Abstract
Salmonella enterica serovar Typhimurium is a common cause of food-borne gastrointestinal illness, but additionally it causes potentially fatal bacteremia in some immunocompromised patients. In mice, systemic spread and replication of the bacteria depend upon infection of and replication within macrophages, but replication in human macrophages is not widely reported or well studied. In order to assess the ability of Salmonella Typhimurium to replicate in human macrophages, we infected primary monocyte-derived macrophages (MDM) that had been differentiated under conditions known to generate different phenotypes. We found that replication in MDM depends greatly upon the phenotype of the cells, as M1-skewed macrophages did not allow replication, while M2a macrophages and macrophages differentiated with macrophage colony-stimulating factor (M-CSF) alone (termed M0) did. We describe how additional conditions that alter the macrophage phenotype or the gene expression of the bacteria affect the outcome of infection. In M0 MDM, the temporal expression of representative genes from Salmonella pathogenicity islands 1 and 2 (SPI1 and SPI2) and the importance of the PhoP/Q two-component regulatory system are similar to what has been shown in mouse macrophages. However, in contrast to mouse macrophages, where replication is SPI2 dependent, we observed early SPI2-independent replication in addition to later SPI2-dependent replication in M0 macrophages. Only SPI2-dependent replication was associated with death of the host cell at later time points. Altogether, our results reveal a very nuanced interaction between Salmonella and human macrophages.
INTRODUCTION
Salmonella enterica serovar Typhimurium is a Gram-negative facultative intracellular pathogen which is estimated to cause over 90 million cases of food-borne illness and 155,000 deaths per year worldwide (1). While most cases in healthy humans consist of a self-limiting gastroenteritis, it can cause life-threatening systemic bacteremia in some patients (2). Although fairly rare in the developed world, in sub-Saharan Africa there is a large population made susceptible due to malaria and advanced AIDS, and nontyphoidal Salmonella enterica (NTS) serovars such as Typhimurium are the most common bacteria isolated from the bloodstream of patients presenting with fever (1). These systemic infections with NTS are difficult to treat and are associated with a 20 to 25% case fatality (1). In HIV-infected patients, recrudescence is common even after effective antibiotic treatment, and it has been postulated that the bacteria persist within the reticuloendothelial system (3, 4).
Salmonella Typhimurium systemic disease has been widely studied in susceptible mice, where it causes a typhoid-like disease (5). In this model, the ability to survive and replicate in macrophages is essential to the systemic spread of the bacteria (6, 7). Two virulence systems that contribute to growth and survival of Salmonella within macrophages are the PhoP/Q two-component regulatory system and the Salmonella pathogenicity island 2 (SPI2)-encoded type III secretion system (T3SS2). Both PhoP/Q and the T3SS2 are induced by intracellular signals and are essential for survival and replication in murine macrophages and for virulence in mice (6, 8–10). Intracellular Salmonella Typhimurium organisms survive and replicate within an acidified, modified phagosome known as the Salmonella-containing vacuole (SCV) (11). In mouse macrophages and macrophage-like cell lines, the T3SS2 and several of its associated effectors are required for survival and replication in the SCV (6, 12). A third virulence system that mediates interactions between Salmonella and host cells is the SPI1-encoded T3SS1. In contrast to the T3SS2 and PhoP/Q, this system is induced in extracellular bacteria and is essential for bacterium-driven entry into nonphagocytic cells (such as intestinal epithelial cells) (13).
In addition to their roles in internalization and intracellular survival, both T3SS can induce cytotoxicity in macrophages. Logarithmic-phase Salmonella Typhimurium, which has high SPI1 expression, induces rapid NLRC4/caspase-1-dependent programmed death (pyroptosis) of mouse macrophages (14–18). Stationary-phase bacteria, which have low SPI1 expression, induce delayed T3SS2-dependent cell death 8 to 17 h postinfection (p.i.) (19, 20). Since the internalization of Salmonella into phagocytic cells does not require T3SS1, studies of Salmonella infection in macrophages generally are done with bacteria grown to stationary phase. However, SPI1-induced Salmonella Typhimurium is released from epithelial cells, suggesting that SPI1-induced bacteria can be encountered by macrophages in vivo (21, 22).
The ability of macrophages to engulf and kill bacteria is largely determined by their activation state. The original classification of macrophages into either “classically activated/M1” or “alternatively activated/M2” was based upon their role in Th1- and Th2-driven immune responses, respectively (23). While these classifications are an oversimplification of the phenotypes macrophages can display, they still provide a useful example of the extremes of macrophage function. The M1 phenotype can be induced in vitro from exposure to the proinflammatory cytokine gamma interferon (IFN-γ) and the stimulation of a toll-like receptor (TLR), such as the stimulation of TLR4 by lipopolysaccharide (LPS). This results in a cell with the antimicrobial and proinflammatory properties essential for fighting bacterial infections. The M2 designation actually comprises several distinct noninflammatory macrophage phenotypes, which are induced in vitro by exposure to different agents (24). One of these, the product of exposure to the Th2 cytokine interleukin-4 (IL-4) (designated M2a), participates in traditional Th2-mediated immune responses, such as fighting extracellular parasites, and contributes to allergic reactions.
In mice, certain subsets of macrophages are the preferred sites of Salmonella Typhimurium survival and replication (25–27); however, its ability to survive and replicate in human macrophages is not well described. In human macrophage-like cell lines (THP-1 and U937), it appears to replicate less than in murine macrophage-like cell lines (RAW 264.7 and J774), and there are few reports describing its replication in human macrophages (28–31). Here, we have revisited the question of whether Salmonella Typhimurium can replicate in human macrophages, and if so, what factors affect its ability to do so. We show that the phenotype of the macrophage is critical in determining whether the bacteria survive and replicate. Additionally, the bacterial determinants involved in survival and replication differ somewhat from what has been described in mouse cells. Future studies using this model system should identify the factors that determine the outcome of Salmonella Typhimurium in human macrophages, potentially providing new therapeutic options.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Strains were maintained on Luria-Bertani Miller (LB-Miller) agar plates containing appropriate antibiotics. Strains and plasmids are listed in Table 1. The 14028 ΔphoP strain was created by P22 transduction from the SL1344 ΔphoP strain into 14028. The promoters for prgH and ssaG were used as reporters for SPI1 and SPI2 regulon activity, respectively. pMPMA3ΔPlac PprgH-gfp [LVA], containing a destabilized variant of green fluorescent protein (GFP) (half-life of approximately 40 min), was described previously (32). To create pMPMA3ΔPlac PssaG-gfp, PssaG-gfp was amplified without the [LVA] tag from pMPMA3ΔPlac PssaG-gfp [LVA] (32), resulting in the original GFPmut3b (33) as the reporter for gene expression. Primers used were PssaG HindIII F (5′-NNNAAGCTTGCATGCCTGCAGGT-3′) and GFP-w/o-LVA HindIII R (5′-NNNAAGCTTTTATTTGTATAGTTCATCCATGCCATGTGTA-3′), and the product was cloned back into pMPMA3ΔPlac. The acid shock response gene asr (STM1485) is induced at pH 4.5 and lower (34). The asr transcriptional reporter (pMPMA3ΔPlac Pasr-gfp) also used GFPmut3b and was made by splicing by overlap extension (SOE) PCR followed by cloning into the ClaI and EcoRV sites of pMPMA3ΔPlac. The 5′ upstream region of asr (403 bp from the translational start site) was amplified from genomic DNA using primers Vector-Pasr F (5′-TTCGATATCAAGCTTGCCATCCTGGTCGGGACG-3′) and GFP-Pasr R (5-TTCTCCTTTGCTCATTTTGATACCCTCGATTTGGT-3′). GFPmut3b was amplified using primers GFPmut3b F (5′-ATGAGTAAAGGAGAAGAACTTTTCA-3′) and vector-GFPmut3b R (5′GTATCGATAAGCTTTTATTTGTATAGTTCATC-3′).
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Genotype/features | Source or reference |
|---|---|---|
| Strains | ||
| SL1344 | hisG46 xyl rpsL | 35 |
| 14028 | Wild-type S. Typhimurium 14028 | ATCC |
| ΔSPI1 | SPI1::kan | 32 |
| ΔSPI2 | SPI2::kan | 36 |
| SL1344 ΔphoP | phoP::kan | 37 |
| 14028 ΔphoP | phoP::kan | This paper |
| Plasmids | ||
| pFPV-mCherry/2 | mCherry constitutive expression | 38 |
| pMPMA3ΔPlac PprgH-gfp [LVA] | PprgH reporter (T3SS1) | 32 |
| pMPMA3ΔPlac PssaG-gfp | PssaG reporter (T3SS2) | 32 and this paper |
| pMPMA3ΔPlac Pasr-gfp | Pasr reporter | This paper |
Macrophages.
