Abstract
Molecular cross-talk between neuronal drebrin A and cofilin is believed to be a part of the activity-dependent cytoskeleton-modulating pathway in dendritic spines. Impairments in this pathway are implicated also in synaptic dysfunction in Alzheimer’s disease, Down syndrome, epilepsy, and normal aging. However, up to now the molecular interplay between cofilin and drebrin has not been elucidated. TIRF microscopy and solution experiments revealed that full length drebrin A or its actin binding core (Drb1-300) inhibits, but do not abolish cofilin-induced severing of actin filaments. Cosedimentation experiments showed that F-actin can be fully occupied with combination of these two proteins. The dependence of cofilin binding on fractional saturation of actin filaments with drebrin suggests direct competition between these two proteins for F-actin binding. This implies that cofilin and drebrin can either overcome or reverse the allosteric changes in F-actin induced by the competitor’s binding. The ability of cofilin to displace drebrin from actin filaments is pH dependent and is facilitated at acidic pH (6.8). Pre-steady state kinetic experiments reveal that both binding and dissociation of drebrin to/from actin filaments is faster than that reported for cooperative binding of cofilin. We found, that drebrin displacement by cofilin is greatly inhibited when actin severing is abolished, which might be linked to the cooperativity of drebrin binding to actin filaments. Our results contribute to molecular understanding of the competitive interactions of drebrin and cofilin with actin filaments.
Keywords: actin, drebrin, cofilin
Introduction
Dendritic spines (DS) are dynamic structures and their morphology and plasticity are modulated by the actin cytoskeleton response to various stimuli [Zhou et al., 2004; Cingolani and Goda, 2008; Cho et al., 2013]. Such plasticity requires switching between stable and dynamic states of actin cytoskeleton in spines and the fine tuning of both actin stabilization and enhanced turnover. Drebrin and actin depolymerizing factors (ADF)/cofilins are key F-actin regulators in spines and their silencing leads to defects in spine morphology and function [Hotulainen et al., 2009; Ivanov et al., 2009b]. Cofilin is an F-actin severing protein the activity of which is tightly regulated by variety of factors, including phosphorylation (Ser 3), small molecules (such as PIP2), and other actin-binding proteins (like some tropomyosin isoforms) [Yonezawa et al., 1990; Arber et al., 1998; Kuhn and Bamburg, 2008]. In contrast to cofilin, which promotes F-actin turnover, drebrin is a filament stabilizing protein that inhibits actin depolymerization [McGough et al., 1997; Mikati et al., 2013]. Accumulating evidence suggests a link between actin remodeling and complex neurological disorders [Kojima and Shirao, 2007; Ivanov et al., 2009a]. Cofilin upregulation followed by the translocation of drebrin A from DS was proposed to be part of a pathological pathway relevant to Alzheimer’s disease (AD), Down syndrome, and epilepsy [Kojima and Shirao, 2007; Ferhat, 2012]. This pathway, which was suggested to lead to spine shrinkage and the impairment of higher order brain function, can be a potential therapeutical target for complex brain disorders [Zhou et al., 2004; Cho et al., 2013]. However, the molecular details of drebrincofilin cross-talk are poorly understood. Thus, it is important to elucidate the mechanism and functional consequences of debrin-cofilin competition for actin binding.
At the molecular level, drebrin-cofilin cross-talk was first suggested based on the results of quantitative immunoprecipitation (IP) experiments [Zhao et al., 2006]. It was shown that less drebrin co-IP with actin in the presence of unphosphorylated cofilin. Moreover, negative correlation between drebrin and cofilin levels in the brains of AD patients has been also reported [Zhao et al., 2006]. It was hypothesized that unphosphorylated cofilin, (at increased levels) displaces drebrin from the filaments, causing destabilization of actin cytoskeleton within the spine, followed by drebrin degradation and spine shrinkage. However, the molecular mechanism of such displacement/competition is not immediately evident. Cooperative binding to actin filaments was documented for both cofilin and drebrin A [Cao et al., 2006; Sharma et al., 2012]. Also, we showed recently the opposite effects of these two proteins on F-actin morphology: cofilin was shown to shorten the helical pitch of actin filaments and drebrin caused an increase in the length of their helical repeats [McGough et al., 1997; Sharma et al., 2010]. Moreover, the propagation of these morphological changes to undecorated filament regions was documented for drebrin by atomic force microscopy (AFM) imaging [Sharma et al., 2012] and was proposed for cofilin based on differential scanning calorimetry (DSC), fluorescence, and cross-linking assays [Bobkov et al., 2006]. Such long range effects can underlie non-competitive (allosteric) inhibition of drebrin-F-actin interaction by cofilin (as was previously reported for Arp2/3 complex [Chan et al., 2009]), and vice versa. However, both proteins contain an ADF homology domain that can provide a structural basis for their direct competition. Taken together, all three possibilities–allosteric mechanism of displacement, direct competition between drebrin-cofilin, and/or their combination appear plausible. Distinguishing among them will be important for better understanding of the state and dynamics of actin-based structures in DS and their availability for interactions with other actin-regulating factors.
In this study, we investigated the effects of drebrin on cofilin-mediated severing and depolymerization employing total internal reflection fluorescence (TIRF) microscopy and equilibrium binding assays. We also determined kinetic parameters of drebrin interaction with actin filaments to clarify the mechanism of its competition with cofilin and other actin binding factors. Our results contribute to a molecular level understanding of the competition of drebrin with cofilin for actin binding, and reveal that it is not driven by allosteric changes in F-actin associated with their respective binding.
Results
Drebrin Effects on Cofilin-Induced Filaments’ Severing and Depolymerization
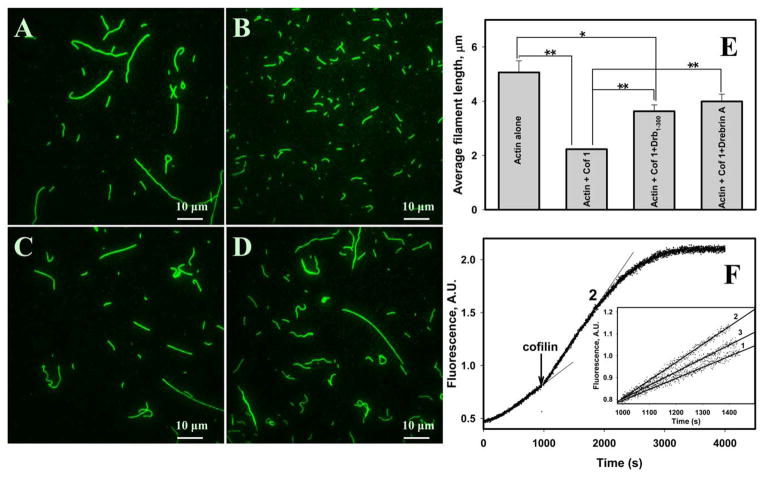
It was previously reported that drebrin does not sever F-actin [Ishikawa et al., 1994]. We confirmed it independently by measuring the average length of bare and drebrin-decorated filament populations. The obtained values were 4.4 μm (n = 154 filaments), 4.2 μm (n = 93 filaments), and 5.2 μm (n = 165 filaments) for bare, Drb1-300 and DrbA-FL-decorated filaments, respectively. The observed increase in average filament length in the presence of DrbA-FL could arise from greater mechanical stability of drebrin-decorated F-actin compared to that of F-actin control. We employed TIRF microscopy (single time point analysis) to probe for the effects of drebrin on cofilin-mediated F-actin severing (Figs. 1A–1D). Optimal cofilin: actin ratio for these experiments, yielding filament fragments sufficiently long for analysis was determined empirically. After incubation of Cy3-labeled F-actin with human cofilin-1 (hCof1) in solution (5:1 molar ratio), the average filament length decreases by ~56%. We observed a reduced severing by hCof1 in the presence of both DrbA-FL (seq. 1–706) and Drb1-300 (seq. 1–300): the average filament length decreased by 21 and 28%, respectively (Fig. 1E). Notably, the original average filaments’ length was not retained in the presence of saturating concentrations (occupancy >0.9) of both drebrin constructs, implying that F-actin decorated with drebrin was still susceptible to cofilin-mediated severing (Figs. 1A–1D).
Fig. 1. Drebrin inhibits cofilin-mediated severing.
