Abstract
CD5 is a pan-T-cell surface marker and is rarely expressed in diffuse large B-cell lymphoma (DLBCL). Large-scale studies of de novo CD5+ DLBCL are lacking in Western countries. In this study by the DLBCL Rituximab-CHOP Consortium, CD5 was expressed in 5.5% of 879 DLBCL patients from Western countries. CD5+ DLBCL was associated with higher frequencies of >1 ECOG performance status, bone marrow involvement, central nervous system relapse, activated B-cell–like subtype, Bcl-2 overexpression, and STAT3 and NF-κB activation, whereas rarely expressed single-stranded DNA-binding protein 2 (SSBP2), CD30 or had MYC mutations. With standard R-CHOP chemotherapy, CD5+ DLBCL patients had significantly worse overall survival (median, 25.3 months vs. not reached, P< .0001) and progression-free survival (median, 21.3 vs. 85.8 months, P< .0001) than CD5− DLBCL patients, which was independent of Bcl-2, STAT3, NF-κB and the International Prognostic Index. Interestingly, SSBP2 expression abolished the prognostic significance of CD5 expression, suggesting a tumor-suppressor role of SSBP2 for CD5 signaling. Gene-expression profiling demonstrated that B-cell receptor signaling dysfunction and microenvironment alterations are the important mechanisms underlying the clinical impact of CD5 expression. This study shows the distinctive clinical and biological features of CD5+ DLBCL patients in Western countries and underscores important pathways with therapeutic implications.
Keywords: ABC, BCL2, CD5, diffuse large B-cell lymphoma, NF-κB
INTRODUCTION
CD5 is a cell surface glycoprotein typically expressed on normal and neoplastic T- cells, as well as on a subset of normal naïve B-cells and lymphoma cells, mainly in chronic lymphocytic leukemia/small lymphocytic lymphoma (CLL/SLL) and mantle cell lymphoma [1-4]. CD5 has an immunoreceptor tyrosine inhibitory motif and functionally inhibits the T-cell response [5, 6] and B-cell receptor (BCR) signaling-mediated apoptosis, probably by recruiting the SH2 domain-containing protein tyrosine phosphatase-1 (SHP-1) after being phosphorylated by Lyn [7, 8]. In lymphocytes, CD5 also inhibits signaling downstream of the BCR pathway, including the calcium response and interleukin-2 (IL2) production whereas augments BCR-mediated IL10 production, an anti-inflammatory cytokine and a survival factor for B-cells [9,10]. In CLL/SLL, CD5 governs the phosphorylation and nuclear translocation of STAT3 and nuclear factor of activated T cells 2 (NFAT2); activated STAT3/NFAT2 in turn leads to excess production of IL10 [11, 12].
CD5 has also been found to be expressed, albeit rarely, in de novo diffuse large B-cell lymphoma (DLBCL). To date, large-scale studies of de novo CD5+ DLBCL have been conducted only in Japan, with a reported frequency of 5 to 22% of all DLBCL [13-20]. Compared with patients with CD5− DLBCL, CD5+ DLBCL patients are reportedly more often elderly, female, and have >1 ECOG performance status, elevated serum lactate dehydrogenase (LDH) level, advanced stage disease, >1 extranodal sites, B-symptoms, and high International Prognostic Index (IPI) at diagnosis [13, 14, 19]. Pathologically, CD5+ DLBCL are associated with centroblastic morphology (rarely immunoblastic), Bcl-2 overexpression, and non-germinal center B-cell (non-GCB) subtype [16,19].
Most studies from Japan have shown that clinical outcomes of CD5+ DLBCL patients treated with standard CHOP (cyclophosphamide, doxorubicin, vincristine, and prednisone) chemotherapy with or without rituximab are poor, although the prognostic significance of CD5 positivity may depend on associated aggressive clinical parameters [13-15, 17, 19]. Bone marrow (BM) involvement (28%) and central nervous system (CNS) relapse (12.7%) are increased in CD5+ DLBCL patients [14, 16, 20]. However, in one study CD5 expression status did not correlate with prognosis by univariate or multivariate analysis, either in all patients or in rituximab-treated patients [18]. The effect of adding rituximab to CHOP on survival of CD5+ DLBCL patients also has been inconsistent in different studies [18-20]. In Western countries, a few cases of de novo CD5+ DLBCL have been reported [21, 22], and a morphologic and immunophenotypic study of 13 cases of de novo CD5+ DLBCL showed heterogeneous features [23]. No large-scale study of CD5+ DLBCL in Western countries has been performed with attention focused on the clinicopathological features and clinical response to R-CHOP.
Biological study of CD5+ DLBCL can enhance understanding of the pathogenesis. Recent gene expression profiling (GEP) analysis by two groups yielded contradictory results. In one study comparison of 11 de novo CD5+ DLBCL and 9 CD5− DLBCL cases showed upregulation of integrin-β 1 and/or CD36 adhesion molecules, which were confirmed by immunohistochemistry (IHC) to be expressed in tumor cells and vascular endothelia, respectively [24]. In another study that compared 22 CD5+ and 26 CD5− DLBCL cases, CD5 positivity was associated with downregulation of extracellular matrix (ECM)-related genes [25].
The purpose of this study is to assess the frequency, clinicopathologic and biological features of de novo CD5+ DLBCL and to evaluate the prognostic significance of CD5 expression in DLBCL treated with rituximab-CHOP (R-CHOP) in Western countries.
RESULTS
Frequency of CD5 expression in DLBCL and associated clinicopathologic features
Figures 1A-B shows representative positive and negative CD5 IHC staining in DLBCL. We observed that thirty (5.6%) DLBCLs in the training set, and eighteen (5.3%) DLBCLs in the validation set were CD5 positive DLBCL. Expression of CD5 was noted on most tumor cells of CD5+ DLBCL; 67% of CD5+ tumors had >80% of the tumor cells positive for CD5. Most (76.7%) CD5+ DLBCL patients were of activated B-cell–like (ABC) subtype (Figure 1C). CD5+ patients DLBCL had significantly higher CD5 mRNA levels compared to CD5− DLBCL patients (P = .0019, Supplemental Figures 1A-B).
Figure 1. Expression and prognostic significance of CD5 in de novo DLBCL.
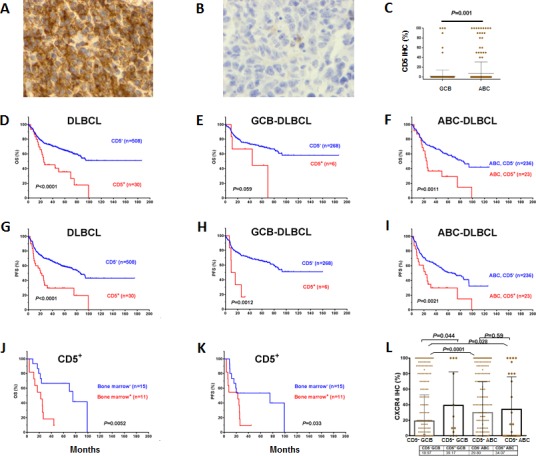
(A-B) Representative of CD5+ and CD5− immunohistochemical staining; (C) Distribution and comparison of CD5 expression between GCB- and ABC-DLBCL; (D-I) CD5 expression was correlated with significantly poor OS and PFS in the overall, GCB-, and ABC- DLBCL cohorts; (J-K) Bone marrow involvement was correlated with significantly poorer OS and PFS in patients with CD5+ DLBCL; (L) Expression of CXCR4 in CD5+ and CD5− GCB- or ABC-DLBCL patients.
Comparison of the clinical characteristics of CD5+ vs. CD5− DLBCL patients in the training set showed that CD5+ DLBCL patients were more frequently elderly (>60 years), and had B-symptoms, high performance status, an IPI score >2, and BM involvement (Table 1). None of the CD5+ patients, compared to eight (1.6%) CD5− DLBCL patients, showed CNS involvement at diagnosis. However, four (8.3%) CD5+ DLBCL patients had CNS relapse during follow-ups.
Table 1: Clinical features of patients between CD5+ and CD5− de novo DLBCL, and patients between CD5+ and CD5− ABC-DLBCL in the training set.
| Variables | CD5+ | CD5− | CD5+ vs. CD5− | CD5+ ABC | CD5− ABC | CD5+ ABC vs. CD5− ABC |
|---|---|---|---|---|---|---|
| N(%) | N(%) | N(%) | N(%) | |||
| Age, years | ||||||
| < 60 | 7(23.3) | 225(44.3) | .024 | 5(21.7) | 83(35.2) | .19 |
| ≥ 60 | 23(76.7) | 283(55.7) | 18(78.3) | 153(64.8) | ||
| Sex | ||||||
| F | 14(45.2) | 210(41.3) | .67 | 10(43.5) | 96(40.7) | .79 |
| M | 17(54.8) | 298(58.7) | 13(56.5) | 140(59.3) | ||
| Stage | ||||||
| I - II | 9(32.1) | 235(47.8) | .11 | 7(31.8) | 91(39.6) | .48 |
| III - IV | 19(67.9) | 257(52.2) | 15(68.2) | 139(60.4) | ||
| B-symptoms | ||||||
| No | 13(46.4) | 319(65.9) | .036 | 10(45.5) | 137(60.6) | .17 |
| Yes | 15(53.6) | 165(34.1) | 12(54.5) | 89(39.4) | ||
| LDH level | ||||||
| Normal | 9(32.1) | 182(38.8) | .48 | 7(30.4) | 79(36.2) | .58 |
| Elevated | 19(67.9) | 287(61.2) | 16(69.6) | 139(63.8) | ||
| No. of extranodal sites | ||||||
| 0 - 1 | 22(76.7) | 369(77.2) | .75 | 17(78.3) | 169(74.2) | .99 |
| ≥ 2 | 8(23.3) | 117(22.8) | 6(21.7) | 60(25.8) | ||
| ECOG performance status | ||||||
| 0 - 1 | 17(60.7) | 387(85.1) | .0007 | 14(60.9) | 179(82.9) | .01 |
| ≥ 2 | 11(39.3) | 68(14.9) | 9(39.1) | 37(17.1) | ||
| Size of largest tumor | ||||||
| < 5cm | 9(45.0) | 224(59.4) | .2 | 7(43.8) | 104(58.1) | .27 |
| ≥ 5cm | 11(55.0) | 153(40.6) | 9(56.3) | 75(41.9) | ||
| IPI score | ||||||
| 0 - 2 | 13(48.3) | 310(63.4) | .05 | 10(43.5) | 127(55.2) | .28 |
| 3 - 5 | 16(51.7) | 182(36.6) | 13(56.5) | 103(44.8) | ||
| Therapy response | ||||||
| CR | 20(66.7) | 396(78) | .15 | 16(69.6) | 192(78) | .35 |
| PR | 6 | 64 | 5 | 34 | ||
| SD | 2 | 20 | 0 | 9 | ||
| PD | 2 | 28 | 2 | 11 | ||
| BM involvement | ||||||
| Yes | 11(42.3) | 39(8.9) | <.0001 | 9(47.4) | 20(9.7) | .0001 |
| No | 15(57.7) | 399(91.1) | 10(52.6) | 187(90.3) | ||
| COO | ||||||
| GCB | 7(23.3) | 268(53.2) | .002 | |||
| ABC | 23(76.7) | 236(46.8) | ||||
Note: LDH, serum lactate dehydrogenase; IPI, International Prognostic Index; BM, bone marrow; COO, cell-or-origin; GCB, germinal center B-like; ABC, activated B-cell like.
