AUTHOR SUMMARY
Staphylococcus aureus is a significant cause of chronic biofilm infections on medical implants. We investigated the biofilm regulatory cascade and discovered that the repressor of toxins (Rot) is part of this pathway. A USA300 community-associated methicillin-resistant S. aureus (CA-MRSA) strain deficient in Rot was unable to form a biofilm using multiple different assays, and we found rot mutants in other strain lineages were also biofilm deficient. By performing a global analysis of transcripts and protein production controlled by Rot, we observed that all the secreted protease genes were upregulated in a rot mutant, and we hypothesized that this regulation could be responsible for the biofilm phenotype. To investigate this question, we determined that Rot bound to the protease promoters, and we observed that activity levels of these enzymes, in particular the cysteine proteases, were increased in a rot mutant. By inactivating these proteases, biofilm capacity was restored to the mutant, demonstrating they are responsible for the biofilm negative phenotype. Finally, we tested the rot mutant in a mouse catheter model of biofilm infection and observed a significant reduction in biofilm burden. Thus S. aureus uses the transcription factor Rot to repress secreted protease levels in order to build a biofilm.
INTRODUCTION
Staphylococcus aureus is an important cause of chronic infections that include osteomyelitis, endocarditis, and growth on medical implants. These types of infections require attachment and growth of S. aureus on a host or foreign body surface, resulting in a community of bacteria at that site, and these communities are encased in a complex matrix and are generally considered biofilms (Kiedrowski et al., 2011a). Biofilms are challenging to treat due to their high level of antibiotic tolerance and resistance to host defenses (Hall-Stoodley et al., 2005, Costerton, 2005, Patel, 2005). Most S. aureus strain types investigated have been described to form biofilms, including community-associated methicillin resistant S. aureus (CA-MRSA), such as the USA300 group of isolates (Lauderdale et al., 2010, Lauderdale et al., 2009). In recent years, there has been tremendous effort on understanding the growing prevalence of USA300 isolates in community and hospital settings across the United States (Chambers et al., 2009), and these strains are increasingly being observed in biofilm infections (Haque et al., 2007, Kourbatova et al., 2005, Seybold et al., 2007).
Regulation of S. aureus biofilm formation involves an intricate network of overlapping circuits. It is generally appreciated that inactivation of the sarA and sigma factor B (SigB) global regulators restricts biofilm formation (Beenken et al., 2003, Lauderdale et al., 2009), while inactivation of the agr quorum-sensing system has the opposite effect (Lauderdale et al., 2010, Boles et al., 2008). In recent years, it has become accepted that a major factor dictating these biofilm phenotypes is the array of extracellular enzymes produced by S. aureus strains, with most reports indicating that the proteases have an important role (Mootz et al., 2013, Beenken et al., 2010, Boles et al., 2010, Lauderdale et al., 2009, Boles & Horswill, 2008, Marti et al., 2010). It has been observed that the regulation of protease transcription through SigB and agr is not direct and instead requires intermediate players (Thoendel et al., 2011), one of the most important of these is the Repressor of toxins (Rot) (McNamara et al., 2000). Rot is a DNA-binding protein from the SarA family of S. aureus regulators that exhibits dual regulation as it promotes the expression of genes that encode for surface proteins and immunomodulators, such as Protein A and the superantigen-like proteins (Benson et al., 2011), and represses genes that encode for exo-toxins and exo-enzymes (Said-Salim et al., 2003, McNamara et al., 2000). Genetic analysis through epistasis studies suggests that Rot functions downstream of agr and represses toxin and exo-enzyme production (McNamara et al., 2000, McNamara et al., 2005). When agr is activated, RNAIII levels rise and prevent translation of Rot protein (Geisinger et al., 2006, Boisset et al., 2007), relieving the Rot repressive effect.
Based on the key role for proteases in S. aureus biofilm development, we hypothesized that inactivation of rot would result in a biofilm phenotype. Indeed, in this study we demonstrate that a rot mutant is defective in biofilm formation using multiple different assays. We find that secreted protease activity is elevated in a rot mutant through global analysis and direct studies, and that genetic and biochemical inhibition of the proteases repairs the biofilm phenotype. The importance of Rot-mediated gene regulation for biofilm-mediated disease is demonstrated using a murine model of catheter infection. Altogether, these data further highlight the complex regulatory networks involved in S. aureus virulence, and identify Rot as a key regulator of biofilm formation.
RESULTS
Rot is essential for biofilm formation by USA300 CA-MRSA
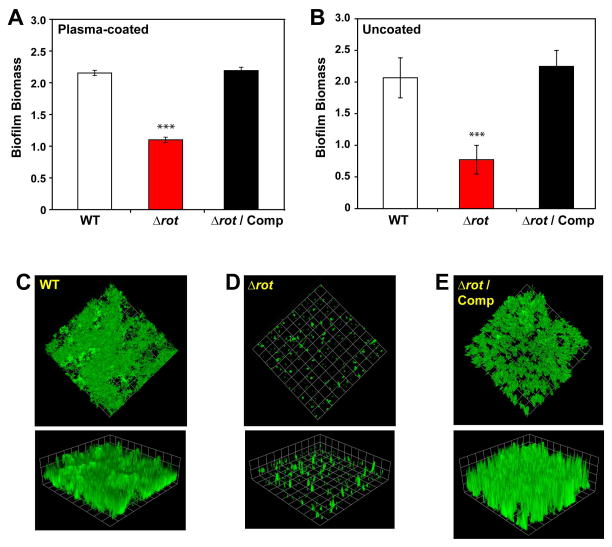
Most studies evaluating the influence of Rot in virulence factor expression have been performed in MSSA-laboratory strains or older clinical isolates such as Newman and COL (Li et al., 2008, Oscarsson et al., 2006, Said-Salim et al., 2003, McNamara et al., 2000). Because of this, the contribution of Rot to the expression of virulence factors in current epidemic strains remains to be fully elucidated. The clinical isolates of the pulse-field gel electrophoresis (PFGE)-type USA300 are the predominant community-associated methicillin-resistant S. aureus (CA-MRSA) strains in the United States and are known to cause chronic biofilm infections (Mootz et al., 2013, Walker et al., 2012, Kaplan et al., 2012, Lauderdale et al., 2010, Thurlow et al., 2011). To define the contribution of Rot to the virulence of this clone, we constructed a Δrot mutant in the USA300 strain LAC. The LAC-WT, Δrot mutant, and complemented strain (Δrot transformed with a complementation plasmid with the rot gene constitutively expressed using the lgt promoter) were compared in both plasma-coated and uncoated biofilm experiments. The Δrot mutant displayed a marked defect in biofilm capacity on both a plasma-coated surface (Fig. 1A) and uncoated surface (Fig. 1B), and in each case, biofilm formation could be restored through complementation of the mutation.
Figure 1. Rot is essential for S. aureus biofilm formation in CA-MRSA USA300.
Biofilm formation by strain LAC (WT), Δrot mutant, and complemented strains in microtiter (A, B) and flow cell (C, D, E) biofilm assays. A. Biofilms on plasma-coated surface. B. Biofilms on uncoated surface. For A and B, color scheme is as follows: white, WT; red, Δrot; black, Δrot Comp (***P<0.001 relative to WT strain as determined by paired T-test). Flow cell biofilms contained C) WT, D) Δrot, and E) Δrot complemented strains. Biofilms were post-stained with SYTO-9 and imaged with confocal microscopy. Representative top-down images and three-dimensional image reconstructions from a z series are shown. Experiments were performed in triplicate.
To extend our observations, the biofilm experiments were performed in a flow cell assay. Biofilms were grown for 2 days on uncoated coverslips, post-stained with SYTO-9, and the biomass of the biofilms visualized with confocal laser scanning microscopy (CLSM). Similar to the microtiter assays, LAC-WT was able to form a thick, confluent biofilm (Fig. 1C). In contrast, the isogenic Δrot mutant was unable to form a biofilm (Fig. 1D), a phenotype that could be restored to WT levels upon rot complementation (Fig. 1E). Taken together, deletion of rot in USA300 strain LAC diminishes the capacity of this strain to form biofilms in standard in vitro assays.
Rot is important for biofilm formation across multiple S. aureus strains
To assess the generality of the Rot requirement for S. aureus biofilm formation, we constructed rot mutants in a number of different genetic backgrounds, including representative USA100-USA800 MRSA strains. To build these strains, a rot::Tn mutant from the Nebraskea Transposon Mutant Library (NTML) was used (Fey et al., 2013), and the resistance was switched to tetracycline (Bose et al., 2013) and transduced into clinical isolates. We confirmed that the transposon in the rot::Tn mutant did not impact other loci by complementing protease phenotypes using the pOS1 plgt-rot plasmid (data not shown). In most backgrounds, the loss of Rot led to a significant drop in biofilm capacity on a plasma-coated surface (Fig. 2). We were unable to move the rot::Tn(Tet) mutant into USA500, but we have observed a significant phenotype using a Δrot::spec mutant in this lineage (data not shown). However, the biofilm requirement for Rot was not consistent in the USA100 and USA400 backgrounds (Fig. 2).
Figure 2. Rot is important for S. aureus biofilm formation in many strain lineages.
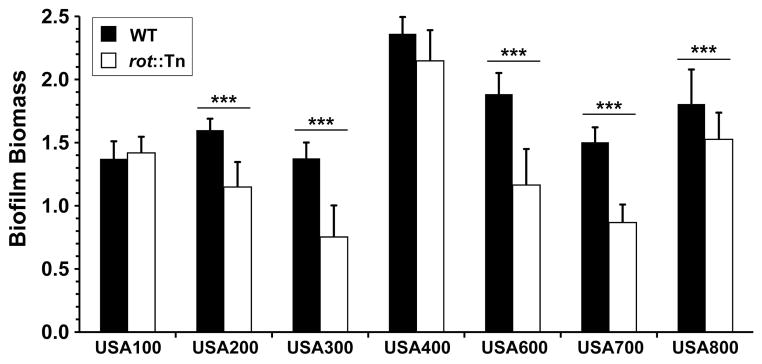
Biofilms of S. aureus WT and rot::Tn(Tet) mutant strains were assessed on a plasma-coated surface (***P<0.005 relative to WT strain as determined by paired T-test).