Representative TIRF images are shown for skeletal F-actin (1.5 μM) alone (A), actin with hCof1 (0.3 μM) (B), actin with hCof1 and 2.5 μM Drb1-300 (C), and actin with hCof1 and 1.5 μM of DrbA-FL (D). TIRF experiments were repeated four times and yielded similar results. Average filaments’ length for all systems tested is shown in (E). Lines connecting the columns show which filament populations are compared. An average result of three independent experiments is presented; bars correspond to the standard error of the mean (n = 3). Student t-test (two-tailed): * P<0.05; ** P<0.01. (F) Acceleration of actin (1.5 μM, 5% pyrene iodoacetamide) polymerization due to hCof1 (0.3 μM) mediated severing. Inset shows initial rates of actin polymerization after cofilin or buffer addition (indicated by arrow), as measured by pyrene fluorescence increase. Trace 1: F-actin (1.5 μM) with Drb1-300 (4.2 μM), buffer added; Trace 2: F-actin alone (1.5 μM), 0.3 μM cofilin added; Trace 3: F-actin (1.5 μM) with Drb1-300 (4.2 μM), 0.3 μM cofilin added. Slopes of the linear fits are: 5.0 × 10−4, 8.2 × 10−4, and 6.1 × 10−4 A.U./s for Traces 1, 2, and 3, respectively.
To confirm this finding, we carried out solution experiments in which bare and drebrin-saturated (occupancy >0.9) filaments were subjected to cofilin-mediated fragmentation (Fig. 1F). At the time of ~20% completion of actin polymerization, cofilin (or buffer) was added to the reactions and mixed gently by inversion. Acceleration of actin elongation was observed with both bare (Fig. 1F, trace 2) and drebrin-saturated filaments (Fig. 1F, trace 3), due to cofilin-induced severing and consequent increase in the number of free filament ends. Consistent with our TIRF experiments, elongation rates after cofilin addition were slower in drebrin containing samples but severing was not abolished (Fig. 1F, trace 3). In contrast to that no acceleration of polymerization was observed in controls (F-actin alone and F-actin-Drb1-300), when buffer was added instead of cofilin (Fig. 1F, trace 1). Notably, these severing experiments yielded similar results at pH 6.8 and at physiological pH (7.5).
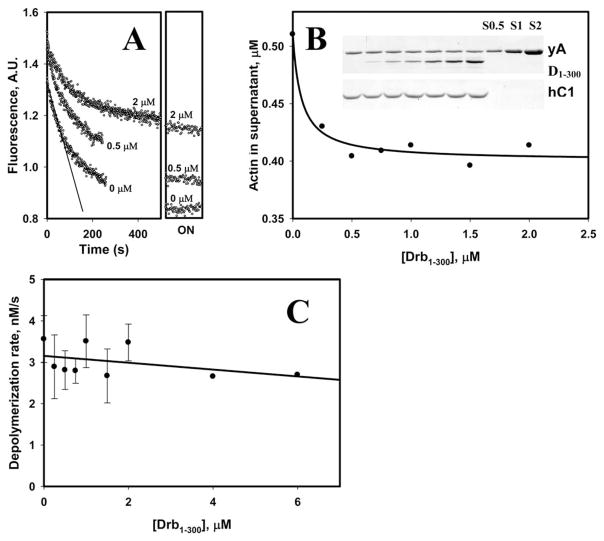
Next, we examined the effect of drebrin on cofilin-induced depolymerization. Cofilin was shown to accelerate pointed end depolymerization of actin filaments [Carlier et al., 1997; Andrianantoandro and Pollard, 2006]. At subsaturating concentrations of cofilin severing events occur at the same time as depolymerization, increasing the number of free filament ends and contributing to the overall increase in bulk depolymerization rates. We showed recently that hCof1 does not sever wild type (WT) yeast actin filaments across a range of cofilin concentrations tested (up to the occupancy of 0.9) [McCullough et al., 2011]. Thus, the number of free filaments’ ends should be the same in WT yeast F-actin samples containing hCof1 alone or both, hCof1, and drebrin. This system allows for uncoupling cofilin-mediated depolymerization from severing, and thus enables examining the effect of drebrin on the depolymerization in solution. As detected by pyrene fluorescence assays, depolymerization curves obtained in the presence of hCof1 alone and with increasing concentrations of Drb1-300 reached different plateau levels (Fig. 2A). According to our data, the main factor contributing to different plateau levels is a decrease (up to ~25%) in total amount of depolymerized actin with increasing concentrations of drebrin (Fig. 2B). To calculate the initial rates of actin cofilin-mediated depolymerization in the presence and absence of Drb1-300, we combined the data obtained in pyrene fluorescence and cosedimentation assays (see Materials and Methods for details). The results shown in Fig. 2C suggest that inhibitory effect of Drb1-300 on the rate of cofilin-induced depolymerization of F-actin is not statistically significant (based on two-tailed Students test). This conclusion is in a good agreement with the previously reported observation [Mikati et al., 2013] that drebrin inhibits barbed-end depolymerization much stronger than that at pointed-end of filaments.
Fig. 2. Depolymerization of yeast WT F-actin in the presence of hCof1 and Drb1-300.
(A) Examples of depolymerization data used to determine the initial rates of F-actin depolymerization. Depolymerization was followed by pyrene fluorescence. As an example, part of the data used to calculate the initial rate is indicated by a solid line. Fluorescence signals of the samples after overnight (ON) incubation, corrected for instrument-related drift of the signal, are shown in the right panel. Conditions: KM2EH7.5 buffer supplemented with 1 mM DTT and 0.2 mM ATP. Final F-actin and cofilin concentrations were 2 and 1.5 μM, respectively. Concentrations of Drb1-300 in the samples are indicated next to the traces. (B) Drb1-300 decreases the extent of actin depolymerization in the presence of hCof1 compared to F-actin-cofilin control. Samples contain increasing concentrations of Drb1-300 (gel insert, left to right: 0; 0.25; 0.5; 0.75; 1; 1.5; 2 μM). Conditions and protein concentrations as in (A.) Gel analysis (SDS PAGE, 12.5%) of the samples [as shown in (A)] after overnight incubation at 4°C. Supernatants and pellets were separated by high speed centrifugation and a representative gel of supernatant samples stained with Coomassie Blue is shown. Protein bands are marked as yA–yeast actin; D1–300–Drb1-300; hC1–human cofilin 1. Amounts of actin in supernatants were quantified using yeast actin standards (labeled as S0.5; S1; S2, numbers (0.5, 1, and 2) correspond to the actin concentration in μM). Dependence of yeast WT F-actin (2 μM) depolymerization rates in the presence of human cofilin-1 (1.5 μM) on the concentration of Drb1-300 construct (average of two independent experiments). Conditions: 1× KM2EH7.5 buffer supplemented with 0.2 mM ATP and 1mM DTT.
Co-binding of Drebrin and Cofilin to Actin Filaments
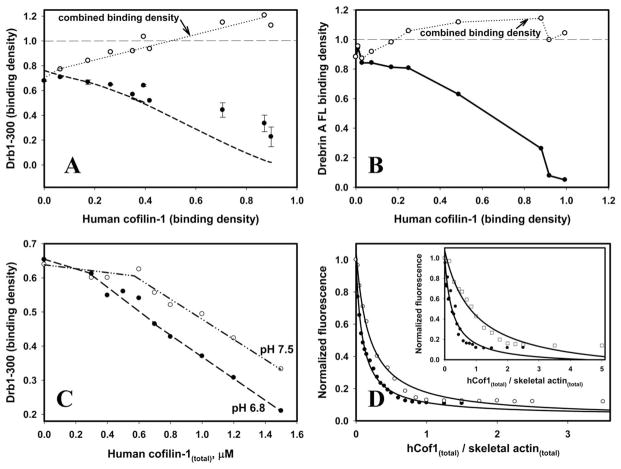
Displacement of drebrin from actin filaments by cofilin was documented before but its mechanism remains unclear [Zhao et al., 2006]. To address this question we conducted cosedimentation experiments using purified hCof1 and drebrin. Increasing concentrations of cofilin were added to drebrin-bound actin filaments, and fractional occupancy of F-actin by drebrin was evaluated and plotted versus fractional occupancy by cofilin (Fig. 3). As evident form Figs. 3A and 3C, at physiological pH, low concentrations of cofilin added to drebrin-decorated F-actin (>0.7 binding density) do not cause significant drebrin displacement until the total combined filaments’ occupancy approaches 1. This indicates that either: (1) in filaments partially decorated with drebrin cofilin-induced intersubunit angular disorder does not propagate over the same range of distances as in undecorated actin, and/or (2) such propagated morphological change in filaments does not displace drebrin from actin filaments.
