2, whereas this response is blunted in the K8−/− colon. Microbiota-produced short-chain fatty acids (SCFAs), substrates in the colonic ketone body pathway, are increased in stool, which correlates with decreased levels of their main transporter, monocarboxylate transporter 1 (MCT1). Microbial populations, including the main SCFA-butyrate producers in the colon, were not altered in the K8−/−. In summary, the regulation of the SCFA-MCT1-HMGCS2 axis is disrupted in K8−/− colonocytes, suggesting a role for keratins in colonocyte energy metabolism and homeostasis.
INTRODUCTION
Keratins (Ks), the intermediate filament (IF) proteins of epithelial cells, are divided into acidic type I (K9–K28) and basic type II (K1–K8 and K71–K80) keratins (Coulombe and Omary, 2002; Schweizer et al., 2006) and make up the largest subgroup of IFs. These two types of keratins form noncovalent obligate heteropolymers in a 1:1 ratio. Keratins are accordingly expressed in specific pairs and in a tissue-specific manner (Quinlan et al., 1984). Intestinal simple epithelial cells express K7, K8, K18–K20, and K23, whereas adult hepatocytes express only K8 and K18 (Omary et al., 2009). One established role of keratins is to protect epithelia from mechanical and nonmechanical stresses; keratins participate in signaling events that regulate processes such as cellular architecture, growth, proliferation, and apoptosis (Omary et al., 2009; Toivola et al., 2010; Pan et al., 2013). The major type II keratin in intestinal epithelia is K8. K8-knockout (K8−/−) mice, which lack almost all colonocyte keratins, develop colonic epithelial hyperproliferation (Baribault et al., 1994), a Th2-type colitis, and an ulcerative colitis–like state that is reversed after antibiotic therapy (Habtezion et al., 2005). K8−/− mice also develop diarrhea due to colonocyte Na and Cl ion transporter mistargeting (Toivola, Krishnan, et al., 2004). The colonic hyperproliferation in K8−/− mice is linked to a decreased capacity of K8−/− colonocytes to undergo normal apoptosis at the upper part of the colonic crypt (Habtezion et al., 2011). Of note, the K8−/− small intestine is spared any major inflammatory response (Baribault et al., 1994; Ameen et al., 2001), which is likely related to the relative lack of bacteria in the small intestine compared to the colon.
Mitochondrial 3-hydroxy-3-methylglutaryl-CoA synthase 2 (HMGCS2) catalyzes the rate-limiting step of ketogenesis, in which ketone bodies (β-hydroxybutyrate, acetoacetate, and acetone) are produced during ketogenic conditions such as starvation, untreated diabetes, and other conditions in which carbohydrate levels are low (Hegardt, 1999). In the ketogenic pathway, acetyl-CoA, mostly derived from the β-oxidation of fatty acids, is converted into ketone bodies through a cascade of enzymatic reactions (Hegardt, 1999). The expression of HMGCS2 is regulated by peroxisome proliferator–activated receptor α (PPARα; Serra et al., 1996a; Meertens et al., 1998), which also regulates many changes in fatty acid and glucose metabolism (Lefebvre et al., 2006) by heterodimerizing with retinoid X receptor and binding to peroxisome proliferator hormone response elements on DNA of target genes (Hegardt, 1999). Ketogenesis occurs mainly in the liver, but also in the colon of adult mammals (Serra et al., 1996a; Hegardt, 1999).
Short-chain fatty acids (SCFAs) are products of intestinal bacterial fermentation of mainly dietary carbohydrates. Butyrate, a major SCFA, constitutes the main energy source for colonocytes and is involved in the regulation of HMGCS2 expression and thus ketone body production (Roediger, 1980; Hegardt, 1999; Cherbuy et al., 2004). Butyrate is thus an important intermediate in maintaining colonic homeostasis and health (Hamer et al., 2008; Bultman, 2014). Absorption of luminal butyrate is mainly mediated by monocarboxylate transporter 1 (MCT1), the expression and function of which are regulated by butyrate (Hadjiagapiou et al., 2000; Cuff et al., 2002; Borthakur et al., 2012) and further upstream by PPARα (König, 2008).
Using a proteomic approach, we identified HMGCS2 as a major down-regulated protein in colonic crypts isolated from K8−/− mice. The decreased HMGCS2 levels correlated with decreased enzyme activity, blunted response to ketogenic conditions, and down-regulation of the upstream MCT1 transporter feeding into the ketogenic pathway, thereby leading to increased luminal SCFA in K8−/− mice.
RESULTS
HMGCS2 is a major down-regulated protein in K8−/− colonocytes
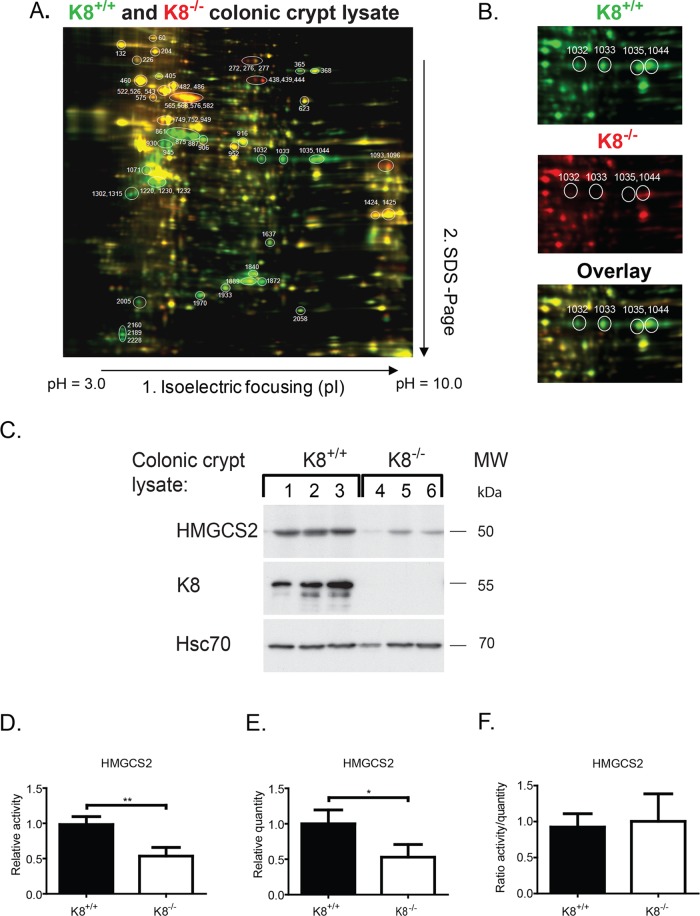
Proteins isolated from colonic crypt lysates of K8 wild-type (K8+/+) and K8−/− mice were compared using two-dimensional differential in-gel electrophoresis (2D DIGE) coupled with mass spectrometry to identify differentially regulated proteins (Figure 1, A and B). In K8−/− colonocytes, the major down-regulated proteins were, as expected, K8, K19, and their fragments (unpublished data). Most differentially expressed proteins are mitochondrial, cytoplasmic or endoplasmic reticulum proteins involved in metabolism and oxidative stress (Supplemental Table S1).
FIGURE 1:
HMGCS is a major down-regulated protein in K8−/− colon. Colonic crypts from K8−/− and K8+/+ mice were isolated and protein lysates labeled with Cy3 (green) for K8+/+ and Cy5 (red) for K8−/− (A). The mixed lysates were separated by 2D DIGE, and differentially expressed proteins were given individual spot numbers and detected by mass spectrometry (Supplemental Table S1). Unchanged protein spots appear yellow. The major down-regulated protein is HMGCS2 (spots 1032, 1033, 1035, and 1044), as shown separately for K8+/+ and K8−/− and as an overlay (B). Lysates of isolated colonic crypts from 3 K8+/+ and 3 K8−/− were analyzed by Western blotting for HMGCS2, K8, and Hsc70 as a loading control (C). Measured HMGCS2 activity values (Supplemental Figure S1) for K8−/− shown as fold decrease relative to HMGCS2 activity in K8+/+ colonocytes (D). HMGCS2 levels were assayed by immunoblotting of the same mitochondrial preparations that were used for measuring HMGCS2 activity. The results were quantified with ImageJ software and normalized to mitochondrial marker prohibitin, which is not altered in K8−/− (E). K8+/+ to K8−/− HMGCS2 enzyme activity and quantity ratios were calculated, and the correlation between HMGCS2 enzyme activity and quantity was analyzed (F). The results are based on three independent experiments (each experiment involving one K8+/+ and one K8−/− mouse) and represent the average ± SD, with significant differences shown as *p < 0.05 and **p < 0.01.
One of the most prominently down-regulated proteins in the K8−/− colon was the rate-limiting enzyme of ketogenesis, HMGCS2. Its four isoforms were, on average, fourfold decreased in K8−/− colonocytes (Figure 1, A and B, spots 1032, 1033, 1035, and 1044) when compared with K8+/+. Western blot analysis of HMGCS2 in isolated K8+/+ and K8−/− colonic crypts (Figure 1C) confirmed the down-regulation of HMGCS2 (see also later discussion of Figures 2A, 4, A–F, and 5C and Supplemental Figures S2 and S3 for total colon lysate and purified mitochondrial samples). Similarly, HMGCS2 was slightly but consistently down-regulated also in K8 small interfering RNA (siRNA)-treated HT-29 and Caco-2 colorectal cancer cells (Supplemental Figure S1). To determine whether HMGCS2 enzyme activity is modulated in K8−/−, we determined HMGCS2 activity and quantity in the same batch of mitochondria isolated from colonic epithelial scrapings (Figure 1D and Supplemental Figure S2). HMGCS2 activity (K8+/+, 0.98 ± 0.11; K8−/−, 0.54 ± 0.12; p < 0.01) and relative protein quantity (K8+/+, 1.00 ± 0.20; K8−/−, 0.53 ± 0.18; p < 0.05) were both significantly decreased in K8−/− colonocytes (Figure 1, D and E). The enzyme activity in K8−/− mice correlated with the decrease in enzyme quantity (HMGCS2 activity/quantity ratio: K8+/+, 0.92 ± 0.19; K8−/−, 1.0 ± 0.38; Figure 1F), and decreased protein levels thus cause the decreased activity.
