Abstract
All insects are colonized by microorganisms on the insect exoskeleton, in the gut and hemocoel, and within insect cells. The insect microbiota is generally different from microorganisms in the external environment, including ingested food. Specifically, certain microbial taxa are favored by the conditions and resources in the insect habitat, by their tolerance of insect immunity, and by specific mechanisms for their transmission. The resident microorganisms can promote insect fitness by contributing to nutrition, especially by providing essential amino acids, B vitamins, and, for fungal partners, sterols. Some microorganisms protect their insect hosts against pathogens, parasitoids, and other parasites by synthesizing specific toxins or modifying the insect immune system. Priorities for future research include elucidation of microbial contributions to detoxification, especially of plant allelochemicals in phytophagous insects, and resistance to pathogens; as well as their role in among-insect communication; and the potential value of manipulation of the microbiota to control insect pests.
Keywords: endosymbiosis, immunity, insect nutrition, microbiota, symbiosis
Introduction
Insects are chronically colonized by microorganisms that are not overtly pathogenic and are often beneficial or even required by the insect host. Most of the cells in a healthy insect are microbial, and the microbiota accounts for up to 1–10% of the insect's biomass. As a result, an insect is fundamentally a multiorganismal entity.
The microbiology of healthy insects has become the focus of intense research interest in recent years. This heightened activity can be attributed to two linked developments: dramatic technical advances in sequencing technologies, enabling microorganisms to be identified and investigated in situ, and large consortial initiatives [e.g., Human Microbiome Project (commonfund.nih.gov/hmp/index), MetaHIT (metahit.eu)] that have successfully applied these technologies to study the resident microorganisms in humans (24) and raised awareness among biologists of the wider importance of animal-associated microbiota. Despite great interest, the study of insect-microbial interactions is still widely regarded as crossing traditional disciplinary boundaries, with the consequence that the literature is scattered among journals of microbiology, ecology, evolution, and molecular biology and physiology, as well as entomology. The purpose of this review is to synthesize this diffuse literature to provide an overview of interactions between insects and their resident microbiota.
Insect Habitats
An insect comprises multiple habitats for microorganisms. The most accessible habitats for microbial colonists are the external cuticle and the gut. Microorganisms that can breach the exoskeleton or gut wall can gain access to the hemocoel and a further set of habitats provided by insect cells.
Cuticle
Although the insect exoskeleton is correctly recognized as a vitally important physical barrier against microbial infections (116), it is also a substrate that can be colonized by various microorganisms. Up to 1,000 culturable bacterial cells are associated with the body surface of Drosophila melanogaster, two orders of magnitude fewer than are borne internally by flies of the same age (92). Factors limiting microbial populations on the insect cuticle can include physical disturbance (e.g., ecdysis and grooming behavior) as well as antimicrobial secretions (e.g., from the meta-pleural glands of ants, Hymenoptera) (122). The extent to which cuticle-associated bacteria can proliferate and form stable communities, as occurs on human skin (46), is largely unknown.
Cuticular structures that promote colonization by specific microorganisms have evolved in many insects. In particular, the mycangia, i.e., cuticular invaginations housing fungi in adult insects, can be considered as culture vessels in which fungi required by the insect's offspring are stored and protected against abiotic factors and contamination by other microorganisms. The defining feature of mycangia—that they house fungi—is somewhat artificial because at least some mycangia additionally bear bacteria (56, 101). Some cuticular modifications house bacteria exclusively. For example, solitary digger wasps of the tribe Philanthini retain Streptomyces spp. in cuticle-lined glandular reservoirs, in each of 5–6 antennal segments (61) (Figure 1a); and attine ants house actinobacteria of the genus Pseudonocardia in similar glandular invaginations, known as crypts or foveae, on the thorax, legs, or other locations of the body, varying with ant species (26).
Figure 1.
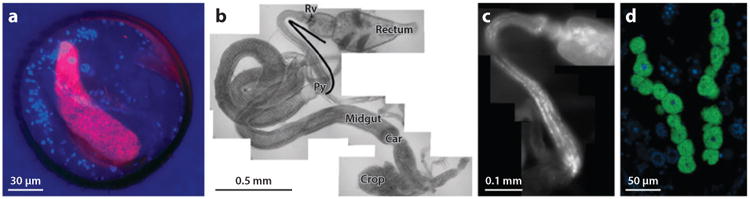
Insect habitats for microorganisms. (a) Antennal gland reservoir of the beewolf Philanthinus quattuordecimpunctatus, with “Candidatus Streptomyces philanthi”–specific probe SPT177-Cy3, the general eubacterial probe EUB784-FAM, and DAPI (blue) counterstain. “Ca. S. philanthi” binds both SPT177 and EUB784 probes, generating yellow fluorescence (note red autofluorescence of chitin at right) (micrograph of M. Kaltenpoth). (b) Dissected gut of Drosophila putrida [Py, pylorus; Rv, rectal valve; Car, cardia from a natural population in Rochester, NY, USA (micrograph by V. Martinson)]. (c) Expanded image of ileum in (b) showing bacteria, false color white fluorescence emission from general bacterial probe EUB338-Cy3 (micrograph by V. Martinson). (d) Section through embryo of black bean aphid Aphis fabae, showing Buchnera symbionts with general eubacterial probe EUB338-FITC (green) in bacteriocytes, and DAPI (blue) counterstain (micrograph by S. Chandler).
Gut
Some attributes of the insect gut are favorable for colonization by microorganisms, including ease of access for food-associated microbial cells, availability of nutrients, and protection from various stresses of the external environment (e.g., desiccation, ultraviolet radiation). Nevertheless, the insect gut poses multiple challenges for microorganisms ingested with the food, including unfavorable physicochemical conditions (e.g., oxygen content, pH, redox potential) in the gut lumen, secreted digestive enzymes and immune-related compounds, physical disturbance caused by peristalsis of gut contents, and loss of habitat at insect molts and metamorphosis. The conditions, resources, and hazards of the gut habitat for microorganisms vary among insect groups and with the life stage of the insect and region within the gut, reflecting the great variation in insect gut anatomy and physiology.
In many insects, the hindgut is the gut region bearing the largest microbial populations (Figure 1b,c). In particular, the ileum (the region between the proximal pylorus and distal rectum) is a relatively benign environment, in that it lacks the digestive enzymes of the midgut and, for many terrestrial insects, the desiccation stress of the distal hindgut, where water is actively resorbed from the lumen into insect tissues. Microbial function and growth may also be favored by the ions and metabolites delivered to the hindgut in the filtrate from the Malpighian tubules. In many insects, the ileum displays no evident morphological or physiological adaptations to maintain microorganisms, but the ileum of some insects (e.g., termites, scarab beetles) is expanded to form an anoxic fermentation chamber in which the microbiota degrade complex plant polysaccharides into products utilizable by aerobic metabolism of the insect (14, 54). In many insect taxa, the cuticle of the hindgut is thrown into spines and plates, and microorganisms can preferentially adhere to these structures (14).
The midgut tends to be a hostile environment for microorganisms. The midgut epithelium secretes an arsenal of enzymes and is immunologically very active. For example, the D. melanogaster midgut produces various antimicrobial peptides (70); a suite of digestive enzymes, including lysozymes (29, 102); and a dual oxidase (DUOX: NADPH oxidase) enzyme that generates microbicidal reactive oxygen species (ROS) (49). It also includes a region of pH < 3 that likely kills many microbial cells (102). However, the strongly acidic region of the midgut in D. melanogaster and other cyclorraphous dipterans is unusual among insects and may be a specific adaptation to bactivory; i.e., utilizing ingested bacteria as food (70). The midgut pH of many insects is mildly acidic to neutral (i.e., 6–7 units), which is suitable for a wide range of microorganisms, but the alkaline midgut (pH 8–12 units) of some insects, including larval lepidopterans, is likely inimical to many microorganisms (51). Compounding the various chemical barriers to the microbial colonization of the insect midgut is the physical barrier posed by the peritrophic matrix (PM), which separates the food bolus from the midgut epithelium. Many ingested microorganisms do not penetrate the PM and transit passively through the midgut with the bulk flow of food. Passage of certain microorganisms across the PM can be facilitated by chitinases of microbial or insect origin (34, 114), and some insects bear apparently benign bacterial communities in the ectoperitrophic space (between the PM and epithelial cells) (14).
For some insects, the dominant foregut habitat for microorganisms is provided by the crop, which can contain microorganisms at densities comparable to, or even exceeding, more distal gut regions (67, 100). However, the crop most commonly functions in the temporary storage of food and is evacuated regularly, raising the possibility that microorganisms may reside in this location for a relatively short period. Unusually, the dipteran olive fly Bactrocera oleae has an esophageal evagination, known as the cephalic bulb, which houses a dense culture of a single bacterium, “Candidatus Erwinia dacicola” (18). In insect vectors of plant or animal pathogens, other regions of the foregut have been identified as sites for microbial adhesion; e.g., the precibarium of the leafhopper Graphocephala atropunctata for the plant pathogen Xylella fastidiosa (85).