Rot expression modulates biofilm formation in USA300
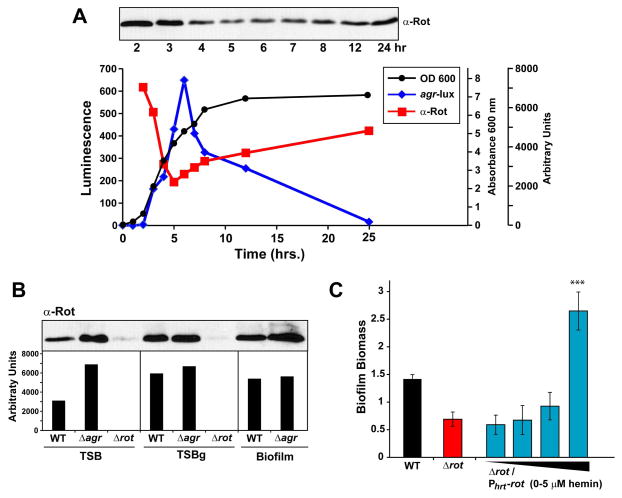
Based on the observation that Rot contributes to biofilm in most MRSA strains, we hypothesized that intracellular levels of Rot would increase under conditions that favored biofilm formation. Glucose is usually added to media to promote S. aureus biofilm formation, and it is well established that non-maintained pH decrease from glucose metabolism represses the agr quorum-sensing system (Regassa et al., 1992b, Boles & Horswill, 2008). With reduced agr function and RNAIII levels, it would be expected that Rot production would increase. To test this hypothesis, we grew an agr P3-lux reporter strain in TSB for 24 hr and collected samples throughout the time course. As shown in Figure 3A, agr P3 promoter activation peaked in late log/early stationary phase and rapidly decreased during stationary phase. In contrast, Rot concentration was inversely correlated with RNAIII expression (as demonstrated by immunoblot in Fig. 3A), consistent with published reports that RNAIII inhibits rot translation (Geisinger et al., 2006, Boisset et al., 2007).
Figure 3. Rot expression correlates with biofilm formation.
A. LAC (WT) containing the agr reporter plasmid pAmiAgrP3 was grown for 24 hr and samples were collected throughout a time course. Culture samples were standardized and whole cell bacterial lysates were used for immunoblot of Rot protein using an Anti-Rot antibody. Optical density measurements at 600 nm (black), luminescence (blue), and arbitrary units of Rot protein levels (red) as measured using densitometry are displayed. B. WT, Δagr and Δrot strains were grown overnight and sub-cultured in either TSB, biofilm media (TSBg) or under static biofilm conditions. Cultures were grown for 24 hr and cell cultures were collected and normalized to an OD600 of 1.5. Protein in whole cell lysates were precipitated with TCA for immunoblot of Rot protein. Arbitrary levels of Rot protein in each sample was measured using densitometry. C. LAC (WT) and Δrot strains containing the hemin-inducible rot expression plasmid were analyzed for biofilm formation in the presence of 0, 0.5, 2, or 5 μM hemin. Color scheme is as follows: black, WT; red, Δrot; light blue, Δrot PhrtAB-rot (***P<0.001 relative to Δrot strain as determined by paired T-test).
To test whether Rot levels increase under biofilm growth, we grew LAC-WT, Δagr and Δrot strains under three separate conditions. Rot production in a biofilm was assessed with either growth in biofilm-promoting media (TSB supplemented with 0.5% glucose and 3% NaCl) or the direct collection of biofilm biomass (except for the Δrot mutant which did not form a biofilm). For comparison, Rot levels were examined under planktonic conditions in TSB. All three conditions were grown for 24 hr, normalized to OD600, and Rot levels were determined with immunoblot and quantified by densitometry. Under planktonic conditions, low levels of Rot were detected in LAC-WT as compared with the isogenic Δagr mutant, indicating that agr was active and rot translation was repressed (Fig. 3B). In contrast, when S. aureus was grown in biofilm media, or when a biofilm was established, Rot was more abundant in both the LAC-WT and Δagr mutant strains as compared to levels in LAC-WT grown in TSB. Thus, as agr is repressed during biofilm formation, Rot levels increase accordingly.
To determine whether the intracellular levels of Rot can be altered to modulate biofilm capacity, a hemin-inducible rot expression plasmid was transformed into a Δrot strain. The ability of this plasmid to control Rot protein levels with hemin induction has been confirmed previously (Benson et al., 2012). Biofilm formation was assessed under varying concentrations of hemin that were sub-inhibitory to S. aureus growth. At low hemin concentrations, the Δrot mutant was unable to form a biofilm as expected compared to LAC-WT (Fig. 3C). As the expression of rot increased with hemin levels, biofilm formation was restored, and at high hemin levels, biofilm capacity continued to increase and exceeded even LAC-WT. As an additional control, a dose-response of hemin was added to Δrot mutant cultures lacking the expression plasmid and no significant changes in biofilm levels were observed (data not shown), demonstrating that hemin addition was not promoting biofilm formation through an independent mechanism. The experiment was also repeated with an empty vector control and only when the rot gene was provided did the biofilm phenotype change (data not shown). Altogether, these data demonstrate that the intracellular concentration of Rot modulates biofilm formation.
Rot is a global regulator of gene expression in USA300 and inhibits protease transcription
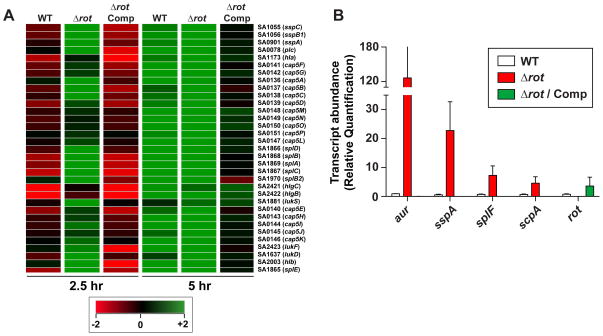
To begin defining mechanism of Rot–mediated biofilm regulation in USA300, microarrays were performed to examine gene expression of LAC Δrot strain compared to isogenic WT and Δrot complemented strain. Rot influences the expression of genes by activating or repressing promoters at both mid-log and late-log phase (Supplementary Table 2). At mid log phase, Rot positively influenced the expression of virulence factors such as ssl11, coa, spa, clfB, and formyl peptide receptor-like 1 inhibitor (Fig. 4A and Supplementary Table 2). In addition to virulence factors, Rot was also a positive regulator of sarS and sarT, two other SarA family members. The number of target genes positively regulated by Rot was found to be much smaller at late log phase, presumably because at this point, agr is activated and Rot levels are decreased due to RNAIII repression of rot translation (Fig 3A). Genes repressed by Rot include: the cytotoxins lukED, lukAB, and hlgCB, in addition to alpha hemolysin (hla), the Cap5 capsular polysaccharide synthesis loci, and the potassium-transporting ABC transporter system Kdp. In addition, we observed that nine of the ten secreted proteases were repressed by Rot (Fig. 4A and Supplementary Table 2), which is supported by a previous microarray study in a laboratory strain (Said-Salim et al., 2003).
Figure 4. Transcription of secreted proteases are upregulated in a rot mutant.
A. Microarray heat map of selected targets from LAC (WT), Δrot, and Δrot complemented strains at mid-log and late-log growth phase. Green coloring indicates increased transcript levels, and red coloring indicates reduced transcript. B. Transcript abundance of proteases and rot in LAC, Δrot, and Δrot complemented strains grown in TSB containing NaCl and glucose as measured by qRT-PCR. Values represent the average of 3 independent experiments ± standard deviation. Color scheme is as follows: white, WT; red, Δrot; green, Δrot Comp
It is well documented that there is strong correlation between protease levels and S. aureus biofilm capacity (Mootz et al., 2013, Lauderdale et al., 2010, Boles et al., 2010, Lauderdale et al., 2009, Zielinska et al., 2012, Beenken et al., 2010, Tsang et al., 2008, Marti et al., 2010, O’Neill et al., 2008). Because of this, we hypothesized that this connection was a major factor in the rot mutant biofilm phenotypes (Fig. 1). To validate the microarray data in regards to the proteases, we performed qRT-PCR analysis to monitor the levels of selected protease transcripts. All the protease transcripts were increased in the Δrot strain compared to WT (Fig. 4B), including aur, which was not seen in the microarray study.
Rot directly binds to secreted protease promoters
Since Rot is a DNA-binding global regulator of virulence factors, we hypothesized that it directly binds protease gene promoters to inhibit promoter activation. To test this hypothesis, we PCR amplified the promoter regions of each of the four secreted protease operons and tested the ability of purified Rot to interact with these promoters by electro-motility shift assays (EMSA). The addition of increasing concentrations of Rot to labeled promoter DNA resulted in a shifted product compared to the no protein control (Fig. 5A). These interactions were found to be specific, as the addition of unlabeled promoter DNA, but not non-promoter DNA, was able to prevent formation of the shifted product (Fig. 5B).
Figure 5. Rot protein binds protease promoters.
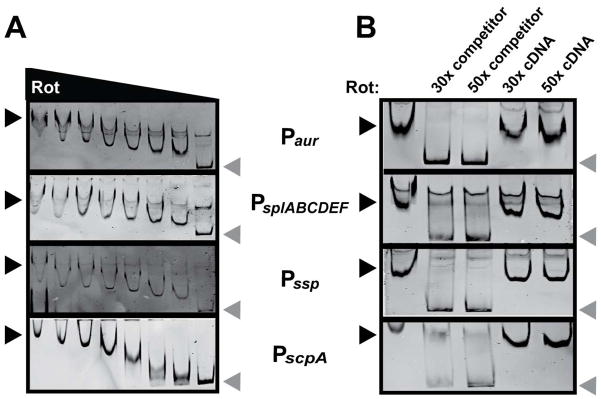
A. EMSAs of purified Rot incubated with protease reporters containing a biotin tag. Two-fold serial dilutions of Rot, starting with 4 pmol, were incubated with 40 fmol DNA. Protein-DNA complexes were separated by PAGE, and DNA probes were visualized using streptavidin DyLight. Black arrows indicate shifted probe, grey arrows indicate free probe. B. EMSA in which 2 pmol of Rot was incubated with 40 fmol of the indicated biotinylated promoter DNA with a 30- or 50-fold molar excess of non-biotinylated promoter DNA or non-biotinylated control DNA. The EMSA reaction was performed and visualized as for panel A. Black arrows indicate shifted probe, grey arrows indicate free probe. The control DNA (cDNA) used for EMSAs is the intragenic DNA between lukA and lukB.