Fig. 3. Drebrin displacement from actin filaments by hCof1.
(A) Equilibrium binding of Drb1-300 to F-actin in the presence of increasing concentrations of hCof1. Solid circles—binding density of Drb1-300; open circles—combined Drb1-300 and hCof1 binding density. Thick dashed line corresponds to the fit of the data to Eq. (1) for competitive binding of a 3-protomer binding protein at different hCof1 occupancy (adopted from Chan et al [Chan et al., 2009], see Materials and Methods). Experiments were repeated four times and yielded similar results. Average of two independent experiments is shown. Error bars correspond to the values of standard deviation; (B) Equilibrium binding of DrbA-FL to F-actin shows a trend similar to that observed with Drb1-300 (solid circles, experimental data points are connected by solid line). Experiments in (B) were done in duplicates and yielded similar results. Conditions (A–B): 1× KM2EH7.5 buffer supplemented with 0.2 mM ATP and 1 mM DTT. (C) Drebrin displacement from actin filaments by hCof1 is facilitated at acidic pH. Observed effects were reproduced 3 times. A representative data set is shown. Conditions: 1× KM2EH7.5 (open circles, dotted, and dashed line) or 1× KM2EI6.8 buffer supplemented with 0.2 mM ATP and 1 mM DTT (solid circles, dashed line) (D) Binding of hCof1 to F-actin (2 μM, 100% pyrene maleimide labeled) in the absence (black symbols) and in the presence (open symbols) of drebrin constructs: Drb1-300 (2.65 μM)—main graph, DrbA-FL (0.4 μM)—inset. Conditions: 1× KM2EI6.8 buffer supplemented with 0.2 mM ATP and 1mM DTT.
We employed previously published simulations, used to analyze Arp2/3-cofilin competition [Chan et al., 2009], to model the obtained results (see Materials and Methods). Drb1-300 (as well as Arp2/3) occupies 3 actin protomers when bound to F-actin. In the case of non-cooperative binding, direct competition will reduce the number of free 3-protomer sites clusters in proportion to (1–c)3, where c is the binding density of cofilin. The power of three reflects the probability of locating any three adjacent unoccupied protomers at the (1–c) concentration of free actin. Stochastic simulations performed by Chan et al [Chan et al., 2009] show that cofilin binding to F-actin reduces its effective binding capacity for actin binding proteins (ABPs) that interact with three protomers (such as Arp2/3 or Drb1-300) by (1–c)1.94. According to the above simulations, factor 1.94 appears to be accurate at least in the range of 0–0.3 cofilin occupancy [Chan et al., 2009]. We applied this approach to fit our Drb1-300–hCof1 binding competition data. As shown in Fig. 3A, the data (for Drb1-300) are in reasonable agreement with a direct competition mechanism in the range of cofilin binding density from 0 to 0.4. At higher cofilin binding densities the amount of drebrin bound exceeds that expected for a direct competition model (Fig. 3A, dashed line), which was in part anticipated due to the cooperativity of its binding to F-actin. Most importantly, the lack of allosteric (indirect) inhibition of binding revealed by this analysis suggests that cofilin and drebrin can either overcome or reverse long-range allosteric changes in F-actin induced by the competitor’s binding. For both drebrin constructs studied here, F-actin decoration with cofilin and drebrin yields a total combined binding density >1 (Figs. 3A and 3B). Several possible scenarios (or their combinations) can account for this: (1) drebrin and cofilin co-binding to the same stretches of actin filaments and the formation of “ternary complexes”; (2) deviation from the assumed binding stoichiometry due to drebrin binding to filaments’ stretches shorter than 3 (for Drb1-300) or 5 (for FL) protomers; (3) non-specific interactions of drebrin and/or cofilin with actin filaments. It should be noted that sedimentation velocity data–obtained for both drebrin constructs used in this study—shows no dimerization or higher order species formation in the absence of F-actin ([Sharma et al., 2010] and data not shown).
We evaluated also the effect of pH on cofilin-drebrin competition for binding to F-actin. As shown in Fig. 3C, at a slightly acidic pH (6.8) cofilin displaces Drb1-300 from actin filaments more efficiently than at pH 7.5. This can be driven by cofilin’s higher affinity to F-actin at lower pH [Ressad et al., 1998].
To quantify the effect of drebrin constructs on cofilin binding, we titrated pyrene-labeled F-actin with increasing amounts of hCof1 in the presence and absence of Drb1-300 and DrbA-FL (Fig. 3D). We documented that Kd(app) of cofilin binding (0.21 ± 0.05 μM) to F-actin increases by a factor of ~2 and ~4 in the presence of Drb1-300 and DrbA-FL (to 0.4 and 0.9 μM, respectively). These results suggest that drebrin decoration decreases only mildly cofilin’s affinity to actin filaments, consistent with drebrin’s partial inhibition of their severing by cofilin.
Kinetics of Drebrin Binding to Actin Filaments
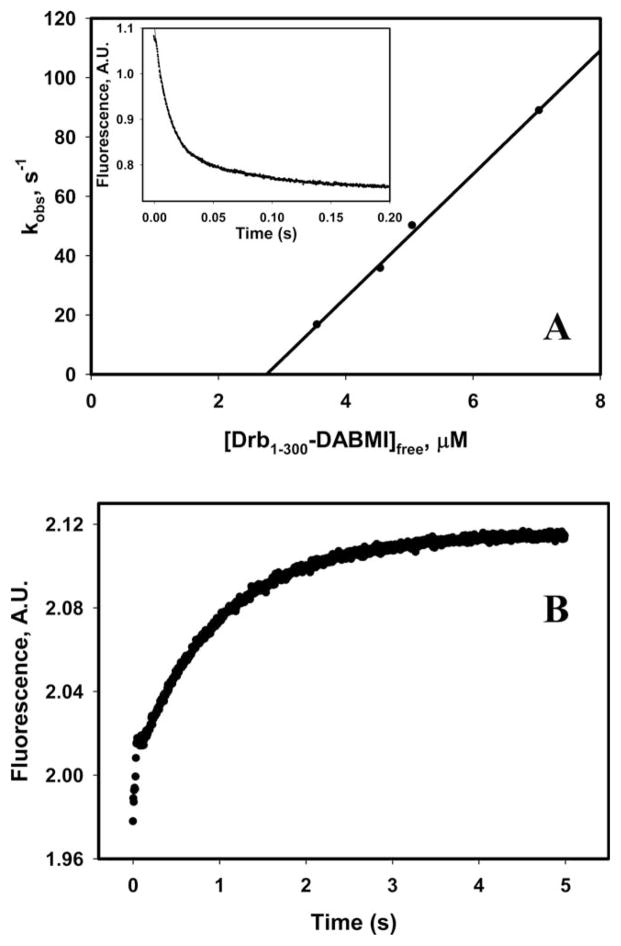
To gain farther insight into the mechanism of drebrincofilin competition, we resolved the kinetics of F-actin-drebrin interaction (Fig. 4) and compared it to the kinetic data reported previously for human cofilin-1 binding to skeletal F-actin. Our use of Drb1-300 in these experiments is based on earlier demonstration that it represents drebrin’s actin-binding core and closely mimics the properties of the FL protein [Grintsevich et al., 2010a; Sharma et al., 2012]. Drb1-300 clustering on actin filaments was also observed by AFM indicating cooperative binding [Sharma et al., 2012]. Here, we employed Förster resonance energy transfer (FRET) between F-actin-IADANSE and DABMI-labeled Drb1-300KCK construct to measure drebrin binding kinetics to actin filaments. Drb1-300DABMI (in large excess over actin’s 3-protomer binding sites; see Materials and Methods) was mixed in stopped flow apparatus with a fixed concentration of IADANSE-labeled filaments and the quenching of IADANSE fluorescence signal with time was recorded. The slope of the linear plot of kobs versus increasing concentrations of Drb1-300 (Fig. 4A) yields cooperative kon of 2 × 107 M−1·s−1 suggesting its fast binding to actin filaments, much faster than that reported for cofilin’s binding to actin (0.08 × 106 M−1·s−1; [Cao et al., 2006]).