Cells
Intracellular microorganisms are widespread or universal in certain insect groups and restricted to cells whose sole function appears to be to maintain and house microorganisms (35). These insect cells are known as bacteriocytes, containing bacteria (Figure 1d), or mycetocytes, containing yeasts. The developmental origin of these insect cells is largely obscure, but the bacteriocytes of the aphid Acyrthosiphon pisum, although morphologically uniform, comprise two populations that differentiate at different stages in embryonic development (10). The dominant bacteria in bacteriocytes (primary symbionts) have no access to the external environment and are transmitted vertically (Table 1), usually by transfer to the ovaries of the female and, thence, to the cytoplasm of the egg. In this way, the host maintains very precise control over the location and abundance of the microorganisms in transit from bacteriocytes to offspring. Many insects with primary symbionts additionally bear other bacteria, known as secondary symbionts, which are associated with the bacteriocytes and are vertically transmitted but differ from primary symbionts in several important traits (Table 1).
Table 1. Characteristics of primary and secondary symbionts: bacteria associated with bacteriocytes of insectsa.
| Primary symbionts | Secondary symbionts |
|---|---|
| Restricted to bacteriocytes | May be located in bacteriocytes, sheath cells bounding bacteriocytes, and hemolymph |
| Present in all individual insects | Intermediate prevalence |
| Vertical transmission only | Vertical and horizontal transmission |
| Required by the insect | Can confer ecologically important traits (e.g., thermal tolerance, resistance to parasitoids or fungi); may reduce or promote insect fitness under laboratory conditions |
Insect-Associated Microorganisms
Taxonomic Diversity
The microbial inhabitants of insects comprise bacterial, archaeal, and eukaryotic (fungi and various unicellular eukaryotes) microorganisms. Viruses are not considered in this article. Although all insects are colonized by microorganisms, most microorganisms are not associated with insects (96). Four phyla of Bacteria (Actinobacteria, Bacteroidetes, Firmicutes, and Proteobacteria) are particularly strongly represented, but other phyla dominate certain insect groups; e.g., Spirochaetae, Fibrobacteres, and candidate phylum TG3 in the hindgut of the wood-feeding termites (Nasu-titermes spp.) (67, 120). Archaea generally are not associated with animals (48), although representatives of the Methanoarchaeota (methanogens) and the nonmethanogenic Thermoplasmatales and Halobacteriales are known in insects and are prevalent in the hindgut of cockroaches (order Blattodea), termites (infraorder Isoptera), and larval scarab beetles (family Scarabaeidae) (5, 15). Most of the eukaryotic microorganisms described in insects are fungi, especially ascomycetes (e.g., Clavicipitaceae, Saccharomycetes). Also well studied are the flagellate protists, apparently restricted to wood roaches and lower termites, and comprising members of the phylum Metamonada (the order Oxymonadida, and trichomonads and hypermastigotes within the class Parabasalia). Anaerobic ciliates of the order Clevelandellida (including Nyctotherus species) are also found in cockroaches and termites (100), and hemipterans, hymenopterans, and dipterans are often infected with trypanosomatids, which are generally benign but can be opportunistic pathogens (20, 74).
Evolutionary History and Ecological Status of Insect-Associated Microorganisms
The duration of relationships between many insects and their associated microorganisms varies across taxa. At one end of the spectrum, some insect-associated microbial taxa maintain substantial free-living populations or are closely allied to free-living microorganisms. In particular, Pantoea spp. (Gammaproteobacteria) are readily isolated from both environmental samples (e.g., water, soil, plant material) and insects, including mosquitoes (Diptera), thrips (Thysanoptera), bees (Hymenoptera), and hemipterans (115); and the Acetobacteraceae (Alphaproteobacteria), found in fruits and fermented foods and beverages, are also found in the guts of insects feeding on sugar-rich diets; e.g., bees, drosophilid fruit flies, and mosquitoes (25). At least some of these microbial populations may transfer regularly between the insect and the external environment, utilizing the insect as a route for dispersal. Other clades of microorganisms are widely distributed across insects and other animals but are unknown or rarely reported in the free-living condition; e.g., some lineages of Rikenellaceae and Porphyromonadaceae (Bacteroidetes), and Clostridiaceae and Ruminococcaceae (Firmi-cutes) (100, 111). At the other end of the scale are insect-specific species of bacteria, including some gut microorganisms [e.g., Snodgrassella alvi (Betaproteobacteria) and Gilliamella apicola (Gammaproteobacteria) in honey bees (Apis mellifera) (42, 69, 73)] and the primary and secondary symbionts associated with insect bacteriocytes (16, 35) (Table 1).
Assembly of Insect-Microbial Associations
The processes shaping the composition of insect microbial communities differ substantially between open associations (i.e., subject to invasion by external microorganisms) and closed associations (isolated by location and host factors from incoming microorganisms). Microbial communities on the cuticle and in the gut are generally open, whereas intracellular symbioses are predominantly closed.
Open Associations
The patterns of assembly of open associations in insects have been studied in gut symbioses, focusing particularly on the variation in the microbial communities with location within the gut. Generally, the microbiota varies longitudinally in the insect gut. For example, the density of microorganisms in the hindgut of the cockroach Shelfordella lateralis is 1–2 × 1010 cells/g, an order of magnitude greater than in the crop, ceca, and midgut (100); members of the bacterial phylum Firmicutes are abundant in the termite Nasutitermes corniger, but the dominant representatives of this phylum vary with gut region (Lactobacillales in the crop, Lachnospiraceae in the midgut, and Ruminococcaceae in the distal hindgut) (67); and the gut microbiota of larval Spodoptera littoralis (Lepidoptera) is dominated by Clostridium species in the midregion of the midgut but by Enterococcus spp. in more proximal and distal gut regions (110). The microbiota can also vary radially, with marked differences between the communities associated with the gut wall and the gut lumen. The gut wall community may dominate some aspects of the interactions with the host because it is persistent (not voided with bulk flow of the food). It can also define metabolite flux across the gut wall and affect the physicochemical conditions in the gut lumen (15, 67).
Diet plays a major role in structuring the gut microbial community. Effects on the microbiota have been observed in comparisons between artificial diets and natural foods, as well as between diets in which the major nutritional classes (protein, lipid, sugar, fiber) are varied (33, 64, 68, 97, 110). These studies raise important and largely unanswered questions about the processes determining the scale and direction of microbial responses to diet. In principle, diet can influence the gut microbiota directly and indirectly. With respect to direct effects, food-associated microorganisms ingested by the insect may vary with the composition of the food, and the microorganisms favored in the gut environment likely include those taxa that can best utilize food-derived nutrients in the gut lumen, including compounds intractable to host digestive enzymes. Indirect effects are mediated through the impact of food on gut anatomy, digestive function, and immunity and may be significant in the many insects where the microbiota in the gut and the food overlap weakly (5, 107, 110). The difference between the microbiota in the gut and that in the food can be exaggerated by behavioral adaptations that further promote the dominant gut microbial taxa, including coprophagy, trophallaxis (transfer of gut fluids by anus-to-mouth or mouth-to-mouth feeding), and maternal smearing of gut microorganisms on the eggshell, which is subsequently consumed by the offspring (11, 14).
The among-individual variation in the composition of the microbiota is substantial for the gut microbiota in some insects. Some of this variation may be driven by intraspecific genetic variation of the host, although the importance of host genotype in these interactions remains to be investigated systematically. Evidence for nongenetic sources of variation come from striking differences in the gut microbiota of single D. melanogaster strains reared on the same food (21, 121), suggesting that the microbiota may not be shaped exclusively by deterministic factors (e.g., gut pH, oxygen tensions). Stochastic processes, including the microorganisms that happen to be ingested, proximity between competing or mutualistic microbial cells in the gut environment, and the gut wall microsite where an individual microbial cell adheres may influence the composition of the gut community and persistence of individual taxa in the gut. Determining the relative contribution of deterministic (niche-based) and stochastic (neutral) processes in the assembly of the gut microbiota is an important challenge for future research.