Production and activity of secreted proteases are increased in a rot mutant
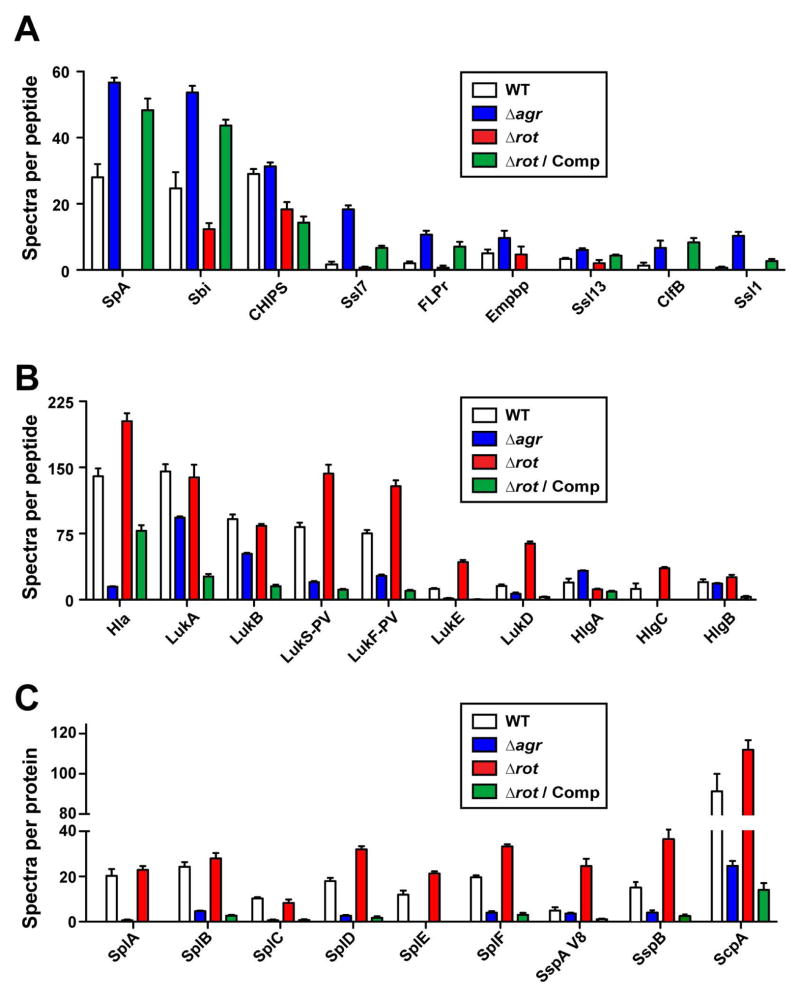
To expand our studies of Rot regulation, we also examined global differences in secreted protein abundance between isogenic WT, Δagr, Δrot, and Δrot complemented strains using LC-MS/MS shotgun proteomics (Fig. 6). We grouped the secreted factors into three categories: immune-modulators (Fig. 6A), cytotoxins (Fig. 6B), and secreted proteases (Fig. 6C). As with the transcriptional analysis (Fig. 4 and Supplementary Table 2), the abundance of immune-modulators such as SpA, FLPr, and the Ssls are influenced in a positive manner by Rot (Fig. 6A), while the abundance of cytotoxins and proteases are negatively influenced by Rot (Fig. 6B & C).
Figure 6. Proteomic analysis of agr and rot mutants.
Exoprotein profiles of USA300 LAC (WT), Δagr, Δrot, and Δrot complemented strains grown in RPMI/cas to late-logarithmic phase. Values represent the average of 3 independent experiments ± standard deviation. A. Immunomodulators. B. Toxins. C. Proteases. Color scheme is as follows: white, WT; blue, Δagr; red, Δrot; green, Δrot Comp.
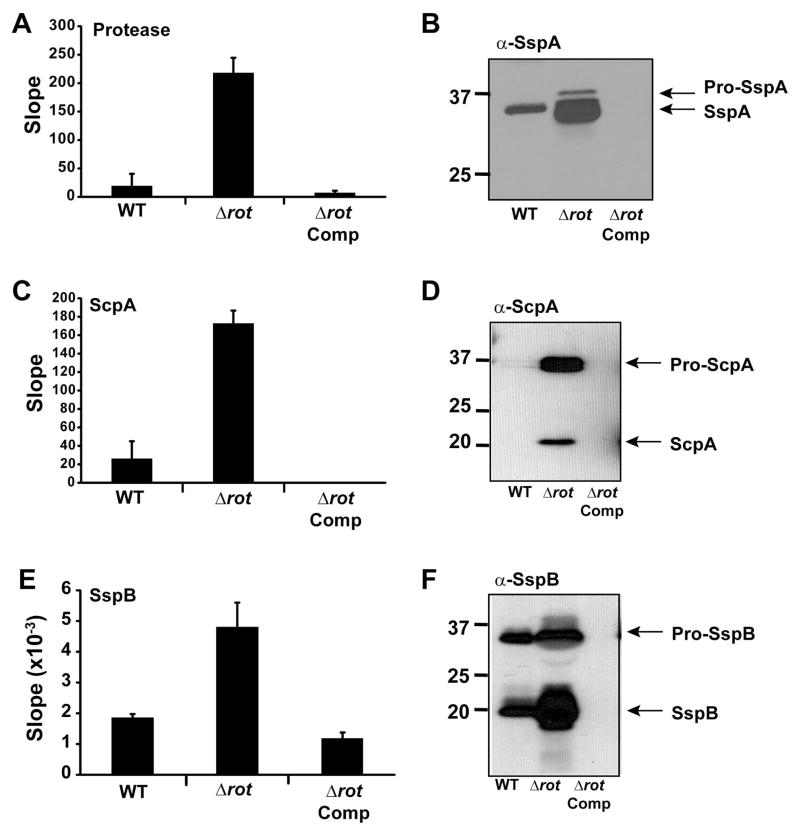
The increased production of secreted proteases detected by proteomics would suggest that there is a higher level of protease activity in S. aureus spent media. As a broad measure of overall protease activity, we utilized a fluorescent-resonance energy transfer (FRET)-based substrate that is susceptible to cleavage by the proteases SspA (V8), Aur, and ScpA. Spent media from a Δrot mutant showed greater protease activity than LAC-WT using this substrate, and this phenotype could be complemented (Fig. 7A). Since SspA is one of the dominant enzymes that cleaves this substrate (data not shown), we also performed immunoblots and observed that indeed SspA production is enhanced in a Δrot mutant (Fig. 7B), supporting the transcriptional (Fig. 4A & B) and proteomic studies (Fig. 6C).
Figure 7. Staphopain activity is upregulated in rot mutant.
Strain LAC (WT), Δrot mutant, and Δrot complemented strains were grown in TSB and samples were collected during late log phase (4 hr) and stationary growth phase (24 hr). Cells were removed by filtration and spent media was saved for enzyme assays and immunoblots. A. Protease activity using a general protease FRET substrate at 24 hr. B. Immunoblot for SspA at 24 hr. C. Measure of ScpA activity using the ScpA-specific FRET substrate at 4 hr. D. Immunoblot for ScpA at 4 hr. E Measure of SspB activity using a pNA substrate at 24 hr. F. Immunoblot for SspB at 24 hr.
Our previous studies with USA300 strains demonstrated that the Staphopain enzymes are important modulators of biofilm formation (Mootz et al., 2013). Staphopain A (ScpA) levels were measured using a specific FRET substrate based on the CXCR2 cleavage site (Laarman et al., 2012), and we observed upregulation of ScpA activity in a Δrot mutant (Fig. 7C). Immunoblots confirmed the increase of processed ScpA enzyme (Fig. 7D). Similarly, Staphopain B (SspB) activity levels increased in Δrot mutant compared to WT, as demonstrated with a specific p-nitroaniline (pNA) substrate (Fig. 7E). Again, immunoblots confirmed the elevated production of the fully processed form of SspB (Fig. 7F). For both Staphopain enzymes, the phenotypes of the Δrot mutant could be complemented.
