Abstract
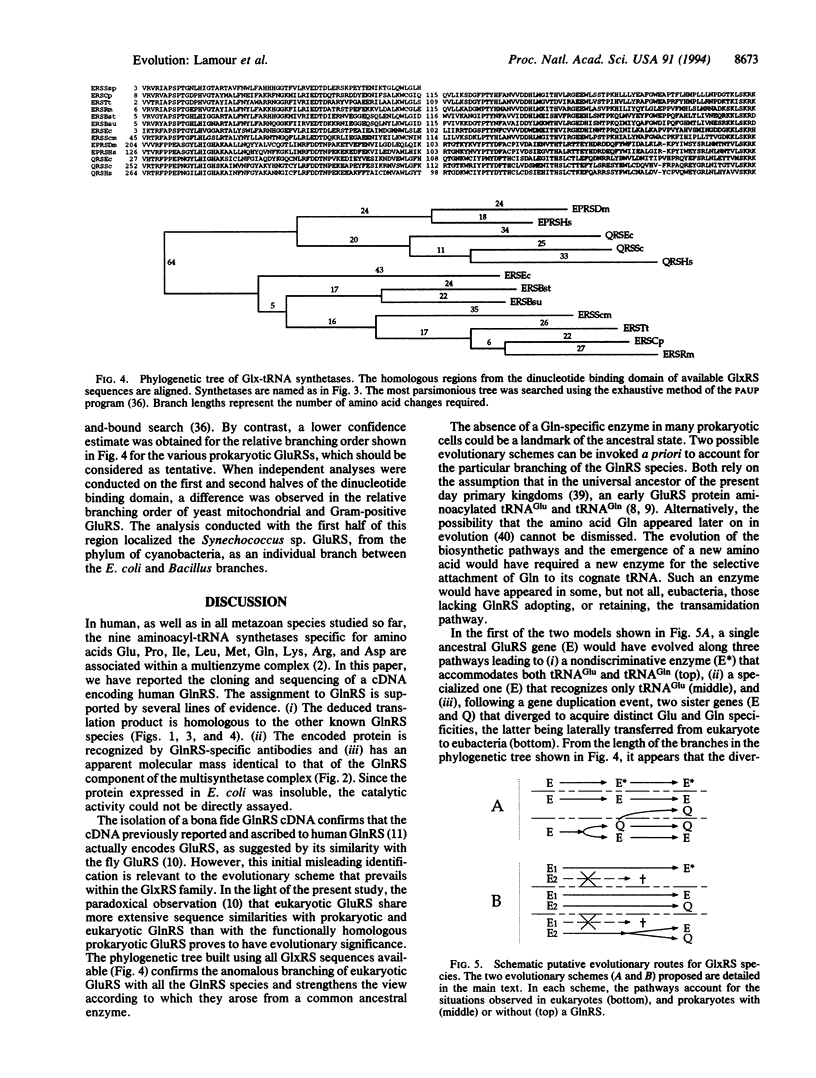
An important step ensuring the fidelity in protein biosynthesis is the aminoacylation of tRNAs by aminoacyl-tRNA synthetases. The accuracy of this process rests on a family of 20 enzymes, one for each amino acid. One exception is the formation of Gln-tRNA(Gln) that can be accomplished by two different pathways: aminoacylation of tRNA(Gln) with Gln by glutaminyl-tRNA synthetase (GlnRS; EC 6.1.1.18) or transamidation of Glu from Glu-tRNA(Gln) mischarged by glutamyl-tRNA synthetase (GluRS; EC 6.1.1.17). The latter pathway is widespread among bacteria and organelles that, accordingly, lack GlnRS. However, some bacterial species, such as Escherichia coli, do possess a GlnRS activity, which is responsible for Gln-tRNA(Gln) formation. In the cytoplasm of eukaryotic cells, both GluRS and GlnRS activities can be detected. To gain more insight into the evolutionary relationship between GluRS and GlnRS enzyme species, we have now isolated and characterized a human cDNA encoding GlnRS. The deduced amino acid sequence shows a strong similarity with other known GlnRSs and with eukaryotic GluRSs. A molecular phylogenetic analysis was conducted on the 14 GlxRS (GluRS or GlnRS) sequences available to date. Our data suggest that bacterial GlnRS has a eukaryotic origin and was acquired by a mechanism of horizontal gene transfer.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. Basic local alignment search tool. J Mol Biol. 1990 Oct 5;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Breton R., Sanfaçon H., Papayannopoulos I., Biemann K., Lapointe J. Glutamyl-tRNA synthetase of Escherichia coli. Isolation and primary structure of the gltX gene and homology with other aminoacyl-tRNA synthetases. J Biol Chem. 1986 Aug 15;261(23):10610–10617. [PubMed] [Google Scholar]
- Breton R., Watson D., Yaguchi M., Lapointe J. Glutamyl-tRNA synthetases of Bacillus subtilis 168T and of Bacillus stearothermophilus. Cloning and sequencing of the gltX genes and comparison with other aminoacyl-tRNA synthetases. J Biol Chem. 1990 Oct 25;265(30):18248–18255. [PubMed] [Google Scholar]
- Burbaum J. J., Schimmel P. Structural relationships and the classification of aminoacyl-tRNA synthetases. J Biol Chem. 1991 Sep 15;266(26):16965–16968. [PubMed] [Google Scholar]
- Cerini C., Kerjan P., Astier M., Gratecos D., Mirande M., Sémériva M. A component of the multisynthetase complex is a multifunctional aminoacyl-tRNA synthetase. EMBO J. 1991 Dec;10(13):4267–4277. doi: 10.1002/j.1460-2075.1991.tb05005.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruzen M. E., Bengtsson U., McMahon J., Wasmuth J. J., Arfin S. M. Assignment of the cysteinyl-tRNA synthetase gene (CARS) to 11p15.5. Genomics. 1993 Mar;15(3):692–693. doi: 10.1006/geno.1993.1128. [DOI] [PubMed] [Google Scholar]
