Abstract
Background
Saccharomyces cerevisiae is the micro-organism of choice for the conversion of fermentable sugars during beverage or bioethanol fermentations. These fermentations are characterised by high osmotic stress on a yeast cell, with selected brewing fermentations beginning at 20–25% fermentable sugars and bioethanol fermentations at 13% fermentable sugars.
Results
RCK2 encodes for a MAPKAP (MAPK-activated protein kinase) enzyme and was identified on a locus by QTL analysis in yeast cells under osmotic stress, RCK2 expression was placed under a tetracycline regulatable vector and rescued glucose, sorbitol or glycerol induced osmotic stress in an rck2 null strain. A strain overexpressing RCK2 had significantly faster fermentation rates when compared with the empty vector control strain.
Conclusions
Presence of RCK2 increased rates of glucose utilisation (~40 g glucose in first 8 h) during a 15% glucose fermentation and concurrent production of ethanol when compared with empty vector controls. Tolerance to osmotic stress using the tetracycline regulatable vectors could be turned off with the addition of tetracycline returning a rck2 null strain back to osmotic sensitivity.
Electronic supplementary material
The online version of this article (doi:10.1186/s12934-015-0276-7) contains supplementary material, which is available to authorized users.
Background
Fermentations, whether for traditional beverage or bioethanol production, impose upon the microbe a variety of stresses. During industrial fermentation yeast strains are exposed to stresses such as oxygen concentration, osmotic pressure, pH, end-product (usually ethanol), nutrient availability and increasing temperature [1]. Osmotic stress can be defined as a situation where there is an imbalance in intracellular and extracellular osmolytes causing an alteration in cellular physiology [2]. In natural habitats, yeast are constantly exposed to fluctuations in osmotic stress which can lead to impaired functioning of the cell [3]. Within the brewing process osmotic stress is encountered upon pitching yeast cells into media (or wort) containing very high concentrations of dissolved fermentable sugars [1, 4]. Thus, resistance to osmotic stress is a desirable phenotypic attribute for improved yeast performance within a fermentation bioreactor.
Using F1 haploid segregants, from clean lineage S. cerevisiae strains, QTL on the yeast chromosome for several stress tolerances, including osmotic stress, were identified [5], genes within the loci have been assessed for their potential role in osmotic tolerance. RCK2, a MAPKAP (MAP activated protein kinase) was significantly up-regulated in yeast cells exposed to osmotic stress. Previous research has revealed that Rck2p is phosphorylated under oxidative or osmotic stress conditions, along with some other components within the high osmolarity glycerol (HOG) pathway [6]. Yeast responds to stress caused by an increase in osmolarity by activating the HOG MAPK cascade [7], and this cascade leads to an elevated synthesis of glycerol [8]. There are a great number of genes responsible for osmotic tolerance and the mechanism for gene regulation through the HOG pathway is still unclear. Around 50 genes are strongly dependant on Hog1 [9], phosphorylation of Hog1p has been shown to influence the activity of metabolic enzymes [10]. Hog1p localization in the nucleus has been shown to be dependent on Rck2p activity [11]. Rck2p also acts on translation elongation factor 2 mediating a transient repression of protein synthesis [12] and regulates the translational expression of osmostress-regulated mRNA [13].
In this article, the importance of RCK2 under osmotic stress was assessed, using phenotypic microarray assays along with performance in fermentation. RCK2 expression was placed under a tetracycline regulatable vector in a Δrck2 null strain and tolerance to osmotic stress inducing chemicals such as d-glucose, sorbitol, glycerol and NaCl determined.
Results
Deletion of RCK2 increases sensitivity to osmotic stress
The metabolic activity of wild type BY4741 and the Δrck2 strain during incubation in the presence of sorbitol (10–30%) was determined by use of a phenotypic microarray as measured by redox signal intensity (redox signal intensity has been defined previously [5]) (Figure 1a). It was observed that Δrck2 was more sensitive to the presence of sorbitol (10–30%) when compared with the background strain. In addition, Δrck2 also displayed increased sensitivity to the presence of increasing glucose and glycerol (Figure 2a, b); however, there was no difference between a Δrck2 strain and BY4741 in the presence of osmotic stress induced by the addition of NaCl (Figure 2c).
Figure 1.
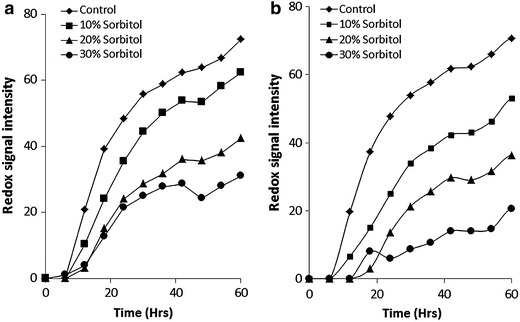
Phenotypic microarray analysis for S. cerevisiae BY4741 or Δrck2 under osmotic stress. a BY4741 under 0–30% sorbitol stress, b Δrck2 under 0–30% sorbitol stress. Mean ± SD (n = 3).
