Abstract
Transcript fusions as a result of chromosomal rearrangements have been a focus of attention in cancer as they provide attractive therapeutic targets. To identify novel fusion transcripts with the potential to be exploited therapeutically, we analyzed RNA sequencing, DNA copy number and gene mutation data from 4,366 primary tumor samples. To avoid false positives, we implemented stringent quality criteria that included filtering of fusions detected in RNAseq data from 364 normal tissue samples. Our analysis identified 7,887 high confidence fusion transcripts across 13 tumor types. Our fusion prediction was validated by evidence of a genomic rearrangement for 78 of 79 fusions in 48 glioma samples where whole genome sequencing data was available. Cancers with higher levels of genomic instability showed a corresponding increase in fusion transcript frequency, whereas tumor samples harboring fusions contained statistically significantly fewer driver gene mutations, suggesting an important role for tumorigenesis. We identified at least one in-frame protein kinase fusion in 324 of 4,366 samples (7.4%). Potentially druggable kinase fusions involving ALK, ROS, RET, NTRK, and FGFR gene families were detected in bladder carcinoma (3.3%), glioblastoma (4.4%), head and neck cancer (1.0%), low grade glioma (1.5%), lung adenocarcinoma (1.6%), lung squamous cell carcinoma (2.3%), and thyroid carcinoma (8.7%), suggesting a potential for application of kinase inhibitors across tumor types. In-frame fusion transcripts involving histone methyltransferase or histone demethylase genes were detected in 111 samples (2.5%) and may additionally be considered as therapeutic targets. In summary, we described the landscape of transcript fusions detected across a large number of tumor samples and revealed fusion events with clinical relevance that have not been previously recognized. Our results support the concept of basket clinical trials where patients are matched with experimental therapies based on their genomic profile rather than the tissue where the tumor originated.
Keywords: Gene fusion, Copy number alteration, Protein kinase, Chromatin modifier, Pan-cancer
Introduction
Transcript fusions resulting from chromosomal rearrangements are an important class of cancer-contributing somatic alteration1. Examples such as BCR-ABL1, first reported in chronic myeloid leukemias2, have led to novel first line therapies with ABL inhibitors such as dasatinib3. Similarly, EML4-ALK fusions were detected in subset of non-small cell lung cancer4 and ALK inhibitors were reported to improve outcome for patients with EML4-ALK positive tumors5. Recent advances in sequencing technology have enabled the comprehensive detection of rearrangements in the cancer genome and transcriptome6, 7. For example, transcriptome sequencing has identified FGFR3-TACC3 fusions in glioblastoma8, bladder cancer9 and head and neck, lung squamous cell carcinoma10, and cell lines expression FGFR3 chimeras were found to be sensitive to the FGFR inhibitors. In addition, recent studies have revealed highly frequent oncogenic fusions in rare tumor types, such as C11orf95-RELA fusion in supratentorial ependymoma11 and DNAJB1-PRKACA fusion in fibrolamellar hepatocellular carcinoma12. Tumor specific fusion gene landscapes of different cancers have been described using genomic and transcriptomic data13–16.
To comprehensively identify fusion transcripts with the potential to be exploited therapeutically across many cancers, we analyzed RNA sequencing and DNA copy number data from 4,366 primary tumor samples and 364 normal samples spanning 13 tumor types. We assessed the significance of fusions per cancer type and evaluated their potential as molecular therapeutic targets by integrating mRNA exon/gene expression, somatic mutations, copy number gains and losses, and protein kinase annotation. Our fusion gene list of TCGA samples is available through a web portal via http://www.tumorfusions.org.
Results
Detection of fusion transcripts
An overview of this study is shown in Supplementary Figure 1. We compiled a mRNA sequencing data set consisting of 4,366 primary tumor samples and 369 normal samples from 13 tissue types (Table 1). Data was generated by The Cancer Genome Atlas and made available through the Cancer Genomics Hub (CGHub, https://cghub.ucsc.edu/). Using supervised hierarchical clustering analysis, we identified five normal samples with a high likelihood of tumor cell contamination and these were excluded from further study (See Supplementary Figure 2 and Methods). We used the Pipeline for RNA sequencing Data Analysis (PRADA)17 to detect 26,995 fusion transcripts supported by at least two discordant read pairs plus one perfect-match junction spanning read, with the other end of the read pair mapping to either of the fusion gene partners. To reduce the number of false positive predictions, we filtered fusion transcripts according to gene homology, transcript allele fraction, and partner gene variety. We used BLASTn to determine homology between partner genes and removed 6,138 fusion pairs consisting of two genes with high similarity. Next, to consider the influence of transcript expression level in the process of fusion detection, we calculated the transcript allele fraction, which is the ratio of junction spanning reads to the total number of reads crossing the junction points in the reference transcripts, and removed fusion candidates with a transcript allele fraction of less than 0.01. Finally, we calculated partner gene variety for each gene and excluded non-specific fusions involving genes showing a large diversity amongst partner genes. After filtering, 9,047 and 192 fusion pairs were identified in 4,366 primary tumor and 364 normal tissue samples, respectively. After removing fusion pairs overlapping between tumor and normal samples, 8,695 tumor specific fusion pairs were identified (Supplementary Table 1). We further classified the final fusion transcript list into four tiers based on level of evidence. Fusions designated as tier 1 were detected through at least three discordant read pairs, two perfect-match junction spanning reads, and gene partner uniqueness within a sample. Tier 2 fusions required at least two discordant read pairs, one perfect-match junction spanning read, plus breakpoints detected in the DNA profile, within 100Kb from predicted junction point. Tier 3 was categorized as fusions with at least two discordant read pairs, one perfect-match junction spanning read, high consistency of predicted junction, and gene partner uniqueness within a sample. The remainder of fusions was directed to tier 4. In total, 6,219 and 1,668 fusion pairs were annotated as tier 1 or tier 2, respectively.
Table 1.
A list of The Cancer Genome Atlas RNAseq data sets
| Tumor type | Tumor* | Normal |
|---|---|---|
| Bladder urothelial carcinoma | 121 | 16 |
| Breast cancer | 1,019 | 110 |
| Glioblastoma multiforme | 158 | - |
| Head and neck squamous cell carcinoma | 300 | 37 |
| Clear cell renal cell carcinoma | 474 | 71 |
| Acute myeloid leukemia | 171 | - |
| Low grade glioma | 266 | - |
| Lung adenocarcinoma | 487 | 57 |
| Lung squamous cell carcinoma | 220 | 17 |
| Ovarian serous cystadenocarcinoma | 400 | - |
| Prostate adenocarcinoma | 178 | - |
| Skin cutaneous melanoma | 78 | - |
| Thyroid carcinoma | 494 | 56 |
| Total | 4,366 | 364 |
Primary tumor samples with both RNAseq and Affymetrix SNP6 array data were analyzed.
Validation of fusion transcript predictions
To verify the reliability of our fusion transcript predictions, we performed BreakDancer18 on whole genome sequencing data from 48 glioma samples and low pass whole genome sequencing from 15 melanoma tumors. A minimum of five supporting read pairs were required for detection of structural variants in whole genome sequencing data and three supporting read pairs in low-pass whole genome sequencing data. Next, we correlated the presence of genomic structural variants with fusion gene predictions from RNA. Structural DNA variants involving both fusion gene partners were considered as high confidence validation, and events involving one of the gene partners were interpreted as medium confidence. As a result, high or medium confidence structural variants were found to support 78 of 79 fusion transcripts detected in 48 glioma samples within 1Mb from the predicted junction points (Supplementary Table 2). As expected, the rate of validation was reduced in low pass sequencing data, where we found support for 31 of 48 fusion transcripts detected in 15 melanoma samples. The validation rate of tier 1 and 2 events in low pass sequencing data was higher compared to tier 3 and 4, and we limited further analysis to 7,887 tier 1 and 2 fusion transcripts.