- Cusack S., Härtlein M., Leberman R. Sequence, structural and evolutionary relationships between class 2 aminoacyl-tRNA synthetases. Nucleic Acids Res. 1991 Jul 11;19(13):3489–3498. doi: 10.1093/nar/19.13.3489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dayhoff M. O., Barker W. C., Hunt L. T. Establishing homologies in protein sequences. Methods Enzymol. 1983;91:524–545. doi: 10.1016/s0076-6879(83)91049-2. [DOI] [PubMed] [Google Scholar]
- Eriani G., Delarue M., Poch O., Gangloff J., Moras D. Partition of tRNA synthetases into two classes based on mutually exclusive sets of sequence motifs. Nature. 1990 Sep 13;347(6289):203–206. doi: 10.1038/347203a0. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Phylogenies from molecular sequences: inference and reliability. Annu Rev Genet. 1988;22:521–565. doi: 10.1146/annurev.ge.22.120188.002513. [DOI] [PubMed] [Google Scholar]
- Fett R., Knippers R. The primary structure of human glutaminyl-tRNA synthetase. A highly conserved core, amino acid repeat regions, and homologies with translation elongation factors. J Biol Chem. 1991 Jan 25;266(3):1448–1455. [PubMed] [Google Scholar]
- Gupta R. Halobacterium volcanii tRNAs. Identification of 41 tRNAs covering all amino acids, and the sequences of 33 class I tRNAs. J Biol Chem. 1984 Aug 10;259(15):9461–9471. [PubMed] [Google Scholar]
- Kanehisa M., Klein P., Greif P., DeLisi C. Computer analysis and structure prediction of nucleic acids and proteins. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):417–428. doi: 10.1093/nar/12.1part1.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. An analysis of vertebrate mRNA sequences: intimations of translational control. J Cell Biol. 1991 Nov;115(4):887–903. doi: 10.1083/jcb.115.4.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunze N., Bittler E., Fett R., Schray B., Hameister H., Wiedorn K. H., Knippers R. The human QARS locus: assignment of the human gene for glutaminyl-tRNA synthetase to chromosome 1q32-42. Hum Genet. 1990 Oct;85(5):527–530. doi: 10.1007/BF00194231. [DOI] [PubMed] [Google Scholar]
- Laberge S., Gagnon Y., Bordeleau L. M., Lapointe J. Cloning and sequencing of the gltX gene, encoding the glutamyl-tRNA synthetase of Rhizobium meliloti A2. J Bacteriol. 1989 Jul;171(7):3926–3932. doi: 10.1128/jb.171.7.3926-3932.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamour V., Lévy N., Desmaze C., Baude V., Lécluse Y., Delattre O., Bernheim A., Thomas G., Aurias A., Lipinski M. Isolation of cosmids and fetal brain cDNAs from the proximal long arm of human chromosome 22. Hum Mol Genet. 1993 May;2(5):535–540. doi: 10.1093/hmg/2.5.535. [DOI] [PubMed] [Google Scholar]