Closed Associations
The bacteriocyte symbioses (Table 1) are predominantly closed systems, raising this question: How is the bacteriocyte protected from colonization by other microorganisms in the insect body while maintaining an intracellular environment that is suitable for the actively dividing primary symbionts? The immunological status of the bacteriocyte may be important, as is suggested by research on antimicrobial peptides (AMPs) in bacteriocytes of the weevil Sitophilus zeamais. AMPs are produced by the insect immune deficiency (IMD) signaling pathway, in response to the bacterial cell wall peptidoglycan (PGN) fragments, but the activity of this pathway is reduced in S. zeamais bacteriocytes by the high expression of an IMD-dependent PGN amidase, PGRP-LB, that degrades the immunogenic PGN fragments (6). Despite this generalized immunological suppression, one AMP (coleoptericin-A) is strongly expressed in the bacteriocytes, and when this AMP is reduced experimentally by RNAi, the symbionts overgrow the bacteriocytes and invade the insect body cavity (71). Downregulation of IMD signaling by PGRP-LB has also been demonstrated in the bacteriocytes of tsetse flies, Glossina spp. (119). However, this mechanism cannot explain the persistence of primary symbionts in all insects. For example, the aphid immune system lacks PGRPs, an intact IMD pathway, and recognizable AMPs expressed in bacteriocytes (47). Other candidate immune effectors may include lysozyme and small, cysteine-rich proteins, both of which are enriched in the transcriptome of aphid bacteriocytes (80, 105).
Phylogenetic analyses of intracellular symbioses in various insects reveal that many of these systems are not perfectly closed. Ancestral symbionts have been displaced by bacteria or yeasts; for example, in dryophthorid weevils (23), cerataphidine aphids (117), and philaenine spittlebugs (65).
Interactions Among Microbial Partners
The field of microbial ecology is replete with examples of interspecific interactions among microorganisms, including multiple mechanisms by which microbes compete for resources or enter into mutualistic consortia that can exploit resources unavailable to consortium members in isolation. This raises the possibility that interactions among microorganisms can affect the composition of the microbiota associated with insects. These interactions have not yet received extensive study but have been demonstrated in several associations.
In the D. melanogaster gut, the prevalence of the various bacteria is generally negatively related (121), suggestive of antagonistic interactions (although the alternative explanation that different individual insects meet the habitat requirements of different bacteria cannot be excluded). However, experimental colonization studies reveal great complexity, with both positive and negative relationships between the abundance of different Acetobacter and Lactobacillus species in the D. melanogaster gut (84). In the gut of the mosquito Aedes albopictus, the prevalence of Asaia and Acinetobacter spp. is positively related (78), but in the desert locust Schistocerca gregaria the abundance of Serratia marcescens is negatively correlated with the abundance of other bacteria (33).
The role of interactions among microorganisms in shaping microbial communities is also evident from the relationship between fungal associates of some insects and antibiotic production by actinobacteria borne on the insect exoskeleton. The dominant fungus partner of the bark beetle Dendroctonus frontalis are Entomocorticium spp., which line the galleries constructed by the insect in the phloem vessels of host trees, but this association is susceptible to invasion by a related fungus, Opisthosoma minus, which supports poor beetle growth. Protection against the antagonistic O. minus is provided by actinobacteria of the genus Streptomyces, which secrete a polyene peroxide antimicrobial that selectively inhibits the growth of O. minus (101). Similarly, antibiotics produced by actinomycete symbionts protect the fungal symbiont of attine ants against the fungal parasites of the genus Escovopsis (27, 28).
Microbial Impacts on Insect Phenotype
Nutrition
Many insect-associated microorganisms promote insect capacity to utilize diets of low or unbalanced nutritional content by providing specific nutrients that the insect cannot synthesize, including essential amino acids and B vitamins and sterols and, for insects feeding on diets rich in plant fiber, by degrading complex plant polysaccharides.
The role of microorganisms in provisioning essential amino acids has been demonstrated most conclusively in hemipteran insects feeding on plant phloem sap. The principal sources of nitrogen in phloem sap are the free amino acids of unbalanced composition, with <20% essential amino acids (the 9/10 of 20 amino acids that contribute to protein that cannot be synthesized by animals) (36). The key evidence that the primary symbiont, Buchnera aphidicola, in aphids synthesizes and releases essential amino acids is threefold: (a) Aphids have no dietary requirement for essential amino acids (unlike most animals) and can synthesize essential amino acids de novo, but they lose these capabilities when the Buchnera bacteria are eliminated by antibiotic treatment (39, 44); (b) isolated Buchnera bacteria release essential amino acids at linear rates for an hour or more (93); and (c) the Buchnera genome has retained the genetic capacity for essential amino acid synthesis, despite massive genome reduction (106). Microbial involvement in essential amino acid provisioning in other plant sap–feeding insects is indicated by the apparently universal incidence of symbioses in these insects (16) and by the retention of essential amino acid biosynthesis genes in all symbionts tested (76). Microbial symbionts have also been implicated in essential amino acid provisioning in ants (45), cockroaches (95), and some wood roaches (113).
Microorganisms associated with insects can gain access to nitrogenous precursors from dietary nitrogen, insect waste nitrogen, and nitrogen fixation. Insect nitrogenous waste is recycled to essential amino acids in the ant-Blochmannia symbiosis, in planthopper (Nilaparvata lugens)-yeast associations, and in cockroaches and termites (45, 90, 95, 98) but apparently not in the aphid-Buchnera symbiosis (72). Persuasive evidence for nitrogen fixation by insect-associated bacteria has been obtained for some termites (87). The microbiota in various other insects includes taxa with the genetic capacity to fix nitrogen and, in some instances, with demonstrable nitrogen fixation or acetylene reduction (which is a valid proxy for nitrogen fixation) (3, 7, 79, 82, 94), but the quantitative contribution of this capability to the nitrogen economy of the insects is largely unexplored.
B vitamins have been inferred to be provided by resident microorganisms, especially in insects feeding throughout the life cycle on vertebrate blood (e.g., the tsetse flies and other Diptera Pupipara, Cimicidae bed bugs, anopluran lice) and some phytophagous and xylophagous insects, including plant sap–feeding hemipterans, and various Coleoptera spp. of the families Anobiidae and Curculionidae (4, 16, 35, 75, 106). Contributions of microbiota to insect sterol nutrition relate exclusively to eukaryotic, particularly yeast, symbionts, because bacteria lack the capacity for sterol synthesis. A fungal source of insect sterols is indicated by the fungal sterol ergosterol and related compounds in the sterol profile of anobiid beetles (Coleoptera, Anobiidae) and planthoppers with yeast symbionts (83, 86). However, sterol analysis of the wood wasp Sirex noctilio suggests that this xylophage derives its sterols from the diet and not the fungal symbiont (112).
Microorganisms make a critical contribution to the degradation of plant cell wall material in insects that feed on sound wood and other plant products with a high lignocellulose content (e.g., termites, wood roaches, scarab beetle larvae). The microorganisms are located in a hindgut fermentation chamber, where they mediate the slow enzymatic degradation of the cellulose and hemicellulose components of the diet to sugars, which are then fermented to short-chain fatty acids and made available to the insect (17). Insects that feed on living plant material are largely independent of microbially mediated degradation of plant cell wall material because they subsist on the soluble carbohydrates and proteins in the plant cell contents and produce midgut glucosyl hydrolases capable of degrading plant cellulose and other plant cell wall polysaccharides (8, 17).
Protection Against Natural Enemies
Resident microorganisms can protect their insect hosts against pathogens and other natural enemies by multiple mechanisms that are not mutually exclusive, including competition for nutrients or space, production of toxins active against the invader, and activation of insect immune system functions that are more deleterious to the invader than the resident. Some of these mechanisms are equivalent to traits of environmental microorganisms that protect a resource patch, for example in soil or the water column, with the implication that protective traits of insect-associated microorganisms are not necessarily specific adaptations to the insect habitat. Microorganisms may defend their insect habitat against competing microorganisms that happen to include insect pathogens. Nevertheless, many protective functions of insect microbiota likely involve adaptations specific to the insect habitat, such as novel microbial chemistries against parasitoids. Coevolutionary interactions between insects and their microbiota are also expected and would lead to selection for reduced toxicity of the microbial agents against the host and coordination of the timing and magnitude of microbial toxin production to optimize protection of particularly vulnerable insect life stages or tissues.
There is now persuasive evidence that resident microorganisms can dictate the outcome of insect interactions with natural enemies, but understanding of the underlying mechanisms is fragmentary. The secondary symbiont Hamiltonella defensa confers pea aphid resistance to the parasitoid Aphidius ervi (88), but not all Hamiltonella spp. are protective. Function has been correlated with a bacteriophage in the Hamiltonella spp., and specifically with phage-encoded genes for toxins, such as Shiga-like toxin, cytolethal distending toxin, and YD-repeat toxins (31). A different group of toxins, polyketides, has been implicated in the Pseudomonas-mediated protection of Paederus rove beetles against predators (89) and in an undefined protective role of Profftella armatura, localized in the bacteriocytes of Asian citrus psyllid Diaphorina citri (81). ROS produced in the insect gut by either microorganisms or the insect gut epithelium can have strong antimicrobial effects. ROS production by prevalent gut bacteria Enterobacter spp. in anopheline mosquitoes inhibits the development of Plasmodium ookinetes into oocysts (22), and Leishmania parasites are sensitive to ROS induced by some members of the gut microbiota in their phlebotomine sand fly vector (Lutzomyia longipalpis) (32).