Diversity of fusion transcripts across 13 tumor types
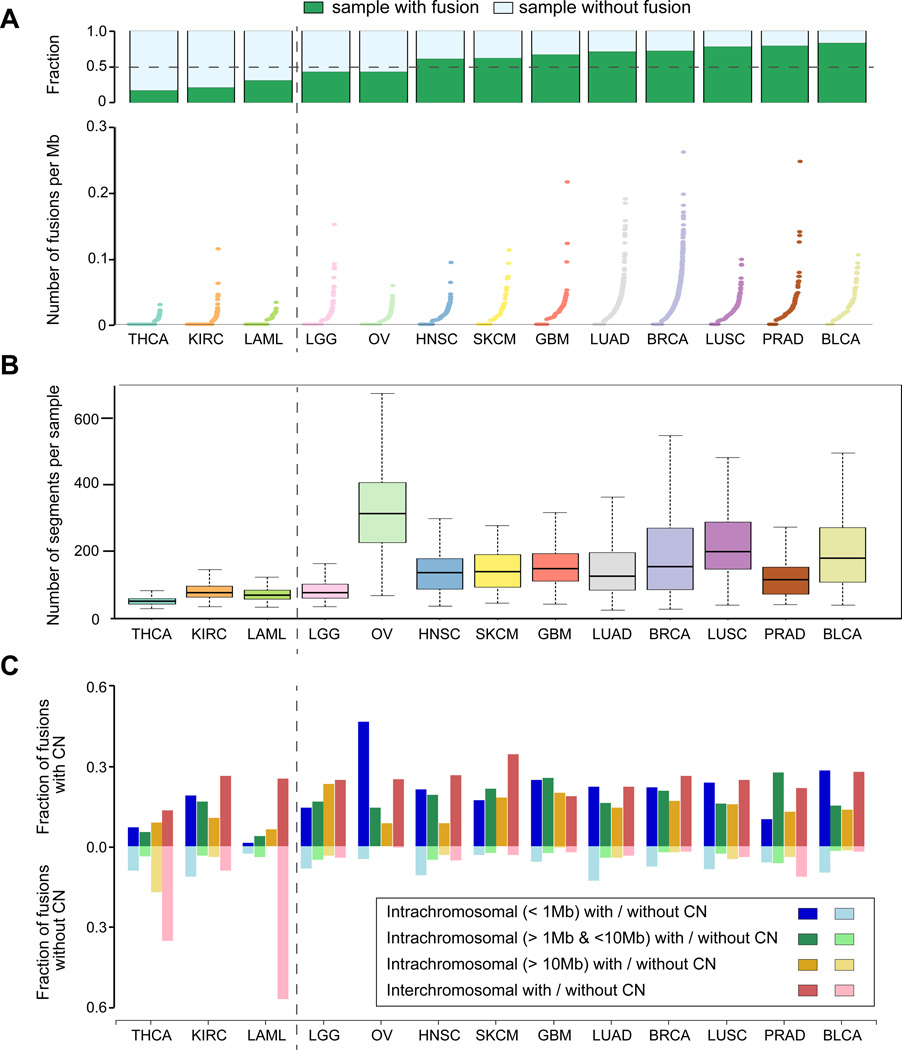
Determining how fusion transcripts promote cancer in various tumor types is an important goal. We categorized fusion transcripts into eight categories based on (i) distance between the two fusion gene partners and (ii) the presence of copy number alterations in proximity to the fusion junction, and examined the distribution of each category for each tumor type. We observed substantial diversity in the frequency of gene fusions, with thyroid carcinoma, clear cell renal cell carcinoma, and acute myeloid leukemia representing the lower end of the spectrum (Figure 1A). A corresponding relative reduction in the frequency of DNA segments was found in these cancer types (Figure 1B). In nine of ten remaining tumor types, the exception being prostate adenocarcinoma, more than 80% of fusion transcripts were associated with DNA amplification or deletion events (Supplementary Figure 3). Acute myeloid leukemia and thyroid carcinoma demonstrated a relatively high frequency of copy-neutral interchromosomal fusions (Figure 1C), suggesting the frequent occurrence of balanced genomic rearrangements1. Fusion transcripts originating from genes within 1 Megabase of each other were dominant in ovarian cancer, which might be related to the high frequency of copy number alteration in ovarian cancer19. Overall, these findings suggest that fusion transcripts resulting from copy number balanced translocations are relatively rare and instead are preferentially derived through genomic instability20, 21.
Figure 1. The distribution of fusion transcripts across twelve tumor types.
(A) Bar plots show the fraction of samples in which at least a single fusion transcript was detected per tumor type (green). The dot plots illustrates the number of detected fusion transcripts per megabase per sample normalized by the sequencing coverage. Tumor types were sorted according to the fraction of samples with fusions. (B) Box-Whisker plots showing the number of DNA segments per sample as a relative measure of genome instability across 13 tumor types. (C) Barplots representing the fraction of different types of fusions classified based on the distance between the genes constituting the fusion and the presence or absence of a DNA copy number alteration within 100Kb of the junction point.
