Immune response regulators IRF3 and ISGF3 bind to and activate an overlapping set of target genes, but IRF3 specificity is critical in activating IFNβ.
Keywords: pathogen response, gene expression, interferon, feed-forward loop, mathematical modeling
Abstract
The innate immune response is largely initiated by pathogen-responsive activation of the transcription factor IRF3. Among other target genes, IRF3 controls the expression of IFN-β, which triggers the activation of the transcription factor ISGF3 via the IFNAR. IRF3 and ISGF3 have been reported to control many of the same target genes and together, control the antimicrobial innate-immune program; however, their respective contributions and specificities remain unclear. Here, we used genomic technologies to characterize their specificity in terms of their physical DNA-binding and genetic function. With the use of ChiP-seq and transcriptomic measurements in WT versus ifnar−/− versus ifnar−/−irf3−/− macrophages responding to intracellular dsRNA, we confirmed the known ISGF3 DNA-binding motif and further specified a distinct IRF3 consensus sequence. The functional specificity of IRF3 is particularly pronounced in cytokine/chemokine regulation; yet, even in the control of IFN-β, that specificity is not absolute. By mathematically modeling IFN-β production within an abstracted tissue layer, we find that IRF3 versus ISGF3 specificity may be critical to limiting IFN-β production and ISGF3 activation, temporally and spatially, but that partial overlap in their specificity is tolerable and may enhance the effectiveness of the innate-immune response.
Introduction
Viral infections are recognized by immune cells via PRRs that bind pathogen-associated molecular patterns, including viral components, such as virus genomic DNA or ss- or dsRNA. More specifically, PRRs that recognize intracellular nucleic acids are known as RLRs, whereas PRRs that recognize extracellular or endosomal nucleic acids are known as TLRs. After binding viral components, RLRs and TLRs initiate effective and appropriate antiviral responses through the activation of many transcription factors, which may act independently or cooperatively to produce a variety of cytokines and induce inflammatory and adaptive immune responses [1–4].
One important transcription factor that is activated after RLR or TLR stimulation is IRF3, which is constitutively expressed and resides primarily in the cytoplasm of uninfected cells [5]. Upon phosphorylation by TNFR-associated factor family member-associated NF-κB activator-binding kinase or inhibitor of NF-κB kinase ε, IRF3 dimerizes and translocates to the nucleus, where it binds consensus IREs to stimulate gene expression [6–9]. The most well-studied IRF3 response gene is type I IFN-β. Type I IFNs, such as IFN-β, are a specialized group of cytokines whose activity is critical to ensure a productive antiviral state [10]. The IFN-β gene is thought to be activated by the coordinated binding of 3 transcription factors: IRF3, NF-κB, and AP-1 [11–13]. IFN-β is secreted from infected cells and stimulates the IFNAR on the same cell or neighboring cells [10]. After binding to IFNAR, receptor-associated tyrosine kinases phosphorylate STAT1 and STAT2, which combine together with IRF9 to form the transcription factor known as ISGF3 [14, 15]; this then translocates to the nucleus, where it binds to ISREs and stimulates the production of many ISGs [15, 16]. Interestingly, IRF3 and ISGF3 have been reported to have similar and overlapping consensus-binding sequences [9, 17, 18]. However, genomic tools have not yet addressed the question of what genes, if any, are distinctly controlled by each transcription factor.
The consensus DNA-binding motif for the IRF family of transcription factors, IRF1–9, is reported as NNGAAANNGAAA [9, 13, 17, 19]. In vitro, IRF3 binding has been shown to be highly dependent on the 5′ GAAA and 3′ GAAA core sequences, whereas other IRFs can tolerate some flexibility in these sequences [20–22]. In addition, a PCR-based DNA-binding site selection experiment with recombinant protein showed that IRF3 may prefer GAAA(G/C)(G/C)GAAAN(T/C) [20]. This sequence completely overlaps with the ISGF3 consensus motif: GAAANNGAAACT [23–25]. There is no crystal structure for ISGF3, so the exact DNA contacts and stoichiometry are still unknown; however, it is proposed that subunits STAT1 and IRF9 make contacts on either side of the GAAA sequence. Interestingly, ISGF3 is not predicted to contact the conserved CT motif following the GAAA core repeats [26, 27]. Surprisingly, there is little in vivo data to characterize the sequence specificity of IRF3 and ISGF3.
The binding of IRF3 and ISGF3 to specific sites within promoters leads to the activation of gene expression. There are hundreds of genes known to be regulated by ISGF3, but what genes are controlled by IRF3 is less clear. This, in part, is a result of the feed-forward loops from IRF3 to ISGF3 and from ISGF3 and IRF7, an IFN-inducible gene, which may exaggerate or mask the loss of IRF3 in a knockout cell [18, 28]. One study aimed to identify IRF3-responsive genes through overexpression of a constitutively active form of IRF3 [29]. However, only a few genes were found to be up-regulated in this system, and that list did not include IFN-β, leading to the conclusion that IRF3 can activate some genes independently but may often function coordinately with other transcription factors. Another approach identified genes that were differentially expressed after viral infection in cell lines that express varying amounts of IRF3 [30]. A few ISGs showed increased expression in cells with overexpressed levels of IRF3; however, the results may not be specific to IRF3, as downstream ISGF3 activity would also be affected. In addition, a microarray study in mouse embryonic fibroblasts was able to identify more IRF3-controlled genes by use of cyclohexamide to inhibit secondary IFN-mediated effects but did not examine the specificity between IRF3 and ISGF3 [31].
Here, we present studies of physical interactions and functional requirement in gene expression to examine the contributions of IRF3 and ISGF3 in the innate-immune response to dsRNA in primary macrophages. Our results confirmed the known ISGF3 DNA-binding motif to be GAAANNGAAACT and further refined the preferred IRF3 consensus sequence: AAATGGAAA. Our transcriptomic studies revealed a functional role for IRF3 in cytokine/chemokine regulation that is distinct from that of ISGF3. However, many IRF3-controlled genes can also be regulated by ISGF3. Even the most IRF3-specific gene, IFN-β, shows a degree of ISGF3 inducibility. To explore the functional consequences of IRF3 versus ISGF3 specificity, we constructed a simple computational model of the IRF3-IFN-β-ISGF3 circuit in an abstracted tissue cell layer. We show that IRF3/ISGF3 specificity is critical to limiting IFN-β-induced ISGF3 activation, spatially and temporally, but that a degree of overlap may be tolerated by the system.
MATERIALS AND METHODS
Animals and cell culture
BMDMs were cultured from C57BL/6, ifnar−/−, and ifnar−/−irf3−/− mice by use of established protocols with L929 medium and as described previously [32]. In brief, a total of 6 × 106 BM cells was cultured in 15 cm suspension dishes in DMEM, supplemented with 20% FBS and 30% L929-conditioned media for 7 days at 37°C with 5% CO2 and replated without L929-conditioned medium on day 7 into smaller cell-culture dishes. All experiments were performed on day 8. BMDMs were transfected with 5 µg/ml dsRNA (poly (I:C) LMW; InvivoGen, San Diego, CA, USA) by use of Lipofectamine RNAiMAX (Invitrogen, Carlsbad, CA, USA), according to the manufacturer's instructions, or stimulated with 100 units/ml IFN-β (PBL Interferon Source, Piscataway, NJ, USA).
Antibodies
ChIP antibodies against IRF3 (#4302) and STAT1 (#4947) were purchased from Cell Signaling Technology (Beverly, MA, USA). Antibodies used in immunoblots were antiphospho-IRF3 (Ser396; #4947; Cell Signaling Technology) and antiactin (sc-1615; Santa Cruz Biotechnology, Santa Cruz, CA, USA).