Human peripheral blood monocytes, enriched by apheresis, were obtained from peripheral blood provided by the Department for Transfusion Medicine and the National Institutes of Health Clinical Center at the National Institutes of Health (Bethesda, MD). Signed, informed consent was obtained from each donor, acknowledging that his or her donation would be used for research purposes by intramural investigators throughout the National Institutes of Health. Monocytes were further enriched by centrifugation with Ficoll-Paque (GE Healthcare) and then resuspended in a freezing medium of 10% dimethyl sulfoxide (DMSO)–90% fetal bovine serum (FBS) at 108 cells/ml and stored in liquid N2. Monocytes were purified with the Dynabeads Untouched human monocytes kit (Life Technologies); the purity of recovered cells was routinely >90% CD14+ by flow cytometry. These cells were plated in complete medium containing RPMI 1640 medium (Gibco), 1 mM sodium pyruvate, 1× MEM nonessential amino acids, 10 mM HEPES buffer, 2 mM glutamate, 5% (vol/vol) heat-inactivated human male AB serum (HuS) (Sigma-Aldrich) or 5% (vol/vol) FBS (Gibco), and 100 ng/ml human recombinant macrophage colony-stimulating factor (M-CSF) (PeproTech). Cells were plated in 8-well optically clear plastic chambered coverslips (Ibidi) at 105 cells/well for all assays except enzyme-linked immunosorbent assay (ELISA) analysis of supernatants (24-well plates, 2 × 105 cells/well), flow cytometry (6-well plates, 106 cells/well), or live cell imaging (optically clear-plastic 35-mm μ-dish [Ibidi], 6 × 105 cells/dish). All substrates were tissue culture treated. Cells were grown in 5% CO2 at 37°C and used for assays on day 7 or 8. On days 3, 5, and 7 (if used on day 8), 50% of the volume of the cultures was replaced with fresh complete RPMI, 5% serum, and 200 ng/ml M-CSF. All data are from 3 to 5 independent experiments with cells prepared on different days from different donors.
Macrophages grown as described above were skewed to an M1 phenotype by the inclusion of 50 ng/ml recombinant human IFN-γ (PeproTech) and 10 ng/ml purified Salmonella enterica serovar Minnesota LPS (Sigma) on days 3 through 7 of culture. For the M2 phenotype, 10 ng/ml of recombinant human IL-4 (PeproTech) was included throughout. Cells grown with M-CSF only were designated M0 for comparison.
Infection of macrophages.
All Salmonella organisms were grown in LB-Miller at 37°C with aeration at 225 rpm. Overnight cultures, started from a single colony, were grown in 2 ml broth containing appropriate antibiotics for 16 to 18 h. Antibiotics used were streptomycin (100 μg/ml), carbenicillin (50 μg/ml), and kanamycin (50 μg/ml). Subcultures were made by diluting the overnight culture 1:33 into fresh LB-Miller broth without antibiotics and grown to late-log phase, which was 3.5 h for wild-type (WT) SL1344 and 3 h for WT 14028 bacteria. Bacteria were pelleted and resuspended in Hanks' balanced salt solution without Ca2+/Mg2+ (HBSS) and then diluted in complete RPMI. For bacteria grown to stationary phase (see Fig. 3A), one colony was inoculated in 10 ml LB-Miller and grown for 18 h. In order to facilitate uptake, the stationary-phase bacteria were opsonized in 50% (vol/vol) human male AB serum (Sigma) in HBSS for 30 min at room temperature (RT) before dilution as described above.
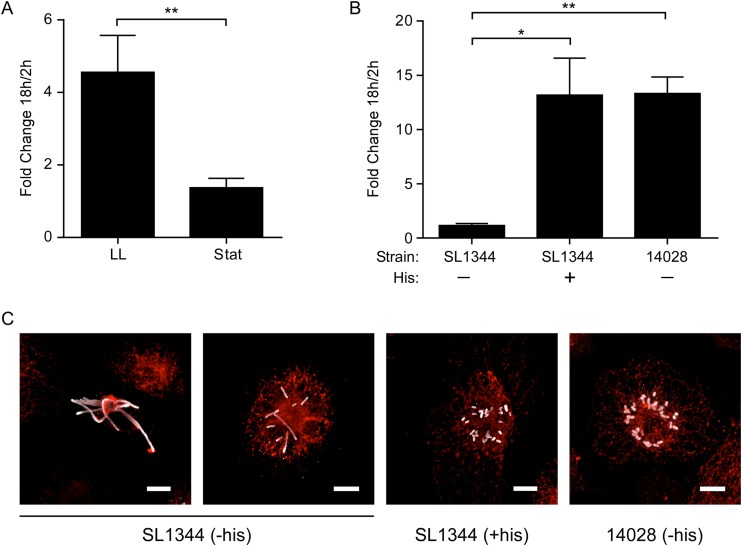
FIG 3.
Replication of Salmonella Typhimurium in monocyte-derived human macrophages is dependent on growth phase and supplemental histidine. (A) Gentamicin protection assay. Shown is the fold change in CFU (at 18 and 2 h p.i.) from M0 cells infected with Salmonella SL1344 grown to late-log phase (LL) or to stationary phase (Stat). Stationary-phase bacteria were opsonized with normal human serum. Results are means ± SD from 3 independent experiments. **, P = 0.008 by paired t test. (B) Gentamicin protection assay. Shown is the fold change in CFU (at 18 and 2 h p.i.) of strain SL1344 (mutation in hisG) with and without supplemental histidine (His) or strain 14028 without histidine. Results are means ± SD from 3 independent experiments. P < 0.05 (*) and P < 0.01 (**) by one-way ANOVA. (C) Representative images of macrophages at 18 h p.i., stained for LPS (gray) and LAMP1 (red). Cells were infected with SL1344 in the absence (−his) or presence (+his) of supplemental histidine. On the far right, strain 14028 is shown in the absence of histidine. Bar, 10 μm.
For all infections, media were prewarmed to 37°C, and the cells were returned to the 37°C incubator between steps. At t = 0, culture medium was removed from the macrophages and replaced with medium containing bacteria at a multiplicity of infection (MOI) of ≈15 bacteria/cell (late-log-phase bacteria). When using opsonized, stationary-phase bacteria (see Fig. 3A), the bacteria were added at an MOI of ≈90:1 in order to achieve similar intracellular numbers at 2 h p.i.
At t = 10 min, cells were washed once with HBSS and fresh medium was added. At t = 30 min, medium was replaced with fresh medium containing 50 μg/ml gentamicin. At t = 45 min, medium was replaced with medium containing 10 μg/ml gentamicin and, for infections using strain SL1344, 500 μg/ml l-histidine (Sigma).
Gentamicin protection assay.
At 2 or 18 h p.i., medium was removed and the cells lysed with 0.02% sodium deoxycholate in phosphate-buffered saline (PBS). Serial 1:10 dilutions in PBS were plated on LB-agar plates in order to assess the number of intracellular bacteria per well. Duplicate wells were averaged for each data point.
Immunofluorescence staining.
Infected cells were fixed with 2.5% (wt/vol) paraformaldehyde (PFA)–PBS for 20 min at 37°C and then washed with PBS and stored at 4°C until staining. All subsequent steps were done at RT. For assays to calculate the number of bacteria per cell, cells were blocked and permeabilized with 0.1% (wt/vol) saponin, 10% (vol/vol) normal goat serum in PBS (SS-PBS) for 30 min. Rabbit anti-Salmonella LPS (Difco) and mouse anti-human LAMP-1 (clone H4A3; Developmental Studies Hybridoma Bank, University of Iowa) were diluted 1:1,000 in SS-PBS and applied for 45 min. Cells were washed once with SS-PBS and then twice with PBS. SS-PBS was added for 15 min, and then the secondary antibodies goat anti-rabbit Alexa 568 and goat anti-mouse Alexa 488 (Life Technologies) diluted 1:800 in SS-PBS were applied for 45 min. Cells were washed once with SS-PBS and twice with PBS. 4′,6-Diamidino-2-phenylindole (DAPI) (300 nM in PBS) was added for 20 min, and cells were washed twice with PBS and then overlaid with 200 μl of mounting medium (Mowiol) (39). For assays using GFP reporter strains (see Fig. 4), cells were stained as described above using the secondary antibodies goat anti-rabbit Alexa 647 and goat anti-mouse Alexa 568 (Life Technologies), each at 1:800.
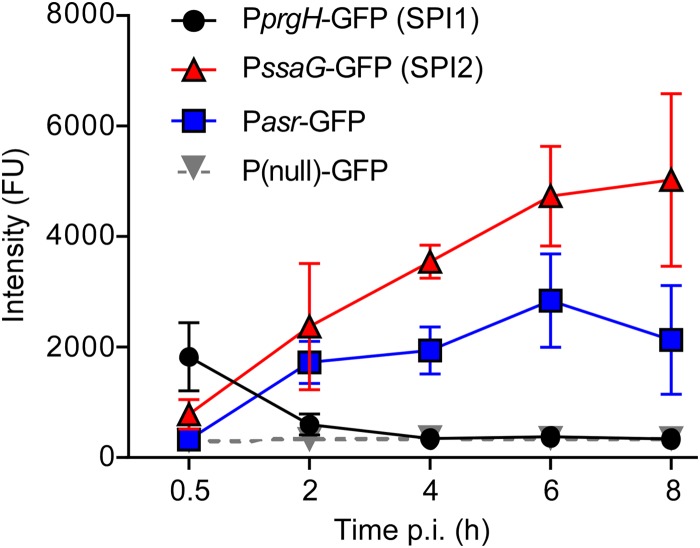
FIG 4.
