Abstract
The Gram-negative bacterium and opportunistic pathogen Serratia marcescens causes ocular infections in healthy individuals. Secreted protease activity was characterized from 44 ocular clinical isolates, and a higher frequency of protease-positive strains was observed among keratitis isolates than among conjunctivitis isolates. A positive correlation between protease activity and cytotoxicity to human corneal epithelial cells in vitro was determined. Deletion of prtS in clinical keratitis isolate K904 reduced, but did not eliminate, cytotoxicity and secreted protease production. This indicated that PrtS is necessary for full cytotoxicity to ocular cells and implied the existence of another secreted protease(s) and cytotoxic factors. Bioinformatic analysis of the S. marcescens Db11 genome revealed three additional open reading frames predicted to code for serralysin-like proteases noted here as slpB, slpC, and slpD. Induced expression of prtS and slpB, but not slpC and slpD, in strain PIC3611 rendered the strain cytotoxic to a lung carcinoma cell line; however, only prtS induction was sufficient for cytotoxicity to a corneal cell line. Strain K904 with deletion of both prtS and slpB genes was defective in secreted protease activity and cytotoxicity to human cell lines. PAGE analysis suggests that SlpB is produced at lower levels than PrtS. Purified SlpB demonstrated calcium-dependent and AprI-inhibited protease activity and cytotoxicity to airway and ocular cell lines in vitro. Lastly, genetic analysis indicated that the type I secretion system gene, lipD, is required for SlpB secretion. These genetic data introduce SlpB as a new cytotoxic protease from S. marcescens.
INTRODUCTION
Microbial keratitis (MK) is a blinding disease with poor visual outcomes even with effective antibiotics and antifungal agents (1–3). In addition to being a major cause of hospital-acquired infections, such as ventilator-associated pneumonia (4, 5), Serratia marcescens is a common cause of MK (1, 2, 6–8), yet the virulence factors involved in this process are poorly understood. In general, bacterial secreted factors, including hemolysins and proteases, contribute to the pathogenesis of bacterial corneal infections (9–12). There are several studies characterizing the importance of proteases from S. marcescens isolates in ocular infections, most recently reviewed by Matsumoto (13). Serralysin is a cytotoxic factor capable of killing mammalian cells in vitro (14, 15). Purified serralysin is sufficient to cause keratitis when injected into rabbit eyes and promotes the spread of bacteria through the corneal stroma (16–18). Additionally, serralysin can degrade components of the human immune system, such as PAR-2, in vitro, and this may impact corneal pathogenesis (13, 19–21). Serralysin, and serralysin family metalloproteases, such as alkaline protease from Pseudomonas aeruginosa, can proteolyze mammalian cell surface proteins, thereby modulating cell physiology. A recent example is the protease-mediated activation of the epithelial sodium channel, leading to a cell surface that is more amenable to bacterial colonization (22, 23). Several bacteria, including Serratia species, invade eukaryotic airway cells in a protease-dependent manner (24–27).
Few ocular clinical isolates of S. marcescens have been characterized for their ability to secrete proteases. These include 3 strains analyzed by Hume et al. in Australia (28) and 3 strains analyzed by Pinna et al. in Italy (29). In this study, we characterized secreted protease activity from more than 40 ocular isolates of S. marcescens collected at a tertiary care hospital in the northeastern United States. Arbitrarily selected isolates were tested for cytotoxicity, and higher levels of cytotoxicity were associated with high-protease-producing strains.
Multiple proteases have been biochemically isolated from a highly virulent keratitis isolate of S. marcescens, but the genes responsible for these proteins were not determined (30). Only the gene for the serralysin protease, prtS, has been cloned and characterized by sequence analysis (31). Here, we report the identification of three uncharacterized serralysin-like proteases in the genome of S. marcescens. To determine which of the metalloproteases mediated cytotoxicity, we cloned and induced expression of each of these genes in trans and in cis in a nearly noncytotoxic laboratory strain of S. marcescens. Only two of the proteases conferred cytotoxicity and elevated secreted protease activity: PrtS (serralysin) and the novel protease named serralysin-like protease B (SlpB). Further genetic analysis using a clinical corneal isolate suggested that SlpB together with PrtS contributes to secreted protease activity by S. marcescens.
MATERIALS AND METHODS
Bacterial growth and strains.
Bacteria were cultured in lysogeny broth (LB) or LB agar (LB broth supplemented with 1.5% [wt/vol] agar) (32) and incubated at 30°C. Milk agar plates were prepared using brain heart infusion agar supplemented with 1% (wt/vol) skimmed milk powder. Liquid cultures were incubated using a New Brunswick TC-7 rotor at speed setting 8 (∼62 rpm). S. marcescens strains used for genetic analysis are listed in Table 1. Ocular clinical isolates were obtained from the Charles T. Campbell Laboratory of Ophthalmic Microbiology at the University of Pittsburgh Medical Center. Escherichia coli strains used in this study were EC100D-pir-116 (Epicentre) and S17-1 λpir (33); Saccharomyces cerevisiae strain InvSc1 (Invitrogen) was used for cloning.
TABLE 1.
S. marcescens strains and plasmids
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| Strains | ||
| CMS376 | WT PIC strain no. 3611 | Presque Isle Cultures |
| C23M13 | CMS786 with lipD::Tn | 37 |
| K904 | Contact lens-associated keratitis isolate | 60 |
| CMS2853 | K904 with prtS deletion mutation | This study |
| CMS3809 | K904 with slpB::pMQ118 | This study |
| CMS3810 | K904 ΔprtS slpB::pMQ118 | This study |
| CMS4063 | CMS376 lipD::Tn | This study |
| CMS4097 | K904 with slpB deletion mutation | This study |
| CMS4098 | K904 ΔprtS ΔslpB | This study |
| Plasmids | ||
| pMQ118 | nptII, rpsL, oriT, oriR6K, URA3, CEN6/ARSH4 | 39 |
| pMQ125 | orip15a, PBAD-lacZa, oripRO1600, oriT, URA3, CEN6/ARSH4 | |
| pMQ131 | oripBBR1, aphA-3, oriT, URA3, CEN6/ARSH4 | 39 |
| pMQ132 | oripBBR1, aacC1, oriT, URA3, CEN6/ARSH4 | 39 |
| pStvZ3 | oriR6K lacZ nptII, oriT, URA3, CEN6/ARSH4 | 60 |
| pMQ200 | oriR6K, nptII, oriT, URA3, CEN6/ARSH4, PBAD-lacZα | 39 |
| pMQ218 | pMQ118 + slpB internal fragment | This study |
| pMQ236 | oriR6K, nptII, rpsL, oriT, URA3, CEN6/ARSH4, I-SceI site | 39 |
| pMQ240 | oripSC101ts, aacC1, oriT, Plac-I-SceI, URA3, CEN6/ARSH4 | 39 |
| pMQ262 | pMQ200 + pigAB′ for induction of prodigiosin | 61 |
| pMQ263 | pMQ236 + prtS | This study |
| pMQ310 | Same as pMQ236 with hygromycin resistance marker hph | 62 |
| pMQ322 | pMQ263 with prtS AgeI-AgeI deletion allele | This study |
| pMQ356 | pMQ125 + His7-prtS | This study |
| pMQ430 | pMQ131 + prtS | This study |
| pMQ431 | pMQ131 + slpB | This study |
| pMQ436 | pMQ125 + His7-slpB | This study |
| pMQ437 | pMQ125 + His7-slpC | This study |
| pMQ444 | pMQ125 + His7-slpD | This study |
| pMQ445 | pMQ310 + lipD::Tn (aacC-1 marker) | This study |
| pMQ460 | pMQ236 + sacB, allelic replacement vector | This study |
| pMQ493 | pMQ460 + slpB | This study |
| pMQ497 | pMQ460 + slpB-PvuI-PvuI deletion allele | This study |
| pMQ510 | pMQ200 + prtS | This study |
| pMQ511 | pMQ200 + slpB | This study |
| pMQ512 | pMQ200 + slpC | This study |
| pMQ513 | pMQ200 + slpD | This study |
Tissue culture and cytotoxicity assays.