ChIP
ChIP was performed as described previously [33] with modifications. In brief, 15 × 106 BMDMs were cross-linked in 2 mM disuccinimidyl glutarate (#20593; Pierce, Rockford, IL, USA) for 30 minutes, followed by 1% formaldehyde/PBS for 15 minutes at room temperature. The reaction was quenched by adding glycine to a final concentration of 125 mM, and cells were pelleted immediately by centrifugation (5 minutes, 700 g, 4°C). Cells were resuspended in hypotonic buffer (10 mM HEPES/KOH, pH 7.9, 85 mM KCl, 1 mM EDTA, 0.5% IGEPAL CA-630) for 5 minutes, spun down, and resuspended in 500 µl lysis buffer (50 mM Tris/HCl, pH 7.4, at 20°C, 1% SDS, 0.5% Empigen BB, 10 mM EDTA). Chromatin was sheared to an average DNA size of 300–500 bp by administering 10 pulses of 15 second duration at 1.5 power output with 30 seconds pause on ice by use of a microtip of a Fisher 450W sonicator. The lysate was cleared by centrifugation (5 minutes, 16,000 g, 4°C), and 500 µl supernatant was diluted 2.5-fold with 750 µl dilution buffer (20 mM Tris/HCl, pH 7.4, at 20°C, 100 mM NaCl, 0.5% Triton X-100, 2 mM EDTA). Magnetic Dynabeads (Life Technologies, Carlsbad, CA, USA) were preincubated with antibody against the protein of interest for 1 hour in 0.5% BSA containing PBS and then added to the lysate with 1% of the lysate reserved as ChIP input. The target protein was immunoprecipitated by rotating lysate with magnetic Dynabeads overnight at 4°C. The beads were magnetically separated and supernatant discarded, and the beads washed 2 times each with 400 µl WB I (20 mM Tris/HCl, pH 7.4, at20°C, 150 mM NaCl, 0.1% SDS, 1% Triton X-100, 2 mM EDTA), WB II (20 mM Tris/HCl, pH 7.4, at 20°C, 500 mM NaCl, 1% Triton X-100, 2 mM EDTA), WB III (10 mM Tris/HCl, pH 7.4, at 20°C, 250 mM LiCl, 1% IGEPAL CA-630, 1% Na-deoxycholate, 1 mM EDTA), and Tris/EDTA. Immunoprecipitated chromatin was eluted twice with 100 µl elution buffer each (100 mM NaHCO3, 1% SDS) into fresh tubes for 20 and 10 minutes, respectively; eluates were pooled, the Na+ concentration was adjusted to 300 mM with 5 M NaCl; and cross-links were reversed overnight at 65°C. The samples were incubated sequentially at 37°C for 2 hours each with 0.33 mg/ml RNase A and 0.5 mg/ml proteinase K. The DNA was isolated by use of the QIAquick PCR purification kit (Qiagen, Valencia, CA, USA), according to the manufacturer's instructions.
ChIP-seq analysis
Nine sample libraries and 9 input libraries were prepared by use of the ChIP-seq Library Prep Kit (Illumina, San Diego, CA, USA). Single-end ChIP-Seq (50 nt) was performed on an Illumina HiSeq, resulting in 7–10 million reads/experimental sample and 20 million reads/input sample. All reads containing adapter sequences were removed, and the remaining reads were aligned to the Mus Musculus genome assembly (Build 38, mm10) by use of Bowtie2 version 2.0.0 [34]. Additional quality-control metrics and peak finding were performed with the HOMER v. 4.3 software [35]. An in-depth description and benchmarking of this software suite can be found at http://homer.salk.edu/homer/. Peaks were filtered for enrichment over the local region and input signal by use of default settings. The enriched peak locations were then used for downstream motif analysis. DNA sequences over-represented within peaks were identified de novo by use of the HOMER v. 4.3 software [35] and reported as sequence logos.
Transcriptomic analysis
BMDMs were treated as indicated, and RNA was extracted with Qiagen RNeasy kit and hybridized to Illumina mouse RefSeq Sentrix-8 V1.1 BeadChips at UCSD, Biogem facility. All WT, ifnar−/− and ifnar−/−irf3−/− samples were normalized to the untreated WT sample and then log2 transformed. Genes for which expression was increased or decreased upon stimulation by 2-fold or more were uploaded into the microarray software suite, Multiple Experiment Viewer [36]. With the use of the Significance Analysis of Microarrays method [37], genes significantly reduced in ifnar−/−irf3−/− compared with ifnar−/− with a FDR of <5% were identified. Motif searches that use JASPAR matrices were performed with the promoter sequences, 1 kb upstream and 0.1 kb downstream of the TSS (National Center for Biotechnology Information 36/mm8) with the motif search program HOMER v. 4.3 software [35]. GO term enrichment was performed by use of DAVID with the entire mouse genome as the background, and P values represent a Bonferroni-corrected modified Fisher’s exact test [38, 39]. The top 2 most enriched GO terms were selected for each case. The contributions of ISGF3 and IRF3 in the control of a specific gene were calculated based on the ΔA and ΔB and displayed as a scatterplot (see Fig. 2D). The phenotype score (see Fig. 3A) was calculated as the geometric mean of the ΔA/ΔB ratios at each time-point, specifically [(ΔA/ΔB)1 hour(ΔA/ΔB)3 hours(ΔA/ΔB)8 hours](1/3).
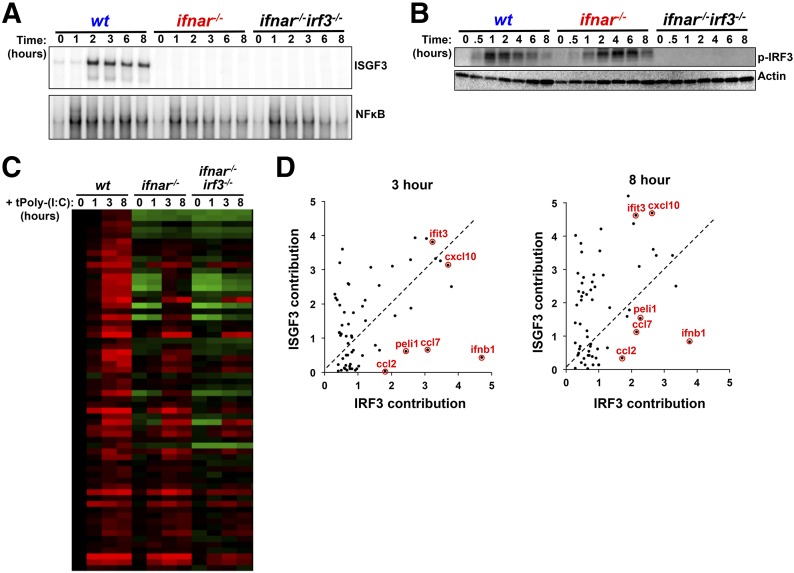
Figure 2. Relative contributions of IRF3 and ISGF3 to the innate-immune response transcriptome.
(A) EMSA of ISGF3 (upper) and NF-κB (lower) activities in BMDMs transfected with dsRNA (5 µg/ml) over 8 hours. Gels shown are representative results from >3 independent EMSAs. (B) Western blot analysis of IRF3 phosphorylation (p-IRF3) in WT, ifnar−/−, and ifnar−/−irf3−/− BMDMs transfected with dsRNA (5 µg/ml) for over 8 hours. Blots shown are representative of >3 independent experiments showing similar results. (C) Microarray mRNA expression data of IRF3-dependent genes from WT, ifnar−/−, and ifnar−/−irf3−/− BMDMs that were left untreated or treated with dsRNA (5 µg/ml) for 1, 3, or 8 hours. All data are normalized to unstimulated WT gene expression. Genes were selected as IRF3 dependent, as described in Materials and Methods. Red represents stimulus-responsive gene induction, whereas green represents repression. (D) Scatterplots indicating the contribution of ISGF3 (y-axis) versus the contribution of IRF3 (x-axis) for ISGF3/IRF3-dependent genes transfected with dsRNA for 3 or 8 hours. The scale indicates the log2-fold contribution of each transcription factor to the overall gene-induction fold.