Fig. 4. Kinetics of Drb1–300 binding to actin filaments.
(A) Dependence of kobs on the concentration of free Drb1-300-DABMI. Inset: Time course of a decrease in FRET signal upon interaction of skeletal F-actin-IAEDANS (2 μM) with 5.5 μM of Drb1-300-DABMI (raw data). (B) Increase in a FRET signal upon Drb1-300-DABMI (1.5 μM) dissociation from IAEDANS-labeled actin filaments due to its competition with unlabeled Drb1-300 (15 μM) (raw data).
The same FRET pair was used to measure the “off” rates of drebrin-F-actin interaction. To this end, DABMI-labeled Drb1-300 was competed off the filaments with 10–20-fold excess of an unlabeled construct. Figure 4B shows two phases of fluorescence increase. The first and very fast phase (completed within ~65 ms) was observed also in control samples (F-actin-Drb1-300 complex mixed with equal volume of buffer). We interpret this phase as representing drebrin dissociation from the ends of filaments upon their shredding in a stopped-flow mixer (see section below). The second phase can be described by a single exponential expression and was interpreted as a fast dissociation of Drb1-300 from the filaments. We did not observe significant dependence of the “off” rates on the nucleotide state of actin or phalloidin addition. In the absence and presence of phosphate (ADP-F-actin supplemented with Na2SO4 and ADP-Pi-F-actin, respectively), the k- was 1 and 0.85 s−1, respectively (Table I). We measured also the “off” rate of Drb1-300DABMI at two different binding densities (0.6 vs. >0.9 as determined by cosedimentation). As shown in Table I, the corresponding values of k- were very close suggesting that in this range of binding densities drebrin bound to F-actin predominantly in clusters and the contribution of the isolated site binding was too low to affect k-.
Table I.
Rates of Drb1–300-DABMI Dissociation from Different Forms of Skeletal F-Actin (2 μM).
| System | koff, s−1 | |
|---|---|---|
| F-actin-ADP + 1-300(1.5) | 0.90 | |
| F-actin-ADP-Ph + 1-300(1.5)a | 0.87 | |
| F-actin-ADP + Na2SO4 + 1-300(1.5)b | 1.02 | |
| F-actin-ADP + Na2SO4 + 1-300(0.5)b | 0.99 | |
| F-actin-ADP-Pi + 1-300(1.5)c | 0.85 | |
|
| ||
| kon, M−1s−1 | ||
|
| ||
| F-actin-ADP | 2.08 × 107 | |
Concentrations of Drb1-300 are given in parentheses (μM). F-actin was aged for at least 2 hours or overnight to obtain F-actin-ADP.
F-actin (aged) was stabilized with phalloidin (1:1 ratio).
Sample was supplemented with 25 mM of Na2SO4 for comparison with the phosphate-containing F-actin sample.
Sample was supplemented with 28 mM phosphate (pH 7.5).
Contribution of Filaments’ Severing to the Competition Between hCof1 and Drebrin
Considering the tighter binding of drebrin constructs to actin filaments (Kd = 0.05–0.2 μM, [Sharma et al., 2010; Grintsevich et al., 2010a], also Fig. 4) versus cofilin’s binding (0.88 μM; cooperative Kd calculated from [Cao et al., 2006]), the observed displacement of the former by the latter in equilibrium binding assays is rather surprising. We hypothesized that cofilin-mediated severing may contribute to this competition. Due to the cooperativity of drebrin binding and its requirement for stretches of 3–5 unoccupied protomers on F-actin this interaction may be sensitive to filament length, as reported previously for tropomyosin (Tm) [Broschat et al., 1989]. To test this possibility experimentally, we employed several actin and cofilin isoforms. First, we compared the displacement of Drb1-300 by hCof1 from skeletal versus yeast actin filaments. As shown in Fig. 5A, displacement of Drb1-300 by human cofilin-1 is greatly inhibited in the system containing yeast F-actin, which is not severed in the chosen range of cofilin concentrations [McCullough et al., 2011]. It should be noted, that similar to yeast cofilin, human cofilin-1 binds to yeast F-actin with high affinity (Kd = 16 nM), as shown previously by fluorescence titrations and cosedimentation assays [McCullough et al., 2011]. To ensure that the observed effect is not due to differences between actin isoforms, we examined the displacement of Drb1-300 from yeast F-actin triggered by additions of hCof1 (no severing) or yeast cofilin (extensive severing). Again, as shown in Fig. 5, much lower concentrations of yeast cofilin (compared to hCof1) are needed to compete off Drb1-300 from yeast actin filaments. These results suggest that the increase in a number of free ends or/and decrease in the filaments’ size contributes to drebrin-cofilin competition. We speculate that cofilin-mediated severing and shortening of the average size of filaments may lower drebrin affinity to such short filaments, as was previously described for tropomyosin [Broschat et al., 1989].
Fig. 5. Cofilin-mediated severing is a contributing factor in drebrin displacement from F-actin.
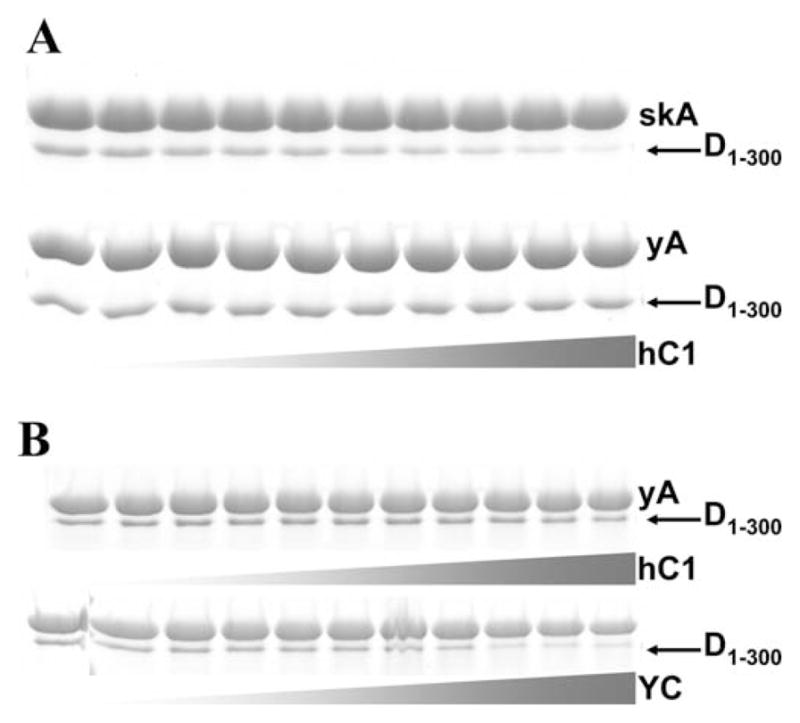
Gel analysis (SDS PAGE, 12.5%) of the samples. Supernatants and pellets were separated by high speed centrifugation. Representative gels stained with Coomassie Blue are shown (pellets only). (A) hCof1 at increasing concentrations (0.25; 0.4; 0.5; 0.6; 0.7; 0.8; 1; 1.5; 2; 2.5 μM) is added to 2 μM skeletal (top panel) or yeast F-actin (bottom panel) supplemented with Drb1-300 (1.5 μM); (B) hCof1 at increasing concentrations (0; 0.25; 0.4; 0.5; 0.6; 0.7; 0.8; 1; 1.5; 2; 2.5 μM) (top panel) or yeast cofilin (at the same concentrations as hCof1, bottom panel) is added to yeast F-actin (2 μM) supplemented with Drb1-300 (0.7 μM). Protein bands are labeled as follows: skA—rabbit skeletal actin; yA–yeast Saccharomyces cerevisiae actin; YC–Saccharomyces cerevisiae cofilin; hC1—human cofilin-1; D1–300—Drb1-300. Gray gradient-colored triangles represent schematically the range of increasing cofilin concentration (0–2.5 μM, from left to right). Conditions: 1× KM2EH7.5 buffer supplemented with 0.2 mM ATP and 1 mM DTT.