Human lung carcinoma line A549 cells (34) were propagated and maintained in tissue culture medium consisting of Dulbecco's modified Eagle's medium (MEM) supplemented with 10% fetal bovine serum (Sigma). Human corneal limbal epithelial (HCLE) cells (35) were maintained in keratinocyte serum-free medium with l-glutamine (Life Technologies), supplemented with bovine pituitary extract (25 μg/ml), embryonic growth factor (0.2 ng/ml), and CaCl2 (0.3 mM). Cytotoxicity was measured using alamarBlue and Presto Blue reagents (Life Technologies) as previously described by Wingard et al. (36). Briefly, bacterial cultures were grown for 18 to 20 h in LB medium with antibiotics if a plasmid was used, and l-arabinose (0.2%) was added to induce expression from the PBAD promoter when desired. Cultures were normalized by adjustment to an optical density at 600 nm (OD600) of 2.0 by addition of LB medium. Culture aliquots were added to microcentrifuge tubes and spun at 14,000 × g for 2 min, followed by filter sterilization of the supernatants using 0.22-μm filters. Two hundred-microliter quantities of filtered bacterial cultures were added to epithelial cell line layers in 24-well dishes with 300 μl of culture medium. After 4 h of incubation, cell layers were washed three times with 0.5 ml of phosphate-buffered saline (PBS) and suspended in culture medium with alamarBlue or Presto Blue viability indicators (Life Technologies) and measured as previously described (36). Alternatively, cell layers were not washed following challenge and were analyzed with Presto Blue. In all cases, cytotoxicity values were determined relative to positive- and negative-control values; i.e., cytotoxicity of the mock treatment (negative control) was set to 0% cytotoxicity, and that of detergent treatment (Triton X-100 at 0.25% [vol/vol]) was set to 100% cytotoxicity. The percent cytotoxicity was determined using the following formula: 100 × [(mock value − experimental value)/(mock value − detergent treatment value)].
Protease assay.
Milk agar plates were used for primary analysis of secreted protease activity. Bacteria were patched on milk agar plates and incubated for 24 to 48 h at 30°C, after which zones of clearance around the colonies were noted. For quantitative analysis, protease activity was measured from normalized spent culture supernatants (OD600 = 2.0) using azocasein (Sigma) as a colorimetric substrate as previously described (37, 38). Normalized cultures were centrifuged and filtered to remove bacteria. A 150-μl aliquot of the filtered cell-free supernatant was mixed with 250 μl of azocasein (2% [wt/vol]) and incubated for 30 min at 37°C. The reaction was stopped by addition of 1.2 ml of 0.6 N trichloroacetic acid, and the mixture was incubated for 15 min on ice. The tubes were centrifuged at 8,000 × g for 10 min. Cold NaOH (1.4 ml of 1 N solution) was added to wells of a 24-well dish, and 1.2 ml of the centrifuged culture mixture was added to the NaOH solution. Colorimetric analysis of liberated azol dye was measured at 440 nm with a plate reader and divided by the original culture OD600.
Molecular biology.
The N-terminal polyhistidine-tagged slpB, slpC, and slpD genes under the control of the PBAD promoter on p15a-based plasmid pMQ125 (39) were generated by replacing prtS on pMQ356 (23) but maintaining the N-terminal His7 tag. This was done by digesting pMQ356 with SalI and SmaI that cut in the prtS gene and then replacing the entire prtS gene using primers that amplify the slp genes and have regions of homology to the His7 tag and vector backbone. The recombination was carried out using Saccharomyces cerevisiae for in vivo recombination (39, 40).
To place the chromosomal candidate protease genes and prtS under the control of PBAD promoter, the open reading frames (ORFs) for prtS, and the candidate proteases were cloned into pMQ200 (39), an oriR6K-based suicide plasmid, using S. cerevisiae recombineering. The plasmid was introduced into the PIC3611 strain by conjugation, and the insertions were verified by PCR.
To generate chromosomal deletions of prtS, the full-length prtS gene was cloned into the pMQ236 (39) allelic replacement vector, generating pMQ263. This plasmid was then digested with AgeI, which cuts in three places in prtS, and then recircularized with T4 ligase. The resulting mutant allele retains 108 bp of the prtS upstream of the AgeI site and 618 bp downstream of the AgeI site. The prtS deletion plasmid was moved into K904, followed by introduction of pMQ337, which has the I-SceI gene under the control of the PBAD promoter. After addition of I-SceI with l-arabinose, chromosomal prtS deletions were screened for kanamycin-susceptible isolates that had lost pMQ236, first by a reduction in protease activity on milk agar plates and then by PCR using primers outside the prtS ORF.
To mutate slpB by plasmid insertion, a 606-bp internal fragment of slpB was cloned into pMQ118 (39). The resulting plasmid, pMQ218, was introduced into K904 by conjugation, and kanamycin-resistant isolates were screened by PCR for the plasmid disruption of the slpB gene. The plasmid inserts after bp 747 out of 1,419 bp, which truncates the slpB gene after codon 249 out of 472 amino acids, and adds on 28 codons from the inserting plasmid sequence until reaching a stop codon. The resulting predicted 277-amino-acid polypeptide has the protease domain but lacks the secretion domain. The insertion was verified by PCR analysis.
To generate the slpB deletion allele, the slpB ORF along with 602 bp of upstream DNA and 523 bp of downstream DNA was cloned into allelic replacement vector pMQ460. A PvuI-PvuI fragment was removed from the slpB gene, removing 657 bp. This in-frame deletion mutation is predicted to remove 219 amino acids from the N-terminal half of the 472-amino-acid protein. The deleted residues from amino acid 49 to 187 include the predicted catalytic site.
LipD mutation.
Strain C23M13 with a transposon mutation in the lipD ORF was identified in a genetic screen for mutations that suppress the elevated protease phenotype of crp mutants in S. marcescens PIC3611 (37). The lipD::Tn allele was cloned into allelic replacement vector pMQ310 (Table 1) to generate pMQ445. The resulting plasmid was moved into PIC3611 by conjugation, and allelic replacement was carried out. The introduced lipD mutation was verified by PCR and loss of lipase and protease activities using plate-based assays as previously described (37).
PAGE analysis of secretomes.
Trichloroacetic acid-precipitated secretomes that had been normalized to an OD600 of 2 were separated on 8 to 16% polyacrylamide gels and stained with Coomassie brilliant blue. Gels were imaged using an infrared imager (Li-Cor; Odyssey) using the 700-nm channel. Band intensities from the digital images were analyzed using ImageJ Software (NIH). Bands of interest were cut out with a clean razor, and proteins were identified using mass spectrometry at the University of Pittsburgh Biomedical Mass Spectrometry Center.
Reverse transcription-PCR (RT-PCR).
RNA was extracted from cultures grown in LB broth to an OD600 of 3.0 using a Qiagen RNA Easy kit, and cDNA preparations were performed as previously described (41). Two rounds of DNase treatment were performed to remove chromosomal DNA. 16S rRNA gene primers were used to normalize signals and to test for chromosomal DNA contamination (see Table S1 in the supplemental material). Control reactions lacking reverse transcriptase were included and verified a lack of chromosomal DNA contamination. The experiment was performed twice with independent RNA samples, with similar results.
SlpB and AprI purification and analysis.
The slpB open reading frame expressed from arabinose regulated expression plasmid pMQ436 in E. coli EC100D pir-116 cells. A culture was grown overnight in the presence of gentamicin, and 10 ml was used as an inoculum for His7-slpB induction in 1-liter cultures. The cultures were grown in LB medium supplemented with gentamicin, and 0.2% (wt/vol) arabinose was added to the culture when it achieved an OD600 of 0.8. Protein was expressed at 37°C for 4 to 6 h. The SlpB protein was purified from the insoluble material under denaturing conditions by following protocols previously described for PrtS and AprA (23). In vitro protein refolding and protease activity were assessed using a fluorescent peptide and casein substrates, as previously described (42). The AprI protease inhibitor was expressed and purified as previously described (23).
Statistical analysis.
Two-tailed Student's t test, one-way analysis of variance (ANOVA) with Tukey's posttest, and Pearson's correlation were performed with Prism 5; significance was set to a P value of <0.05.
RESULTS
Secreted protease activity from ocular clinical isolates.
Protease activity, secreted by ocular clinical isolates of S. marcescens, was analyzed. Isolates were obtained from patients with conjunctivitis (n = 7), endophthalmitis (n = 2), and keratitis (n = 35). Bacteria were patched onto milk agar plates, and after 24 h at 30°C, zones of clearance around the bacteria were counted as a positive indication of secreted protease activity, with any zone of ≥1 mm being considered positive. A total of 38 were positive (86%) for clearance at 24 h, although all produced at least a small zone of clearance by 48 to 72 h. Similar results were observed at 37°C; for example, clearing zones on milk plates around patches of strain K904 were 3.6 ± 0.3 mm at 30°C and 3.6 ± 0.4 mm at 37°C (n ≥ 7; P = 0.96). By 48 h the zones increased to 4.9 ± 0.8 mm at 30°C and 5.5 ± 0.7 mm at 37°C (n = 21; P = 0.02).