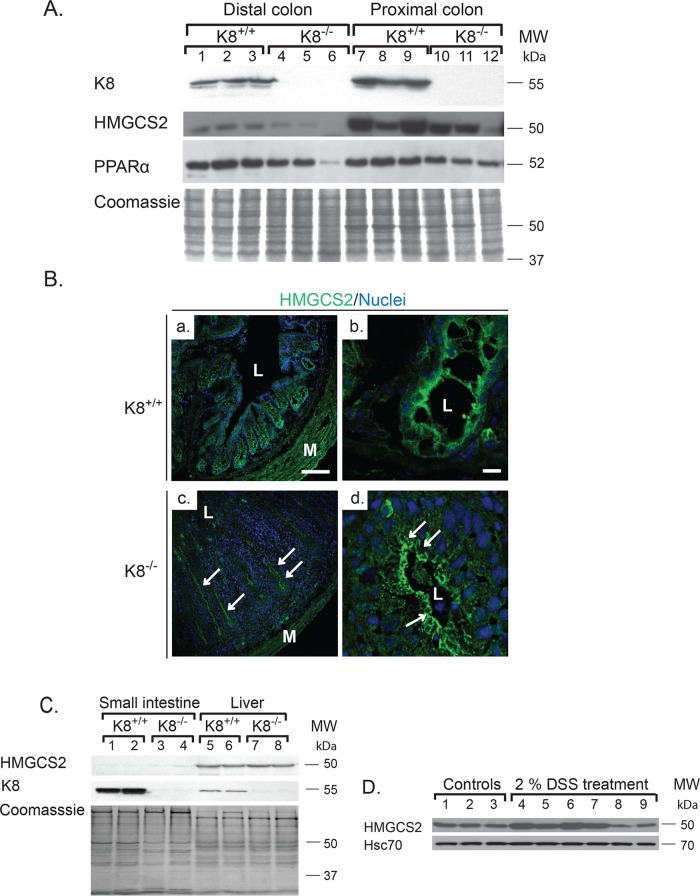
FIGURE 2:
HMGCS2 and PPARα are decreased and HMGCS2 is mislocalized in adult colon but not in small intestine or liver of K8−/− mice. Lysates of distal and proximal colon (A) and small intestine and liver (C) were obtained from K8+/+ and K8−/− mice, and the lysates were normalized by protein assay. Protein levels of HMGCS2, PPARα, and K8 were analyzed by immunoblotting, and equal loading was confirmed by Coomassie brilliant blue staining (A, C). Immunostaining of colonic HMGCS2 (B) showed decreased cytoplasmic pools of HMGCS2 (green) in crypts, with the remaining HMGCS2 located apically in the colonocytes (arrows). Scale bars, 100 μm (a, c), 10 μm (b, d); L, lumen; M, muscles. (D) Proximal colon lysates were obtained from 2.5-mo-old BALB/c mice treated with 2% DSS or without DSS (control) for 8 d. Samples were analyzed by immunoblotting for HMGCS2. Equal loading is shown by Hsc70.
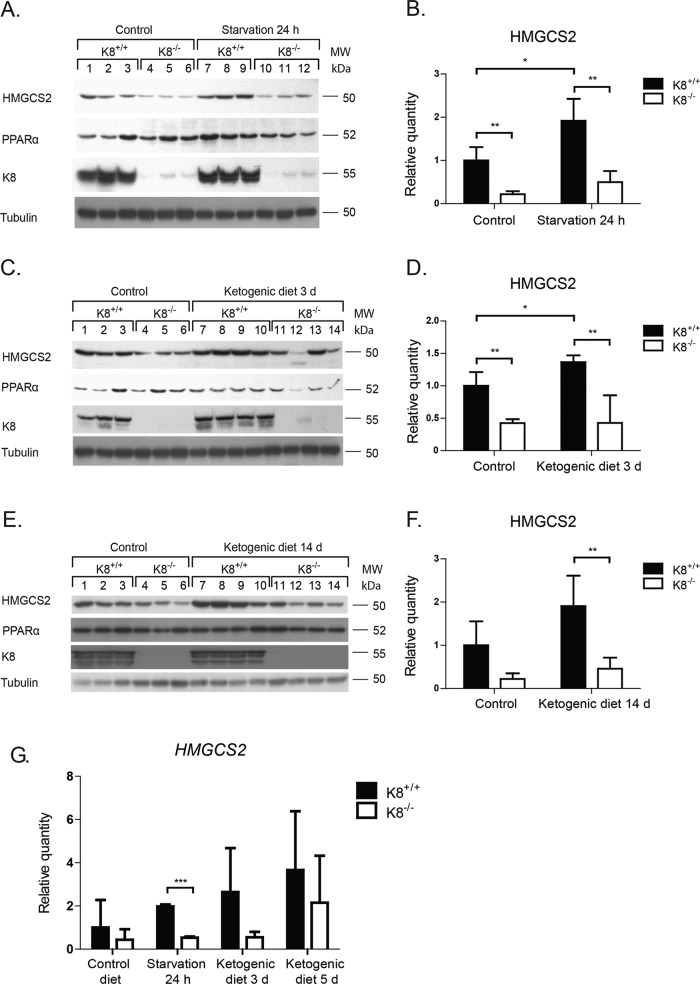
FIGURE 4:
Onset of ketogenic conditions fails to increase colonic HMGCS2 in K8−/− to the same levels as in K8+/+. Total colon lysates from K8+/+ and K8−/− mice subjected to normal and ketogenic conditions (24 h of starvation (A) or 3 d (C) or 14 d (E) of a ketogenic diet) were analyzed by Western blot for levels of HMGCS2, PPARα, and K8. Equal amounts of proteins were loaded after normalization by protein assay, as shown by tubulin protein levels (A, C, E). HMGCS2 levels (B, D, F) were quantified with ImageJ and normalized to tubulin (A, C, E, respectively; average ± SD, *p < 0.05, **p < 0.01). HMGCS2 mRNA levels (G) were examined by RT-PCR and normalized to actin and are shown as fold change of HMGCS2 mRNA levels between the different genotypes and treatment regimens (average ± SD, ***p < 0.001).
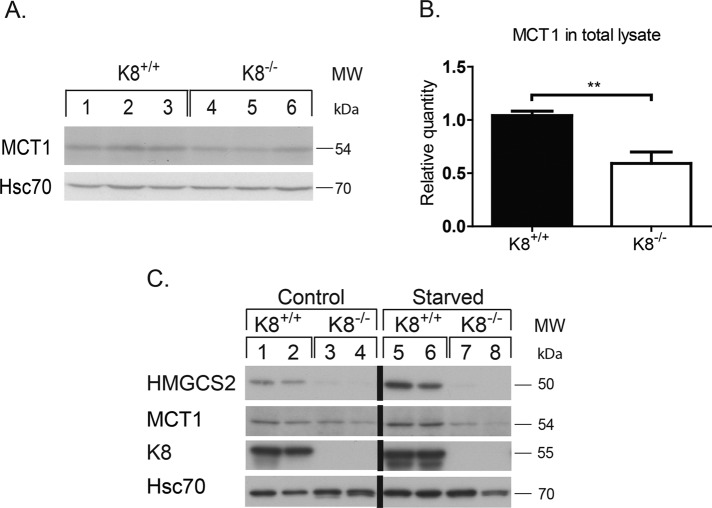
FIGURE 5:
MCT1 and HMGCS2 are down-regulated in K8−/− under normal and ketogenic conditions. Whole lysates of colon from K8+/+ and K8−/− mice were analyzed by immunoblotting for MCT1. Equal loading is shown by Hsc70 (A). The immunoblots were quantified, and MCT1 was normalized against Hsc70 (B). The results represent the average ± SD, with significant differences shown as **p < 0.01. Two- to 2.5-mo-old K8+/+ and K8−/− male mice were subjected to normal conditions or 18 h of starvation, and lysates of isolated colonic epithelium were prepared. The samples were analyzed by immunoblotting for HMGCS2 and MCT1 (C). Equal loading is shown by Hsc70, and the absence of keratin filaments is shown by K8.
K8-knockout–induced HMGCS2 down-regulation is a distinct noninflammatory phenotype unique for the colon and is associated with a decrease in PPARα
To investigate whether the K8-related down-regulation of HMGCS2 in the colon also occurs in other digestive organs, we analyzed HMGCS2 levels in K8−/− distal and proximal colon, small intestine, and liver. HMGCS2 down-regulation was clearly observed in both parts of the colon, and the levels of HMGCS2 in the proximal colon were in general higher than in the distal colon in both K8+/+ and K8−/− genotypes (Figure 2A and Supplemental Figure S3). This is expected because HMGCS2 levels correlate with luminal fermenting bacteria, which are more numerous in the proximal than the distal colon, therefore producing more SCFA (Topping and Clifton, 2001; Hamer et al., 2008). PPARα was also significantly decreased 1.3- to 1.8-fold (p = 0.03 for distal colon) in K8−/− (Figure 2A). Confocal immunofluorescence microscopy analysis confirmed that HMGCS2 levels are decreased in K8−/− colonocytes and that the remaining HMGCS2 accumulated in the apical part of the colonocytes (Figure 2B). HMGCS2 was undetectable in the small intestine (Figure 2C), as previously described (Hegardt, 1999). The liver, which is the main ketogenic organ, contains more HMGCS2 than the proximal colon (Supplemental Figure S3) but did not reveal any major difference in HMGCS2 levels between the genotypes (Figure 2C and Supplemental Figure S3). Our results were confirmed using two independent HMGCS2 antibodies (Supplemental Figure S3). Furthermore, HMGCS2 down-regulation is likely not caused by colitis in general since HMGCS2 levels were unchanged or increased after experimental dextran sulfate sodium (DSS)–induced colitis (Figure 2D).