Inhibition of Staphopains restores biofilm capacity to rot mutants
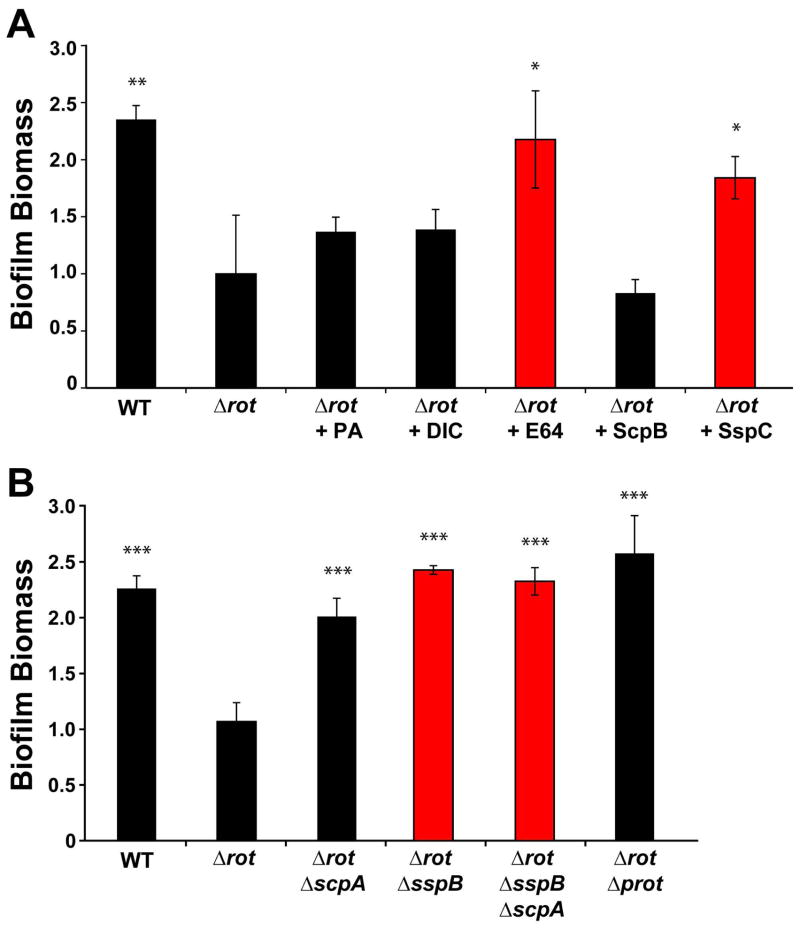
We have demonstrated that reduced biofilm formation in a USA300-LAC Δrot mutant correlates with the upregulation of secreted proteases. To further validate these findings, and to elucidate the individual contribution of each protease to biofilm inhibition, protease inhibitors were utilized. The inhibitors selected were the metalloprotease inhibitor 1,10-Phenanthroline (PA), serine protease inhibitor Dichloroisocoumarin (DIC), the cysteine protease inhibitor E-64, and the Staphostatin inhibitory proteins (Mootz et al., 2013). Each inhibitor was added at concentrations sub-inhibitory to growth of the Δrot mutant at time zero and biofilm formation was assessed after 24 hours. As shown in Figure 8A, only the general cysteine protease inhibitor E-64 and the SspC Staphostatin (specific inhibitor of SspB) were able to restore biofilm capacity. These observations suggest that the Staphopains, and in particular SspB, are detrimental to biofilm formation in the Δrot mutant.
Figure 8. Biochemical and genetic inhibition of Staphopains restores rot mutant biofilm capacity.
Biofilm formation by strain LAC (WT), Δrot, and Δrot Δprotease combination mutants were performed in a plasma-coated biofilm assay. A. Δrot biofilm formation was assessed in the presence of protease inhibitors at sub-inhibitory concentrations [10 μM] of either the metalloprotease inhibitor 1,10-phenanthroline, serine protease inhibitor 3,4-Dichloroisocoumarin (DIC) or cysteine protease inhibitor E-64. Δrot biofilm formation was also assessed in the presence of Staphostatin SspC or ScpB at 400 nM concentration. Red colored bars indicate conditions that include an inhibitor of SspB (*P<0.05, **P<0.01 relative to Δrot strain as determined by paired T-test). B. Biofilm formation of WT, Δrot and Δrot protease mutants was similarly assessed. Red colored bars indicate strains with sspB mutations (***P<0.001 relative to Δrot strain as determined by paired T-test).
To further validate the inhibitor findings, we used a genetic approach to introduce protease mutations into the Δrot background and tested the effect of these mutations on biofilm capacity. Based on the inhibitor analysis, we initially focused on mutations that removed the expression of the Staphopains. As shown in Figure 8B, deletion of the sspB and scpA genes restored Δrot mutant biofilm formation. Mutation of both Staphopain genes (scpA and sspB), or inactivation of all protease genes, completely restored Δrot biofilm formation to similar biomass levels of LAC-WT levels (Fig 8B). In additional experiments, the deletion of the Staphopain genes, or all the secreted protease genes, also restored Δrot biofilm formation on uncoated surfaces (data not shown), demonstrating that anti-biofilm effects of the proteases are self-targeting and not specific to the plasma coating. Thus, the genetic and biochemical protease inhibition results indicate that proteases are the predominant players in the Δrot biofilm phenotype, with Staphopain B (SspB) being the most important contributor.
Rot is important for USA300 pathogenesis in a mouse model of catheter-associated biofilm infection
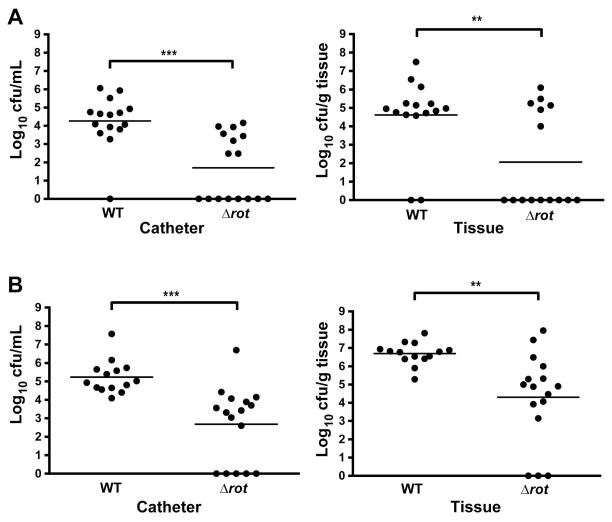
S. aureus readily adheres to catheter material in vivo resulting in chronic biofilm infections. Biofilms provide inherent resistance to innate immune defenses, as well as antibiotics, and therefore make it difficult for the host to clear an established infection (Thurlow et al., 2011, Heim et al., 2014). As Rot is important for biofilm formation in vitro, we speculated that Δrot would be attenuated in a mouse model of biofilm infection. We utilized a well-established model of S. aureus catheter-associated biofilm infection, where mice were challenged with a low infectious inoculum (103 CFUs) of WT or Δrot. Beginning 24 hours post-infection, animals received low-dose antibiotic treatment twice daily for the duration of the experiment to provide selective pressure. Importantly, both WT and Δrot were equally susceptible to the antibiotic combination utilized in these experiments (daptomycin and rifampicin). Animals were sacrificed three and seven days post-infection, whereupon bacterial burdens on the catheter and surrounding tissue were quantified. Compared with LAC-WT, the isogenic Δrot mutant exhibited significantly decreased bacterial burdens in both the catheter and surrounding tissue at both time points examined (Fig. 9), suggesting that Rot plays a key role in S. aureus biofilm formation and persistence.
Figure 9. Rot is essential for biofilm formation in a murine model of catheter-associated biofilm infection.
Biofilm infections were established in C57BL/6 mice following the inoculation of 103 CFU of WT or Δrot S. aureus into the lumen of subcutaneous implanted catheters. Mice were sacrificed at 3 (A) and 7 (B) days post-infection, whereupon bacterial burdens on the catheter and surrounding tissue were assessed. Statistical analysis was performed using a Student’s t-test with Welch’s correction for unequal variances. For all analyses, a p-value of less than 0.05 was considered statistically significant.
DISCUSSION
Since the establishment of S. aureus chronic infection is often due to biofilm formation, there is increasing interest in how regulatory factors coordinate the biofilm developmental process. Studies examining S. aureus biofilm regulation have pointed to several key regulators including sarA, sigB, and agr (O’Gara, 2007, Archer et al., 2011, Boles et al., 2011, Kiedrowski & Horswill, 2011a). Often these regulators modulate each other’s expression to impact biofilm formation, as is the case with SigB inhibition of agr function (Lauderdale et al., 2009). When activated, agr induces expression of secreted proteases to inhibit biofilms, however, the molecular details of this cascade are not fully elucidated (Boles & Horswill, 2008, Thoendel et al., 2011). Based on previous studies (Boisset et al., 2007, Geisinger et al., 2006, McNamara et al., 2000, Said-Salim et al., 2003), the agr effector molecule RNAIII acts through the regulatory intermediate Rot, a master regulator of exoprotein expression. In this report, we endeavored to connect all these regulatory components together to assess the role of Rot in biofilm formation.
Our current study demonstrates that Rot is important for S. aureus biofilm formation (Figs. 1 & 2) and that Rot intracellular levels modulate the biofilm forming capacity of S. aureus (Fig. 3C). As anticipated, these levels are dictated in large part by agr, where RNAIII expression levels are inversely correlated with Rot protein concentration (Fig. 3A). Interestingly, when S. aureus cells were collected from a biofilm or when cells were grown under conditions that promote biofilm formation, Rot levels in these cells were higher than in cells grown under planktonic conditions (Fig. 3B). These findings mimic those of the essential biofilm regulator SarA, which is present at higher concentrations during biofilm growth (Valle et al., 2003). In contrast, agr is significantly repressed in biofilms due to the drop in extracellular pH that results from excretion of acidic metabolites (Boles & Horswill, 2008, Regassa et al., 1992a). Thus, the repression of agr and simultaneous accumulation of Rot protein is a critical regulatory event that is essential for maintaining S. aureus biofilm integrity. However, we did find a couple S. aureus clinical isolates where Rot is not as important for biofilm formation (Fig. 2), suggesting that the cascade might be more complicated in some strains.
To understand the biofilm phenotype of a USA300 rot mutant, we investigated the role of Rot in secreted protease regulation. The proteases are known to exhibit anti-biofilm effects (Mootz et al., 2013, Zielinska et al., 2012, Lauderdale et al., 2010, Lauderdale et al., 2009, Boles & Horswill, 2008, Marti et al., 2010, O’Neill et al., 2008), and there are previous reports that protease activity is up-regulated in rot mutants constructed in laboratory strains (Oscarsson et al., 2006, Said-Salim et al., 2003, McNamara et al., 2000). In a USA300 strain, microarray studies confirmed that the transcription of the secreted protease genes was up-regulated (Fig. 4A), which was validated by qRT-PCR analysis (Fig. 4B). Proteomic (Fig. 6C), immunoblot (Fig. 7B, D, F), and enzyme activity (Fig. 7A, C, E) studies all verified that these secreted proteases are produced at higher levels in a rot mutant. In addition, we found that Rot directly binds all four secreted protease promoters (Fig. 5), which is in line with previous work by Oscarsson et al., which demonstrated that Rot binds to the aur and ssp promoters (Oscarsson et al., 2006). Taken together, these findings are significant in that they provide a direct link between agr and protease expression, facilitating the generation of a model of the regulatory network (Fig. 10). Briefly, agr activation results in increased RNAIII expression (further elevated with sigB mutation), and RNAIII then prevents Rot translation. This in turn relieves the Rot-mediated repression of protease gene transcription, which limits biofilm formation due to high activity of the secreted proteases. We hypothesize these proteases are targeting the surface and matrix proteins that serve as bridges to encase the S. aureus biofilm. In contrast, when agr is inhibited, RNAIII levels are low and Rot protein levels stabilize, resulting in repression of protease production and an enhancement in biofilm formation. By directly modulating Rot levels in the studies described herein, we further defined the role of Rot to solidify the final piece of this biofilm regulatory model.