Three of the seven conjunctivitis isolates had low levels of proteolysis on milk agar plates (no zone by 24 h), whereas only 3 of the 35 keratitis isolates were protease deficient at 24 h (P = 0.0478, Fisher's exact test). As previously reported, we observed that the laboratory strain PIC3611 used in this study did not produce a zone of clearing at 24 h but did exhibit a small zone of protease activity on milk plates by 48 h (37).
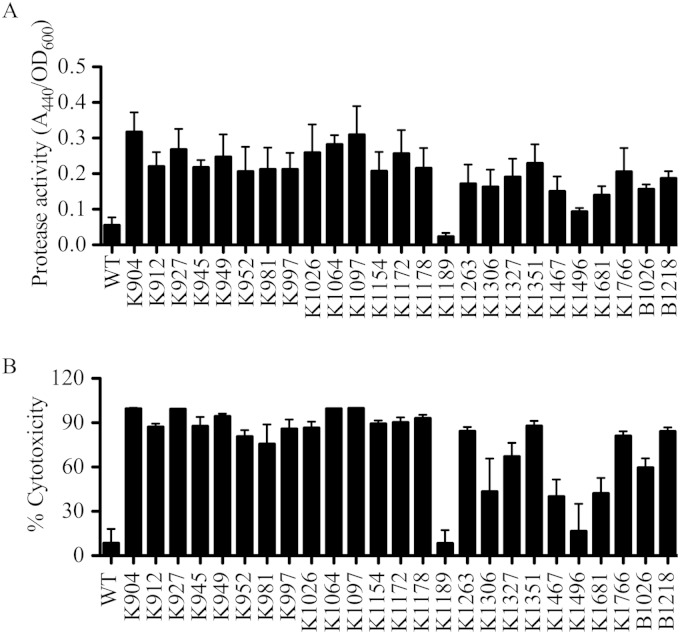
Secreted protease activity was measured from culture supernatants of a subset of the strains analyzed with milk agar plates. Since keratitis is the most frequent ocular infection caused by S. marcescens, further analysis primarily used keratitis isolates. Protease activity is shown in Fig. 1A and followed a strain-dependent pattern. Of the 21 tested keratitis strains, all but 2 (K1189 and K1496) demonstrated elevated levels of proteolysis relative to that of laboratory strain PIC3611 (P < 0.05, ANOVA with Dunnett's multiple-comparison test).
FIG 1.
Secreted protease activity by ocular clinical isolates of S. marcescens correlates with cytotoxicity to human corneal cells. (A) Secreted protease activity from strain PIC3611 (WT) and ocular clinical isolates of S. marcescens measured using azocasein. (B) Cytotoxicity of S. marcescens secreted factors to a human ocular epithelial cell line (HCLE).
Cytotoxicity of ocular clinical isolates to a corneal cell line in vitro is strain dependent.
Cytotoxicity of secreted components produced by ocular clinical isolates and laboratory strain PIC3611 to HCLE cells was investigated. In contrast to the laboratory strain, many of the clinical isolates were highly toxic to the HCLE cell layers (Fig. 1B). Cytotoxicity and protease activity of supernatants from PIC3611 and clinical conjunctivitis and keratitis isolates were found to have a positive correlation (Pearson's r = 0.925; P < 0.001 (see Fig. S1 in the supplemental material).
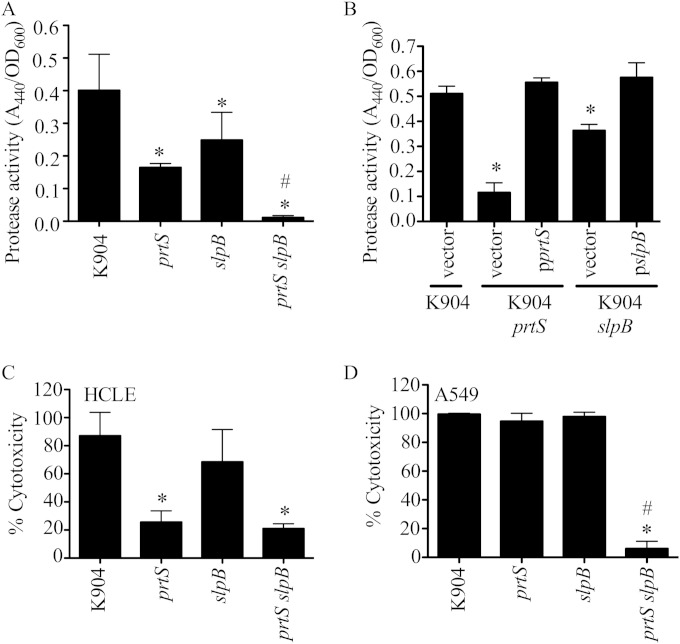
Mutation of prtS in clinical isolate K904 confers loss of secreted protease and cytotoxicity to human epithelial cells.
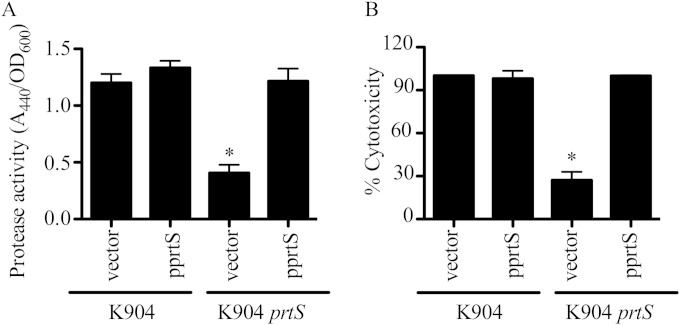
To test whether PrtS contributes to the cytotoxicity of S. marcescens keratitis isolates to corneal cells in vitro, which has not previously been reported, a deletion allele of the prtS gene was imparted to the chromosome of strain K904 by allelic replacement. This in-frame deletion allele is missing amino acids 37 to 297 of the predicted protein, which includes the zinc-dependent metalloprotease domain. Secreted fractions from the K904 ΔprtS mutant strain were defective in protease activity, with a >50% reduction, and this deficit could be complemented by the intact prtS gene on a plasmid (Fig. 2A). Cytotoxicity to human corneal cells was likewise reduced (Fig. 2B). It was of interest that the secreted protease activity and cytotoxicity were not completely eliminated in supernatants of the K904 ΔprtS mutant (mock-treated cells had 0% cytotoxicity, and LB medium had no detectable protease activity [data not shown]), suggesting a contribution by another secreted protease(s).
FIG 2.
Serralysin (PrtS) is required for the majority of secreted protease activity and cytotoxic power of supernatants from a keratitis isolate of S. marcescens. (A) Secreted protease activity from bacterial supernatants measured by azocasein (n = 7). (B) Cytotoxicity of secreted supernatants to HCLE cells (n = 6). Vector = pMQ125, pprtS = pMQ356. Mock-treated cells had 0% cytotoxicity, and detergent-treated cells had 100% cytotoxicity. Means and standard deviations are shown. The asterisk indicates a difference from K904 plus vector by ANOVA with Tukey's posttest.
Predicted serralysin-like proteins were identified following in silico analysis of S. marcescens genomes.
In search of additional proteases, we scanned the S. marcescens Db11 genome (Wellcome-Trust Sanger Center) using the PrtS amino acid sequence as a query. We identified 3 ORFs with high similarity to serralysin that were also found in the genomes of more recently sequenced S. marcescens strains (43, 44) using an NCBI BLAST search (45). These predicted proteins are here noted as serralysin-like proteases B, C, and D (see Table S2 in the supplemental material).
The amino acid identities of the predicted proteins to serralysin were 52.6, 50.3, and 45%, respectively. Phylogenetic analysis indicates that of the three predicted proteases, SlpB is closest by sequence identity to serralysin, and it is also most similar to alkaline protease from Pseudomonas aeruginosa, AprA (see Fig. S2 in the supplemental material).
Serralysins are peptidases, a subset of the larger M10 metalloprotease group that includes mammalian matrix metallopeptidases (MMPs). Serralysin family proteins have an N-terminal HEXXHXXGXXH sequence involved in Zn2+ coordination in their N termini and catalytic activity in addition to repeats in toxin (RTX) motifs (GGXGXD) in the C terminus involved in binding to calcium and secretion via type I secretion systems (T1SSs) (46). The C-terminal domain is noted by the Protein Family Database (47) designation pfam00353. Predicted SlpB, SlpC, and SlpD proteins have the N-terminal HEXXHXXGXXH domain and RTX repeats, as well as a highly conserved M-turn methionine residue proximal to the active site (see Table S2). Notably, serralysin and SlpB have four GGXGXD motifs, whereas SlpC and SlpD have two.