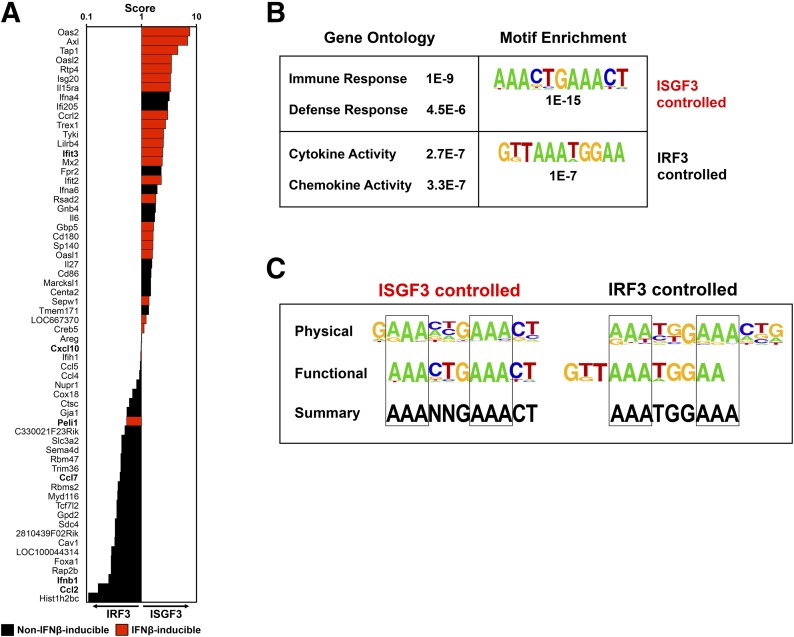
Figure 3. Genes that are primarily IRF3 dependent following dsRNA treatment are functionally distinct from those that are primarily ISGF3 dependent.
(A) Phenotype score (x-axis), calculated as described in Materials and Methods for all genes with reduced expression in the absence of IRF3 (listed along y-axis) in response to dsRNA. Genes that are IFN inducible by 2-fold or more (Supplemental Fig. 2) are red. Genes with a score >1.00 are classified as primarily ISGF3 controlled, whereas genes with a score ≤1.00 are classified as primarily IRF3 controlled. Red bars represent genes that were also found to the induced by IFN-β (Supplemental Fig. 2). (B) GO analysis of genes controlled by ISGF3 and IRF3 in response to transfected dsRNA (5 µg/ml) determined by the DAVID bioinformatics database tool (left) and the most highly enriched IRE-like motifs identified de novo within −1.0 to +0.1 kb of the transcriptional start sites for ISGF3- and IRF3-dependent genes with P values for statistical significance (right). (C) Physical, functional, and a summary thereof for consensus-binding sequences for primarily ISGF3 (left)- or primarily IRF3 (right)-controlled genes.
Biochemical assays
Immunoblotting, EMSAs, and ELISA were conducted with standard methods, as described previously [40]. ELISA was performed, according to the manufacturer’s instructions (#KMC4041; Invitrogen). Total RNA was isolated by use of Qiagen RNAeasy kit from BMDMs treated as indicated. RNA was reverse transcribed with iScript RT (Bio-Rad Laboratories, Hercules, CA, USA), and resulted cDNA was used for real-time qPCR analysis (SYBR Green; Bio-Rad Laboratories). qRT-PCR reactions without the RT were performed as control for genomic contamination. qRT-PCR primers include: GAPDH forward AACTTTGGCATTGTGGAAGG , GAPDH reverse GGATGCAGGGATGATGTTCT ; IFN-β forward GGTCCGAGCAGAGATCTTCA , IFN-β reverse CTGAGGCATCAACTGACAGG . EMSA probes include: κB, GCTACAAGGGACTTTCCGCTGGGGACTTTCCAGGGAGG ; ISRE, GATCCTCGGGAAAGGGAAACCTAAACTGAAGCC ; IFN-γ activation sequence, TACAACAGCCTGATTTCCCCGAAATGACGC .
Mathematical modeling
The model was developed by use of CompuCell3D simulation package [41]. The simulation is stochastic and agent based with each cell a unique agent. Activation of ISGF3 followed a simple Hill activation function with a Hill coefficient of 2. In the simulation, a single cell is infected initially with virus, and as a result, it secretes a constant amount of IFN-β and dies after 10 hours. In all other noninfected cells, activation of IRF3 and ISGF3 was dependent on the level of positive feedback in the model. The strength of the positive feedback was scaled as a function of the initial viral-dependent secretion of IFN-β.
In the model, cell shape and cohesiveness of the tissue were maintained dynamically through the addition of a phenomenological “energy” term:
where A(t) is the current area of each cell, Atarget is a target area, C is a cohesion factor, and is a δ function that is 1 when 2 adjacent pixels are from 2 different cells and 0 otherwise. During the simulation, pixel identity (i.e., cells or media) was allowed to flip between neighboring pixels randomly by use of a metropolis-like step; such a pixel flip was accepted if it reduced the energy-term expression above or was within an exponentially decreasing probability that is based on the possible increase in the effective energy term above. The spatiotemporal dynamic of IFN-β was simulated simultaneously on the same grid by use of a finite difference method for the simulation of IFN-β diffusion and uptake by cells. The equation for IFN-β dynamics was: , where D is the diffusion coefficient for IFN-β.
Simulation was done on a grid of cells that included >1000 cells. The simulation used a nonadaptive time step of 1 second. At each time-point, the concentration of IFN-β was updated according to the diffusion equation shown above; cells secreted additional IFN-β depending on their internal state of IRF3 and ISGF3, and cell shapes were updated according to the phenomenological energy minimization.