Figure 10. Schematic of Rot role in the S. aureus biofilm formation pathway.
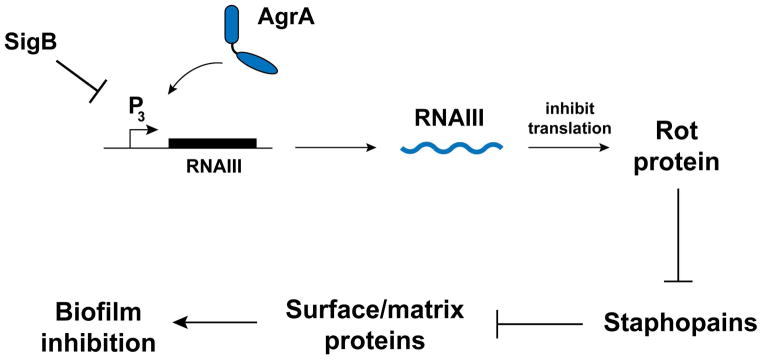
Sigma B inhibits the agr system by an unknown mechanism. RNAIII, the primary agr effecter molecule, binds to rot mRNA and prevents translation. Rot, a DNA-binding transcriptional regulator, directly binds protease gene promoters and inhibits transcription. Secreted proteases are destructive towards biofilms due to cleavage of biofilm matrix constituents.
While we demonstrated that Rot represses protease gene expression, and subsequently protease production, it was still unclear which of the secreted proteases were responsible for the anti-biofilm properties of the rot mutant. We observed that biofilm formation could be restored to a rot mutant using the chemical protease inhibitor E-64, the biochemical inhibitor Staphostatin B (SspC), or through specific protease mutations (Fig. 8), all seeming to point toward the cysteine proteases, and in particular Staphopain B (SspB), as being the primary biofilm inhibitor. This finding is supported by the fact that SspB protein levels and activity are markedly enhanced in a rot mutant (Fig 7C & D). We recently performed a study to investigate protease impact on USA300 biofilm formation using sigB mutants (Mootz et al., 2013), and we observed that Staphopain A (ScpA) was the most significant anti-biofilm protease. The rot mutant impact on ScpA regulation was significant (Fig. 7C&D) and removal of ScpA from a rot mutant did partially repair biofilm capacity (Fig. 8B). Thus, both Staphopain enzymes appear to be important contributors to the rot mutant biofilm phenotype.
Finally, we observed that the rot mutant was attenuated in a murine catheter model of biofilm infection (Fig. 9). This finding demonstrates that the in vitro biofilm phenotype is relevant in vivo during infection. However, the mechanism of the in vivo phenotype is complicated by the fact that Rot is an important regulator of immunomodulatory proteins (Benson et al., 2011), and these may have a role in protecting the organism during catheter infection. Additionally, the secreted proteases themselves are virulence factors that are known to tailor the secreted proteome of S. aureus (Kolar et al., 2013). Altered production of Aureolysin in particular has been shown to modulate proteome levels and in turn influence infection outcomes (Cassat et al., 2013, Zielinska et al., 2011). The data presented in this study identify exciting new roles for the transcriptional regulator Rot in maintenance of biofilm integrity, and further elucidate the individual roles of each of the proteases during biofilm formation.
EXPERIMENTAL PROCEDURES
Ethics statement
This study was conducted in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The protocol was approved by the Institutional Animal Care and Use Committee of the University of Nebraska Medical Center (Protocol number 09-049-06-EP).
Strains and growth conditions
S. aureus cultures were grown in Tryptic Soy Broth (TSB), Tryptic Soy Agar (TSA), or Roswell Park Memorial Institute medium (RPMI) supplemented with 1% casamino acids. The bacterial strains and plasmids used in this study are described in Table 1. Escherichia coli cultures were grown in LB broth or on LB agar plates supplemented with 100 μg/mL ampicillin (Amp) as required for plasmid maintenance. S. aureus chromosomal markers or plasmids were selected for, or maintained in, 10 μg/mL of chloramphenicol (Cam) or erythromycin (Erm), 0.625 μg/mL tetracycline (Tet), and 100 μg/mL spectinomycin (Spec). Unless otherwise stated, all broth cultures were grown at 37°C with shaking at 200 rpm.
Table 1.
Strain and Plasmid List
| Strain or Plasmid | Description | Source or Reference |
|---|---|---|
| Escherichia coli | ||
| ER2566 | Cloning Strain | New England Biolabs |
| Staphylococcus aureus | ||
| RN4220 | Restriction modification deficient cloning host | (Nair et al., 2011) |
| AH1263 | LAC* USA300 CA-MRSA ErmS | (Boles et al., 2010) |
| AH1292 | AH1263 Δagr::tetM | (Kiedrowski et al., 2011b) |
| AH1825 | AH1263 ΔsspAB | (Wormann et al., 2011) |
| AH1825 | AH1263 ΔscpA | (Wormann et al., 2011) |
| AH1892 | AH1263 ΔsspAB ΔscpA | (Wormann et al., 2011) |
| AH1919 | AH1263 Δaur ΔsspAB ΔscpA Δspl::erm | (Wormann et al., 2011) |
| AH2184 | AH1263 Δrot::spec | This work |
| AH2544 | AH1263 Δrot::spec Δagr::tet | This work |
| AH2546 | AH1263 Δrot::spec Δaur ΔsspAB ΔscpA Δspl::erm | This work |
| AH2595 | AH1263 ΔsspB::pSMUT ΔscpA | (Mootz et al., 2013) |
| AH2553 | AH1263 Δrot::spec ΔsspAB | This work |
| AH2554 | AH1263 Δrot::spec ΔscpA | This work |
| AH2557 | AH1263 Δrot::spec ΔsspAB ΔscpA | This work |
| AH2664 | AH1263 Δrot::spec ΔsspB::pSMUT | This work |
| AH2665 | AH1263 Δrot::spec ΔsspB::pSMUT ΔscpA | This work |
| AH3545 | AH1263 rot::tetM | This work |
| Newman | MSSA | (Baba et al., 2008) |
| AH3546 | Newman rot::tetM | This work |
| MN8 | USA200 toxic shock isolate | (Maira-Litran et al., 2002) |
| AH3548 | MN8 rot::tetM | This work |
| MW2 | USA400 CA-MRSA | (Baba et al., 2002) |
| AH3606 | MW2 rot::tetM | This work |
| AH2623 | USA100 blood isolate #209 | D. Diekema |
| AH3575 | AH2623 rot::tetM | This work |
| BK22950 | USA600 | B. Kreiswirth |
| AH3587 | BK22950 rot::tetM | This work |
| AH2652 | USA700 | NARSA collection |
| AH3573 | USA700 rot::tetM | This work |
| AH2654 | USA800 | NARSA collection |
| AH3563 | USA800 rot::tetM | This work |
| Plasmids | ||
| pAmiAgrP3 | agrP3-lux | (Subrt et al., 2011) |
| pJM01 | pOS1 plgt with new MCS | This work |
| pOS1 phrtAB rot | rot gene cloned into pOS1 phrtAB | This work |
| pOS1 plgt rot | rot gene cloned into pOS1 plgt | (Benson et al., 2012) |
Recombinant DNA and genetic techniques
Oligonucleotides were synthesized by Integrated DNA Technologies (Coralville, IA) and are listed in Supplementary Table 1. Restriction modification enzymes were purchased from New England Biolabs and used according to the manufacturer’s instructions. Plasmids were first electroporated into S. aureus RN4220 as previously described (Schenk et al., 1992) and subsequently transformed into select strains. As needed, chromosomal mutations were transduced into select strains using bacteriophage 11 or 80α (Novick, 1991). Non-radioactive sequencing was performed at the University of Iowa or New York University DNA Facility to confirm constructs.
Plasmid construction
To constitutively express rot we used a complementation plasmid where rot expression was controlled by the lgt promoter (Benson et al., 2011). The rot allele was also cloned into the pOS-PhrtAB plasmid where rot expression was controlled by the hrtAB heme-inducible promoter (Benson et al., 2012). Lastly, an empty vector control with a new multiple cloning site (MCS) was developed from the rot complementation plasmid. Oligonucleotides JMM032 and JMM033 were allowed to anneal and ligated into NdeI and XhoI digested pOS1 plgt rot resulting in pJM01. Plasmid construction was confirmed by PCR and enzymatic digestion.
Strain construction
S. aureus Δrot::spec mutant strains were generated using the pKOR-1 allelic replacement strategy previously described (Bae et al., 2006). Briefly, sequences flanking rot were PCR amplified with primers VJT423 and VJT424 for the upstream fragment and primers VJT425 and VJT426 for a downstream fragment. The PCR amplicons were digested with XmaI and PstI and assembled into pGEMT-T (Promega). To generate the plasmid containing the rot::spec construct, the spectinomycin resistance cassette (aad9) was amplified from plasmid pBT-S (kindly provided to us by Dr. Anthony Richardson) and cloned into the pGEM-T containing the Δrot construct as described previously (Benson et al., 2012). A PCR amplicon of the resultant Δrot::spec construct was then recombined into pKOR-1 resulting in the pKOR-1Δrot::spec plasmid. Allelic replacement was carried out in strain LAC according to previously described methods.