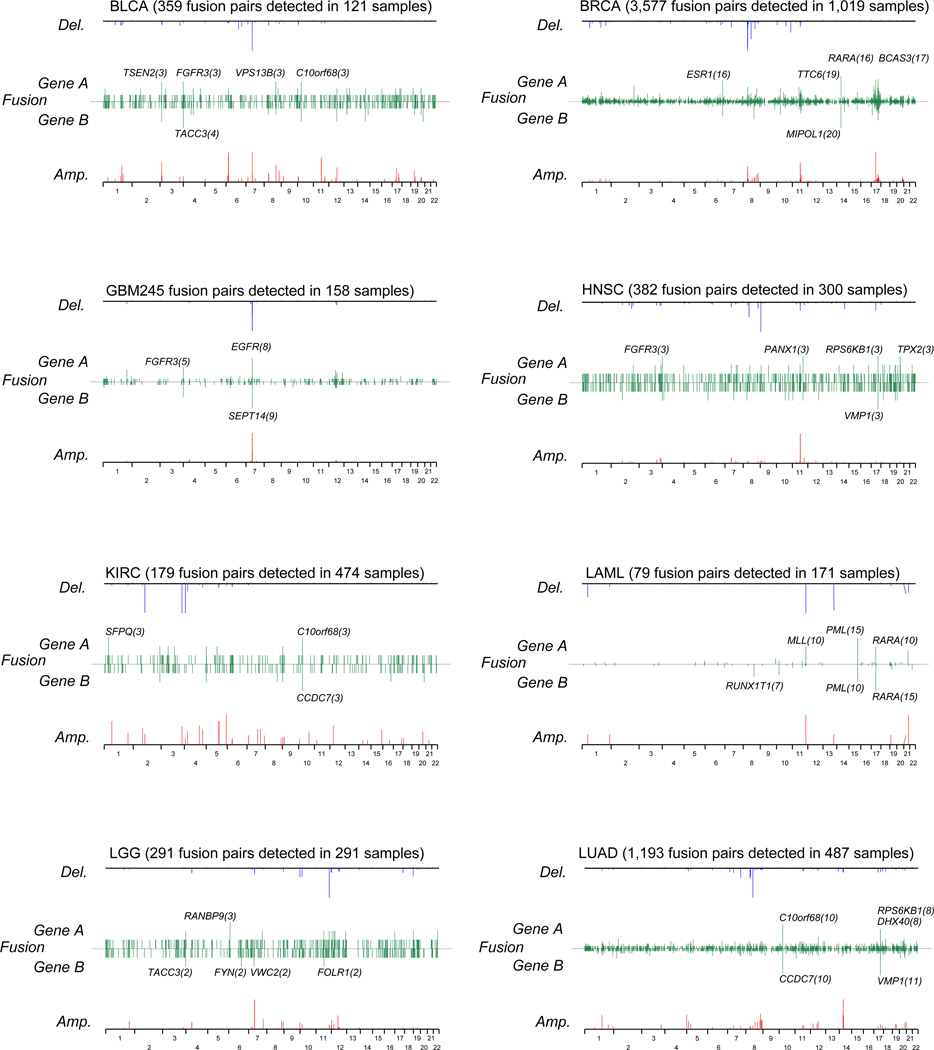
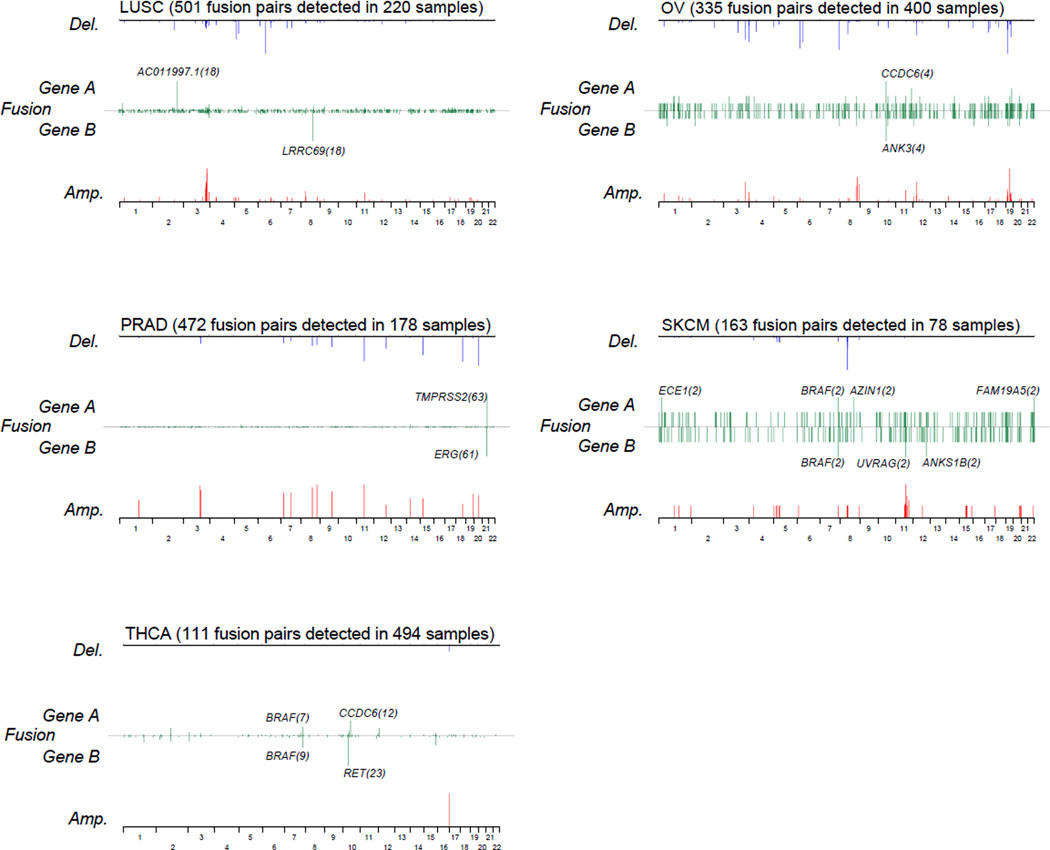
Next, we generated a summary of recurrent fusion transcripts across 13 tumor types (Supplementary Table 3) which included 263 fusions occurring at least twice. Of these, 24 recurrent fusions have been previously reported22, 23. Furthermore, we focused on fusions with the same gene fused to multiple different partners (Figure 2 and Supplementary Table 4). Perhaps the most prominent and novel recurrent gene was the estrogen receptor 1 (ESR1). We identified 16 ESR1 associated fusions in breast cancer, which represents 1.5%) of the entire breast cancer cohort. Only one of these was predicted to be in frame and these fusions may thus be disruptive events rather than activating (Supplementary Figure 4). On the basis of this result, we extracted 221 fusions involving a tumor suppressor gene (TSG) that have a potential to result in loss of function (Supplementary Table 5). All samples harboring TSG fusions were called wild type except one low grade glioma sample.
Figure 2. The chromosomal location of recurrent fusion genes for each tumor types.
Line plots representing the frequency of fusion gene A and B across the genome (green), the negative log (q-value) of DNA amplifications (red) and deletions (blue) per tumor type.
DNA copy number alterations with q-value less than 0.05 as determined by GISTIC are shown.
Approximately 36% of detected fusion transcripts were predicted to be in-frame and thus may result in a functional protein, with acute myeloid leukemia and thyroid carcinoma showing relatively high fractions of in-frame fusions (78.5% and 70.3%) compared to other tumor types (Supplementary Figure 5A). When we re-evaluated the distribution of eight fusion categories only using 2,811 in-frame fusions, the distribution of eight categories for each tumor type was generally similar with those based on 7,887 fusion transcripts (Supplementary Figure 5B, Supplementary Table 6).
In total, 80 of 2,811 in-frame fusion transcripts were detected in at least two samples across the entire cohort (Supplementary Table 7), including well known fusions such as TMPRSS2-ERG24, PML-RARA25, FGFR3-TACC38, EGFR-SEPT1426, 27. Interestingly, we observed reduced frequencies of significant gene mutation in samples with recurrent in frame fusion transcripts compared to those without recurrent in frame fusion transcripts. The difference was statistically significant in bladder carcinoma, breast cancer, head and neck squamous cell carcinoma, clear cell renal cell carcinoma, acute myeloid leukemia, and thyroid cancer (Welch’s t-test, P = 0.0067, 0.022, 0.030, 0.063, 4.8-e15, and 8.3e–88, respectively), suggesting that fusions in these cancer types could be functioning as incidental cancer driving events (Supplementary Figure 6).