RESULTS
IRF3 and ISGF3 bind similar consensus sequences in response to dsRNA
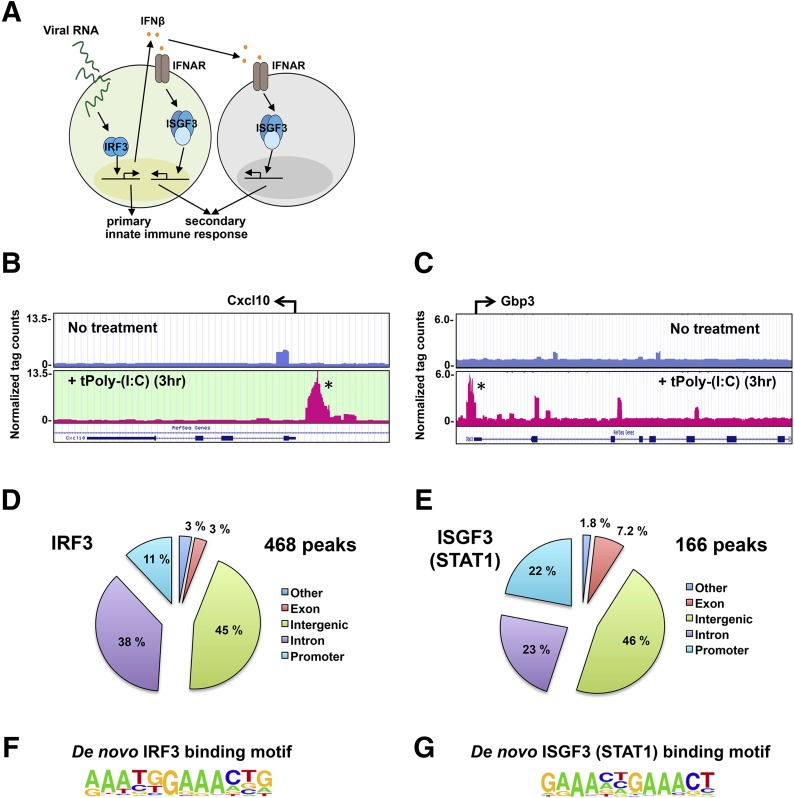
To understand the contributions of IRF3 and ISGF3 in the generation of primary and secondary innate immune responses to viral RNA (Fig. 1A), we first set out to determine the consensus-binding sequences of IRF3 and ISGF3 within the native chromatin environment. To that end, we performed ChIP, followed by ChIP-seq experiments for these factors after dsRNA treatment. ISGF3 is a complex of STAT1, STAT2, and IRF9, and we used antibodies against STAT1 to pull down the entire ISGF3-DNA complex. Although STAT1 is also a component of the IFN-γ-inducible STAT1 homodimer transcription factor, we found substantially less STAT1 homodimer activation following dsRNA transfection over 8 hours than in response to IFN-γ (Supplemental Fig. 1). Thus, the majority of the STAT1 signal obtained by ChIP in response to dsRNA was expected to derive from ISGF3. Following immunoprecipitation and sequencing, we used Bowtie2 [34] to identify peaks that were induced in the presence of dsRNA compared with the untreated sample. Browser tracks are shown for the Cxcl10 gene after IRF3 pull-down in the presence and absence of stimulus, and the peak that was identified is highlighted with an asterisk (Fig 1B). Likewise, browser tracks are shown for the Gbp3 gene after ISGF3 pull-down in the presence and absence of stimulus, and the peak that was identified is highlighted with an asterisk (Fig. 1C). There were 468 peaks and 166 peaks total identified for IRF3 and ISGF3, respectively (Fig. 1D and E), which presumably constitutes representative subsets of actual binding events, as the proportion of transcription factor-binding events identified by ChIP-seq is a function of antibody quality, background, stringency of software settings to avoid false positives, and chosen time-points. The majority of peaks for both transcription factors was found in intergenic and intron regions, whereas 11% and 22% of peaks were found in promoter regions for IRF3 and ISGF3, respectively (Fig. 1D and E). With the use of de novo HOMER [35] analysis, we determined the DNA-binding motif of IRF3 and ISGF3 (Fig. 1F and G). Our results confirm the ISGF3 consensus sequence as GAAANNGAAACT. Interestingly, for IRF3, our results show that IRF3 prefers a -TG- sequence following the first -AAA-. This represents a novel finding in IRF3 DNA-binding specificity, which had been addressed previously, exclusively in cell-free experimental systems.
Figure 1. ChIP-seq analysis of IRF3 and ISGF3.
(A) Schematic diagram of primary and secondary innate-immune responses to viral RNA stimulation. (B, D, and F) IRF3 ChIP-seq analysis of murine BMDMs transfected with dsRNA (5 µg/ml) for 3 hours. (C, E, and G) ISGF3 (STAT1) ChIP-seq analysis of BMDMs transfected with dsRNA (5 µg/ml) for 3 hours. (B and C) Example genome browser tracks of unstimulated BMDMs (upper) and BMDMs transfected with dsRNA (5 µg/ml) for 3 hours (lower) for Cxcl10 and Gbp3 example genes, respectively. tPoly-(I:C), transfected poly (I:C). (D and E) Location analysis of IRF3 (D)- and ISGF3 (E)-binding sites (peaks). IRF3- and ISGF3-binding peaks were mapped relative to their nearest RefSeq genes. The promoter region was defined as <1 kb upstream from the TSS. (F and G) Consensus motif position weight matricies generated by a de novo motif search of IRF3- and ISGF3-binding sites. Motifs were analyzed within ±50 bp from the center of a given peak.
The identification of relative contributions of IRF3 and ISGF3 in the innate-immune response transcriptome
To examine the role of IRF3 and ISGF3 in gene expression, we compared WT BMDMs, ifnar−/− BMDMs, and ifnar−/−irf3−/− BMDMs following treatment with dsRNA. To identify IRF3-dependent genes, we used a genetic background, in which type I IFN signaling is absent to avoid possible feed-forward loops of ISGF3 or IRF7 that would complicate interpretation of results, as irf3−/− alone leads to a decrease in ISGF3 and compensation by other family members, such as IRF7. First, we characterized transcription factor activation in ifnar−/− and ifnar−/−irf3−/− double-knockout BMDMs. The ifnar−/− BMDMs showed no activation of ISGF3, as expected (Fig. 2A, upper), although NF-κB activation remained largely unaffected (Fig. 2A, lower). In contrast, IRF3 activation, measured by IRF3 phosphorylation, remained intact (Fig. 2B). Furthermore, ifnar−/−irf3−/− cells were defective in ISGF3 and IRF3 activation but had a similar NF-κB response, as observed in ifnar−/− cells (Fig. 2A and B). Following microarray analysis in response to transfected dsRNA, we first identified dsRNA-induced genes in WT cells and then determined which of these were reduced significantly, limited to a FDR of <5% in the ifnar−/−irf3−/− compared with ifnar−/− BMDMs. This analysis yielded 61 genes that were affected by the loss of IRF3 (Fig. 2C). When comparing the WT expression profiles to the ifnar−/− profiles, we noticed that some of these 61 genes were also greatly affected by the initial loss of type I IFN signaling (Fig. 2C).
To determine the relative contribution of IFNAR/ISGF3 and IRF3 in the expression of each gene, we calculated the difference in log2 fold change from the WT expression profile to the ifnar−/− expression profile and the difference from the ifnar−/− expression profile to the ifnar−/−irf3−/− expression profile, which we termed the ISGF3 contribution and the IRF3 contribution, respectively, and plotted as a scatterplot (Fig. 2D). We restricted the analysis to 3 and 8 hour time-points, as there was little induction at 1 hour in WT BMDMs. The results are remarkably consistent for most genes at these 2 time-points and reveal a notable diversity of expression phenotypes: whereas most genes are induced with contributions from IRF3 and ISGFR3, there are genes that show more dependence on ISGF3 and others that show more dependence on IRF3. As expected, our data show IFN-β1 to be one of the genes that depends most highly on IRF3, but other genes, such as ccl2, that are less highly induced are also largely IRF3 controlled.
Functional distinctions between IRF3 and ISGF3
To classify genes as being primarily IRF3 or ISGF3 controlled , we calculated an overall phenotype score for each gene as the geometric mean of the ratio of ISGF3 versus IRF3 contributions at each time-point (see Materials and Methods). Genes were ranked in score order (Fig. 3A). By this measure, genes with a score of >1.00 were classified as primarily ISGF3 controlled (right of the vertical axis), whereas genes with a score of <1.00 were classified as primarily IRF3 controlled (left of the vertical axis). Of note, the bars are not a measure of expression level, but rather, they indicate the degree to which the induction of the gene is skewed for contribution from IRF3 (left) or ISGF3 (right). In principle, our analysis predicts that the set of ISGF3-controlled genes should be inducible by exogenous IFN-β stimulation, whereas IRF3-controlled genes should not. To test this prediction, we treated BMDMs with IFN-β alone, performed microarray analysis, and identified induced genes at 1, 3, or 8 hour time-points (Supplemental Fig. 2). With the cross-referencing to the aforementioned gene classification, we highlighted IFN-β-inducible genes (Fig. 3A, red). As expected, the majority of genes classified as primarily ISGF3 dependent for dsRNA-induced activation were also responsive to IFN-β stimulation, indicating that ISGF3 is indeed sufficient for their activation. In contrast, dsRNA-inducible genes classified as primarily IRF3 controlled, with the exception of Peli1, were generally not IFN-β inducible.
