Abstract
Voltage-gated sodium channels initiate action potentials in nerve, muscle, and other electrically excitable cells. Voltage-gated calcium channels are activated by depolarization during action potentials, and calcium influx through them is the key second messenger of electrical signaling, initiating secretion, contraction, neurotransmission, gene transcription, and many other intracellular processes. Drugs that block sodium channels are used in local anesthesia and the treatment of epilepsy, bipolar disorder, chronic pain, and cardiac arrhythmia. Drugs that block calcium channels are used in the treatment of epilepsy, chronic pain, and cardiovascular disorders, including hypertension, angina pectoris, and cardiac arrhythmia. The principal pore-forming subunits of voltage-gated sodium and calcium channels are structurally related and likely to have evolved from ancestral voltage-gated sodium channels that are widely expressed in prokaryotes. Determination of the structure of a bacterial ancestor of voltage-gated sodium and calcium channels at high resolution now provides a three-dimensional view of the binding sites for drugs acting on sodium and calcium channels. In this minireview, we outline the different classes of sodium and calcium channel drugs, review studies that have identified amino acid residues that are required for their binding and therapeutic actions, and illustrate how the analogs of those key amino acid residues may form drug-binding sites in three-dimensional models derived from bacterial channels.

Introduction
Introduction to the Pharmacology of Voltage-Gated Sodium and Calcium Channels
Sodium Channels.
Local anesthetics prevent pain by blocking initiation and propagation of the action potential in sensory nerves through blockage of voltage-gated sodium channels (Catterall and Mackie, 2011). Some antiepileptic drugs, including diphenylhydantoin, carbamazepine, and lamotrigine, prevent seizures by blocking brain sodium channels (McNamara, 2011). Similarly, some antiarrhythmic drugs, including quinidine, procainamide, lidocaine, and flecainide, interrupt and prevent cardiac arrhythmias by blocking cardiac sodium channels (Sampson and Kass, 2011). Thus, reduction of sodium channel activity is a major mechanism in pharmacology.
Complete block of sodium channels would have major unwanted side effects, including complete loss of sensation in sensory nerves, sedation or coma in the brain, and cardiac arrest in the heart. Moreover, sodium channel–blocking drugs in current clinical use are not selective among the nine voltage-gated sodium channels in mammals (Catterall et al., 2005a) so unwanted side effects of complete sodium channel block would be observed in multiple tissues. Sodium channel blockers are only usable in therapy because the frequency and voltage dependence of their action allows them to inhibit action potential generation in the depolarized and rapidly firing cells that are responsible for pain, epilepsy, and arrhythmia without complete block of action potential generation in normally functioning cells.
Frequency- and voltage-dependent block of sodium channels is currently understood in terms of the modulated receptor hypothesis (Hille, 1977). According to this concept, sodium channel–blocking drugs can gain access to their receptor site in the lumen of the pore more rapidly when the intracellular activation gate of the channel is open during an action potential, and these drugs bind with higher affinity to inactivated sodium channels that accumulate during high-frequency firing of healthy cells and/or steady depolarization of damaged cells. Understanding the structural basis for frequency- and voltage-dependent block of sodium channels is an important goal in sodium channel pharmacology and will provide the foundation for future structure-based design of safer and more efficacious drugs.
Calcium Channels.
Voltage-gated calcium channels are also important drug targets. Phenylalkylamine calcium channel blockers like verapamil are used in the treatment of atrial arrhythmias, as are benzothiazepines like diltiazem (Sampson and Kass, 2011). These drugs are thought to act at receptor site(s) in the pore of the calcium channels and block them, much like sodium channel–blocking drugs block sodium channels. The action of verapamil and diltiazem is frequency- and voltage-dependent, and these effects are currently understood in terms of the modulated receptor hypothesis (Hondeghem and Katzung, 1984). Effective use of verapamil and diltiazem in the treatment of atrial arrhythmias is thought to depend on this state-dependent block.