Gene expression in the course of infection. M0 macrophages were infected with SL1344 bacteria carrying a plasmid-borne reporter of the indicated genes. The GFP intensities in individual bacteria were measured as fluorescence units (FU) and the geometric means calculated. Shown are the geometric means ± SD from 3 independent experiments.
ELISA.
Supernatants were collected 12 h p.i. and stored at −80°C. ELISAs were performed according to the manufacturer's directions using DuoSet ELISA kits (R&D Systems) to detect the human cytokines IL-6, IL-1β, tumor necrosis factor alpha (TNF-α), IL-12p40, IL-8/CXCL8, IL-10, and monocyte chemoattractant protein 1 (MCP-1). Samples were assayed in duplicate and the values averaged.
Flow cytometry.
Macrophages were washed once with PBS and then incubated with TrypLE Select (Life Technologies) for 20 min at 37°C. The loosened cells were removed from the plastic with a cell lifter (Costar), and the well was rinsed once with PBS. These cells were collected into tubes on ice, centrifuged at 250 × g for 5 min at 4°C to pellet, and then resuspended in ice-cold PBS containing 2% (vol/vol) fetal calf serum and 0.01% (wt/vol) sodium azide (fluorescence-activated cell sorter [FACS] buffer). Between 2 × 104 and 2 × 105 cells were placed per well into a round-bottom 96-well plate, cells were spun down, and supernatant removed. Fc receptors were blocked by incubation with 10 μg of purified whole human IgG (ChromPure; Jackson ImmunoResearch) for 10 min in 10 μl FACS buffer. Cells then were stained by addition of one or more fluorophore-conjugated antibodies in 100 μl of FACS buffer for 30 min on ice. Antibodies were used at the manufacturer's recommended dilution. From BioLegend we obtained HLA-DR fluorescein isothiocyanate (FITC) (L243), CD163 phycoerythrin (PE) (GHI/61), CD80 PE (2D10), CD11b peridinin chlorophyll protein (PerCP)/Cy5.5 (ICRF44), and CD36 allophycocyanin (APC)/Cy7 (5-271). From eBioscience we obtained CD33 PE-Cy7 (WM-53), CD16 APC (eBioCB16), and CD14 eFluor450 (61D3). From BD Pharmingen we obtained CD206 APC (19.2). Cells were washed twice with FACS buffer and then resuspended in 1% PFA–PBS (wt/vol) and stored at 4°C in the dark until analysis (up to 24 h). Cells were analyzed on a BD LSRII flow cytometer (BD Biosciences) using FACSDiva software, and the data were analyzed using FlowJo software (v. 9.8). A compensation matrix was established using single-color stained cells. For quantification, the macrophages were gated on CD33 and CD11b, and then the geometric mean fluorescence intensity of this population in each channel was measured. Unstained cells of the same phenotype/treatment were used to measure the background fluorescence in each channel, which then was subtracted from the values obtained on the stained samples.
Assessment of cell death.
Cells plated in 8-well μ-slides (Ibidi number 80826) were infected as described above, using approximate MOIs of 10 and 100 for WT bacteria or 30 and 300 for ΔSPI-1 bacteria (to compensate for lower internalization efficiency). Mock-infected cells underwent the identical infection protocol but with plain media in place of diluted bacteria. One hour after the start of the infection, FLICA reagent (ImmunoChemistry Technologies), which binds specifically to active caspase-1, was added to the cells. At 2 h, cells were washed to remove unbound FLICA reagent, and then propidium iodide (PI) was added. Cells were analyzed immediately by immunofluorescence microscopy for the percentage of cells staining green (FLICA+, active caspase-1) and red (PI+, compromised cell membrane).
Phagocytosis assay.
The phagocytic capacity of macrophages was assessed by incubation with Alexa 488-labeled killed Escherichia coli BioParticles (Life Technologies). Approximately 2.5 × 105 BioParticles were added per well in prewarmed complete medium, and cells were incubated for 10 min. Cells were washed once with warm HBSS and then incubated with prewarmed medium for an additional 20 min. Cells then were washed twice with PBS and fixed in 2.5% (wt/vol) PFA–PBS for 15 min at 37°C. Cells were stained with wheat germ agglutinin conjugated to Alexa 647 (Life Technologies) for 20 min at RT as a way to visualize the macrophages. Cells were fixed again with 2.5% PFA–PBS for 10 min, and then extracellular BioParticles were stained with rabbit anti-Alexa 488 antibody (Life Technologies), followed by goat anti-rabbit Alexa 564. The total number of cells and the number of intracellular particles per cell were counted by microscopy, and random fields were counted in each of two duplicate wells. At least 100 (FBS grown) or 290 (HuS grown) macrophages were counted per experiment.
Imaging.
For live cell imaging, macrophages were infected with mCherry-expressing WT or ΔSPI2 bacteria as described above. Imaging was initiated at approximately 1 h p.i. and continued for approximately 21 h using a TiE microscope with a Perfect Focus system (Nikon), a CSU10 spinning disk (Yokogawa), and a custom laser launch (Prairie Technologies). A stage-top incubation chamber (Pathology Devices) was used to maintain temperature and humidity. mCherry and differential interference contrast (DIC) images were taken every 15 min using a Plan Fluor 40×/1.3 numeric-aperture (n.a.) oil DIC objective (Nikon) and either a Cascade II (Photometrics) or an iXon (Andor) electron-multiplying charge-coupled device (EMCCD) camera.
Phase-contrast photos shown in Fig. 2 and Fig. S1 in the supplemental material were taken using a Nikon Eclipse TS100 microscope with a 10×/0.25-n.a. Ph1 ADL objective (Nikon) and captured on a Nikon Sight DS-Fi1 camera.
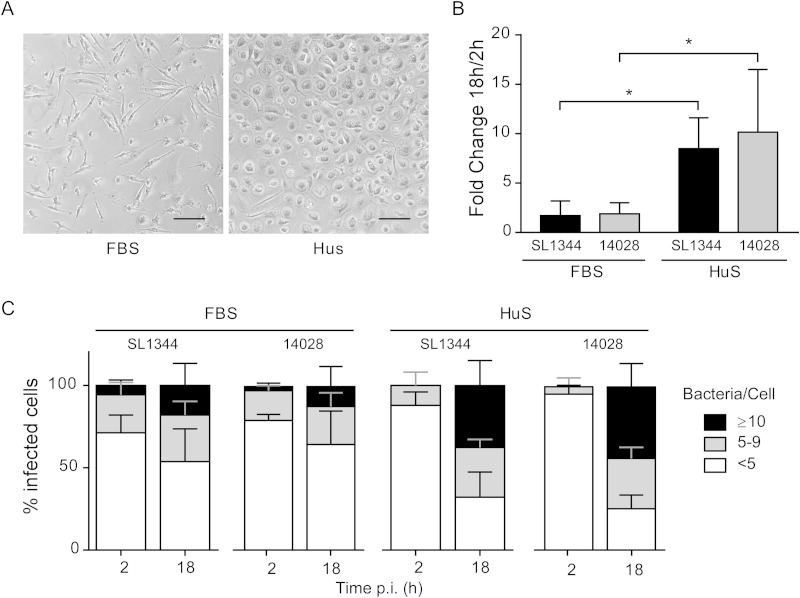
FIG 2.
Differentiation of monocyte-derived human macrophages in fetal bovine serum versus human serum affects the outcome of Salmonella Typhimurium infection. Cells were cultured in media containing FBS or HuS for 7 days as described in the text and then infected with Salmonella WT strains SL1344 and14028. (A) Phase-contrast images of uninfected cells. Bar, 100 μm. (B) Gentamicin protection assay. Shown is the fold change in CFU (at 18 and 2 h p.i.). Results are means ± SD from 4 independent experiments. *, P < 0.05 by two-way ANOVA. (C) The percentages of infected cells containing <5, 5 to 9, or ≥10 bacteria were assessed by microscopy at 2 and 18 h p.i. Results are means ± SD from 4 independent experiments.
Fluorescent images in Fig. 1B and 3C are maximum intensity projections of laser-scanning confocal images taken with an LSM710 using Zen 2010 software and a Plan-Apochromat 63×/1.40-n.a. oil DIC M27 objective (Carl Zeiss).
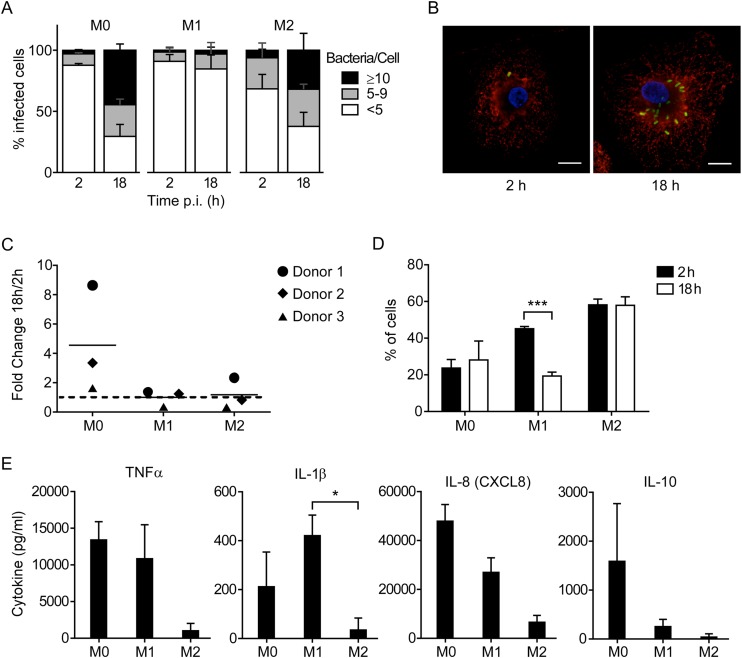
FIG 1.
