Abstract
Protein prenylation is a posttranslational modification that is indispensable for translocation of membrane GTPases like Ras, Rho, Ras etc. Proteins of Ras family undergo farnesylation by FTase while Rho family goes through geranylgeranylation by GGTase1. There is only an infinitesimal difference in signal recognition between FTase and GGTase1. FTase inhibitors mostly end up selecting the cells with mutated Ras proteins that have acquired affinity towards GGTase1 in cancer microcosms. Therefore, it is of interest to identify GGTase1 and FTase dual inhibitors using the docking tool AutoDock Vina. Docking data show that curcumin (from turmeric) has higher binding affinity to GGTase1 than that of established peptidomimetic GGTase1 inhibitors (GGTI) such as GGTI-297, GGTI-298, CHEMBL525185. Curcumin also interacts with FTase with binding energy comparable to co-crystalized compound 2-[3-(3-ethyl-1-methyl-2-oxo-azepan-3-yl)-phenoxy]-4-[1-amino-1-(1-methyl-1h-imidizol-5-yl)-ethyl]-benzonitrile (BNE). The docked complex was further simulated for 10 ns using molecular dynamics simulation for stability. Thus, the molecular basis for curcumin binding to GGTase1 and FTase is reported.
Keywords: Prenylation, GGTase1, Rho, AutoDock Vina, Curcumin, Molecular dynamics simulations, FTase
Background
Protein prenylation is an important posttranslational modification through which naïve protein molecules are targeted to membranes. It also helps in protein-protein interactions and reversible binding of some transport proteins to membranes [1]. Protein prenylation is the addition of either Farnesyl or geranylgeranyl moieties to proteins. Many proteins including Ras superfamily of proteins require prenylation for their proper function [2]. Three independent prenylating enzymes namely protein farnesyl transferase (FTase) and two protein geranylgeranyl transferases (GGTase1 and 2) are responsible for addition of respective isoprenoids. The subtle change in amino acid recognition sequence by these three enzymes confirms that any given protein is prenylated with only one of them. Both FTase and GGTase1 recognize CAAX motif present in C-terminal of the proteins to be prenylated, where C is cysteine, A can be any aliphatic amino acid. FTase binds to the protein if X is serine, methionine or glutamine. If X is leucine, GGTase1 binds to it resulting in geranylgeranylation of the protein. In a stark contrast GGTase2 recognizes proteins with C-C or CXC domain and prenylate them. To date, Rab family of proteins are the only known candidates to possess CC or CXC domain [3, 4].
Ras farnesylation was targeted soon after its discovery to combat tumor malignancy [5, 6]. Farnesylation was effectively stopped by inhibiting FTase through peptidomimetic compounds. Though proven to be very good drugs with amazingly nil side effects, FTase inhibitors (FTI) failed to prevent tumor proliferation completely as some Ras isoforms like K-Ras-4B bind the enzyme more avidly than FTIs like L- 744,832, and FTI-277 or undergo alternative prenylation i.e. geranylgeranylation [7]. Rho family of GTPases (about eight members) belongs to Ras superfamily of proteins that are geranylgeranylated by GGTase1. Members of Rho family especially RhoA and Rac1 play a vital role in Ras mediated transformation of NIH 3T3 cells [8]. A combination of FTI and GGTI was required for deterring K-Ras processing in A549 and Calu-1 cells [9]. The importance of GGTase1 and FTase inhibitors in anti-cancer therapy was described elsewhere [5, 10, 11]. Hence, it becomes clear that an FTase-GGTase1 dual inhibitor may be of certain interest.
FTase and GGTase1 are structurally very similar with a common α-subunit [12]. The β-subunit, with precisely two amino acids, W102 and Y365 (FTase) T49 and F324 (GGTase-I) recognizes their respective CAAX peptide [13]. Previously, we have identified curcumin as an inhibitor of GGTase1 by docking simulations using AutoDock Vina [14]. Curcumin (C21H20O6) is an active ingredient of Turmeric (Figure 1) is known to inhibit FTase but the molecular mechanism and structural basis of this activity is unknown [15, 16]. In the present study, we have evaluated the molecular dynamics of the enzyme inhibitor complex using Desmond software [17]. Additionally, molecular dynamics of curcumin binding to FTase and enzyme–inhibitor interactions were studied using Desmond software.