Ketogenesis is blunted in the K8−/− colon
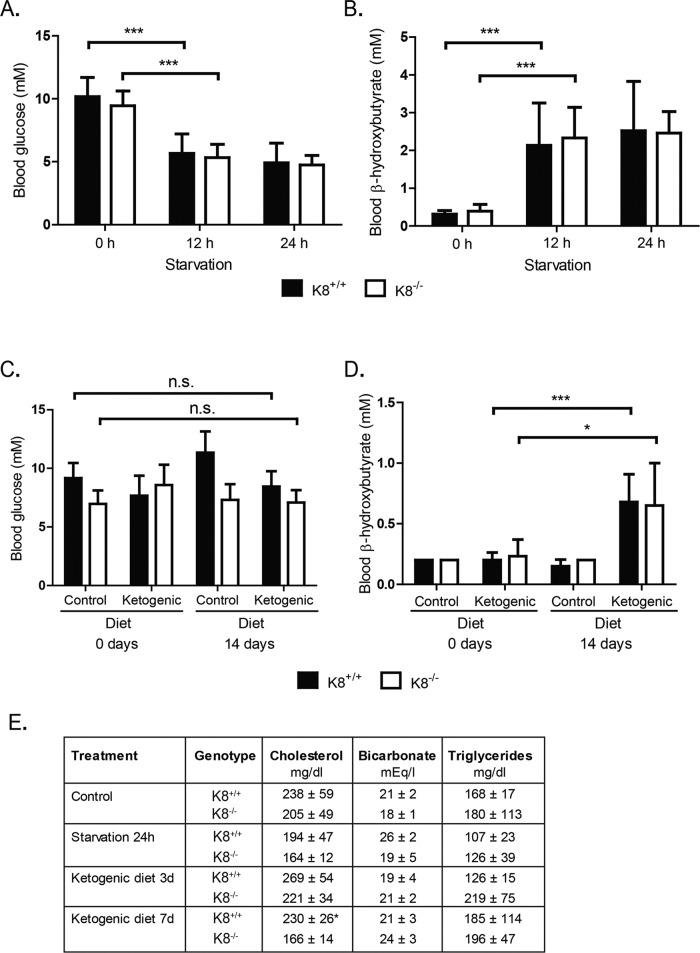
To study whether the decreased levels of HMGCS2 in K8−/− colonocytes are enough to respond to ketogenic conditions, mice were starved for 12 and 24 h or given a ketogenic diet for 3 or 14 d, after which HMGCS2, blood glucose, and blood β-hydroxybutyrate levels were assayed. Ketogenesis was readily induced, since K8+/+ and K8−/– blood glucose levels decreased consistently with starvation at 12 h and remained low at 24 h (Figure 3A), whereas blood β-hydroxybutyrate, which is the most prominent ketone body, simultaneously increased threefold to fourfold (Figure 3B). The ketogenic diet did not dramatically affect blood glucose levels, but β-hydroxybutyrate was moderately increased in both genotypes 14 d after onset of the ketogenic diet (Figure 3, C and D). Serum cholesterol, bicarbonate, and triglycerides were not markedly different between K8+/+ and K8−/−, except for a slight decrease of cholesterol in K8−/− 7 d after onset of ketogenic diet (Figure 3E). No differences in food ingestion or weight gain were detected between genotypes (Supplemental Figure S4). Because the liver is the main ketogenic organ controlling blood ketone levels, these baseline data suggest that liver ketogenesis remains unchanged despite K8 deletion and that a ketogenic diet and starvation should be useful models for assessing colonocyte ketogenesis.
FIGURE 3:
Blood and serum parameters are similar in K8+/+ and K8−/− mice under baseline and ketogenic conditions. Mice were starved for 24 h (A, B, E) or fed with a control or ketogenic diet (C, D, E) as outlined in Materials and Methods. Blood samples from male and female K8+/+ and K8−/− mice were taken from the submandibular vein 0, 12, and 24 h after onset of starvation (A, B) and after 0 and 14 d of a ketogenic diet (C, D) and were assayed for the levels of blood glucose (A, C), blood β-hydroxybutyrate (B, D), and a panel of blood markers (E). Serum samples from mice subjected to 24 h of starvation or to 3 and 7 d of a ketogenic diet were analyzed for cholesterol, bicarbonate, and triglycerides (E). The results represent the average ± SD; shown as n.s., p > 0.05, not significant; *p < 0.05 and ***p < 0.001, statistically significant.
Western blot analysis showed, as expected, that both starvation and 3 d of ketogenic diet significantly increased HMGCS2 levels in K8+/+, whereas no significant changes in HMGCS2 levels were observed in K8−/− at any time after starvation (Figure 4, A and B) or ketogenic diet (Figure 4, C–F). The decrease of K8−/− HMGCS2 was, in comparison to K8+/+, on average threefold during normal conditions and fourfold during ketogenic conditions (Figure 4, A–F). The corresponding decrease of HMGCS2 mRNA levels was 3.8-fold during normal diet conditions and on average 3.7-fold after starvation and 3 d of ketogenic diet (Figure 4G). The protein levels of PPARα, in general, follow the same pattern as HMGCS2 levels (Figure 4, A, C, and E). A slight up-regulation of PPARα was observed after ketogenic conditions in K8+/+ mice, whereas a slight down-regulation in K8−/− was seen after both normal and ketogenic conditions. K8 knockdown thus clearly leads to a blunted colonic ketogenic response.
Normal colonic mitochondrial ultrastructure and adenosine monophosphate–activated protein kinase signaling in K8−/− colon
We next assessed whether the K8−/− colon ketogenic phenotype could stem from abnormalities in mitochondria and common energy metabolism pathways. In contrast to K8−/− hepatocyte mitochondria, which are smaller than K8+/+ mitochondria (Tao et al., 2009), no differences in mitochondrial size were noted in K8−/− colonocytes, even if there were fewer cristae per mitochondrial section area compared with K8+/+ (Supplemental Figure S5, B and C). The protein levels of mitochondrial prohibitin and cytochrome c were comparable in K8−/− and K8+/+ (Supplemental Figure S2), suggesting that the mitochondrial function is unaltered in the K8−/− colonocytes. To analyze whether there are additional changes in K8−/− colon energy metabolism, we analyzed a panel of proteins involved in energy metabolism pathways under baseline and ketogenic conditions. Acetyl-CoA carboxylase (ACC), a central enzyme in the biosynthesis of fatty acids, converts acetyl-CoA into malonyl-CoA and functions as an inhibitor of β-oxidation (Lopaschuk et al., 2010). Ketogenic conditions induce β-oxidation of fatty acids into acetyl-CoA (Hegardt, 1999), whereas ACC activity remains low (Lopaschuk et al., 2010) in order to maintain the availability of acetyl-CoA for conversion into ketone bodies through ketogenesis. The levels of ACC in K8+/+ and K8−/− colons were unaltered under basal conditions but remained high under ketogenic conditions in K8−/− but not K8+/+ colon (Supplemental Figure S6, A–C; a significant 2.5-fold decrease in K8+/+ [p = 0.0005], whereas it was only 1.4-fold [not significant] in K8−/− after 3 d of ketogenic diet when quantified from Western blots), thus supporting the notion of a blunted ketogenic pathway after K8 inactivation. No changes were observed in the expression patterns of ACC inhibitors adenosine monophosphate–activated protein kinase α (AMPKα) and AMPKβ and their inactive phosphorylated forms in K8+/+ and K8−/− colons (Supplemental Figure S6, A–C). Independent of basal or active ketogenesis, the expression levels of glucose transporter 4 (GLUT4) and cytochrome c oxidase (COX IV; a mitochondrial enzyme of the respiratory chain; Supplemental Figure S6, A–C) were marginally decreased in total colon lysates of K8−/− mice and unchanged in colonic epithelium isolated from K8−/− mice in comparison to K8+/+ (unpublished data). Analysis of ADP/ATP and NAD/NADH ratios in epithelial scrapings did not reveal any major differences between the genotypes (unpublished data). Together these results confirm that ketogenesis is activated under the ketogenic conditions used here and blunted in K8−/−, and that mitochondrial ultrastructure and energy intermediates are largely unchanged in the absence of K8.