Pathological features of CD5+ vs. CD5− DLBCL patients were characterized by comparing their protein expression profiles (Table 2 showed the results for most but not all the biomarkers in 879 patients). CD5+ DLBCL, as compared with CD5− DLBCL, were more often positive for Bcl-2, FOXP1, pSTAT3, c-Rel and CXCR4, and less often expressed GCET, CD10, CD30, and SSBP2 (single-stranded DNA binding protein 2), or had MYC nonsilent mutations (Table 2, Supplemental Figures 1C-E). REL amplification or BCL2 translocation, which was found in only one CD5+ GCB-DLBCL DLBCL, did not account for the increased c-Rel or Bcl-2 level. CD5+ DLBCL also had no association with BCL2 amplifications, unlike one earlier study [26]. When comparison was restricted to the ABC subtype, CD5+ DLBCL were associated with significantly higher frequencies of Bcl-2+ and pSTAT3+ and lower frequencies of MYC mutations, CD30+, SSBP2+, and NF-κB1/p50+ (Table 2).
Table 2: Pathological features of patients with CD5+ and CD5− de novo DLBCL (combined training and validation sets).
| Variables | CD5+ | CD5− | CD5+ vs. CD5− | CD5+ ABC | CD5− ABC | CD5+ ABC vs. CD5− ABC |
|---|---|---|---|---|---|---|
| N(%) | N(%) | N(%) | N(%) | |||
| Bcl-2 overexpression | ||||||
| < 70% | 12(25.5) | 369(47.2) | .0039 | 5(15.2) | 145(39.2) | .0076 |
| ≥ 70% | 35(74.5) | 412(52.8) | 28(84.8) | 225(60.8) | ||
| BCL2 gene | ||||||
| Normal | 31(83.8) | 520(78.4) | .13 | 19(79.2) | 248(84.3) | 0.38 |
| Translocation | 1(2.7) | 86(13) | 0(0) | 9(3.1) | ||
| Amplification or polysomy | 5(13.5) | 57(8.6) | 5(20.8) | 37(12.6) | ||
| MYC nonsilent CDS mutations | ||||||
| Mut | 1(2.6) | 97(17.5) | .04 | 0(0) | 39(15.2) | .034 |
| WT | 38(97.4) | 458(82.5) | 27(100) | 218(84.8) | ||
| CXCR4 expression | ||||||
| < 20% | 21(52.5) | 461(66) | .089 | 15(53.6) | 194(59.1) | .56 |
| ≥ 20% | 19(47.5) | 237(34) | 13(46.4) | 134(40.9) | ||
| GCET overexpression | ||||||
| < 50% | 40(85.1) | 577(70.8) | .044 | 31(93.9) | 339(87.8) | .40 |
| ≥ 50% | 7(14.9) | 238(29.2) | 2(6.1) | 47(12.2) | ||
| FOXP1 overexpression | ||||||
| < 60% | 13(27.7) | 352(43.2) | .047 | 4(12.1) | 78(20.1) | .36 |
| ≥ 60% | 34(72.3) | 463(56.8) | 29(87.9) | 310(79.9) | ||
| CD10 overexpression | ||||||
| < 40% | 34(59.6) | 518(63) | .67 | 28(84.8) | 375(95.9) | .017 |
| ≥ 40% | 23(40.4) | 304(37) | 5(15.2) | 16(4.1) | ||
| CD30 | ||||||
| Negative | 44(95.7) | 674(81.8) | .015 | 33(100) | 311(79.9) | .0016 |
| Positive | 2(4.3) | 150(18.2) | 0(0) | 78(20.1) | ||
| Blimp-1 expression | ||||||
| < 10% | 32(68.1) | 492(66.8) | 1.0 | 21(63.6) | 205(59.8) | .71 |
| ≥ 10% | 15(31.9) | 244(33.2) | 12(36.4) | 138(40.2) | ||
| pStat3 overexpression | ||||||
| < 50% | 25(61) | 611(85.3) | .0002 | 16(55.2) | 279(82.1) | .0014 |
| ≥ 50% | 16(39) | 105(14.7) | 13(44.8) | 61(17.9) | ||
| Cyclin D3 | ||||||
| Negative | 38(90.5) | 526(77.5) | .05 | 28(93.3) | 248(76.8) | .037 |
| Positive | 4(9.5) | 153(22.5) | 2(6.7) | 75(23.2) | ||
| p50 nuclear expression | ||||||
| Negative | 23(53.5) | 337(45.4) | .35 | 17(54.8) | 129(36.6) | .05 |
| Positive | 20(46.5) | 405(54.6) | 14(45.2) | 223(63.4) | ||
| p52 nuclear expression | ||||||
| Negative | 36(80) | 537(76) | .59 | 27(84.4) | 254(76.3) | .38 |
| Positive | 9(20) | 170(24) | 5(15.6) | 79(23.7) | ||
| p65 nuclear expression | ||||||
| Negative | 16(36.4) | 376(49.2) | .12 | 11(35.5) | 194(52.9) | .09 |
| Positive | 28(63.6) | 389(50.8) | 20(64.5) | 173(47.1) | ||
| RelB nuclear expression | ||||||
| Negative | 25(96.2) | 378(85.3) | .15 | 19(95) | 172(83.5) | .33 |
| Positive | 1(3.8) | 65(14.7) | 1(5) | 34(16.5) | ||
| c-Rel nuclear expression | ||||||
| Negative | 23(56.1) | 521(72.3) | .033 | 18(62.1) | 255(74.6) | .19 |
| Positive | 18(43.9) | 200(27.7) | 11(37.9) | 87(25.4) | ||
| SSBP2 expression | ||||||
| ≤ 5% | 36(76.6) | 283(37.7) | < .0001 | 27(81.8) | 182(52.6) | .0015 |
| > 5% | 11(23.4) | 468(62.3) | 6(18.4) | 164(47.4) | ||
Note: LDH, serum lactate dehydrogenase; IPI, International Prognostic Index; BM, bone marrow; COO, cell-of-origin; GCB, germinal center B-like; ABC, activated B-cell like. Due to tissue exhaustion, immunohistochemical staining analysis for some markers was not successful in few cases.
CD5 expression is associated with significantly poorer survival in DLBCL
CD5+ DLBCL patients had significantly poorer overall survival (OS) (median OS: 25.3 months vs. not reached, hazard ratio [HR]: 3.87, 95% confidence interval [CI] of rate: 1.99-7.51, P < .0001) and progression-free survival (PFS) (median PFS: 21.3 vs. 85.8 months, HR: 4.31, 95% CI: 2.26-8.23, P < .0001) in the training set, regardless of cell-of-origin (COO) (Figures 1D-I). The 5-year OS rates for patients with CD5+ vs. CD5− DLBCL were 35.5% vs. 64.8%, and the 5-year PFS rates for patients with CD5+ vs. CD5− DLBCL were 29.6% vs. 59%, respectively. Between CD5+ patients with GCB- and ABC-DLBCL there was no significant difference in OS or PFS (P = .76 for OS, and P = .51 for PFS).
BM involvement significantly impacted OS (P = .0052) and PFS (P = .033) of CD5+ DLBCL patients (Figures 1J-K), and CD5 expression appeared to impact nodal DLBCL more than extranodal DLBCL (Supplemental Figures 1F-I) in the training set. CXCR4, a chemokine receptor involved in tumor cell homing to bone marrow and lymph node [27-29] was expressed at higher levels in CD5+ compared with CD5− DLBCL (P= .05, Supplemental Figure 1E). However, only in GCB-DLBCL was expression of CXCR4 significantly upregulated (P = .044). Expression levels of CXCR4 in CD5+ were similar to that in CD5−ABC-DLBCL (P = .59), although significantly higher than in CD5− patients with GCB-DLBCL (P = .028, Figure 1L).
Effect on Bcl-2 expression and prognostic independence of CD5 expression
Almost 75% of CD5+ DLBCL patients had concurrent overexpression (≥70% of the tumor cells) of antiapoptotic Bcl-2, an unfavorable biomarker [30, 31]. This frequency was significantly higher than that in CD5− DLBCL patients (53%, P = .0039, Table 2). Moreover, the correlation of CD5+ and Bcl-2 overexpression was observed at both the mRNA and protein levels and remained significant in the comparison of CD5+ with CD5− DLBCL within the ABC subtype, which commonly overexpressed Bcl-2 (Figures 2A-D).