Figure 2.
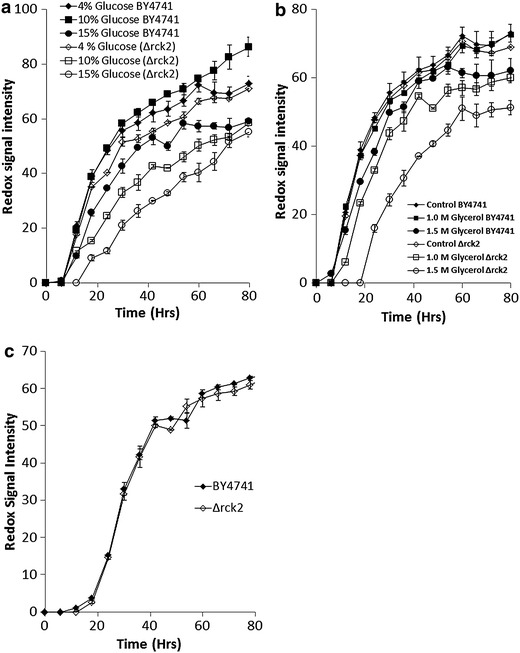
Phenotypic microarray analysis for S. cerevisiae BY4741 or Δrck2 under osmotic stress. a BY4741 or Δrck2 under 4,10, or 15% glucose stress, and b BY4741 or Δrck2 under control, 1.0 or 1.5 M glycerol stress. c BY4741 or Δrck2 under 2 M NaCl. Mean ± SD (n = 3).
Expression of RCK2 in the Δrck2 strain recovers osmotic tolerance
Insertion of a tetracycline regulatable vector (pCM161:RCK2) into a Δrck2 strain was assessed for impact on sensitivity to osmotic stress and compared with a strain carrying an empty vector (pCM161) as control. qPCR confirmed that expression of RCK2 in the Δrck2pCM161(RCK2), was 32-fold higher when compared with Δrck2pCM161 (data not shown). Transformation with the pCM161 empty vector had no impact on metabolic output when under osmotic stress (10% sorbitol) compared with a Δrck2 strain (p = 0.9102), however, a Δrck2 strain containing a pCM161(RCK2) had significantly higher metabolic output (p = 0.0001) under this stress condition (Figure 3a). We also assessed for performance under increasing concentrations of glucose and observed that assays using a strain with an empty vector were identical to the Δrck2 strain, however, assays with a Δrck2 strain carrying pCM161(RCK2) revealed that there was a significant increase in metabolic output at all glucose concentrations (p = 0.0001) (Figure 3b), and metabolic output in the presence of 15% glucose was no different to that at 4% (p = 0.844) (Additional file 1: Figure S1). Assays using 20% glucose displayed a reduction in metabolic output in the Δrck2pCM161(RCK2) strain compared with this strains output at 4–15% glucose (p = 0.0432), however, metabolic output was still significantly higher than a Δrck2(pCM161) in the presence of 20% glucose (p = 0.0005) (Additional file 1: Figure S1).
Figure 3.
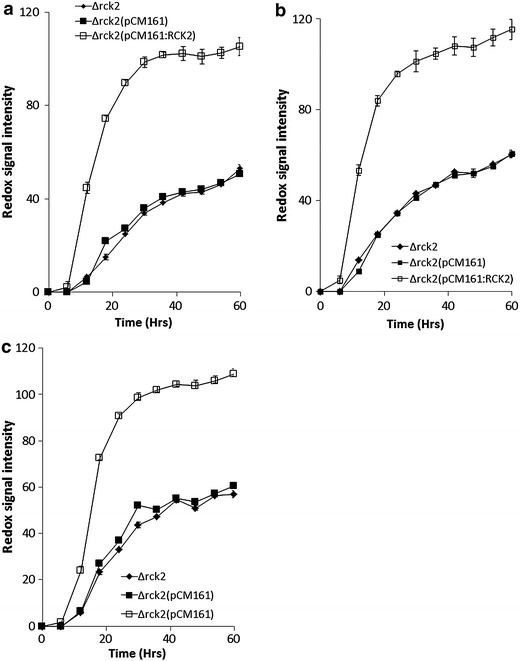
Phenotypic microarray analysis for S. cerevisiae Δrck2, S. cerevisiae Δrck2pCM161 and S. cerevisiae Δrck2pCM161:RCK2 under osmotic stress a 10% sorbitol stress, b 4% glucose stress, c 1.0 M glycerol stress. Mean ± SD (n = 3).
Confirmation of phenotypic microarray strain assessments using mini-fermentation analysis
The fermentation profiles of the strains using 40 g/L glucose were assessed in terms of glucose utilisation and ethanol production (Figure 4). It was observed that a strain with a pCM161(RCK2) vector utilised glucose and produced ethanol significantly faster than the empty vector control (p = 0.03) (Figure 4a, b). Addition of 1 µg/mL tetracycline reduced glucose utilisation and ethanol production when compared with absence of tetracycline (p = 0.03) (Figure 4a, b), however, glucose utilisation and ethanol production was significantly higher than in the pCM161 empty vector control (p = 0.03) (Figure 4a, b).