Calcium antagonist drugs are also used in the treatment of hypertension and angina pectoris; however, dihydropyridines are the drugs of choice for these indications (Michel and Hoffmann, 2011). Dihydropyridines can either enhance or inhibit activation of voltage-gated calcium channels in heart and vascular smooth muscle by acting at an allosteric modulatory site located outside of the pore (Hess et al., 1984; Kokubun and Reuter, 1984; Kokubun et al., 1986). Their binding is highly voltage dependent, and the calcium antagonist drugs used in therapy bind with high affinity to the inactivated state of calcium channels (Bean, 1984; Kokubun and Reuter, 1984; Kokubun et al., 1986). It is thought that their efficacy in hypertension and angina pectoris depends on the voltage-dependent block of calcium channels in depolarized, continuously contracting vascular smooth muscle cells. They have lesser effects on calcium channels in the heart because the membrane potential of cardiac myocytes returns to negative values in the range of −85 mV during each cardiac cycle, allowing dissociation of the drugs from the resting state of calcium channels. Understanding the structural basis for the frequency- and voltage-dependent block of calcium channels would allow structure-based design and the development of safer and more effective drugs for therapy of cardiovascular diseases.
New generations of calcium channel drugs have emerged for the treatment of epilepsy and chronic pain. ω-Conotoxins are potent blockers of presynaptic calcium channels in neurons. The synthetic derivative ziconitide (Prialt) is administered intrathecally for the control of severe pain in advanced cancer patients and others with intractable pain conditions (Miljanich and Ramachandran, 1995; Lewis et al., 2012). Gabapentin (Neurontin) and pregabalin (Lyrica) were originally designed as GABA-related drugs (hence the names), but these compounds actually act on calcium channels by altering their membrane trafficking (Davies et al., 2007). These drugs act on the auxiliary α2δ subunits of calcium channels, so recent advances on the structure of prokaryotic channels have not given insight into the sites and mechanisms of action of these drugs.
Structures of Sodium Channels
Sodium Channel Protein.
Voltage-gated sodium channels are complexes of a large, pore-forming α subunit of ∼260 kDa and one or two β subunits of 30–40 kDa (Catterall et al., 2005b). The α subunit of ∼2000 amino acid residues is organized in four homologous domains that each contain six transmembrane segments (S1–S6) and an additional membrane re-entrant segment (Fig. 1A). Segments S1–S4 form the voltage-sensing module, whereas segments S5 and S6 and the P loop between them form the pore. Fast inactivation is mediated by the short intracellular loop between domains III and IV. The receptor site for local anesthetics and related drugs is formed by amino acid residues in the IS6, IIS6, and IVS6 segments, which contribute to a single drug-binding site in the pore (Fig. 1B). This two-dimensional map of the sodium channel has provided the template for detailed structure-function studies, but the large mammalian sodium channels have resisted efforts at high-resolution three-dimensional structure determination. The discovery of bacterial sodium channels that are analogous in structure to one domain of mammalian sodium channels and function as homotetramers (Ren et al., 2001) has opened the way to the structural biology of sodium channels.
Fig. 1.
Structure of voltage-gated sodium channels. (A) A transmembrane folding diagram of the NaV1.2 channel. Cylinders represent α-helical segments. Bold lines represent the polypeptide chains of each subunit, with a length approximately proportional to the number of amino acid residues in the brain sodium channel subtypes. The extracellular domains of the β1 and β2 subunits are shown as immunoglobulin-like folds. Ψ, sites of probable N-linked glycosylation; P in red, sites of demonstrated protein phosphorylation by protein kinase A (circles) and protein kinase C (diamonds); blue, pore-lining segments; yellow circles, the outer (EEEE) and inner (DEKA) rings of amino residues that form the tetrodotoxin-binding site and ion selectivity filter; green, S1–S4 voltage sensors; h in blue circle, inactivation particle in the inactivation gate loop; blue circles, sites implicated in forming the inactivation gate receptor. ScTx, scorpion toxin. (B) Model of the local anesthetic receptor site in mammalian NaV1.2 channels. (C) Side view of NaVAb channels colored according to (A): voltage-sensing module (green); pore module (blue); S4-S5 linker (red). (D) Side view of the ion selectivity filter. Glu177 (purple) interactions with Gln172, Ser178, and the backbone of Ser180 are shown in the far subunit. Fo-Fc omit map, 4.75 σ (blue); putative cations or water molecules (red spheres, ionEX). Electron-density around Leu176 (gray; Fo-Fc omit map at 1.75 σ) and a putative water molecule is shown (gray sphere). Na+-coordination sites: siteHFS, siteCEN, and siteIN. (E) Architecture of the NaVAb pore: Glu177 side chains (purple) and pore volume (gray). The S5 and S6 segments and P loop from two lateral subunits are shown.