Figure 2. Correlation between CD5 and Bcl-2 overexpression and the prognostic significance of CD5 expression in DLBCL independent of Bcl-2 overexpression.
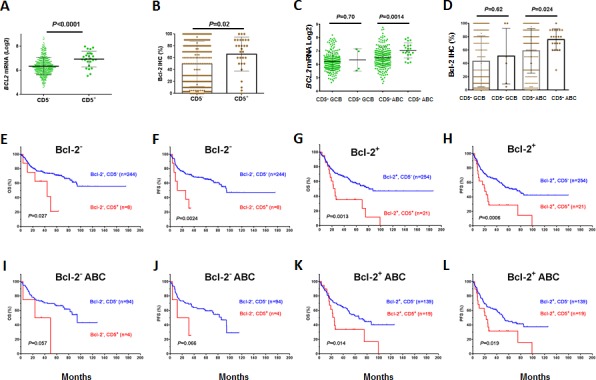
(A-D) CD5 expression was correlated with Bcl-2 overexpression and upregulated mRNA levels, in both GCB and ABC subtypes; (E-H) The prognostic significance of CD5+ was independent of Bcl-2 overexpression in DLBCL; (I-L) The prognostic significance of CD5+ was independent of Bcl-2 overexpression in ABC-DLBCL.
However, CD5 expression predicted unfavorable clinical outcomes independent of Bcl-2+ status in the overall or ABC-DLBCL (Figures 2E-L), and vice versa.
Effect on NF-κB and STAT3 activation and prognostic independence of CD5 expression
CD5 expression was associated with nuclear expression of the NF-κB subunits c-Rel and p65, but not mRNA levels of REL or RELA (Figures 3A-D, Table 2, Supplemental Figures 1J-K). However, nuclear expression of p50, which is also indicative of canonical NF-κB pathway activation [32], was significantly decreased in CD5+ ABC-DLBCL (Figure 3E, Table 2), which was not due to the NFKB1 downregulation at the mRNA level (Supplemental Figure 1L). There was no correlation between CD5+ and nuclear expression of p52 or RelB, two subunits involved in noncanonical/alternative NF-κB pathway activation [32] (although showing trends of downregulation in CD5+ DLBCL, Figures 3F-G).
Figure 3. Association between CD5 and NF-κB/STAT3 activation in DLBCL and the independent prognostic significance of CD5 expression.
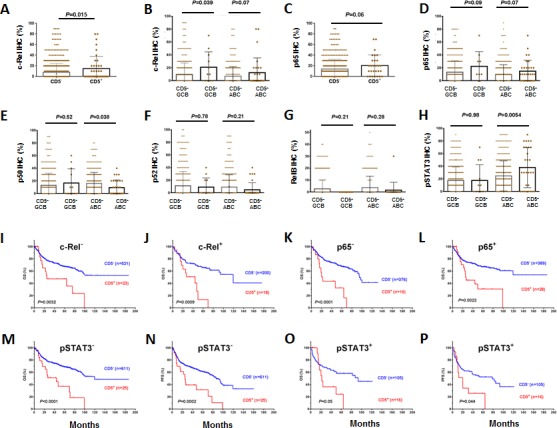
(A-D) Association of CD5 expression with nuclear expression of c-Rel and p65; (E) CD5 expression was associated with decreased nuclear expression of p50 in ABC-DLBCL; (F-G) Comparison of nuclear expression of p52 and RelB between CD5+ and CD5− patients; (H) CD5 expression was associated with STAT3 activation in ABC-DLBCL; (I-P) The prognostic significance of CD5+ was independent of c-Rel, p65 or STAT3 activation.
CD5+ was also associated significantly with nuclear expression of phosphorylated/activated STAT3 but not upreglulated STAT3 mRNA in the ABC-DLBCL subtype (Figure 3H, Supplemental Figure 1O). The nuclear expression of pSTAT3 has been associated with poorer survival [33].
However, the adverse effect of CD5 expression on prognosis did not depend on c-Rel, p65 or STAT3 activation. In both c-Rel− and c-Rel+, p65− and p65+, and pSTAT3− and pSTAT3+ patients, CD5 expression correlated with significantly poorer OS and PFS (Figures 3I-P, showing analysis in the combined training and validation sets due to the limited CD5+ cases).
Frequent loss of SSBP2 expression in CD5+ DLBCL
CD5 positivity in DLBCL was frequently associated with lack of SSBP2 expression, a tumor suppressor protein for lymphoma and leukemia (Figures 4A-D, Table 2) [34-36]. Furthermore, the prognostic significance of CD5 expression was restricted to DLBCLs without or with very low SSBP2 expression. In SSBP2+ patients, CD5 expression in DLBCL was not associated with prognosis (Figures 4E-H).
Figure 4. Correlation between CD5 and SSBP2 expression in DLBCL.
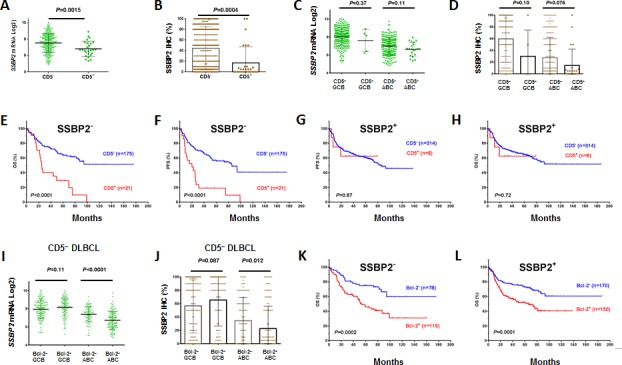
(A-D) CD5 expression was correlated with decreased SSBP2 expression; (E-F) The prognostic significance of CD5+ was present only in patients without SSBP2 expression; (G-H) CD5+ was not prognostic in patients with SSBP2 expression; (I-J) Bcl-2+ compared with Bcl-2− patients had decreased SSBP2 mRNA and protein levels only in the ABC-DLBCL subset but not in the GCB-DLBCL subset; (K-L) The prognostic significance of Bcl-2+ was independent of SSBP2 expression.
In ABC-DLBCL, Bcl-2+ vs. Bcl-2− also had decreased SSBP2 mRNA and protein expression independent of CD5 expression (Figures 4I-J, Supplemental Figures 1P-Q). However, the prognostic significance of Bcl-2 was independent of SSBP2 expression (Figures 4K-L).
Multivariate survival analysis
Multivariate analysis of clinical and pathological factors including IPI (defined by age, stage, serum LDH, performance status, and extranodal sites), sex, B-symptoms and tumor size, CD5+, Bcl-2+, and COO confirmed that CD5 expression independently predicted significantly poorer OS (P = .005) and PFS (P = .014) in DLBCL, in addition to the known unfavorable prognostic factors IPI >2 and Bcl-2+ (Table 3). COO classification did not reach statistical significance to be an independent factor in this multivariate analysis, but did remain as an independent prognostic factor after removal of CD5 as a factor in the survival analysis, suggesting that CD5 expression significantly contributed to the poor prognosis of ABC-DLBCL.
Table 3: Multivariate analysis in the overall and ABC-DLBCL cohort of patients.
| OS | PFS | |||||
|---|---|---|---|---|---|---|
| Variables | HR | 95% CI | p | HR | 95% CI | p |
| Overall DLBCL | ||||||
| IPI > 2 | 2.40 | 1.70-3.39 | <.0001 | 2.24 | 1.63-3.09 | <.0001 |
| CD5+ | 2.12 | 1.25-3.5 | .005 | 1. 93 | 1.14-3.24 | .014 |
| Bcl-2+ | 2.04 | 1.45-2.87 | <.0001 | 1.92 | 1.39-2.63 | <.0001 |
| ABC | 1.27 | .89-1.81 | .19 | 1.33 | .95-1.85 | .094 |
| Female | .94 | .73-1.36 | .73 | 1.0 | .67-1.30 | .99 |
| Tumor size>5cm | 1.33 | .97-1.83 | .078 | 1.26 | .93-1.70 | .14 |
| B-symptoms | 1.27 | .90-1.78 | .17 | 1.23 | .89-1.70 | .20 |
| ABC-DLBCL | ||||||
| IPI > 2 | 2.22 | 1.44-3.40 | <.0001 | 2.08 | 1.38-3.13 | <.0001 |
| CD5+ | 1. 96 | 1.06-3.62 | .032 | 1. 62 | 0.88-2.99 | .12 |
| Bcl-2+ | 1.92 | 1.26-2.97 | .005 | 1.93 | 1.26-3.88 | .003 |
| Female | 1.13 | .72-1.78 | .58 | 1.10 | 0.72-1.70 | .64 |
| Tumor size>5cm | 1.14 | .75-1.74 | .54 | 1.05 | 0.69-1.57 | .83 |
| B-symptoms | 1.14 | .72-1.81 | .57 | 1.28 | 0.82-1.98 | .27 |
Note: OS, overall survival; PFS, progression-free survival; HR, hazard ratio; CI, confidence interval; IPI, International Prognostic Index.
The prognostic significance of these variables (except COO) was further analyzed in the ABC-DLBCL subset. CD5 positivity remained as an independent prognostic factor predicting poorer OS (P = .032). However, the P value for PFS did not reach statistical significance (P = .12). IPI>2 and Bcl-2+ remained independent prognostic factors for both OS and PFS (Table 3).
Prognostic and biological impact of CD5 expression in the validation set
In an independent validation set from multiple medical centers, we validated the prognostic significance (Figures 5A-B) as well biological impact of CD5+ discovered in the training set. In the validation set, CD5+ DLBCL were also associated with elevated Bcl-2 and decreased SSBP2 expression (Figures 5C-D), and its prognosis was independent of Bcl-2+ (Figures 5E-F) but depended on SSBP2− (Figures 5G-H).
Figure 5. Biological and prognostic impact of CD5 expression in the validation DLBCL cohort.
