Abstract
Cell migration is a cellular response and results in various biological processes such as cancer metastasis, that is, the primary cause of death for cancer patients. Quantitative investigation of the correlation between cell migration and extracellular stimulation is essential for developing effective therapeutic strategies for controlling invasive cancer cells. The conventional method to determine cell migration rate based on comparison of successive images may not be an objective approach. In this work, a microfluidic chip embedded with measurement electrodes has been developed to quantitatively monitor the cell migration activity based on the impedimetric measurement technique. A no-damage wound was constructed by microfluidic phenomenon and cell migration activity under the stimulation of cytokine and an anti-cancer drug, i.e., interleukin-6 and doxorubicin, were, respectively, investigated. Impedance measurement was concurrently performed during the cell migration process. The impedance change was directly correlated to the cell migration activity; therefore, the migration rate could be calculated. In addition, a good match was found between impedance measurement and conventional imaging analysis. But the impedimetric measurement technique provides an objective and quantitative measurement. Based on our technique, cell migration rates were calculated to be 8.5, 19.1, and 34.9 μm/h under the stimulation of cytokine at concentrations of 0 (control), 5, and 10 ng/ml. This technique has high potential to be developed into a powerful analytical platform for cancer research.
I. INTRODUCTION
Cell migration is a cellular response after a highly integrated multistep molecular process.1,2 It results various biological processes such as orchestration of embryonic morphogenesis, cancer metastasis, tissue repair and regeneration, wound healing, and inflammation.3–7 Because cells can be stimulated to promote or inhibit cell migration in response to particular factors, study of cell migration is critical for understanding the underlying molecular mechanisms. For example, increase of interleukin-6 (IL-6) cytokine can promote cancer cell migration and cancer metastasis.8–10 When IL-6 engages its receptor in cells, STAT3 transcription factor is activated and regulates the target genes.11 STAT3 controls a wide range of cellular processes including cell proliferation, oncogenesis, and cancer metastasis. On the other hand, an anti-cancer drug, e.g., doxorubicin, can inhibit cancer cell migration and induce cell apoptosis.12,13 Doxorubicin is an effective anti-cancer drug used to treat a wide range of cancers; but serious side effects are also induced.14 Control of the dosage of doxorubicin is important in cancer chemotherapy. Knowledge of the correlation between cell migration and the extracellular stimulation, e.g., cytokine and anti-cancer drugs, is essential for understanding the cancer pathogenesis and thus developing effective therapeutic strategies for controlling invasive cancer cells.
In biological laboratory, one of the widely used assays for the study of cell migration is scratched wound healing assay.15,16 This assay is initiated by physically introducing a wound by scratching cells in a confluent monolayer. Cells migrate into the artificially generated space to close this wound. This conventional assay has the advantage of simplicity; however, it also has inherent limitations. Physical scratching by razor blades or scalpels often damages the cells on the wound edges. For the observation of cell migration activity, cells near the wound edges are very important and such uncertain cell damage may induce influence of the assay. Therefore, alternative method of creating wound edges is necessary for the investigation of cell migration. In the past decades, microfluidic systems have been rapidly developed and various biomedical applications have been implemented to these miniaturized systems.17,18 One of the key features of microfluidic systems is that fluid flow in microchannels exhibits laminar flow. Multiple parallel fluid streams can be created and this feature was first utilized to pattern cells and their environments.19 Based on this development, constructing a wound edge in a cultured cell monolayer using microfluidic technique has been reported to study the cell migration.20–22 The wound edge was constructed by partially digesting a confluent cell monolayer inside a microchannel using multiple laminar flows with and without trypsin. Previous studies showed that the effect of the shear stress on the cells can be ignored due to the slow flow and no-damage wound in microchannels can be prepared by this technique.20,21 By capturing and comparing the images at the beginning and at regular intervals during cell migration, cell migration rate can be estimated. However, comparison of images to determine migration rate may not be an objective method because the leading edge becomes blur after a certain time and observation is influenced by different human perceptions. To tackle the technical hurdles, in this work, electrical impedimetric monitoring of cell migration activity in a microfluidic system is proposed to conduct a real-time, non-invasive, objective, and quantitative investigation.
Impedance measurement across a pair of coplanar electrodes has been widely demonstrated to quantify cellular activities.23,24 Because cellular membrane has a membrane capacitance of 0.5–1.3 μF/cm2 and a membrane resistance of 102–105 Ω/cm2,25 change of cellular behaviors can be monitored by the change of total impedance across the electrodes. For example, if cells adhere and proliferate on the electrode surface, the effective surface area of the electrode is reduced and thus the total impedance increased. Conversely, cell death leads to the release of cells from the electrode surface, which induces the decrease of the total impedance. Based on this rationale, cell proliferation and viability were, respectively, monitored by the change of total impedance.26,27 It showed that cells do not behave significant change in their viability, motility, anchorage, or cell-cycle time when exposed to electric field generated by coplanar electrodes.28 Moreover, a microfluidic device was developed for trapping and measuring the dielectric properties of microtumors over time using electrical impedance spectroscopy.29 These excellent demonstrations showed that the impedance measurement provides a convenient and reliable technique for real-time monitoring of cellular activity.