Antimicrobial compounds are of particular importance to insects living in enclosed, humid environments, where opportunistic fungal or bacterial infections can develop rapidly. Adult females of the solitary digger wasp Philanthus triangulum smear the ceiling of each subterranean brood cell with an antennal secretion containing antibiotic-producing Streptomyces spp. (Figure 1a); and the larva subsequently transfers the secretion to the surface of the cocoon. Survival is reduced from 80% to 10% if the Streptomyces bacteria are removed (60). Similarly, adults of the spruce bark beetle Dendroctonus rufipennis smear oral secretions containing bacteria onto the gallery walls of the trees they infest, likely conferring protection against antagonistic fungi, such as Aspergillus spp. (19).
From an evolutionary perspective, these beneficial effects of resident microorganisms in insects challenged by natural enemies can be attributed to strong selective overlap between the micro-biota and their insect host: Persistence of the insect habitat is advantageous to the microbiota. Some microbial partners may, however, respond to pathogen/parasitoid-mediated reduction in insect fitness by increased proliferation and dissemination from the failing insect. These microbial residents of insects are opportunistic pathogens. One possible instance of this response comes from the reduced virulence of baculovirus infecting Spodoptera exigua treated with antibiotic to eliminate the gut microbiota, relative to untreated caterpillars (59).
Detoxification of Toxins: Plant Allelochemicals and Insecticides
Most described instances of detoxification in insects are intrinsic. They are mediated by capabilities encoded by the insect genome, including cytochrome P450 monooxygenases, glutathione S-transferases, and esterases. Resident microorganisms have, however, been implicated in a few systems. Elimination of the yeast-like symbiont Symbiotaphrina kochi from Lasioderma serricorne beetles depresses larval development on diets containing allelochemicals that cultured S. kochi can degrade (40, 104). The capacity of the mountain pine beetle Dendroctonus ponderosae to utilize terpene-rich trees may be facilitated by species of Pseudomonas, Rahnella, and other resident gut bacteria that have the genetic capacity to degrade terpenes (1). A laccase enzyme produced by the fungal symbiont of attine ants also mediates the detoxification of plant material brought to the nest by the worker ants (30).
Resident microorganisms have repeatedly been proposed as a source of insecticide resistance, but most claims lack proper validation. Exceptionally, the resistance of the alydid stink bug Riptortus pedestris to the organophosphate fenitrothion is mediated by fenitrothion-degrading Burkholderia bacteria that are acquired from the soil by the insects (63). Further research is required to establish whether other insects benefit from microorganisms that can utilize both the insect habitat and the wider environment in this way.
A Source of Cues and Signals
Microorganisms associated with insects have been invoked as the source of chemicals that alter the behavior of conspecifics or other organisms (43), to the benefit or disadvantage of the insect host. To illustrate: The phenolic guaiacol in the aggregation pheromone of the desert locust Schistocerca gregaria is synthesized by Pantoea agglomerans and other Enterobacteriaceae in the insect gut (33); Drosophila prefer to mate with conspecifics that have a similar gut microbiota, and this preference is probably linked to microbiota-dependent variation in the cuticle hydrocarbon profile (103); and parasitic wasps of the bark beetle Dendroctonus ponderosae are attracted to logs containing the fungal partners (Grosmannia clavigera and Ophiostoma montium) of the beetle, suggesting that these para-sitoids use fungal volatiles as cues to locate beetle larvae and pupae (2). Most experimental studies, however, lack definitive evidence (38), and establishing precisely the role of the microorganisms in the synthesis of insect semiochemicals is a priority for future research in insect chemical ecology.
Resident Microorganisms in Economically Important Insects
The resident microbiota offers great potential for improved methods to manage economically important insects. Three primary opportunities are to predict the traits of insect pests, and hence efficacy of control strategies, from the composition of the microbiota, to target the microbiota for insect pest control, and to manipulate the microbiota to depress the vector competence of insects.
Predictor of Insect Pest Traits
Traits crucial to the management of certain insect pests are dictated by their possession of particular microorganisms. Examples include the secondary symbionts that determine the resistance of aphids to parasitoids and fungal pathogens used as biological control agents (88, 99); the pesticide-resistant Burkholderia strains that confer pesticide resistance to Riptortus pedestris (63); and bacteria that enable Megacopta stink bugs to utilize soybean crops (53). In these systems, the prevalence of the critical microorganisms in insect populations can be used to monitor the pest status and identify preferred control strategies. Regular monitoring would be required because the micro-biota in insect populations can change rapidly (52, 58). Monitoring may be particularly valuable for exotic insect species, whose invasiveness can depend on interactions with microorganisms in the introduced range. For example, the US turpentine beetle Dendroctonus valens (Scolytinae) is a minor forestry pest in its native range, but it has been causing high mortality to Chinese pines since its introduction to China in the 1980s (108), partly because of its acquisition of fungi from local Chinese Scolytinae (109). By contrast, Megacopta cribraria, introduced from eastern Asia to the eastern United States in 2009, appears to have retained the ancestral Ishikawaella symbiont; and its rapid transfer to soybean crops in the United States is not linked to symbiont switching (12).
A Target of Novel Insect-Pest-Control Strategies
The potential of insect pest control with the microbiota as the primary target is greatest for insects dependent on vertically transmitted microorganisms because the insect has no opportunity to acquire equivalent microorganisms from the environment. Many insects with vertically transmitted bacteriocyte symbioses (Table 1) are agricultural or medically important pests: aphids (super-family Aphidoidea), whiteflies (family Aleyrodidae), planthoppers (infraorder Fulgoroidea), and sharpshooters (tribe Proconiini of the family Cicadellidae)feeding on plant sap; anopluran lice (suborder Anoplura), bed bugs (family Cimicidae), and tsetse flies (family Glossinidae) feeding on vertebrate blood; pests of stored products and timber (e.g., various beetles of the families Curculionidae and Anobiidae); and cockroaches (order Blattodea). The population increase of these insects is abrogated by antibiotics that eliminate the microbial symbionts. The key priority is to identify alternatives to antibiotics that are cost-effective and specific. Recent advances in understanding the cellular processes underlying vertical transmission (66) and nutrient translocation between the insect and microbial partners (91, 93) are providing candidate molecular targets for disruption of these symbioses.
Microbially Mediated Manipulation of Insect Traits
The insects of greatest interest for microbially mediated manipulation are vectors of disease agents, especially mosquitoes. The goal is to introduce microorganisms that both suppress vector competence and promote their own dissemination through the insect population. Bacteria of the genus Wolbachia have long been identified as candidate microorganisms for this application. Most mosquito vector species are not infected with Wolbachia naturally, and the stable introductions of Wolbachia from Drosophila into Aedes aegypti (77) and Anopheles stephensi (9) are major breakthroughs. The insects bearing Wolbachia display enhanced resistance to dengue and chikungunya viruses and Plasmodium parasites, probably through heightened immunological function (9, 55, 62, 118). Field trials are investigating the fate of introduced Wolbachia-infected mosquitoes, with the long-term goal to release these insects to reduce disease transmission.
An alternative route for microbial manipulation of insect pests is to exploit members of the native microbiota of the insect vector by introducing gene(s) deleterious to the disease agent into a bacterial symbiont. Proof of principle has been obtained for Trypanosoma cruzi, the agent of Chagas disease, vectored by Rhodnius prolixus; when the gut symbiont, Rhodococcus rhodnii, was genetically modified to express the antimicrobial peptide cecropin A and then introduced to R. prolixus, transmission of T. cruzi was suppressed (41). Furthermore, R. prolixus populations in domestic environments are readily infected when they feed on mock fecal pellets containing the genetically modified bacteria, a formulation that has been developed as Cruzigard. Analogous approaches are under development to suppress transmission of the Leishmania parasite by sand flies Phlebotomus argentipes (57).
In principle, multiple opportunities are available to modify the pest status of insects by promoting members of the insect's native microbiota that influence vector competence or other traits of interest (e.g., plant range of crop pests, capacity for dispersal, mate choice, oviposition preference). Effective manipulation of native microbiota in pest management, however, depends on a detailed understanding of the function of insect-associated microorganisms and the interplay of factors that shape their abundance within an insect and dissemination through insect populations.
Perspectives and Future Directions
Microorganisms are ubiquitous in insects and have pervasive impacts on multiple aspects of insect biology. Consequently, microorganisms should be included as candidate factors affecting virtually any aspect of insect biology. Fortunately, the tools to study these associations are increasingly available, including methods to identify and quantify microorganisms and their functions, to manipulate the composition of the microbiota, and to investigate their interactions with the nutrition, immunity, and other physiological systems of the insect.