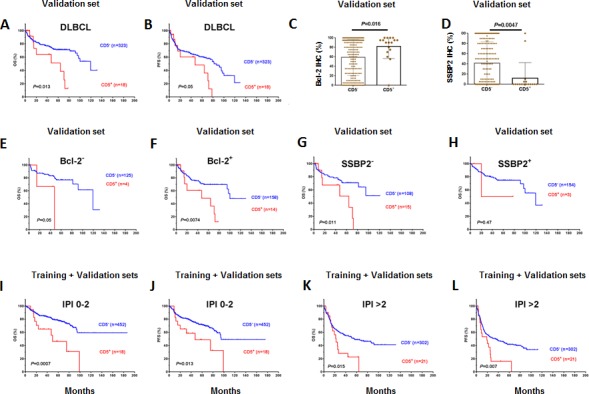
(A-B) CD5 expression was correlated with significantly poorer OS and PFS in the overall DLBCL cohort; (C-D) CD5 expression was correlated with Bcl-2 overexpression and decreased SSBP2 expression; (E-F) The prognostic significance of CD5+ was independent of Bcl-2 overexpression; (G-H) The prognostic significance of CD5+ in the validation DLBCL cohort depended on loss of SSBP2 expression; (I-L) The prognostic significance of CD5+ was independent of the IPI.
In both the training and validation sets, the prognostic significance of CD5+ was independent of the IPI. Figures 5I-L shows analysis in the combined training and validation sets.
Effect of CD5 expression on BCR/TCR signaling
CD5+ DLBCL had significantly higher levels of CARD11/CARM1 mRNA compared with CD5− DLBCL (Figure 6A). CARD11 is a scaffold protein downstream of BCR signaling that activates NF-κB via interaction with BCL10 [32, 37, 38]. A20, a negative regulator of NF-κB signaling [37], and IL10, which mediates immune inhibition and cell survival downstream the CD5 signaling [10, 11], did not show differential expression of mRNAs between CD5+ and CD5− DLBCL patients. (P= .30 and P= .36). In addition, TCL1A (which modulates AKT activation downstream of the TCR signaling [39]) and BCL11A (proto-oncogene with a critical role in lymphoid development [40]), which was often co-amplified with REL in lymphoid malignancies [41, 42], were significantly upregulated in CD5+ ABC-DLBC patients (Figures 6B-C). PTEN encoding a tumor suppressor antagonizing the PI3K/AKT signaling was significantly downregulated in CD5+ GCB-DLBCL (Figure 6D).
Figure 6. mRNA expression of BCR signaling-related genes between CD5+ and CD5− DLBCL patients, or Bcl-2+ and Bcl-2− DLBCL patients.
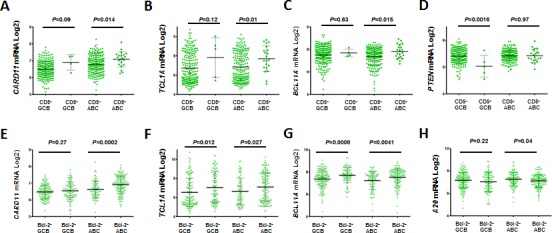
(A-B) Association of CD5 expression with upregulated CARD11 and TCL1 mRNA levels; (C) CD5 expression was associated with increased BCL11A mRNA in ABC-DLBCL; (D) CD5 expression was associated with decreased PTEN mRNA in GCB-DLBCL; (E-H) Differential expression of CARD11, TCL1, BCL11A and A20 between Bcl-2+ and Bcl-2− patients resembled the patterns between CD5+ and CD5− DLBCL patients.
Differential expression of CARD11, TCL1A, and BCL11A between Bcl-2+ and Bcl-2− DLBCL patients resembled the effect of CD5 signaling (Figures 6E-G), suggesting that there are similar signaling pathways potentially underlying the association of CD5 and Bcl-2 expression. In contrast, regulation of other genes such as A20 (Figure 6H), PTEN, CD10, PIK3CA, MYC, CXCL12, PRDM1, STAT3 showed different patterns in Bcl-2+ (vs. Bcl-2−) and CD5+ (vs. CD5−) patients.
CD5 expression signatures in de novo DLBCL
Gene expression profiles of CD5+ and CD5− DLBCL were compared to identify CD5 gene signatures in de novo DLBCL, which showed 86 differentially expressed genes (DEGs) by comparison within the overall cohort, and 39 DEGs by comparison within the ABC-DLBCL subcohort (Figures 5A-B, Table 4). Comparison within the GCB-DLBCL subcohort with limited CD5+ cases found 17 DEGs, most of which are related to cytoskeleton, microtubule or nervous system function (Figure 5C). Only a few genes were found expressed differentially between CD5+ GCB- and ABC-DLBCLs (Table 5).
Table 4: Differentially expressed genes between patients with CD5+ and CD5− DLBCL.
| Function categories | Numbers | Upregulated gene names | Numbers | Downregulated gene names |
|---|---|---|---|---|
| Antiapoptosis | 2 | BCL2, TNFAIP8 | ||
| T-cell and B-cell receptor signaling | 7 | CARD11, CLECL1, IGHM, LYN, PTPN2, SIT1, SH3BP5ⱡ | ||
| Hematopoietic development, immune response, central nervous system, signaling including NF-kB, MAPK, Wnt and calcium-dependent pathways | 10 | ARHGAP17, BSPRY, C2orf34ⱡ, CXXC5ⱡ, SIPA1L3ⱡ, OGT, MPEG1, OAS1ⱡ, TMEM149, GLYR1* | 14 | APCDD1ⱡ, CLEC11A, DZIP1, INHBA#x2C61;, PI15ⱡ, PDE4D, SGK1, SOBP, CAMK2N1*, FGF1*, FGF17*, PPM1L*, PTPRR*, SLC6A9* |
| Cell cycle progression | 3 | CCND2, CDK3, TLK1 | 1 | NEK6ⱡ |
| Transcriptional activator/repressor | 11 | CREB3L2, ETV6, FOXP1, IRF2BP2, JARID2, TCF4ⱡ, ZNF589, RBM39*, ANKRD12*, ZNF614*, ZNF814* | 7 | LMO2, LMO4, MAML3, TFEC, MYBL1, ZNF709, LHX2* |
| mRNA stability, RNA regulation | 2 | LUC7L, SERBP1 | 1 | SREK1 |
| Protein modification, processing, degradation, transporter | 3 | ERP29, FAM175Bⱡ, SLC38A2 | 2 | CTSK, UGGT2ⱡ |
| Cytoskeletal protein, extracellular matrix, cell adhesion, migration | 1 | PDLIM1 | 14 | CCBE1, COL5A1, COL6A3, ENPP3, FAM198Bⱡ, FN1, LTBP1, PCDH9, PTK2, PTPRB, RAPH1, BAIAP2L1*, ITGBL1*, MYO7A* |
| Metabolism | 1 | ST3GAL2* | ||
| IncRNA, unknown function and others | 12 | C13orf18ⱡ, CXorf21, FAM129Cⱡ, LOC100506168, LOC202781, KLHL14, KIAA0040ⱡ, LIC7L, MAPS8, RRP7A, NPIP*, NAPSB* | 11 | CRNDE,CCDC144B, C15orf48, FRMD6, GLT1D1, LOC100288271ⱡ, LOC100506457ⱡ, LOC100133790ⱡ, LOC440864, KIAA1211, SAMD12, LOC100130815* |
Note: Genes marked by
are those identified in comparison of CD5+ vs. CD5− ABC-DLBCLs; genes marked by
are those identified in both CD5+ vs. CD5− DLBCL and CD5+ vs. CD5− ABC-DLBCL. Bolded genes are involved in nervous system development or function.
Table 5: Differentially expressed genes between patients with CD5+ and CD5− GCB-DLBCL, and between patients with CD5+ GCB-DLBCL and CD5+ ABC-DLBCL (protein function is from NCBI http://www.ncbi.nlm.nih.gov and UniProtKB http://www.uniprot.org).
| CD5+ vs. CD5− GCB-DLBCL | CD5+ GCB vs. CD5+ ABC-DLBCL | |||
|---|---|---|---|---|
| Function categories | Up | Down | Up | Down |
| Ion-channel | CHRNA3, KCNK5, | TRPM6 | KCNJ2 | |
| Nervous system function | SERPINE2, SLC38A2 | DIO2, SSTR1, ADAM22 | SERPINE2 | |
| Immune response | LILRA1 | |||
| Protein folding, transportation | UGGT2 | |||
| Transcription | TRPS1 | |||
| Cytoskeleton, microtubule | WDR69 | FGD6, CCIN, MAP6 | WDR69 | COL28A1 |
| Metabolism | CYP1A1, CROT | GALT | ||
| Unknown function | KIAA0125, GLT1D1, RBFA | C4orf34 | ||
CD5 expression signatures included both activators and inhibitors of TCR/BCR signaling. Activation of BCR/TCR was suggested by upregulation of CARD11, CLECL1 (encoding a T-cell costimulatory molecule), and IGHM. However, inhibition of BCR/TCR and increased threshold for activation in CD5+ DLBCL was also suggested by upregulation of PTPN2 and SIT1 (which negatively regulate TCR signaling), LYN (which has roles in inhibiting BCR), and SH3BP5 (which inhibits BTK signaling), and downregulation of PDE4D (which hydrolyzes c-AMP, thereby removing the c-AMP constraint for TCR) in CD5+ DLBCL [43].
DEGs downstream of TCR/BCR also suggested activated BCR signaling with negative feedback in CD5+ DLBCL. Activation of the NF-κB, MAPK, and Wnt pathways was indicated by upregulation of CXXC5 (activating NF-κB and MAPK) and GLYR1 (activating MAPK) and downregulation of APCDD1 (negative regulator of Wnt signaling), PI15 (trypsin inhibitor), CAMK2N1 (CAMK2 inhibitor), PPM1L (dephosphorylating MAPKs), and PTPRR (sequester and inhibitor of MAPKs). In turn, antiapoptotic BCL2 and TNFAIP8 downstream of the NF-κB pathway were significantly upregulated. On the other hand, inhibition of the calcium-dependent signaling, which is required for the activation of proteins downstream BCR including NF-κB and NFAT [44, 45], was indicated by upregulation of BSPRY (inhibiting calcium influx) and downregulation of SGK1, which activates ion channels and calcium entry.