The S. aureus rot::tetM mutant was generated from strain NE386 (JE2 rot::φNΣ) (Fey et al., 2013). Briefly, allelic replacement was used to change the mariner transposon-encoded erythromycin resistance cassette to a tetracycline resistance cassette (tetM), as previously described (Bose et al., 2013). The rot::tetM allele was transduced into a variety of S. aureus backgrounds using phage 11 and mutants were confirmed by PCR.
Transduction of Δrot::spec into AH1292 resulted in the double mutant Δrot::spec Δagr::tet (designated AH2544). Protease mutant strains were previously constructed (Mootz et al., 2013). For construction of rot/protease mutant combinations, Δrot::spec was transduced into select protease mutants. Clones were selected for on TSA with Spec and mutations were confirmed using PCR.
Biofilm assays
Microtiter plate biofilms
Biofilm formation was assessed on both coated and uncoated surfaces. Plasma-coated static microtiter plate assays were performed as previously described (Mootz et al., 2013). Briefly, lyophilized human plasma (Sigma) was resuspended to 20% in Carbonate-Bicarbonate buffer, filtered through a 0.22 μm membrane (Millipore), and added to the wells of a 96-well microtiter plate (Corning). Coated plates were incubated overnight at 4°C before the plasma was removed by aspiration. Immediately following aspiration, bacteria diluted 1:200 in biofilm media (TSB + 3% NaCl + 0.5% glucose) were added to coated wells and statically incubated for 24 hr at 37°C. Spent supernate was removed, and biofilms were washed 3 times in 1X PBS, fixed with 100% ethanol, stained with crystal violet and again washed 3 times with PBS. Biomass was assessed by resuspending biofilms in isopropanol and measuring absorbance at 595 nm using a Tecan plate reader. Uncoated microtiter plate biofilm assays were performed as previously described (Kiedrowski et al., 2011b). All biofilm additives were added at time zero at concentrations that did not inhibit growth (data not shown). Antibiotics were used to maintain plasmids as necessary. pOS1-PhrtAB rot was induced with 0, 0.5, 2 and 5 μM hemin (Sigma). 1,10-Phenathroline monohydrate (1,10-PA), 3,4-Dichloro-isocoumarin (DIC) and E-64 (Sigma) were each used at a concentration of 10 μM. Purified MBP, MBP-ScpB and MBP-SspC were used at a final concentration of 400 nM.
Flow cell biofilms
Flow cell biofilm formation was assessed as previously described (Kiedrowski et al., 2011b, Boles & Horswill, 2008). Bacteria were grown in 2% TSB supplemented with 0.2% glucose in flow cell chambers for 48 hr. When required for plasmid maintenance, 5 μg/mL Cam was added to the growth media. Biofilms were post-stained with SYTO-9 to detect biomass and confocal laser scanning microscopy (CLSM) was performed on a Nikon Eclipse E600 microscope using the Radiance 2100 image capturing system (Biorad). Image acquisition was performed with the Laser Sharp 2000 software (Zeiss) and images were processed using Volocity software (Improvision).
RNAIII reporter assay
A S. aureus LAC reporter strain containing pAmiAgrP3 (agrP3-lux) (Subrt et al., 2011) was grown overnight in TSB supplemented with Cam and sub-cultured in 25 mL to an OD600 of 0.1. Two-hundred μL samples were collected throughout a 24 hr time course and measured for luminescence in a microtiter plate (Corning) using a Tecan plate reader.
Protein preparation and Western blotting
The Rot immunoblots were performed using a rabbit polyclonal antibody described previously (Benson et al., 2012). S. aureus Rot protein was derived from whole cell bacterial lysates. To determine relative quantities of Rot protein in bacterial cultures grown under differing conditions, cells were collected after 24 hr growth and standardized to an OD600 of 1.5. For cells collected from mature biofilms, spent supernate was removed from microtiter wells and the biofilm was subsequently resuspended in 200 μL of fresh biofilm media. Multiple biofilm cultures from separate wells were pooled prior to measuring light absorbance. Protein was collected from whole cell lysates by Trichloroacetic acid (TCA) precipitation. To analyze protease protein content, select strains were grown overnight and sub-cultured to an OD600 of 0.05. These cultures were grown for 24 hrs and spent supernates were collected at 4 hr (late log phase) and 24 hr (stationary phase) time points and TCA precipitated. TCA precipitated samples were mixed with SDS-PAGE loading buffer and 5 μL of each sample was electrophoresed on a 12% polyacrylamide gel. The proteins were transferred to Immobilon-P PVDF membranes (Millipore) using a Protean II device (Bio-Rad Laboratories, Hercules, CA). Membranes were blocked overnight at 4°C with 5% milk in Tris-buffered saline (20 mM Tris-HCl, pH 7.0, with 137 mM NaCl) containing 0.1% Tween 20 (TBST). Primary antibodies were diluted in 5% milk in TBST and incubated with the membranes at room temperature for 2 hr. Membranes were rinsed 3 times in TBST and washed with agitation for 15 min, and subsequently two more times for 5 min. Secondary HRP-conjugated goat anti-chicken IgG (or goat anti-rabbit) was diluted 1:20,000 in 5% milk in TBST and incubated at room temperature for 1 hr. The membrane was rinsed and washed again as above. SuperSignal West Pico chemiluminescent substrate was added for 5 min at room temperature followed by exposure to X-ray film. For quantification of Rot protein the X-ray film was scanned and densitometry analysis performed using ImageJ software. Polyclonal rabbit anti-SspA was generously provided by Dr. Martin McGavin and used at 1:5,000, and immunoblots were performed as described above. Polyclonal chicken anti-SspB and anti-ScpA was used at concentrations of 1:2,000 and immunoblots were performed as previously described (Mootz et al., 2013).
RNA isolation
RNA from S. aureus was isolated as described previously (Benson et al., 2011). Briefly, S. aureus cultures were grown in RPMI/CAS or TSB + 0.5% glucose + 3% NaCl for five hours (stationary phase) and then normalized to the same OD600. Cultures were then mixed with an equal volume of 1:1 ethanol:acetone mixture and frozen at −80°C. For RNA extraction, frozen samples were allowed to thaw on ice; cells were then sedimented and washed two times with TE buffer. Cells were broken mechanically using fastprep bead beater (MP Biomedicals, Solon, OH), and the total RNA was isolated using the RNeasy mini kit (Qiagen, Valencia, CA) according to the manufacturer’s recommendation. On-column DNase digestion was performed and after RNA elution, residual DNA contamination was removed using the RNeasy mini kit (Qiagen, Valencia, CA).
Microarray
Microarrays were performed as described previously (Morrison et al., 2012b, Morrison et al., 2012a). Briefly, ten micrograms of total bacterial RNA from cultures grown in RPMI/CAS was labeled and hybridized to S. aureus GeneChip arrays by following the manufacturer’s recommendations for antisense prokaryotic arrays (Affymetrix, Santa Clara, CA). Average GeneChip signal intensities for biological replicates (two) for each condition were obtained from values normalized to the total GeneChip values. Differentially expressed genes exhibiting an average 2-fold difference in expression in LAC wild-type and Δrot pOS1plgt rot cells in comparison with that in LAC Δrot cells were determined using GeneSpring 7.2 software (Agilent Technologies, Redwood City, CA) as previously described (Anderson et al., 2010, Anderson et al., 2006, Beenken et al., 2004).
qRT-PCR analysis
100 ng of RNA purified as described above from cultures grown in TSB + 0.5% glucose + 3% NaCl was used to perform qRT-PCR using SYBER Green Master Mix (Qiagen, Valencia, CA) in a 7300 Real Time PCR System (Applied Biosystems). Primers used to detect specific transcripts are listed in Supplemental Table 1. 10 ng of 16S RNA was used as an endogenous control. Analysis was performed using 7300 Real Time PCR System Software.
Purification of Rot protein and Electrophoretic Mobility Shift Assays
These experiments were performed as described previously (Benson et al., 2012). As done previously, the Rot-biotinylated probe complex was competed with 30–50 fold excess of nonbiotinylated promoter DNA (probe) or control DNA (lukAB intragenic DNA). It includes the end of the lukA gene, the short intragenic region, and the beginning of the lukB coding sequence
Proteomics
Briefly, S. aureus LAC WT, Δrot, and Δrot pOS1plgt rot were grow overnight in RPMI/CAS (+ CM for strain containing plasmid). Overnight cultures were subcultured 1:100 into fresh RPMI/CAS and grown for 5 hrs. to late exponential phase. Cultures were OD600 normalized and sedimented by centrifugation. Supernatants were filtered and precipitated with 10% TCA. Proteins were resuspended in 1X SDS-digestion buffer and loaded into 12% SDS-PAGE gel (without a stacking gel) and electrophoresed 2 cm into the gel. The gel was stained with Colloidal Blue (Invitrogen, Carlsbad, CA) then destained with distilled water over night. Proteins were then subjected to in-gel trypsin digestion and peptide extraction and the resulting peptides were then analyzed as described previously (Attia et al., 2010).
Protease activity assays
Protease activity was monitored using three unique peptide substrates. To prepare samples to measure protease activity using peptide substrates, overnight cultures were sub-cultured to an OD600 of 0.05 and allowed to grow for an additional 4 hr or 24 hr as indicated. Spent media was collected and filtered through 0.22 μm Costar Spin-X centrifuge tube filters (Corning, NY). Fluorescence measurements (excitation 490 nm, emission 520 nm) were obtained at 37°C in a Tecan plate reader. A Fluorescence Resonance Energy Transfer (FRET) assay was used to examine protease activity. The FRET substrate (5-FAM-Lys-Lys-Ala-Ala-Glu-Ala-Ser-Lys-(QXL520)-OH; AnaSpec, Fremont, CA) is based on a known SspA peptide substrate but is also cleaved to a lesser extent by Aur and ScpA (Mootz et al., 2013). Similarly, a FRET substrate assay was developed to measure ScpA activity. The FRET substrate (5-FAM-Lys-Leu-Leu-Asp-Ala-Ala-Pro-Lys(QXL-520)-OH; AnaSpec) is based on the known ScpA cleavage substrate CXCR2 (Laarman et al., 2012). The substrate was resuspended to 50 μM using 20 mM Tris pH 7.4. Spent media samples were mixed 1:1 with the FRET substrate and fluorescence measurements (excitation 490 nm, emission 520 nm) were obtained at 37°C in a microtiter plate reader. Substrate specificity was measured using a combination of purified enzymes and protease mutational analysis. Purified Aureolysin, SspB and ScpA were purchased from BioCentrum (Krakow, Poland), and purified SspA was purchased from Worthington Biochemical Corporation (Lakewood, NJ). Our findings indicate that among the S. aureus secreted proteases only ScpA readily cleaves the CXCR2-based substrate (data not shown). SspB protease activity was measured using the synthetic chromogenic substrate Bz-Pro-Phe-Arg-pNA (Bachem, Torrance, CA) as described (Mootz et al., 2013).