Cell phenotype affects the ability of Salmonella Typhimurium to survive and replicate in human MDM. (A) M1, M2, and M0 macrophages were infected with wild-type strain SL1344, fixed, and stained for immunofluorescence microscopy at the indicated times. Infected cells were scored as containing ≥10, 5 to 9, or <5 bacteria. The means ± standard deviations (SD) from 3 independent experiments using three different donors are shown. (B) Representative images of infected M0 macrophages. Green, LPS; red, LAMP1; blue, DAPI. Bar, 10 μm. (C) Gentamicin protection assays were performed using cells infected as described for panel A. At 2 and 18 h p.i., the cells were lysed and recoverable CFU were assessed by plating. Each point represents the fold change in CFU (18 versus 2 h p.i.) obtained in one experiment, and lines represent the means from 3 independent experiments. (D) Cells were infected, fixed, and stained for microscopy as described for panel A. The percentage of cells containing bacteria at each time point is shown. Bars represent means ± SD from 3 independent experiments. ***, P = 0.0003 by two-way ANOVA. (E) Cytokine levels in cell culture supernatants at 12 h p.i. Results are means ± SD from 3 independent experiments. *, P = 0.027 by one-way ANOVA.
For quantification of cells per field, percentage of infected cells, number of bacteria per cell, and fluorescence of bacteria containing GFP reporters (see Fig. 4) or BioParticles per cell, a Nikon Eclipse TE2000 epifluorescence microscope with a Plan Apo 60×/1.40-n.a. oil Ph3 DM objective (Nikon) was used. Images were captured on a Photometrics CoolSnap HQ CCD camera.
All postacquisition image analysis and figure preparation was done using ImageJ software (version 1.4.8; W. S. Rasband, National Institutes of Health, Bethesda, MD) and Adobe Photoshop (CS5, v12.1; Adobe).
Statistics.
The statistical significance of comparisons was assessed using paired t tests, one-way analysis of variance (ANOVA), or two-way ANOVA, as stated in the figure legends. P values for one-way ANOVA were calculated using Tukey's multiple-comparison test and for two-way ANOVA using Bonferroni's multiple-comparison test. These analyses were done using GraphPad Prism (v6.0). Comparisons left unmarked do not represent statistically significant differences (P > 0.05).
RESULTS
Macrophage phenotype affects the ability of Salmonella to replicate.
To study the ability of Salmonella Typhimurium to survive and replicate within human macrophages, we chose monocyte-derived macrophages (MDM) differentiated with macrophage colony-stimulating factor (M-CSF) that have been skewed to an M1 or M2a (here designated M2) phenotype by the addition of IFN-γ and LPS or IL-4, respectively. These cells were compared to macrophages differentiated without any additions besides M-CSF (designated M0).
Cell morphology, as viewed by phase-contrast microscopy, and analysis by flow cytometry confirmed that these culturing conditions resulted in cells with different phenotypes (see Fig. S1A and B in the supplemental material). On M2 cells, the surface expression of CD206 (macrophage mannose receptor) was high and the level of CD14 was low, as has been reported previously (40). On M1 cells the levels of HLA-DR and CD14 were higher. Although cells differentiated in M-CSF alone are sometimes described as M2 (40–42), the M0 cells were clearly distinct from the IL-4-derived M2 cells.
M1, M2, and M0 macrophages were infected with Salmonella Typhimurium to assess the ability of the bacteria to survive and replicate in these cells (Fig. 1). For these experiments, an MOI of ≈15 was used, and cells were incubated with bacteria for 10 min and then washed. Gentamicin was added to the cells at 30 min p.i. to prevent growth of extracellular bacteria. At 2 and 18 h p.i., cells were either fixed for analysis by microscopy or lysed to release intracellular bacteria for CFU enumeration. We selected 18 h as the latest time point, because we have observed significant macrophage death at later time points. Fixed cells were immunolabeled for Salmonella (with an anti-LPS antibody) and the SCV (with an anti-LAMP1 antibody) and analyzed by immunofluorescence microscopy. At 2 h p.i. the vast majority of infected cells contained <5 bacteria (Fig. 1A). To facilitate comparisons between the three cell types, we categorized the infected cells according to whether they contained a low (<5), intermediate (5 to 9), or high (≥10) number of intracellular bacteria. This revealed an increase over time in the number of bacteria per cell for M0 and M2 cells (Fig. 1A and B), so that at 18 h the majority of infected cells contained >5 bacteria, with 40 to 50% of infected M0 cells and 30 to 40% of infected M2 cells containing ≥10 bacteria. In contrast, the number of intracellular bacteria per cell did not change significantly in M1 cells.
In agreement with the microscopy analysis, the number of recoverable intracellular Salmonella organisms in M0 cells increased between 2 and 18 h p.i., although the extent of this increase appeared to be donor dependent (Fig. 1C). In contrast, there was no detectable increase in recoverable intracellular bacteria in M2 cells, even though the microscopy data suggested that intracellular replication did occur. This discrepancy may be due to death of M2 macrophages following infection (data not shown) and underscores the importance of performing single-cell analysis by microscopy when assessing bacterial replication.
A comparison of the percentages of infected cells at 2 and 18 h p.i. also reveals significant differences between the different cell types. At 2 h p.i. more M2 cells (≈58%) contained bacteria than M1 (≈40%) or M0 cells (≈24%) (Fig. 1D). At 18 h p.i., the numbers of infected cells remained the same for M0 and M2 cells, whereas in M1 cells it decreased by ≈50% (P = 0.0003), suggesting that either a portion of these cells cleared the bacteria or infected cells were lost/detached. Since no difference in the total number of cells per field between 2 and 18 h p.i. was detected for any of the cell types, it is likely that M1 cells cleared bacteria more efficiently than M0 or M2 cells.
Given the observed differences in survival/replication of intracellular Salmonella, we hypothesized that there also would be measurable differences in the responses of the differentiated macrophages to infection as indicated by cytokine secretion. As expected, M1 cells responded to Salmonella Typhimurium infection with increased production of the proinflammatory cytokines TNF-α, IL-1β, and IL-6 and the chemokine IL-8 (CXCL8) (Fig. 1E; also see Fig. S1C in the supplemental material). In comparison, M2 cells produced low levels of these cytokines, fitting with their reported role as a noninflammatory cell type. Interestingly, M0 cells also produced the proinflammatory cytokines TNF-α, IL-1β, and IL-6 while also producing the anti-inflammatory cytokine IL-10. Given the ability of Salmonella to replicate in M0 cells while also stimulating a robust cytokine response, we chose to further characterize this interaction.
For the above-described experiments, Salmonella organisms were grown under SPI1-inducing conditions (32). The SPI1-encoded T3SS1 can cause rapid caspase-1-mediated cell death (pyroptosis) in macrophages (14, 15); however, we could detect no loss of infected cells at 2 h p.i. (see Fig. S2A in the supplemental material). One possibility is that the relatively low MOI and short infection time used here result in the internalization of lower numbers of bacteria than required for induction of pyroptosis. To investigate this possibility, we labeled M0 cells at 2 h p.i. with the reagent FLICA, which specifically binds active caspase-1, and propidium iodide (PI) to indicate membrane permeability (see Fig. S2B). Accordingly, significant caspase-1 activation and cell death were induced when cells were infected with a high MOI of ≈100 (>40% of infected cells were PI+/FLICA+) but not at the lower MOI of ≈10. In comparison, an SPI1-deficient strain (ΔSPI1) did not induce cell death even when added at an MOI of ≈300.
Differentiation of MDM with fetal bovine serum decreases Salmonella replication.
In this study, MDM were cultured in medium containing human serum (HuS), although some protocols for human MDM differentiation use fetal bovine serum (FBS). To determine how much of an effect this would have on M0 cells and their interaction with Salmonella, we directly compared cells differentiated in FBS versus HuS. We observed marked differences in morphology and cell surface marker expression. The predominant shape of cells grown in HuS was amoeboid, whereas that of cells grown in FBS was elongated or spindle-like (Fig. 2A). The levels of several surface markers (CD14, CD80, CD16, and CD163) were higher in FBS-grown cells, whereas the level of CD36 was higher in HuS-grown cells (see Fig. S3A in the supplemental material), and HLA-DR, CD11b, CD33, and CD206 levels were similar.