CD5 expression signatures also included 18 transcription factors, suggesting distinct transcription programs in CD5+ DLBCL patients. Upregulated transcription factors included CREB3L2 (a transcription activator binding to the cAMP response element), ETV6 (a transcriptional repressor), FOXP1 (an essential transcriptional repressor of B-cell development), IRF2BP2 (a transcription corepressor repressing the NFAT target genes including IL2/IL4), JARID2 (a transcriptional repressor), TCF4 (a transcription factor binding to the immunoglobulin enhancer), and ZNF589 (a transcriptional repressor). Downregulated transcription factors included LIM factors LMO2, LMO4, and LHX2 (which have roles in proliferation, differentiation and hematopoietic development), MAML3 (a transcriptional coactivator for NOTCH proteins), TFEC (which binds to the immunoglobulin heavy-chain/IGH gene enhancer), and MYBL1 (a strong transcriptional activator). Altogether it appears that transcription factors that repress TCR/BCR signaling and proliferation outnumbered those that enhance TCR/BCR signaling.
Cell cycle and proliferation genes also appeared under concurrent positive and negative regulations in CD5+ DLBCL. Three genes promoting cell cycle progression were upregulated, including CCND2 (a cyclin important for G1/S transition), CDK3 (a cyclin-dependent protein kinase involved in G0-G1 and G1-S transitions), and TLK1 (a kinase involved in the regulation of chromatin assembly), whereas NEK6 (playing an important role in mitotic cell cycle progression) was downregulated. JARID2 (which negatively regulates cell proliferation signaling) was upregulated, and CLEC11A, PTK2, MYBL1, and SGK2 genes (which stimulate proliferation) were downregulated.
Supporting a previous study [25], another distinctive feature of the CD5 expression signatures were the downregulation of genes related to cell adhesion, ECM remodeling, and migration (CCBE1, COL5A1, COL6A3, ENPP3, FAM198B, FN1, ITGBL1, PCDH9, PTK2, RAPH1, and MYO7A). LTBP1 (functioning in the assembly, secretion, and targeting of TGFβ1 to ECM), INHBA (encoding a TGFβ family member), and PTPRB (involved in blood vessel remodeling and angiogenesis) were downregulated in CD5+ DLBCL.
Gene set enrichment analysis (GSEA) was performed to enrich the relevant pathways. Downregulated “ECM Receptor Interaction and upregulated “Nitrogen Metabolism” had nominal P-values < 1% (.006 and .0039 respectively) although no gene sets were enriched by an FDR threshold of 25% (probably due to the small number of CD5+ cases, and the highly heterogeneous nature of CD5− DLBCL patients). In addition, “Focal Adhesion” had a nominal P-value of .026 with an FDR of 30%. These results reinforced the notion that downregulation of genes involved in ECM and cell adhesion is a prominent feature of the CD5 expression signature (Figures 5D-F). Pathway analysis by the Ingenuity Pathway Analysis (IPA) software showed CD5 expression signatures were associated with functional networks of Hematopoiesis, Nervous System Development and Function, Cellular Growth and Proliferation (Supplemental Figure 2).
DISCUSSION
De novo CD5+ DLBCL is a unique subset of DLBCL [1] and has not been studied on a large scale in Western countries. The current study of 879 patients with de novo DLBCL identified 48 (5.5%) CD5+ patients, associated with higher frequencies of >1 ECOG performance status, BM involvement, CNS relapse, ABC subtype, Bcl-2+, and STAT3 activation whereas with lower frequencies of CD30+, SSBP2+, and MYC mutations. CD5 signaling appears to differentially regulate NF-κB subunits, activating c-Rel and p65 but decreasing p50 activation. Other features associated with CD5+ DLBCL in this study, such as elderly age, B-symptoms, IPI, GCET, CD10, FOXP1 and CXCR4, were probably due to the predominance of ABC subtype of CD5+ DLBCL patients. Compared with previous studies conducted in Japan, CD5+ DLBCL in Western countries had lower prevalence, shared common features of performance status, ABC subtype, Bcl-2 overexpression, BM involvement, and development of CNS recurrence, but lacked features of female predominance, extranodal involvement, elevated serum LDH, and higher disease stage [13-15, 17-19]. These differences may reflect ethnic and genetic variation, the heterogeneity of CD5+ DLBCL, the larger number of CD5+ DLBCLs (n = 109) in the Japanese cohort and cohort-specific features.
We further assessed the prognostic impact of CD5 expression and found that CD5+ independently correlated with poorer survival in DLBCL with R-CHOP treatment. Moreover, the 48 CD5+ DLBCL patients treated by R-CHOP in this study cohort did not show significant improvement in OS (P= .66) or PFS (P= .81) compared to the 14 CD5+ DLBCL patients treated with CHOP from an independent CHOP-treated DLBCL cohort (results not shown). Furthermore, our attempt to understand the biology of CD5+ DLBCL suggested that molecular pathways downstream BCR signaling which promote cell proliferation and survival (such as Bcl-2 [but not Myc] overexpression, and activation of c-Rel, p65, and STAT3) were likely relevant for the pathogenesis of CD5+ DLBCL; however, the adverse impact of CD5 expression did not depend on any of these factors alone.
BM involvement also appeared to impact prognosis of CD5+ DLBCL significantly in the training set (Figures 1J-K, which however was not confirmed in the validation set), and development of CNS relapse (0% at diagnosis, 8.3% after treatment) was remarkable for CD5+ DLBCL. A role of CXCR4/CXCL12 axis in BM involvement and CNS relapse of CD5+ DLBCL was suggested by the higher CXCR4 expression in the studied CD5+ DLBCL patients (Supplemental Figure 1E) [28, 46]. However, restricting within ABC-DLBCL, CD5+ compared to CD5− patients had similar levels of CXCR4 but had a higher incidence of BM involvement (34.5% vs. 7.2%, P < .0001, Table 2), suggesting that CXCR4/CXCL12 axis was not sufficient to explain for the BM involvement. A previous study in CLL/SLL also suggested that other factors in addition to the CXCR4/CXCL12 axis may account for marrow infiltration of neoplastic cells [46]. In this study, downregulation of genes involved in ECM and cell adhesion, which was a prominent feature of the CD5+ signature revealed by GEP analysis, likely contributed to the BM involvement and development of CNS relapse in CD5+ DLBCL.
Interestingly, CD30 and SSBP2 (a tumor-suppressor [34, 36, 47]) expression was frequently negative in CD5+ DLBCL patients. SSBP2 has a critical regulatory role in the transcriptional program of hematopoietic stem and progenitor cells in vivo, via modulating the abundance and function of multiple transcription cofactors including LIM domain-binding protein 1 (LDB1), LMO, and LHX. In mouse models, loss of SSBP2 resulted in hypoplastic hematopoietic tissues and impaired hematopoiesis and was associated with shortened lifespan and greater susceptibility to B-cell lymphomas [34, 35, 48]. Prognostic significance of CD5 expression in SSBP2+ and SSBP2− DLBCL patients also suggests a tumor-suppressor function of SSBP2 for CD5 signaling.
GEP analysis suggested both positive and negative regulation of TCR/BCR in CD5+ DLBCL patients, and differential regulation of BCR downstream pathways (activation of NF-κB, MAPK, and Wnt pathways and inhibition of calcium influx). This may suggest the activated but reprogrammed BCR signaling in CD5+ DLBCL patients, and the role of CD5 expression in mitigating BCR signaling, and promoting tumor cell survival by previous studies [7-10]. Likewise, both positive and negative regulations of proliferation, growth, and cell cycle were suggested by CD5 expression signatures. Therefore it appears that CD5 signaling contributes to survival yet an anergic-like state of B-cells.
It is also possible that the unique characteristics of the CD5 expression signature in antiapoptosis, proliferation, signaling, and transcription reflects the COO and the differentiation stages of the lymphoma cells. For example, downregulated LMO2, LMO4, LHX2, and CLEC11A as well as upregulated FOXP1, ZNF589, LYN, and BCL2 are hematopoietic mediators and have distinct expression patterns during B-cell development. In addition, IGHM expressed in naïve B-cells and plasma cells was upregulated in CD5+ patients, whereas MYC mutations, which may arise from an aberrant hypermutation process in DLBCL [49], were almost absent in CD5+ DLBCL patients (only one CD5+ GCB-DLBCL case had MYC mutations). Previous studies indicated that CD5+ DLBCL and CD5+ CLL had higher frequencies of germline vs. somatically hypermutated IGHV genes compared with CD5− DLBCL [50], suggesting the COO of CD5+ DLBCL might be distinct from that of CD5− DLBCL and yet similar to that of CD5+ CLL [51]. Our collective results of GEP, MYC mutations, and Blimp-1 expression (indicating commitment to plasma cell differentiation) [52-54] in CD5+ and CD5− DLBCL (Tables 2, 4) suggest that subsets of CD5+ DLBCL may originate from pre-GC, memory B-cells, or neoplasms differentiated into plasma cells yet never through GC reaction [60, 61].
In summary, in this study we show that de novo CD5+ DLBCL, which occurs at a low frequency (5.5%) in Western countries, was associated with unfavorable clinicopathologic variables and with inferior survival following R-CHOP treatment. Although heterogeneity still exists in this disease subset, dysregulated BCR signaling is significantly implicated in lymphoma cell survival and disease dissemination. Bcl-2 inhibitors, STAT3 inhibitors, and therapeutic strategies modulating BCR signaling, tumor microenvironment and cytokine/chemokine axes may help in the management of CD5+ DLBCL patients.
Figure 7. Gene expression profiling and gene set enrichment analysis of CD5DLBCL.