In this work, we have implemented the impedance measurement scheme in a microfluidic chip to investigate cell migration activity. Influence of cell migration rate under the respective stimulation of cytokine and an anti-cancer drug, i.e., IL-6 and doxorubicin, was demonstrated to quantitatively study the correlation between the extracellular stimulation and cell migration. A microfluidic chip with a Y-shaped microchannel was developed and 16 electrodes were embedded along the microchannel for the impedance measurement. Nasopharyngeal carcinoma (NPC) cell line (NPC-BM1) derived from a bone marrow metastatic lesion was used because it is highly metastatic. After a cell monolayer was cultured in the microchannel, a wound edge was constructed by partially digesting the monolayer using multiple laminar flows with and without trypsin. Quantitative impedimetric monitoring of cell migration was demonstrated under the respective stimulation of cytokine and an anti-cancer drug. A good correlation was found between impedance measurement and conventional imaging analysis. In conclusion, this technique provides an efficient, objective, and quantitative measurement to study the correlation between the cell migration activity and the extracellular stimulation.
II. MATERIALS AND METHODS
A. Cell culture
NPC cell line (NPC-BM1) derived from a bone marrow metastatic lesion was kindly provided by Dr. Jenny Liu, Chang Gung University. Culture medium was Dulbecco's modified eagle medium (DMEM; Invitrogen, USA) supplemented with 10% fetal bovine serum (FBS; Invitrogen, USA) and antibiotic/antimycotic (100 U/ml of penicillin G sodium, 100 mg/ml of streptomycin, and 0.25 mg/ml of amphotericin B; Gibco-BRL Life Technologies, USA). Cells were amplified by standard cell culture technique and trypsinized using 0.05% trypsin (Gibco-BRL Life Technologies, USA) for 3 min, centrifuged at 1200 rpm for 5 min, and re-suspended in the medium for further experiments.
B. Design and fabrication of the microfluidic system
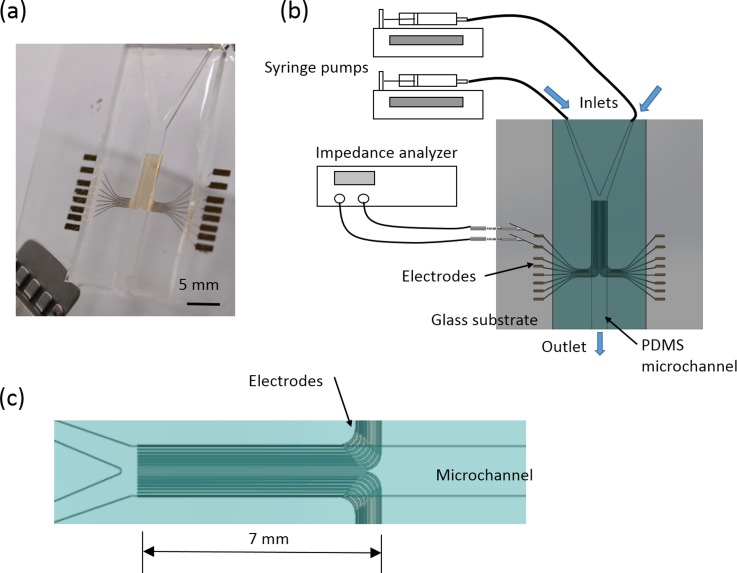
A microfluidic chip has been developed for the quantitative study of the correlation between the cell migration activity and the extracellular stimulation. The chip consisted of a polydimethylsiloxane (PDMS) layer with a Y-shaped microchannel and a glass substrate embedded with 16 electrodes. The electrodes were aligned along the microchannel and used for the impedance measurement, as shown in Fig. 1(a). The dimension of the mainstream was 25 mm in length, 3 mm in width, and 500 μm in height. The width of each electrode was 100 μm and the gap between electrodes was 100 μm. The total length of the electrodes was 7 mm and the electrodes were aligned along the microchannel, as shown in Fig. 1(c). In the impedance measurement, cells attach on the electrode surface and the total impedance across a pair of electrodes increases to determine the presence of cells. In this study, cells migrated across the microchannel and the impedance change of each pair of electrodes represented the location and number of cells. Cell migration activity could be quantitatively monitored by, respectively, measuring each pair of electrodes. Such that cell migration rate was calculated by continuous impedance measurement of each pair of electrodes at different fixed distances. Illustration of the experimental setup is shown in Fig. 1(b). Therefore, the correlation between the cell migration activity and the extracellular stimulation (in this case, cytokine and an anti-cancer drug) could be objectively studied. Such design of the microfluidic chip provides well-defined environment for quantitative study of cell migration activity.
FIG. 1.
Microfluidic chip for quantitative impedimetric monitoring of cell migration activity. (a) Photographic image of the microfluidic chip. (b) Schematic illustration of the experiment setup. (c) Illustration of the electrode structure inside the microchannel.
The microfluidic chip was composed of a PDMS (Model: Sylgard® 184; Dow Corning, USA) layer with microchannel and a glass substrate embedded with electrodes. The PDMS layer was fabricated by soft lithography. Briefly, a poly(methyl methacrylate) (PMMA) mold with a negative pattern of microchannels was machined by a CNC engraving machine (Model: EGX-400; Roland, USA). Next, PDMS pre-polymer and curing agents were mixed at 10:1 ratio under the instructions from the manufacturer. After being stirred thoroughly and degassed in a vacuum chamber, the prepared PDMS mixture was poured on the mold and cured at 70 °C for 1 h. The cured PDMS layer was peeled off from the mold and ready for use. For the fabrication of the Cr/Au (200 Å/1000 Å) electrodes on the glass substrate, a standard micro-fabrication process including thermal evaporation and lift-off, respectively, was performed. Finally, the microfluidic chip was assembled by bonding two layers together using oxygen plasma treatment. Because the bonding of the PDMS layer and glass substrate was manually handled, electrodes on most sides were used for the alignment of the sidewalls of the microchannel. Before performing the experiment, the microfluidic chip was sterilized using 70% (w/v) ethanol for 6 h, washed by phosphate-buffered saline (PBS), and kept under ultraviolet light overnight.