Protein kinase fusions across 13 tumor types
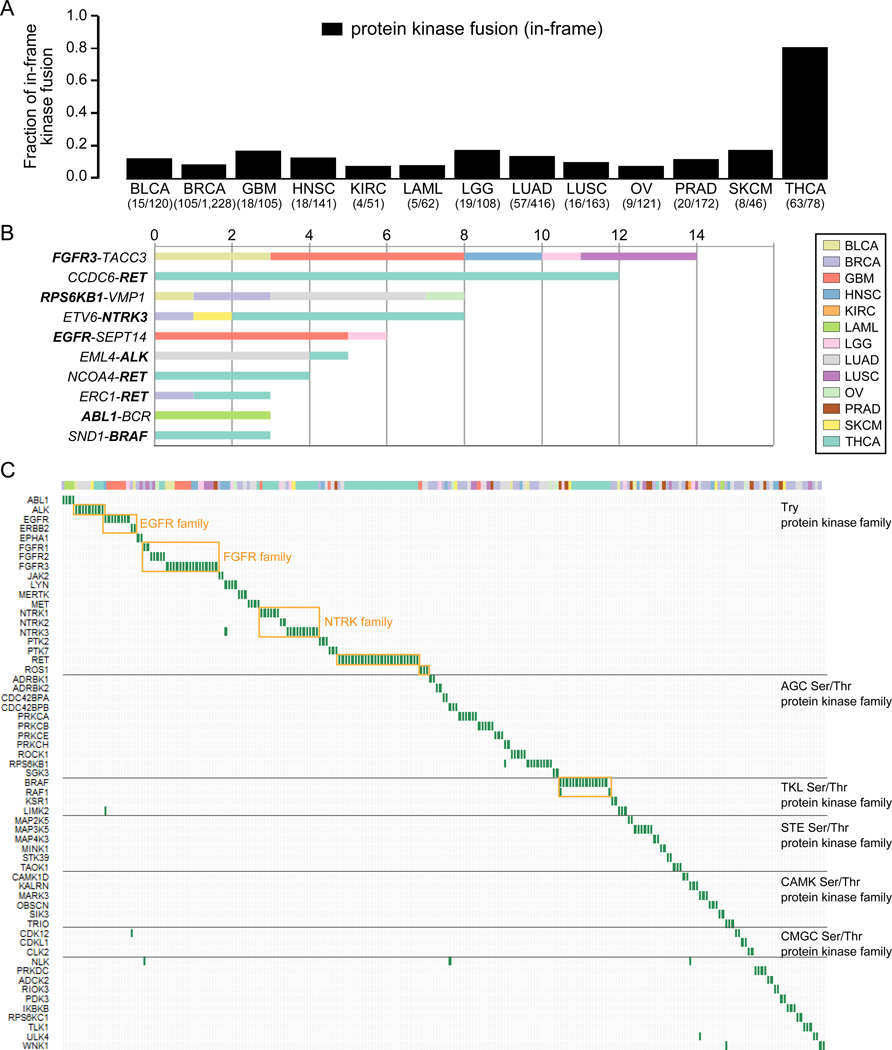
Fusion genes with oncogenic kinase activation have been identified in many cancers1, 2, 4 and cancer cells harboring these types of fusions are frequently highly susceptible to kinase inhibitors28. To discover fusion candidates with therapeutic potential, we focused on fusion transcripts involving a protein kinase gene. An in-frame protein kinase fusion was detected in 324 (7.4%) of 4,366 samples (minimum, 0.8% in clear cell renal cell carcinoma; maximum, 11.6% in bladder carcinoma) (Supplementary Table 8). The majority of in frame kinase fusions belonged to the tyrosine kinase family (36.1%), the AGC serine/threonine protein kinases (14.8%) and the tyrosine kinase-like serine/threonine protein kinase group (10.1%) (Supplementary Table 9).
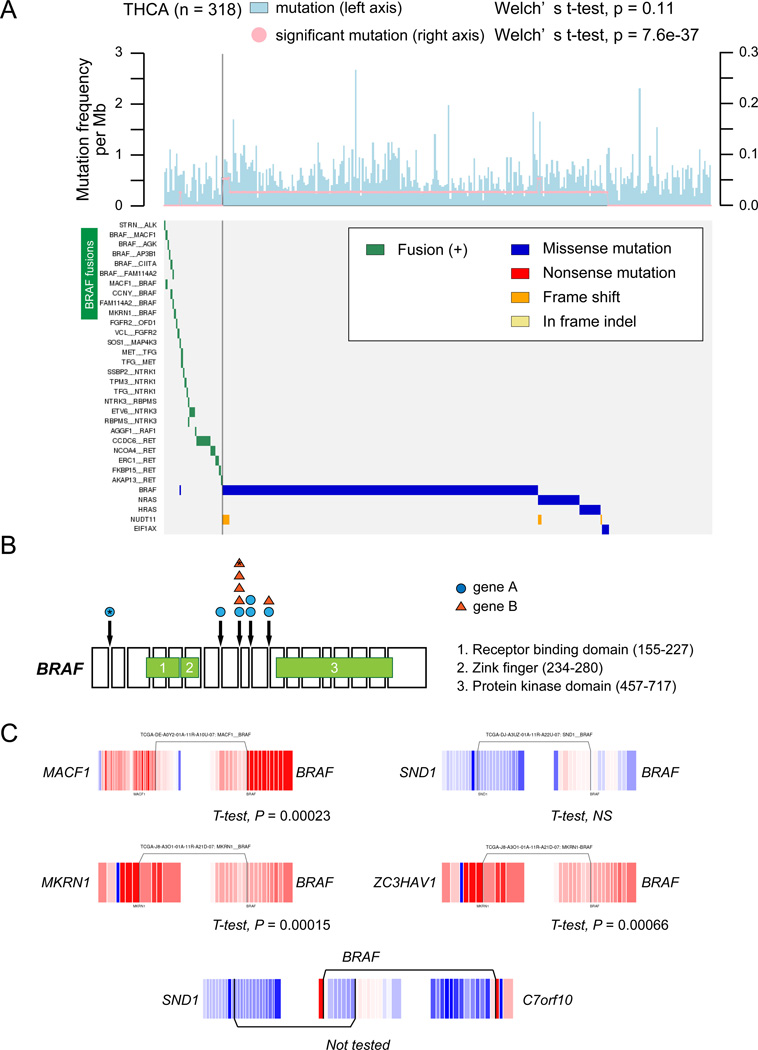
The fraction of protein kinase fusions was significantly higher in thyroid carcinoma compared to other tumor types, involving genes such as RET (n=24), NTRK3 (n=9) and BRAF (n=16) (Fisher’s exact test, P = 2.2e–16) (Figure 3A). BRAF fusions were also detected in two prostate adenocarcinoma, two melanoma and one low grade glioma samples (Supplementary Table 10). BRAF fusions are notable because of mutually exclusivity with BRAF mutation (Figure 4) as well as the life-prolonging effects of RAF and MEK inhibitors for patients with melanoma harboring BRAF V600E mutations29. RET is frequently activated by mutations in medullary thyroid cancer and inhibitors of multiple tyrosine kinases including RET has been approved for medullary thyroid cancer by the Food and Drug Administration (FDA), while treatment of NTRK1 fusion positive lung cancer cells with a kinase inhibitor led to suppression of cell growth30. Our findings suggest that kinase inhibition may have broad applicability for treatment of thyroid cancers31, 32.
Figure 3. An overview of protein kinase fusions across 13 tumor types.
(A) Bar plots show the fraction of in-frame protein kinase fusions relative to the total number of in frame fusions per tumor type. (B) Recurrent in-frame protein kinase fusion across 13 tumor types (n≥2). Color represents tumor type. (C) The landscape of protein kinase fusions across cancer. The horizontal and vertical axes represent tumor samples and kinase genes, respectively. Genes were ordered based on kinase family annotation. Color bar depicts tumor type.
Figure 4. Significance of RAF family fusions in thyroid cancer.