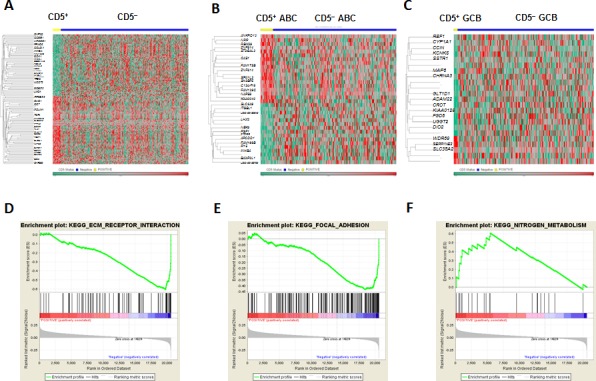
(A-C) Gene expression profiling of CD5+ vs. CD5− patients in the overall, ABC-, and GCB -DLBCL cohorts; (D-F) Gene set enrichment analysis of CD5 expression signatures.
MATERIALS AND METHODS
Patients
This study was part of the International DLBCL Rituximab-CHOP Consortium Program, comprising a total of 879 patients (538 patients in the training set and 341 patients in the validation set) with de novo DLBCL who were treated with R-CHOP chemotherapy. Patients who had a history of low-grade B-cell lymphoma, primary mediastinal, cutaneous, or central nervous system DLBCL, or human immunodeficiency virus infection were excluded. All patients were reviewed by a group of hematopathologists and were diagnosed according to the World Health Organization classification criteria. The study was conducted in accordance with the Declaration of Helsinki, and the protocol was reviewed and approved by the Institutional Review Boards of each participating center, and the comprehensive collaborative study was approved by the Institutional Review Board at The University of Texas MD Anderson Cancer Center.
Tissue microarray and biomarkers
Immunohistochemical analysis for CD5 expression using a monoclonal CD5 antibody (Novocastra Labs, UK) was performed on 879 DLBCL biopsy specimens using formalin-fixed, paraffin-embedded tissue microarrays as described previously [55, 56]. CD5 expression was scored by three pathologists independently on 400 cells in each of cases under a microscope at 40× magnification. Expression of other markers such as CD10, GCET1, MUM1, FOXP1, Bcl-6, Bcl-2, Myc, Ki-67, p53, MDM2, NF-κB subunits, pSTAT3, CD30, CXCR4, and SSBP2 [31, 33, 55-59] was also assessed using respective antibodies. SSBP2 antibody was kindly provided by Dr. Lalitha Nagarajan, PhD from the Department of Genetics, MD Anderson Cancer Center [34,36]. Due to tissue exhaustion, IHC analysis for some markers other than CD5 was not successful in few cases. Gene translocations and amplifications were detected using the methods described previously [30, 31, 59]. MYC mutation was detected using the Sanger sequencing method.
Cell-of-origin classification
Cell-of-origin classification as either GCB or ABC DLBCLs was determined by GEP for patients in the training set and by IHC according to the Visco-Young algorithm and/or Choi algorithms [55] for all the patients in the training and validation sets.
Gene expression profiling
GEP were achieved in 488 DLBCL (27 CD5+ and 461 CD5−) patients of the training set using total RNAs extracted from each formalin-fixed, paraffin-embedded tissue sample and Affymetrix GeneChips array as described previously [30, 55, 57, 58]. The microarray data were quantified and normalized, and the DEGs between CD5+ and CD5− DLBCL patients at false discovery rate of .01 were identified using multiple t-tests. Gene set enrichment analysis was performed on the KEGG pathway gene sets. Pathway analysis for the DEGs was performed using the Ingenuity Pathway Analysis software program (IPA, http://www.qiagen.com/ingenuity).
Statistical analysis
Clinicopathologic differences between different DLBCL subgroups were assessed using Fisher's exact test and the Spearman rank correlation. The mRNA expression levels of affected genes were also retrieved from the GEP data and compared between CD5+ and CD5− DLBCL patients using unpaired t tests. Overall survival was calculated from the time of diagnosis to death from any cause or last follow-up. Progression-free survival was calculated from the time of diagnosis to disease progression, relapse, or death from any cause. Patients who were alive and/or had no disease progression were censored at last follow-up. Survival analysis was performed using the Kaplan–Meier method with GraphPad Prism 6 (GraphPad Software, San Diego, CA), and differences were compared using the log-rank test. Multivariate survival analysis was performed using the Cox proportional hazards regression model with the SPSS statistics software program (version 19.0; IBM Corporation, Armonk, NY). All differences with P ≤ .05 were considered statistically significant.
SUPPLEMENTARY MATERIAL FIGURES
Acknowledgments
This work is supported by The University of Texas MD Anderson Cancer Center Institutional Research Grant Award, an MD Anderson Lymphoma Specialized Programs of Research Excellence (SPORE) Research Development Program Award, an MD Anderson Myeloma SPORE Research Development Program Award, MD Anderson Collaborative Research Funds with High-Throughput Molecular Diagnostics, Gilead Pharmaceutical, Adaptive Biotechnology, and Roche Molecular Systems, and National Cancer Institute and National Institutes of Health grants (R01CA138688 and R01CA187415) to K.H.Y. This work was also partially supported by National Cancer Institute and National Institutes of Health grants (P50CA136411 and P50CA142509), and by the MD Anderson Cancer Center Support Grant CA016672. Dr. Xu-Monette, PhD is the recipient of the Harold C. and Mary L. Daily Endowment Fellowships and Shannon Timmins Fellowship for Leukemia Research Award. Dr. Kausar J. Jabbar, MD is the recipient of the Pathology Division Biomarker Fellowship Award.
Footnotes
Author contributions
Conception and design: ZYXM, KHY; Research performance: ZYXM, KHY; Provision of study materials, key reagents and technology: ZYXM, MT, KJJ, XC, AT, CV, QC, SMM, YA, KD, AC, AO, YZ, GB, KLR, EDH, WWLC, JHK, MP, AJMF, XZ, MAP, JPF, JNW, RNM, LJM, KHY; Collection and assembly of data under approved IRB and MTA: ZYXM, KJJ, XC, AZ, CV, SMM, KD, AC, AO, YZ, GB, KLR, EDH, WWLC, JHK, MP, AJMF, XZ, MAP, JPF, JNW, KHY; Data analysis and interpretation: ZYXM, KHY; Manuscript writing: ZYXM, MT, KHY; Final approval of manuscript: All authors.
Editorial note
This paper has been accepted based in part on peerreview conducted by another journal and the authors' response and revisions as well as expedited peer-review in Oncotarget.
CONFLICTS OF INTEREST DISCLOSURE
The authors declare no conflicts of interest.
REFERENCES
- 1.Young KH, Medeiros LJ, Chan WC. Diffuse large B-cell lymphoma. In: Orazi A, Weiss LM, Foucar K, Knowles DM, editors. Neoplastic Hematopathology. Philadelphia, PA, USA: Lippincott Willaims & Wilkins; 2014. pp. 502–565. [Google Scholar]
- 2.Stein H, Warnke RA, Chan WC, Jaffe ES, Chan JKC, Gatter KC, Campo E, Swerdlow SH, Campo E. Diffuse large B-cell lymphoma, not otherwise specified. In: Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri SA, Stein H, Thiele J, Vardiman JW, et al., editors. WHO classification of tumours of haematopoetic and lymphoid tissues in. 4. Lyon, France: International Agency for Research on Cancer (IARC); 2008. pp. 233–261. [Google Scholar]
- 3.Jain P, Fayad LE, Rosenwald A, Young KH, O'Brien S. Recent advances in de novo CD5+ diffuse large B cell lymphoma. Am J Hematol. 2013;88:798–802. doi: 10.1002/ajh.23467. [DOI] [PubMed] [Google Scholar]
- 4.Berland R, Wortis HH. Origins and functions of B-1 cells with notes on the role of CD5. Annu Rev Immunol. 2002;20:253–300. doi: 10.1146/annurev.immunol.20.100301.064833. [DOI] [PubMed] [Google Scholar]
- 5.Brossard C, Semichon M, Trautmann A, Bismuth G. CD5 inhibits signaling at the immunological synapse without impairing its formation. J Immunol. 2003;170:4623–9. doi: 10.4049/jimmunol.170.9.4623. [DOI] [PubMed] [Google Scholar]
- 6.Bamberger M, Santos AM, Goncalves CM, Oliveira MI, James JR, Moreira A, Lozano F, Davis SJ, Carmo AM. A new pathway of CD5 glycoprotein-mediated T cell inhibition dependent on inhibitory phosphorylation of Fyn kinase. J Biol Chem. 2011;286:30324–36. doi: 10.1074/jbc.M111.230102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jevremovic D, Dronca RS, Morice WG, McPhail ED, Kurtin PJ, Zent CS, Hanson CA. CD5+ B-cell lymphoproliferative disorders: Beyond chronic lymphocytic leukemia and mantle cell lymphoma. Leuk Res. 2010;34:1235–8. doi: 10.1016/j.leukres.2010.03.020. [DOI] [PubMed] [Google Scholar]
- 8.Tibaldi E, Brunati AM, Zonta F, Frezzato F, Gattazzo C, Zambello R, Gringeri E, Semenzato G, Pagano MA, Trentin L. Lyn-mediated SHP-1 recruitment to CD5 contributes to resistance to apoptosis of B-cell chronic lymphocytic leukemia cells. Leukemia. 2011;25:1768–81. doi: 10.1038/leu.2011.152. [DOI] [PubMed] [Google Scholar]