C. Experimental procedures
In order to promote cell adhesion to the surface in the microchannel, solution with collagen in the concentration of 1 μg/ml was introduced to the microchannel and incubated for 30 min. Collagen-coated surface was created after washing by PBS (50 mM phosphate, 150 mM NaCl, and 10 mM EDTA; pH 7.6). Cells suspended solution with cell concentration of 5 × 107 cells/ml were then applied to the microchannel. Cells spread and formed a monolayer on the bottom surface of the microchannel after 1 day culture in a 37 °C, humidified atmosphere, and 5% CO2 incubator (Model: 370; Thermoscientific, USA). Then, a wound edge was constructed by partially digesting the cell monolayer. Culture mediums with and without 0.25% trypsin were simultaneously applied from two inlets of the microfluidic chip at the flow rate of 0.08 ml/min for 10 min and then 0.15 ml/min until cells that were in contact with trypsin were washed away under the observation using an upright microscope (Model: BX51M; Olympus, Japan). Because of low Reynolds number, the flow was laminar flow and two parallel fluid streams were created. Cells that were in contact with trypsin were trypsinized and detached from the surface. Next, the microchannel was washed by applying medium at the flow rate of 0.15 ml/min. However, in some situations, cells without contacting with trypsin were also washed away because of shear force. In order to confirm the initial cell concentration before cell migration experiment, electrodes on the portion of without trypsin treatment were used for the impedance measurement to estimate the cell concentration. If the impedance value was in the acceptable range (in this case, >600 Ω at the measurement frequency of 20 kHz), the wound edge was successfully constructed. Then, the entire microfluidic chip was placed in the incubator during the cell migration experiment. In this study, investigation of cell migration without extracellular stimulation was first carried out for up to 6 days. The result of cell migration rate was defined as control to compare with the results under the stimulation of cytokine and an anti-cancer drug, i.e., IL-6 (Invitrogen, USA) and doxorubicin (Sigma, USA), in different concentrations. For these experiments, impedimetric monitoring was concurrently conducted by using an impedance analyzer (Model: VersaSTAT 4; Princeton Applied Research, USA) to provide objective and quantitative results in a real-time and non-invasive manner.
D. Impedance measurement of cell migration activity
Impedance measurement was performed every 12 h during the cell migration activity. In order to confirm the conditions of temperature and pH of the medium, fresh 37 °C medium was applied to the microchannel before each measurement. Potential of 0.1 Vrms was applied across a pair of electrodes, and the impedance was measured from 50 Hz to 1 MHz. Only seven electrodes on the portion of treating with trypsin, i.e., half of the microchannel, were used for the impedance measurement during the cell migration activity. They were, respectively, located at 0, 200, 400, 600, 800, 1000, and 1200 μm to the center line of the microchannel. Six measurements were, respectively, conducted by six pairs of electrodes located at 0 and 200 μm, 200 and 400 μm, 400 and 600 μm, 600 and 800 μm, 800 and 1000 μm, and 1000 and 1200 μm. Therefore, six impedance values were measured for each successive time point. In order to eliminate the variation between electrodes, background impedance value of each pair of electrodes was also measured at the beginning of the cell migration experiment, i.e., impedance value at 0 h. Impedance change was defined as the subtraction of the impedance value at the successive time point and the background impedance value.
III. RESULTS AND DISCUSSION
A. Optimization of the impedance measurement conditions
Impedance measurement has been widely used for monitoring cellular behaviors such as cell proliferation and viability.23,24 Analysis of the sensitivity and frequency characteristics of coplanar electrodes for monitoring cells was discussed, in detail.30 When cells attach onto the electrode surface, the total impedance across the electrodes can be modeled as capacitances and resistances in series, which reflects the biophysical properties of the insulating cell membrane. When the frequency is sufficiently low or high, sensitivity of the cell impedance measurement becomes extremely low. It was suggested that the most appropriate frequency for cell impedance measurement is 20 kHz, at which the sensor will be at the highest sensitivity.30
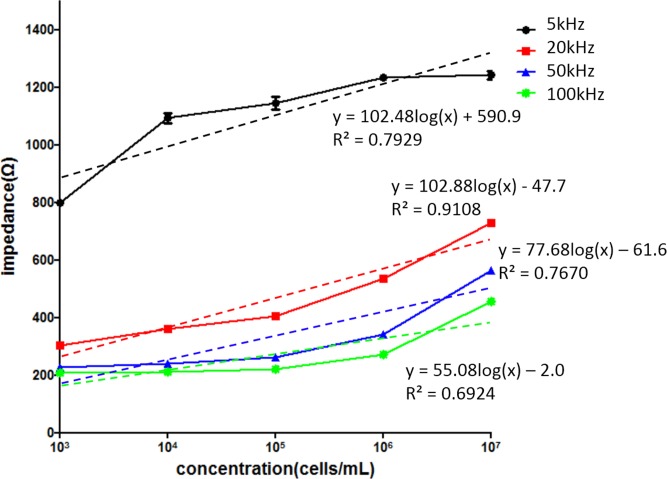
In our microfluidic chip, the optimized measurement frequency was investigated in order to find out the highest sensitivity and linear correlation between the impedance magnitude and cell concentration. Cells at different concentrations, i.e., 103, 104, 105, 106, and 107 cells/ml, were, respectively, introduced to the microchannel and incubated for 8 h for cell attachment before measurement. Then, impedance measurement was performed to investigate the electrical characteristics. Relationship between impedance magnitude and cell concentration under different measurement frequencies of 5, 20, 50, 100 kHz is shown in Fig. 2. The cell concentration was found to be proportional to the impedance magnitude at all measurement frequencies. However, the sensitivity and the linear correlation were different. The sensitivities were 102.48, 102.88, 77.68, and 55.08, respectively, and the R-squared values were 0.7929, 0.9108, 0.7670, and 0.6924, respectively. Measurement at 20 kHz showed the highest sensitivity and linear correlation to describe cell concentration using impedance magnitude, which is as the same as in the previous work.30 Therefore, measurement frequency of 20 kHz was used for the following experiments.