The SCFA transporter MCT1 is down-regulated in K8−/− and luminal SCFA acid levels are increased upstream of HMGCS2
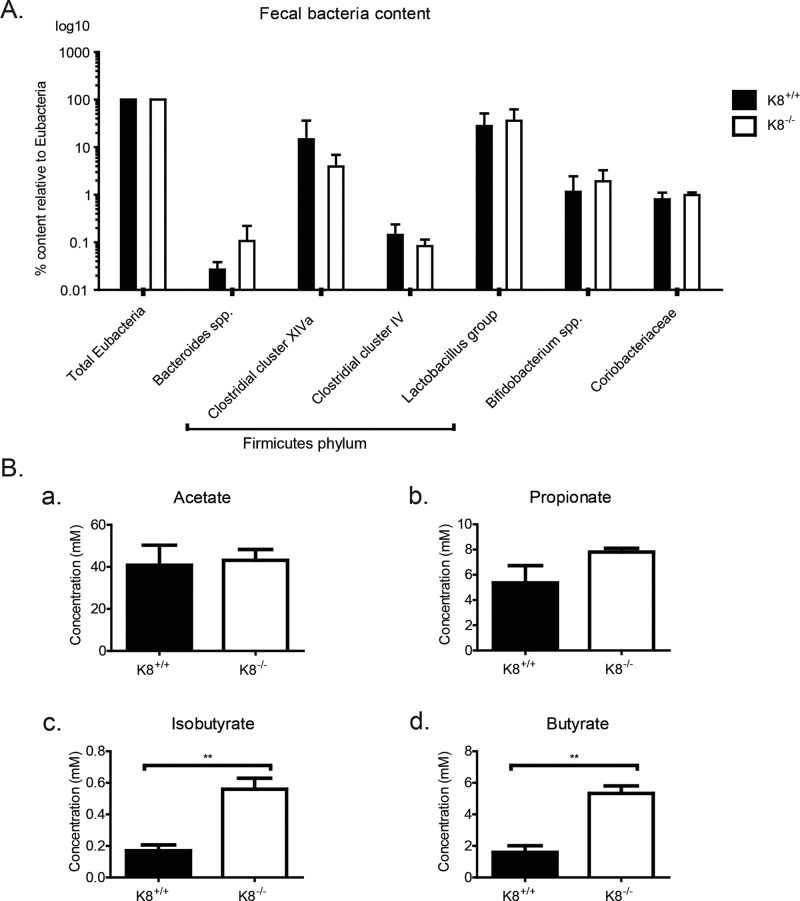
Absorption of luminal SCFA is mainly mediated by MCT1 (Hadjiagapiou et al., 2000). To assess whether expression of MCT1, and consequently the absorption of SCFA, could be perturbed in the absence of keratins, we assayed K8−/− colon MCT1 levels by immunoblotting. Significant down-regulation of MCT1 was observed in total lysates of K8−/− (0.59 ± 0.10; p < 0.005) in comparison to K8+/+ (1.04 ± 0.04; Figure 5, A and B) and in isolated K8−/− colonic epithelium (0.40 ± 0.11) compared with K8+/+ (1.00 ± 0.11; Figure 5C) under both control and starved conditions. The decrease in MCT1 levels was also confirmed in HT-29 and Caco-2 cells in which K8 has been down-regulated (Supplemental Figure S1). This decrease in MCT1 could also be appreciated in MCT1 immunostainings in K8−/− colon and in Caco-2 cells treated with K8 siRNA (Supplemental Figure S7). However, K8 down-regulation or absence did not lead to any differences in MCT1 localization, which is seen as a tight plasma membrane staining in Caco-2 cells and mostly laterally between the epithelial cells in the colon in vivo (Supplemental Figure S7). Because MCT1 expression and function are up-regulated by butyrate (Cuff et al., 2002), the ratio of butyrate-producing bacteria to total Eubacteria in K8+/+ and K8−/− was assayed. No differences in K8−/− and K8+/+ stool microbial populations were seen, including the two clusters of butyrate-producing bacteria of the Firmicutes phylum, Clostridium clusters XIVa and IV (Pryde et al., 2002; Figure 6A). Therefore the metabolic state of the K8−/− phenotype is not caused by a difference in the microbial balance and butyrate-producing bacteria. The SCFA levels in K8+/+ and K8−/− stool were furthermore profiled to determine whether the observed down-regulation of MCT1 could be due to diminished production of butyrate by the microflora in K8−/−. Of interest, a significant increase in the levels of butyric acid (K8+/+, 1.59 ± 0.72 mM; K8−/−, 5.33 ± 0.83 mM; p < 0.005) and isobutyric acid (K8+/+, 0.17 ± 0.062 mM; K8−/−, 0.56 ± 0.12 mM; p < 0.01) was detected in K8−/− stool in comparison to K8+/+ (Figure 6B), indicating that bacterial production of SCFA is not perturbed, whereas the uptake may be compromised due to decreased levels of MCT1.
FIGURE 6:
Ratios of the butyrate-producing bacteria from the Firmicutes phylum are unaltered in K8−/− colons, whereas K8−/− stool contains increased levels of butyrate. Colonic contents (fecal stool) were collected from K8+/+ and K8−/− mice and analyzed by quantitative PCR for detection and quantification of the total amount of Eubacteria and a panel of bacterial groups and clusters. (A) No significant differences in the levels of butyrate-producing bacteria in Clostridium clusters XIVa and IV were observed in the colon of K8−/− and K8+/+ mice. Levels are normalized to total eubacteria for both genotypes. (B) Levels of the SCFAs acetate, propionate, isobutyrate, and butyrate were assessed from freshly collected K8+/+ and K8−/− stool by gas chromatography profiling. The results represent the mean ± SD, with significant differences shown as **p < 0.01.
DISCUSSION
The molecular functions of keratins in the colonic epithelium are largely unclear, even if it is well established that K8 ablation in mice leads to early and chronic colitis (Baribault et al., 1994; Toivola, Krishnan, et al., 2004; Habtezion et al., 2005, 2011). To understand how keratins may help maintain intestinal homeostasis, we used a proteomic approach to identify differentially expressed proteins in K8−/− colonocytes. We show that 1) HMGCS2 is decreased and the reduced HMGCS2 levels in K8−/− colonocytes correlate with decreased enzyme activity in mitochondria; 2) the HMGCS2 down-regulation phenotype is colon-specific and not related to an acute inflammation per se, as determined using the DSS colitis model; and 3) the decrease of HMGCS2 leads to blunted ketogenesis in K8−/− colon. In addition, by studying upstream regulation of HMGCS2, we find that 4) the HMGCS2 transcriptional regulator PPARα and the SCFA (butyrate) transporter MCT1 are down-regulated in the K8−/– colon, whereas 5) luminal butyrate levels are increased 6) without changes in the composition of the butyrate-producing bacteria. To our knowledge, this is the first study describing IF keratins as modulators of colonocyte energy metabolism.
HMGCS2 is an important and unique enzyme in the colonic epithelium, since ketogenesis only occurs in a few other organs apart from the liver (Serra et al., 1996b; Hegardt, 1999). Colonic HMGCS2 is regulated by bacterially produced SCFA (mainly butyrate), which constitute the substrate for ketogenesis and promote colonic health, for example, by inhibiting inflammatory responses and strengthening the colonic defense barrier (Hamer et al., 2008). Adult small intestine, on the other hand, does not express HMGCS2 and is fueled by glucose and glutamine in the fed state and liver-derived ketone bodies during starvation (Ardawi and Majzoub, 1988). Moreover, there is no major disease phenotype in the K8−/− small intestine (Baribault et al., 1994; Ameen et al., 2001), whereas the colitis phenotype is prominent especially in the proximal colon in these mice (Habtezion et al., 2005). HMGCS2 is highly expressed in the proximal colon, where also the vast majority of the colonic microflora resides. Because K8−/− mice have fewer luminal bacteria and the colitis is reversible with antibiotics (Habtezion et al., 2005, 2011), it is conceivable that HMGCS2 plays a contributory role in the development of colitis.
Because the decreased HMGCS2 protein amount correlates with decreased enzyme activity (Hegardt, 1999) in the K8−/− colon, it is unlikely that keratins directly regulate HMGCS2 activity. The management of energy stores during ketogenic conditions is regulated by PPARα (Kersten et al., 1999), which has been shown to have a stimulatory effect on hepatic HMGCS2 expression (Rodriguez et al., 1994). We also noted down-regulation of colonic PPARα under baseline and ketogenic conditions in K8−/− mice, in line with the expression of HMGCS2 (Figures 2 and 4). Clinically, low HMGCS2 levels are also observed in colon cancer (Camarero et al., 2006), and mutations in HMGCS2 lead to decreased protein and hypoketotic hypoglycemia upon prolonged starvation (Aledo et al., 2006). HMGCS2 is highly expressed in the liver, where keratins have established cytoprotective roles (Omary et al., 2009; Toivola et al., 2010). However, no major change in HMGCS2 expression was found in K8−/− liver, despite the fact that these hepatocyte mitochondria are smaller than wild-type mitochondria (Tao et al., 2009), and consequently no differences were seen in serum ketone body levels between genotypes. In the K8−/− liver, mitochondrial and cytoplasmic proteins were also the two groups of proteins most affected, of which transferrin and aldehyde dehydrogenase were altered in both organs (Tao et al., 2009). Because the liver is lacking the microbial component, these findings further suggest the central involvement of microflora and/or SCFA in the down-regulation of K8−/− colonic HMGCS2. Moreover, the modulation of HMGCS2 described here is likely specifically due to the ablation of K8 and not due to the inflammatory status of the K8−/− colon phenotype, since the expression of HMGCS2 was increased rather than decreased in DSS colitis (Naito et al., 2010).