- 9.Gary-Gouy H, Bruhns P, Schmitt C, Dalloul A, Daeron M, Bismuth G. The pseudo-immunoreceptor tyrosine-based activation motif of CD5 mediates its inhibitory action on B-cell receptor signaling. J Biol Chem. 2000;275:548–56. doi: 10.1074/jbc.275.1.548. [DOI] [PubMed] [Google Scholar]
- 10.Gary-Gouy H, Harriague J, Bismuth G, Platzer C, Schmitt C, Dalloul AH. Human CD5 promotes B-cell survival through stimulation of autocrine IL-10 production. Blood. 2002;100:4537–43. doi: 10.1182/blood-2002-05-1525. [DOI] [PubMed] [Google Scholar]
- 11.Garaud S, Morva A, Lemoine S, Hillion S, Bordron A, Pers JO, Berthou C, Mageed RA, Renaudineau Y, Youinou P. CD5 promotes IL-10 production in chronic lymphocytic leukemia B cells through STAT3 and NFAT2 activation. J Immunol. 2011;186:4835–44. doi: 10.4049/jimmunol.1003050. [DOI] [PubMed] [Google Scholar]
- 12.Challagundla P, Jorgensen JL, Kanagal-Shamanna R, Gurevich I, Pierson DM, Ferrajoli A, Reyes SR, Medeiros LJ, Miranda RN. Utility of quantitative flow cytometry immunophenotypic analysis of CD5 expression in small B-cell neoplasms. Arch Pathol Lab Med. 2014;138:903–9. doi: 10.5858/arpa.2013-0367-OA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yamaguchi M, Ohno T, Oka K, Taniguchi M, Ito M, Kita K, Shiku H. De novo CD5-positive diffuse large B-cell lymphoma: clinical characteristics and therapeutic outcome. Br J Haematol. 1999;105:1133–9. doi: 10.1046/j.1365-2141.1999.01513.x. [DOI] [PubMed] [Google Scholar]
- 14.Yamaguchi M, Seto M, Okamoto M, Ichinohasama R, Nakamura N, Yoshino T, Suzumiya J, Murase T, Miura I, Akasaka T, Tamaru J, Suzuki R, Kagami Y, et al. De novo CD5+ diffuse large B-cell lymphoma: a clinicopathologic study of 109 patients. Blood. 2002;99:815–21. doi: 10.1182/blood.v99.3.815. [DOI] [PubMed] [Google Scholar]
- 15.Harada S, Suzuki R, Uehira K, Yatabe Y, Kagami Y, Ogura M, Suzuki H, Oyama A, Kodera Y, Ueda R, Morishima Y, Nakamura S, Seto M. Molecular and immunological dissection of diffuse large B cell lymphoma: CD5+, and CD5- with CD10+ groups may constitute clinically relevant subtypes. Leukemia. 1999;13:1441–7. doi: 10.1038/sj.leu.2401487. [DOI] [PubMed] [Google Scholar]
- 16.Yamaguchi M, Nakamura N, Suzuki R, Kagami Y, Okamoto M, Ichinohasama R, Yoshino T, Suzumiya J, Murase T, Miura I, Ohshima K, Nishikori M, Tamaru J, et al. De novo CD5+ diffuse large B-cell lymphoma: results of a detailed clinicopathological review in 120 patients. Haematologica. 2008;93:1195–202. doi: 10.3324/haematol.12810. [DOI] [PubMed] [Google Scholar]
- 17.Ennishi D, Takeuchi K, Yokoyama M, Asai H, Mishima Y, Terui Y, Takahashi S, Komatsu H, Ikeda K, Yamaguchi M, Suzuki R, Tanimoto M, Hatake K. CD5 expression is potentially predictive of poor outcome among biomarkers in patients with diffuse large B-cell lymphoma receiving rituximab plus CHOP therapy. Ann Oncol. 2008;19:1921–6. doi: 10.1093/annonc/mdn392. [DOI] [PubMed] [Google Scholar]
- 18.Hyo R, Tomita N, Takeuchi K, Aoshima T, Fujita A, Kuwabara H, Hashimoto C, Takemura S, Taguchi J, Sakai R, Fujita H, Fujisawa S, Ogawa K, et al. The therapeutic effect of rituximab on CD5-positive and CD5-negative diffuse large B-cell lymphoma. Hematol Oncol. 2010;28:27–32. doi: 10.1002/hon.896. [DOI] [PubMed] [Google Scholar]
- 19.Niitsu N, Okamoto M, Tamaru JI, Yoshino T, Nakamura N, Nakamura S, Ohshima K, Nakamine H, Hirano M. Clinicopathologic characteristics and treatment outcome of the addition of rituximab to chemotherapy for CD5-positive in comparison with CD5-negative diffuse large B-cell lymphoma. Ann Oncol. 2010;21:2069–74. doi: 10.1093/annonc/mdq057. [DOI] [PubMed] [Google Scholar]
- 20.Miyazaki K, Yamaguchi M, Suzuki R, Kobayashi Y, Maeshima AM, Niitsu N, Ennishi D, Tamaru JI, Ishizawa K, Kashimura M, Kagami Y, Sunami K, Yamane H, et al. CD5-positive diffuse large B-cell lymphoma: a retrospective study in 337 patients treated by chemotherapy with or without rituximab. Ann Oncol. 2011;22:1601–7. doi: 10.1093/annonc/mdq627. [DOI] [PubMed] [Google Scholar]
- 21.Davidson-Moncada JK, McDuffee E, Roschewski M. CD5+ diffuse large B-cell lymphoma with hemophagocytosis. J Clin Oncol. 2013;31:e76–9. doi: 10.1200/JCO.2012.44.2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Westin J, McLaughlin P. De novo CD5+ diffuse large B-cell lymphoma: a distinct subset with adverse features, poor failure-free survival and outcome with conventional therapy. Leuk Lymphoma. 2010;51:161–3. doi: 10.3109/10428190903324244. [DOI] [PubMed] [Google Scholar]
- 23.Kroft SH, Howard MS, Picker LJ, Ansari MQ, Aquino DB, McKenna RW. De novo CD5+ diffuse large B-cell lymphomas. A heterogeneous group containing an unusual form of splenic lymphoma. Am J Clin Pathol. 2000;114:523–33. doi: 10.1309/RM1Q-1T0B-WKQB-AF5A. [DOI] [PubMed] [Google Scholar]
- 24.Kobayashi T, Yamaguchi M, Kim S, Morikawa J, Ogawa S, Ueno S, Suh E, Dougherty E, Shmulevich I, Shiku H, Zhang W. Microarray reveals differences in both tumors and vascular specific gene expression in de novo CD5+ and CD5- diffuse large B-cell lymphomas. Cancer Res. 2003;63:60–6. [PubMed] [Google Scholar]
- 25.Suguro M, Tagawa H, Kagami Y, Okamoto M, Ohshima K, Shiku H, Morishima Y, Nakamura S, Seto M. Expression profiling analysis of the CD5+ diffuse large B-cell lymphoma subgroup: development of a CD5 signature. Cancer Sci. 2006;97:868–74. doi: 10.1111/j.1349-7006.2006.00267.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yamamoto K, Okamura A, Yakushijin K, Hayashi Y, Matsuoka H, Minami H. Tandem triplication of the BCL2 gene in CD5-positive intravascular large B cell lymphoma with bone marrow involvement. Ann Hematol. 2014;93:1791–3. doi: 10.1007/s00277-014-2035-y. [DOI] [PubMed] [Google Scholar]
- 27.Azab AK, Runnels JM, Pitsillides C, Moreau AS, Azab F, Leleu X, Jia X, Wright R, Ospina B, Carlson AL, Alt C, Burwick N, Roccaro AM, et al. CXCR4 inhibitor AMD3100 disrupts the interaction of multiple myeloma cells with the bone marrow microenvironment and enhances their sensitivity to therapy. Blood. 2009;113:4341–51. doi: 10.1182/blood-2008-10-186668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Balkwill F. Cancer and the chemokine network. Nat Rev Cancer. 2004;4:540–50. doi: 10.1038/nrc1388. [DOI] [PubMed] [Google Scholar]
- 29.Chang BY, Francesco M, De Rooij MF, Magadala P, Steggerda SM, Huang MM, Kuil A, Herman SE, Chang S, Pals ST, Wilson W, Wiestner A, Spaargaren M, et al. Egress of CD19(+)CD5(+) cells into peripheral blood following treatment with the Bruton tyrosine kinase inhibitor ibrutinib in mantle cell lymphoma patients. Blood. 2013;122:2412–24. doi: 10.1182/blood-2013-02-482125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Visco C, Tzankov A, Xu-Monette ZY, Miranda RN, Tai YC, Li Y, Liu WM, d'Amore ES, Li Y, Montes-Moreno S, Dybkaer K, Chiu A, Orazi A, et al. Patients with diffuse large B-cell lymphoma of germinal center origin with BCL2 translocations have poor outcome, irrespective of MYC status: a report from an International DLBCL rituximab-CHOP Consortium Program Study. Haematologica. 2013;98:255–63. doi: 10.3324/haematol.2012.066209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hu S, Xu-Monette ZY, Tzankov A, Green T, Wu L, Balasubramanyam A, Liu WM, Visco C, Li Y, Miranda RN, Montes-Moreno S, Dybkaer K, Chiu A, et al. MYC/BCL2 protein coexpression contributes to the inferior survival of activated B-cell subtype of diffuse large B-cell lymphoma and demonstrates high-risk gene expression signatures: a report from The International DLBCL Rituximab-CHOP Consortium Program. Blood. 2013;121:4021–31. doi: 10.1182/blood-2012-10-460063. quiz 4250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shaffer AL, 3rd, Young RM, Staudt LM. Pathogenesis of human B cell lymphomas. Annu Rev Immunol. 2012;30:565–610. doi: 10.1146/annurev-immunol-020711-075027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ok CY, Chen J, Xu-Monette ZY, Tzankov A, Manyam GC, Li L, Visco C, Montes-Moreno S, Dybkaer K, Chiu A, Orazi A, Zu Y, Bhagat G, et al. Clinical Implications of Phosphorylated STAT3 Expression in De Novo Diffuse Large B-cell Lymphoma. Clin Cancer Res. 2014;20:5113–23. doi: 10.1158/1078-0432.CCR-14-0683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang Y, Klumpp S, Amin HM, Liang H, Li J, Estrov Z, Zweidler-McKay P, Brandt SJ, Agulnick A, Nagarajan L. SSBP2 is an in vivo tumor suppressor and regulator of LDB1 stability. Oncogene. 2010;29:3044–53. doi: 10.1038/onc.2010.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li J, Kurasawa Y, Wang Y, Clise-Dwyer K, Klumpp SA, Liang H, Tailor RC, Raymond AC, Estrov Z, Brandt SJ, Davis RE, Zweidler-McKay P, Amin HM, et al. Requirement for ssbp2 in hematopoietic stem cell maintenance and stress response. J Immunol. 2014;193:4654–62. doi: 10.4049/jimmunol.1300337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liang H, Samanta S, Nagarajan L. SSBP2, a candidate tumor suppressor gene, induces growth arrest and differentiation of myeloid leukemia cells. Oncogene. 2005;24:2625–34. doi: 10.1038/sj.onc.1208167. [DOI] [PubMed] [Google Scholar]