FIG. 2.
Relationship between impedance magnitude and cell concentration under different measurement frequencies of 5, 20, 50, and 100 kHz. Error bars represent the standard deviations of three repeated experiments.
B. Construction of a wound edge
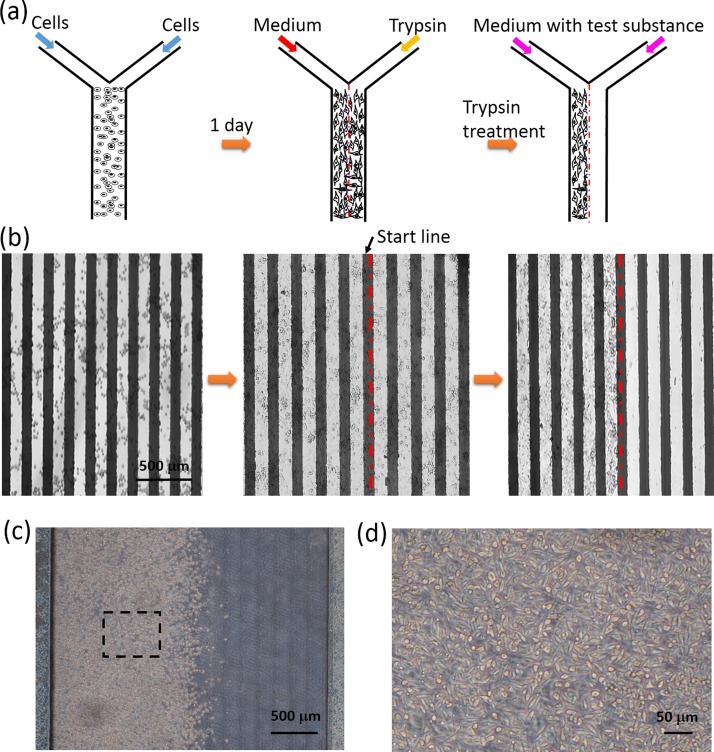
Before observing the cell migration activity, construction of a wound edge in the microchannel was performed and the procedures are illustrated in Fig. 3(a). Based on the formation of two parallel laminar flows in the microchannel, only cells that were in contact with trypsin were detached from the surface, while the untreated portion of the cell monolayer remained intact and spread to form a clear wound edge. Because the electrodes were not transparent, cells were observed using an upright microscope. A sequence of photographic images was captured and is shown in Fig. 3(b). Cells were shown to spread and form a monolayer on both glass and electrode surfaces after 1 day culture. After the treatment of trypsin, the resultant wound edge of cell monolayer was linear, and there was no cell remaining on the cell-denuded portion. A start line of cell migration could be defined on the wound edge. In addition, a microfluidic chip without embedding electrodes was used to repeat the experiment of constructing the wound edge. Such that the wound edge and the cell monolayer could be clearly observed using an inverted microscope. Photographic images are, respectively, shown in Figs. 3(c) and 3(d). The resultant wound edge was shown to be linear and cells achieved a good morphology and monolayer growth status.
FIG. 3.
Construction of a wound edge in the microchannel. (a) Schematic illustration of the procedures of constructing the wound edge based on the formation of two parallel laminar flows in the microchannel. (b) A sequence of photographic images showing the process of constructing the wound edge in the microchannel. The red dashed line represents the start line of the cell migration process. Images were captured under an upright microscope because the electrodes were not transparent. (c) Photographic image of the wound edge constructing in the microchannel without embedding electrodes. Images were captured under an inverted microscope. (d) Photographic image of the cell monolayer corresponding to the dashed line square in (c).