(A) The top panel indicates frequencies of somatic mutations (lightblue) and significant mutations (pink). To compare the frequency between samples with and without recurrent fusions (n≥2), a Welch’ s t-test was performed. The bottom panel shows a heatmap of fusions and significant gene mutations in 312 thyroid cancers. (B) Position of each domain in BRAF gene and junction points of BRAF fusions. (C) Exon expression plots demonstrated Z-normalized exon expression for each exon in thyroid cancers. Red and blue represent relatively high and low exon expression.
Amongst 357 kinase fusions, the ALK-ROS1-RET lineage, FGFR, and NTRK family kinase fusions have previously been considered as druggable28 and were commonly detected in tumor types including bladder carcinoma (3.3%), glioblastoma (4.4%), head and neck cancer (1.0%), low grade glioma (1.5%), lung adenocarcinoma (1.6%), lung squamous cell carcinoma (2.3%), prostate adenocarcinoma (1.7%), and thyroid carcinoma (8.7%)(Figure 3B and Supplementary Table 10), suggesting a potential for application of kinase inhibitors across tumor types (Figure 3C and Supplementary Table 11). ALK fusions can be targeted by ALK inhibitors and have been reported in non-small cell lung cancer as well as breast, colorectal, esophageal, renal cell, and renal medullary cancers33. We detected ALK fusions in lung adenocarcinoma (0.8%), bladder (0.8%), melanoma (1,3%), and thyroid cancer (0.6%), suggesting that ALK fusions are rare but occur across different tumor lineages.
Chromatin modifier fusions across 13 tumor types
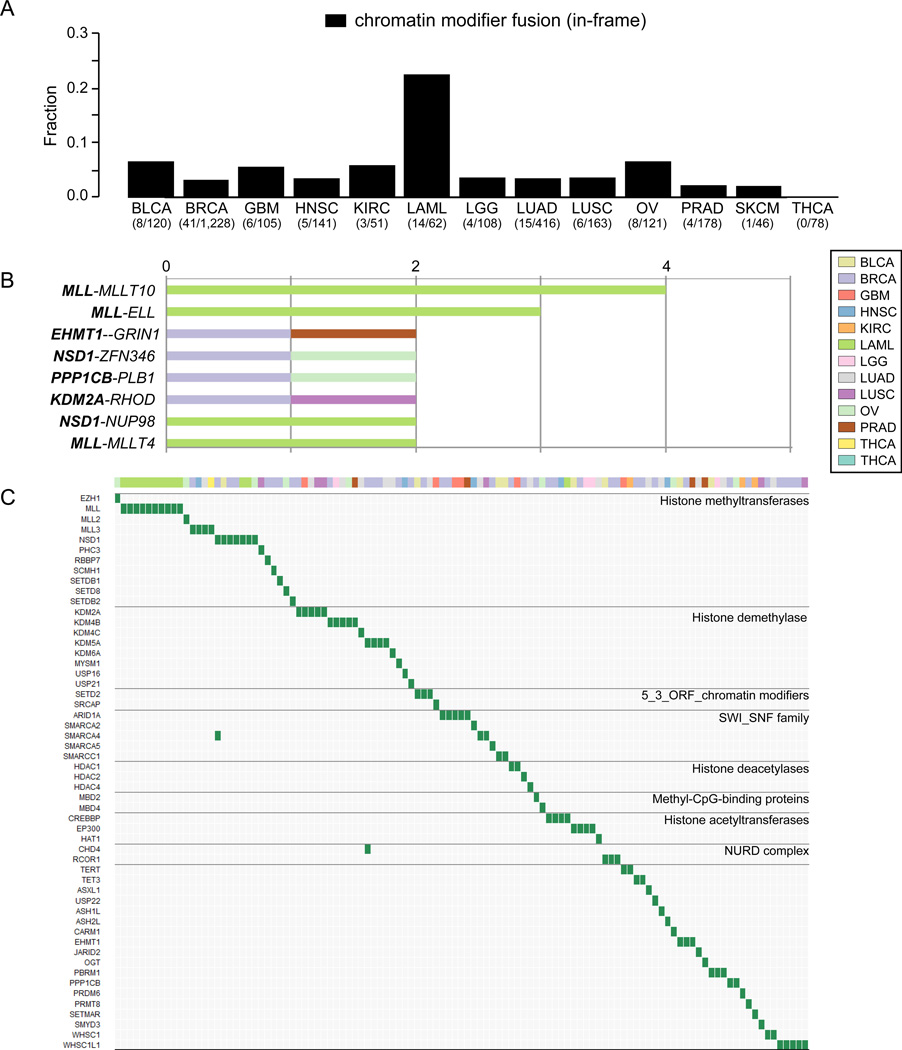
Recent studies demonstrated that genes associated with chromatin modification are frequently mutated and drive many types of cancers, leading to developing new drugs for epigenetic protein families. Inhibitors of DNA methylation and histone deacetylates (HDAC) show antitumor activity34, 35, and have been approved for the treatment of myelodysplastic syndrome36 and cutaneous T cell lymphoma37 by the FDA. In-frame gene fusions involving a chromatin modifier gene were detected in 115 (2.6%) of 4,366 samples (Supplementary Table 12 and 13) and were mutually exclusive with protein kinase fusions (Fisher’s exact test, P = 0.031). The fraction of chromatin modifier fusions in acute myeloid leukemia was higher than other tumor types (Figure 5A), and included four samples with MLL fusions (5.8%) which may be druggable by DOT1L inhibitors38. Although there were only seven recurrent chromatin modifier fusions across 13 tumor types (Figure 5B), fusions related to histone methyltransferases and demethylase families with potential as a target for anticancer therapy were detected in 48 (1.1%) of 4,366 samples (Figure 5C)35. For example, an association of the lysine-specific demethylase 5A gene (KDM5A/JARID1A/RBP2) overexpression with tumorigenesis or metastasis has been previously reported in lung cancer39. The KDM5A JmjC domain plays an important role in demethylating lysine 4 of histone 3 and upregulated of this domain was observed in three of four samples harboring KDM5A fusions (Supplementary Figure 7).
Figure 5. A survey of chromatin modifier fusions across 13 tumor types.
(A) Bar plots show the fraction of in-frame chromatin modifier fusions relative to the total number of in frame fusions per tumor type. (B) Recurrent in-frame chromatin modifier fusions across 13 tumor types (n≥2). Color represents tumor type. (C) The landscape of chromatin modifier fusions across cancer. The horizontal and vertical axes represent tumor samples and chromatin modifier genes, respectively. Genes were ordered based on chromatin modifier class. Color bar depicts tumor type.
A resource of fusion transcripts from The Cancer Genome Atlas
To allow integration of structural transcript variations with other types of molecular data generated by The Cancer Genome Atlas, we developed a user-friendly fusion gene database which is accessible at http://54.84.12.177/PanCanFusV2/. Through a user-friendly web interface, this portal enables users to search fusion transcripts by gene, by fusion, by TCGA patient ID and tumor type.
Discussion
This study presents a bona-fide catalog of fusion transcripts through analysis of 4,730 paired-end RNA sequencing data sets. We comprehensively identified the diversity of fusion transcripts across 13 tumor types, including the association of fusion transcripts with somatic mutation and DNA double strand breaks.