The ramifications of low HMGCS2 levels in K8−/− are noticeable after exposing mice to ketogenic conditions, upon which K8−/− mice were not able to increase colonic HMGCS2, in contrast to wild-type mice. Ketogenic conditions, such as the intake of a high-fat, low-carbohydrate diet or starvation, activates β-oxidation of fatty acids and generates an excess of acetyl-CoA that is converted into ketone bodies (Cullingford, 2004). The blunted ketogenic response in K8−/− was supported by our observation that ACC levels remained relatively unchanged in K8−/− colon under ketogenic conditions. ACC catalyzes the conversion of acetyl-CoA into malonyl-CoA, which is an inhibitor of carnitine palmitoyltransferase I (CPTI), the rate-limiting enzyme of fatty acid β-oxidation (McGarry and Brown, 1997). Because the main function of ACC in liver is to provide malonyl-CoA and therefore inhibit β-oxidation (McGarry and Brown, 1997), lower levels of ACC under ketogenic conditions are expected due to decrease levels of acetyl-CoA (Guzman et al., 2000). These results also suggest diminished β-oxidation in K8−/− colon. How keratins modulate this pathway needs further study but could be linked to the observation that K8/K18 are inhibitory of hepatic CPTI activity (Velasco et al., 1998). Changes in proteins involved in ketone body metabolism (decreased acetoacetyl-CoA thiolase in the first step of ketogenesis) and β-oxidation have also been described in desmin-null mouse muscle (Fountoulakis, Soumaka, Rapti, et al., 2005). In contrast to hepatocyte mitochondria, which are smaller after K8 knockdown (Tao et al., 2009), colon mitochondrial size appears normal, even if alterations in cristae can be seen (Supplemental Figure S5). Hence mitochondrial morphology changes are unrelated to the observed changes in colonocyte HMGCS2.
Mice raised in a germ-free environment have diminished metabolic processes, since the butyrate-producing bacteria are nonexistent (Donohoe et al., 2011). HMGCS2 expression is dependent on colonic microflora and SCFA (Cherbuy et al., 2004), and the K8−/− colonic microflora consist of fewer microorganisms than K8+/+ microflora (Habtezion et al., 2011). These data, together with the fact that HMGCS2 is not altered in K8−/− hepatocytes (Figure 2C), suggest that the decreased HMGCS2 in the absence of K8 could be due to diminished availability of luminal SCFA or aberrant transport of SCFA across the cell membrane. SCFAs are transported into colonocytes mainly through MCT1, the expression and function of which are up-regulated by butyrate (Cuff et al., 2002) and PPARα (König, 2008). Because butyrate is the primary source of colonocyte energy and inhibits inflammation and carcinogenesis, it is feasible that diminished availability of luminal butyrate or the inability of colonocytes to oxidize butyrate could contribute to the onset of inflammation in the colon (Roediger, 1990; Hamer et al., 2008). We show here that the absence of K8 correlates with lower epithelial levels of MCT1 and higher levels of K8−/− fecal butyrate, which, however, are not caused by changes in the ratios of butyrate-producing bacterial species in the microbiome (Figures 5 and 6). These results suggest that the K8−/− microbiome is capable of producing SCFAs, which, however, may not enter colonocytes properly and, hence, affect the fuel for ketogenesis. A similar association between keratins and butyrate levels was also seen in a human colorectal cancer study, in which higher levels of fecal butyrate corresponded to decreased K8 levels (Khan et al., 2011). Besides the down-regulation of MCT1, down-regulation of PPARα is also seen when K8 is absent in colonocytes, confirming that PPARα up-regulates genes involved in fatty acid oxidation and ketogenesis (König B, 2008).
Keratins could participate in the entry of SCFAs to epithelial cells by allowing proper targeting of MCT1 to the cell membrane. This hypothesis is supported by the generalized mistargeting of membrane proteins in the K8−/− colon, including ion transporters AE1/2 and ENACγ (Toivola, Krishnan, et al., 2004). Lack or mutation of keratins and other IF proteins leads to altered distribution of macromolecules in several contexts, including GLUT transporters in embryonic epithelia (Vijayaraj, Kröger, et al., 2009) and the endocrine pancreas (Alam et al., 2013) and cholesterol in the adrenal gland (Shen et al., 2012). In this study, however, we could not detect any major mistargeting of MCT1 in the absence of K8, indicating that keratins may affect MCT1 stability in the cell. Of interest, butyrate is an inhibitor of histone deacetylases, and it has been shown that colonic K8 lysines also are acetylated and this is regulated by butyrate levels (Drake, Griffiths, et al., 2009; Kilner et al., 2012).
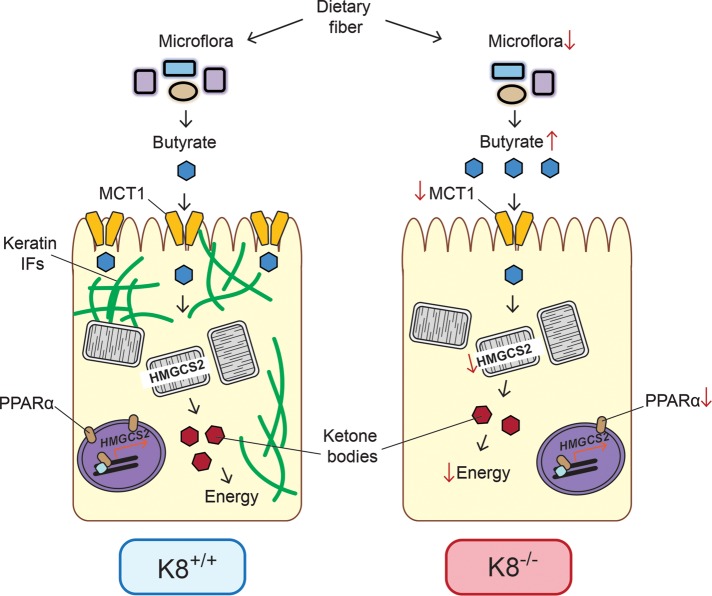
Taken together, these results suggest a model in which absorption of SCFAs is decreased in K8−/− colonocytes due to reduced MCT1 levels, leading to elevated levels of butyrate in the lumen, as well as diminish HMGCS2 levels in colonocytes. These perturbations likely contribute to the inflammatory phenotype observed in the K8−/− mouse (Figure 7).
FIGURE 7:
The role of keratins in colon energy metabolism. The SCFA butyrate is the primary source of energy for colonic epithelium and is involved in the maintenance of epithelial homeostasis. Butyrate and other SCFAs are produced by the colonic microflora via fermentation of, for example, dietary fiber. The amount of luminal bacteria is slightly decreased (Habtezion et al., 2011) and while the ratio of butyrate-producing species is unaltered, butyrate levels are increased in K8−/− compared with K8+/+ colonic lumen. SCFAs are absorbed primarily by MCT1, which is down-regulated in K8−/− colon. No evidence of MCT1 mistargeting as a consequence of K8-down-regulation could be detected. The localization of MCT1 in the cell varies depending on the nutritional state of the colon and changes from a lateral to a presumably functional apical location. SCFAs are transported to mitochondria, where they are converted to acetyl-CoA through β-oxidation. Acetyl-CoA is then used in the citric acid cycle and for ketone body production, which serves as energy in colonocytes under normal conditions. The absence of keratin filaments leads to decreased energy metabolism in the colon of K8−/− mice, which may be central in the observed K8−/− colitis phenotype.
MATERIALS AND METHODS
Mice
K8−/− and K8+/+ mice in the FVB/n background were generated by interbreeding K8 heterozygote (K8+/−) mice and genotyped as described (Baribault et al., 1994). BALB/c mice were purchased from the Turku Central Animal Facility, where all of the mice were housed. Animals were treated according to the animal study protocol approved by the State Provincial Office of South Finland.
Antibodies
Primary antibodies used for Western blotting and immunostaining were chicken anti-HMGCS2 (Genway, San Diego, CA), rabbit anti-HMGCS2 (AVIVA, San Diego, CA; epitope different from the Genway antibody), chicken anti-MCT1 (Chemicon, Temecula, CA), rabbit anti-MCT1, goat anti-PPARα (Santa Cruz Biotechnology, Dallas, TX), rabbit anti–α-smooth muscle actin, rabbit anti-prohibitin (Abcam, Cambridge, UK), rabbit anti–cytochrome c, rabbit anti-Glut4, rabbit anti–COX IV, rabbit anti-ACC, rabbit anti–phospho-ACC, rabbit anti-AMPKα, rabbit anti–P-AMPKα, rabbit anti-AMPKβ1, rabbit anti–P-AMPKβ1 (Cell Signaling Technology, Danvers, MA), rat anti-Hsc70 (Enzo Life Sciences, Helsinki, Finland), rat anti-K8 (Troma I Developmental Studies Hybridoma Bank, Iowa City, IA), and mouse anti-tubulin (Sigma-Aldrich, Munich, Germany). Secondary antibodies used for Western blotting were anti-mouse immunoglobulin G (IgG)–horseradish peroxidase (HRP), anti-rat IgG-HRP (GE Healthcare, Buckinghamshire, UK), anti-rabbit IgG-HRP (Cell Signaling Technology), anti-chicken IgG-HRP (Genway), and anti-goat IgG-HRP (Cell Signaling Technology). Secondary antibodies used for immunostaining were anti-rabbit Alexa Fluor 488 and anti-rat Alexa Fluor 546 (Life Technologies, Carlsbad, CA). Nuclei were stained with Toto-3 (Life Technologies) or DRAQ5 (Cell Signaling Technology).
Isolation of colonic crypts and enrichment of the colonic epithelium
Colonic crypts used for the proteomics study were isolated using MatriSperse Cell Recovery Solution (BD Biosciences, Bedford, MA) as described (Perreault and Beaulieu, 1998). For enrichment of colonic epithelium also used, for example, for ATP+NAD, the colon was excised, placed on an ice-cold glass surface, cut open longitudinally, and rinsed with ice-cold phosphate-buffered saline, after which the colonic epithelium was collected by scraping with a glass slide.