Figure 4.
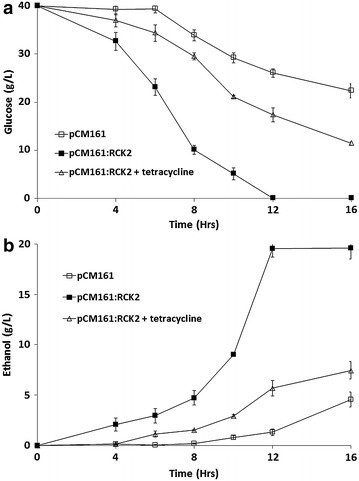
Performance of Δrck2(pCM161) and Δrck2(pCM161:RCK2) during a 40 g/L fermentation in the presence and absence of tetracycline a glucose utilisation b ethanol production. Mean + SD (n = 3).
This data can be used to assess the efficiency of the conversion of glucose into ethanol. Under control conditions, the empty vector Δrck2 control strain had a 0.09 ± 0.003 g ethanol/g glucose conversion efficiency whilst the pCM161:RCK2 strain had an efficiency of 0.48 ± 0.001 ethanol/g glucose conversion after 12 h. Addition of tetracycline reduced conversion efficiency to 0.24 ± 0.012. The theoretical maxima is 0.511 g ethanol per g of glucose consumed [14], therefore the pCM161:RCK2 strain was converting glucose into ethanol at near theoretical maximum during a fermentation in the presence of 40 g/L glucose.
Overexpression of RCK2 improved fermentation in the presence of 15% glucose
Weight loss experiments using Δrck2pCM161(RCK2) and Δrck2pCM161 revealed that presence of RCK2 significantly improved rates of fermentation in the presence of 15% glucose (Additional file 2: Figure S2). There have been a number of studies where ethanol fermentations by S. cerevisiae have been started with initial glucose concentration of around 100 g/L [15] so we tested these strains in 2 L fermentation in the presence of 150 g/L glucose and monitored glucose utilisation, ethanol production, and production of metabolites such as acetic acid and glycerol; we also determined the concentration of stress response storage carbohydrates in the cell such as trehalose and glycogen. Results revealed that presence of RCK2 significantly speeded up utilisation of glucose and production of ethanol when compared with the empty vector controls (Figure 5a, b), fermentations containing RCK2 also produced more acetic acid than the empty vector control (Figure 5c), however, concentrations of glycerol were significantly lower than with the empty vector control (Figure 5c).
Figure 5.
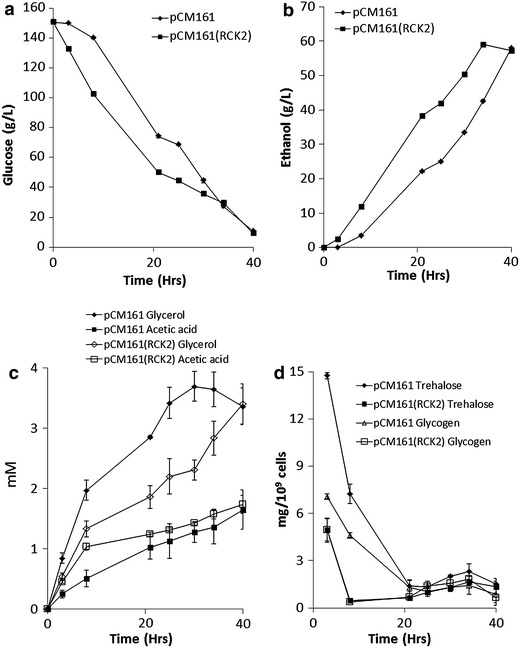
Performance of Δrck2(pCM161) and Δrck2(pCM161:RCK2) during a 150 g/L fermentation. a glucose utilisation b ethanol production c acetic acid (mM) and glycerol (mM) concentrations d trehalose and glycogen concentrations (mg/109 cells). Mean + SD (n = 3).
An assessment of accumulation of storage carbohydrates such as trehalose and glycogen revealed that there was initially higher concentrations of these polymers in the empty vector controls when compared with the strain containing RCK2 (Figure 5d). Trehalose and glycogen concentrations dropped rapidly at the start of the fermentation and after 20 h there was no difference between the yeast types (Figure 5d).
Carbon partitioning during fermentations using Δrck2pCM161 or Δrck2pCM161(RCK2)
Carbon balance analysis was carried out for yeast strains Δrck2pCM161 and Δrck2pCM161(RCK2) during the 150 g/L glucose fermentations (Table 1). For carbon analysis, the cultivation period was divided into two phases, I (0–21 h) and II (21–40 h). The two strains differed in consumption of glucose and ethanol formation in each phase with Δrck2pCM161 assimilating 429.1 mM and Δrck2pCM161(RCK2) assimilating 559.4 mM glucose, respectively in phase I. The difference became more significant with respect to ethanol production, Δrck2pCM161(RCK2) strain generated about 73% more ethanol (831.1 mM) in comparison with Δrck2pCM161 (481.1 mM) in the first 21 h, this trend was reversed in the second phase of the fermentation (21–40 h) where consumption 354.1 and 228.2 mM glucose by Δrck2pCM161 and Δrck2pCM161(RCK2) resulted in 781.3 and 411.6 mM ethanol, respectively.