We next compared the ability of two Salmonella Typhimurium wild-type strains, SL1344 and 14028, to survive and replicate in M0 MDM differentiated in FBS or HuS. Neither strain showed significant replication in FBS-grown cells (Fig. 2B and C), whereas in HuS-grown cells the recovered CFU increased by ≈10-fold from 2 to 18 h p.i., and by microscopy there was a corresponding increase in the number of bacteria per infected cell (Fig. 2B and C). A comparison of the percentages of infected cells also revealed differences. The FBS-grown cells showed the highest percentage of infection with SL1344 (50 to 70%) and 14028 (5 to 45%), while the levels in HuS-grown cells infected with either strain were lower (10 to 25%) (see Fig. S3B in the supplemental material). The distribution was similar at both 2 and 18 h p.i., suggesting that there is no significant clearance of bacteria. To examine whether the difference in numbers of infected cells could be attributed to differences in phagocytic uptake, we compared their capacity to internalize killed E. coli. After 10 min of internalization, followed by a 20-min chase, approximately 60% of FBS-grown cells contained E. coli, compared to less than 10% of HuS-grown cells (see Fig. S3C). Thus, FBS-grown cells appeared more phagocytic than the HuS-grown cells. Altogether, these experiments highlight the influence of culture conditions on MDM phenotype and function and, in turn, on the outcome of infection.
Bacterial growth conditions affect replication in macrophages.
Here, we have used late-log-phase, SPI1-induced bacteria to infect macrophages, whereas many other studies use stationary-phase bacteria, which are not SPI1 induced (32, 43). Non-SPI1-induced Salmonella bacteria often are opsonized to allow efficient uptake via receptor-mediated phagocytosis, while late-log-phase bacteria can enter by either SPI1-mediated or macrophage-driven uptake. To compare the consequences of infection with Salmonella Typhimurium grown to express SPI-1 or not, M0 cells (differentiated in HuS) were infected with either stationary-phase bacteria (opsonized with human serum) or late-log-phase bacteria. By gentamicin assay, replication of stationary-phase bacteria was greatly reduced compared to that of SPI1-induced bacteria (mean of 1.4-fold versus 4.6-fold increase from 2 to 18 h p.i.; P = 0.008) (Fig. 3A). Single-cell analysis by microscopy confirmed that this difference was due to replication, since neither the percentage of infected cells nor the total number of cells changed significantly between 2 and 18 h p.i. (not shown). Thus, the ability of Salmonella Typhimurium to replicate in M0 cells is dependent on the transcriptome state and/or the mechanism of entry.
Salmonella Typhimurium SL1344, commonly used as a wild-type strain, is a histidine auxotroph due to a mutation in hisG (35). Although this does not appear to affect its virulence in mice, one study showed that intracellular replication is reduced in the absence of supplemental histidine in some cell types (44). In our hands, replication of SL1344 in M0 MDM was completely eliminated in the absence of supplemental histidine (Fig. 3B), and fluorescence microscopy revealed SL1344 bacteria with a filamentous morphology (Fig. 3C). In comparison, supplemental histidine was not required for intracellular replication of strain 14028, which has a functional hisG gene, and these bacteria exhibited a normal rod-shaped morphology indistinguishable from that of SL1344 in the presence of supplemental histidine. Thus, SL1344 requires supplemental histidine to replicate in human MDMs.
Salmonella gene expression during infection of human MDM.
The expression of the T3SSs encoded by SPI1 and SPI2 is strictly regulated during the infection of both epithelial cells and macrophages. After entry into a host cell, the T3SS1 is rapidly downregulated, followed by the upregulation of the T3SS2 and PhoP/Q concurrently with SCV acidification (32, 45, 46). To confirm that this sequence of events also occurs during the infection of M0 human macrophages, we assessed the transcriptional activity of representative SPI1 (PprgH) and SPI2 (PssaG) promoters in intracellular bacteria using green fluorescent protein (GFP) as a reporter. Cells infected with SPI1-induced, late-log-phase bacteria were fixed at various time points and immunostained with anti-LPS (to label all bacteria) and anti-LAMP1 (to label the SCV) antibodies for analysis by immunofluorescence microscopy (Fig. 4). At 30 min p.i., the earliest time point examined, only intracellular bacteria carrying the SPI1 reporter plasmid, PprgH-gfp, had detectable GFP fluorescence. This decreased significantly by 2 h p.i. and was undetectable at 4 and 8 h p.i., consistent with rapid downregulation of the SPI1 regulon after internalization. In contrast, bacteria carrying the SPI2 reporter plasmid PssaG-gfp were not fluorescent at 30 min p.i. but thereafter showed steadily increasing fluorescence, with maximum fluorescence at 6 to 8 h p.i. A similar response was seen for the promoter of the acid-responsive gene asr. These results suggest that the bacteria respond to their intracellular environment in M0 cells similarly to what has been described in murine macrophages and macrophage-like cells (45, 46).
Requirement for the PhoP/Q two-component regulatory system and SPI2 for intracellular replication.
Survival of Salmonella Typhimurium within mouse macrophages is dependent on both the SPI2-encoded T3SS2 and the PhoP/Q two-component regulatory system. The PhoP/Q system controls a number of virulence genes, including those involved in resisting the innate immune system, and is essential for replication in mouse macrophages and macrophage-like cell lines (47). To confirm the role of PhoP/Q in human macrophages, we compared infection with WT SL1344 and the isogenic ΔphoP mutant (Fig. 5). At both 8 and 18 h p.i., the mutant displayed a profound defect in replication compared to the WT by both gentamicin assay and microscopic analysis. Thus, replication in M0 MDM is dependent on the PhoP/Q regulatory system.
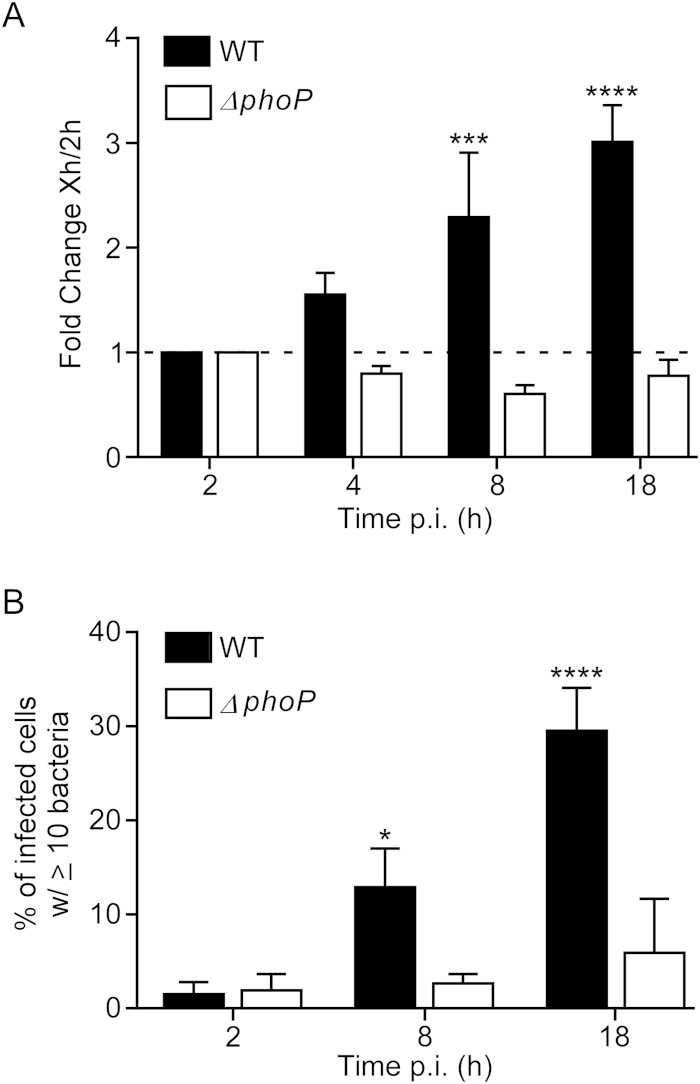
FIG 5.
Salmonella Typhimurium organisms lacking the PhoP/Q two-component regulatory system are defective for replication in human MDM. M0 macrophages were infected with WT SL1344 or an isogenic ΔphoP strain. A gentamicin assay was performed. Shown is the fold change in recoverable CFU compared to the level at 2 h p.i. (A) and the percentage of cells containing ≥10 bacteria as assessed by microscopy (B). Results are means ± SD from 3 independent experiments. P < 0.05 (*), P < 0.01 (***), and P < 0.0001 (****) by two-way ANOVA of comparisons to 2-h p.i. values.
In contrast to the PhoP mutant, an isogenic ΔSPI2 mutant (36) showed no replication defect at 4 and 8 h p.i. However, this was no longer the case by 18 h p.i., when the increase compared to the level at 2 h was 2.6-fold for the mutant and 5.5-fold for the WT (P = 0.03) (Fig. 6A). Single-cell analysis by fluorescence microscopy confirmed that at 8 h p.i. the mutant was indistinguishable from the WT, but at 18 h p.i. there were significantly fewer cells containing ≥10 bacteria (Fig. 6B).