- Lapointe J., Duplain L., Proulx M. A single glutamyl-tRNA synthetase aminoacylates tRNAGlu and tRNAGln in Bacillus subtilis and efficiently misacylates Escherichia coli tRNAGln1 in vitro. J Bacteriol. 1986 Jan;165(1):88–93. doi: 10.1128/jb.165.1.88-93.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazard M., Mirande M. Cloning and analysis of a cDNA encoding mammalian arginyl-tRNA synthetase, a component of the multisynthetase complex with a hydrophobic N-terminal extension. Gene. 1993 Oct 15;132(2):237–245. doi: 10.1016/0378-1119(93)90201-d. [DOI] [PubMed] [Google Scholar]
- Ludmerer S. W., Schimmel P. Gene for yeast glutamine tRNA synthetase encodes a large amino-terminal extension and provides a strong confirmation of the signature sequence for a group of the aminoacyl-tRNA synthetases. J Biol Chem. 1987 Aug 5;262(22):10801–10806. [PubMed] [Google Scholar]
- Ludmerer S. W., Wright D. J., Schimmel P. Purification of glutamine tRNA synthetase from Saccharomyces cerevisiae. A monomeric aminoacyl-tRNA synthetase with a large and dispensable NH2-terminal domain. J Biol Chem. 1993 Mar 15;268(8):5519–5523. [PubMed] [Google Scholar]
- Mirande M. Aminoacyl-tRNA synthetase family from prokaryotes and eukaryotes: structural domains and their implications. Prog Nucleic Acid Res Mol Biol. 1991;40:95–142. doi: 10.1016/s0079-6603(08)60840-5. [DOI] [PubMed] [Google Scholar]
- Mirande M., Cirakoğlu B., Waller J. P. Macromolecular complexes from sheep and rabbit containing seven aminoacyl-tRNA synthetases. III. Assignment of aminoacyl-tRNA synthetase activities to the polypeptide components of the complexes. J Biol Chem. 1982 Sep 25;257(18):11056–11063. [PubMed] [Google Scholar]
- Mirande M., Lazard M., Martinez R., Latreille M. T. Engineering mammalian aspartyl-tRNA synthetase to probe structural features mediating its association with the multisynthetase complex. Eur J Biochem. 1992 Feb 1;203(3):459–466. doi: 10.1111/j.1432-1033.1992.tb16570.x. [DOI] [PubMed] [Google Scholar]
- Mirande M., Le Corre D., Waller J. P. A complex from cultured Chinese hamster ovary cells containing nine aminoacyl-tRNA synthetases. Thermolabile leucyl-tRNA synthetase from the tsH1 mutant cell line is an integral component of this complex. Eur J Biochem. 1985 Mar 1;147(2):281–289. doi: 10.1111/j.1432-1033.1985.tb08748.x. [DOI] [PubMed] [Google Scholar]
- Nagel G. M., Doolittle R. F. Evolution and relatedness in two aminoacyl-tRNA synthetase families. Proc Natl Acad Sci U S A. 1991 Sep 15;88(18):8121–8125. doi: 10.1073/pnas.88.18.8121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M., Stephens J. C., Saitou N. Methods for computing the standard errors of branching points in an evolutionary tree and their application to molecular data from humans and apes. Mol Biol Evol. 1985 Jan;2(1):66–85. doi: 10.1093/oxfordjournals.molbev.a040333. [DOI] [PubMed] [Google Scholar]