Native expression of slpB, slpC, and slpD is detectable in a laboratory and a clinical isolate of S. marcescens.
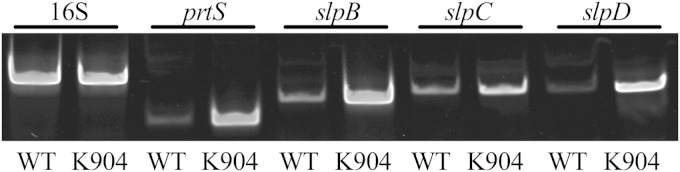
We tested whether the genes noted above from the Db11 genome were present in the genomes of strains PIC3611 and K904 used in this study. PCR analysis revealed that the prtS, slpB, slpC, and slpD genes were present in both genomes (data not shown). This does not, however, prove that these putative metalloprotease genes are transcribed. To test whether slpB, slpC, and slpD were expressed under laboratory conditions, RNA was harvested and RT-PCR was performed. For both PIC3611 and K904, amplicons corresponding to the slpB, slpC, and slpD genes and 16S and prtS controls were detectable from RT-treated RNA (Fig. 3). Furthermore, expression was relatively higher for each of the protease genes in the clinical isolate K904 than for the laboratory strain PIC3611 (Fig. 3).
FIG 3.
Elevated metalloprotease gene transcription in keratitis isolate K904. Semiquantitative RT-PCR analysis of gene expression of control and putative metalloprotease genes from Serratia marcescens strain PIC3611 (WT) and clinical ocular isolate K904 was performed. RNA was collected from cells at an OD600 of 3, purified, and found to be free of contaminating chromosomal DNA. The 16S ribosomal gene was used a control for input cDNA. Expression of prtS and three putative metalloprotease genes was elevated in the clinical strain K904 relative to that in the laboratory strain PIC3611. The experiment was performed four times using RNA from three independent experiments. All experiments demonstrated the same trend for each gene. Results from a representative experiment are shown.
Induced expression of serralysin and slpB, but not slpC and slpD increased extracellular protease activity.
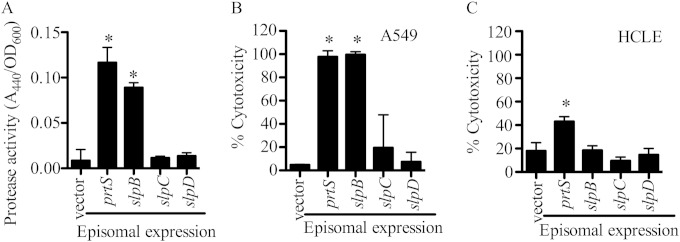
The putative chromosomal protease genes for prtS, slpB, slpC, and slpD were placed under the control of an arabinose-inducible promoter in the laboratory strain PIC3611. This strain was chosen because of a relatively low basal level of extracellular protease activity compared to those for the clinical isolates (Fig. 1A), and this low level would facilitate measuring increases in extracellular protease levels. Arabinose-induction of polyhistidine (His7)-tagged versions of prtS or slpB from an episomal plasmid resulted in a clear increase in extracellular protease activity, whereas no increases in protease levels were detected from supernatants of cells expressing the His7-tagged versions of slpC and slpD genes or a negative vector control (Fig. 4A). As a second test, the chromosomal prtS, slpB, slpC, and slpD genes were expressed under the control of the PBAD promoter integrated in front of each gene in the PIC3611 strain. Similar to episomal induction of these genes, chromosomal expression of prtS and slpB conferred increased secreted protease activity compared to the empty vector control, whereas slpC and slpD expression did not (see Fig. S3A). Induction of a control nonprotease gene, pigA pigment biosynthetic gene, showed little background protease activity (see Fig. S3A).
FIG 4.
Secreted protease activity from strain PIC3611 with induced expression of putative metalloprotease genes. (A to C) Induction of His7-tagged putative metalloprotease genes from a p15a-based plasmid in strain PIC3611 was followed by measuring protease activity and cytotoxic capacity in the filtered supernatants from cultures grown overnight. The negative-control was episomal plasmid pMQ125 (vector). Means and standard deviations are shown, and the asterisks indicate a significant difference from the vector control by ANOVA with Tukey's posttest (n = 6). (A) Protease production measured with azocasein. (B) Cytotoxicity to A549 airway cell line measured with Presto blue. (C) Cytotoxicity to HCLE cell line measured with Presto blue.
Induced expression of serralysin and SlpB increased cytotoxicity to human epithelial cell lines in vitro.
We tested whether induced expression of serralysin and the other putative proteases in PIC3611 could influence the cytotoxicity of bacterial secretomes to human epithelial cells in vitro. Unlike the vector negative control, expression of prtS induced bacterial cytotoxicity to A549 airway cells (Fig. 4B). Induced expression of slpB, but not slpC or slpD, was similar to prtS induction with cytotoxicity to A549 cells (Fig. 4B). A similar pattern was observed when prtS, slpB, slpC, and slpD were expressed from an episomal plasmid (Fig. S3B). Interestingly, unlike cell line A549, the human ocular cell line HCLE was less susceptible to secreted proteases induced in PIC3611, where only prtS induction caused significant levels of cytotoxicity (Fig. 4C).
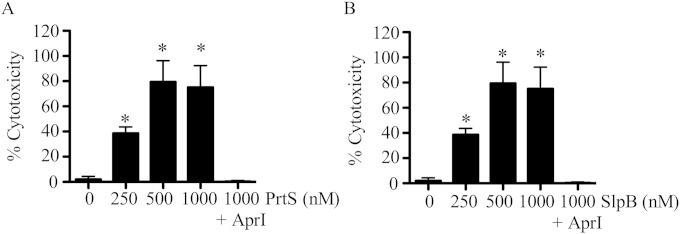
Alkaline protease inhibitor (AprI) from P. aeruginosa inhibits PrtS and SlpB-mediated proteolysis and cytotoxicity.
AprI was recently shown to be able to inhibit purified PrtS protease activity in vitro (23). This was confirmed in our model system, where induced PrtS was inhibited by AprI in a dose-dependent manner (see Fig. S4A in the supplemental material). A similar dose-dependent inhibition of SlpB activity by AprI was observed (see Fig. S4A).
Since AprI was able to inhibit both PrtS and SlpB, it was used as another way to assess whether protease activity was responsible for the observed cytotoxicity to epithelial cells. When AprI was added at 250 nM, cytotoxicity to A549 cells was significantly reduced (see Fig. S4A and B). AprI was also able to inhibit secreted cytotoxicity capability from strain K904 to A549 cells (Fig. S4C). Together, these data support the notion that protease activity is responsible for the observed cytotoxic effects and increased the scope of proteases known to be inhibited by AprI.
Mutation of slpB in clinical isolate K904 confers loss of secreted protease activity and epithelial cell type-specific cytotoxicity.
To test the role of SlpB in secreted protease activity and cytotoxicity, allelic replacement of the slpB gene on the chromosome of K904 with a deletion allele of slpB was performed. The mutation results in an in-frame deletion of 219 amino acids, removing the active site and most of the N-terminal protease domain. The ΔslpB mutant was defective in secreted proteolysis (Fig. 5A and B), and the defect could be complemented by slpB on a plasmid (Fig. 5B). Mutation of prtS caused a more severe reduction in protease activity than deletion of slpB (Fig. 5A and B) (P < 0.05). Consistent phenotypes were recorded with directed insertional mutation of the slpB gene in strain K904 (data not shown). This mutation was achieved by recombination of an internal fragment of slpB on a suicide plasmid into the chromosome. The plasmid inserts after bp 747 out of 1,419 bp. Mutation of both prtS and slpB in the same strain conferred a further reduction in secreted protease activity, suggesting an independent and additive contribution from PrtS and SlpB (Fig. 5A).
FIG 5.
Deletion of prtS and slpB confers a protease and cytotoxicity defective phenotype. (A) Protease activity of stationary-phase bacterial supernatants using azocasein as a substrate and normalized by culture densities. Data for 5 independent replicates from 3 separate experiments are shown. (B) Protease assay of complemented strains as described above, with 4 independent replicates. Vector = pMQ125, pprtS = pMQ356, pslpB = pMQ436. (C) Cytotoxicity of normalized bacterial supernatants to HCLE cells, with 6 independent biological replicates from 5 separate experiments. (D) Cytotoxicity of normalized bacterial supernatants to A549 cells (n = 3). In all graphs the means and standard deviations are shown. The asterisk indicates statistical difference from K904 by ANOVA with Tukey's posttest. Pound signs indicate a significant difference between K904 prtS and K904 prtS slpB by ANOVA with Tukey's posttest.