Architecture of the Pore.
Determination of the crystal structure of the bacterial sodium channel NavAb at high resolution (2.7 Å) has given new insights into the structural basis for many aspects of sodium channel function (Fig. 1, B and D) (Payandeh et al., 2011). As viewed from the side, NavAb has a central bundle of four S5 and four S6 segments that form the pore (Fig. 1C, blue). As viewed from the top, the central pore is surrounded by four pore-forming modules composed of S5 and S6 segments and the intervening P loop (Fig. 1C, blue). Four voltage-sensing modules composed of S1–S4 segments are symmetrically associated with the outer rim of the pore module (Fig. 1C, green), which is connected by the S4-S5 linkers in each domain (Fig. 1C, red). The transmembrane architecture of NavAb shows that adjacent subunits have swapped functional modules, such that each voltage-sensing module is most closely associated with the pore-forming module of its neighbor (Fig. 1C) (Payandeh et al., 2011). It is likely that this module-swapping arrangement enforces concerted gating of the four subunits of homotetrameric bacterial sodium channels and possibly also has the same function in the four homologous domains of the more complex pseudotetrameric mammalian sodium channels.
Amino acid residues in the short α-helical segments between S5 and S6 (Fig. 1A) form the receptor site for the pore blocker tetrodotoxin (Fig. 1A, yellow circles) (Noda et al., 1989; Terlau et al., 1991), and mutations of these amino acid residues alter ion selectivity (Heinemann et al., 1992). The crystal structure of the NavAb channel reveals a narrow ion selectivity filter with three ion interaction sites: SiteHFS formed by the negatively charged carboxylate groups of Glu177, SiteCEN formed by the backbone carbonyls of Leu176, and SiteIN formed by the backbone carbonyls of Thr175 (Fig. 1D). This selectivity filter is set in a transmembrane pore with a large external vestibule (Fig. 1E), the narrow ion selectivity filter containing the amino acid residues that determine ion selectivity (Fig. 1E), a large central cavity lined by the S6 segments (Fig. 1E), and an intracellular activation gate formed at the crossing of the S6 segments at the intracellular surface of the membrane (Fig. 1E). The activation gate is tightly closed in the NavAb structure (Payandeh et al., 2011). A high-resolution structure of a pore-only construct of NaVMs (McCusker et al., 2012) and a cryo–electron microscopy structure of NaVCt at a resolution of 9 Å (Tsai et al., 2013) reveal two possible open-gate conformations.
The NavAb ion selectivity filter has a high field-strength site at its extracellular end (Fig. 1D) formed by the side chains of four glutamate residues (Payandeh et al., 2011), which are highly conserved and key determinants of ion selectivity in vertebrate sodium and calcium channels (Heinemann et al., 1992). Considering selectivity-filter dimensions of approximately 4.6 Å2, Na+ with two planar waters of hydration could fit in this high field–strength site. This outer site is followed on its intracellular side by two ion coordination sites formed by backbone carbonyls (Fig. 1D) (Payandeh et al., 2011). These two carbonyl sites are perfectly designed to bind Na+ with four planar waters of hydration but would be much too large to bind Na+ directly. Thus, Na+ is selected and conducted as a hydrated ion interacting with the pore through its inner shell of bound water molecules.
Structural Basis for Voltage-Dependent Gating.