Although the frequency of recurrent fusion transcripts is generally substantially less compared to somatic mutation events40 such as TP53, PIK3CA, or PTEN, the detection of specific events such as the EML4-ALK protein kinase fusion in non-small cell lung cancer has led to development of treatment effectively targeted this lesion28. Importantly, we showed that in-frame and potentially activating NTRK1, and ALK rearrangements are not limited to breast, thyroid, and lung cancer respectively28, 41, but can be detected across cancer at low frequency. Similarly, FGFR related fusions with therapeutic potential have been reported across tumor types10, which was confirmed by our study. Similar to kinase fusions, our cross-sectional fusion list suggests that there may be opportunity for sporadic application of DNA methylation and histone deacetylase inhibitors, such as have been approved for clinical use in hematological malignant tumors35, 42. For instance, Cadot et al. have reported that suppressing HDAC4 causes chromosome segregation defects in p53-deficient tumor cells43 and one lung adenocarcinoma sample harboring HDAC4-SNX18 fusion showed HDAC4 mRNA overexpression and TP53 somatic mutation, suggesting a possible beneficial effect of HDAC inhibitors. We observed a significant anti-correlation between the presence of a transcript fusion and significant gene mutations in most tumor types, which suggested that driver genome and transcriptome rearrangements may occur infrequently but with high relevance to the tumor in which they are detected. Out findings provide a strong rationale for unbiased clinical testing of targetable fusion events. Basket clinical trials in which patients are treated on the basis of gene abnormalities, instead of tumor type tissue of origin, have the potential to overcome the infrequency of druggable events and may particularly evaluated in the context of transcript fusions
Our fusion database (http://54.84.12.177/PanCanFusV2/) would be a largest database of fusion transcripts obtained from pair-end RNA sequencing data based on unified criteria, demonstrating that druggable fusions are not so frequent but relevant across many tumor types. A comprehensive understanding of fusion transcripts across tumor types could facilitate development of new therapeutic strategies for various tumor types based on fusion events.
Methods
Data preparation
TCGA RNA sequencing data were downloaded from Cancer Genome Hub (CGHub, https://cghub.ucsc.edu). In this study, we used RNA sequencing data obtained from 4,730 TCGA samples (4,366 primary tumor and 364 normal tissues) consisting of 13 tumor types (Table 1). To exclude the possibility of tumor cell contamination in normal tissue, we compared gene expression profiles between primary tumor and normal samples by using SAM algorithm44 (Fold change >2 and p < 0.0001) and performed supervised hierarchical clustering using differentially expressed genes for each tumor type. Of 369 normal samples, five samples (one clear cell renal cell cancer, one lung adenocarcinoma, and three thyroid carcinoma) belonging to tumor cluster were excluded in this study (Supplementary Figure 2).
Identification of fusion transcripts
We used the pipeline for RNA sequencing Data Analysis (PRADA, http://bioinformatics.mdanderson.org/Software/PRADA/)17. Briefly, PRADA extracts all best alignments per read from the dual (genome and transcriptome) reference file using BWA45. After initial mapping, the alignments of reads that map to both genome and transcriptome are collapsed into single genome coordinates. Once mapped, reads are filtered out if their best placements are not mapped to multiple genomic coordinates. Quality scores are recalibrated using the Genome Analysis Toolkit (GATK)46. Index files are generated using Samtools47 and duplicate reads are flagged using Picard (http://picard.sourceforge.net). The PRADA fusion module detects fusion transcripts through identification of discordant read pairs and junction spanning reads. Discordant read pairs are paired read ends that map uniquely to different protein-coding genes with orientation consistent with formation of a sense-sense chimera. Junction spanning reads are detected by the construction of a sequence database that holds all possible exon-exon junctions that match the 3’ end of one gene fused to 5’ end of a second gene. Unmapped reads aligned to the database of all hypothetical exon junctions created by using the Ensembl transcriptome reference. Only reads of which the mate pair maps to either of the two fusion partner genes are considered as fusion transcripts. In this study, we extracted fusions (1) with at least two discordant read pairs, (2) at least one junction spanning reads, and (3) without high gene homology between each fusion gene partner (E-value > 0.001). Next, we applied the concept of mutation allele fraction to RNA sequencing data, and calculated the ratio of junction spanning reads to the total number of reads crossing over the junction point in the reference transcript (Supplementary Figure 8). We used the transcript allele fraction (TAF) to exclude artifacts depending on highly expressed transcripts. We included fusion transcripts showing more than 0.01 in TAF of both genes in our fusion list. In addition, we assessed a variety of partner genes for each gene. The partner genes variety was defined as the kinds of chromosome arms in which partner genes were located (Supplementary Figure 9). We calculated random distribution of partner gene variety (permutation: 100,000 times) per number of fusions comprising one specific gene with consideration of gene frequency for each chromosomal arm, and excluded fusions in which partner genes were randomly distributed to various chromosome arms (p < 0.00001).
Next we utilized TCGA level 3 copy number data to scan the existence of breakpoints within 100 Kb from predicted junction point26. We set copy number threshold value as 0.3. We applied fusion transcripts to a four-tier system as follow; Tier 1: fusions harboring at least three discordant read pairs, at least two junction spanning reads, and gene partner uniqueness within a sample48. Tier 2: fusions having at least two discordant read pairs and at least one junction spanning reads, plus breakpoints within 100Kb from predicted junction point. Tier 3: fusions showing high consistency of predicted junction and gene partner uniqueness within a sample as well as having at least two discordant reads pairs and at least one junction spanning reads. Tier 4: other than tier 1 to 3. We used total 7,415 tier 1 and tier 2 fusion transcripts in this study. Fusions that have never been reported were annotated as “novel” based on Mitelman Database of Chromosome Aberrations and Gene Fusions in Cancer (http://cgap.nci.nih.gov/Chromosomes/Mitelman), Cancer genome project (http://www.sanger.ac.uk/research/projects/cancergenome/) and ChimerDB 2.0 (http://biome.ewha.ac.kr:8080/FusionGene/). We included genes overlapping between TSGene: Tumor Suppressor Gene Database (http://bioinfo.mc.vanderbilt.edu/TSGene/)49 and Cancer Gene census (http://cancer.sanger.ac.uk/cancergenome/projects/census/)50 in a list of tumor suppressor gene.
Validation of fusion transcripts
We obtained TCGA whole genome sequence data on 28 glioblastoma, 20 low-grade glioma, 18 melanoma (low pass), and matched normal samples from CGHub. We applied BreakDancer (version 1.12)18 to whole genome sequencing data and identified somatic rearrangements that had 5 or more supporting reads in whole genome sequencing data, or 3 or more supporting reads in low-pass whole genome sequencing data, and were not in matched normal samples. To validate fusion transcripts by using whole genome sequencing data, we set two confidence level (high and medium) and two window size (100Kb and 1Mb). When Break Dancer predicts structural variant involving connecting both gene partners of fusion transcripts or involving one of the gene partners, we defined “high confidence” and “medium confidence”, respectively (Supplementary Figure 9).
Exon expression analysis
TCGA level 3 RNA sequence exon level expression data was obtained from TCGA Data Portal (https://tcga-data.nci.nih.gov/tcga/). The Generic Annotation files (GAF) including annotations for all exon was downloaded from https://tcga-data.nci.nih.gov/tcgafiles/ftp_auth/distro_ftpusers/anonymous/other/GAF/GAF_bundle/outputs/TCGA.Sept2010.09202010.gaf. We used exon quantification text file to perform Z-normalization for each exon expression in each tumor type. To examine the association of fusion events with gene expression, we performed Welch’s t-test score between exons before and after junction point for each gene.