The ΔslpB mutant was slightly less cytotoxic to HCLE cells than the parental strain, but the reduction was not significant (Fig. 5C). Secretomes from the ΔprtS mutant were significantly less cytotoxic to HCLE cells than from the ΔslpB mutant but produced equal cytotoxicity to secretomes from the double mutant (Fig. 5C).
A clear difference to HCLE cells was observed when A549 cells were challenged with bacterial supernatants, where neither mutation of prtS or slpB reduced cytotoxicity of K904 (Fig. 5D). However, the ΔprtS ΔslpB double mutant was almost completely defective in cytotoxicity to HCLE cells (Fig. 5D). This suggests that either PrtS or SlpB alone is sufficient for cytotoxicity to A549 cells.
The cytotoxicity assays used previously in this study measure both loss of cells due to proteolytic removal of cells (cell detachment) and cell death after 4 h of challenge with bacterial supernatants. As an alternative approach, the cytotoxicity assays were performed without washing away detached HCLE cells after 4 h and 24 h of exposure to K904 and mutant secretomes. After 4 h, the majority of HCLE cells exposed to K904 supernatants were dead, supporting the notion that the K904 secretomes led to HCLE cell death (Fig. 6A). K904 secretomes produced 92% cytotoxicity after 24 h of exposure (Fig. 6B). The prtS mutant was less cytotoxic than the parental strain (77%) (P < 0.05). As observed at 4 h, the slpB mutant was as cytotoxic as the parental strain (90%) after 24 h. Strikingly, the prtS slpB double mutant was the least cytotoxic, at 59% (Fig. 6B). These experiments further support the notion that the S. marcescens secretomes are cytotoxic to HCLE cells and reveal HCLE cytotoxicity requiring SlpB that was evident after a 24-h challenge.
FIG 6.
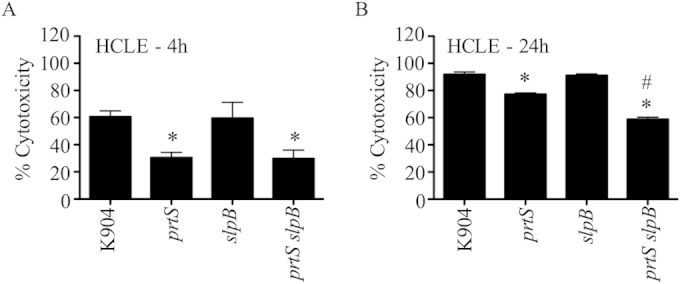
Prolonged exposure of HCLE cells to supernatants of K904 and protease mutants suggest a role for SlpB in cytotoxocity to ocular cells. Cytotoxicity of normalized bacterial supernatants to HCLE cells was measured using Presto blue (n = 3). These experiments were done without a washing step so that detached HCLE cells were included in the assay. Means and standard deviations are shown. The asterisk indicates statistical difference from K904 by ANOVA with Tukey's posttest. Pound signs indicate a significant difference between K904 prtS and K904 prtS slpB by ANOVA with Tukey's posttest. (A) 4-h exposure; (B) 24-h exposure.
Secretion of SlpB requires type I secretion protein LipD.
PrtS is secreted through a type I secretion system (T1SS) composed of three proteins LipB, LipC, and LipD (48, 49). In this study, a genetic approach was used to assess whether SlpB is also secreted through the LipBCD T1SS. To do so, slpB expression was induced in the wild-type (WT) PIC3611 strain and an isogenic strain with a transposon mutation in lipD. Secreted protease activity should be highly reduced in the lipD mutant compared to the wild-type strain with a functional LipBCD system if this T1SS is used to secrete SlpB. A prtS expression plasmid (pprtS) was included as a control to verify the approach. Compared to a vector control, elevated secreted protease activity was measured from the wild-type strain (PIC3611) expressing prtS and slpB, but not the lipD::Tn strain expressing prtS and slpB (see Fig. S5 in the supplemental material). These data indicate that like PrtS, SlpB requires LipBCD for secretion.
SlpB encodes an active, Ca2+-regulated protease.
Recombinant SlpB was expressed and purified from E. coli to characterize its biochemical and functional properties in vitro. The protein was expressed robustly in E. coli and could be purified to homogeneity under denaturing conditions in guanidinium hydrochloride. The protein was then refolded using protocols previously described for other similar serralysin metalloproteases (23).
The protein was refolded by rapid dilution into buffers in the presence and absence of Ca2+, a structural cofactor that has been shown to regulate the folding of RTX proteins and other members of the serralysin protease family (42). The purified protein was first diluted into buffers containing 2 mM Ca2+, which is predicted to saturate the binding sites in the C-terminal RTX domain, and activity was monitored using a fluorescent substrate. Rapid dilution into buffers containing millimolar Ca2+ concentrations resulted in robust protease activity, as monitored by the cleavage of the fluorescent peptide substrate (see Fig. S6). This activity was not seen in the buffer controls or in refolding reactions that lacked Ca2+, consistent with the activation of other serralysin family proteases (23).
To further characterize these Ca2+-regulated activities, Ca2+ titrations were performed and protease activity was assessed using this fluorescent peptide (see Fig. S6B). The purified SlpB protease showed no observable protease activity when refolded into buffers containing less than 15 µM Ca2+. Between 15 and 40 µM Ca2+, protease activity increased with increasing Ca2+. At and above 50 µM Ca2+, the protease activity appeared saturated and did not increase with further Ca2+ addition. The apparent affinity of the Ca2+-induced activity was 22 µM and was cooperative with a Hill coefficient of 5.5.
Given the apparent sequence and putative structural similarity between SlpB and other members of the serralysin protease family, the AprI protease inhibitor from P. aeruginosa was evaluated for its ability to block SlpB activity. Previous studies have shown that the AprI inhibitor binds with high-affinity to P. aeruginosa AprA and other serralysin proteases (23, 50). AprI was purified and coincubated in reaction mixtures containing refolded SlpB in the presence and absence of saturating Ca2+. When incubated at stoichiometric concentrations, the AprI protease inhibitor effectively blocked the SlpB protease activity (see Fig. S6C). This binding and protease inhibition suggests that the structure of the folded SlpB protease is likely similar to that of other serralysin family members. Further, the Ca2+-regulated activation and AprI inhibition demonstrate that SlpB functions as a bona fide protease.
Purified PrtS and SlpB were cytotoxic to HCLE cells in vitro, and SlpB is found at lower concentrations than PrtS in K904 secretomes.
Having purified SlpB, we tested whether recombinant PrtS and SlpB were cytotoxic to HCLE cells. Both PrtS and SlpB were cytotoxic to HCLE cells in a dose-dependent manner and could be inhibited by equimolar AprI (Fig. 7). This suggests that rather than being less cytotoxic than PrtS to HCLE cells, extracellular SlpB is produced at lower levels or is less stable.
FIG 7.
Purified PrtS and SlpB are cytotoxic to HCLE cells. Cytotoxicity of purified proteins to HCLE cells was measured with Presto blue. Mean values (n = 8) and standard deviations are shown. The asterisk equals significant difference from no protease control by ANOVA with Tukey's posttest. AprI was added at 1,000 nM. (A) Experiments performed with PrtS; (B) experiments performed with SlpB.
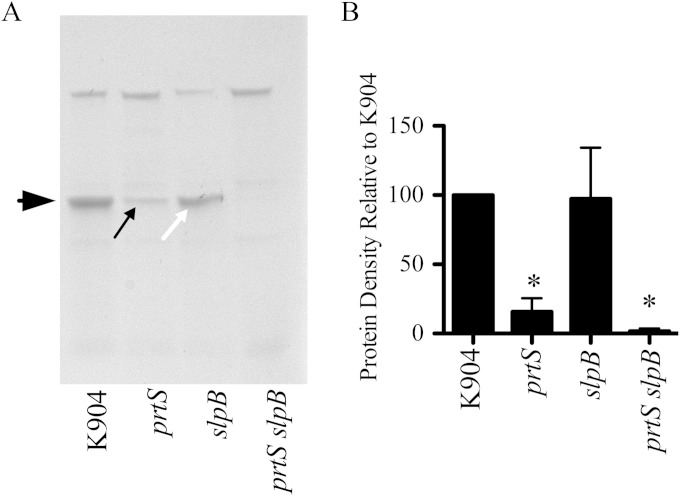
To investigate relative amounts of PrtS and SlpB in K904 secretomes, PAGE analysis was performed. The major band in the K904 secretome was ∼50 kDa (Fig. 8). The 50-kDa band was highly reduced in the ΔprtS strain (Fig. 8). The ΔslpB mutant secretomes had the 50-kDa band at levels similar to that of the parental K904 strain, and the band was completely absent in the double mutant, suggesting that PrtS and SlpB together constitute this band. Mass spectrometry analysis confirms that the band in the ΔprtS secretomes is SlpB and the band in the ΔslpB secretomes is PrtS. These data suggest that PrtS is made at higher levels or is more stable than SlpB, leading to its relative importance in cytotoxocity to corneal cells.