Voltage-gated sodium channels are activated in response to depolarization by the outward movement of electrically charged particles across the cell membrane through the transmembrane electric field (Hodgkin and Huxley, 1952b). The transmembrane movement of these gating charges was detected as a small capacitive gating current in a squid giant axon (Armstrong and Bezanilla, 1973). The S4 segments of sodium channels, which contain four to eight repeated motifs of a positively charged amino acid residue (usually arginine) flanked by two hydrophobic residues, were proposed to span the membrane (Fig. 1A) and carry the gating charges in the sliding helix or helical screw model of voltage sensing (Catterall, 1986; Guy and Seetharamulu, 1986; Yarov-Yarovoy et al., 2006, 2012). In this model, the S4 segment is in a transmembrane position in both the resting and activated states, the gating charges are stabilized by forming ion pairs with neighboring negatively charged and hydrophilic residues in other transmembrane segments, and their outward movement is catalyzed by exchange of ion pair partners (Catterall, 1986; Guy and Seetharamulu, 1986; Yarov-Yarovoy et al., 2006, 2012). Extensive structure-function studies now provide strong support for all of the elements of this model [reviewed by Catterall (2010)], and a consensus mechanism for voltage sensor function based on the sliding helix model has been presented by several leading investigators (Vargas et al., 2012). The voltage-driven outward movement of the gating charges in the S4 segment initiates the conformational changes that lead to pore opening and inactivation of sodium channels.
Sodium channels in vertebrates open in response to depolarization and then inactivate within 1–2 milliseconds (Hodgkin and Huxley, 1952a). This fast inactivation process is required for repetitive firing of action potentials in neural circuits and for control of excitability in nerve and muscle cells. The short intracellular loop connecting homologous domains III and IV of the sodium channel α subunit is responsible for fast inactivation by folding into the intracellular mouth of the pore and blocking it (Fig. 1A), as shown by blockage of fast inactivation by antibodies directed against this peptide segment (Vassilev et al., 1988). Fast inactivation is also greatly slowed by cutting this loop by expression of the sodium channel as two pieces: one containing domains I–III and a second containing domain IV (Stuhmer et al., 1989). The key amino acid motif IFM is required to maintain closure of the inactivation gate (West et al., 1992), and peptides containing this inactivation gate sequence motif can restore fast inactivation to mutant sodium channels (Eaholtz et al., 1994). Analysis of the structure of the inactivation gate by NMR showed that it contains a rigid α helix preceded by two loops of protein that optimally array the IFM motif and a neighboring Thr residue for interaction with and block of the open pore of the channel (Rohl et al., 1999). The fast inactivation gate is not present in homotetrameric bacterial sodium channels, so further structural analysis must await determination of the three-dimensional structure of a vertebrate sodium channel.
Bacterial sodium channels do not have a fast inactivation process because they lack the equivalent of the linker between domains III and IV of vertebrate sodium channels. However, bacterial sodium channels have a slow inactivation process that does not involve a cytosolic inactivation gate (Pavlov et al., 2005). Like vertebrate sodium channels (Vilin and Ruben, 2001), slow inactivation of bacterial sodium channels involves amino acid residues in the P loop and S6 segment (Zhao et al., 2004a,b; Pavlov et al., 2005). In crystallographic studies of two different bacterial sodium channels, the nearly square 4-fold symmetric structure of the pore in the preopen state of NavAb is collapsed to a parallelogram in a dimer-of-dimers symmetry by movement of two S6 segments toward the central axis of the pore and corresponding movement of the other two S6 segments away from the central axis (Payandeh et al., 2012). This movement is more pronounced in NavRh than in NavAb, suggesting a sequence of conformational changes during the multistep slow inactivation process (Zhang et al., 2012).
Drug Receptor Sites in Sodium Channels
Sodium channels are blocked by drugs used clinically as local anesthetics, antiarrhythmics, and antiepileptics as described above. Site-directed mutagenesis studies revealed the receptor site for local anesthetics and related drugs, which is formed by amino acid residues in the S6 segments in domains I, III, and IV (Fig. 1, A and B) (Ragsdale et al., 1994, 1996; Wang et al., 1998; Yarov-Yarovoy et al., 2001, 2002). These drugs bind to a common receptor site in the pore and impede ion permeation. The amino acid residues that form the receptor sites for sodium channel blockers line the inner surface of the S6 segments and create a three-dimensional drug receptor site whose occupancy would block the pore (Fig. 1B) (Ragsdale et al., 1994; Catterall, 2000; Yarov-Yarovoy et al., 2002; Payandeh et al., 2011). Access to this receptor site by large or hydrophilic drugs would require opening of the intracellular activation gate, which is tightly closed in the NavAb structure (Fig. 1E) (Payandeh et al., 2011). This tight closure of the activation gate provides a structural basis for use-dependent block of sodium channels by local anesthetics and related drugs (Hille, 1977), as they would bind much more rapidly when the channel is frequently opened.