Figure 1.
2-Dimensional structure of curcumin shown in one of its tautomeric (keto) forms.
Methodology
GGTase1 target structure and molecular docking:
A homology based high-resolution structure of GGTase1 was prepared and docking was done as described elsewhere [14].
FTase target structure and molecular docking:
2Å resolution crystal structure of human FTase (PDB ID 1MZC). 2-[3-(3-ethyl-1-methyl-2-oxo-azepan-3-yl)-phenoxy]-4- [1-amino-1-(1-methyl-1h-imidizol-5-yl)-ethyl]-benzonitrile (BNE) were downloaded from protein data bank (PDB) [18]. This structure was chosen due to its human origin, better resolution and co-crystallization with an inhibitor. The structures were minimized and prepared as mentioned elsewhere using chimera [19]. Autogrid was constructed around the active site defined in the crystal structure. All docking simulations were done on a PyRx GUI v0.8 [20] containing AutoDock Vina tool.
Molecular dynamics simulations:
MDS were run on Desmond [17] software of Maestro 9.9 GUI (academic versions) study with OPLS 2005 force field [21]. Docked complexes were solvated using the SPC explicit water model that is most suitable for a cytosol proteins. The complexes were neutralized by addition of Na+ ions. System builder tool of Desmond software imposed topology and force field parameters. A Nose-Hoover chain thermostat method [22] and Martyna-Tobias-Klein barostat method [23] were used to maintain the water bath at 300K and a constant isotropic pressure of 1atm with 2ps relaxation time respectively. Smooth particle mesh Ewald method [24] with a cut off radius of 9Å was used to study the Coulombic and a tolerance of 1e-09 was used for long range interactions. The setup was then simulated for 10ns using above said conditions.
Results
Virtual screening and molecular docking:
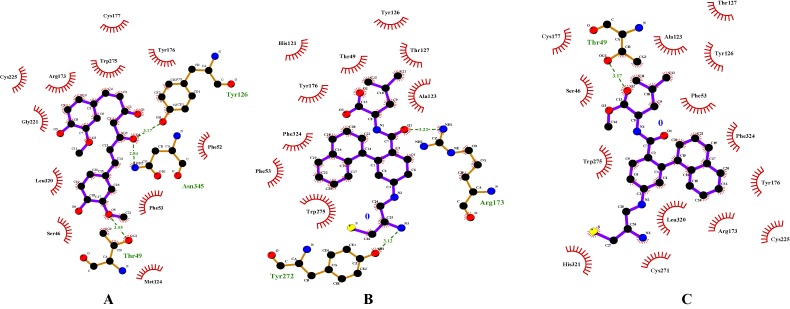
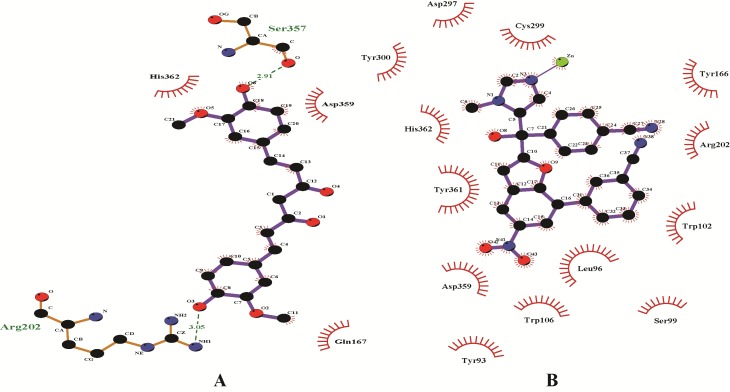
Enzyme inhibitory site-compound library screening using AutoDock Vina yielded 10 highest score enzyme-inhibitor conformations. Based on zero RMSD value, the best enzymesubstrate conformation was selected. The best score for the compounds were Curcumin -7.3 as compared to GGTI-297 -7.5 and GGTI-298 -7.5. All these compounds formed hydrogen bond with T49 which is a key amino acid for GGTase1 specificity (Figure 2). All the hydrogen bonds were observed within a range of 2.1-2.5 Å. Curcumin and 2-[3-(3-ethyl-1- methyl-2-oxo-azepan-3-yl)-phenoxy]-4-[1-amino-1-(1-methyl- 1h-imidizol-5-yl)-ethyl]-benzonitrile (BNE) with score of -7.9 and -9.5 respectively, bind FTase in the inhibitory site. Curcumin formed hydrogen bonds with R202 and S357 but BNE did not form any hydrogen bond with the enzyme (Figure 3). The critical residues S99B and D359 were also shown to interact with the inhibitors but through non-covalent interactions other than hydrogen bonds. Table 1 & Table 2 (see supplementary material) shows the docking scores of various compounds with GGTase1 and FTase respectively.