- Nguyen Van Cong, Weil D., Finaz C., Cohen-Haguenauer O., Gross M. S., Jegou-Foubert C., de Tand M. F., Cochet C., de Grouchy J., Frezal J. Panel of twenty-five independent man-rodent hybrids for human genetic marker mapping. Ann Genet. 1986;29(1):20–26. [PubMed] [Google Scholar]
- Nureki O., Suzuki K., Hara-Yokoyama M., Kohno T., Matsuzawa H., Ohta T., Shimizu T., Morikawa K., Miyazawa T., Yokoyama S. Glutamyl-tRNA synthetase from Thermus thermophilus HB8. Molecular cloning of the gltX gene and crystallization of the overproduced protein. Eur J Biochem. 1992 Mar 1;204(2):465–472. doi: 10.1111/j.1432-1033.1992.tb16656.x. [DOI] [PubMed] [Google Scholar]
- Rould M. A., Perona J. J., Söll D., Steitz T. A. Structure of E. coli glutaminyl-tRNA synthetase complexed with tRNA(Gln) and ATP at 2.8 A resolution. Science. 1989 Dec 1;246(4934):1135–1142. doi: 10.1126/science.2479982. [DOI] [PubMed] [Google Scholar]
- Schimmel P. Aminoacyl tRNA synthetases: general scheme of structure-function relationships in the polypeptides and recognition of transfer RNAs. Annu Rev Biochem. 1987;56:125–158. doi: 10.1146/annurev.bi.56.070187.001013. [DOI] [PubMed] [Google Scholar]
- Schuler G. D., Altschul S. F., Lipman D. J. A workbench for multiple alignment construction and analysis. Proteins. 1991;9(3):180–190. doi: 10.1002/prot.340090304. [DOI] [PubMed] [Google Scholar]
- Schön A., Kannangara C. G., Gough S., Söll D. Protein biosynthesis in organelles requires misaminoacylation of tRNA. Nature. 1988 Jan 14;331(6152):187–190. doi: 10.1038/331187a0. [DOI] [PubMed] [Google Scholar]
- Singleton C. K., Manning S. S., Ken R. Primary structure and regulation of vegetative specific genes of Dictyostelium discoideum. Nucleic Acids Res. 1989 Dec 11;17(23):9679–9692. doi: 10.1093/nar/17.23.9679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith M. W., Feng D. F., Doolittle R. F. Evolution by acquisition: the case for horizontal gene transfers. Trends Biochem Sci. 1992 Dec;17(12):489–493. doi: 10.1016/0968-0004(92)90335-7. [DOI] [PubMed] [Google Scholar]
- Sprague G. F., Jr Genetic exchange between kingdoms. Curr Opin Genet Dev. 1991 Dec;1(4):530–533. doi: 10.1016/s0959-437x(05)80203-5. [DOI] [PubMed] [Google Scholar]
- Wichlan D. G., Hatch T. P. Identification of an early-stage gene of Chlamydia psittaci 6BC. J Bacteriol. 1993 May;175(10):2936–2942. doi: 10.1128/jb.175.10.2936-2942.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese C. R. Bacterial evolution. Microbiol Rev. 1987 Jun;51(2):221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong J. T. A co-evolution theory of the genetic code. Proc Natl Acad Sci U S A. 1975 May;72(5):1909–1912. doi: 10.1073/pnas.72.5.1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamao F., Inokuchi H., Cheung A., Ozeki H., Söll D. Escherichia coli glutaminyl-tRNA synthetase. I. Isolation and DNA sequence of the glnS gene. J Biol Chem. 1982 Oct 10;257(19):11639–11643. [PubMed] [Google Scholar]