Remarkably, as also predicted by the modulated receptor hypothesis (Hille, 1977), fenestrations lead from the lipid phase of the membrane sideways into the drug receptor site, providing a specific hydrophobic access pathway for binding of small hydrophobic drugs in the resting state of the channel (Fig. 2, A and B, pore portals) (Payandeh et al., 2011). Access to the drug-binding site in NavAb channels is controlled by the side chain of a single amino acid residue Phe203 (Fig. 2, A and B) (Payandeh et al., 2011), which is homologous to amino acid residues identified in previous structure-function studies that control drug access and egress from the local anesthetic receptor site in mammalian cardiac and brain sodium channels (Ragsdale et al., 1994; Qu et al., 1995).
Fig. 2.
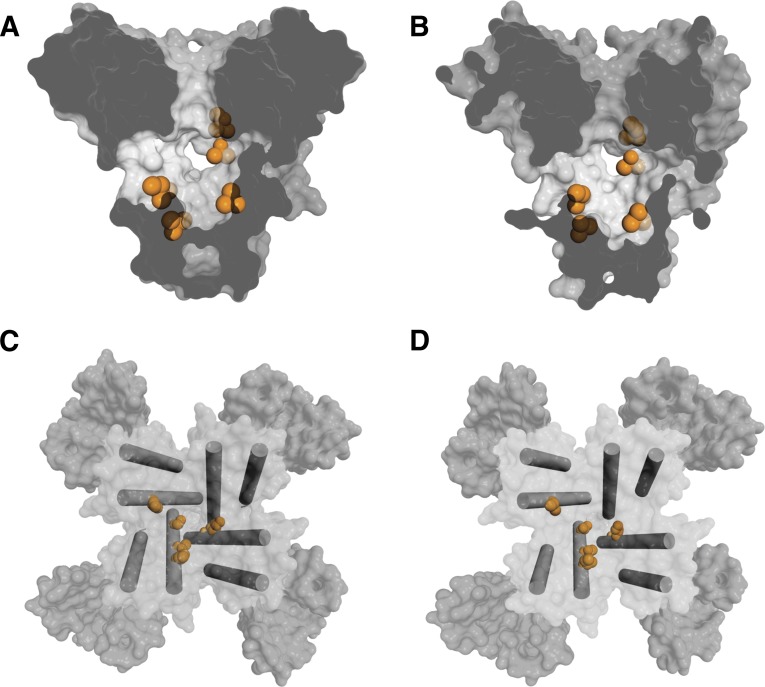
Drug-binding sites and fenestrations in NavAb. (A) Side view through the pore module illustrating fenestrations (portals) and hydrophobic access to central cavity. Phe203 side chains are yellow sticks. Surface representations of NaVAb residues aligning with those implicated in drug binding and block: Thr206 (blue), Met209 (green), and Val213 (orange). Membrane boundaries are gray lines. Electron-density from an Fo-Fc omit map is contoured at 2.0 σ. (B) Top view sectioned below the selectivity filter colored as in (A). (C) Structure of the drug-binding site in the slow-inactivated state in NaVAb.
The conformational changes in the pore during slow inactivation cause striking rearrangements of the drug-binding sites and alter the size and orientation of the fenestrations (Fig. 2C) (Payandeh et al., 2012), which would be expected to alter both drug-binding affinity and drug access to the binding site. State-dependent drug access and drug affinity are both hallmarks of sodium channel pharmacology, whose molecular basis is revealed in a preliminary form in these crystal structures of bacterial sodium channels. The conformational changes that take place during slow inactivation likely contribute to the increased affinity for drug binding to inactivated sodium channels.