FIG 8.
S. marcescens strain K904 produces more PrtS than SlpB. (A) PAGE analysis of secretomes of K904 and noted isogenic mutants. Secretomes were normalized by optical density and trichloroacetic acid precipitated, and equal volumes were loaded. The large black arrow indicates an ∼50-kDa band. The small black arrow indicates SlpB. The small white arrow indicates PrtS. (B) Quantification of the 50-kDa band from 4 gels using independent samples. Values were normalized to the 50-kDa band from the K904 secretomes. Means and standard deviations are shown. The asterisk equals significant difference from K904 by ANOVA with Tukey's posttest.
DISCUSSION
Proteolysis of host immune components and eukaryotic surface proteins plays an important role in the pathogenic process of a number of pathogens (51). A variety of model systems have demonstrated that secreted proteases are key virulence factors, facilitating invasion of bacteria into mammalian cells, cleavage of host innate immune factors such as immunoglobulins and surfactant protein D, and disruption of tight junctions and epithelial cell integrity (12, 21, 26, 51). In polymicrobial infections, proteases could potentiate virulence by other microbes, not only through enabling bacterial invasion but also through degrading host immune components and activating host proteins, such as matrix metalloproteases (52).
Published data implicate the S. marcescens secreted metalloprotease PrtS as a virulence factor (13). Nevertheless, previous studies have assessed protease production by a very limited number of clinical isolates, and its role in cytotoxicity to ocular cells has not been tested in vitro. In this study, we tested a much larger number of ocular isolates from the three major eye infections caused by S. marcescens: conjunctivitis, endophthalmitis, and keratitis. S. marcescens is most prevalent as an agent of keratitis, with many studies listing it as the second most common cause of contact lens-associated keratitis, behind P. aeruginosa (2, 6, 53–56). S. marcescens is associated with common contact lens-associated inflammation complications (57) and activates inflammation through both the TLR4/MD-2/MyD88 and MyD-88/IL-1R1 pathways (58). Serralysin itself is sufficient to induce an immune response (21).
In this study, all tested ocular isolates produced some secreted protease activity if allowed to incubate on protease detection agar for 72 h. However, a range of secreted proteolysis was observed, and interestingly, isolates with delayed protease activity were more prevalent among conjunctivitis strains that are associated with a less severe infection. The reasons for the different levels of secreted protease activity between strains are not at this point fully understood, but the differences in expression levels of prtS and slpB were much lower in the low-protease-producing PIC3611 than in the highly protease-producing K904, suggesting that differential regulation is behind the different levels of protease production. However, amino acid differences influencing protease activity could be responsible for strain differences in secreted protease activity.
Using strain K904 as a representative keratitis isolate that secreted more cytotoxic and proteolytic activity than a wild-type laboratory strain, PIC3611, we obtained evidence suggesting that there may be proteases other than serralysin secreted by the clinical isolate K904. First, a previous study describes multiple secreted proteases purified from a particularly virulent keratitis isolate of S. marcescens (30). Although biochemical data were obtained, the identities of the proteins were not determined. Second, deletion of the prtS gene was not sufficient to completely eliminate secreted protease activity by clinical isolate K904. These observations suggested the existence of other protease genes in the genome of S. marcescens, a prediction that was validated by searching the published genome of strain Db11 (44). Transcripts from each of the three ORFs could be detected in both a clinical and laboratory strain and were much higher in the clinical isolate, supporting the conclusion that they were not cryptic ORFs.
To obtain insight into the function of these putative proteases, the putative metalloprotease ORFs and positive-control prtS were placed under transcriptional control of the inducible PBAD promoter on the chromosome of a largely nonproteolytic strain. Of slpB, slpC, and slpD, only slpB and the positive-control prtS conferred measurable protease activity. Each of the independently cloned genes was also induced from a low- to medium-copy-number plasmid, and the identical trend was observed. The potential reasons why slpC and slpD expression did not generate detectable secreted protease activity include that our assays used only casein as a substrate, and SlpC and SlpD may have specificities that exclude casein. Similarly, SlpC and SlpD may have pH optima or other requirements not met under our assay conditions. Additionally, these putative proteases may not be efficiently secreted under the tested conditions or are not stable.
One of the outcomes of this study is that PrtS was apparently more important for toxicity to HCLE cells than SlpB, since induced expression of prtS from a noncytotoxic strain conferred much higher levels of cytotoxicity to HCLE cells than expression of slpB. Additionally, mutation of prtS had a much more deleterious effect on cytotoxicity from clinical isolate K904 than did mutation of slpB. However, given that purified SlpB was similarly cytotoxic to PrtS, it may be that PrtS is more efficiently secreted or more stable than SlpB. PAGE analysis using single and double mutants is consistent with this model, but further biochemical analysis will be required to determine whether the difference is due to reduced secretion, lower stability of SlpB than of PrtS, and other mechanisms. Nevertheless, cytotoxicity from purified SlpB and reduced cytotoxicity from a K904 prtS slpB double mutant compared to that from the K904 prtS mutant support a potential role for SlpB in cytotoxicity to ocular cells.
The serralysin protease may have differential cytotoxicity by host cell type. A study by Ishii et al. demonstrated that S. marcescens supernatants enhanced the release of phagocytic hemocytes into silkworm hemolymph but did not kill the silkworm (59). Further elegant biochemical analysis revealed that serralysin contributed to the loss of hemocyte adhesion through degradation of adhesion molecules on the hemocyte membrane. The lack of cytotoxicity seen in that study may have to do with differences in bacterial strains, differences in susceptibilities of silkworm hemocytes to human epithelial cell lines, or differences in the timing of the experimental analysis which were not explicitly stated in the insect study (59). Conversely, other groups found that serralysin was cytotoxic to HeLa cells (14) and human embryonic lung fibroblasts (15), and protease-deficient mutants of Serratia sp. strain SCBI were defective in cytotoxicity to a buffalo green monkey kidney cell line. In this study, we observed that A549 airway cells were more susceptible than HCLE corneal cells to S. marcescens proteases.
This study identifies SlpB as a new cytotoxic factor secreted by S. marcescens and supports that under the experimental conditions used, PrtS and SlpB are the major secreted cytotoxic proteins of S. marcescens. Lastly, the metal-dependent nature of S. marcescens metalloproteases suggests that nontoxic metal chelators could be of use in prevention of protease-associated tissue damage.
Supplementary Material
ACKNOWLEDGMENTS
We thank James Fender, Dayna Helvick, Eric Kalivoda, and Christina Medaglia for technical assistance, Regis Kowalski for providing clinical isolates, and the P30 EY085570 grant-supported hybridoma and molecular biology modules for support.
This study was supported by NIH grants AI085570 to R.M.Q.S. and DK083284 to P.H.T., with additional funding supplied by the Eye and Ear Foundation of Pittsburgh and unrestricted funds from Research to Prevent Blindness. K.M.B. was supported by NIH training grant 2T32 EY017271. This project used the UPCI Cancer Biomarkers Facility, which is supported in part by award P30CA047904.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.03096-14.