Discussion
Drebrin versus Cofilin Binding to Actin Filaments
Recent studies revealed several important aspects of F-actin-drebrin interaction: (i) drebrin (FL and Drb1-300) induces morphological changes in actin filaments that result in an increase in actin’s helical pitch from 36 to 40 nm; (ii) these changes propagate into undecorated F-actin regions (over up to two helical crossovers or 26 protomers); (iii) drebrin binding to F-actin is cooperative [Sharma et al., 2012]. The same set of features–cooperative binding, morphological changes in F-actin (albeit resulting in a decreased pitch) and their conformational spread into bare filaments’ regions—was reported previously for cofilin [McGough et al., 1997; Bobkov et al., 2006; Cao et al., 2006; De La Cruz and Sept, 2010].
Based on the propagation of drebrin-induced morphological changes in F-actin, we expected to see indirect (allosteric) inhibition of cofilin binding to actin filaments in the presence of drebrin. One example of such allosteric inhibition is the competition between cofilin and Arp2/3 for F-actin binding. In this case, the spread of conformational change from cofilin-decorated regions lowers dramatically the affinity of Arp2/3 to actin filaments, resulting in <1 total combined occupancy (= binding density) of these two proteins on F-actin [Chan et al., 2009]. Our study revealed that this is not the case for drebrin and cofilin competition. Our equilibrium binding assays suggest that direct (non-allosteric) competition between these two proteins is predominant; as total binding density of 1 can be achieved upon filaments’ co-decoration with cofilin and drebrin (Figs. 3A, 3B, and 6). This implies that cofilin can bind in immediate proximity to drebrin clusters and vice versa. Moreover, we found that cofilin binding to drebrin-decorated F-actin is weaker by a factor of 2–4 (for Drb1-300 and FL, respectively) compared to the bare actin control (Fig. 3D). For comparison, the affinity of Arp2/3 to F-actin decreases ~34-fold upon cofilin decoration [Chan et al., 2009].
Fig. 6. Different scenarios of drebrin-cofilin competition for F-actin binding.
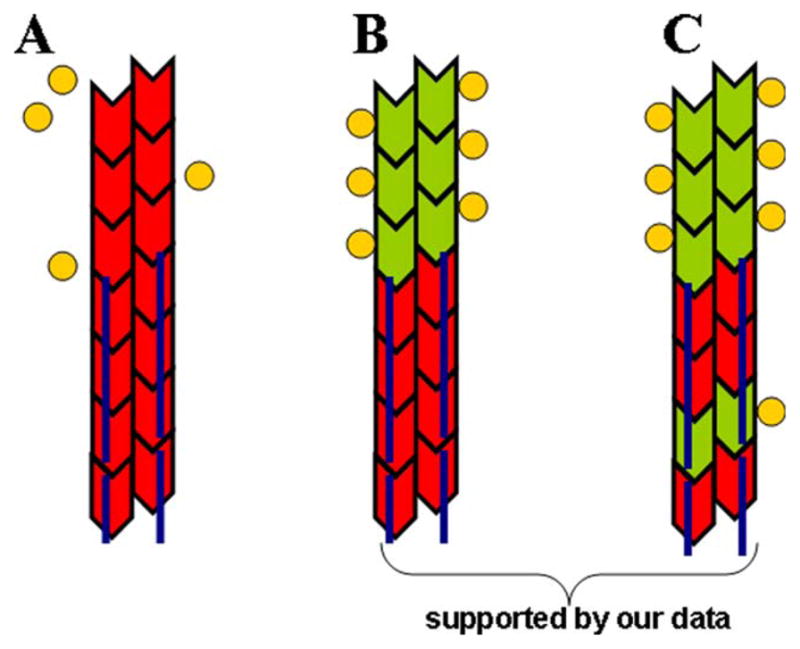
In red—F-actin sites unavailable for cofilin binding either due to their protection by drebrin or due to the propagation of 40 nm change in helical repeat [Sharma et al., 2010; Sharma et al., 2012] from a drebrin cluster; green—F-actin sites available for cofilin binding; yellow circles—cofilin; dark blue line—Drb1-300 (interacts with 3 actin protomers). (A) Direct and allosteric inhibition of cofilin binding by drebrin. Undecorated actin region next to the drebrin cluster is unavailable for cofilin binding due to the propagation of morphological changes in actin filaments. Such allosteric effect, together with direct competition for F-actin binding, would lead to a dramatic decrease in cofilin’s affinity to drebrin-decorated filaments (not observed); (B) Direct competition. Drebrin-decorated actin region is protected from cofilin binding due to their competition for binding sites on F-actin. Cofilin affinity to F-actin is only mildly reduced in the presence of saturating concentrations of drebrin allowing it to bind to actin sites adjacent to drebrin clusters (consistent with our data); (C) Direct competition with the possibility of cofilin binding to F-actin within drebrin clusters. Total combined occupancy of drebrin and cofilin >1 suggests the possibility of drebrin and cofilin co-binding to F-actin, leading to a ternary complex formation (suggested by our data).
To investigate further the competition between drebrin and cofilin, we determined the kinetic parameters of drebrin binding to actin filaments. Based on equilibrium binding assays with acrylodan-labeled A167C/C374A yeast actin mutant [Sharma et al., 2012], drebrin binding to F-actin is not significantly affected by the presence of phosphate or F-actin stabilizing drug phalloidin (data not shown). To rule out the possibility that both “on” and “off” rates of drebrin binding to actin filaments are changed in the presence of these ligands, we examined their effects on drebrin dissociation from actin filaments (Table I). It should be noted that both phalloidin and phosphate significantly decrease the affinity of cofilin to F-actin. Such effect was not observed with Drb1-300 for which phalloidin and phosphate did not change its “off” rates from actin filaments more than 25% compared to control filaments (Table I). Our data suggests that—unlike cofilin–drebrin binding to actin filaments is neither phalloidin nor nucleotide-sensitive. This implies that in vivo ADP-Pi caps of F-actin-based neuronal structures will be decorated with drebrin but free of cofilin which has low affinity to ADP-Pi F-actin. The lack of phalloidin effect on equilibrium binding and dissociation kinetics of drebrin suggests that in contrast to cofilin, a wide range of dynamic motions in F-actin is not required for drebrin binding.
A comparison of kinetic parameters for cooperative binding of drebrin and cofilin to F-actin reveals several important differences. First, the association rate of Drb1-300 to skeletal F-actin is ~2 orders of magnitude faster than that reported for human cofilin-1 (2 × 107 and 0.08 × 106 M−1s−1, Fig. 4 and [Cao et al., 2006], respectively). Second, cofilin dissociation rate from F-actin is also slower (~10-fold, [Cao et al., 2006]) than that of Drb1-300. These differences in the association rates between cofilin and drebrin can be rationalized by structural polymorphism of actin filaments and the consequent heterogeneity of its binding sites available for actin-binding proteins [Galkin et al., 2010]. The faster binding of drebrin (compared to cofilin) can arise from a higher probability for transient occurrence of “undertwisted” (40 nm helical pitch) F-actin states, versus “overtwisted” states (~27–30 nm) that favor cofilin binding. It is also possible that the binding of the first of drebrin’s actin-interacting domains induces/stabilizes conformational changes in F-actin and facilitates the binding of other domains. Based on our data we speculate that drebrin binding interface on F-actin is more exposed than that of cofilin. The overall result is faster association and dissociation of drebrin from actin filaments. The kinetic parameters of drebrin interaction with F-actin determined here can be useful for future examination of its interrelations with other actin-binding proteins.