NavAb and other bacterial sodium channels are blocked by local anesthetics and related drugs in a voltage- and frequency-dependent manner, similar to mammalian NaV channels, but with lower affinity (Ren et al., 2001; Zhao et al., 2004a; Lee et al., 2012; Bagneris et al., 2014). Nevertheless, despite their lower affinity, the amino acid residues in the bacterial channels that are in analogous positions to those that bind local anesthetics and related drugs in mammalian sodium channels form a functional receptor site and mediate drug block. The locations of the side chains of these amino acid residues are illustrated in Figs. 3A and C for NavAb in the preopen state and in Figs. 3B and D in the inactivated state. When viewed in a transverse section through the pore module, amino acid residues that are analogous to those in transmembrane segments IIIS6 and IVS6 form a distributed binding site along the walls of the cavity in three of the S6 segments that line the pore (Fig. 3, A and C). When viewed from the extracellular side via a section through the structure above the activation gate, these side chains form a potential drug-binding site on the wall of the cavity, with amino acid residues from three domains contributing side chains to the site (Fig. 3, C and D). The positions of these amino acid side chains are altered in the slow inactivated state (Fig. 3, B and D; orange), potentially contributing to state-dependent binding of local anesthetic drugs. In Fig. 3D, the inner ends of two of the S6 segments have moved away from the pore axis, two have moved closer in the slow inactivated state, and all four S6 segments have twisted with respect to the pore axis. This movement of S6 segments changes the geometry of the receptor site by moving the amino acid side chain in pseudo-domain I (upper left, orange) away from those in pseudo-domains III and IV (lower left and lower right, orange) and by moving the amino acid side chains in pseudo-domains III and IV closer together. Higher resolution structures of NavAb and chimeras with substitutions of amino acid residues from mammalian sodium channels should provide crucial information on the mechanism of state-dependent block and the design of highly state-dependent sodium channel blockers.
Fig. 3.
Amino acid side chains in the local anesthetic–binding site of NavAb. (A) Amino acid residues identified by mutagenesis of mammalian sodium channels and shown to be important in the drug block are illustrated as orange spheres. Helices S5 and S6 and the pore domain are depicted in a side view of the preopen state of NavAb. (B) Similar view to (A) showing the slow-inactivated state of NavAb. (C) NavAb is shown from the extracellular side. The S5 and S6 segments are shown as cylinders. Amino acid residues identified by mutagenesis of mammalian sodium channels and shown to be important in the drug block are illustrated as orange spheres. (D) Similar view to (C) showing the inactivated state of NavAb.
A combination of structural modeling and ligand-docking approaches has led to predictions of the location and pose of classic local anesthetics bound in mammalian NaV channels, whose structures have been modeled based on the crystal structure of KcsA and NaV1.5 (Bruhova et al., 2008). Surprisingly, tetracaine is predicted to bind in a pose that lies horizontally across the central cavity, with its positively charged ammonium group in the center of the cavity near the focus of the partially negatively charged ends of the four P helices and its aromatic group in the crevice between domains III and IV (Bruhova et al., 2008). In this position, the aromatic group makes π-π interactions with the highly conserved Phe residue, which was previously implicated in drug binding by site-directed mutagenesis with substitutions of natural and unnatural amino acid residues (Ragsdale et al., 1994; Pless et al., 2011). Further structural work with bacterial and mammalian NaV channels with bound drugs should give a more detailed picture of this important drug-receptor interaction.
Subtype-Selective Sodium Channel Blockers
Sodium channels are encoded by 10 different genes, which are differentially expressed in the major excitable tissues, yet all of the sodium channel drugs used clinically are nonselective, preventing systemic administration in doses sufficient to saturate their binding sites. The value of tissue selectivity is illustrated by successful systemic administration of a neurospecific peptide snail toxin, μ-conotoxin KIIIA, to prevent pain in a mouse model of inflammatory pain without cardiac and neuromuscular side effects (Zhang et al., 2007). Great interest has arisen in the discovery of selective, high-affinity blockers of the NaV1.7 and NaV1.8 channels, which are highly expressed in sensory neurons (Priest and Kaczorowski, 2007; Krafte and Bannon, 2008). Pore blockers that are selective for NaV1.8 channels and gating modifiers that bind selectively and modulate the voltage sensors in NaV1.7 have been described (Jarvis et al., 2007; McCormack et al., 2013). Moreover, the first structural evidence for the location of a drug receptor site in the sodium channel has come from studies of a novel brominated drug, PI1, which selectively blocks mammalian NaV1.8 channels and retains binding to a pore-only construct of the ancestral bacterial sodium channel NaVMs (Bagneris et al., 2014). The anomalous scattering signal from the Br atom was located in four symmetrically disposed positions near the fenestrations on the sides of the central cavity in the pore. No electron density was observed for the bound drug itself. However, molecular modeling suggested that the bound drug would extend from the fenestration toward and into the inner mouth of the ion selectivity filter. This pose of the bound drug is generally consistent with previous site-directed mutagenesis studies of classic, nonselective sodium channel blockers in mammalian NaV channels. Similarly, mutagenesis studies of the amino acid residues in NaVMs that are required for high-affinity binding of PI1, suggest a similar binding site to the classic channel blockers, despite the selective high affinity of PI1 for NaV1.8 channels (Bagneris et al., 2014). However, the binding pose of PI1 differs significantly from the predicted binding pose of tetracaine. PI1 extends farther upward into the ion selectivity filter to make interactions with the innermost ion coordination site (siteIN) (Payandeh et al., 2011) in the ion selectivity filter. Structural studies in which the pose of the bound drug is observed directly in electron density profiles will be needed to determine whether this apparent difference in drug binding, which is inferred from ligand docking algorithms, is confirmed.