2D DIGE
Colonic crypts from two pairs of K8+/+ and K8−/− mice were isolated. K8+/+ and K8−/− colonic protein samples were labeled with Cy3 and Cy5, respectively, pooled, and then separated by 2D DIGE (Figure 1). Differentially expressed proteins were excised from gels, identified, and quantified by mass spectrometry and numbered (Figure 1 and Supplemental Table S1).
Ketogenic conditions
K8+/+ mice (nine females, five males) and K8−/− mice (nine females, six males) 3 mo of age were subjected to 24 h of starvation with unlimited access to water. Weight, as well as blood glucose and β-hydroxybutyrate levels, was measured at 0, 12, and 24 h after the beginning of food starvation. Blood glucose and β-hydroxybutyrate levels were measured with blood obtained from the submandibular vein using goldenrod lancets (Medipoint, Mineola, NY) and a glucose/β-hydroxybutyrate meter (Precision Xceed; Abbott Diabetes Care, Alameda, CA). Mice were killed by CO2 asphyxiation at the end of 24 h of starvation.
K8+/+ mice (three females, two males) and K8−/− mice (three females, three males) 3–6 mo of age were subjected to ketogenic diet (TD.96355; Harlan Teklad Laboratories, Madison, WI) for 3, 7, or 14 d. Corresponding K8+/+ and K8−/− mice were subjected to control diet (TD.00606; Harlan Teklad Laboratories). The macronutrient composition of the ketogenic diet (by weight) was 15.3% protein, 0.5% carbohydrate, and 67.4% fat, with an energy content of 6.7 kcal/g. The corresponding composition of the control diet was 9.2% protein, 70.9% carbohydrate, and 5.1% fat, with an energy content of 3.7 kcal/g. Because the ketogenic diet contains twice as many calories, mice eat less of it than the control diet (manufacturer's suggestion). Therefore the intake of protein, minerals, and vitamins is halved in the ketogenic diet. Cages, water, and the control diet were changed on day 7, and the ketogenic diet was changed on days 1, 2, 4, 6, 7, 9, and 11. The control diet consisted of pellets administered with normal feeder systems, and the ketogenic diet, which has a butter-like texture, was administered by a feeder designed for powdered food. Daily measurements of mouse and food weight, as well as of blood glucose and β-hydroxybutyrate levels, were done. Blood glucose and blood β-hydroxybutyrate were measured at days 0, 3, 7, and 14 with a glucose/β-hydroxybutyrate meter (Precision Xceed). Mice were killed by CO2 inhalation on day 14. Liver, small intestine, and distal and proximal colon samples were excised and stored in liquid nitrogen (for protein analysis), in optimum cutting temperature compound for immunofluorescence (Sakura, Alphen, Netherlands), in RNAlater for RNA quantification (Qiagen, Valencia, CA), and in collidine–glutaraldehyde buffer for electron microscopy.
DSS-induced colitis
DSS 2% (40,000 Da; TdB Consultancy, Uppsala, Sweden) was administered in autoclaved drinking water to 2.5-mo-old BALB/c mice for 8 d to induce acute colitis (Wirtz et al., 2007). Control BALB/c mice were given normal drinking water, and mice were killed on day 8 of the experiment. Distal and proximal colon samples were excised and stored in liquid nitrogen for protein analysis.
Mitochondrion isolation, HMGCS2 activity assay, and ADP/ATP and NAD/NADH assays
For HMGCS2 enzyme activity measurements, mitochondria were purified from colonic epithelium scrapings isolated from K8+/+ and K8−/− 5- to 6-mo-old mice starved overnight for 18 h, by a modification of a method described by Nonn et al. (2003). Colonic scrapings were suspended in MSH buffer (250 mM mannitol, 75 mM sucrose, 5 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid, pH 7.4) with 1 mM EDTA directly after isolation and homogenized manually with a Dounce homogenizer (75 strokes), following by pelleting (10 min, 600 × g, 4°C). The supernatants were recentrifuged for 15 min at 6800 × g and 4°C, and the pellets containing mitochondria were resuspended in ice-cold MSH buffer. The protein content of the isolated mitochondria was determined with a Pierce BCA Protein Assay Kit (Thermo Scientific, Rockford, IL). HMGCS2 activity in the isolated mitochondria was assayed by a modification of a method described by Patel et al. (2007). The activity of HMGCS2 was determined as the difference in the rate of disappearance of acetoacetyl-CoA at 303 nm before and after the addition of acetyl-CoA and phosphotransacetylase (PTA) in a pairwise analysis of K8+/+ and K8−/− samples with equal amounts of mitochondrial proteins as input. Reagents for the enzyme assay were obtained from Sigma-Aldrich (Munich, Germany). Briefly, a 1-ml quartz cuvette containing 50 mM Tris-HCl (pH 8.0), 10 mM MgCl2, 0.2 mM dithiothreitol, and 5 mM acetyl phosphate to a total volume of 827 μl was prewarmed to 30°C. A 75-μg amount of isolated mitochondria was lysed by incubation with Triton X-100 (1.4% [vol/vol]) for 4 min at 30°C. After addition of 10 μM acetoacetyl-CoA (AcAcCoA) and the lysed mitochondria to the cuvette, the absorbance was measured at 303 nm for 1 min after 2 min of incubation. Acetyl-CoA (AcCoA; 100 μM) and PTA (10 U) were added to the cuvette and the change in absorbance was measured. HMGCS2 activity was then calculated as follows:
HMG-CoA synthase activity = [activity after addition of AcCoA + regeneration system (AcAcCoA hydrolase + HMG-CoA synthase activity)] − [activity (2–3 min) before addition of AcCoA + regeneration system (AcAcCoA hydrolase activity)]
HMGCS2 activities were related to HMGCS2 protein levels, which in turn were determined by immunoblotting of the same mitochondrial preparations, and normalized to the levels of the mitochondrial marker prohibitin.
The intracellular ADP/ATP ratios in colonocytes isolated from K8+/+ and K8−/− mice were measured using a bioluminescent ADP/ATP ratio assay kit (Abnova, Taipei, Taiwan) according to the manufacturer's instructions. The luminescence readings were performed using a Hidex Sense microplate reader (Hidex, Turku, Finland). The intracellular levels of NAD and NADH were measured using a colorimetric NAD/NADH assay kit (Abcam, Taipei, Taiwan) according to the manufacturer's instructions. Total NAD and NADH levels were determined through colorimetric detection of absorbance at 450 nm with a Wallac 1420 VICTOR2 multilabel counter (PerkinElmer, Boston, MA).
Immunoblotting and immunostaining
Samples for protein analysis were homogenized in a homogenization buffer (0.187 M Tris-HCl, pH 6.8, 3% SDS, 5 mM EDTA) to obtain total tissue lysates. Equal parts of the central distal and proximal colons were mixed for colon lysates unless otherwise stated. Protein concentrations were measured with a Pierce BCA protein assay kit (Thermo Scientific, Waltham, MA); the samples were normalized and separated by SDS–PAGE, transferred to a polyvinylidene fluoride membrane, and analyzed by immunoblotting. Western blot bands were quantified with ImageJ software (National Institutes of Health, Bethesda, MD) and normalized to loading control (tubulin, Hsc70, or prohibitin). Fresh-frozen K8+/+ and K8−/− colon samples were cryosectioned (6 μm) and fixed in −20°C acetone for 10 min, Caco-2 cells were fixed in 1% paraformaldehyde for 10 min, and the fixed tissue and cell samples were immunostained as described (Ku et al., 2004). Samples were analyzed with a Leica TCS SP5 (Leica, Mannhein, Germany) confocal microscope.
Transmission electron microscopy and cristae quantification
For transmission electron microscopy, K8+/+ and K8−/− mice were starved for 18 h before killing, and the colon was excised and fixed directly in 5% glutaraldehyde in 0.16 M collidine buffer (pH 7.4). After dehydration and embedding, 70-nm sections were cut and stained with 12.5% uranyl acetate in methanol with 0.001% acetone and 0.25% lead citrate and analyzed with a JEM-100S transmission electron microscope (Jeol, Tokyo, Japan) and JEM-1400 Plus transmission electron microscope. For quantifying cristae, the number of cristae per mitochondria section area was counted using ImageJ. The fold change with SD was calculated on three mice, with six mitochondria per genotype.
RNA quantification
Samples for RNA quantification were homogenized with an Ultra-Turrax T8 (IKA-Analysentechnik) homogenizer, and RNA was isolated with a RNeasy kit (Qiagen, Hilden, Germany). After normalization of the RNA concentrations, DNase treatment (Promega, Madison, WI) and cDNA synthesis (Promega) were performed. Real-time (RT) PCR analysis by a Taqman 7900HT (Applied Biosystems, Foster City, CA) was done for HMGCS2 with forward and reverse primers (forward, CCGAGTGTCCAAGGATGC; reverse, TGGGCAGATCTGACACACTAGA; Oligomer, Helsinki, Finland) and probes (#80; Universal Probe Library, Roche, Basel, Switzerland). Actin was used as an endogenous control. Fold change differences were analyzed with RQ Manager (Applied Biosystems) and Excel (Microsoft, Redmond, WA).
Profiling of fecal SCFA and bacterial levels
Approximately 200 mg of fresh mouse stool was collected immediately after defecation from K8+/+ and K8−/− mice (three per genotype) and profiled for SCFA by gas chromatography (Alimetrics, Espoo, Finland) and for a panel of butyrate-producing and other bacteria by quantitative PCR (Alimetrics).