Functional Consequences of the Competition Between Drebrin and Cofilin for F-Actin Binding
We documented that in the presence of near saturating concentrations of drebrin cofilin-mediated severing is inhibited but not abolished (Fig. 1A–1F). According to a model—accepted currently in the field, cofilin-induced severing occurs at the boundaries between undecorated and cofilin-bound F-actin regions [McCullough et al., 2008; McCullough et al., 2011; Suarez et al., 2011]. This model proposes that F-actin partially decorated with cofilin accumulates stress from thermally driven fluctuations at the boundaries between bare (stiff, with persistence length (Lp) of 9.4 μm) and cofilin-decorated (flexible, Lp = 3 μm) segments, resulting in filaments’ fragmentation. Reduction of severing at saturating concentrations of drebrin suggests that cofilin occupancy on F-actin and/or the number of boundaries is decreased, but it still can bind and sever actin filaments. These results were unexpected considering that drebrin binds to actin filaments stronger than human cofilin-1 ([Cao et al., 2006] and Fig. 4). The following scenarios (or their combination) offer possible explanations for the observed effects. (1) It is possible that low binding stoichiometry of drebrin to actin filaments (1:3 and 1:5 for Drb1-300 and FL, respectively) results in the occurrence of small gaps (2–4 protomers) between drebrin-bound clusters on F-actin which are too short to accommodate a whole drebrin molecule. Cofilin binding within these gaps would create a boundary between flexible (cofilin-decorated) and stiff (drebrin-bound, Lp = 10.9 μm) actin regions and allow for severing. (2) Our equilibrium binding data support the possibility of actin filaments’ codecoration with cofilin and drebrin, with a maximum combined binding density >1 (Figs. 3A, 3B, and 6). We hypothesize that at least part of the F-actin-drebrin interacting interface might be compatible with cofilin binding and potentially allow for their interaction with the same segments of F-actin. The presence of cofilin-compatible binding sites on drebrin-decorated F-actin combined with very fast drebrin binding and dissociation rates, could explain why cofilin can occasionally sever filaments in the presence of near saturating concentrations of drebrin.
Preferential binding of cofilin to an “overtwisted” (27 nm helical pitch) F-actin conformation would appear to disfavor its interaction with drebrin-decorated filaments with their propagated increase in the length of helical repeats (40 nm). However, it should be noted that in contrast to cofilin, the structural basis of drebrin-induced change in the helical pitch from 36 to 40 nm remains unknown. It was previously documented that, similar to cofilin, the strongest actin-binding module of drebrin molecule (DrbABD) reduces the efficiency of cross-linking between Cys 374 (subdomain 1 of actin, SD1) and residue 41 (subdomain 2 of actin, SD2) on an adjacent protomer, which can be interpreted as an increase in the distance between SD1 and SD2 [Bobkov et al., 2006; Mikati et al., 2013]. In the case of drebrin such actin conformer could be stabilized by its short- and long-range effects on lateral and longitudinal interfaces in F-actin [Sharma et al., 2012; Mikati et al., 2013]. We speculate that such structural changes in F-actin may partially expose cofilin-binding interface on F-actin and do not inhibit (or can even facilitate) cofilin binding to the sites where steric clashes with drebrin molecules do not occur [De La Cruz and Sept, 2010].
Biological Implications
Both cofilin and drebrin are enriched in DS. Our data show that drebrin inhibits but does not block cofilin function. It appears that cellular concentrations of drebrin and cofilin should be fine-tuned for normal spine function. According to previous reports, cofilin silencing and drebrin overexpression lead to similar defects in spine morphology— spines’ elongation and filopodia like protrusions have been observed [Hayashi and Shirao, 1999; Hotulainen et al., 2009; Ivanov et al., 2009b; Mizui et al., 2005]. This suggests that filaments’ fragmentation is essential for proper spine morphology and function. It is possible that drebrin-stabilized short filaments serve as seeds for F-actin repolymerization during activity-dependent spine remodeling or as mother filaments for Arp2/3-mediated branching.
Earlier work defines drebrin as “neuronal tropomyosin” due to its F-actin stabilizing effects and enrichment in neurons [Sekino et al., 2007]. One of the intriguing questions is why drebrin is needed in neurons together with the large variety of Tm isoforms. It was reported previously that filaments saturated with the mixture of neuronal Tm isoforms are even more ADF/cofilin-resistant than the those ones decorated with skeletal muscle Tm [Kuhn and Bamburg, 2008]. It is possible that such a strong effect of neuronal Tm is too restrictive for dynamic actin structures present in DS. Our data suggests that drebrin might be more suitable (compared to Tm) for the fast transition between stable and dynamic states of actin cytoskeleton in spines upon ADF/cofilin activation.
Materials and Methods
Proteins
Skeletal actin [Spudich and Watt, 1971], WT yeast actin [Grintsevich et al., 2008], yeast cofilin [Grintsevich et al., 2008], FL drebrin [Sharma et al., 2010], and Drb1-300 construct [Grintsevich et al., 2010a] were expressed and/or purified as described previously. Human cofilin 1 (hCof1) plasmid was a kind gift form Prof. E. M. De La Cruz. hCof1 expression in BL21(DE3)pLysS cells (4 hr, 37°C) was induced by 1 mM IPTG. HCof1 was purified employing ion exchange chromatography (strong cation exchanger, SP Sepharose FF, GE Healthcare) followed by gel-filtration on HiLoad 16/60 Superdex 75 (Amersham Biosciences) column.
TIRF Microscopy Assays and Analysis
In-solution severing experiments were conducted at pH 6.8 in order to inhibit cofilin-mediated actin depolymerization [McCullough et al., 2011]. Rabbit skeletal Mg-ATP-G-actin (30% Cy3-maleimide-labeled) was polymerized overnight, at 4°C, by addition of 10× KM2EI6.8 buffer (final concentrations: 50 mM KCl, 2 mM MgCl2, 0.2 mM ethylene glycol tetraacetic acid (EGTA), 20 mM imidazole, pH 6.8) supplemented with 0.2 mM ATP and 1 mM DTT. Cofilin severing was induced by mixing F-actin (1.5 μM) or F-actin-drebrin complexes with cofilin at 5:1 molar ratio. After allowing reactions to proceed for 3 min at RT, reaction mixtures were diluted to 2.1 nM in the 1× KM2EI6.8 buffer supplemented with 0.2 mM ATP, 1 μM phalloidin and 100 mM DTT and applied on the polylysine-coated coverslips. Longer incubation times (70 min) and higher ATP concentrations (1 mM) resulted in similar F-actin fragmentation upon cofilin severing (Grintsevich EE, unpublished observations). Filaments were imaged using DMI6000 TIRF microscope (Leica, Wetzlar, Germany). For each experimental condition 4–9 randomly selected fields were imaged per slide. On average ~180–350 filaments were analyzed per condition in each repeat. Filaments’ length was analyzed using a custom script provided kindly by Dr. Orkun Akin.
F-Actin Depolymerization Assays
Yeast WT Mg-ATP-G-actin (5% pyrene-maleimide) was polymerized for at least 2 hr (or overnight) with the addition of 10× KM2EH7.5 buffer (final concentrations: 50 mM KCl, 2 mM MgCl2, 0.4 mM EGTA, 10 mM HEPES, pH 7.5). Filaments (2 μM, final concentration) were preincubated with Drb1-300 for ~3 min, at 25°C, and then gently mixed with hCof1 (1.5 μM) in the same buffer. Mixing time (~35 s on average) was noted for each sample. Changes in pyrene fluorescence signal were recorded for few minutes. Initial rates of fluorescence change were calculated using data collected within the first 60 s of depolymerization. Linear fit of this data yielded values of F0 (at the intersection with Y axes). For each set of samples several depolymerization reaction were monitored till completion, to use as reference points and to correct the plateau levels for instrument-related drift of fluorescence signal. To determine the plateau levels, samples were incubated overnight at 4°C and the fluorescence signal was recorded the next day. Samples were subjected then to high speed ultracentrifugation (TLA100, 90,000 rpm, 4°C, 20 min) and supernatants and pellets were analyzed separately by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS PAGE). After determining WT actin concentration in supernatants and total changes in F (F0-F[plateau]), changes in F were converted into μM/fluorescence count. Using these values and the initial rates, depolymerization rates were calculated and expressed in nM/s.
Fluorescence Titration Assays
Equilibrium binding of cofilin to drebrin-decorated and bare actin filaments (100% pyrene-maleimide labeled) was monitored by measuring their fluorescence at 25°C in KM2EI6.8 buffer supplemented with 0.2 mM ATP and 1 mM DTT using a Photon Technologies Intl. (South Brunswick, NJ) fluorimeter. Samples were excited at 365 nm and the fluorescence emission signal was recorded at 407 nm and used for analysis. Data was normalized using values of fluorescence signal in the absence of cofilin (F0). Normalized curves were fitted to Hyperbolic Decay using SigmaPlot software and yielded the values of Kd(app).