C. Impedimetric monitoring of cell migration
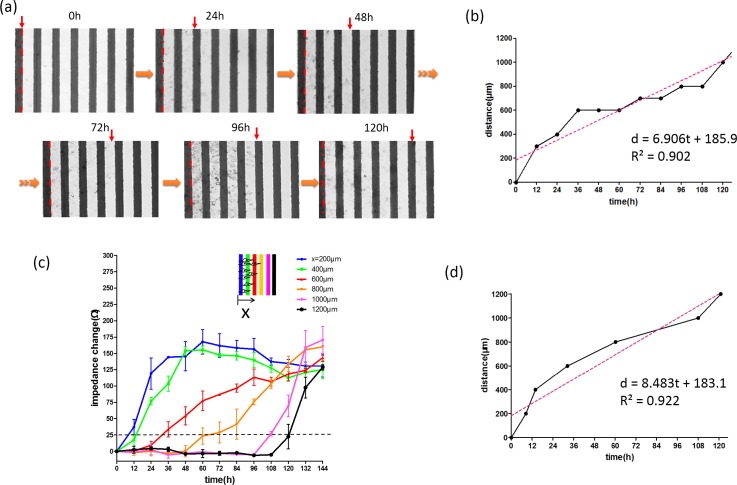
After the wound was obtained in the microchannel, cells were incubated with culture medium in the incubator and the cell migration activity started. For every 12 h during the cell migration process, photographic images were captured and impedance values across the electrodes were concurrently measured. Comparison of the successive images is conventionally used to determine the cell migration rate, as shown in Fig. 4(a). Distances of the leading edge to the start line at different successive time points were identified from the images and plotted in Fig. 4(b). Based on the linear regressive approximation, cell migration rate was estimated to be 6.9 μm/h. This conventional method is convenient but subjective. Determination of the distance was difficult because the leading edge became blur after a certain time. On the other hand, result of impedance measurements across six pairs of electrodes at different successive time points is shown in Fig. 4(c). Increase of impedance change represented increase of cells attaching on the electrode. Results quantitatively showed cells progressively migrated from the start line until the distance of 1200 μm. In order to calculate the cell migration rate, we defined the impedance change of 25 Ω as a threshold of cells reaching the electrodes. Because environmental noise may influence the impedance measurement, but we found that impedance change over 25 Ω indicated the presence of cells on the electrode surface in our microfluidic chip (data not shown). At this threshold, six time points for cells reaching six electrodes at the distances of 200, 400, 600, 800, 1000, and 1200 μm were calculated by linear interpolation. Relationship between the migration distance and time was plotted and is shown in Fig. 4(d). Based on the linear regressive approximation, cell migration rate was calculated to be 8.5 μm/h, which reasonably matches with the conventional method.
FIG. 4.
Analyses of cell migration activity. (a) Successive images of cell migration process. The red dashed line represents the start line of the cell migration process. The red arrow indicates the leading edge. (b) Relationship between the migration distance and time identified from the images is shown in (a). (c) Impedance measurements across six pairs of electrodes at distances of 200, 400, 600, 800, 1000, and 1200 μm at different successive time points. Error bars represent the standard deviations of three repeated experiments. (d) Relationship between the migration distance and time calculated from the impedance data is shown in (c).
D. Cell migration under the stimulation of cytokine and anti-cancer drug
In order to confirm that cell migration could be examined within such a microfluidic chip, analyses of cell migration under the stimulation of cytokine and an anti-cancer drug were, respectively, performed to either promote or inhibit cell migration. The migration ability of cancer cells represents the probability of cancer metastasis that is the primary cause of death for cancer patients. Hence, understanding the underlying molecular mechanisms is critical for the development of effective therapeutic strategies for controlling invasive cancer cells.
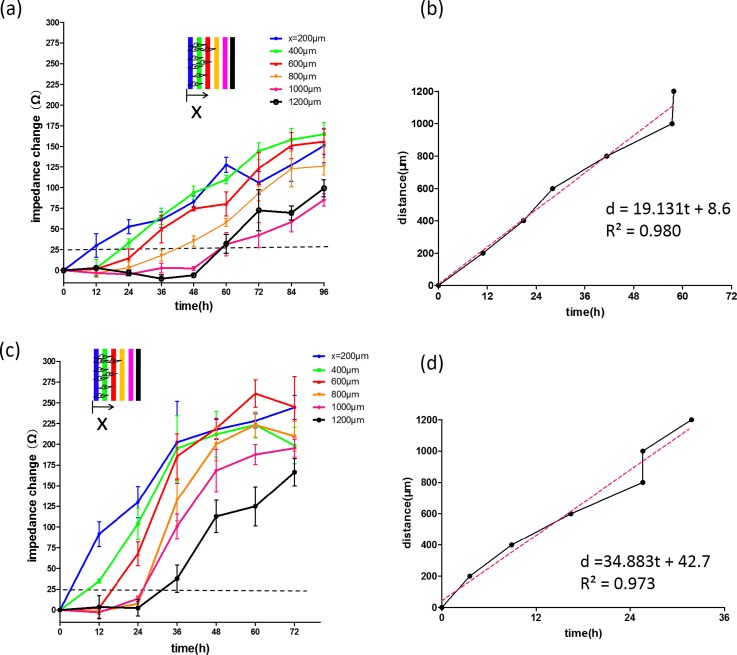
Studying the role of cytokine is one of the interesting topics of the investigation of cancer pathogenesis.31–33 It was reported that cells infected by Epstein-Barr virus lead to the oncogenic transformation34,35 and induce the production of cytokines such as IL-6 and IL-10.36,37 The production of IL-6 from virus-transformed cells affects the neighboring cells for aberrant cell proliferation and migration.8–10 In this work, cell migration rate under the stimulation of IL-6 at different concentrations of 5 and 10 ng/ml was quantitatively studied using the microfluidic chip and the results are shown in Fig. 5. Cells were stimulated by IL-6 and their migration ability was regulated. Based on our technique, cell migration rates were, respectively, calculated to be 19.1 and 34.9 μm/h, as shown in Figs. 5(b) and 5(d). Cell migration rate was directly related to the IL-6 concentration because higher activation level of the transcription factors was triggered by higher concentration of IL-6.38 Also, the invasiveness of cells was found to be significantly enhanced by IL-6 ranging from 0 to 10 ng/ml. This result was similar to those that had been reported previously using a conventional method.39 But our microfluidic chip could quantitatively monitor the entire cell migration process.