With the inspection of the genes in detail, we identified several immune-defense genes within the ISGF3-controlled set, such as the transporter associated with antigen processing 1 gene and various immune-signaling receptors, such as IL-15Rα and IFN-α4. Likewise, several chemokines within the IRF3-controlled set were identified (e.g., CCL2, CCL4, CCL5, CCL7, and CXCL10) that are involved in the recruitment of monocytes, macrophages, and other immune cells to areas of infection. Taken together, these data suggest a complimentary, albeit somewhat distinct, role for each factor in the innate-immune response. Furthermore, GO analysis of genes identified as primarily ISGF3 controlled is in agreement with these findings, with most classified as involved in immune and defense responses, the 2 highest scoring (lowest P values) of the biologic process terms (Fig. 3B, upper left). In contrast to the ISGF3 set, for the primarily IRF3-controlled genes, the GO terms that were most highly enriched were cytokine and chemokine activity (Fig. 3B, lower left).
With the use of de novo sequence motif analysis, we determined the most highly enriched sequence motifs within a range of −1.0 to +0.1 kb from the transcriptional start site of ISGF3- or IRF3-controlled genes (Fig. 3B, right). This analysis revealed sequences that resemble the ISRE, but as in our ChIP-seq analysis, we found different preferences for predominantly IRF3- versus ISGF3-controlled genes (Fig. 3B, right). After examining the sequences from our physical binding data and functional gene-expression data, we confirmed the ISGF3 consensus sequence and further characterized the IRF3 consensus as being AAATGGAAA with less variability in the first half-site and a preference for TGG in the connecting region between the half-sites (Fig. 3C).
Functional specificity is critical to limiting a potential “IFN storm”
Our in vivo physical binding and functional genetic requirement data indicate that IRF3 and ISGF3 have overlapping sequence preferences, with IRF3 being more restrictive, showing increased stringency for a particular half-site and stronger selection for bases between the half-sites. Despite the limited sequence specificity, gene-expression studies revealed a substantial specificity in the genetic ISGF3 versus IRF3 requirement and show that IRF3-dependent genes are enriched for secreted cytokine and chemokine proteins, with IFN-β being a particularly prominent IRF3-specific gene. As IRF3 and ISGF3 are connected by IFN-β, we decided to explore the issue of specificity by mathematically modeling this gene circuit in a 2-dimensional layer of cells following infection of a single cell. To parameterize the model, we confirmed the microarray results by qPCR and indeed, find that IFN-β mRNA production depends highly on IRF3 (Fig. 4A). We measured ifnb1 expression over time following dsRNA transfection and found that the ifnar−/−irf3−/− BMDMs displayed almost no ifnb1 mRNA expression, whereas ifnar−/− cells were only slightly less responsive to dsRNA treatment at early time-points than WT cells (Fig. 4A). Secreted IFN-β profiles, determined by ELISA, followed a similar pattern (Fig. 4B), with ifnar−/− cells having a reduced production level, especially at early time-points, and ifnar−/−irf3−/− BMDMs generating levels of IFN-β below the detectable limit of the assay (Fig. 4B).
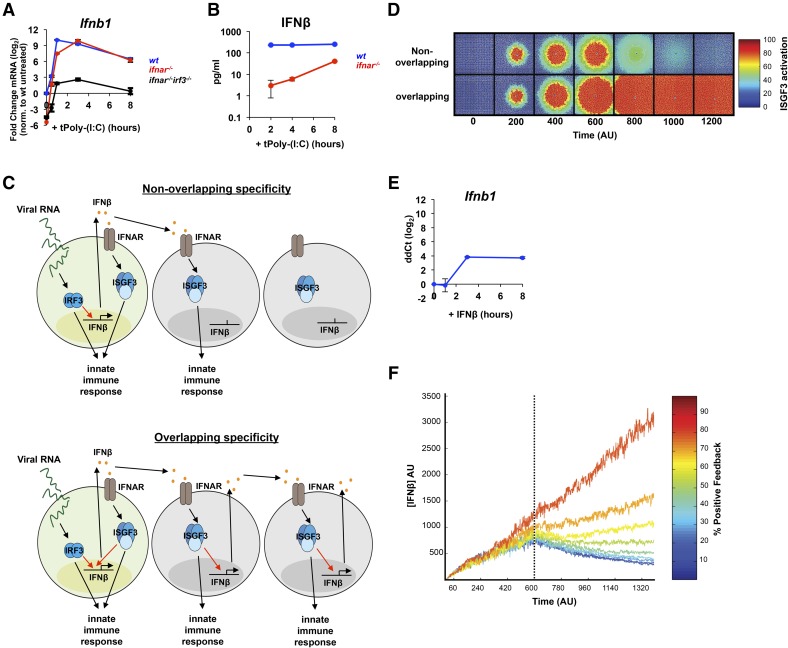
Figure 4. IRF3 specificity in the expression of IFN-β prevents an IFN storm.
(A) qRT-PCR) analysis of ifnb mRNA expression in WT (blue), ifnar−/− (red), and ifnar−/− irf3−/− (black) BMDMs transfected with dsRNA (5 µg/ml). All data points are normalized to unstimulated WT mRNA levels and reported as fold change on a log2 scale. Error bars represent PCR triplicates. Data are representative of 3 experiments that gave consistent results. (B) ELISA analysis of secreted IFN-β in the media following dsRNA transfection (5 µg/ml) of WT BMDMs. Error bars represent triplicate wells, and the results shown are representative of 2 experiments. (C) Two models to illustrate the following scenarios. (Upper) A scenario in which IRF3 and ISGF3 have nonoverlapping specificities, and only IRF3 can activate IFN-β expression. (Lower) A scenario in which IRF3 and ISGF3 can activate IFN-β expression to the same degree. (D) Model simulation of ISGF3 activation across a 2-dimensional layer of cells by use of mathematical models representing the scenarios depicted in C. (E) qRT-PCR analysis of IFN-β mRNA expression in WT BMDMs following treatment with IFN-β (100 units/ml). ddCt, ΔΔ Cycle threshold. (F) Model simulation of total IFN-β concentration over time with feedback strength from 10 to 90%.
We used these data to inform parameter selection for 2 models to explore what role IRF3 and ISGF3 specificity plays in the spatial and temporal activation of innate-immune responses. The first model describes a scenario in which IRF3 and ISGF3 have nonoverlapping specificities and ISGF3 cannot produce more IFN-β in the infected cell or in neighboring cells (Fig. 4C, upper). The second model is just the opposite, in which IRF3 and ISGF3 have completely overlapping specificities, and ISGF3 can activate IFN-β to the same degree as IRF3 in the infected cell and the neighboring cells forming a positive-feedback loop (Fig. 4C, lower). We used these models to simulate how the IFN response is propagated across a layer of cells in each case. First, IRF3 is activated only in the centermost cell to simulate infection, which then activates IFN-β production. IFN-β diffuses outwardly to activate ISGF3 in neighboring cells. After a period of time (600 AU), the initial “infected” cell ceases to exist, akin to virus-infected cells that may undergo cell death as a result of virulence factors or prolonged IFN-β exposure. Our results show that when there is ISGF3, feedback is absent, and IFN-β production depends solely on IRF3, the activation of ISGF3 in neighboring cells increases but decreases quickly after the initial cell dies (Fig. 4D, upper). However, when ISGF3 can feed back into the system to activate IFN-β at 100%, our simulation indicates that ISGF3 can be activated in cells far distal from the initially infected cell and that ISGF3 activity is not terminated after the infected cell dies (Fig. 4D, lower). These results demonstrate that positive ISGF3 feedback can lead to a bistable system, in which an initial signal spreads rapidly to all cells within a simulated tissue and cannot return to the resting state. Conversely, in the absence of ISGF3 feedback, neighboring cells are able to return to their initial state, and the spatial spread of ISGF3 activation is limited by the IFN-β diffusion rate, and the lifespan of the initially infected cell (Fig. 4D, upper).