REFERENCES
- 1.Cheng KH, Leung SL, Hoekman HW, Beekhuis WH, Mulder PG, Geerards AJ, Kijlstra A. 1999. Incidence of contact-lens-associated microbial keratitis and its related morbidity. Lancet 354:181–185. doi: 10.1016/S0140-6736(98)09385-4. [DOI] [PubMed] [Google Scholar]
- 2.Mah-Sadorra JH, Najjar DM, Rapuano CJ, Laibson PR, Cohen EJ. 2005. Serratia corneal ulcers: a retrospective clinical study. Cornea 24:793–800. doi: 10.1097/01.ico.0000159738.06167.88. [DOI] [PubMed] [Google Scholar]
- 3.Keay L, Edwards K, Naduvilath T, Taylor HR, Snibson GR, Forde K, Stapleton F. 2006. Microbial keratitis predisposing factors and morbidity. Ophthalmology 113:109–116. doi: 10.1016/j.ophtha.2005.08.013. [DOI] [PubMed] [Google Scholar]
- 4.Richards MJ, Edwards JR, Culver DH, Gaynes RP. 2000. Nosocomial infections in combined medical-surgical intensive care units in the United States. Infect Control Hosp Epidemiol 21:510–515. doi: 10.1086/501795. [DOI] [PubMed] [Google Scholar]
- 5.Mahlen SD. 2011. Serratia infections: from military experiments to current practice. Clin Microbiol Rev 24:755–791. doi: 10.1128/CMR.00017-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alexandrakis G, Alfonso EC, Miller D. 2000. Shifting trends in bacterial keratitis in south Florida and emerging resistance to fluoroquinolones. Ophthalmology 107:1497–1502. doi: 10.1016/S0161-6420(00)00179-2. [DOI] [PubMed] [Google Scholar]
- 7.Lavinsky F, Avni-Zauberman N, Barequet IS. 2013. Clinical characteristics and outcomes of patients admitted with presumed microbial keratitis to a tertiary medical center in Israel. Arq Bras Oftalmol 76:175–179. doi: 10.1590/S0004-27492013000300009. [DOI] [PubMed] [Google Scholar]
- 8.Kowalski RP, Kowalski TA, Shanks RM, Romanowski EG, Karenchak LM, Mah FS. 2013. In vitro comparison of combination and monotherapy for the empiric and optimal coverage of bacterial keratitis based on incidence of infection. Cornea 32:830–834. doi: 10.1097/ICO.0b013e318268d6f4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Engel LS, Hill JM, Moreau JM, Green LC, Hobden JA, O'Callaghan RJ. 1998. Pseudomonas aeruginosa protease IV produces corneal damage and contributes to bacterial virulence. Invest Ophthalmol Vis Sci 39:662–665. [PubMed] [Google Scholar]
- 10.Girgis DO, Sloop GD, Reed JM, O'Callaghan RJ. 2005. Effects of toxin production in a murine model of Staphylococcus aureus keratitis. Invest Ophthalmol Vis Sci 46:2064–2070. doi: 10.1167/iovs.04-0897. [DOI] [PubMed] [Google Scholar]
- 11.Green SN, Sanders M, Moore QC III, Norcross EW, Monds KS, Caballero AR, McDaniel LS, Robinson SA, Onwubiko C, O'Callaghan RJ, Marquart ME. 2008. Protection from Streptococcus pneumoniae keratitis by passive immunization with pneumolysin antiserum. Invest Ophthalmol Vis Sci 49:290–294. doi: 10.1167/iovs.07-0492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mun JJ, Tam C, Kowbel D, Hawgood S, Barnett MJ, Evans DJ, Fleiszig SM. 2009. Clearance of Pseudomonas aeruginosa from a healthy ocular surface involves surfactant protein D and is compromised by bacterial elastase in a murine null-infection model. Infect Immun 77:2392–2398. doi: 10.1128/IAI.00173-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matsumoto K. 2004. Role of bacterial proteases in pseudomonal and serratial keratitis. Biol Chem 385:1007–1016. [DOI] [PubMed] [Google Scholar]
- 14.Marty KB, Williams CL, Guynn LJ, Benedik MJ, Blanke SR. 2002. Characterization of a cytotoxic factor in culture filtrates of Serratia marcescens. Infect Immun 70:1121–1128. doi: 10.1128/IAI.70.3.1121-1128.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Molla A, Matsumoto K, Oyamada I, Katsuki T, Maeda H. 1986. Degradation of protease inhibitors, immunoglobulins, and other serum proteins by Serratia protease and its toxicity to fibroblast in culture. Infect Immun 53:522–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kreger AS, Griffin OK. 1975. Cornea-damaging proteases of Serratia marcescens. Invest Ophthalmol 14:190–198. [PubMed] [Google Scholar]
- 17.Lyerly D, Gray L, Kreger A. 1981. Characterization of rabbit corneal damage produced by Serratia keratitis and by a serratia protease. Infect Immun 33:927–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miyagawa S, Matsumoto K, Kamata R, Okamura R, Maeda H. 1991. Spreading of Serratia marcescens in experimental keratitis and growth suppression by chicken egg white ovomacroglobulin. Jpn J Ophthalmol 35:402–410. [PubMed] [Google Scholar]
- 19.Molla A, Oda T, Maeda H. 1987. Different binding kinetics of Serratia 56K protease with plasma alpha 2-macroglobulin and chicken egg white ovomacroglobulin. J Biochem 101:199–205. [DOI] [PubMed] [Google Scholar]
- 20.Molla A, Kagimoto T, Maeda H. 1988. Cleavage of immunoglobulin G (IgG) and IgA around the hinge region by proteases from Serratia marcescens. Infect Immun 56:916–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kida Y, Inoue H, Shimizu T, Kuwano K. 2007. Serratia marcescens serralysin induces inflammatory responses through protease-activated receptor 2. Infect Immun 75:164–174. doi: 10.1128/IAI.01239-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Butterworth MB, Zhang L, Heidrich EM, Myerburg MM, Thibodeau PH. 2012. Activation of the epithelial sodium channel (ENaC) by the alkaline protease from Pseudomonas aeruginosa. J Biol Chem 287:32556–32565. doi: 10.1074/jbc.M112.369520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Butterworth MB, Zhang L, Liu X, Shanks RM, Thibodeau PH. 2014. Modulation of the epithelial sodium channel (ENaC) by bacterial metalloproteases and protease inhibitors. PLoS One 9:e100313. doi: 10.1371/journal.pone.0100313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Norqvist A, Norrman B, Wolf-Watz H. 1990. Identification and characterization of a zinc metalloprotease associated with invasion by the fish pathogen Vibrio anguillarum. Infect Immun 58:3731–3736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cowell BA, Twining SS, Hobden JA, Kwong MS, Fleiszig SM. 2003. Mutation of lasA and lasB reduces Pseudomonas aeruginosa invasion of epithelial cells. Microbiology 149:2291–2299. doi: 10.1099/mic.0.26280-0. [DOI] [PubMed] [Google Scholar]
- 26.Bozhokina ES, Tsaplina OA, Efremova TN, Kever LV, Demidyuk IV, Kostrov SV, Adam T, Komissarchik YY, Khaitlina SY. 2011. Bacterial invasion of eukaryotic cells can be mediated by actin-hydrolysing metalloproteases grimelysin and protealysin. Cell Biol Int 35:111–118. doi: 10.1042/CBI20100314. [DOI] [PubMed] [Google Scholar]
- 27.Tonry JH, McNichol BA, Ramarao N, Chertow DS, Kim KS, Stibitz S, Schneewind O, Kashanchi F, Bailey CL, Popov S, Chung MC. 2012. Bacillus anthracis protease InhA regulates BslA-mediated adhesion in human endothelial cells. Cell Microbiol 14:1219–1230. [DOI] [PubMed] [Google Scholar]
- 28.Hume EB, Conerly LL, Moreau JM, Cannon BM, Engel LS, Stroman DW, Hill JM, O'Callaghan RJ. 1999. Serratia marcescens keratitis: strain-specific corneal pathogenesis in rabbits. Curr Eye Res 19:525–532. doi: 10.1076/ceyr.19.6.525.5283. [DOI] [PubMed] [Google Scholar]
- 29.Pinna A, Usai D, Sechi LA, Carta A, Zanetti S. 2011. Detection of virulence factors in Serratia strains isolated from contact lens-associated corneal ulcers. Acta Ophthalmol 89:382–387. doi: 10.1111/j.1755-3768.2009.01689.x. [DOI] [PubMed] [Google Scholar]
- 30.Matsumoto K, Maeda H, Takata K, Kamata R, Okamura R. 1984. Purification and characterization of four proteases from a clinical isolate of Serratia marcescens kums 3958. J Bacteriol 157:225–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Létoffé S, Delepelaire P, Wandersman C. 1991. Cloning and expression in Escherichia coli of the Serratia marcescens metalloprotease gene: secretion of the protease from E. coli in the presence of the Erwinia chrysanthemi protease secretion functions. J Bacteriol 173:2160–2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bertani G. 1951. Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J Bacteriol 62:293–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miller VL, Mekalanos JJ. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J Bacteriol 170:2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Giard DJ, Aaronson SA, Todaro GJ, Arnstein P, Kersey JH, Dosik H, Parks WP. 1973. In vitro cultivation of human tumors: establishment of cell lines derived from a series of solid tumors. J Natl Cancer Inst 51:1417–1423. [DOI] [PubMed] [Google Scholar]