FIG. 5.
Impedimetric monitoring of cell migration under the stimulation of IL-6 cytokine at concentrations of (a) 5 and (c) 10 ng/ml. Error bars represent the standard deviations of three repeated experiments. (b) and (d) Relationship between the migration distance and time for the calculation of cell migration rate.
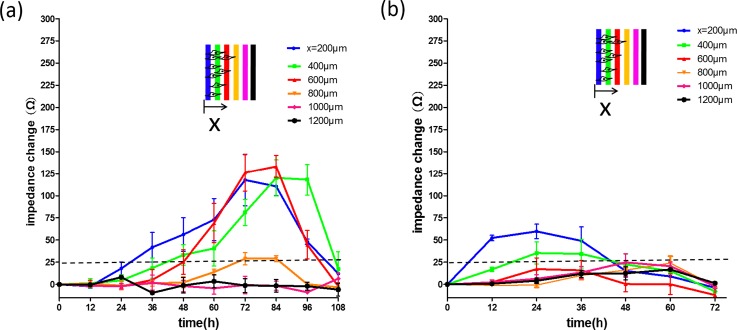
In cancer chemotherapy, the use of anti-cancer drugs can inhibit cell migration and induce cell apoptosis;12,13 but serious side effects are also induced by a high dosage of drugs.14 Because cell migration ability reflects the cell vitality to a certain extent, investigation of cell migration under the stimulation of anti-cancer drugs is essential to understand the trade-off between cell vitality and side effects. In this study, impedimetric monitoring of cell migration activity was investigated under the stimulation of doxorubicin at different concentrations of 0.5 and 1 μg/ml, as shown in Fig. 6. Results revealed the drug's effective time and the longest cell migration distance. Under the drug concentration of 0.5 μg/ml (Fig. 6(a)), cells migrated until 72 h and cell apoptosis was then induced. The longest migration distance was shown to be 800 μm. In addition, for the drug concentration of 1 μg/ml (Fig. 6(b)), cell apoptosis occurred after 24 h of drug application and the longest migration distance was 400 μm. Reliable assessment of cell vitality is very important for the development of effective and safe cancer chemotherapy. Our microfluidic chip provides quantitative results to investigate cell vitality under the stimulation of anti-cancer drugs.
FIG. 6.
Impedimetric monitoring of cell migration under the stimulation of doxorubicin at concentrations of (a) 0.5 and (b) 1 μg/ml. Error bars represent the standard deviations of three repeated experiments.
IV. CONCLUSION
Impedimetric monitoring of cell migration activity has been demonstrated in a microfluidic chip. Based on the microfluidic feature, a no-damage wound was constructed by partially digesting the monolayer using multiple laminar flows with and without trypsin. Quantitative correlations between cell migration activity and the extracellular stimulation, i.e., IL-6 and doxorubicin, were, respectively, investigated. The impedance change was directly correlated to the cell migration activity; therefore, migration rate could be calculated. In addition, a good match was found between impedance measurement and conventional imaging analysis. Results showed that this microfluidic chip provides a promising tool to study cell migration activity in a real-time, non-invasive, objective, and quantitative manner. This technique has high potential to be developed into a powerful analytical platform for cancer research.
ACKNOWLEDGMENTS
This work was partially supported by the Ministry of Science and Technology, Taiwan (Project No. MOST103-2221-E-182-004-MY3).