Given the overlap in IRF3 and ISGF3 consensus-binding sequences, we thought it prudent to explore the possibility that ISGF3 may be able to activate IFN-β to some degree. To test this, we stimulated wt BMDMs with rIFN-β to activate ISGF3 and indeed, found a small degree of IFN-β mRNA induction (Fig. 4E) relative to dsRNA (24 compared with 210 peak-fold change relative to dsRNA in Fig. 4A), indicating that even in the control of the IFN-β enhancer, there is functional overlap in the specificity of IRF3 and ISGF3 and also suggesting that a low level of positive feedback may operate in the control of IFN-β. We examined the functional consequence of this finding in our tissue layer model by varying the positive-feedback strength of ISGF3 in our simulation from 10 to 90% and plotted the total amount of IFN-β in our system over time (Fig. 4F). Our results indicate that low amounts of ISGF3 feedback (<40% for the specific parameter sets chosen) can be tolerated by the system, as the levels of IFN-β can be reduced after the loss of the initial signal. However, at greater feedback strengths from greater overlap in IRF3/ISGF3 specificity, the IFN-β levels continue to increase even after the loss of the initial signal, generating a bistable system in which the second state represents a runaway IFN-β-expressing state that will lead to cell death in an increasing number of cells.
DISCUSSION
Following pathogen invasion, it is critical for a single infected cell not only to induce an effective innate-immune response within itself but also to warn surrounding uninfected cells of impending danger [10]. IRF3 is activated in response to pathogen exposure and was reported to be genetically required for IFN-β production in response to stimulation with LPS but not with poly I:C [42–45], largely as a result of compensating family member IRF7. The IRFs are thus critical for triggering a protective response in neighboring cells by controlling the secretion of IFN-β. Once secreted, IFN-β binds to its cognitive receptor (IFNAR) to induce the expression of immune-response genes through the transcriptional activity of ISGF3 [14, 16]. Whereas it is clear that IRF3 and ISGF3 functions are coordinated, it remains unclear to what degree they activate the same or distinct transcriptional programs. Here, we have combined physical interaction and functional gene expression analyses to define the relative contributions of IRF3 and ISGF3 in the innate-immune-responsive transcriptome to dsRNA stimulation.
With the use of a genetic background of IFNAR−/−, in which IRF7 expression is reduced, our results indicate that IRF3 plays a critical role in the acute response to dsRNA. We found that many IRF3-controlled genes encode cytokines and chemokines that can be secreted from infected cells to warn uninfected cells of infection or recruit other immune cells necessary to clear the pathogen. This correlates with fast-activation dynamics of IRF3, as early as 30 minutes following dsRNA treatment. On the other hand, ISGF3 is activated only after IFN-β is produced, and thus, its dynamics are delayed, with activation appearing ∼45 minutes poststimulation. Our analysis shows that genes that are controlled primarily by ISGF3 are involved in immune and defense responses. Therefore, it is likely that ISGF3 is the key determinant of the innate defenses in the infected cell and—prophylactically—in neighboring, uninfected cells. Our data do not preclude that IRF3 contributes to the defense within the infected cell; however, our genetic analysis shows that by and large, it is not sufficient, as ISGF3 is required. Thus, we did not obtain evidence that the intrinsic acute defense within infected cells is distinct from the prophylactic defense preparation of neighboring, uninfected cells. It is important to note that the expression of many genes appears to be controlled by IRF3 and ISGF3 to some degree. This redundancy and overlap may ensure that innate-immune defenses are initiated rapidly and in a sustained manner.
ChIP-seq analyses, combined with genetic functional analyses, led us to define the consensus sequences for IRF3 and ISGF3. In the case of ISGF3, we confirmed the previously reported sequence, GAAANNGAAACT. Interestingly, the functional sequence for genes primarily controlled by ISGF3 was enriched for AAACTGAAACT, which is slightly different from the ChIP result. These genes are also controlled by IRF3, to some degree, according to our analysis, so it is possible that genes that are bound by ISGF3 and IRF3 only need a single, complete GAAACT half-site for function in vivo. In the case of IRF3, we defined a novel consensus sequence: AAATGGAAA. Our data suggest a preference for the -TG- dinucleotide between the GAAA core repeat sequences, which vary slightly from the previously reported sequence GAAA(G/C)(G/C)GAAAN(T/C) based on cell-free systems studies [20]. The crystal structure of IRF3 bound to the IFN-β enhancer shows that dimeric IRF3 binds to 2 overlapping stretches of AANNGAAA, with the 2 IRF3 molecules occupying opposite sides of the DNA double helix, making minor groove contacts with the first 2 A bases and major groove contacts with the GAAA sequence [13]. The dinucleotide sequence in between the core repeats does not contact the DNA directly but may play a crucial role in determining specificity through very small conformational changes that cannot be detected in the crystal structure. Whereas our data address responses to dsRNA exclusively, the physical recognition by IRF3 or ISGF3 of their respective binding sites is likely determined by these physical considerations and may apply to other stimuli.
Our work was motivated by the goal to account for the apparent gene-expression specificity by IRF3 and ISGF3 in terms of their in vivo DNA-binding specificities. Whereas we were able to identify some sequence preferences for these 2 transcription factors, the specificities were quite limited; sequences identified by binding and genetics were largely overlapping. Thus, our data suggest that IRF3 and ISGF3 specificity may not be mediated by sequence specificity alone but are likely codetermined by protein factors within enhancer assemblies.
The enhancer of IFN-β may be an example in this regard, with several early studies identifying intricate protein–protein interactions within an enhanceosome complex to mediate stimulus-specificity specificity [46]. Indeed, we recently identified the activation-incompetent NF-κB family member p50:p50 as a specificity factor for IFN-β expression [47]. Notably, our findings identify IFN-β as one of the most strongly skewed genes for IRF3 control; however, the IRE within the positive regulatory domain III (PRDIII) of the ifnb1 gene more closely conforms to the consensus identified for ISGF3 than that for IRF3, supporting the notion that protein factors likely play a critical role in determining specificity. Of note, we were not able to demonstrate ISGF3 presence at the ifnb1 enhancer by ChIP; however, this does not preclude ISGF3 presence at distant enhancers of IFN-β production. Indeed, recent work has identified such long-range enhancers for IFN-β that conform to ISGF3- and IRF3-binding motifs [48].
Our modeling efforts illustrate how the functional specificity of IRF3 and ISGF3 combines to control IFN-β within a simulated tissue. We started by examining how ISGF3 activity spreads from a single, infected cell, both spatially and temporally. We discovered that distinct specificities of IRF3 and ISGF3 are critical for preventing an IFN storm via bistable control that would be catastrophic for the tissue and perhaps subsequently for the organism as well. However, whereas specificities must be distinct, they may be partially overlapping; by varying the amount of positive-feedback strength from 10 to 90% in our simulations, we found that the system can tolerate moderate levels of feedback from ISGF3. Experimentally, we found that whereas ISGF3 is able to activate IFN-β to some degree, the almost 2 orders of magnitude difference in induction observed with poly (I:C) are unlikely to result in a positive-feedback loop that will lead to a detrimental bistable system. As the IFN-β enhancer is not only controlled by IRF3, the functional specificity described here may not be a function of IRF3 and ISGF3 DNA-binding specificity alone but likely involves protein–protein interactions as well. We may imagine that the larger ISGF3 is sterically impeded from binding the AP-1 and NF-κB-bound IFN-β enhanceosome or that it fails to synergize with the coordinated factors, whereas IRF3 is not hindered in this way. Future studies may address the mechanism of ISGF3 versus IRF3 specificity in the expression of IFN-β.