- 37.Staudt LM. Oncogenic activation of NF-kappaB. Cold Spring Harb Perspect Biol. 2010;2:a000109. doi: 10.1101/cshperspect.a000109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thome M, Charton JE, Pelzer C, Hailfinger S. Antigen receptor signaling to NF-kappaB via CARMA1, BCL10, and MALT1. Cold Spring Harb Perspect Biol. 2010;2:a003004. doi: 10.1101/cshperspect.a003004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rodig SJ, Vergilio JA, Shahsafaei A, Dorfman DM. Characteristic expression patterns of TCL1, CD38, and CD44 identify aggressive lymphomas harboring a MYC translocation. Am J Surg Pathol. 2008;32:113–22. doi: 10.1097/PAS.0b013e3180959e09. [DOI] [PubMed] [Google Scholar]
- 40.Weniger MA, Pulford K, Gesk S, Ehrlich S, Banham AH, Lyne L, Martin-Subero JI, Siebert R, Dyer MJ, Moller P, Barth TF. Gains of the proto-oncogene BCL11A and nuclear accumulation of BCL11A(XL) protein are frequent in primary mediastinal B-cell lymphoma. Leukemia. 2006;20:1880–2. doi: 10.1038/sj.leu.2404324. [DOI] [PubMed] [Google Scholar]
- 41.Satterwhite E, Sonoki T, Willis TG, Harder L, Nowak R, Arriola EL, Liu H, Price HP, Gesk S, Steinemann D, Schlegelberger B, Oscier DG, Siebert R, et al. The BCL11 gene family: involvement of BCL11A in lymphoid malignancies. Blood. 2001;98:3413–20. doi: 10.1182/blood.v98.12.3413. [DOI] [PubMed] [Google Scholar]
- 42.Deambrogi C, De Paoli L, Fangazio M, Cresta S, Rasi S, Spina V, Gattei V, Gaidano G, Rossi D. Analysis of the REL, BCL11A, and MYCN proto-oncogenes belonging to the 2p amplicon in chronic lymphocytic leukemia. Am J Hematol. 2010;85:541–4. doi: 10.1002/ajh.21742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Peter D, Jin SL, Conti M, Hatzelmann A, Zitt C. Differential expression and function of phosphodiesterase 4 (PDE4) subtypes in human primary CD4+ T cells: predominant role of PDE4D. J Immunol. 2007;178:4820–31. doi: 10.4049/jimmunol.178.8.4820. [DOI] [PubMed] [Google Scholar]
- 44.Woyach JA, Johnson AJ, Byrd JC. The B-cell receptor signaling pathway as a therapeutic target in CLL. Blood. 2012;120:1175–84. doi: 10.1182/blood-2012-02-362624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Feske S. Calcium signalling in lymphocyte activation and disease. Nat Rev Immunol. 2007;7:690–702. doi: 10.1038/nri2152. [DOI] [PubMed] [Google Scholar]
- 46.Barretina J, Junca J, Llano A, Gutierrez A, Flores A, Blanco J, Clotet B, Este JA. CXCR4 and SDF-1 expression in B-cell chronic lymphocytic leukemia and stage of the disease. Ann Hematol. 2003;82:500–5. doi: 10.1007/s00277-003-0679-0. [DOI] [PubMed] [Google Scholar]
- 47.Fleisig HB, Orazio NI, Liang H, Tyler AF, Adams HP, Weitzman MD, Nagarajan L. Adenoviral E1B55K oncoprotein sequesters candidate leukemia suppressor sequence-specific single-stranded DNA-binding protein 2 into aggresomes. Oncogene. 2007;26:4797–805. doi: 10.1038/sj.onc.1210281. [DOI] [PubMed] [Google Scholar]
- 48.Xu Z, Meng X, Cai Y, Liang H, Nagarajan L, Brandt SJ. Single-stranded DNA-binding proteins regulate the abundance of LIM domain and LIM domain-binding proteins. Genes Dev. 2007;21:942–55. doi: 10.1101/gad.1528507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pasqualucci L, Neumeister P, Goossens T, Nanjangud G, Chaganti RS, Kuppers R, Dalla-Favera R. Hypermutation of multiple proto-oncogenes in B-cell diffuse large-cell lymphomas. Nature. 2001;412:341–6. doi: 10.1038/35085588. [DOI] [PubMed] [Google Scholar]
- 50.Nakamura N, Abe M. Histogenesis of CD5-positive and CD5-negative B-cell neoplasms on the aspect of somatic mutation of immunoglobulin heavy chain gene variable region. Fukushima J Med Sci. 2003;49:55–67. doi: 10.5387/fms.49.55. [DOI] [PubMed] [Google Scholar]
- 51.Nakamura N, Kuze T, Hashimoto Y, Tasaki K, Hojo H, Sasaki Y, Sato M, Abe M. Analysis of the immunoglobulin heavy chain gene variable region of 101 cases with peripheral B cell neoplasms and B cell chronic lymphocytic leukemia in the japanese population. Pathol. Int. 1999;49:595–600. doi: 10.1046/j.1440-1827.1999.00911.x. [DOI] [PubMed] [Google Scholar]
- 52.Angelin-Duclos C, Cattoretti G, Lin KI, Calame K. Commitment of B lymphocytes to a plasma cell fate is associated with Blimp-1 expression in vivo. J Immunol. 2000;165:5462–71. doi: 10.4049/jimmunol.165.10.5462. [DOI] [PubMed] [Google Scholar]
- 53.Nutt SL, Fairfax KA, Kallies A. BLIMP1 guides the fate of effector B and T cells. Nat Rev Immunol. 2007;7:923–7. doi: 10.1038/nri2204. [DOI] [PubMed] [Google Scholar]
- 54.Shaffer AL, Lin KI, Kuo TC, Yu X, Hurt EM, Rosenwald A, Giltnane JM, Yang L, Zhao H, Calame K, Staudt LM. Blimp-1 orchestrates plasma cell differentiation by extinguishing the mature B cell gene expression program. Immunity. 2002;17:51–62. doi: 10.1016/s1074-7613(02)00335-7. [DOI] [PubMed] [Google Scholar]
- 55.Visco C, Li Y, Xu-Monette ZY, Miranda RN, Green TM, Li Y, Tzankov A, Wen W, Liu WM, Kahl BS, d'Amore ES, Montes-Moreno S, Dybkaer K, et al. Comprehensive gene expression profiling and immunohistochemical studies support application of immunophenotypic algorithm for molecular subtype classification in diffuse large B-cell lymphoma: a report from the International DLBCL Rituximab-CHOP Consortium Program Study. Leukemia. 2012;26:2103–13. doi: 10.1038/leu.2012.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hu S, Xu-Monette ZY, Balasubramanyam A, Manyam GC, Visco C, Tzankov A, Liu WM, Miranda RN, Zhang L, Montes-Moreno S, Dybkaer K, Chiu A, Orazi A, et al. CD30 expression defines a novel subgroup of diffuse large B-cell lymphoma with favorable prognosis and distinct gene expression signature: a report from the International DLBCL Rituximab-CHOP Consortium Program Study. Blood. 2013;121:2715–24. doi: 10.1182/blood-2012-10-461848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xu-Monette ZY, Wu L, Visco C, Tai YC, Tzankov A, Liu WM, Montes-Moreno S, Dybkaer K, Chiu A, Orazi A, Zu Y, Bhagat G, Richards KL, et al. Mutational profile and prognostic significance of TP53 in diffuse large B-cell lymphoma patients treated with R-CHOP: report from an International DLBCL Rituximab-CHOP Consortium Program Study. Blood. 2012;120:3986–96. doi: 10.1182/blood-2012-05-433334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xu-Monette ZY, Moller MB, Tzankov A, Montes-Moreno S, Hu W, Manyam GC, Kristensen L, Fan L, Visco C, Dybkaer K, Chiu A, Tam W, Zu Y, et al. MDM2 phenotypic and genotypic profiling, respective to TP53 genetic status, in diffuse large B-cell lymphoma patients treated with rituximab-CHOP immunochemotherapy: a report from the International DLBCL Rituximab-CHOP Consortium Program. Blood. 2013;122:2630–40. doi: 10.1182/blood-2012-12-473702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tzankov A, Xu-Monette ZY, Gerhard M, Visco C, Dirnhofer S, Gisin N, Dybkaer K, Orazi A, Bhagat G, Richards KL, Hsi ED, Choi WW, van Krieken JH, et al. Rearrangements of MYC gene facilitate risk stratification in diffuse large B-cell lymphoma patients treated with rituximab-CHOP. Mod Pathol. 2014;27:958–71. doi: 10.1038/modpathol.2013.214. [DOI] [PubMed] [Google Scholar]
- 60.Dybkær K, Bøgsted M, Falgreen S, Bødker JS, Kjeldsen MK, Schmitz A, et al. A diffuse large B-cell lymphoma classification system that associates normal B-cell subset phenotypes with prognosis. J Clin Oncol. 2015 doi: 10.1200/JCO.2014.57.7080. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Testoni M, Zucca E, Young KH, Bertoni F. Genetic lesions in diffuse large B-cell lymphomas. Ann Oncol. 2015 doi: 10.1093/annonc/mdv019. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