Copy number alteration analysis
TCGA level 3 copy number data based on Affymetrix SNP 6.0 array was obtained from TCGA Data Portal. We calculated DNA segments per sample as a measure of genome instability. To detect high frequent region of copy number alterations and copy number status for each gene for each tumor type, we used the genome identification of significant targets in cancer (GISTIC) algorithm (version 2)51. Copy number levels were categorized into five levels (high and low-level amplification, high and low-level deletion, and no alteration).
Mutation data analysis
We downloaded somatic mutation data (syn1710680) from Synapse (https://www.synapse.org/#) and determined significant mutated genes per tumor type by MutSigCV40. Of 13 tumor types, melanoma samples with recurrent fusion had no mutation data. For each tumor type, we extracted overlapped samples between fusion and mutation data sets to compare mutation rate and significant mutation frequency between samples with and without recurrent fusions. Low grade glioma, prostate adenocarcinoma and melanoma data sets in which no sample with recurrent fusions was detected in the overlapped data set were excluded in this analysis.
Protein expression data analysis
We downloaded reverse phase protein array (RPPA) data in TCGA breast cancer from The Cancer Protein Atlas (TCPA) website (http://app1.bioinformatics.mdanderson.org/tcpa/_design/basic/index.html)52. We focused on ER alpha protein expression in ESR1 fusion positive breast cancer samples and compared ER alpha and phosphorylated ER alpha expression between samples with ESR1 fusion positive and negative breast cancer samples.
Statistical analysis
We conduced all computations with R 3.0.153 and used standard statistical tests as appropriate.
Supplementary Material
Acknowledgments
The results published here are in whole or part based upon data generated by The Cancer Genome Atlas project established by the NCI and NHGRI. Information about TCGA and the investigators and institutions who constitute the TCGA research network can be found at “http://cancergenome.nih.gov”. This work was supported in part by the Mary K. Chapman Foundation, the Michael & Susan Dell Foundation (honoring Lorraine Dell), U.S. National Cancer Institute (NCI; MD Anderson TCGA Genome Data Analysis Center) grant numbers CA143883 and CA083639, and MD Anderson Cancer Center Support Grant P30 CA016672 (the Bioinformatics Shared Resource).
Footnotes
Conflict of interest
The authors declare no conflict of interest.
Supplementary Information
Supplementary Information accompanies the paper on the Oncogene website (http://www.nature.com/onc).
References
- 1.Mitelman F, Johansson B, Mertens F. The impact of translocations and gene fusions on cancer causation. Nat Rev Cancer. 2007;7:233–245. doi: 10.1038/nrc2091. [DOI] [PubMed] [Google Scholar]
- 2.Nowell PC, Hungerford DA. A minute chromosome in human chronic granulocytic leukemia. Science. 1960;132:1488–1501. [Google Scholar]
- 3.Kantarjian H, Shah NP, Hochhaus A, Cortes J, Shah S, Ayala M, et al. Dasatinib versus imatinib in newly diagnosed chronic-phase chronic myeloid leukemia. N Engl J Med. 2010;362:2260–2270. doi: 10.1056/NEJMoa1002315. [DOI] [PubMed] [Google Scholar]
- 4.Soda M, Choi YL, Enomoto M, Takada S, Yamashita Y, Ishikawa S, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature. 2007;448:561–566. doi: 10.1038/nature05945. [DOI] [PubMed] [Google Scholar]
- 5.Shaw AT, Kim DW, Nakagawa K, Seto T, Crino L, Ahn MJ, et al. Crizotinib versus chemotherapy in advanced ALK-positive lung cancer. N Engl J Med. 2013;368:2385–2394. doi: 10.1056/NEJMoa1214886. [DOI] [PubMed] [Google Scholar]
- 6.Meyerson M, Gabriel S, Getz G. Advances in understanding cancer genomes through second-generation sequencing. Nature reviews Genetics. 2010;11:685–696. doi: 10.1038/nrg2841. [DOI] [PubMed] [Google Scholar]
- 7.Watson IR, Takahashi K, Futreal PA, Chin L. Emerging patterns of somatic mutations in cancer. Nature reviews Genetics. 2013;14:703–718. doi: 10.1038/nrg3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Singh D, Chan JM, Zoppoli P, Niola F, Sullivan R, Castano A, et al. Transforming fusions of FGFR and TACC genes in human glioblastoma. Science. 2012;337:1231–1235. doi: 10.1126/science.1220834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guo G, Sun X, Chen C, Wu S, Huang P, Li Z, et al. Whole-genome and whole-exome sequencing of bladder cancer identifies frequent alterations in genes involved in sister chromatid cohesion and segregation. Nat Genet. 2013;45:1459–1463. doi: 10.1038/ng.2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu YM, Su F, Kalyana-Sundaram S, Khazanov N, Ateeq B, Cao X, et al. Identification of targetable FGFR gene fusions in diverse cancers. Cancer Discov. 2013;3:636–647. doi: 10.1158/2159-8290.CD-13-0050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Parker M, Mohankumar KM, Punchihewa C, Weinlich R, Dalton JD, Li Y, et al. C11orf95-RELA fusions drive oncogenic NF-kappaB signalling in ependymoma. Nature. 2014 doi: 10.1038/nature13109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Honeyman JN, Simon EP, Robine N, Chiaroni-Clarke R, Darcy DG, Lim II, et al. Detection of a recurrent DNAJB1-PRKACA chimeric transcript in fibrolamellar hepatocellular carcinoma. Science. 2014;343:1010–1014. doi: 10.1126/science.1249484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shah N, Lankerovich M, Lee H, Yoon JG, Schroeder B, Foltz G. Exploration of the gene fusion landscape of glioblastoma using transcriptome sequencing and copy number data. BMC Genomics. 2013;14:818. doi: 10.1186/1471-2164-14-818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shern JF, Chen L, Chmielecki J, Wei JS, Patidar R, Rosenberg M, et al. Comprehensive genomic analysis of rhabdomyosarcoma reveals a landscape of alterations affecting a common genetic axis in fusion-positive and fusion-negative tumors. Cancer Discov. 2014;4:216–231. doi: 10.1158/2159-8290.CD-13-0639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seo JS, Ju YS, Lee WC, Shin JY, Lee JK, Bleazard T, et al. The transcriptional landscape and mutational profile of lung adenocarcinoma. Genome Res. 2012;22:2109–2119. doi: 10.1101/gr.145144.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu C, Wyatt AW, Lapuk AV, McPherson A, McConeghy BJ, Bell RH, et al. Integrated genome and transcriptome sequencing identifies a novel form of hybrid and aggressive prostate cancer. J Pathol. 2012;227:53–61. doi: 10.1002/path.3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Torres-Garcia W, Zheng S, Sivachenko A, Vegesna R, Wang Q, Yao R, et al. PRADA: Pipeline for RNA sequencing Data Analysis. Bioinformatics. 2014 doi: 10.1093/bioinformatics/btu169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen K, Wallis JW, McLellan MD, Larson DE, Kalicki JM, Pohl CS, et al. Break Dancer: an algorithm for high-resolution mapping of genomic structural variation. Nature methods. 2009;6:677–681. doi: 10.1038/nmeth.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zack TI, Schumacher SE, Carter SL, Cherniack AD, Saksena G, Tabak B, et al. Pan-cancer patterns of somatic copy number alteration. Nat Genet. 2013;45:1134–1140. doi: 10.1038/ng.2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kalyana-Sundaram S, Shankar S, Deroo S, Iyer MK, Palanisamy N, Chinnaiyan AM, et al. Gene fusions associated with recurrent amplicons represent a class of passenger aberrations in breast cancer. Neoplasia. 2012;14:702–708. doi: 10.1593/neo.12914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Edgren H, Murumagi A, Kangaspeska S, Nicorici D, Hongisto V, Kleivi K, et al. Identification of fusion genes in breast cancer by paired-end RNA-sequencing. Genome Biol. 2011;12:R6. doi: 10.1186/gb-2011-12-1-r6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim P, Yoon S, Kim N, Lee S, Ko M, Lee H, et al. Chimer DB 2.0--a knowledgebase for fusion genes updated. Nucleic Acids Res. 2010;38:D81–D85. doi: 10.1093/nar/gkp982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mitelman F, Johansson B, Mertens F. Fusion genes and rearranged genes as a linear function of chromosome aberrations in cancer. Nat Genet. 2004;36:331–334. doi: 10.1038/ng1335. [DOI] [PubMed] [Google Scholar]