We note that the IRF family of transcription factors not only comprises IRF3 and ISGF3 but also other members, such as IRF1 and IRF7, which have overlapping functions in the innate-immune response. Like IRF3, these are cell-intrinsic response factors (not activated in neighboring, uninfected cells), and their role in this system should be addressed in future work. Furthermore, the IRF7 gene is induced by ISGF3 and may play a role in basal expression in resting cells and in sustaining expression in stimulated cells. Quantitative information about consensus sequences, functional specificity, and dynamics is critical for a predictive understanding of the IRF signaling system and the control of innate-immune responses.
AUTHORSHIP
D.R.O. and A.H. designed the study. D.R.O. undertook experimental work and data analysis. H.B. assisted in the bioinformatic analysis. N.O. and R.W. did the model simulations. D.R.O., J.D.V., and A.H. wrote the manuscript.
Supplementary Material
Acknowledgments
This study was supported by the U.S. National Institutes of Health Grants R01 GM071573 and R01 AI083453 (to A.H.). The authors thank Michael David (UCSD) for advice throughout this project and members of the A.H. lab for critical comments.
Glossary
- −/−
deficient
- ΔA
differences in log2-induction folds in wild-type versus type I IFNR-deficient
- ΔB
differences in log2-induction folds in type I IFNR-deficient versus type I IFNR-deficient IFN regulatory factor 3-deficient
- AU
arbitrary units
- BM
bone marrow
- BMDM
bone marrow-derived macrophage
- ChIP
chromatin immunoprecipitation
- ChIP-seq
chromatin immunoprecipitation massively parallel DNA sequencing
- DAVID
Database for Annotation, Visualization, and Integrated Discovery
- FDR
false discovery rate
- gbp3
guanylate-binding protein 3
- GO
gene ontology
- HOMER
Hypergeometric Optimization of Motif Enrichment
- IFNAR
type I IFNR
- IRE
IFN response element
- IRF
IFN regulatory factor
- ISG
IFN-stimulated gene
- ISGF3
IFN-stimulated gene factor 3
- ISRE
IFN-stimulated response element
- Peli1
pellino homolog 1
- poly (I:C)
polyinosinic:polycytidylic acid
- PRR
pattern-recognition receptor
- qRT-PCR
quantitative RT-PCR
- RefSeq
references sequence
- RLR
retinoic acid-inducible gene 1-like receptor
- TSS
transcription start site
- UCSD
University of California, San Diego
- WB
wash buffer
- WT
wild-type
Footnotes
The online version of this paper, found at www.jleukbio.org, includes supplemental information.
DISCLOSURES
The authors declare no conflicts of interest.
REFERENCES
- 1.Kawai T., Akira S. (2006) Innate immune recognition of viral infection. Nat. Immunol. 7, 131–137. [DOI] [PubMed] [Google Scholar]
- 2.Kawai T., Akira S. (2008) Toll-like receptor and RIG-I-like receptor signaling. Ann. N. Y. Acad. Sci. 1143, 1–20. [DOI] [PubMed] [Google Scholar]
- 3.Wathelet M. G., Lin C. H., Parekh B. S., Ronco L. V., Howley P. M., Maniatis T. (1998) Virus infection induces the assembly of coordinately activated transcription factors on the IFN-beta enhancer in vivo. Mol. Cell 1, 507–518. [DOI] [PubMed] [Google Scholar]
- 4.Doyle S., Vaidya S., O’Connell R., Dadgostar H., Dempsey P., Wu T., Rao G., Sun R., Haberland M., Modlin R., Cheng G. (2002) IRF3 mediates a TLR3/TLR4-specific antiviral gene program. Immunity 17, 251–263. [DOI] [PubMed] [Google Scholar]
- 5.Kumar K. P., McBride K. M., Weaver B. K., Dingwall C., Reich N. C. (2000) Regulated nuclear-cytoplasmic localization of interferon regulatory factor 3, a subunit of double-stranded RNA-activated factor 1. Mol. Cell. Biol. 20, 4159–4168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fitzgerald K. A., McWhirter S. M., Faia K. L., Rowe D. C., Latz E., Golenbock D. T., Coyle A. J., Liao S.-M., Maniatis T. (2003) IKKepsilon and TBK1 are essential components of the IRF3 signaling pathway. Nat. Immunol. 4, 491–496. [DOI] [PubMed] [Google Scholar]
- 7.Sharma S., tenOever B. R., Grandvaux N., Zhou G.-P., Lin R., Hiscott J. (2003) Triggering the interferon antiviral response through an IKK-related pathway. Science 300, 1148–1151. [DOI] [PubMed] [Google Scholar]
- 8.Lin R., Heylbroeck C., Pitha P. M., Hiscott J. (1998) Virus-dependent phosphorylation of the IRF-3 transcription factor regulates nuclear translocation, transactivation potential, and proteasome-mediated degradation. Mol. Cell. Biol. 18, 2986–2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fujii Y., Shimizu T., Kusumoto M., Kyogoku Y., Taniguchi T., Hakoshima T. (1999) Crystal structure of an IRF-DNA complex reveals novel DNA recognition and cooperative binding to a tandem repeat of core sequences. EMBO J. 18, 5028–5041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stetson D. B., Medzhitov R. (2006) Type I interferons in host defense. Immunity 25, 373–381. [DOI] [PubMed] [Google Scholar]
- 11.Agalioti T., Lomvardas S., Parekh B., Yie J., Maniatis T., Thanos D. (2000) Ordered recruitment of chromatin modifying and general transcription factors to the IFN-beta promoter. Cell 103, 667–678. [DOI] [PubMed] [Google Scholar]
- 12.Ford E., Thanos D. (2010) The transcriptional code of human IFN-beta gene expression. Biochim. Biophys. Acta 1799, 328–336. [DOI] [PubMed] [Google Scholar]
- 13.Panne D., Maniatis T., Harrison S. C. (2004) Crystal structure of ATF-2/c-Jun and IRF-3 bound to the interferon-beta enhancer. EMBO J. 23, 4384–4393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Darnell J. E. Jr., Kerr I. M., Stark G. R. (1994) Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science 264, 1415–1421. [DOI] [PubMed] [Google Scholar]
- 15.Qureshi S. A., Salditt-Georgieff M., Darnell J. E. Jr. (1995) Tyrosine-phosphorylated Stat1 and Stat2 plus a 48-kDa protein all contact DNA in forming interferon-stimulated-gene factor 3. Proc. Natl. Acad. Sci. USA 92, 3829–3833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.O’Shea J. J., Gadina M., Schreiber R. D. (2002) Cytokine signaling in 2002: new surprises in the Jak/Stat pathway. Cell 109 (Suppl), S121–S131. [DOI] [PubMed] [Google Scholar]
- 17.Tanaka N., Kawakami T., Taniguchi T. (1993) Recognition DNA sequences of interferon regulatory factor 1 (IRF-1) and IRF-2, regulators of cell growth and the interferon system. Mol. Cell. Biol. 13, 4531–4538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sato M., Suemori H., Hata N., Asagiri M., Ogasawara K., Nakao K., Nakaya T., Katsuki M., Noguchi S., Tanaka N., Taniguchi T. (2000) Distinct and essential roles of transcription factors IRF-3 and IRF-7 in response to viruses for IFN-alpha/beta gene induction. Immunity 13, 539–548. [DOI] [PubMed] [Google Scholar]
- 19.Escalante C. R., Nistal-Villán E., Shen L., García-Sastre A., Aggarwal A. K. (2007) Structure of IRF-3 bound to the PRDIII-I regulatory element of the human interferon-beta enhancer. Mol. Cell 26, 703–716. [DOI] [PubMed] [Google Scholar]