Some aspects of insect-microbe interactions now have a firm experimental foundation, but others remain contentious. Of particular interest for future research are the mechanisms by which resident microorganisms influence insect susceptibility to pathogens, insect capacity to degrade phytotoxins, and insect capacity to vector plant viruses and medically important disease agents. Careful experimental analyses are required to assess the generality of microorganisms as determinants of insect communication (38), plant range (50), resistance to insecticides (63), and insect speciation events (13).
Summary Points.
An insect represents multiple habitats, including the exoskeleton, gut lumen, and cells, that are colonized by microorganisms.
The composition and abundance of insect-associated microorganisms are shaped by the physicochemical conditions in the insect habitat, insect immune function, interactions among microorganisms, and transmission mechanisms of the insect.
Some microorganisms contribute to insect nutrition, by providing nutrients or degrading plant material intractable to insect digestion.
Resident microorganisms protect their insect hosts against natural enemies, including viruses, bacteria, and parasitoids, by synthesizing toxins or modulating the insect immune system.
Microorganisms have been implicated in the detoxification of dietary compounds and insecticides, and as the source of signals and cues important to insect communication, but the incidence and general significance of these functions are largely unexplored.
The resident microbiota of insects has great potential in promoting effective management of insect pests, as biomarkers for insect traits, as modulators of insect vector competence, and as targets for novel strategies to control pests and manipulate their traits.
Acknowledgments
I acknowledge the very many studies on insect microbiota that could not be included in this review because of space restrictions. I thank Dr. Martin Kaltenpoth, Dr. Vincent Martinson and Dr. Simon Chandler for providing micrographs, and Dr. Peter Newell and Dr. Rob Raguso for helpful comments on the manuscript. Financial support was provided by RO1 GM095372 (NIH), NYW-2011-04650 (AFRI-NIFA) and BIO 1241099 (NSF).
Footnotes
Disclosure Statement: The author is not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
Literature Cited
- 1.Adams AS, Aylward FO, Adams SM, Erbilgin N, Aukema BH, et al. Mountain pine beetles colonizing historical and naive host trees are associated with a bacterial community highly enriched in genes contributing to terpene metabolism. Appl Environ Microbiol. 2013;79:3468–75. doi: 10.1128/AEM.00068-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adams AS, Six DL. Detection of host habitat by parasitoids using cues associated with mycangial fungi of the mountain pine beetle, Dendroctonua ponderosae. Can Entomol. 2008;140:124–27. [Google Scholar]
- 3.Aharon Y, Pasternak Z, Ben Yosef M, Behar A, Lauzon C, et al. Phylogenetic, metabolic, and taxonomic diversities shape Mediterranean fruit fly microbiotas during ontogeny. Appl Environ Microbiol. 2013;79:303–13. doi: 10.1128/AEM.02761-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Akman L, Yamashita A, Watanabe H, Oshima K, Shiba T, et al. Genome sequence of the endocellular obligate symbiont of tsetse flies, Wigglesworthia glossinidia. Nat Genet. 2002;32:402–7. doi: 10.1038/ng986. [DOI] [PubMed] [Google Scholar]
- 5.Andert J, Marten A, Brandl R, Brune A. Inter- and intraspecific comparison of the bacterial assemblages in the hindgut of humivorous scarab beetle larvae (Pachnoda spp.) FEMS Microbiol Ecol. 2010;74:439–49. doi: 10.1111/j.1574-6941.2010.00950.x. [DOI] [PubMed] [Google Scholar]
- 6.Anselme C, Vallier A, Balmand S, Fauvarque MO, Heddi A. Host PGRP gene expression and bacterial release in endosymbiosis of the weevil Sitophilus zeamais. Appl Environ Microbiol. 2006;72:6766–72. doi: 10.1128/AEM.00942-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bansal R, Hulbert S, Schemerhorn B, Reese JC, Whitworth RJ, et al. Hessian fly-associated bacteria: transmission, essentiality, and composition. PLOS ONE. 2011;6:e23170. doi: 10.1371/journal.pone.0023170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berenbaum M. Adaptive significance of midgut pH in larval Lepidoptera. Am Nat. 1980;115:138–46. [Google Scholar]
- 9.Bian G, Joshi D, Dong Y, Lu P, Zhou G, et al. Wolbachia invades Anopheles stephensi populations and induces refractoriness to Plasmodium infection. Science. 2013;340:748–51. doi: 10.1126/science.1236192. [DOI] [PubMed] [Google Scholar]
- 10.Braendle C, Miura T, Bickel R, Shingleton AW, Kambhampati S, Stern DL. Developmental origin and evolution of bacteriocytes in the aphid-Buchnera symbiosis. PLoS Biol. 2003;1:E21. doi: 10.1371/journal.pbio.0000021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bright M, Bulgheresi S. A complex journey: transmission of microbial symbionts. Nat Rev Mi-crobiol. 2010;8:218–30. doi: 10.1038/nrmicro2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brown AM, Huynh LY, Bolender CM, Nelson KG, McCutcheon JP. Population genomics of a symbiont in the early stages of a pest invasion. Mol Ecol. 2014;23:1516–30. doi: 10.1111/mec.12366. [DOI] [PubMed] [Google Scholar]
- 13.Brucker RM, Bordenstein SR. The roles of host evolutionary relationships (genus: Nasonia) and development in structuring microbial communities. Evolution. 2012;66:349–62. doi: 10.1111/j.1558-5646.2011.01454.x. [DOI] [PubMed] [Google Scholar]
- 14.Brune A. Symbiotic associations between termites and prokaryotes. In: Dworkin M, Falkow S, Rosenberg E, Schleifer KH, Stackebrandt E, editors. The Prokaryotes. Vol. 1. New York: Springer; 2006. pp. 439–74. [Google Scholar]
- 15.Brune A. Methanogenesis in the digestive tracts of insects. In: Timmis KN, editor. Handbook of Hydrocarbon and Lipid Microbiology. Berlin: Springer-Verlag; 2010. pp. 706–28. [Google Scholar]
- 16.Buchner P. Endosymbioses of Animals with Plant Microorganisms. Chichester, UK: Wiley; 1965. [Google Scholar]
- 17.Calderon-Cortes N, Quesada M, Watanabe H, Cano-Camacho H, Oyama K. Endogenous plant cell wall digestion: a key mechanism in insect evolution. Annu Rev Ecol Evol Syst. 2012;43:45–71. [Google Scholar]
- 18.Capuzzo C, Firrao G, Mazzon L, Squartini A, Girolami V. ‘Candidatus Erwinia dacicola’, a co-evolved symbiotic bacterium of the olive fly Bactrocera oleae (Gmelin) Int J Syst Evol Microbiol. 2005;55:1641–47. doi: 10.1099/ijs.0.63653-0. [DOI] [PubMed] [Google Scholar]
- 19.Cardoza YJ, Klepzig KD, Raffa KF. Bacteria in oral secretions of an endophytic insect inhibit antagonistic fungi. Ecol Entomol. 2006;31:636–45. [Google Scholar]
- 20.Chandler JA, James PM. Discovery of trypanosomatid parasites in globally distributed Drosophila species. PLOS ONE. 2013;8:e61937. doi: 10.1371/journal.pone.0061937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chandler JA, Lang JM, Bhatnagar S, Eisen JA, Kopp A. Bacterial communities of diverse Drosophila species: ecological context of a host-microbe model system. PLOS Genet. 2011;7:e1002272. doi: 10.1371/journal.pgen.1002272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cirimotich CM, Dong Y, Clayton AM, Sandiford SL, Souza-Neto JA, et al. Natural microbemediated refractoriness to Plasmodium infection in Anopheles gambiae. Science. 2011;332:855–58. doi: 10.1126/science.1201618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Conrad C, Despres L, Vallier A, Balmand S, Miquel C, et al. Long-term evolutionary stability of bacterial endosymbiosis in Curculionoidea: additional evidence of symbiont replacement in the Dryophthoridae family. Mol Biol Evol. 2008;25:859–68. doi: 10.1093/molbev/msn027. [DOI] [PubMed] [Google Scholar]