Figure 2.
Interaction diagrams of GGTase1 with A) Curcumin B) GGTI-297 and C) GGTI-298. Hydrogen bonds between the enzyme-inhibitor complex is shown as green dashed lines. Other amino acids shown here form the binding pocket which may interact with the inhibitors through non-covalent interactions other than hydrogen bonding.
Figure 3.
Interaction diagrams of FTase1 with A) Curcumin and B) BNE. Hydrogen bonds between the enzyme-inhibitor complex is shown as green dashed lines. Other amino acids shown here form the binding pocket which may interact with the inhibitors through non-covalent interactions other than hydrogen bonding.
Molecular dynamics simulations:
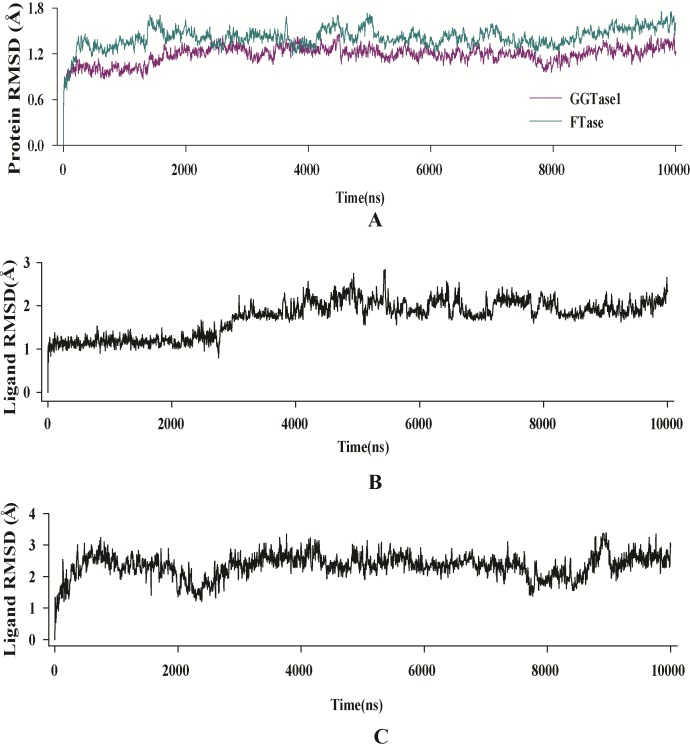
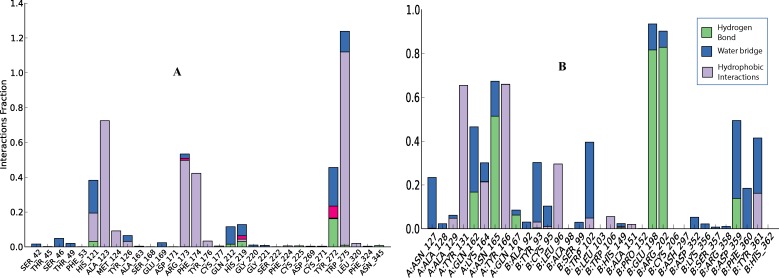
RMSD of enzyme C-α and inhibitor fit to enzyme of both complexes (GGTase1 and FTase) were found to be in equilibrium after 3 ns and fluctuations were not observed till the end of simulation time (10 ns) (Figure 4). While GGTase1 interacted with curcumin mainly through hydrophobic interactions, FTase did so with hydrogen bonding. W275 of GGTase1 showed strong hydrophobic interaction with curcumin almost 120% of the time (due to multiple interactions other than that of hydrophobic). Hydrogen bonds with T49 and F324, the key amino acids which give substrate specificity for GGTase1, perished during the course of simulation time and showed only a little interaction. In the case of FTase E198 and R202 of the B chain from was found to hydrogen bond with curcumin for more than 80% of the time. Of this hydrogen bond with R202 was realized during docking studies. Interestingly, hydrogen bond with S357 was lost throughout the simulation time. W102 which gives FTase its substrate specificity, interacts with the inhibitor for about 40% of the time. Enzyme-inhibitor interactions throughout the simulation time is shown in Figure 5.