- 24.Tomlins SA, Rhodes DR, Perner S, Dhanasekaran SM, Mehra R, Sun XW, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–648. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 25.The Cancer Genome Atlas Research N. Genomic and Epigenomic Landscapes of Adult De Novo Acute Myeloid Leukemia. N Engl J Med. 2013 doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zheng S, Fu J, Vegesna R, Mao Y, Heathcock LE, Torres-Garcia W, et al. A survey of intragenic breakpoints in glioblastoma identifies a distinct subset associated with poor survival. Genes Dev. 2013 doi: 10.1101/gad.213686.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brennan CW, Verhaak RG, McKenna A, Campos B, Noushmehr H, Salama SR, et al. The somatic genomic landscape of glioblastoma. Cell. 2013;155:462–477. doi: 10.1016/j.cell.2013.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shaw AT, Hsu PP, Awad MM, Engelman JA. Tyrosine kinase gene rearrangements in epithelial malignancies. Nat Rev Cancer. 2013;13:772–787. doi: 10.1038/nrc3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Flaherty KT, Infante JR, Daud A, Gonzalez R, Kefford RF, Sosman J, et al. Combined BRAF and MEK inhibition in melanoma with BRAF V600 mutations. N Engl J Med. 2012;367:1694–1703. doi: 10.1056/NEJMoa1210093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vaishnavi A, Capelletti M, Le AT, Kako S, Butaney M, Ercan D, et al. Oncogenic and drug-sensitive NTRK1 rearrangements in lung cancer. Nat Med. 2013 doi: 10.1038/nm.3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wells SA, Jr, Gosnell JE, Gagel RF, Moley J, Pfister D, Sosa JA, et al. Vandetanib for the treatment of patients with locally advanced or metastatic hereditary medullary thyroid cancer. J Clin Oncol. 2010;28:767–772. doi: 10.1200/JCO.2009.23.6604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lam ET, Ringel MD, Kloos RT, Prior TW, Knopp MV, Liang J, et al. Phase II clinical trial of sorafenib in metastatic medullary thyroid cancer. J Clin Oncol. 2010;28:2323–2330. doi: 10.1200/JCO.2009.25.0068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hallberg B, Palmer RH. Mechanistic insight into ALK receptor tyrosine kinase in human cancer biology. Nat Rev Cancer. 2013;13:685–700. doi: 10.1038/nrc3580. [DOI] [PubMed] [Google Scholar]
- 34.Hojfeldt JW, Agger K, Helin K. Histone lysine demethylases as targets for anticancer therapy. Nature reviews Drug discovery. 2013;12:917–930. doi: 10.1038/nrd4154. [DOI] [PubMed] [Google Scholar]
- 35.Arrowsmith CH, Bountra C, Fish PV, Lee K, Schapira M. Epigenetic protein families: a new frontier for drug discovery. Nature reviews Drug discovery. 2012;11:384–400. doi: 10.1038/nrd3674. [DOI] [PubMed] [Google Scholar]
- 36.Kantarjian H, Issa JP, Rosenfeld CS, Bennett JM, Albitar M, DiPersio J, et al. Decitabine improves patient outcomes in myelodysplastic syndromes: results of a phase III randomized study. Cancer. 2006;106:1794–1803. doi: 10.1002/cncr.21792. [DOI] [PubMed] [Google Scholar]
- 37.Olsen EA, Kim YH, Kuzel TM, Pacheco TR, Foss FM, Parker S, et al. Phase IIb multicenter trial of vorinostat in patients with persistent, progressive, or treatment refractory cutaneous T-cell lymphoma. J Clin Oncol. 2007;25:3109–3115. doi: 10.1200/JCO.2006.10.2434. [DOI] [PubMed] [Google Scholar]
- 38.Daigle SR, Olhava EJ, Therkelsen CA, Majer CR, Sneeringer CJ, Song J, et al. Selective killing of mixed lineage leukemia cells by a potent small-molecule DOT1L inhibitor. Cancer Cell. 2011;20:53–65. doi: 10.1016/j.ccr.2011.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Teng YC, Lee CF, Li YS, Chen YR, Hsiao PW, Chan MY, et al. Histone demethylase RBP2 promotes lung tumorigenesis and cancer metastasis. Cancer Res. 2013;73:4711–4721. doi: 10.1158/0008-5472.CAN-12-3165. [DOI] [PubMed] [Google Scholar]
- 40.Lawrence MS, Stojanov P, Polak P, Kryukov GV, Cibulskis K, Sivachenko A, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013 doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Asmann YW, Hossain A, Necela BM, Middha S, Kalari KR, Sun Z, et al. A novel bioinformatics pipeline for identification and characterization of fusion transcripts in breast cancer and normal cell lines. Nucleic Acids Res. 2011;39:e100. doi: 10.1093/nar/gkr362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gough SM, Lee F, Yang F, Walker RL, Zhu YJ, Pineda M, et al. NUP98-PHF23 is a chromatin modifying oncoprotein that causes a wide array of leukemias sensitive to inhibition of PHD domain histone reader function. Cancer Discov. 2014 doi: 10.1158/2159-8290.CD-13-0419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cadot B, Brunetti M, Coppari S, Fedeli S, de Rinaldis E, Dello Russo C, et al. Loss of histone deacetylase 4 causes segregation defects during mitosis of p53-deficient human tumor cells. Cancer Res. 2009;69:6074–6082. doi: 10.1158/0008-5472.CAN-08-2796. [DOI] [PubMed] [Google Scholar]
- 44.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The Sequence Alignment/Map format and SAM tools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cancer Genome Atlas Research N. Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature. 2013;499:43–49. doi: 10.1038/nature12222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhao M, Sun J, Zhao Z. TS Gene: a web resource for tumor suppressor genes. Nucleic Acids Res. 2013;41:D970–D976. doi: 10.1093/nar/gks937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Futreal PA, Coin L, Marshall M, Down T, Hubbard T, Wooster R, et al. A census of human cancer genes. Nat Rev Cancer. 2004;4:177–183. doi: 10.1038/nrc1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mermel CH, Schumacher SE, Hill B, Meyerson ML, Beroukhim R, Getz G. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol. 2011;12:R41. doi: 10.1186/gb-2011-12-4-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li J, Lu Y, Akbani R, Ju Z, Roebuck PL, Liu W, et al. TCPA: a resource for cancer functional proteomics data. Nature methods. 2013;10:1046–1047. doi: 10.1038/nmeth.2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.R Core Team. R: A language and environment for statistical computing. 2013 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.