Table 1.
Carbon balance for batch cultivation of Δrck2pCM161(RCK2) and Δrck2pCM161(RCK2) strains during a 15% glucose fermentation
| Substrate and products | Phase I (0–21 h) | Phase II (21–40 h) | ||||
|---|---|---|---|---|---|---|
| Amount (mM) | Carbon (mM) | Carbon (%) | Amount (mM) | Carbon (mM) | Carbon (%) | |
| Δrck2pCM161 | ||||||
| Glucose | 426.1 | 2556.6 | 100.0 | 354.1 | 2124.6 | 100.0 |
| Ethanol | 481.1 | 962.2 | 37.6 | 781.3 | 1562.6 | 73.5 |
| Glycerol | 2.9 | 8.7 | 0.3 | 0.5 | 1.5 | 0.1 |
| Acetic acid | 1.0 | 2.0 | 0.1 | 0.6 | 1.2 | 0.1 |
| Trehalose | 1.2 | 7.2 | 0.3 | 0.0 | 0.0 | 0.0 |
| Glycogen | 1.1 | 6.6 | 0.3 | 0.4 | 2.4 | 0.1 |
| CO2 | 481.1 | 481.1 | 18.8 | 781.3 | 781.3 | 36.8 |
| Carbon recovery | 57.6 | 110.6 | ||||
| Δrck2pCM161(RCK2) | ||||||
| Glucose | 559.4 | 3356.4 | 100.0 | 228.4 | 1370.4 | 100.0 |
| Ethanol | 831.1 | 1662.2 | 49.5 | 411.6 | 823.2 | 60.1 |
| Glycerol | 1.9 | 5.7 | 0.2 | 1.5 | 4.5 | 0.3 |
| Acetic acid | 1.2 | 2.4 | 0.1 | 0.5 | 1.0 | 0.1 |
| Trehalose | 1.0 | 6.0 | 0.2 | 0.4 | 2.4 | 0.2 |
| Glycogen | 1.1 | 6.6 | 0.2 | 0.4 | 2.4 | 0.2 |
| CO2 | 831.1 | 831.1 | 24.8 | 411.6 | 411.6 | 30.0 |
| Carbon recovery | 74.9 | 90.9 | ||||
Discussion
The conversion of sugars into ethanol is the prime function of yeast during a brewing or bioethanol fermentation, the osmotic effect of unfermented sugars and ethanol inhibition are major stresses imposed on the yeast cell which inevitably affect their growth and rates of fermentation [4, 16]. Adaptation to hyperosmotic stress via the high osmolarity glycerol (HOG) transduction pathway is well defined in yeast [17].
Rck2p is a serine/threonine protein kinase homologous to mammalian calmodulin kinases, which requires phosphorylation for activation, phosphorylation is transiently increased during osmotic stress or in strains overexpressing the HOG pathway [18]. A QTL analysis for yeast revealed that on chromosome XII there were genes responsible for osmotolerance, upon further examination of the QTL, RCK2 was found to be up-regulated in yeast cells under osmotic stress [5, 19].
Placing RCK2 expression under a tetracycline regulatable vector, we measured performance under osmotic stress and compared this with the performance of a Δrck2 strain. Performance of the Δrck2 strain under osmotic stress (glucose, sorbitol, or glycerol) confirmed that absence of RCK2 was characterised by osmosensitivity. Expression of RCK2 in a rck2 null background conferred resistance to osmotic stress in terms of both increased metabolic output and rates of fermentation. Addition of tetracycline reduced RCK2 expression and reduced the rates of fermentation.
Overexpression of RCK2 has been observed previously to confer osmotic tolerance in hog1 null yeast cells when induced by the addition of NaCl [6], tolerance as measured by inhibition was believed to be downstream of Hog1 phosphorylation. Rck2p along with other protein kinases forms a sub-family of MAPK enzymes which incorporates a characteristic glycine loop in their C-terminal domain [20]. In general, there is a down-regulation in translation in cells under osmotic stress, however, this down-regulation is mitigated in hog1 or rck2 mutants [18] and Rck2p has been shown to phosphorylate elongation factor 2 a component of translation regulation [12, 18] indicating an importance of Rck2p in maintaining translation under osmotic stress conditions.
Conclusion
Results reported here are the first example of how overexpressing RCK2 conferred resistance to osmotic stress when induced by the presence of reducing sugars during a fermentation, results revealed that the improved rates of fermentation in a cell overexpressing RCK2 was not characterised by an increase in glycerol when compared with an empty vector control indicating that the role of Rck2p in osmotically stressed cells is not solely through an up-regulation of the HOG pathway which is characterised by an increased in glycerol production [8]. Future work will be to determine the cellular role of RCK2 in osmotic stressed yeast cells and to track RCK2 expression during fermentations.