Figure 4.
RMSD evolution of the A) enzymes, B) curcumin fit into GGTase1 and C) curcumin fit into FTase. All protein frames are first aligned on the reference frame backbone, and then the C-α RMSD is calculated. Changes in C-α RMSD lie within acceptable limit of 1-1.4 Å. Ligand (inhibitor) RMSD shows the RMSD of the inhibitor when the enzyme-inhibitor complex is first aligned on the protein backbone of the reference and then the RMSD of the inhibitor heavy atoms is measured. The values observed were not significantly larger than the RMSD of the enzyme.
Figure 5.
Amino acid residues that interacted non-covalently with the inhibitor during the simulation period. Values on yaxes represent the percentage of time the inhibitor interacted with that amino acids. This percentage can be more than 100% due to the multiple interactions. A. Interaction of curcumin with GGTase1. W275 is shown to have hydrophobic interactions with curcumin almost throughout the simulation period. B. Interaction of curcumin with FTase. Both the A chain and B chain interact with curcumin. E198 and R202 of the B chain form hydrogen bond with curcumin for more than 80% of the simulation time.
Discussion
Ras mediated transformation is manifested in many cancers. As mentioned earlier, a combined administration of FTase and GGTase1 inhibitors has been shown to be effective in treatment of cancers facilitated by Ras. Peptidomimic compounds, the only proven drugs that act against GGTase1 which is structurally similar to CAAX tetrapeptide. Molecular docking is a fast track method for screening drugs with low cost but high efficiency and rapidity. Many compounds are already available in market that were initially screened by molecular docking. But there are no inhibitors for GGTase1 other than peptidomimic compounds reported. Taylor et al., [25] concluded that amino acid at 49th position which ascertains GGTase1 specificity can be exploited for structure based drug design. Our findings indicate that curcumin can act as dual inhibitor of GGTase1 and FTase. It specifically binds T49 which may confer inhibitory effect of GGTase1. Curcumin, a pleiotropic molecule is shown to ameliorate cancer progression by a number of ways [26]. In a previous study with piperidine based FTase inhibitors, the synthesized compounds were shown to interact with S99B and D359B through molecular modelling approach [27]. In this study, despite curcumin was not shown to interact with either of these amino acids after docking, MDS studies show that D359 and S99 significantly interacted with curcumin for about 40% of the total simulation time (10 ns). Hence, we hypothesize that anti-cancer activity of these compounds are in part may be due to FTase and GGTase1 inhibition. Extensive studies need to decipher the complete mechanism of curcumin׳s action on FTase and GGTase1 inhibition.
Conclusion
Curcumin is a highly pleiotropic molecule with excellent anticancer activity. Molecular docking and dynamics simulation studies indicate that, part of curcumin׳s anticancer activity is attributable to its binding of two of the most important prenylating enzymes. In conclusion, curcumin exhibits its anticancer activity at least in part due to the inhibition of FTase and GGTase1 and in future, more rigorous studies are required to prove this fact.