- 35.Gipson IK, Spurr-Michaud S, Argüeso P, Tisdale A, Ng TF, Russo CL. 2003. Mucin gene expression in immortalized human conreal-limbal and conjunctival epithelial cell lines. Invest Ophthalmol Vis Sci 44:2496–2506. doi: 10.1167/iovs.02-0851. [DOI] [PubMed] [Google Scholar]
- 36.Wingard JB, Romanowski EG, Kowalski RP, Mah FS, Ling Y, Bilonick RA, Shanks RM. 2011. A novel cell-associated protection assay demonstrates the ability of certain antibiotics to protect ocular surface cell lines from subsequent clinical Staphylococcus aureus challenge. Antimicrob Agents Chemother 55:3788–3794. doi: 10.1128/AAC.01828-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shanks RM, Stella NA, Arena KE, Fender JE. 2013. Mutation of crp mediates Serratia marcescens serralysin and global secreted protein production. Res Microbiol 164:38–45. doi: 10.1016/j.resmic.2012.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cruz-Romero M, Kelly AL, Kerry JP. 2008. Influence of packaging strategy on microbiological and biochemical changes in high-pressure-treated oysters. J Sci Food Agric 88:2713–2723. doi: 10.1002/jsfa.3398. [DOI] [Google Scholar]
- 39.Shanks RM, Kadouri DE, MacEachran DP, O'Toole GA. 2009. New yeast recombineering tools for bacteria. Plasmid 62:88–97. doi: 10.1016/j.plasmid.2009.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shanks RM, Caiazza NC, Hinsa SM, Toutain CM, O'Toole GA. 2006. Saccharomyces cerevisiae-based molecular tool kit for manipulation of genes from gram-negative bacteria. Appl Environ Microbiol 72:5027–5036. doi: 10.1128/AEM.00682-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shanks RM, Lahr RM, Stella NA, Arena KE, Brothers KM, Kwak DH, Liu X, Kalivoda EJ. 2013. A Serratia marcescens PigP homolog controls prodigiosin biosynthesis, swarming motility and hemolysis and is regulated by cAMP-CRP and HexS. PLoS One 8:e57634. doi: 10.1371/journal.pone.0057634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang L, Conway JF, Thibodeau PH. 2012. Calcium-induced folding and stabilization of the Pseudomonas aeruginosa alkaline protease. J Biol Chem 287:4311–4322. doi: 10.1074/jbc.M111.310300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chung WC, Chen LL, Lo WS, Kuo PA, Tu J, Kuo CH. 2013. Complete genome sequence of Serratia marcescens WW4. Genome Announc 1(2):e0012613. doi: 10.1128/genomeA.00126-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Iguchi A, Nagaya Y, Pradel E, Ooka T, Ogura Y, Katsura K, Kurokawa K, Oshima K, Hattori M, Parkhill J, Sebaihia M, Coulthurst SJ, Gotoh N, Thomson NR, Ewbank JJ, Hayashi T. 2014. Genome evolution and plasticity of Serratia marcescens, an important multidrug-resistant nosocomial pathogen. Genome Biol Evol 6:2096–2110. doi: 10.1093/gbe/evu160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stöcker W, Grams F, Baumann U, Reinemer P, Gomis-Ruth FX, McKay DB, Bode W. 1995. The metzincins—topological and sequential relations between the astacins, adamalysins, serralysins, and matrixins (collagenases) define a superfamily of zinc-peptidases. Protein Sci 4:823–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Punta M, Coggill PC, Eberhardt RY, Mistry J, Tate J, Boursnell C, Pang N, Forslund K, Ceric G, Clements J, Heger A, Holm L, Sonnhammer EL, Eddy SR, Bateman A, Finn RD. 2012. The Pfam protein families database. Nucleic Acids Res 40:D290–D301. doi: 10.1093/nar/gkr1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Akatsuka H, Kawai E, Omori K, Shibatani T. 1995. The three genes lipB, lipC, and lipD involved in the extracellular secretion of the Serratia marcescens lipase which lacks an N-terminal signal peptide. J Bacteriol 177:6381–6389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Binet R, Letoffe S, Ghigo JM, Delepelaire P, Wandersman C. 1997. Protein secretion by Gram-negative bacterial ABC exporters—a review. Gene 192:7–11. doi: 10.1016/S0378-1119(96)00829-3. [DOI] [PubMed] [Google Scholar]
- 50.Hege T, Feltzer RE, Gray RD, Baumann U. 2001. Crystal structure of a complex between Pseudomonas aeruginosa alkaline protease and its cognate inhibitor: inhibition by a zinc-NH2 coordinative bond. J Biol Chem 276:35087–35092. doi: 10.1074/jbc.M104020200. [DOI] [PubMed] [Google Scholar]
- 51.Hoge R, Pelzer A, Rosenau F, Wilhelm S. 2010. Weapons of a pathogen: proteases and their role in virulence of Pseudomonas aeruginosa. Curr Res Technol Educ Top Appl Microbiol Microb Biotechnol 2:383–395. http://www.formatex.info/microbiology2/383-395.pdf. [Google Scholar]
- 52.Miyajima S, Akaike T, Matsumoto K, Okamoto T, Yoshitake J, Hayashida K, Negi A, Maeda H. 2001. Matrix metalloproteinases induction by pseudomonal virulence factors and inflammatory cytokines in vitro. Microb Pathog 31:271–281. doi: 10.1006/mpat.2001.0470. [DOI] [PubMed] [Google Scholar]
- 53.Das S, Sheorey H, Taylor HR, Vajpayee RB. 2007. Association between cultures of contact lens and corneal scraping in contact lens related microbial keratitis. Arch Ophthalmol 125:1182–1185. doi: 10.1001/archopht.125.9.1182. [DOI] [PubMed] [Google Scholar]
- 54.Mayo MS, Schlitzer RL, Ward MA, Wilson LA, Ahearn DG. 1987. Association of Pseudomonas and Serratia corneal ulcers with use of contaminated solutions. J Clin Microbiol 25:1398–1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schaefer F, Bruttin O, Zografos L, Guex-Crosier Y. 2001. Bacterial keratitis: a prospective clinical and microbiological study. Br J Ophthalmol 85:842–847. doi: 10.1136/bjo.85.7.842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Verhelst D, Koppen C, Van Looveren J, Meheus A, Tassignon MJ. 2006. Contact lens-related corneal ulcers requiring hospitalization: a 7-year retrospective study in Belgium. Acta Ophthalmol Scand 84:522–526. doi: 10.1111/j.1600-0420.2006.00681.x. [DOI] [PubMed] [Google Scholar]
- 57.Holden BA, La Hood D, Grant T, Newton-Howes J, Baleriola-Lucas C, Willcox MD, Sweeney DF. 1996. Gram-negative bacteria can induce contact lens related acute red eye (CLARE) responses. CLAO J 22:47–52. [PubMed] [Google Scholar]
- 58.Zhou R, Zhang R, Sun Y, Platt S, Szczotka-Flynn L, Pearlman E. 2012. Innate immune regulation of Serratia marcescens-induced corneal inflammation and infection. Invest Ophthalmol Vis Sci 53:7382–7388. doi: 10.1167/iovs.12-10238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ishii K, Adachi T, Hamamoto H, Sekimizu K. 2014. Serratia marcescens suppresses host cellular immunity via the production of an adhesion-inhibitory factor against immunosurveillance cells. J Biol Chem 289:5876–5888. doi: 10.1074/jbc.M113.544536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kalivoda EJ, Stella NA, Aston MA, Fender JE, Thompson PP, Kowalski RP, Shanks RM. 2010. Cyclic AMP negatively regulates prodigiosin production by Serratia marcescens. Res Microbiol 161:158–167. doi: 10.1016/j.resmic.2009.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kadouri DE, Shanks RM. 2013. Identification of a methicillin-resistant Staphylococcus aureus inhibitory compound isolated from Serratia marcescens. Res Microbiol 164:821–826. doi: 10.1016/j.resmic.2013.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kalivoda EJ, Horzempa J, Stella NA, Sadaf A, Kowalski RP, Nau GJ, Shanks RM. 2011. New vector tools with a hygromycin resistance marker for use with opportunistic pathogens. Mol Biotechnol 48:7–14. doi: 10.1007/s12033-010-9342-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.