- 24.Consort. Hum. Microb. A framework for human microbiome research. Nature. 2012;486:215–21. doi: 10.1038/nature11209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Crotti E, Rizzi A, Chouaia B, Ricci I, Favia G, et al. Acetic acid bacteria, newly emerging symbionts of insects. Appl Environ Microbiol. 2010;76:6963–70. doi: 10.1128/AEM.01336-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Currie CR, Poulsen M, Mendenhall J, Boomsma JJ, Billen J. Coevolved crypts and exocrine glands support mutualistic bacteria in fungus-growing ants. Science. 2006;311:81–83. doi: 10.1126/science.1119744. [DOI] [PubMed] [Google Scholar]
- 27.Currie CR, Scott JA, Summerbell RC, Malloch D. Fungus-growing ants use antibiotic-producing bacteria to control garden parasites. Nature. 1999;398:701–4. [Google Scholar]
- 28.Currie CR, Wong B, Stuart AE, Schultz TR, Rehner SA, et al. Ancient tripartite coevolution in the attine ant-microbe symbiosis. Science. 2003;299:386–88. doi: 10.1126/science.1078155. [DOI] [PubMed] [Google Scholar]
- 29.Daffre S, Kylsten P, Samakovlis C, Hultmark D. The lysozyme locus in Drosophila melanogaster: an expanded gene family adapted for expression in the digestive tract. Mol Gen Genet. 1994;242:152–62. doi: 10.1007/BF00391008. [DOI] [PubMed] [Google Scholar]
- 30.De Fine Licht HH, Schiøtt M, Rogowska-Wrzesinska A, Nygaard S, Roepstorff P, Boomsma JJ. Laccase detoxification mediates the nutritional alliance between leaf-cutting ants and fungus-garden symbionts. Proc Natl Acad Sci USA. 2013;110:583–87. doi: 10.1073/pnas.1212709110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Degnan PH, Moran NA. Diverse phage-encoded toxins in a protective insect endosymbiont. Appl Environ Microbiol. 2008;74:6782–91. doi: 10.1128/AEM.01285-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Diaz-Albiter H, Sant'Anna MR, Genta FA, Dillon RJ. Reactive oxygen species-mediated immunity against Leishmania mexicana and Serratia marcescens in the sand phlebotomine fly Lutzomyia longipalpis. J Biol Chem. 2012;287:23995–4003. doi: 10.1074/jbc.M112.376095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dillon R, Charnley K. Mutualism between the desert locust Schistocerca gregaria and its gut microbiota. Res Microbiol. 2002;153:503–9. doi: 10.1016/s0923-2508(02)01361-x. [DOI] [PubMed] [Google Scholar]
- 34.Dostálová A, Volf P. Leishmania development in sand flies: parasite-vector interactions overview. Parasites Vectors. 2012;5:276. doi: 10.1186/1756-3305-5-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Douglas AE. Mycetocyte symbiosis in insects. Biol Rev. 1989;64:409–34. doi: 10.1111/j.1469-185x.1989.tb00682.x. [DOI] [PubMed] [Google Scholar]
- 36.Douglas AE. Phloem-sap feeding by animals: problems and solutions. J Exp Bot. 2006;57:747–54. doi: 10.1093/jxb/erj067. [DOI] [PubMed] [Google Scholar]
- 37.Douglas AE. The microbial dimension in insect nutritional ecology. Funct Ecol. 2009;23:38–47. [Google Scholar]
- 38.Douglas AE, Dobson AJ. New synthesis: animal communication mediated by microbes: fact or fantasy? J Chem Ecol. 2013;39:1149. doi: 10.1007/s10886-013-0343-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Douglas AE, Minto LB, Wilkinson TL. Quantifying nutrient production by the microbial symbionts in an aphid. J Exp Biol. 2001;204:349–58. doi: 10.1242/jeb.204.2.349. [DOI] [PubMed] [Google Scholar]
- 40.Dowd PF, Shen SK. The contribution of symbiotic yeast to toxin resistance of the cigarette beetle (Lasioderma serricorne) Entomol Exp Appl. 1990;56:241–48. [Google Scholar]
- 41.Durvasula RV, Gumbs A, Panackal A, Kruglov O, Aksoy S, et al. Prevention of insect-borne disease: an approach using transgenic symbiotic bacteria. Proc Natl Acad Sci USA. 1997;94:3274–78. doi: 10.1073/pnas.94.7.3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Engel P, Martinson VG, Moran NA. Functional diversity within the simple gut microbiota of the honey bee. Proc Natl Acad Sci USA. 2012;109:11002–7. doi: 10.1073/pnas.1202970109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ezenwa VO, Gerardo NM, Inouye DW, Medina M, Xavier JB. Microbiology: animal behavior and the microbiome. Science. 2012;338:198–99. doi: 10.1126/science.1227412. [DOI] [PubMed] [Google Scholar]
- 44.Febvay G, Rahbe Y, Rynkiewicz M, Guillaud J, Bonnot G. Fate of dietary sucrose and neosynthesis of amino acids in the pea aphid, Acyrthosiphon pisum, reared on different diets. J Exp Biol. 1999;202:2639–52. doi: 10.1242/jeb.202.19.2639. [DOI] [PubMed] [Google Scholar]
- 45.Feldhaar H, Straka J, Krischke M, Berthold K, Stoll S, et al. Nutritional upgrading for omnivorous carpenter ants by the endosymbiont Blochmannia. BMC Biol. 2007;5:48. doi: 10.1186/1741-7007-5-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Findley K, Yang J, Conlan S, Deming C, et al. Topographic diversity of fungal and bacterial communities in human skin. Nature. 2013;498:367–70. doi: 10.1038/nature12171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gerardo NM, Altincicek B, Anselme C, Atamian H, Barribeau SM, et al. Immunity and other defenses in pea aphids, Acyrthosiphon pisum. Genome Biol. 2010;11:R21. doi: 10.1186/gb-2010-11-2-r21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gill EE, Brinkman FS. The proportional lack of archaeal pathogens: Do viruses/phages hold the key? BioEssays. 2011;33:248–54. doi: 10.1002/bies.201000091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ha EM, Oh CT, Bae YS, Lee WJ. A direct role for dual oxidase in Drosophila gut immunity. Science. 2005;310:847–50. doi: 10.1126/science.1117311. [DOI] [PubMed] [Google Scholar]
- 50.Hansen AK, Moran NA. The impact of microbial symbionts on host plant utilization by herbivorous insects. Mol Ecol. 2014;23:1473–96. doi: 10.1111/mec.12421. [DOI] [PubMed] [Google Scholar]
- 51.Harrison JF. Insect acid-base physiology. Annu Rev Entomol. 2001;46:221–50. doi: 10.1146/annurev.ento.46.1.221. [DOI] [PubMed] [Google Scholar]
- 52.Himler AG, Adachi-Hagimori T, Bergen JE, Kozuch A, Kelly SE, et al. Rapid spread of a bacterial symbiont in an invasive whitefly is driven by fitness benefits and female bias. Science. 2011;332:254–56. doi: 10.1126/science.1199410. [DOI] [PubMed] [Google Scholar]
- 53.Hosokawa T, Kikuchi Y, Meng XY, Fukatsu T. The making of symbiont capsule in the plataspid stinkbug Megacopta punctatissima. FEMS Microbiol Ecol. 2005;54:471–77. doi: 10.1016/j.femsec.2005.06.002. [DOI] [PubMed] [Google Scholar]
- 54.Huang SW, Zhang HY, Marshall S, Jackson TA. The scarab gut: a potential bioreactor for bio-fuel production. Insect Sci. 2010;17:175–83. [Google Scholar]
- 55.Hughes GL, Koga R, Xue P, Fukatsu T, Rasgon JL. Wolbachia infections are virulent and inhibit the human malaria parasite Plasmodium falciparum in Anopheles gambiae. PLoS Pathog. 2011;7:e1002043. doi: 10.1371/journal.ppat.1002043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hulcr J, Rountree NR, Diamond SE, Stelinski LL, Fierer N, Dunn RR. Mycangia of ambrosia beetles host communities of bacteria. Microb Ecol. 2012;64:784–93. doi: 10.1007/s00248-012-0055-5. [DOI] [PubMed] [Google Scholar]
- 57.Hurwitz I, Fieck A, Read A, Hillesland H, Klein N, et al. Paratransgenic control of vector borne diseases. Int J Biol Sci. 2011;7:1334–44. doi: 10.7150/ijbs.7.1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jaenike J, Unckless R, Cockburn SN, Boelio LM, Perlman SJ. Adaptation via symbiosis: recent spread of a Drosophila defensive symbiont. Science. 2010;329:212–15. doi: 10.1126/science.1188235. [DOI] [PubMed] [Google Scholar]
- 59.Jakubowska AK, Vogel H, Herrero S. Increase in gut microbiota after immune suppression in baculovirus-infected larvae. PLOS Pathog. 2013;9:e1003379. doi: 10.1371/journal.ppat.1003379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kaltenpoth M, Gottler W, Herzner G, Strohm E. Symbiotic bacteria protect wasp larvae from fungal infestation. Curr Biol. 2005;15:475–79. doi: 10.1016/j.cub.2004.12.084. [DOI] [PubMed] [Google Scholar]