Cosedimentation Assays
Reaction mixtures (120 μl) were incubated at RT for 20 min and then subjected to high speed ultracentrifugation (TLA100, 90,000 rpm, 4°C, 20 min). Pellets were concentrated 5-fold for gel analysis. Supernatants and pellets were analyzed by SDS PAGE. Known concentrations of G-actin, drebrin, and cofilin (three dilutions for each protein) were loaded on the gels as standards. The amounts of G-actin, F-actin, cofilin, and drebrin were determined by densitometry analysis of protein bands on the gels. Binding densities were calculated based on the following F-actin binding stoichiometries: 1:1 for hCof1 [McGough et al., 1997]; 1:3 for Drb1-300 [Grintsevich et al., 2010a]; 1:5 for DrbA-FL [Ishikawa et al., 1994; Sharma et al., 2010]. Occupancy of Drb1-300 on actin filaments in the presence of increasing concentrations of cofilin was modeled with an equation describing competitive binding (modified from Chan et al [Chan et al., 2009]):
| (1) |
where Kd = 0.2 μM (binding affinity of Drb1-300 to F-actin) [Grintsevich et al., 2010a]; Amax = maximum number of Drb1-300 binding sites on F-actin ([F-actin]/3); c—cofilin binding density; 1.94—reflects the probability of locating any three adjacent unoccupied protomers [Chan et al., 2009] at the (1–c) concentration of free actin. Factor 1.94 was calculated for F-actin binding protein interacting noncooperatively with 3-protomer sites on F-actin (Arp2/3). Since the cooperativity of cofilin and Drb1-300 binding to F-actin is relatively low, this simplified approach allowed for estimation of Drb1-300 occupancy at increasing binding densities of cofilin and ruled out non-competitive (allosteric) drebrin displacement through cofilin-induced long-range conformational changes in the filaments. Data points falling below the calculated curve would indicate that free 3-protomer sites—although available–are structurally altered due to long-range (allosteric) effects of cofilin, and Drb1-300 binding to such sites is decreased or abolished (not observed). Data points above the calculated curve would reflect a contribution of the cooperative binding of both, Drb1-300 and cofilin to actin filaments and would be consistent with their direct competition (observed). Total combined binding density of both proteins will report on the possibility of the “ternary” complex formation (actin-Drb1-300-cofilin).
Pre-steady State Kinetic Measurements
Drb1-300-KCK construct was expressed, purified and labeled with 4-dimethylaminophenylazophenyl-4′-maleimide (DABMI) using previously described procedure [Sharma et al., 2012] with the following modifications. DABMI solution (in 100% DMF) was added to the protein at 1:3 molar ratio (Drb1-300KCK:DABMI). After the labeling (10 min at RT), preparations were passed through 0.5 ml Zeba™ Spin Desalting Column (Thermo Scientific) and spun down at 21,000 g, for 20 min, at 4°C. The resulting supernatants were used in the assays. The extent of DABMI incorporation was determined under denaturing conditions (5.2 M guanidinium HCl) using ε 472 = 14,226 M−1·cm−1 corrected for the solvent conditions, as previously described [Grintsevich et al., 2010b]. Skeletal actin labeling with 5-[[((2-iodoacetyl)amino)ethyl] amino]naphthalene-1-sulfonic acid (IADANSE) was performed as previously described [Bobkov et al., 2006]. Binding of Drb1-300KCK-DABMI to IADANSE-F-actin was confirmed by cosedimentation. Transient kinetics measurements were carried out at 25°C in Applied Photophysics SX·18MV (Leatherhead, UK) stopped-flow apparatus. Excitation wavelength was set to 340 nm and the emission was monitored through a 435 nm direct filter. Stopped flow experiments were performed in 1×KM2EH7.5 supplemented with 0.2 mM ATP and 1 mM DTT. Samples supplemented with equimolar concentration of Na2SO4 were used as controls for reaction mixtures containing phosphate.
For association rate constants (k+) measurements total actin concentration was 1.5 μM and the critical concentration was assumed to be 0.1 μM. Given the 1:3 Drb1-300:F-actin binding stoichiometry and effective F-actin concentration of 1.4 μM (1.5–0.1 μM), the maximum concentration of 3-protomer binding sites was 0.47 μM (equals to maximum concentration of bound Drb1-300). Since we employed Drb1-300 concentrations in the range of 4–7.5 μM (i.e., at least 9-fold higher than the concentration of 3-protomer binding sites), pseudo-first order conditions were assumed in the analysis of “on” rates. The data were fitted to single exponential expression by non-linear least-squares in SigmaPlot, from which the kobs values were calculated. Concentrations of Drb1-300-DABMI(free) were calculated as [Drb1-300-DABMI(total)-Drb1-300-DABMI(bound)] based on equilibrium Kd = 0.2 μM, binding stoichiometry 1:3, and maximum concentration of 3-protomer binding sites. The slope of linear dependence of kobs on Drb1-300-DABMI(free) yields cooperative k+ composed of an unknown association rate constant for the isolated site on F-actin (kss+) and a cooperativity parameter for Drb1-300 binding to actin filaments (ω+) [Cao et al., 2006]:
Acknowledgments
The authors thank Prof. Margot Quinlan for the access to the TIRF microscope and valuable comments on the manuscript. The authors also thank Dr. Orkun Akin for providing his custom script for the filaments’ length analysis and Dr. Justin Bois for valuable discussions. This work was supported by the US Public Health Service through grant GM 077190 (to E.R.).
Abbreviations used
- AD
Alzheimer’s disease
- Drb1-300
drebrin a.a.1-300
- DrbABD
drebrin actin binding domain, a.a. 233-317
- DrbA-FL
drebrin A full length (Mus Musculus)
- hCof1
human cofilin-1
- TIRF microscopy
total internal reflection fluorescence microscopy
Footnotes
The Authors declare no conflict of interest.