Experimental procedures
Yeast strains and growth conditions
Yeast strains employed in this work derive from S. cerevisiae BY4741 (Table 2), all strains were grown in YPD [1% (w/v) yeast extract (Oxoid); 2% (w/v) Bacto-peptone (Oxoid); 2% (w/v) glucose]. Strains were deleted for tryptophan biosynthesis (trp1::URA3) using pAG60 (Euroscarf, Frankfurt, Germany) with a URA3 selectable marker using primers (Table 3). A Δrck2 null mutant was obtained from Euroscarf (Frankfurt, Germany) and TRP1 was also deleted from this strain as above for BY4741.
Table 2.
Strains used in this study, all strains are derived from BY4741 (MATa his3∆0 leu2∆0 met15∆0 ura3∆0), with the additional TRP1 gene knocked out to allow for selection with pCM plasmids
| Strains used in this study | Integrative plasmida |
|---|---|
| BY4741Δtrp1 (MATa his3∆0 leu2∆0 met15∆0 ura3∆0 trp1Δ0) | – |
| BY4741Δtrp1Δrck2 | – |
| BY4741Δtrp1Δrck2 | pCM161 |
| BY4741Δtrp1Δrck2 | pCM161 (RCK2) |
aIntegrative plasmids were constructed as indicated in “Plasmid construction”.
Table 3.
Primers used in this study for the knockout of TRP1 and insertion of pCM vectors into yeast strains
| Primers | Sequence |
|---|---|
| TRP1 knockout forward | cgccagatggcagtagtggaagatattctttattgaaaaatagcttgtcaATGACAGTCAACACTAAGACCTATA |
| TRP1 knockout reverse | ttttatgcttgcttttcaaaaggcctgcaggcaagtgcacaaacaatactTTATAATTGGCCAGTCTTTTTC |
| pCM161 forward | ATCGATATGCTTAAAATAAAGGCC |
| pCM161 reverse | GGATCCCTATTCCCTGATAGTGGC |
Plasmid construction
Plasmid pCM161 is a centromeric yeast plasmid, marker TRP1, tetracycline repressed expression of lacZ under the control of the tetO2 promoter. For the construction of the pCM161(RCK2), a PCR product encoding for RCK2 with relevant restriction enzyme sites appropriate for ligation into the vectors was prepared. For cloning into pCM161, Cla1 and BamH1 restriction enzymes (NEB) were used and the relevant digest site added to the forward and reverse primers (Table 3).
PCR of RCK2 from genomic DNA
S. cerevisiae BY4741 was grown to stationary phase, cells harvested and broken with glass beads using a MagNalyser (Roche, Burges Hill, UK) bead beater for 30 s at 4°C, before incubating on ice for 15 min to precipitate proteins. Cell debris and proteins were harvested by centrifugation for 15 min (17,000×g at 4°C). The cell-free supernatant was used for the extraction of total DNA using an isolation kit from. PCR was performed using primers (pCM161F—ATCGATATGCTTAAAATAAAGGCC and pCM161R—GGATCCCTATTCCCTGATAGTGGC–GGATCCCTATTCCCTGATAGTGGC). PCR products using above primers were digested and linearized digested plasmids with appropriate restriction enzymes were ligated using the Quick ligation mix (New England Biosciences, US,) and the ligation mixture used in a standard lithium acetate transformation [21] and plated out on appropriate selection agar plates.
qPCR
Strains BY4741, Δrck2 and Δrck2 containing plasmids were grown to the mid-logarithmic stage of growth in 4% SD-Trp media 30°C and stressed by the addition of 20% sorbitol for 15 min, rotated at 150 rpm. Cells were broken with glass beads using a MagNalyser (Roche, Burges Hill, UK) bead-beater for 30 s at 4°C, before incubating on ice for 15 min to precipitate proteins. Cell debris and proteins were harvested by centrifugation for 15 min (17,000×g at 4°C). The cell-free supernatant was used for the extraction of total RNA using an isolation kit from Qiagen (Hilden, Germany) and cDNA prepared using a first strand cDNA synthesis kit (GE Healthcare, Bucks, UK). Transcriptional levels were determined by qPCR using the following conditions follows: 0.5 ng/µl cDNA, 6.25 µM forward primer, 6.25 µM reverse primer, 5 µL of 2× SYBR Green master mix (Applied Bio Systems) and made up to 20 µL using molecular grade water.
All data was compared against ACT1 which encodes for actin a structural protein in yeast as an internal normaliser and expression data from genes within the relevant loci were presented as fold-change in comparison to ACT1 transcript levels in control and stress conditions.
Phenotypic microarray analysis
For phenotypic microarray (PM) analysis, medium was prepared as described previously [5]. Osmotic stress was induced by addition of 10–30% sorbitol, 1–2 M glycerol, or 1–2 M NaCl using 80% sorbitol, 5 M glycerol or 10 M NaCl as stock solution as appropriate. Appropriate amounts from these stocks were added to the wells of the phenotypic microarray assay displacing water to maintain a final volume of 120 µL in the assay.