Supplementary material
Acknowledgments
The financial support provided by the University Grants Commission, New Delhi [UGC F. No: 42-176/2013(SR)], India is gratefully acknowledged.
Footnotes
Citation:Subramani et al, Bioinformation 11(5): 248-253 (2015)
References
- 1.Zhang FL, et al. Annu Rev Biochem. 1996;65:241. doi: 10.1146/annurev.bi.65.070196.001325. [DOI] [PubMed] [Google Scholar]
- 2.Casey PJ. J Lipid Res. 1992;33:1731. [PubMed] [Google Scholar]
- 3.Casey PJ, et al. Proc Natl Acad Sci U S A. 1991;88:8631. doi: 10.1073/pnas.88.19.8631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cox AD, et al. Small GTPases. 2010;1:2. doi: 10.4161/sgtp.1.1.12178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sebti SM, et al. Oncogene. 2000;19:6584. doi: 10.1038/sj.onc.1204146. [DOI] [PubMed] [Google Scholar]
- 6.Zimmermann G, et al. Nature. 2013;497:638. doi: 10.1038/nature12205. [DOI] [PubMed] [Google Scholar]
- 7.Fiordalisi JJ, et al. J Biol Chem. 2003;278:41718. doi: 10.1074/jbc.M305733200. [DOI] [PubMed] [Google Scholar]
- 8.Khosravi-Far R, et al. Mol Cell Biol. 1995;15:6443. doi: 10.1128/mcb.15.11.6443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sun J, et al. Oncogene. 1998;16:1467. doi: 10.1038/sj.onc.1201656. [DOI] [PubMed] [Google Scholar]
- 10.Lobell RB, et al. Cancer Res. 2001;61:8758. [PubMed] [Google Scholar]
- 11.Kelloff GJ, et al. Cancer Epidemiol Biomarkers Prev. 1997;6:267. [PubMed] [Google Scholar]
- 12.Seabra MC, et al. Cell. 1991;65:429. doi: 10.1016/0092-8674(91)90460-g. [DOI] [PubMed] [Google Scholar]
- 13.Terry KL, et al. Biochemistry. 2006;45:9746. doi: 10.1021/bi060295e. [DOI] [PubMed] [Google Scholar]
- 14.Thippanna M, et al. Bioinformation. 2013;9:973. doi: 10.6026/97320630009973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen X, et al. Anticancer Res. 1997;17:2555. [PubMed] [Google Scholar]
- 16.Kang HM, et al. Nat Prod Res. 2004;18:295. doi: 10.1080/14786410310001620691. [DOI] [PubMed] [Google Scholar]
- 17.Guo Z, et al. Chem Biol Drug Des. 2010;75:348. doi: 10.1111/j.1747-0285.2010.00951.x. [DOI] [PubMed] [Google Scholar]
- 18.Berman HM, et al. Nucleic Acids Res. 2000;28:235. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pettersen EF, et al. J Comput Chem. 2004;25:1605. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 20.Wolf LK, et al. Chemical & Engineering News Archive. 2009;87:32. [Google Scholar]
- 21.Banks JL, et al. J Comput Chem. 2005;26:1752. doi: 10.1002/jcc.20292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Evans DJ, et al. The Journal of Chemical Physics. 1985;83:4069. [Google Scholar]
- 23.Martyna GJ, et al. The Journal of Chemical Physics. 1994;101:4177. [Google Scholar]
- 24.Essmann U, et al. The Journal of Chemical Physics. 1995;103:8577. [Google Scholar]
- 25.Taylor JS, et al. EMBO J. 2003;22:5963. doi: 10.1093/emboj/cdg571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Subramani PA, et al. Nat Prod Commun. 2013;8:121. [PubMed] [Google Scholar]
- 27.Tanaka R, et al. Bioorg Med Chem. 2007;15:1363. doi: 10.1016/j.bmc.2006.11.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.