K8 and K18 knockdown in HT-29 and Caco-2 cells
HT-29 colon cancer cells were transfected for 8 h and overnight for 24 h with Pre-design Chimera RNAi (Abnova, Taipei, Taiwan) designed for KRT8 (H00003856-R01), KRT18 (H00003875-R01), and scrambled negative control Naito1 (R0017). Transfections of 30% confluent HT-29 cells were performed with 400 nM RNA interference in Lipofectamine2000 (Invitrogen, Carlsbad, CA) and diluted in Opti-MEM medium (Gibco). Cells were harvested after 72 h.
Caco-2 cells were transfected with siMAX siRNA (Eurofins Genomics, Ebersberg, Germany) designed for KRT8 (5′-GCCUCCUUCAUAGACAAGGUA(dTdT)-3′). The sequence for the K8 siRNA (clone ID TRCN0000062384) was obtained from the TRC Library Database, and nontarget (scr) siRNA (5′-CCUACAUCCCGAUCGAUGAUG(dTdT)-3′) was used as a control. Transfections of 20–30% confluent Caco-2 cells were performed with 50 pmol of siRNA in Lipofectamine2000 and diluted in Opti-MEM. Cells were harvested after 72 h.
Statistical analysis
Statistical calculations were performed using Excel and GraphPad Prism with Student's t test.
Supplementary Material
Acknowledgments
We thank Catharina Alam (Åbo Akademi University, Turku, Finland) for help with statistical analysis; Helena Saarento, Mary Mohlin, Lijiao Yang, Jolanta Lundgren, Bianca Forsell, Cecilia Antman, and Carl-Gustaf Stenvall (Åbo Akademi University), Kaija Ollikainen (Stanford University, Stanford, CA), the personnel at the Central Animal Laboratory, Turku Centre for Disease Modeling, and the Cell Imaging Core at the Turku Centre for Biotechnology, and members of the Toivola lab for skillful technical assistance; Héléne Baribault (Amgen, South San Francisco, CA) for providing the K8−/− mouse strain; J. Peter Slotte (Åbo Akademi University) for discussions on enzyme activity measurements; and Margaretha Gustafsson (Åbo Akademi University) for advice and assistance in analyzing the mitochondrial ultrastructure. This work was supported by the Academy of Finland, Sigrid Juselius Foundation, Stiftelsen Liv och Hälsa, EU FP7 Marie Curie IRG, and Åbo Akademi University Center of Excellence on Cell Stress and Molecular Aging (D.T.); Victoriastiftelsen (T.H.); the Turku Doctoral Programme for Biomedical Sciences (J.M.); Svenska Kulturfonden; makarna Agneta och Carl-Erik Olins fond (J.N.); National Institutes of Health Grant DK47918 and the Department of Veterans Affairs (M.B.O.); and the Department of Veterans Affairs Office of Research and Development Medical Service and National Institutes of Health Grant HL92473 (S.A.).
Abbreviations used:
- ACC
acetyl CoA carboxylase
- AMPK
adenosine monophosphate–activated protein kinase
- COX IV
cytochrome c oxidase
- HMGCS2
mitochondrial 3-hydroxy-3-methylglutaryl CoA synthase 2
- IF
intermediate filament
- K
keratin
- K8+/+
keratin 8 wild type
- K8−/−
keratin 8-knockout
- MCT1
monocarboxylate transporter 1
- PPARα
peroxisome proliferator–activated receptor α.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E14-02-0736) on April 22, 2015.
*These authors contributed equally and should be regarded as joint first authors.
REFERENCES
Boldface names denote co–first authors.
- Alam CM, Silvander JS, Daniel EN, Tao GZ, Kvarnstrom SM, Alam P, Omary MB, Hanninen A, Toivola DM. Keratin 8 modulates beta-cell stress responses and normoglycaemia. J Cell Sci. 2013;126:5635–5644. doi: 10.1242/jcs.132795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aledo R, Mir C, Dalton RN, Turner C, Pie J, Hegardt FG, Casals N, Champion MP. Refining the diagnosis of mitochondrial HMG-CoA synthase deficiency. J Inherit Metab Dis. 2006;29:207–211. doi: 10.1007/s10545-006-0214-2. [DOI] [PubMed] [Google Scholar]
- Ameen NA, Figueroa Y, Salas PJ. Anomalous apical plasma membrane phenotype in CK8-deficient mice indicates a novel role for intermediate filaments in the polarization of simple epithelia. J Cell Sci. 2001;114:563–575. doi: 10.1242/jcs.114.3.563. [DOI] [PubMed] [Google Scholar]
- Ardawi MS, Majzoub MF. Glutamine and ketone-body metabolism in the small intestine of starved peak-lactating rats. Biochimie. 1988;70:749–755. doi: 10.1016/0300-9084(88)90104-6. [DOI] [PubMed] [Google Scholar]
- Baribault H, Penner J, Iozzo RV, Wilson-Heiner M. Colorectal hyperplasia and inflammation in keratin 8-deficient FVB/N mice. Genes Dev. 1994;8:2964–2973. doi: 10.1101/gad.8.24.2964. [DOI] [PubMed] [Google Scholar]
- Borthakur A, Priyamvada S, Kumar A, Natarajan AA, Gill RK, Alrefai WA, Dudeja PK. A novel nutrient sensing mechanism underlies substrate-induced regulation of monocarboxylate transporter-1. Am J Physiol Gastrointest Liver Physiol. 2012;303:G1126–1133. doi: 10.1152/ajpgi.00308.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bultman SJ. Molecular pathways: gene-environment interactions regulating dietary fiber induction of proliferation and apoptosis via butyrate for cancer prevention. Clin Cancer Res. 2014;20:799–803. doi: 10.1158/1078-0432.CCR-13-2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camarero N, Mascaro C, Mayordomo C, Vilardell F, Haro D, Marrero PF. Ketogenic HMGCS2 Is a c-Myc target gene expressed in differentiated cells of human colonic epithelium and down-regulated in colon cancer. Mol Cancer Res. 2006;4:645–653. doi: 10.1158/1541-7786.MCR-05-0267. [DOI] [PubMed] [Google Scholar]
- Cherbuy C, Andrieux C, Honvo-Houeto E, Thomas M, Ide C, Druesne N, Chaumontet C, Darcy-Vrillon B, Duee PH. Expression of mitochondrial HMGCoA synthase and glutaminase in the colonic mucosa is modulated by bacterial species. Eur J Biochem. 2004;271:87–95. doi: 10.1046/j.1432-1033.2003.03908.x. [DOI] [PubMed] [Google Scholar]
- Coulombe PA, Omary MB. “Hard” and “soft” principles defining the structure, function and regulation of keratin intermediate filaments. Curr Opin Cell Biol. 2002;14:110–122. doi: 10.1016/s0955-0674(01)00301-5. [DOI] [PubMed] [Google Scholar]
- Cuff MA, Lambert DW, Shirazi-Beechey SP. Substrate-induced regulation of the human colonic monocarboxylate transporter, MCT1. J Physiol. 2002;539:361–371. doi: 10.1113/jphysiol.2001.014241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullingford TE. The ketogenic diet; fatty acids, fatty acid-activated receptors and neurological disorders. Prostaglandins Leukot Essent Fatty Acids. 2004;70:253–264. doi: 10.1016/j.plefa.2003.09.008. [DOI] [PubMed] [Google Scholar]
- Donohoe DR, Garge N, Zhang X, Sun W, O'Connell TM, Bunger MK, Bultman SJ. The microbiome and butyrate regulate energy metabolism and autophagy in the mammalian colon. Cell Metab. 2011;13:517–526. doi: 10.1016/j.cmet.2011.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake PJ, Griffiths GJ, Shaw L, Benson RP, Corfe BM. Application of high-content analysis to the study of post-translational modifications of the cytoskeleton. J Proteome Res. 2009;8:28–34. doi: 10.1021/pr8006396. [DOI] [PubMed] [Google Scholar]
- Fountoulakis M, Soumaka E, Rapti K, Mavroidis M, Tsangaris G, Maris A, Weisleder N, Capetanaki Y. Alterations in the heart mitochondrial proteome in a desmin null heart failure model. J Mol Cell Cardiol. 2005;38:461–474. doi: 10.1016/j.yjmcc.2004.12.008. [DOI] [PubMed] [Google Scholar]
- Guzman M, Velasco G, Geelen MJ. Do cytoskeletal components control fatty acid translocation into liver mitochondria. Trends Endocrinol Metab. 2000;11:49–53. doi: 10.1016/s1043-2760(99)00223-4. [DOI] [PubMed] [Google Scholar]
- Habtezion A, Toivola DM, Asghar MN, Kronmal GS, Brooks JD, Butcher EC, Omary MB. Absence of keratin 8 confers a paradoxical microflora-dependent resistance to apoptosis in the colon. Proc Natl Acad Sci USA. 2011;108:1445–1450. doi: 10.1073/pnas.1010833108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habtezion A, Toivola DM, Butcher EC, Omary MB. Keratin-8-deficient mice develop chronic spontaneous Th2 colitis amenable to antibiotic treatment. J Cell Sci. 2005;118:1971–1980. doi: 10.1242/jcs.02316. [DOI] [PubMed] [Google Scholar]
- Hadjiagapiou C, Schmidt L, Dudeja PK, Layden TJ, Ramaswamy K. Mechanism(s) of butyrate transport in Caco-2 cells: role of monocarboxylate transporter 1. Am J Physiol Gastrointest Liver Physiol. 2000;279:G775–780. doi: 10.1152/ajpgi.2000.279.4.G775. [DOI] [PubMed] [Google Scholar]