References
- 1. Ridley A. J., Schwartz M. A., Burridge K., Firtel R. A., Ginsberg M. H. et al. , “ Cell migration: Integrating signals from front to back,” Science 302, 1704–1709 (2003). 10.1126/science.1092053 [DOI] [PubMed] [Google Scholar]
- 2. Lauffenburger D. A. and Horwitz A. F., “ Cell migration: A physically integrated molecular process,” Cell 84, 359–369 (1996). 10.1016/S0092-8674(00)81280-5 [DOI] [PubMed] [Google Scholar]
- 3. Montell D. J., “ Morphogenetic cell movements: diversity from modular mechanical properties,” Science 322, 1502–1505 (2008). 10.1126/science.1164073 [DOI] [PubMed] [Google Scholar]
- 4. Liotta L. A., Steeg P. S., and Stetler-Stevenson W. G., “ Cancer metastasis and angiogenesis: An imbalance of positive and negative regulation,” Cell 64, 327–336 (1991). 10.1016/0092-8674(91)90642-C [DOI] [PubMed] [Google Scholar]
- 5. Karnoub A. E., Dash A. B., Vo A. P., Sullivan A., Brooks M. W. et al. , “ Mesenchymal stem cells within tumour stroma promote breast cancer metastasis,” Nature 449, 557–563 (2007). 10.1038/nature06188 [DOI] [PubMed] [Google Scholar]
- 6. Martin P. and Leibovich S. J., “ Inflammatory cells during wound repair: The good, the bad and the ugly,” Trends Cell Biol. 15, 599–607 (2005). 10.1016/j.tcb.2005.09.002 [DOI] [PubMed] [Google Scholar]
- 7. Stramer B., Wood W., Galko M. J., Redd M. J., Jacinto A. et al. , “ Live imaging of wound inflammation in Drosophila embryos reveals key roles for small GTPases during in vivo cell migration,” J. Cell Biol. 168, 567–573 (2005). 10.1083/jcb.200405120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Snyder M., Huang X. Y., and Zhang J. J., “ Signal transducers and activators of transcription 3 (STAT3) directly regulates cytokine-induced fascin expression and is required for breast cancer cell migration,” J. Biol. Chem. 286, 38886–38893 (2011). 10.1074/jbc.M111.286245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Graness A., Chwieralski C. E., Reinhold D., Thim L., and Hofmann W., “ Protein kinase C and ERK activation are required for TFF-peptide-stimulated bronchial epithelial cell migration and tumor necrosis factor-α-induced interleukin-6 (IL-6) and IL-8 secretion,” J. Biol. Chem. 277, 18440–18446 (2002). 10.1074/jbc.M200468200 [DOI] [PubMed] [Google Scholar]
- 10. McLoughlin R. M., Jenkins B. J., Grail D., Williams A. S., Fielding C. A. et al. , “ IL-6 trans-signaling via STAT3 directs T cell infiltration in acute inflammation,” Proc. Natl. Acad. Sci. U.S.A. 102, 9589–9594 (2005). 10.1073/pnas.0501794102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ogata A., Chauhan D., Teoh G., Treon S. P., Urashima M. et al. , “ IL-6 triggers cell growth via the ras-dependent mitogen-activated protein kinase cascade,” J. Immunol. 159, 2212–2221 (1997); available at http://www.jimmunol.org/content/159/5/2212. [PubMed] [Google Scholar]
- 12. Tanaka T., Yamaguchi J., Shoji K., and Nangaku M., “ Anthracycline inhibits recruitment of hypoxia-inducible transcription factors and suppresses tumor cell migration and cardiac angiogenic response in the host,” J. Biol. Chem. 287, 34866–34882 (2012). 10.1074/jbc.M112.374587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chen M. B., Wu X. Y., Gu J. H., Guo Q. T., Shen W. X., and Lu P. H., “ Activation of AMP-activated protein kinase contributes to doxorubicin-induced cell death and apoptosis in cultured myocardial H9c2 cells,” Cell Biochem. Biophys. 60, 311–322 (2011). 10.1007/s12013-011-9153-0 [DOI] [PubMed] [Google Scholar]
- 14. Tao R., Karliner J. S., Simonis U., Zheng J., Zhang J. et al. , “ Pyrroloquinoline quinone preserves mitochondrial function and prevents oxidative injury in adult rat cardiac myocytes,” Biochem. Biophys. Res. Commun. 363, 257–262 (2007). 10.1016/j.bbrc.2007.08.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Liang C. C., Park A. Y., and Guan J. L., “ In vitro scratch assay: A convenient and inexpensive method for analysis of cell migration in vitro,” Nat. Protoc. 2, 329–333 (2007). 10.1038/nprot.2007.30 [DOI] [PubMed] [Google Scholar]
- 16. Linderholm P., Braschler T., Vannod J., Barrandon Y., Brouard M., and Renaud P., “ Two-dimensional impedance imaging of cell migration and epithelial stratification,” Lab Chip 6, 1155–1162 (2006). 10.1039/b603856e [DOI] [PubMed] [Google Scholar]
- 17. Sackmann E. K., Fulton A. L., and Beebe D. J., “ The present and future role of microfluidics in biomedical research,” Nature 507, 181–189 (2014). 10.1038/nature13118 [DOI] [PubMed] [Google Scholar]
- 18. Lei K. F., “ Microfluidic systems for diagnostic applications: A review,” J. Lab. Autom. 17, 330–347 (2012). 10.1177/2211068212454853 [DOI] [PubMed] [Google Scholar]
- 19. Takayama S., McDonald J. C., Ostuni E., Liang M. N., Kenis P. J. A. et al. , “ Patterning cells and their environments using multiple laminar fluid flows in capillary networks,” Proc. Natl. Acad. Sci. U.S.A. 96, 5545–5548 (1999). 10.1073/pnas.96.10.5545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nie F. Q., Yamada M., Kobayashi J., Yamato M., Kikuchi A. et al. , “ On-chip cell migration assay using microfluidic channels,” Biomaterials 28, 4017–4022 (2007). 10.1016/j.biomaterials.2007.05.037 [DOI] [PubMed] [Google Scholar]