Confirmation of phenotypic microarray results using mini fermentation vessels
Fermentations were conducted in 180 mL mini-fermentation vessels (FV). Cryopreserved yeast colonies were streaked onto YPD plates and incubated at 30°C for 48 h. Colonies of yeast strains were used to inoculate 20 mL of YPD broth and incubated in an orbital shaker at 30°C for 24 h. These were then transferred to 200 mL of YPD and grown for 48 h in a 500 mL conical flask shaking at 30°C. Cells were harvested and washed three times with sterile RO water and then re-suspended in 5 mL of sterile water. For control conditions, 1.5 × 107 cells mL−1 were inoculated in 99.6 mL of medium containing 40 g/L (4%) glucose, 2% YNB-trp with 0.4 mL reverse osmosis (RO) water. For stress conditions, 1.5 × 107 cells mL−1 were incubated in 99.6 mL of medium containing 40 g/L glucose, 2% YNB-trp with 75 mM acetic acid, Volumes of media were adjusted to account for the addition of the inhibitory compounds (~400 μL) to ensure that all fermentations began with the same carbon load. Fermentations with 80, 100 or 150 g/L glucose had a starting carbon load of 80, 100 or 150 g/L in a final volume of 100 mL with pitching levels identical to fermentations with 40 g/L glucose.
Anaerobic conditions were prepared using a sealed butyl plug (Fisher, Loughborough, UK) and aluminium caps (Fisher Scientific). A hypodermic needle attached with a Bunsen valve was purged through rubber septum to facilitate the release of CO2. All experiments were performed in triplicate and weight loss was measured at each time point. Mini-fermentations were conducted at 30°C, with orbital shaking at 200 rpm.
15% glucose fermentation in 2 L fermentation vessels
After growing the yeast strains in 180 mL mini-fermentation vessels, batch fermentations of two strains (pCM161 and pCM161(RCK2)) were carried out in 2 L vessels with a working volume of 1.5 L SD-trp medium. The initial glucose concentration was 150 g/L. The temperature and agitation was controlled to 30°C and 250 rpm, respectively. The stirring was achieved using magnetic beads. The pH was not controlled during the fermentation but initial pH was adjusted to pH 4.0–4.5 using phosphoric acid. These vessels have provisions for sparging of air and the experiments were conducted under microaerobic conditions which were achieved by sparging air at an aeration rate of 0.1 vvm.
Detection of glucose ethanol, acetic acid and glycerol from FV experiments via HPLC
Glucose, ethanol, acetic acid and glycerol were quantified by HPLC. The HPLC system included a Jasco AS-2055 Intelligent auto sampler (Jasco, Tokyo, Japan) and a Jasco PU-1580 Intelligent pump (Jasco). The chromatographic separation was performed on a Rezex ROA H+ organic acid column, 5 μm, 7.8 mm × 300 mm, (Phenomenex, Macclesfield, UK) at ambient temperature. The mobile phase was 0.005 N H2SO4 with a flow rate of 0.5 mL/min. For detection a Jasco RI-2031 Intelligent refractive index detector (Jasco) was employed. Data acquisition was via the Azur software (version 4.6.0.0, Datalys, St Martin D’heres, France) and concentrations were determined by peak area comparison with injections of authentic standards. The injected volume was 10 μL and analysis was completed in 40 min. All chemicals used were analytical grade (>95% purity, Sigma-Aldrich, UK).
Determination of intercellular glycogen and trehalose concentrations
Intracellular glycogen and trehalose were estimated based on the method of Parrou and Francois [22]. Frozen yeast samples were thawed on ice and an appropriate cell suspension volume containing 1 × 109 cells was centrifuged at 3,000 rpm for 5 min at 4°C. The pellet was washed three times with distilled water. Cells were lysed by resuspension of the pellet in sodium carbonate (0.25 mL; 0.25 M) and were incubated at 95°C for 2 h followed by the addition of sodium acetate (0.6 mL; 0.2 M) and acetic acid (0.15 mL; 1 M). 0.5 mL aliquots were then assessed for glycogen and trehalose concentrations. Glycogen and trehalose were broken down into glucose by adding 10 μL of α-amyloglucosidase (10 mg/mL; 59.9 Units/mg; Fluka Biochemika, Steinheim) or 10 μL trehalase (3 m Units; Sigma-Aldrich, UK) respectively followed by incubation at 57 and 37°C for 14 h. Post incubation, samples were centrifuged (3,500 rpm; 5 min) and the supernatant (0.1 mL) containing liberated glucose was quantified using the Megazyme Glucose Assay kit (GOPOD, Megazyme, Ireland) at an optical density of 510 nm. Analysis for each time point was conducted in triplicates and results were expressed in concentration of glucose as a function of cell number.