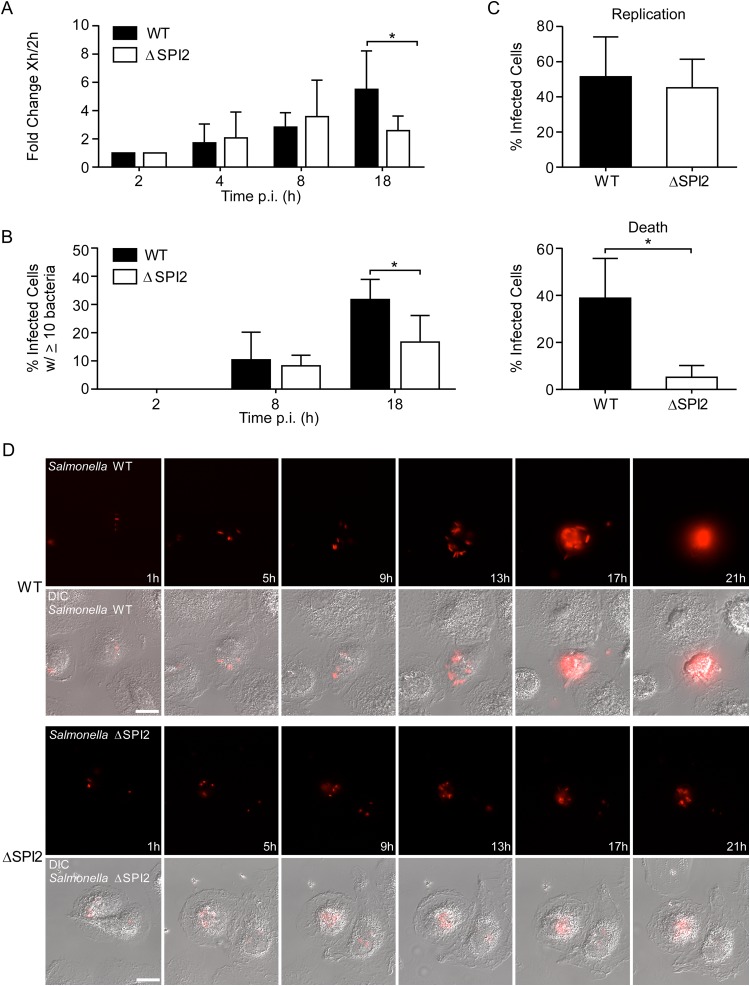
FIG 6.
SPI2-dependent and -independent replication of Salmonella Typhimurium in M0 macrophages. (A) Gentamicin assay. Shown is the fold change in recoverable CFU compared to the values at 2 h p.i. Results are means ± SD from 5 independent experiments. (B) The percentage of infected cells containing the indicated number of bacteria was assessed by microscopy. Results are means ± SD from 3 independent experiments. (C and D) Intracellular replication was assessed by live cell imaging. Macrophages infected with mCherry-expressing WT or ΔSPI2 bacteria were imaged from 1 to 20 h p.i. every 15 min. (C) Cells that contained bacteria at 1 h were assessed for bacterial replication (top) and the appearance of cell death by morphological changes (bottom) over the time course. Macrophages from 3 donors were analyzed for each strain. A total of 62 cells infected with the WT and 59 infected with the ΔSPI2 strain were analyzed. (D) Stills from representative movies showing the replication of mCherry-expressing WT (top) or ΔSPI2 (bottom) bacteria in macrophages. Also see Movies S1 and S2 in the supplemental material. Bar, 10 μm. *, P < 0.05 by two-way ANOVA (A and B) and by paired t test (C).
Since several studies have shown that SPI2 is essential for replication in macrophages, we performed a more detailed analysis of the fate of intracellular Salmonella Typhimurium in M0 MDM using live cell imaging, which allows individual cells to be monitored through time and can provide a much more detailed picture of the interaction between host and pathogen. We have previously used a spinning-disc confocal imaging system to image Salmonella-infected HeLa cells for up to 20 h p.i. (48). Here, we have used the same approach with M0 cells infected with Salmonella constitutively expressing the red fluorescent protein mCherry. These experiments revealed that while both WT and ΔSPI2 bacteria do replicate within M0 cells, there are significant differences. The extent of replication was consistently higher for WT bacteria, and this was associated with rounding up and membrane blebbing of the host cell by ≈20 h p.i. (Fig. 6C and D; also see Movie S1 in the supplemental material). Replication of the ΔSPI2 mutant was observed in more than half of infected cells, but it did not reach the level of the WT, and very few infected cells died by 20 h p.i. (Fig. 6C and D; also see Movie S2). Thus, both SPI2-independent and SPI2-dependent replication of Salmonella Typhimurium is observed in human M0 MDM.
DISCUSSION
In mice, the ability of Salmonella Typhimurium to survive and replicate within macrophages is essential for systemic disease, whereas in humans the role of macrophages has not been well studied. In vivo, macrophage phenotypes are extremely heterogeneous and are determined by local cues in different tissue microenvironments, which act upon populations of macrophages at different stages of differentiation (49). Such phenotypic complexity cannot be reproduced in vitro, because many of the environmental signals are not understood. However, given the extremely limited availability of human tissues, monocyte-derived cultures differentiated under defined conditions are a valid option.
Here, we used human monocyte-derived macrophages as a model to study the intracellular survival and replication of Salmonella Typhimurium. We found that the bacteria could replicate in macrophages with an alternatively activated, or M2, phenotype or a nonactivated, or M0, phenotype, while no net replication was detected in macrophages with a classically activated, or M1, phenotype (Fig. 1). Additionally, the M1 macrophages appeared to be able to kill the bacteria, in agreement with previous data showing that priming of MDM with IFN-γ enhances killing of Salmonella Typhimurium (50). Macrophage polarization has been very well characterized in mice, but many of these findings do not extend to human macrophages (51), making it difficult to draw conclusions based on mouse studies. However, these results do correlate with a recent finding that Salmonella preferentially replicates within IL-4-treated M2 bone marrow-derived mouse macrophages but not within IFN-γ-treated M1 cells (52). In the same report, using an in vivo mouse model of persistent infection, it was shown that Salmonella persisted in certain subsets of macrophages expressing M2 markers. It is reasonable to expect that during human systemic Salmonella infections the bacteria persist and replicate within a subset of macrophages, similar to what has been seen in mice.
The importance of the macrophage phenotype in the clearance of the bacteria may help to explain why people with certain diseases are much more susceptible to the development of systemic bacteremia. Individuals with mutations in IL-12 or IFN-γ are extremely susceptible to systemic nontyphoidal Salmonella infections (53), and the resulting lack of IFN-γ would impair the skewing of macrophages to the bactericidal M1-like phenotype. The default pathway in the absence of IFN-γ may result in the development of a replication-permissive M2-like phenotype (54). Similarly, in patients with advanced HIV, the lowered CD4+ T cell counts and resulting lack of CD4+-derived cytokines may result in a macrophage phenotype that contributes to disease.
The amount of replication that we and others (29) have observed in human MDMs is relatively modest, generally 2- to 10-fold between 2 and 18 h, and less than that reported for most murine macrophages and macrophage-like cell lines, where it is generally 10- to 100-fold. Aside from species-specific differences, multiple factors, including donor variation and the growth conditions of both the host cell and the bacterium, can affect intracellular survival/replication. The comparison here of cells grown in human versus fetal bovine serum clearly demonstrates how different protocols can have profound impacts on the results. We also confirmed the findings of Henry et al. (44), who showed that supplemental histidine is required for growth of the histidine auxotroph SL1344 in several cell types. To our knowledge, histidine is not routinely added during infection of murine macrophages or macrophage-like cell lines with SL1344. Finally, although this has not been systematically addressed, differences in bacterial growth conditions and/or different mechanisms of internalization can have significant impacts on the ability of Salmonella Typhimurium to survive and replicate in human macrophages.
It has been well documented that Salmonella Typhimurium expressing T3SS1 triggers pyroptosis shortly after entry (55–57). However, under the conditions used here, including a relatively low MOI and a short internalization of 10 min, the vast majority of infected cells contained less than 5 bacteria at 2 h p.i., and pyroptosis was not observed (see Fig. S2 in the supplemental material). When a 10× higher MOI was used, we did observe significant early SPI1-dependent cell death, indicating that this is a concentration-dependent process. In vivo it is likely that macrophages will encounter bacteria with various transcriptional profiles, often at low MOIs. For example, SPI1-induced bacteria are released from infected epithelial cells in vivo (21, 22). Further study is required to determine what role the number of intracellular bacteria plays in the induction of pyroptosis both in vitro and in vivo.
Interestingly, we found that the requirement for the SPI2-encoded T3SS2 may not be as clear-cut as has been described for mouse macrophages, particularly at early time points. In vitro studies using mouse macrophages or macrophage-like cell lines report a strong dependence on T3SS2 for replication (6, 12), although these studies used stationary-phase opsonized bacteria and looked only at late (16 to 20 h) time points. Here, we used log-phase bacteria internalized by invasion and observed early SPI2-independent replication followed by later SPI2-dependent replication, so that there were approximately 50% fewer viable intracellular SPI2 mutant bacteria than WT bacteria at 18 h p.i. Host cell death was clearly associated with SPI2-dependent replication at ≈20 h p.i., since death was observed in cells containing replicating WT but not ΔSPI2 bacteria. SPI2-dependent late apoptosis of infected macrophages in vitro has been described previously in both mouse and human macrophages (20, 31) and may be an important mechanism for the spread of the bacteria to neighboring cells in vivo (58).