- 21. Van der Meer A. D., Vermeul K., Poot A. A., Feijen J., and Vermes I., “ A microfluidic wound-healing assay for quantifying endothelial cell migration,” Am. J. Physiol. 298, H719–H725 (2010) 10.1152/ajpheart.00933.2009. [DOI] [PubMed] [Google Scholar]
- 22. Huang X., Li L., Tu Q., Wang J., Liu W. et al. , “ On-chip cell migration assay for quantifying the effect of ethanol on MCF-7 human breast cancer cells,” Microfluid. Nanofluid. 10, 1333–1341 (2011). 10.1007/s10404-011-0766-9 [DOI] [Google Scholar]
- 23. Lei K. F., “ Review on impedance detection of cellular responses in micro/nano environment,” Micromachines 5, 1–12 (2014). 10.3390/mi5010001 [DOI] [Google Scholar]
- 24. Hong J., Kandasamy K., Marimuthu M., Choi C. S., and Kim S., “ Electrical cell-substrate impedance sensing as a non-invasive tool for cancer cell study,” Analyst 136, 237–245 (2011). 10.1039/C0AN00560F [DOI] [PubMed] [Google Scholar]
- 25. Pethig R., Dielectric and Electronic Properties of Biological Materials ( Wiley, New York, 1979). [Google Scholar]
- 26. Ehret R., Baumann W., Brischwein M., Schwinde A., Stegbauer K. et al. , “ Monitoring of cellular behavior by impedance measurements on interdigitated electrode structures,” Biosens. Bioelectron. 12, 29–41 (1997). 10.1016/0956-5663(96)89087-7 [DOI] [PubMed] [Google Scholar]
- 27. Thakur M., Mergel K., Weng A., Frech S., Gilabert-Oriol R. et al. , “ Real time monitoring of the cell viability during treatment with tumor-targeted toxins and saponins using impedance measurement,” Biosens. Bioelectron. 35, 503–506 (2012). 10.1016/j.bios.2012.03.024 [DOI] [PubMed] [Google Scholar]
- 28. Fuhr G., Glasser H., Muller T., and Schnelle T., “ Cell manipulation and cultivation under a.c. electric field influence in highly conductive culture media,” Biochim. Biophys. Acta 1201, 353–360 (1994). 10.1016/0304-4165(94)90062-0 [DOI] [PubMed] [Google Scholar]
- 29. Luongo K., Holton A., Kaushik A., Spence P., Ng B. et al. , “ Microfluidic device for trapping and monitoring three dimensional multicell spheroids using electrical impedance spectroscopy,” Biomicrofluidics 7, 034108 (2013). 10.1063/1.4809590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wang L., Wang H., Wang L., Mitchelson K., Yu Z. et al. , “ Analysis of the sensitivity and frequency characteristics of coplanar electrical cell-substrate impedance sensors,” Biosens. Bioelectron. 24, 14–21 (2008). 10.1016/j.bios.2008.03.018 [DOI] [PubMed] [Google Scholar]
- 31. Johnson C., Han Y., Hughart N., McCarra J., Alpini G. et al. , “ Interleukin-6 and its receptor, key players in hepatobiliary inflammation and cancer,” Transl. Gastrointest Cancer 1, 58–70 (2012) 10.3978/j.issn.2224-4778.2011.11.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tang Y., Kitisin K., Jogunoori W., Li C., Deng C. X. et al. , “ Progenitor/stem cells give rise to liver cancer due to aberrant TGF-β and IL-6 signaling,” Proc. Natl. Acad. Sci. U.S.A. 105, 2445–2450 (2008). 10.1073/pnas.0705395105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Liu Y., Li P. K., Li C., and Lin J., “ Inhibition of STAT3 signaling blocks the anti-apoptotic activity of IL-6 in human liver cancer cells,” J. Biol. Chem. 285, 27429–27439 (2010). 10.1074/jbc.M110.142752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wang D., Liebowitz D., and Kieff E., “ An EBV membrane protein expressed in immortalized lymphocytes transforms established rodent cells,” Cell 43, 831–840 (1985). 10.1016/0092-8674(85)90256-9 [DOI] [PubMed] [Google Scholar]
- 35. Baichwal V. R. and Sudgen B., “ Transformation of Balb 3T3 cells by the BNLF-1 gene of Epstein-Barr virus,” Oncogene 2, 461–467 (1988); available at http://europepmc.org/abstract/med/2836780. [PubMed] [Google Scholar]
- 36. Eliopoulos A. G., Stack M., Dawson C. W., Kaye K. M., Hodgkin L. et al. , “ Epstein-Barr virus-encoded LMP1 and CD40 mediate IL-6 production in epithelial cells via an NF-kappaB pathway involving TNF receptor-associated factors,” Oncogene 14, 2899–2916 (1997). 10.1038/sj.onc.1201258 [DOI] [PubMed] [Google Scholar]
- 37. Nakagomi H., Dolcetti R., Bejarano M. T., Pisa P., Kiessling R., and Masucci M., “ The Epstein-Barr virus latent membrane protein-1 (LMP2) induces interleukin-10 production in burkitt lymphoma lines,” Int. J. Cancer 57, 240–244 (1994). 10.1002/ijc.2910570218 [DOI] [PubMed] [Google Scholar]
- 38. Lei K. F. and Huang C. H., “ Paper-based microreactor integrating cell culture and subsequent immunoassay for the investigation of cellular phosphorylation,” ACS Appl. Mater. Interfaces 6, 22423–22429 (2014). 10.1021/am506388q [DOI] [PubMed] [Google Scholar]
- 39. Obata N. H., Tamakoshi K., Shibata K., Kikkawa F., and Tomoda Y., “ Effects of interleukin-6 on in vitro cell attachment, migration and invasion of human ovarian carcinoma,” Anticancer Res. 17, 337–342 (1997); available at http://europepmc.org/abstract/med/9066674. [PubMed] [Google Scholar]