- 20.Lin R., Génin P., Mamane Y., Hiscott J. (2000) Selective DNA binding and association with the CREB binding protein coactivator contribute to differential activation of alpha/beta interferon genes by interferon regulatory factors 3 and 7. Mol. Cell. Biol. 20, 6342–6353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schmid S., Mordstein M., Kochs G., García-Sastre A., Tenoever B. R. (2010) Transcription factor redundancy ensures induction of the antiviral state. J. Biol. Chem. 285, 42013–42022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morin P., Bragança J., Bandu M.-T., Lin R., Hiscott J., Doly J., Civas A. (2002) Preferential binding sites for interferon regulatory factors 3 and 7 involved in interferon-A gene transcription. J. Mol. Biol. 316, 1009–1022. [DOI] [PubMed] [Google Scholar]
- 23.Kessler D. S., Levy D. E., Darnell J. E. Jr. (1988) Two interferon-induced nuclear factors bind a single promoter element in interferon-stimulated genes. Proc. Natl. Acad. Sci. USA 85, 8521–8525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reich N., Evans B., Levy D., Fahey D., Knight E. Jr., Darnell J. E. Jr. (1987) Interferon-induced transcription of a gene encoding a 15-kDa protein depends on an upstream enhancer element. Proc. Natl. Acad. Sci. USA 84, 6394–6398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rutherford M. N., Hannigan G. E., Williams B. R. (1988) Interferon-induced binding of nuclear factors to promoter elements of the 2-5A synthetase gene. EMBO J. 7, 751–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tenoever B. R., Ng S.-L., Chua M. A., McWhirter S. M., García-Sastre A., Maniatis T. (2007) Multiple functions of the IKK-related kinase IKKepsilon in interferon-mediated antiviral immunity. Science 315, 1274–1278. [DOI] [PubMed] [Google Scholar]
- 27.Seeman N. C., Rosenberg J. M., Rich A. (1976) Sequence-specific recognition of double helical nucleic acids by proteins. Proc. Natl. Acad. Sci. USA 73, 804–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sato M., Hata N., Asagiri M., Nakaya T., Taniguchi T., Tanaka N. (1998) Positive feedback regulation of type I IFN genes by the IFN-inducible transcription factor IRF-7. FEBS Lett. 441, 106–110. [DOI] [PubMed] [Google Scholar]
- 29.Grandvaux N., Servant M. J., tenOever B., Sen G. C., Balachandran S., Barber G. N., Lin R., Hiscott J. (2002) Transcriptional profiling of interferon regulatory factor 3 target genes: direct involvement in the regulation of interferon-stimulated genes. J. Virol. 76, 5532–5539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Elco C. P., Guenther J. M., Williams B. R. G., Sen G. C. (2005) Analysis of genes induced by Sendai virus infection of mutant cell lines reveals essential roles of interferon regulatory factor 3, NF-kappaB, and interferon but not Toll-like receptor 3. J. Virol. 79, 3920–3929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Andersen J., VanScoy S., Cheng T.-F., Gomez D., Reich N. C. (2008) IRF-3-dependent and augmented target genes during viral infection. Genes Immun. 9, 168–175. [DOI] [PubMed] [Google Scholar]
- 32.Caldwell A. B., Cheng Z., Vargas J. D., Birnbaum H. A., Hoffmann A. (2014) Network dynamics determine the autocrine and paracrine signaling functions of TNF. Genes Dev. 28, 2120–2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Métivier R., Penot G., Hübner M. R., Reid G., Brand H., Kos M., Gannon F. (2003) Estrogen receptor-alpha directs ordered, cyclical, and combinatorial recruitment of cofactors on a natural target promoter. Cell 115, 751–763. [DOI] [PubMed] [Google Scholar]
- 34.Langmead B., Salzberg S. L. (2012) Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Heinz S., Benner C., Spann N., Bertolino E., Lin Y. C., Laslo P., Cheng J. X., Murre C., Singh H., Glass C. K. (2010) Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saeed A. I., Sharov V., White J., Li J., Liang W., Bhagabati N., Braisted J., Klapa M., Currier T., Thiagarajan M., Sturn A., Snuffin M., Rezantsev A., Popov D., Ryltsov A., Kostukovich E., Borisovsky I., Liu Z., Vinsavich A., Trush V., Quackenbush J. (2003) TM4: a free, open-source system for microarray data management and analysis. Biotechniques 34, 374–378. [DOI] [PubMed] [Google Scholar]
- 37.Tusher V. G., Tibshirani R., Chu G. (2001) Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA 98, 5116–5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang W., Sherman B. T., Lempicki R. A. (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57. [DOI] [PubMed] [Google Scholar]
- 39.Huang W., Sherman B. T., Lempicki R. A. (2009) Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Werner S. L., Barken D., Hoffmann A. (2005) Stimulus specificity of gene expression programs determined by temporal control of IKK activity. Science 309, 1857–1861. [DOI] [PubMed] [Google Scholar]
- 41.Swat M. H., Thomas G. L., Belmonte J. M., Shirinifard A., Hmeljak D., Glazier J. A. (2012) Multi-scale modeling of tissues using CompuCell3D. Methods Cell Biol. 110, 325–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sakaguchi S., Negishi H., Asagiri M., Nakajima C., Mizutani T., Takaoka A., Honda K., Taniguchi T. (2003) Essential role of IRF-3 in lipopolysaccharide-induced interferon-beta gene expression and endotoxin shock. Biochem. Biophys. Res. Commun. 306, 860–866. [DOI] [PubMed] [Google Scholar]
- 43.Honda K., Yanai H., Negishi H., Asagiri M., Sato M., Mizutani T., Shimada N., Ohba Y., Takaoka A., Yoshida N., Taniguchi T. (2005) IRF-7 is the master regulator of type-I interferon-dependent immune responses. Nature 434, 772–777. [DOI] [PubMed] [Google Scholar]
- 44.Fujita T., Sakakibara J., Sudo Y., Miyamoto M., Kimura Y., Taniguchi T. (1988) Evidence for a nuclear factor(s), IRF-1, mediating induction and silencing properties to human IFN-beta gene regulatory elements. EMBO J. 7, 3397–3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weiss G., Maaetoft-Udsen K., Stifter S. A., Hertzog P., Goriely S., Thomsen A. R., Paludan S. R., Frøkiær H. (2012) MyD88 drives the IFN-β response to Lactobacillus acidophilus in dendritic cells through a mechanism involving IRF1, IRF3, and IRF7. J. Immunol. 189, 2860–2868. [DOI] [PubMed] [Google Scholar]
- 46.Thanos D., Maniatis T. (1995) Virus induction of human IFN beta gene expression requires the assembly of an enhanceosome. Cell 83, 1091–1100. [DOI] [PubMed] [Google Scholar]
- 47.Cheng C. S., Feldman K. E., Lee J., Verma S., Huang D.-B., Huynh K., Chang M., Ponomarenko J. V., Sun S.-C., Benedict C. A., Ghosh G., Hoffmann A. (2011) The specificity of innate immune responses is enforced by repression of interferon response elements by NF-κB p50. Sci. Signal. 4, ra11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Banerjee A. R., Kim Y. J., Kim T. H. (2014) A novel virus-inducible enhancer of the interferon-β gene with tightly linked promoter and enhancer activities. Nucleic Acids Res. 42, 12537–12554. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.