Our data confirm that Salmonella Typhimurium is capable of survival and replication in some subsets of human macrophages. We have shown that multiple factors, on both the host and pathogen side, can profoundly affect the outcome of infection. Further studies, in particular a systematic comparison of different in vitro-generated macrophage phenotypes, and a thorough comparison of primary mouse and human macrophages are needed to better understand the interactions critical to human infections.
Supplementary Material
ACKNOWLEDGMENTS
We thank the members of the Steele-Mortimer laboratory, specifically Timothy Bauler, Ciaran Finn, and Sushmita Sridhar, and Scott A. Wetzel (University of Montana) for critical review of the manuscript.
This work was supported by the Intramural Research Program of the NIH, National Institute of Allergy and Infectious Diseases.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.00033-15.
REFERENCES
- 1.Feasey NA, Dougan G, Kingsley RA, Heyderman RS, Gordon MA. 2012. Invasive non-typhoidal salmonella disease: an emerging and neglected tropical disease in Africa. Lancet 379:2489–2499. doi: 10.1016/S0140-6736(11)61752-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gordon MA. 2011. Invasive nontyphoidal Salmonella disease: epidemiology, pathogenesis and diagnosis. Curr Opin Infect Dis 24:484–489. doi: 10.1097/QCO.0b013e32834a9980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gordon MA, Banda HT, Gondwe M, Gordon SB, Boeree MJ, Walsh AL, Corkill JE, Hart CA, Gilks CF, Molyneux ME. 2002. Non-typhoidal salmonella bacteraemia among HIV-infected Malawian adults: high mortality and frequent recrudescence. AIDS 16:1633–1641. doi: 10.1097/00002030-200208160-00009. [DOI] [PubMed] [Google Scholar]
- 4.Gordon MA, Kankwatira AMK, Mwafulirwa G, Walsh AL, Hopkins MJ, Parry CM, Faragher EB, Zijlstra EE, Heyderman RS, Molyneux ME. 2010. Invasive non-typhoid salmonellae establish systemic intracellular infection in HIV-infected adults: an emerging disease pathogenesis. Clin Infect Dis 50:953–962. doi: 10.1086/651080. [DOI] [PubMed] [Google Scholar]
- 5.Tsolis RM, Xavier MN, Santos RL, Bäumler AJ. 2011. How to become a top model: impact of animal experimentation on human Salmonella disease research. Infect Immun 79:1806–1814. doi: 10.1128/IAI.01369-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cirillo DM, Valdivia RH, Monack DM, Falkow S. 1998. Macrophage-dependent induction of the Salmonella pathogenicity island 2 type III secretion system and its role in intracellular survival. Mol Microbiol 30:175–188. doi: 10.1046/j.1365-2958.1998.01048.x. [DOI] [PubMed] [Google Scholar]
- 7.Fields PI, Swanson RV, Haidaris CG, Heffron F. 1986. Mutants of Salmonella typhimurium that cannot survive within the macrophage are avirulent. Proc Natl Acad Sci U S A 83:5189–5193. doi: 10.1073/pnas.83.14.5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Groisman EA. 2001. The pleiotropic two-component regulatory system PhoP-PhoQ. J Bacteriol 183:1835–1842. doi: 10.1128/JB.183.6.1835-1842.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ochman H, Soncini FC, Solomon F, Groisman EA. 1996. Identification of a pathogenicity island required for Salmonella survival in host cells. Proc Natl Acad Sci U S A 93:7800–7804. doi: 10.1073/pnas.93.15.7800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thompson JA, Liu M, Helaine S, Holden DW. 2011. Contribution of the PhoP/Q regulon to survival and replication of Salmonella enterica serovar Typhimurium in macrophages. Microbiology 157:2084–2093. doi: 10.1099/mic.0.048926-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Steele-Mortimer O. 2008. The Salmonella-containing vacuole: moving with the times. Curr Opin Microbiol 11:38–45. doi: 10.1016/j.mib.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hensel M, Shea JE, Waterman SR, Mundy R, Nikolaus T, Banks G, Vázquez-Torres A, Gleeson C, Fang FC, Holden DW. 1998. Genes encoding putative effector proteins of the type III secretion system of Salmonella pathogenicity island 2 are required for bacterial virulence and proliferation in macrophages. Mol Microbiol 30:163–174. doi: 10.1046/j.1365-2958.1998.01047.x. [DOI] [PubMed] [Google Scholar]
- 13.Moest TP, Méresse S. 2013. Salmonella T3SSs: successful mission of the secret(ion) agents. Curr Opin Microbiol 16:38–44. doi: 10.1016/j.mib.2012.11.006. [DOI] [PubMed] [Google Scholar]
- 14.Monack DM, Navarre WW, Falkow S. 2001. Salmonella-induced macrophage death: the role of caspase-1 in death and inflammation. Microbes Infect 3:1201–1212. doi: 10.1016/S1286-4579(01)01480-0. [DOI] [PubMed] [Google Scholar]
- 15.Lundberg U, Vinatzer U, Berdnik D, von Gabain A, Baccarini M. 1999. Growth phase-regulated induction of Salmonella-induced macrophage apoptosis correlates with transient expression of SPI-1 genes. J Bacteriol 181:3433–3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brennan MA, Cookson BT. 2000. Salmonella induces macrophage death by caspase-1-dependent necrosis. Mol Microbiol 38:31–40. doi: 10.1046/j.1365-2958.2000.02103.x. [DOI] [PubMed] [Google Scholar]
- 17.Miao EA, Leaf IA, Treuting PM, Mao DP, Dors M, Sarkar A, Warren SE, Wewers MD, Aderem A. 2010. Caspase-1-induced pyroptosis is an innate immune effector mechanism against intracellular bacteria. Nat Immunol 11:1136–1142. doi: 10.1038/ni.1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miao EA, Warren SE. 2010. Innate immune detection of bacterial virulence factors via the NLRC4 inflammasome. J Clin Immunol 30:502–506. doi: 10.1007/s10875-010-9386-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Libby SJ, Lesnick M, Hasegawa P, Weidenhammer E, Guiney DG. 2000. The Salmonella virulence plasmid spv genes are required for cytopathology in human monocyte-derived macrophages. Cell Microbiol 2:49–58. doi: 10.1046/j.1462-5822.2000.00030.x. [DOI] [PubMed] [Google Scholar]
- 20.van der Velden AW, Lindgren SW, Worley MJ, Heffron F. 2000. Salmonella pathogenicity island 1-independent induction of apoptosis in infected macrophages by Salmonella enterica serotype Typhimurium. Infect Immun 68:5702–5709. doi: 10.1128/IAI.68.10.5702-5709.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Knodler LA, Vallance BA, Celli J, Winfree S, Hansen B, Montero M, Steele-Mortimer O. 2010. Dissemination of invasive Salmonella via bacterial-induced extrusion of mucosal epithelia. Proc Natl Acad Sci U S A 107:17733–17738. doi: 10.1073/pnas.1006098107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Laughlin RC, Knodler LA, Barhoumi R, Payne HR, Wu J, Gomez G, Pugh R, Lawhon SD, Bäumler AJ, Steele-Mortimer O, Adams LG. 2014. Spatial segregation of virulence gene expression during acute enteric infection with Salmonella enterica serovar Typhimurium. mBio 5:e00946-13. doi: 10.1128/mBio.00946-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mills CD, Kincaid K, Alt JM, Heilman MJ, Hill AM. 2000. M-1/M-2 macrophages and the Th1/Th2 paradigm. J Immunol 164:6166–6173. doi: 10.4049/jimmunol.164.12.6166. [DOI] [PubMed] [Google Scholar]
- 24.Mantovani A, Sica A, Sozzani S, Allavena P, Vecchi A, Locati M. 2004. The chemokine system in diverse forms of macrophage activation and polarization. Trends Immunol 25:677–686. doi: 10.1016/j.it.2004.09.015. [DOI] [PubMed] [Google Scholar]
- 25.Richter-Dahlfors A, Buchan AM, Finlay BB. 1997. Murine salmonellosis studied by confocal microscopy: Salmonella typhimurium resides intracellularly inside macrophages and exerts a cytotoxic effect on phagocytes in vivo. J Exp Med 186:569–580. doi: 10.1084/jem.186.4.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McCoy MW, Moreland SM, Detweiler CS. 2012. Hemophagocytic macrophages in murine typhoid fever have an anti-inflammatory phenotype. Infect Immun 80:3642–3649. doi: 10.1128/IAI.00656-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Salcedo SP, Noursadeghi M, Cohen J, Holden DW. 2001. Intracellular replication of Salmonella typhimurium strains in specific subsets of splenic macrophages in vivo. Cell Microbiol 3:587–597. doi: 10.1046/j.1462-5822.2001.00137.x. [DOI] [PubMed] [Google Scholar]