Purification and testing of MBP fusion proteins
Plasmid constructs were previously generated for expression of MBP-Staphostatin fusion proteins (Mootz et al., 2013). Purification of MBP-ScpB and MBP-SspC was performed according to the manufacturer’s instructions (New England Biolabs). Proteins were purified over amylose resin and eluted fractions analyzed via SDS-PAGE and concentrated using an Amicon Ultra-15 with a 10K cut-off (Millipore). Protein concentration was measured using the Bio-Rad protein assay (Hercules, CA). Activity of MBP-ScpB and MBP-SspC was measured as previously described (Mootz et al., 2013).
Mouse model of S. aureus catheter-associated biofilm infection
S. aureus catheter-associated biofilm infections were established as previously described (Cassat et al., 2007, Thurlow et al., 2011). Briefly, male C57BL/6 mice (8 weeks of age) were obtained from Charles River Laboratories (Frederick, MD), anesthetized with tribromoethanol (Avertin) and the skin was shaved and disinfected with povidone-iodine. A small s.c. incision was made in the flank and a blunt probe was used to create a pocket for insertion of a sterile 14-gauge teflon i.v. catheter 1 cm in length (Excel International, St. Petersburg, FL). Incisions were sealed using Vetbond Tissue Adhesive (3M, St. Paul, MN), whereupon 103 CFU of LAC, or its isogenic mutant LAC Δrot::spec were slowly injected through the skin, directly into the catheter lumen in a volume of 20 μL sterile PBS. Beginning on day 1 post-infection, animals received antibiotic treatment (rifampicin and daptomycin; 25 and 5 μg/kg, respectively) twice per day until animals were sacrificed at the appropriate time points for determination of bacterial burdens.
Recovery of catheters and surrounding tissues for S. aureus enumeration
Mice were euthanized at days 3 and 7 post-infection with an overdose of inhaled isoflurane. Catheters were removed and sonicated in 1 mL PBS to dissociate bacteria from the catheter surface. The heart, kidney, and tissues surrounding the infected catheters were collected, weighed, and disrupted in 500 μL homogenization buffer (PBS supplemented with 100 μL RNasin and a protease inhibitor tablet [Roche Diagnostics, Indianapolis, IN]) using a Bullet Blender (Next Advance, Averill Park, NY). Bacterial titers associated with catheters and surrounding tissues as well as the heart and kidney were quantified on TSA plates supplemented with 5% sheep blood (HemoStat Laboratories, Dixon, CA) and are expressed as Log10 CFU per milliliter for catheters or Log10 CFU per gram wet tissue weight.
Statistical analysis
Significant differences between experimental groups were determined using Student’s t-test with Welch’s correction for unequal variances (GraphPad Prism 4.02, GraphPad Software, Inc., La Jolla, CA). For all analyses, a p-value of less than 0.05 was considered statistically significant.
Supplementary Material
Acknowledgments
We thank Dr. Mark McGavin for providing SspA antisera, Drs. Barry Kreiswirth and Daniel Diekema for strains, and Dr. W. Hayes McDonald and the Mass Spectrometry Research Center at Vanderbilt University School of Medicine, for the mass spectrometry analyses. Research reported in this publication was supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health (NIH) under award numbers AI07511 T32 training grant (to JM), AI078921 (to ARH), AI083211 (to ARH and TK), and AI101533 (to VJT).
Footnotes
The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
References
- Anderson KL, et al. Characterization of the Staphylococcus aureus heat shock, cold shock, stringent, and SOS responses and their effects on log-phase mRNA turnover. J Bacteriol. 2006;188:6739–6756. doi: 10.1128/JB.00609-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson KL, Roux CM, Olson MW, Luong TT, Lee CY, Olson R, Dunman PM. Characterizing the effects of inorganic acid and alkaline shock on the Staphylococcus aureus transcriptome and messenger RNA turnover. FEMS Immunol Med Microbiol. 2010;60:208–250. doi: 10.1111/j.1574-695X.2010.00736.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archer NK, Mazaitis MJ, Costerton JW, Leid JG, Powers ME, Shirtliff ME. Staphylococcus aureus biofilms: properties, regulation, and roles in human disease. Virulence. 2011;2:445–459. doi: 10.4161/viru.2.5.17724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attia AS, Benson MA, Stauff DL, Torres VJ, Skaar EP. Membrane damage elicits an immunomodulatory program in Staphylococcus aureus. PLoS Pathog. 2010;6:e1000802. doi: 10.1371/journal.ppat.1000802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba T, Bae T, Schneewind O, Takeuchi F, Hiramatsu K. Genome sequence of Staphylococcus aureus strain Newman and comparative analysis of staphylococcal genomes: polymorphism and evolution of two major pathogenicity islands. J Bacteriol. 2008;190:300–310. doi: 10.1128/JB.01000-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba T, et al. Genome and virulence determinants of high virulence community-acquired MRSA. Lancet. 2002;359:1819–1827. doi: 10.1016/s0140-6736(02)08713-5. [DOI] [PubMed] [Google Scholar]
- Bae T, Schneewind O. Allelic replacement in Staphylococcus aureus with inducible counter-selection. Plasmid. 2006;55:58–63. doi: 10.1016/j.plasmid.2005.05.005. [DOI] [PubMed] [Google Scholar]
- Beenken KE, Blevins JS, Smeltzer MS. Mutation of sarA in Staphylococcus aureus limits biofilm formation. Infect Immun. 2003;71:4206–4211. doi: 10.1128/IAI.71.7.4206-4211.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beenken KE, et al. Global gene expression in Staphylococcus aureus biofilms. J Bacteriol. 2004;186:4665–4684. doi: 10.1128/JB.186.14.4665-4684.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beenken KE, et al. Epistatic relationships between sarA and agr in Staphylococcus aureus biofilm formation. PLoS ONE. 2010;5:e10790. doi: 10.1371/journal.pone.0010790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson MA, Lilo S, Nygaard T, Voyich JM, Torres VJ. Rot and SaeRS cooperate to activate expression of the staphylococcal superantigen-like exoproteins. J Bacteriol. 2012;194:4355–4365. doi: 10.1128/JB.00706-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson MA, et al. Staphylococcus aureus regulates the expression and production of the staphylococcal superantigen-like secreted proteins in a Rot-dependent manner. Mol Microbiol. 2011;81:659–675. doi: 10.1111/j.1365-2958.2011.07720.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisset S, et al. Staphylococcus aureus RNAIII coordinately represses the synthesis of virulence factors and the transcription regulator Rot by an antisense mechanism. Genes & Development. 2007;21:1353–1366. doi: 10.1101/gad.423507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boles BR, Horswill AR. agr-mediated dispersal of Staphylococcus aureus biofilms. PLoS Pathogens. 2008;4:e1000053. doi: 10.1371/journal.ppat.1000052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boles BR, Horswill AR. Staphylococcal biofilm disassembly. Trends in microbiology. 2011;19:449–455. doi: 10.1016/j.tim.2011.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boles BR, Thoendel M, Roth AJ, Horswill AR. Identification of genes involved in polysaccharide-independent Staphylococcus aureus biofilm formation. PLoS ONE. 2010;5:e10146. doi: 10.1371/journal.pone.0010146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bose JL, Fey PD, Bayles KW. Genetic tools to enhance the study of gene function and regulation in Staphylococcus aureus. Appl Environ Microbiol. 2013;79:2218–2224. doi: 10.1128/AEM.00136-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassat JE, et al. A secreted bacterial protease tailors the Staphylococcus aureus virulence repertoire to modulate bone remodeling during osteomyelitis. Cell Host Microbe. 2013;13:759–772. doi: 10.1016/j.chom.2013.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassat JE, Lee CY, Smeltzer MS. Investigation of biofilm formation in clinical isolates of Staphylococcus aureus. Methods Mol Biol. 2007;391:127–144. doi: 10.1007/978-1-59745-468-1_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers HF, Deleo FR. Waves of resistance: Staphylococcus aureus in the antibiotic era. Nat Rev Microbiol. 2009;7:629–641. doi: 10.1038/nrmicro2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costerton JW. Biofilm theory can guide the treatment of device-related orthopaedic infections. Clin Orthop Relat Res. 2005;437:7–11. doi: 10.1097/00003086-200508000-00003. [DOI] [PubMed] [Google Scholar]
- Fey PD, Endres JL, Yajjala VK, Widhelm TJ, Boissy RJ, Bose JL, Bayles KW. A genetic resource for rapid and comprehensive phenotype screening of nonessential Staphylococcus aureus genes. MBio. 2013;4:e00537–00512. doi: 10.1128/mBio.00537-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisinger E, Adhikari RP, Jin R, Ross HF, Novick RP. Inhibition of rot translation by RNAIII, a key feature of agr function. Mol Microbiol. 2006;61:1038–1048. doi: 10.1111/j.1365-2958.2006.05292.x. [DOI] [PubMed] [Google Scholar]
- Hall-Stoodley L, Stoodley P. Biofilm formation and dispersal and the transmission of human pathogens. Trends Microbiol. 2005;13:7–10. doi: 10.1016/j.tim.2004.11.004. [DOI] [PubMed] [Google Scholar]
- Haque NZ, et al. Infective endocarditis caused by USA300 methicillin-resistant Staphylococcus aureus (MRSA) Int J Antimicrob Agents. 2007;30:72–77. doi: 10.1016/j.ijantimicag.2007.02.007. [DOI] [PubMed] [Google Scholar]