Carbon analysis
Carbon analysis was calculated based on the consumed substrate and accumulated metabolites, glucose, ethanol, glycerol, acetic acid, trehalose, glycogen and carbon dioxide [23]. The evolution of carbon dioxide was estimated using the following equation: C6H12O6 → 2C2H5OH + 2CO2 and CO2 coming from other sources such as the TCA cycle was not included into the calculations. The carbon used for biomass formation was not taken into account due to lack of information on biomass composition of yeast cells.
Statistical analysis
Data derived from phenotypic microarrays was analysed for analysis of variance (ANOVA) using ezANOVA (http://www.cabiatl.com/mricro/ezanova), with statistical significance signified by use of, * = 0.05% significance, ** = 0.01% significance and *** 0.001% significance.
Authors’ contribution
VK and AJH performed the experiments, AJH and TTW constructed the pCM vectors containing RCK2, GAT proofread the document and DG designed the experiments and wrote the manuscript. All authors read and approved the final manuscript.
Acknowledgements
The research reported here was supported (in full or in part) by the Biotechnology and Biological Sciences Research Council (BBSRC) Sustainable Bioenergy Centre (BSBEC), under the programme for ‘Lignocellulosic Conversion To Ethanol’ (LACE) (Grant Ref: BB/G01616X/1). This is a large interdisciplinary programme and the views expressed in this paper are those of the authors alone, and do not necessarily reflect the views of the collaborators or the policies of the funding bodies. This project is part financed by the European Regional Development Fund project EMX05568.
Compliance with ethical guidelines
Competing interests The authors declare that they have no competing interests and all authors have agreed to submit the paper to the journal of microbial cell factories.
Additional files
Additional file 1: Figure S1. Phenotypic microarray analysis for S. cerevisiae Δrck2(pCM161:RCK2) under 4 and 15% glucose. Mean + SD (n = 3).
Additional file 2: Figure S2. Fermentation profiles for S. cerevisiae Δrck2 pCM161 or S. cerevisiae Δrck2 pCM161:RCK2 under osmotic stress (A) performance of S. cerevisiae Δrck2 pCM161 and S. cerevisiae Δrck2 pCM161:RCK2 in 15% glucose. Mean + SD (n = 3).
Footnotes
V Kumar and A J Hart contributed equally
Contributor Information
V Kumar, Email: vinod.kumar@nottingham.ac.uk.
A J Hart, Email: andrew.hart@nottingham.ac.uk.
T T Wimalasena, Email: tithira.wimalasena@gmail.com.
G A Tucker, Email: gregory.tucker@nottingham.ac.uk.
D Greetham, Email: darren.greetham@nottingham.ac.uk.
References
- 1.Briggs DE, Boulton CA, Brookes PA, Stevens R. Brewing: science and practice. Amsterdam: Elsevier; 2004. [Google Scholar]
- 2.Csonka LN, Hanson AD. Prokaryotic osmoregulation: genetics and physiology. Ann Rev Microbiol. 1991;45:569–606. doi: 10.1146/annurev.mi.45.100191.003033. [DOI] [PubMed] [Google Scholar]
- 3.Beney L, Martinez de Maranon I, Marechal PA, Gervais P. Influence of thermal and osmotic stresses on the viability of the yeast Saccharomyces cerevisiae. Int J Food Microbiol. 2000;55(1–3):275–279. doi: 10.1016/S0168-1605(00)00203-8. [DOI] [PubMed] [Google Scholar]
- 4.Devantier R, Scheithauer B, Villas-Boas SG, Pedersen S, Olsson L. Metabolite profiling for analysis of yeast stress response during very high gravity ethanol fermentations. Biotechnol Bioeng. 2005;90(6):703–714. doi: 10.1002/bit.20457. [DOI] [PubMed] [Google Scholar]
- 5.Greetham D, Wimalasena T, Kerruish DW, Brindley S, Ibbett RN, Linforth RL, et al. Development of a phenotypic assay for characterisation of ethanologenic yeast strain sensitivity to inhibitors released from lignocellulosic feedstocks. J Ind Microbiol Biotechnol. 2014;41(6):931–945. doi: 10.1007/s10295-014-1431-6. [DOI] [PubMed] [Google Scholar]
- 6.Bilsland-Marchesan E, Arino J, Saito H, Sunnerhagen P, Posas F. Rck2 kinase is a substrate for the osmotic stress-activated mitogen-activated protein kinase Hog1. Mol Cell Biol. 2000;20(11):3887–3895. doi: 10.1128/MCB.20.11.3887-3895.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boguslawski G. PBS2, a yeast gene encoding a putative protein kinase, interacts with the RAS2 pathway and affects osmotic sensitivity of Saccharomyces cerevisiae. J Gen Microbiol. 1992;138(11):2425–2432. doi: 10.1099/00221287-138-11-2425. [DOI] [PubMed] [Google Scholar]