- 28.Schwan WR, Huang X-Z, Hu L, Kopecko DJ. 2000. Differential bacterial survival, replication, and apoptosis-inducing ability of Salmonella serovars within human and murine macrophages. Infect Immun 68:1005–1013. doi: 10.1128/IAI.68.3.1005-1013.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Strandberg KL, Richards SM, Gunn JS. 2012. Cathelicidin antimicrobial peptide expression is not induced or required for bacterial clearance during Salmonella enterica infection of human monocyte-derived macrophages. Infect Immun 80:3930–3938. doi: 10.1128/IAI.00672-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gallois A, Klein J, Allen L-AH, Jones BD, Nauseef WM. 2001. Salmonella pathogenicity island 2-encoded type III secretion system mediates exclusion of NADPH oxidase assembly from the phagosomal membrane. J Immunol 166:5741–5748. doi: 10.4049/jimmunol.166.9.5741. [DOI] [PubMed] [Google Scholar]
- 31.Browne SH, Lesnick ML, Guiney DG. 2002. Genetic requirements for Salmonella-induced cytopathology in human monocyte-derived macrophages. Infect Immun 70:7126–7135. doi: 10.1128/IAI.70.12.7126-7135.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ibarra JA, Knodler LA, Sturdevant DE, Virtaneva K, Carmody AB, Fischer ER, Porcella SF, Steele-Mortimer O. 2010. Induction of Salmonella pathogenicity island 1 under different growth conditions can affect Salmonella-host cell interactions in vitro. Microbiology 156:1120–1133. doi: 10.1099/mic.0.032896-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cormack BP, Valdivia RH, Falkow S. 1996. FACS-optimized mutants of the green fluorescent protein (GFP). Gene 173:33–38. doi: 10.1016/0378-1119(95)00685-0. [DOI] [PubMed] [Google Scholar]
- 34.Allam US, Krishna MG, Sen M, Thomas R, Lahiri A, Gnanadhas DP, Chakravortty D. 2014. Acidic pH induced STM1485 gene is essential for intracellular replication of Salmonella. Virulence 3:122–135. doi: 10.4161/viru.19029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hoiseth SK, Stocker BAD. 1981. Aromatic-dependent Salmonella typhimurium are non-virulent and effective as live vaccines. Nature 291:238–239. doi: 10.1038/291238a0. [DOI] [PubMed] [Google Scholar]
- 36.Knodler LA, Vallance BA, Hensel M, Jäckel D, Finlay BB, Steele-Mortimer O. 2003. Salmonella type III effectors PipB and PipB2 are targeted to detergent-resistant microdomains on internal host cell membranes. Mol Microbiol 49:685–704. doi: 10.1046/j.1365-2958.2003.03598.x. [DOI] [PubMed] [Google Scholar]
- 37.Bustamante VH, Martínez LC, Santana FJ, Knodler LA, Steele-Mortimer O, Puente JL. 2008. HilD-mediated transcriptional cross-talk between SPI-1 and SPI-2. Proc Natl Acad Sci U S A 105:14591–14596. doi: 10.1073/pnas.0801205105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Drecktrah D, Levine-Wilkinson S, Dam T, Winfree S, Knodler LA, Schroer TA, Steele-Mortimer O. 2008. Dynamic behavior of Salmonella-induced membrane tubules in epithelial cells. Traffic 9:2117–2129. doi: 10.1111/j.1600-0854.2008.00830.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Valnes K, Brandtzaeg P. 1985. Retardation of immunofluorescence fading during microscopy. J Histochem Cytochem 33:755–761. doi: 10.1177/33.8.3926864. [DOI] [PubMed] [Google Scholar]
- 40.Ambarus CA, Krausz S, van Eijk M, Hamann J, Radstake TRDJ, Reedquist KA, Tak PP, Baeten DLP. 2012. Systematic validation of specific phenotypic markers for in vitro polarized human macrophages. J Immunol Methods 375:196–206. doi: 10.1016/j.jim.2011.10.013. [DOI] [PubMed] [Google Scholar]
- 41.Verreck FAW, de Boer T, Langenberg DML, Hoeve MA, Kramer M, Vaisberg E, Kastelein R, Kolk A, de Waal-Malefyt R, Ottenhoff THM. 2004. Human IL-23-producing type 1 macrophages promote but IL-10-producing type 2 macrophages subvert immunity to (myco)bacteria. Proc Natl Acad Sci U S A 101:4560–4565. doi: 10.1073/pnas.0400983101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Verreck FAW, de Boer T, Langenberg DML, van der Zanden L, Ottenhoff THM. 2006. Phenotypic and functional profiling of human proinflammatory type-1 and anti-inflammatory type-2 macrophages in response to microbial antigens and IFN-gamma- and CD40L-mediated costimulation. J Leukoc Biol 79:285–293. doi: 10.1189/jlb.0105015. [DOI] [PubMed] [Google Scholar]
- 43.Kröger C, Colgan A, Srikumar S, Händler K, Sivasankaran SK, Hammarlöf DL, Canals R, Grissom JE, Conway T, Hokamp K, Hinton JCD. 2013. An infection-relevant transcriptomic compendium for Salmonella enterica serovar Typhimurium. Cell Host Microbe 14:683–695. doi: 10.1016/j.chom.2013.11.010. [DOI] [PubMed] [Google Scholar]
- 44.Henry T, García-del Portillo F, Gorvel JP. 2005. Identification of Salmonella functions critical for bacterial cell division within eukaryotic cells. Mol Microbiol 56:252–267. doi: 10.1111/j.1365-2958.2005.04540.x. [DOI] [PubMed] [Google Scholar]
- 45.Alpuche-Aranda CM, Swanson JA, Loomis WP, Miller SI. 1992. Salmonella typhimurium activates virulence gene transcription within acidified macrophage phagosomes. Proc Natl Acad Sci U S A 89:10079–10083. doi: 10.1073/pnas.89.21.10079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Martin-Orozco N, Touret N, Zaharik ML, Park E, Kopelman R, Miller S, Finlay BB, Gros P, Grinstein S. 2006. Visualization of vacuolar acidification-induced transcription of genes of pathogens inside macrophages. Mol Biol Cell 17:498–510. doi: 10.1091/mbc.E04-12-1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dalebroux ZD, Miller SI. 2014. Salmonellae PhoPQ regulation of the outer membrane to resist innate immunity. Curr Opin Microbiol 17:106–113. doi: 10.1016/j.mib.2013.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Malik-Kale P, Winfree S, Steele-Mortimer O. 2012. The bimodal lifestyle of intracellular Salmonella in epithelial cells: replication in the cytosol obscures defects in vacuolar replication. PLoS One 7:e38732. doi: 10.1371/journal.pone.0038732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gordon S, Plüddemann A. 2013. Tissue macrophage heterogeneity: issues and prospects. Semin Immunopathol 35:533–540. doi: 10.1007/s00281-013-0386-4. [DOI] [PubMed] [Google Scholar]
- 50.Gordon MA, Jack DL, Dockrell DH, Lee ME, Read RC. 2005. Gamma interferon enhances internalization and early nonoxidative killing of Salmonella enterica serovar Typhimurium by human macrophages and modifies cytokine responses. Infect Immun 73:3445–3452. doi: 10.1128/IAI.73.6.3445-3452.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Raes G, Van den Bergh R, De Baetselier P, Ghassabeh GH, Scotton C, Locati M, Mantovani A, Sozzani S. 2005. Arginase-1 and Ym1 are markers for murine, but not human, alternatively activated myeloid cells. J Immunol 174:6561–6562. doi: 10.4049/jimmunol.174.11.6561. [DOI] [PubMed] [Google Scholar]
- 52.Eisele NA, Ruby T, Jacobson A, Manzanillo PS, Cox JS, Lam L, Mukundan L, Chawla A, Monack DM. 2013. Salmonella require the fatty acid regulator PPARδ for the establishment of a metabolic environment essential for long-term persistence. Cell Host Microbe 14:171–182. doi: 10.1016/j.chom.2013.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jouanguy E, Doffinger R, Dupuis S, Pallier A, Altare F, Casanova JL. 1999. IL-12 and IFN-gamma in host defense against mycobacteria and salmonella in mice and men. Curr Opin Immunol 11:346–351. doi: 10.1016/S0952-7915(99)80055-7. [DOI] [PubMed] [Google Scholar]
- 54.Martinez FO, Gordon S, Locati M, Mantovani A. 2006. Transcriptional profiling of the human monocyte-to-macrophage differentiation and polarization: new molecules and patterns of gene expression. J Immunol 177:7303–7311. doi: 10.4049/jimmunol.177.10.7303. [DOI] [PubMed] [Google Scholar]
- 55.Miao EA, Alpuche-Aranda CM, Dors M, Clark AE, Bader MW, Miller SI, Aderem A. 2006. Cytoplasmic flagellin activates caspase-1 and secretion of interleukin 1β via Ipaf. Nat Immunol 7:569–575. doi: 10.1038/ni1344. [DOI] [PubMed] [Google Scholar]
- 56.Miao EA, Rajan JV. 2011. Salmonella and caspase-1: a complex interplay of detection and evasion. Front Microbiol 2:85. doi: 10.3389/fmicb.2011.00085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Broz P, von Moltke J, Jones JW, Vance RE, Monack DM. 2010. Differential requirement for caspase-1 autoproteolysis in pathogen-induced cell death and cytokine processing. Cell Host Microbe 8:471–483. doi: 10.1016/j.chom.2010.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Grant AJ, Morgan FJE, McKinley TJ, Foster GL, Maskell DJ, Mastroeni P. 2012. Attenuated Salmonella Typhimurium lacking the pathogenicity island-2 type 3 secretion system grow to high bacterial numbers inside phagocytes in mice. PLoS Pathog 8:e1003070. doi: 10.1371/journal.ppat.1003070. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.