- 61.Kaltenpoth M, Yildirim E, Gu¨rbu¨z MF, Herzner G, Strohm E. Refining the roots of the beewolf-Streptomyces symbiosis: antennal symbionts in the rare genus Philanthinus (Hymenoptera, Crabronidae) Appl Environ Microbiol. 2012;78:822–27. doi: 10.1128/AEM.06809-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kambris Z, Blagborough AM, Pinto SB, Blagrove MS, Godfray HC, et al. Wolbachia stimulates immune gene expression and inhibits Plasmodium development in Anopheles gambiae. PLoS Pathog. 2010;6:e1001143. doi: 10.1371/journal.ppat.1001143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kikuchi Y, Hayatsu M, Hosokawa T, Nagayama A, Tago K, Fukatsu T. Symbiont-mediated insecticide resistance. Proc Natl Acad Sci USA. 2012;109:8618–22. doi: 10.1073/pnas.1200231109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Koch H, Cisarovsky G, Schmid-Hempel P. Ecological effects on gut bacterial communities in wild bumblebee colonies. J Anim Ecol. 2012;81:1202–10. doi: 10.1111/j.1365-2656.2012.02004.x. [DOI] [PubMed] [Google Scholar]
- 65.Koga R, Bennett GM, Cryan JR, Moran NA. Evolutionary replacement of obligate symbionts in an ancient and diverse insect lineage. Environ Microbiol. 2013;15:2073–81. doi: 10.1111/1462-2920.12121. [DOI] [PubMed] [Google Scholar]
- 66.Koga R, Meng XY, Tsuchida T, Fukatsu T. Cellular mechanism for selective vertical transmission of an obligate insect symbiont at the bacteriocyte-embryo interface. Proc Natl Acad Sci USA. 2012;109:E1230–37. doi: 10.1073/pnas.1119212109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Köhler T, Dietrich C, Scheffrahn RH, Brune A. High-resolution analysis of gut environment and bacterial microbiota reveals functional compartmentation of the gut in wood-feeding higher termites (Nasutitermes spp.) Appl Environ Microbiol. 2012;78:4691–701. doi: 10.1128/AEM.00683-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kuechler SM, Dettner K, Kehl S. Characterization of an obligate intracellular bacterium in the midgut epithelium of the bulrush bug Chilacis typhae (Heteroptera, Lygaeidae, Artheneinae) Appl Environ Microbiol. 2011;77:2869–76. doi: 10.1128/AEM.02983-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kwong WK, Moran NA. Cultivation and characterization of the gut symbionts of honey bees and bumble bees: description of Snodgrassella alvi gen. nov., sp nov., a member of the family Neisseriaceae of the Betaproteobacteria, and Gilliamella apicola gen nov., sp nov., a member of Orbaceae fam nov., Orbales ord nov., a sister taxon to the order ‘Enterobacteriales’ of the Gammaproteobacteria. Int J Syst Evol Microbiol. 2013;63:2008–18. doi: 10.1099/ijs.0.044875-0. [DOI] [PubMed] [Google Scholar]
- 70.Lemaitre B, Miguel-Aliaga I. The digestive tract of Drosophila melanogaster. Annu Rev Genet. 2013;47:377–404. doi: 10.1146/annurev-genet-111212-133343. [DOI] [PubMed] [Google Scholar]
- 71.Login FH, Balmand S, Vallier A, Vincent-Monegat C, Vigneron A, et al. Antimicrobial peptides keep insect endosymbionts under control. Science. 2011;334:362–65. doi: 10.1126/science.1209728. [DOI] [PubMed] [Google Scholar]
- 72.Macdonald SJ, Lin GG, Russell CW, Thomas GH, Douglas AE. The central role of the host cell in symbiotic nitrogen metabolism. Proc R Soc Lond B. 2012;279:2965–73. doi: 10.1098/rspb.2012.0414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Martinson VG, Danforth BN, Minckley RL, Rueppell O, Tingek S, Moran NA. A simple and distinctive microbiota associated with honey bees and bumble bees. Mol Ecol. 2011;20:619–28. doi: 10.1111/j.1365-294X.2010.04959.x. [DOI] [PubMed] [Google Scholar]
- 74.Maslov DA, Votypka J, Yurchenko V, Lukes J. Diversity and phylogeny of insect trypanosomatids: All that is hidden shall be revealed. Trends Parasitol. 2013;29:43–52. doi: 10.1016/j.pt.2012.11.001. [DOI] [PubMed] [Google Scholar]
- 75.McCutcheon JP, Moran NA. Parallel genomic evolution and metabolic interdependence in an ancient symbiosis. Proc Natl Acad Sci USA. 2007;104:19392–97. doi: 10.1073/pnas.0708855104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.McCutcheon JP, Moran NA. Extreme genome reduction in symbiotic bacteria. Nat Rev Microbiol. 2012;10:13–26. doi: 10.1038/nrmicro2670. [DOI] [PubMed] [Google Scholar]
- 77.McMeniman CJ, Lane RV, Cass BN, Fong AW, Sidhu M, et al. Stable introduction of a life-shortening Wolbachia infection into the mosquito Aedes aegypti. Science. 2009;323:141–44. doi: 10.1126/science.1165326. [DOI] [PubMed] [Google Scholar]
- 78.Minard G, Tran FH, Raharimalala FN, Hellard E, Ravelonandro P, et al. Prevalence, genomic and metabolic profiles of Acinetobacter and Asaia associated with field-caught Aedes albopictus from Madagascar. FEMS Microbiol Ecol. 2013;83:63–73. doi: 10.1111/j.1574-6941.2012.01455.x. [DOI] [PubMed] [Google Scholar]
- 79.Morales-Jiménez J, Zúñiga G, Villa-Tanaca L, Hernández-Rodríguez C. Bacterial community and nitrogen fixation in the red turpentine beetle, Dendroctonus valens LeConte (Coleoptera: Curculionidae: Scolytinae) Microb Ecol. 2009;58:879–91. doi: 10.1007/s00248-009-9548-2. [DOI] [PubMed] [Google Scholar]
- 80.Nakabachi A, Shigenobu S, Sakazume N, Shiraki T, Hayashizaki Y, et al. Transcriptome analysis of the aphid bacteriocyte, the symbiotic host cell that harbors an endocellular mutualistic bacterium, Buchnera. Proc Natl Acad Sci USA. 2005;102:5477–82. doi: 10.1073/pnas.0409034102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Nakabachi A, Ueoka R, Oshima K, Teta R, Mangoni A, et al. Defensive bacteriome symbiont with a drastically reduced genome. Curr Biol. 2013;23:1478–84. doi: 10.1016/j.cub.2013.06.027. [DOI] [PubMed] [Google Scholar]
- 82.Nardi JB, Mackie RI, Dawson JO. Could microbial symbionts of arthropod guts contribute significantly to nitrogen fixation in terrestrial ecosystems? J Insect Physiol. 2002;48:751–63. doi: 10.1016/s0022-1910(02)00105-1. [DOI] [PubMed] [Google Scholar]
- 83.Nasir H, Noda H. Yeast-like symbiotes as a sterol source in anobiid beetles (Coleoptera, Anobiidae): possible metabolic pathways from fungal sterols to 7-dehydrocholesterol. Arch Insect Biochem Physiol. 2003;52:175–82. doi: 10.1002/arch.10079. [DOI] [PubMed] [Google Scholar]
- 84.Newell PD, Douglas AE. Interspecies interactions determine the impact of the gut microbiota on nutrient allocation in Drosophila melanogaster. Appl Environ Microbiol. 2014;80:788–96. doi: 10.1128/AEM.02742-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Newman KL, Almeida RP, Purcell AH, Lindow SE. Cell-cell signaling controls Xylella fastidiosa interactions with both insects and plants. Proc Natl Acad Sci USA. 2004;101:1737–42. doi: 10.1073/pnas.0308399100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Noda H, Koizumi Y. Sterol biosynthesis by symbiotes: cytochrome P450 sterol C-22 desaturase genes from yeastlike symbiotes of rice planthoppers and anobiid beetles. Insect Biochem Mol Biol. 2003;33:649–58. doi: 10.1016/s0965-1748(03)00056-0. [DOI] [PubMed] [Google Scholar]