- 8.Albertyn J, Hohmann S, Thevelein JM, Prior BA. GPD1, which encodes glycerol-3-phosphate dehydrogenase, is essential for growth under osmotic stress in Saccharomyces cerevisiae, and its expression is regulated by the high-osmolarity glycerol response pathway. Mol Cell Biol. 1994;14(6):4135–4144. doi: 10.1128/mcb.14.6.4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Posas F, Chambers JR, Heyman JA, Hoeffler JP, de Nadal E, Arino J. The transcriptional response of yeast to saline stress. J Biol Chem. 2000;275(23):17249–17255. doi: 10.1074/jbc.M910016199. [DOI] [PubMed] [Google Scholar]
- 10.Dihazi H, Kessler R, Eschrich K. High osmolarity glycerol (HOG) pathway-induced phosphorylation and activation of 6-phosphofructo-2-kinase are essential for glycerol accumulation and yeast cell proliferation under hyperosmotic stress. J Biol Chem. 2004;279(23):23961–23968. doi: 10.1074/jbc.M312974200. [DOI] [PubMed] [Google Scholar]
- 11.Bilsland E, Molin C, Swaminathan S, Ramne A, Sunnerhagen P. Rck1 and Rck2 MAPKAP kinases and the HOG pathway are required for oxidative stress resistance. Mol Microbiol. 2004;53(6):1743–1756. doi: 10.1111/j.1365-2958.2004.04238.x. [DOI] [PubMed] [Google Scholar]
- 12.Melcher ML, Thorner J. Identification and characterization of the CLK1 gene product, a novel CaM kinase-like protein kinase from the yeast Saccharomyces cerevisiae. J Biol Chem. 1996;271(47):29958–29968. doi: 10.1074/jbc.271.47.29958. [DOI] [PubMed] [Google Scholar]
- 13.Warringer J, Hult M, Regot S, Posas F, Sunnerhagen P. The HOG pathway dictates the short-term translational response after hyperosmotic shock. Mol Biol Cell. 2010;21(17):3080–3092. doi: 10.1091/mbc.E10-01-0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krishnan MS, Ho NW, Tsao GT. Fermentation kinetics of ethanol production from glucose and xylose by recombinant Saccharomyces 1400(pLNH33) Appl Biochem Biotechnol. 1999;77–79:373–388. doi: 10.1385/ABAB:78:1-3:373. [DOI] [PubMed] [Google Scholar]
- 15.Alfenore S, Molina-Jouve C, Guillouet SE, Uribelarrea JL, Goma G, Benbadis L. Improving ethanol production and viability of Saccharomyces cerevisiae by a vitamin feeding strategy during fed-batch process. Appl Microbiol Biotechnol. 2002;60(1–2):67–72. doi: 10.1007/s00253-002-1092-7. [DOI] [PubMed] [Google Scholar]
- 16.Zheng DQ, Wu XC, Tao XL, Wang PM, Li P, Chi XQ, et al. Screening and construction of Saccharomyces cerevisiae strains with improved multi-tolerance and bioethanol fermentation performance. Bioresource Technol. 2011;102(3):3020–3027. doi: 10.1016/j.biortech.2010.09.122. [DOI] [PubMed] [Google Scholar]
- 17.Hohmann S. Osmotic stress signaling and osmoadaptation in yeasts. Microbiol Mol Biol Rev. 2002;66(2):300–372. doi: 10.1128/MMBR.66.2.300-372.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Teige M, Scheikl E, Reiser V, Ruis H, Ammerer G. Rck2, a member of the calmodulin-protein kinase family, links protein synthesis to high osmolarity MAP kinase signaling in budding yeast. Proc Natl Acad Sci USA. 2001;98(10):5625–5630. doi: 10.1073/pnas.091610798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Greetham D, Wimalasena TT, Leung K, Marvin ME, Chandelia Y, Hart AJ, et al. The genetic basis of variation in clean lineages of Saccharomyces cerevisiae in response to stresses encountered during bioethanol fermentations. PLoS One. 2014;9(8):e103233. doi: 10.1371/journal.pone.0103233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Asp E, Sunnerhagen P. Mkp1 and Mkp2, two MAPKAP-kinase homologues in Schizosaccharomyces pombe, interact with the MAP kinase Sty1. Mol Genet Genomics. 2003;268(5):585–597. doi: 10.1007/s00438-002-0786-y. [DOI] [PubMed] [Google Scholar]
- 21.Gietz RD, Schiestl RH. Large-scale high-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat Protoc. 2007;2(1):38–41. doi: 10.1038/nprot.2007.15. [DOI] [PubMed] [Google Scholar]
- 22.Parrou JL, Francois J. A simplified procedure for a rapid and reliable assay of both glycogen and trehalose in whole yeast cells. Anal Biochem. 1997;248(1):186–188. doi: 10.1006/abio.1997.2138. [DOI] [PubMed] [Google Scholar]
- 23.Durgapal M, Kumar V, Yang TH, Lee HJ, Seung D, Park S. Production of 1,3-propanediol from glycerol using the newly isolated Klebsiella pneumoniae J2B. Bioresource Tech. 2014;159:223–231. doi: 10.1016/j.biortech.2014.01.126. [DOI] [PubMed] [Google Scholar]