References
- Andrianantoandro E, Pollard TD. Mechanism of actin filament turnover by severing and nucleation at different concentrations of ADF/Cofilin. Mol Cell. 2006;24(1):13–23. doi: 10.1016/j.molcel.2006.08.006. [DOI] [PubMed] [Google Scholar]
- Arber S, Barbayannis FA, Hanser H, Schneider C, Stanyon CA, Bernard O, Caroni P. Regulation of actin dynamics through phosphorylation of cofilin by LIM-kinase. Nature. 1998;393(6687):805–809. doi: 10.1038/31729. [DOI] [PubMed] [Google Scholar]
- Bobkov AA, Muhlrad A, Pavlov DA, Kokabi K, Yilmaz A, Reisler E. Cooperative effects of cofilin (ADF) on actin structure suggest allosteric mechanism of cofilin function. J Mol Biol. 2006;356(2):325–334. doi: 10.1016/j.jmb.2005.11.072. [DOI] [PubMed] [Google Scholar]
- Broschat KO, Weber A, Burgess DR. Tropomyosin stabilizes the pointed end of actin filaments by slowing depolymerization. Biochemistry. 1989;28(21):8501–8506. doi: 10.1021/bi00447a035. [DOI] [PubMed] [Google Scholar]
- Cao W, Goodarzi JP, De La Cruz EM. Energetics and kinetics of cooperative cofilin-actin filament interactions. J Mol Biol. 2006;361(2):257–267. doi: 10.1016/j.jmb.2006.06.019. [DOI] [PubMed] [Google Scholar]
- Carlier MF, Laurent V, Santolini J, Melki R, Didry D, Xia GX, Hong Y, Chua NH, Pantaloni D. Actin depolymerizing factor (ADF/Cofilin) enhances the rate of filament turnover: Implication in Actin-based motility. J Cell Biol. 1997;136(6):1307–1322. doi: 10.1083/jcb.136.6.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan C, Beltzner CC, Pollard TD. Cofilin dissociates Arp2/ 3 complex and branches from actin filaments. Curr Biol. 2009;19(7):537–545. doi: 10.1016/j.cub.2009.02.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho IH, Lee MJ, Kim DH, Kim B, Bae J, Choi KY, Kim SM, Huh YH, Lee KH, Kim CH, Song WK. SPIN90 dephosphorylation is required for cofilin-mediated actin depolymerization in NMDA-stimulated hippocampal neurons. Cell Mol Life Sci. 2013;70(22):4369–4383. doi: 10.1007/s00018-013-1391-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cingolani LA, Goda Y. Actin in action: The interplay between the actin cytoskeleton and synaptic efficacy. Nat Rev Neurosci. 2008;9(5):344–356. doi: 10.1038/nrn2373. [DOI] [PubMed] [Google Scholar]
- De La Cruz EM, Sept D. The kinetics of cooperative cofilin binding reveals two states of the cofilin-actin filament. Biophys J. 2010;98(9):1893–1901. doi: 10.1016/j.bpj.2010.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferhat L. Potential role of drebrin A, an F-actin binding protein, in reactive synaptic plasticity after pilocarpine-induced seizures: Functional implications in epilepsy. Int J Cell Biol. 2012;2012:1–12. doi: 10.1155/2012/474351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galkin VE, Orlova A, Schroder GF, Egelman EH. Structural polymorphism in F-actin. Nat Struct Mol Biol. 2010;17(11):1318–1323. doi: 10.1038/nsmb.1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grintsevich EE, Benchaar SA, Warshaviak D, Boontheung P, Halgand F, Whitelegge JP, Faull KF, Ogorzalek Loo RR, Sept D, Loo JA, Reisler E. Mapping the cofilin binding site on yeast G-actin by chemical cross-linking. J Mol Biol. 2008;377(2):395–409. doi: 10.1016/j.jmb.2007.12.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grintsevich EE, Galkin VE, Orlova A, Ytterberg AJ, Mikati MM, Kudryashov DS, Loo JA, Egelman EH, Reisler E. Mapping of drebrin binding site on F-actin. J Mol Biol. 2010a;398(4):542–554. doi: 10.1016/j.jmb.2010.03.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grintsevich EE, Phillips M, Pavlov D, Phan M, Reisler E, Muhlrad A. Antiparallel dimer and actin assembly. Biochemistry. 2010b;49(18):3919–3927. doi: 10.1021/bi1002663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi K, Shirao T. Change in the shape of dendritic spines caused by overexpression of drebrin in cultured cortical neurons. J Neurosci. 1999;19(10):3918–3925. doi: 10.1523/JNEUROSCI.19-10-03918.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotulainen P, Llano O, Smirnov S, Tanhuanpää K, Faix J, Rivera C, Lappalainen P. Defining mechanisms of actin polymerization and depolymerization during dendritic spine morphogenesis. J Cell Biol. 2009;185(2):323–339. doi: 10.1083/jcb.200809046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa R, Hayashi K, Shirao T, Xue Y, Takagi T, Sasaki Y, Kohama K. Drebrin, a development-associated brain protein from rat embryo, causes the dissociation of tropomyosin from actin filaments. J Biol Chem. 1994;269(47):29928–29933. [PubMed] [Google Scholar]
- Ivanov A, Esclapez M, Ferhat L. Role of drebrin A in dendritic spine plasticity and synaptic function. Commun Integr Biol. 2009a;2(3):268–270. doi: 10.4161/cib.2.3.8166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov A, Esclapez M, Pellegrino C, Shirao T, Ferhat L. Drebrin A regulates dendritic spine plasticity and synaptic function in mature cultured hippocampal neurons. J Cell Sci. 2009b;122(4):524–534. doi: 10.1242/jcs.033464. [DOI] [PubMed] [Google Scholar]
- Kojima N, Shirao T. Synaptic dysfunction and disruption of postsynaptic drebrin-actin complex: A study of neurological disorders accompanied by cognitive deficits. Neurosci Res. 2007;58(1):1–5. doi: 10.1016/j.neures.2007.02.003. [DOI] [PubMed] [Google Scholar]
- Kuhn TB, Bamburg JR. Tropomyosin and ADF/cofilin as collaborators and competitors. Adv Exp Med Biol. 2008;644:232–249. doi: 10.1007/978-0-387-85766-4_18. [DOI] [PubMed] [Google Scholar]
- McCullough BR, Blanchoin L, Martiel JL, De La Cruz EM. Cofilin increases the bending flexibility of actin filaments: Implications for severing and cell mechanics. J Mol Biol. 2008;381(3):550–558. doi: 10.1016/j.jmb.2008.05.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullough BR, Grintsevich EE, Chen CK, Kang H, Hutchison AL, Henn A, Cao W, Suarez C, Martiel JL, Blanchoin L, Reisler E, De La Cruz EM. Cofilin-linked changes in actin filament flexibility promote severing. Biophys J. 2011;101(1):151–159. doi: 10.1016/j.bpj.2011.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGough A, Pope B, Chiu W, Weeds A. Cofilin changes the twist of F-actin: Implications for actin filament dynamics and cellular function. J Cell Biol. 1997;138(4):771–781. doi: 10.1083/jcb.138.4.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikati MA, Grintsevich EE, Reisler E. Drebrin-induced stabilization of actin filaments. J Biol Chem. 2013;288(27):19926–19938. doi: 10.1074/jbc.M113.472647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizui T, Takahashi H, Sekino Y, Shirao T. Overexpression of drebrin A in immature neurons induces the accumulation of Factin and PSD-95 into dendritic filopodia, and the formation of large abnormal protrusions. Mol Cell Neurosci. 2005;30(1):149–157. doi: 10.1016/j.mcn.2005.06.008. [DOI] [PubMed] [Google Scholar]
- Ressad F, Didry D, Xia GX, Hong Y, Chua NH, Pantaloni D, Carlier MF. Kinetic analysis of the interaction of actin-depolymerizing factor (ADF)/Cofilin with G- and F-Actins. J Biol Chem. 1998;273(33):20894–20902. doi: 10.1074/jbc.273.33.20894. [DOI] [PubMed] [Google Scholar]
- Sekino Y, Kojima N, Shirao T. Role of actin cytoskeleton in dendritic spine morphogenesis. Neurochem Int. 2007;51(2–4):92–104. doi: 10.1016/j.neuint.2007.04.029. [DOI] [PubMed] [Google Scholar]
- Sharma S, Grintsevich EE, Phillips ML, Reisler E, Gimzewski JK. Atomic force microscopy reveals drebrin induced remodeling of F-actin with subnanometer resolution. Nano Lett. 2010;11(2):825–827. doi: 10.1021/nl104159v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S, Grintsevich E, Hsueh C, Reisler E, Gimzewski J. Molecular cooperativity of drebrin1–300 binding and structural remodeling of F-Actin. Biophys J. 2012;103(2):275–283. doi: 10.1016/j.bpj.2012.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spudich JA, Watt S. The regulation of rabbit skeletal muscle contraction. I. Biochemical studies of the interaction of the tropomyosin-troponin complex with actin and the proteolytic fragments of myosin. J Biol Chem. 1971;246(15):4866–4871. [PubMed] [Google Scholar]
- Suarez C, Roland J, Boujemaa-Paterski R, Kang H, McCullough BR, Reymann AC, Guérin C, Martiel JL, De La Cruz EM, Blanchoin L. Cofilin tunes the nucleotide state of actin filaments and severs at bare and decorated segment boundaries. Curr Biol. 2011;21(10):862–868. doi: 10.1016/j.cub.2011.03.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonezawa N, Nishida E, Iida K, Yahara I, Sakai H. Inhibition of the interactions of cofilin, destrin, and deoxyribonuclease I with actin by phosphoinositides. J Biol Chem. 1990;265(15):8382–8386. [PubMed] [Google Scholar]
- Zhao L, Ma QL, Calon F, Harris-White ME, Yang F, Lim GP, Morihara T, Ubeda OJ, Ambegaokar S, Hansen JE, Weisbart RH, Teter B, Frautschy SA, Cole GM. Role of p21-activated kinase pathway defects in the cognitive deficits of Alzheimer disease. Nat Neurosci. 2006;9(2):234–242. doi: 10.1038/nn1630. [DOI] [PubMed] [Google Scholar]
- Zhou Q, Homma KJ, Poo Mm. Shrinkage of dendritic spines associated with long-term depression of hippocampal synapses. Neuron. 2004;44(5):749–757. doi: 10.1016/j.neuron.2004.11.011. [DOI] [PubMed] [Google Scholar]