- 87.Ohkuma M, Noda S, Kudo T. Phylogenetic diversity of nitrogen fixation genes in the symbiotic microbial community in the gut of diverse termites. Appl Environ Microbiol. 1999;65:4926–34. doi: 10.1128/aem.65.11.4926-4934.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Oliver KM, Moran NA, Hunter MS. Variation in resistance to parasitism in aphids is due to symbionts not host genotype. Proc Natl Acad Sci USA. 2005;102:12795–800. doi: 10.1073/pnas.0506131102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Piel J. A polyketide synthase-peptide synthetase gene cluster from an uncultured bacterial symbiont of Paederus beetles. Proc Natl Acad Sci USA. 2002;99:14002–7. doi: 10.1073/pnas.222481399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Potrikus CJ, Breznak JA. Gut bacteria recycle uric acid nitrogen in termites: a strategy for nutrient conservation. Proc Natl Acad Sci USA. 1981;78:4601–5. doi: 10.1073/pnas.78.7.4601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Price DR, Feng H, Baker JD, Bavan S, Luetje CW, Wilson AC. Aphid amino acid transporter regulates glutamine supply to intracellular bacterial symbionts. Proc Natl Acad Sci USA. 2014;111:320–25. doi: 10.1073/pnas.1306068111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ren C, Webster P, Finkel SE, Tower J. Increased internal and external bacterial load during Drosophila aging without life-span trade-off. Cell Metab. 2007;6:144–52. doi: 10.1016/j.cmet.2007.06.006. [DOI] [PubMed] [Google Scholar]
- 93.Russell CW, Bouvaine S, Newell PD, Douglas AE. Shared metabolic pathways in a coevolved insect-bacterial symbiosis. Appl Environ Microbiol. 2013;79:6117–23. doi: 10.1128/AEM.01543-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Russell JA, Moreau CS, Goldman-Huertas B, Fujiwara M, Lohman DJ, Pierce NE. Bacterial gut symbionts are tightly linked with the evolution of herbivory in ants. Proc Natl Acad Sci USA. 2009;106:21236–41. doi: 10.1073/pnas.0907926106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sabree ZL, Kambhampati S, Moran NA. Nitrogen recycling and nutritional provisioning by Blattabacterium, the cockroach endosymbiont. Proc Natl Acad Sci USA. 2009;106:19521–26. doi: 10.1073/pnas.0907504106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sachs JL, Skophammer RG, Regus JU. Evolutionary transitions in bacterial symbiosis. Proc Natl Acad Sci USA. 2011;108(Suppl. 2):10800–7. doi: 10.1073/pnas.1100304108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Santo Domingo JW, Kaufman MG, Klug MJ, Holben WE, Harris D, Tiedje JM. Influence of diet on the structure and function of the bacterial hindgut community of crickets. Mol Ecol. 1998;7:761–67. [Google Scholar]
- 98.Sasaki T, Kawamura M, Ishikawa H. Nitrogen recycling in the brown planthopper, Nilaparvata lugens: involvement of yeast-like endosymbionts in uric acid metabolism. J Insect Physiol. 1996;42:125–29. [Google Scholar]
- 99.Scarborough CL, Ferrari J, Godfray HC. Aphid protected from pathogen by endosymbiont. Science. 2005;310:1781. doi: 10.1126/science.1120180. [DOI] [PubMed] [Google Scholar]
- 100.Schauer C, Thompson CL, Brune A. The bacterial community in the gut of the cockroach Shelfordella lateralis reflects the close evolutionary relatedness of cockroaches and termites. Appl Environ Microbiol. 2012;78:2758–67. doi: 10.1128/AEM.07788-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Scott JJ, Oh DC, Yuceer MC, Klepzig KD, Clardy J, Currie CR. Bacterial protection of beetlefungus mutualism. Science. 2008;322:63. doi: 10.1126/science.1160423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Shanbhag S, Tripathi S. Epithelial ultrastructure and cellular mechanisms of acid and base transport in the Drosophila midgut. J Exp Biol. 2009;212:1731–44. doi: 10.1242/jeb.029306. [DOI] [PubMed] [Google Scholar]
- 103.Sharon G, Segal D, Ringo JM, Hefetz A, Zilber-Rosenberg I, Rosenberg E. Commensal bacteria play a role in mating preference of Drosophila melanogaster. Proc Natl Acad Sci USA. 2010;107:20051–56. doi: 10.1073/pnas.1009906107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Shen SK, Dowd PF. Detoxification spectrum of the cigarette beetle symbiont Symbiotaphrina kochii in culture. Entomol Exp Appl. 1991;60:51–59. [Google Scholar]
- 105.Shigenobu S, Stern DL. Aphids evolved novel secreted proteins for symbiosis with bacterial endosymbiont. Proc R Soc Lond B. 2013;280:20121952. doi: 10.1098/rspb.2012.1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Shigenobu S, Watanabe H, Hattori M, Sakaki Y, Ishikawa H. Genome sequence of the endocellular bacterial symbiont of aphids Buchnera sp. APS. Nature. 2000;407:81–86. doi: 10.1038/35024074. [DOI] [PubMed] [Google Scholar]
- 107.Sudakaran S, Salem H, Kost C, Kaltenpoth M. Geographical and ecological stability of the symbiotic mid-gut microbiota in European firebugs, Pyrrhocoris apterus (Hemiptera, Pyrrhocoridae) Mol Ecol. 2012;21:6134–51. doi: 10.1111/mec.12027. [DOI] [PubMed] [Google Scholar]
- 108.Sun J, Lu M, Gillette NE, Wingfield MJ. Red turpentine beetle: innocuous native becomes invasive tree killer in China. Annu Rev Entomol. 2013;58:293–311. doi: 10.1146/annurev-ento-120811-153624. [DOI] [PubMed] [Google Scholar]
- 109.Taerum SJ, Duong TA, de Beer ZW, Gillette N, Sun JH, et al. Large shift in symbiont assemblage in the invasive red turpentine beetle. PLOS ONE. 2013;8:e78126. doi: 10.1371/journal.pone.0078126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Tang X, Freitak D, Vogel H, Ping L, Shao Y, et al. Complexity and variability of gut commensal microbiota in polyphagous lepidopteran larvae. PLOS ONE. 2012;7:e36978. doi: 10.1371/journal.pone.0036978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Thomas F, Hehemann JH, Rebuffet E, Czjzek M, Michel G. Environmental and gut Bacteroidetes: the food connection. Front Microbiol. 2011;2:93. doi: 10.3389/fmicb.2011.00093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Thompson BM, Grebenok RJ, Behmer ST, Gruner DS. Microbial symbionts shape the sterol profile of the xylem-feeding woodwasp, Sirex noctilio. J Chem Ecol. 2013;39:129–39. doi: 10.1007/s10886-012-0222-7. [DOI] [PubMed] [Google Scholar]
- 113.Tokuda G, Elbourne LD, Kinjo Y, Saitoh S, Sabree Z, et al. Maintenance of essential amino acid synthesis pathways in the Blattabacterium cuenoti symbiont of a wood-feeding cockroach. Biol Lett. 2013;9:20121153. doi: 10.1098/rsbl.2012.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Tsai YL, Hayward RE, Langer RC, Fidock DA, Vinetz JM. Disruption of Plasmodium falciparum chitinase markedly impairs parasite invasion of mosquito midgut. Infect Immun. 2001;69:4048–54. doi: 10.1128/IAI.69.6.4048-4054.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Valiente Moro C, Tran FH, Raharimalala FN, Ravelonandro P, Mavingui P. Diversity of culturable bacteria including Pantoea in wild mosquito Aedes albopictus. BMC Microbiol. 2013;13:70. doi: 10.1186/1471-2180-13-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Vallet-Gely I, Lemaitre B, Boccard F. Bacterial strategies to overcome insect defences. Nat Rev Microbiol. 2008;6:302–13. doi: 10.1038/nrmicro1870. [DOI] [PubMed] [Google Scholar]
- 117.Vogel KJ, Moran NA. Functional and evolutionary analysis of the genome of an obligate fungal symbiont. Genome Biol Evol. 2013;5:891–904. doi: 10.1093/gbe/evt054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Walker T, Johnson PH, Moreira LA, Iturbe-Ormaetxe I, Frentiu FD, et al. The wMel Wolbachia strain blocks dengue and invades caged Aedes aegypti populations. Nature. 2011;476:450–53. doi: 10.1038/nature10355. [DOI] [PubMed] [Google Scholar]
- 119.Wang J, Wu Y, Yang G, Aksoy S. Interactions between mutualist Wigglesworthia and tsetse peptidoglycan recognition protein (PGRP-LB) influence trypanosome transmission. Proc Natl Acad Sci USA. 2009;106:12133–38. doi: 10.1073/pnas.0901226106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Warnecke F, Luginbuhl P, Ivanova N, Ghassemian M, Richardson TH, et al. Metagenomic and functional analysis of hindgut microbiota of a wood-feeding higher termite. Nature. 2007;450:560–65. doi: 10.1038/nature06269. [DOI] [PubMed] [Google Scholar]
- 121.Wong AC, Chaston JM, Douglas AE. The inconstant gut microbiota of Drosophila species revealed by 16S rRNA gene analysis. ISME J. 2013;7:1922–32. doi: 10.1038/ismej.2013.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Yek SH, Mueller UG. The metapleural gland of ants. Biol Rev. 2011;86:774–91. doi: 10.1111/j.1469-185X.2010.00170.x. [DOI] [PubMed] [Google Scholar]