- Hamer HM, Jonkers D, Venema K, Vanhoutvin S, Troost FJ, Brummer RJ. Review article: the role of butyrate on colonic function. Aliment Pharmacol Ther. 2008;27:104–119. doi: 10.1111/j.1365-2036.2007.03562.x. [DOI] [PubMed] [Google Scholar]
- Hegardt FG. Mitochondrial 3-hydroxy-3-methylglutaryl-CoA synthase: a control enzyme in ketogenesis. Biochem J. 1999;338:569–582. [PMC free article] [PubMed] [Google Scholar]
- Kersten S, Seydoux J, Peters JM, Gonzalez FJ, Desvergne B, Wahli W. Peroxisome proliferator-activated receptor alpha mediates the adaptive response to fasting. J Clin Invest. 1999;103:1489–1498. doi: 10.1172/JCI6223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan AQ, Bury JP, Brown SR, Riley SA, Corfe BM. Keratin 8 expression in colon cancer associates with low faecal butyrate levels. BMC Gastroenterol. 2011;11:2. doi: 10.1186/1471-230X-11-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilner J, Waby JS, Chowdry J, Khan AQ, Noirel J, Wright PC, Corfe BM, Evans CA. A proteomic analysis of differential cellular responses to the short-chain fatty acids butyrate, valerate and propionate in colon epithelial cancer cells. Mol Biosystems. 2012;8:1146–1156. doi: 10.1039/c1mb05219e. [DOI] [PubMed] [Google Scholar]
- König B, Koch A, Giggel K, Dordschbal B, Eder K, Stangl GI. Monocarboxylate transporter (MCT)-1 is up-regulated by PPARalpha. Biochim Biophys Acta. 2008;6:899–904. doi: 10.1016/j.bbagen.2008.03.002. [DOI] [PubMed] [Google Scholar]
- Ku NO, Toivola DM, Zhou Q, Tao GZ, Zhong B, Omary MB. Studying simple epithelial keratins in cells and tissues. Methods Cell Biol. 2004;78:489–517. doi: 10.1016/s0091-679x(04)78017-6. [DOI] [PubMed] [Google Scholar]
- Lefebvre P, Chinetti G, Fruchart JC, Staels B. Sorting out the roles of PPAR alpha in energy metabolism and vascular homeostasis. J Clin Invest. 2006;116:571–580. doi: 10.1172/JCI27989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopaschuk GD, Ussher JR, Folmes CD, Jaswal JS, Stanley WC. Myocardial fatty acid metabolism in health and disease. Physiol Rev. 2010;90:207–258. doi: 10.1152/physrev.00015.2009. [DOI] [PubMed] [Google Scholar]
- McGarry JD, Brown NF. The mitochondrial carnitine palmitoyltransferase system. From concept to molecular analysis. Eur J Biochem. 1997;244:1–14. doi: 10.1111/j.1432-1033.1997.00001.x. [DOI] [PubMed] [Google Scholar]
- Meertens LM, Miyata KS, Cechetto JD, Rachubinski RA, Capone JP. A mitochondrial ketogenic enzyme regulates its gene expression by association with the nuclear hormone receptor PPARalpha. EMBO J. 1998;17:6972–6978. doi: 10.1093/emboj/17.23.6972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naito Y, Takagi T, Okada H, Omatsu T, Mizushima K, Handa O, Kokura S, Ichikawa H, Fujiwake H, Yoshikawa T. Identification of inflammation-related proteins in a murine colitis model by 2D fluorescence difference gel electrophoresis and mass spectrometry. J Gastroenterol Hepatol. 2010;25(Suppl 1):S144–S148. doi: 10.1111/j.1440-1746.2009.06219.x. [DOI] [PubMed] [Google Scholar]
- Nonn L, Williams RR, Erickson RP, Powis G. The absence of mitochondrial thioredoxin 2 causes massive apoptosis, exencephaly, and early embryonic lethality in homozygous mice. Mol Cell Biol. 2003;23:916–922. doi: 10.1128/MCB.23.3.916-922.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omary MB, Ku NO, Strnad P, Hanada S. Toward unraveling the complexity of simple epithelial keratins in human disease. J Clin Invest. 2009;119:1794–1805. doi: 10.1172/JCI37762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X, Hobbs RP, Coulombe PA. The expanding significance of keratin intermediate filaments in normal and diseased epithelia. Curr Opin Cell Biol. 2013;25:47–56. doi: 10.1016/j.ceb.2012.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel VB, Spencer CH, Young TA, Lively MO, Cunningham CC. Effects of 4-hydroxynonenal on mitochondrial 3-hydroxy-3-methylglutaryl (HMG-CoA) synthase. Free Radic Biol Med. 2007;43:1499–1507. doi: 10.1016/j.freeradbiomed.2007.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perreault N, Beaulieu JF. Primary cultures of fully differentiated and pure human intestinal epithelial cells. Exp Cell Res. 1998;245:34–42. doi: 10.1006/excr.1998.4221. [DOI] [PubMed] [Google Scholar]
- Pryde SE, Duncan SH, Hold GL, Stewart CS, Flint HJ. The microbiology of butyrate formation in the human colon. FEMS Microbiol Lett. 2002;217:133–139. doi: 10.1111/j.1574-6968.2002.tb11467.x. [DOI] [PubMed] [Google Scholar]
- Quinlan RA, Cohlberg JA, Schiller DL, Hatzfeld M, Franke WW. Heterotypic tetramer (A2D2) complexes of non-epidermal keratins isolated from cytoskeletons of rat hepatocytes and hepatoma cells. J Mol Biol. 1984;178:365–388. doi: 10.1016/0022-2836(84)90149-9. [DOI] [PubMed] [Google Scholar]
- Rodriguez JC, Gil-Gomez G, Hegardt FG, Haro D. Peroxisome proliferator-activated receptor mediates induction of the mitochondrial 3-hydroxy-3-methylglutaryl-CoA synthase gene by fatty acids. J Biol Chem. 1994;269:18767–18772. [PubMed] [Google Scholar]
- Roediger WE. Role of anaerobic bacteria in the metabolic welfare of the colonic mucosa in man. Gut. 1980;21:793–798. doi: 10.1136/gut.21.9.793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roediger WE. The starved colon–diminished mucosal nutrition, diminished absorption, and colitis. Dis Colon Rectum. 1990;33:858–862. doi: 10.1007/BF02051922. [DOI] [PubMed] [Google Scholar]
- Schweizer J, Bowden PE, Coulombe PA, Langbein L, Lane EB, Magin TM, Maltais L, Omary MB, Parry DA, Rogers MA, Wright MW. New consensus nomenclature for mammalian keratins. J Cell Biol. 2006;174:169–174. doi: 10.1083/jcb.200603161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serra D, Bellido D, Asins G, Arias G, Vilaro S, Hegardt FG. The expression of mitochondrial 3-hydroxy-3-methylglutaryl-coenzyme-A synthase in neonatal rat intestine and liver is under transcriptional control. Eur J Biochem. 1996a;237:16–24. doi: 10.1111/j.1432-1033.1996.0016n.x. [DOI] [PubMed] [Google Scholar]
- Serra D, Fillat C, Matas R, Bosch F, Hegardt FG. Tissue-specific expression and dietary regulation of chimeric mitochondrial 3-hydroxy-3-methylglutaryl coenzyme A synthase/human growth hormone gene in transgenic mice. J Biol Chem. 1996b;271:7529–7534. doi: 10.1074/jbc.271.13.7529. [DOI] [PubMed] [Google Scholar]
- Shen WJ, Zaidi SK, Patel S, Cortez Y, Ueno M, Azhar R, Azhar S, Kraemer FB. Ablation of vimentin results in defective steroidogenesis. Endocrinology. 2012;153:3249–3257. doi: 10.1210/en.2012-1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao GZ, Looi KS, Toivola DM, Strnad P, Zhou Q, Liao J, Wei Y, Habtezion A, Omary MB. Keratins modulate the shape and function of hepatocyte mitochondria: a mechanism for protection from apoptosis. J Cell Sci. 2009;122:3851–3855. doi: 10.1242/jcs.051862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toivola DM, Krishnan S, Binder HJ, Singh SK, Omary MB. Keratins modulate colonocyte electrolyte transport via protein mistargeting. J Cell Biol. 2004;164:911–921. doi: 10.1083/jcb.200308103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toivola DM, Strnad P, Habtezion A, Omary MB. Intermediate filaments take the heat as stress proteins. Trends Cell Biol. 2010;20:79–91. doi: 10.1016/j.tcb.2009.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topping DL, Clifton PM. Short-chain fatty acids and human colonic function: roles of resistant starch and nonstarch polysaccharides. Physiol Rev. 2001;81:1031–1064. doi: 10.1152/physrev.2001.81.3.1031. [DOI] [PubMed] [Google Scholar]
- Velasco G, Geelen MJ, Gomez del Pulgar T, Guzman M. Malonyl-CoA-independent acute control of hepatic carnitine palmitoyltransferase I activity. Role of Ca2+/calmodulin-dependent protein kinase II and cytoskeletal components. J Biol Chem. 1998;273:21497–21504. doi: 10.1074/jbc.273.34.21497. [DOI] [PubMed] [Google Scholar]
- Vijayaraj P, Kroger C, Reuter U, Windoffer R, Leube RE, Magin TM. Keratins regulate protein biosynthesis through localization of GLUT1 and -3 upstream of AMP kinase and Raptor. J Cell Biol. 2009;187:175–184. doi: 10.1083/jcb.200906094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirtz S, Neufert C, Weigmann B, Neurath MF. Chemically induced mouse models of intestinal inflammation. Nat Protoc. 2007;2:541–546. doi: 10.1038/nprot.2007.41. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.