- Heim CE, Vidlak D, Scherr TD, Kozel JA, Holzapfel M, Muirhead DE, Kielian T. Myeloid-Derived Suppressor Cells Contribute to Staphylococcus aureus Orthopedic Biofilm Infection. J Immunology. 2014;192:3778–92. doi: 10.4049/jimmunol.1303408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan JB, et al. Low levels of beta-lactam antibiotics induce extracellular DNA release and biofilm formation in Staphylococcus aureus. MBio. 2012;3:e00198–00112. doi: 10.1128/mBio.00198-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiedrowski MR, Horswill AR. New approaches for treating staphylococcal biofilm infections. Annal New York Acad Sci. 2011a;1241:104–121. doi: 10.1111/j.1749-6632.2011.06281.x. [DOI] [PubMed] [Google Scholar]
- Kiedrowski MR, et al. Nuclease modulates biofilm formation in community-associated methicillin-resistant Staphylococcus aureus. PLoS ONE. 2011b;6:e26714. doi: 10.1371/journal.pone.0026714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolar SL, et al. Extracellular proteases are key mediators of Staphylococcus aureus virulence via the global modulation of virulence-determinant stability. Microbiologyopen. 2013;2:18–34. doi: 10.1002/mbo3.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kourbatova EV, Halvosa JS, King MD, Ray SM, White N, Blumberg HM. Emergence of community-associated methicillin-resistant Staphylococcus aureus USA300 clone as a cause of health care-associated infections among patients with prosthetic joint infections. Am J Infect Control. 2005;33:385–391. doi: 10.1016/j.ajic.2005.06.006. [DOI] [PubMed] [Google Scholar]
- Laarman AJ, et al. Staphylococcus aureus Staphopain A inhibits CXCR2-dependent neutrophil activation and chemotaxis. EMBO J. 2012;31:3607–3619. doi: 10.1038/emboj.2012.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauderdale KJ, Boles BR, Cheung AL, Horswill AR. Interconnections between Sigma B, agr, and proteolytic activity in Staphylococcus aureus biofilm maturation. Infect Immun. 2009;77:1623–1635. doi: 10.1128/IAI.01036-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauderdale KJ, Malone CL, Boles BR, Morcuende J, Horswill AR. Biofilm dispersal of community-associated methicillin-resistant Staphylococcus aureus on orthopedic implant material. J Orthop Res. 2010;28:55–61. doi: 10.1002/jor.20943. [DOI] [PubMed] [Google Scholar]
- Li D, Cheung A. Repression of hla by rot is dependent on sae in Staphylococcus aureus. Infect Immun. 2008;76:1068–1075. doi: 10.1128/IAI.01069-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maira-Litran T, Kropec A, Abeygunawardana C, Joyce J, Mark G, 3rd, Goldmann DA, Pier GB. Immunochemical properties of the staphylococcal poly-N-acetylglucosamine surface polysaccharide. Infect Immun. 2002;70:4433–4440. doi: 10.1128/IAI.70.8.4433-4440.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marti M, Trotonda MP, Tormo-Mas MA, Vergara-Irigaray M, Cheung AL, Lasa I, Penades JR. Extracellular proteases inhibit protein-dependent biofilm formation in Staphylococcus aureus. Microbes Infect. 2010;12:55–64. doi: 10.1016/j.micinf.2009.10.005. [DOI] [PubMed] [Google Scholar]
- McNamara PJ, Bayer AS. A rot mutation restores parental virulence to an agr-null Staphylococcus aureus strain in a rabbit model of endocarditis. Infect Immun. 2005;73:3806–3809. doi: 10.1128/IAI.73.6.3806-3809.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNamara PJ, Milligan-Monroe KC, Khalili S, Proctor RA. Identification, cloning, and initial characterization of rot, a locus encoding a regulator of virulence factor expression in Staphylococcus aureus. J Bacteriol. 2000;182:3197–3203. doi: 10.1128/jb.182.11.3197-3203.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mootz JM, Malone CL, Shaw LN, Horswill AR. Staphopains modulate Staphylococcus aureus biofilm integrity. Infect Immun. 2013;81:3227–3238. doi: 10.1128/IAI.00377-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison JM, Anderson KL, Beenken KE, Smeltzer MS, Dunman PM. The staphylococcal accessory regulator, SarA, is an RNA-binding protein that modulates the mRNA turnover properties of late-exponential and stationary phase Staphylococcus aureus cells. Front Cell Infect Microbiol. 2012a;2:26. doi: 10.3389/fcimb.2012.00026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison JM, et al. Characterization of SSR42, a novel virulence factor regulatory RNA that contributes to the pathogenesis of a Staphylococcus aureus USA300 representative. J Bacteriol. 2012b;194:2924–2938. doi: 10.1128/JB.06708-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair D, et al. Whole-genome sequencing of Staphylococcus aureus strain RN4220, a key laboratory strain used in virulence research, identifies mutations that affect not only virulence factors but also the fitness of the strain. J Bacteriol. 2011;193:2332–2335. doi: 10.1128/JB.00027-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick RP. Genetic systems in staphylococci. Methods Enzymol. 1991;204:587–636. doi: 10.1016/0076-6879(91)04029-n. [DOI] [PubMed] [Google Scholar]
- O’Gara JP. ica and beyond: biofilm mechanisms and regulation in Staphylococcus epidermidis and Staphylococcus aureus. FEMS Microbiol Lett. 2007;270:179–188. doi: 10.1111/j.1574-6968.2007.00688.x. [DOI] [PubMed] [Google Scholar]
- O’Neill E, et al. A novel Staphylococcus aureus biofilm phenotype mediated by the fibronectin-binding proteins, FnBPA and FnBPB. J Bacteriol. 2008;190:3835–3850. doi: 10.1128/JB.00167-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oscarsson J, Tegmark-Wisell K, Arvidson S. Coordinated and differential control of aureolysin (aur) and serine protease (sspA) transcription in Staphylococcus aureus by sarA, rot and agr (RNAIII) I J Med Micro. 2006;296:365–380. doi: 10.1016/j.ijmm.2006.02.019. [DOI] [PubMed] [Google Scholar]
- Patel R. Biofilms and antimicrobial resistance. Clin Ortho Rel Res. 2005;437:41–47. doi: 10.1097/01.blo.0000175714.68624.74. [DOI] [PubMed] [Google Scholar]
- Regassa LB, Betley MJ. Alkaline pH decreases expression of the accessory gene regulator (agr) in Staphylococcus aureus. J Bacteriol. 1992a;174:5095–5100. doi: 10.1128/jb.174.15.5095-5100.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regassa LB, Novick RP, Betley MJ. Glucose and nonmaintained pH decrease expression of the accessory gene regulator (agr) in Staphylococcus aureus. Infect Immun. 1992b;60:3381–3388. doi: 10.1128/iai.60.8.3381-3388.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Said-Salim B, et al. Global regulation of Staphylococcus aureus genes by Rot. J Bacteriol. 2003;185:610–619. doi: 10.1128/JB.185.2.610-619.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenk S, Laddaga RA. Improved method for electroporation of Staphylococcus aureus. FEMS Microbiol Lett. 1992;73:133–138. doi: 10.1016/0378-1097(92)90596-g. [DOI] [PubMed] [Google Scholar]
- Seybold U, Talati NJ, Kizilbash Q, Shah M, Blumberg HM, Franco-Paredes C. Hematogenous osteomyelitis mimicking osteosarcoma due to Community Associated Methicillin-Resistant Staphylococcus aureus. Infection. 2007;35:190–193. doi: 10.1007/s15010-007-6058-3. [DOI] [PubMed] [Google Scholar]
- Subrt N, Mesak LR, Davies J. Modulation of virulence gene expression by cell wall active antibiotics in Staphylococcus aureus. J Antimicrob Chemother. 2011;66:979–984. doi: 10.1093/jac/dkr043. [DOI] [PubMed] [Google Scholar]
- Thoendel M, Kavanaugh JS, Flack CE, Horswill AR. Peptide signaling in the staphylococci. Chem Rev. 2011;111:117–151. doi: 10.1021/cr100370n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurlow LR, et al. Staphylococcus aureus biofilms prevent macrophage phagocytosis and attenuate inflammation in vivo. Journal Immunology. 2011;186:6585–6596. doi: 10.4049/jimmunol.1002794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang LH, Cassat JE, Shaw LN, Beenken KE, Smeltzer MS. Factors contributing to the biofilm-deficient phenotype of Staphylococcus aureus sarA mutants. PLoS ONE. 2008;3:e3361. doi: 10.1371/journal.pone.0003361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valle J, Toledo-Arana A, Berasain C, Ghigo JM, Amorena B, Penades JR, Lasa I. SarA and not sigmaB is essential for biofilm development by Staphylococcus aureus. Mol Microbiol. 2003;48:1075–1087. doi: 10.1046/j.1365-2958.2003.03493.x. [DOI] [PubMed] [Google Scholar]
- Walker JN, Horswill AR. A coverslip-based technique for evaluating Staphylococcus aureus biofilm formation on human plasma. Front Cell Infect Microbiol. 2012;2:39. doi: 10.3389/fcimb.2012.00039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wormann ME, Reichmann NT, Malone CL, Horswill AR, Grundling A. Proteolytic cleavage inactivates the Staphylococcus aureus lipoteichoic acid synthase. J Bacteriol. 2011;193:5279–5291. doi: 10.1128/JB.00369-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zielinska AK, et al. Defining the strain-dependent impact of the Staphylococcal accessory regulator (sarA) on the alpha-toxin phenotype of Staphylococcus aureus. J Bacteriol. 2011;193:2948–2958. doi: 10.1128/JB.01517-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zielinska AK, et al. sarA-mediated repression of protease production plays a key role in the pathogenesis of Staphylococcus aureus USA300 isolates. Mol Microbiol. 2012;86:1183–1196. doi: 10.1111/mmi.12048. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.