Structure of Calcium Channels
Calcium Channel Protein.
Like sodium channels, voltage-gated calcium channels have a large, pore-forming subunit (designated α1) that is composed of four homologous domains, with six transmembrane segments and a pore loop in each (Catterall et al., 2005b). However, calcium channels have a different set of auxiliary subunits, including CaV β, γ, and α2δ subunits (Catterall et al., 2005b). The members of the bacterial sodium channel family, including NavAb, are thought to have been the ancestors of both mammalian sodium channels and calcium channels based on their amino acid sequence similarity to both mammalian channel families (Ren et al., 2001; Yu and Catterall, 2004). Therefore, the general architectures of the voltage-sensing modules and pore modules of CaV channels are likely to resemble those of NaV channels (Fig. 1). The structure of NavAb will give important insights into the structural basis for calcium channel function based on this strong similarity in the amino acid sequence.
Structural Basis for Calcium Selectivity.
As a first step toward understanding the structural basis for calcium channel function, site-directed mutations were made in the bacterial sodium channels that introduce new negatively charged residues on the extracellular side of the high field-strength site (Yue et al., 2002; Shaya et al., 2014; Tang et al., 2014). Substitution of Asp residues located one and four positions on the extracellular side of the high field-strength site plus substitution of Asp for the key Glu at the high field-strength site in the selectivity filter confer high calcium selectivity on NaChBac and NavAb, with PCa:PNa ∼400, which is similar to mammalian calcium channels (Yue et al., 2002; Tang et al., 2014). Structural and functional analyses revealed that the key substitution of Asp for Ser in the position just on the extracellular side of the Glu at the high field-strength site (Glu177 in NavAb) forms a second calcium-binding site in the pore. Remarkably, this single substitution is sufficient to increase calcium selectivity ∼1000-fold from PCa:PNa ∼0.03 to PCa:PNa ∼30 (Tang et al., 2014). Structural analysis revealed a series of five binding sites for Ca2+, leading from the entry to the outer vestibule through the selectivity filter and into the central cavity (Tang et al., 2014). These results are consistent with a knock-off mechanism in which binding of one calcium ion blocks the pore and prevents monovalent cation entry, whereas binding of a second calcium ion to the newly introduced calcium-binding site at the position on the extracellular side of the high field-strength site induces electrostatic repulsion that pushes the first bound calcium ion through the selectivity filter and eventually into the cytosol (Tang et al., 2014). This mechanism explains how the calcium channel can achieve high selectivity against monovalent cation permeation and still have high conductance for calcium, even though the atomic diameters of sodium and calcium are nearly identical.
Receptor Sites for Calcium Antagonist Drugs
Calcium channel antagonist drugs used in the therapy of cardiovascular diseases fall into three structural classes that act at separate sites. They all preferentially target the CaV1.2 channels that initiate excitation-contraction coupling in cardiac and vascular smooth muscle. The polypeptide conotoxins from cone snails used in the treatment of chronic extreme pain block presynaptic CaV2.2 channels by binding at a fourth site in the outer pore of calcium channels. The gabapentinoid drugs, which are also used in the treatment of chronic pain, bind to a separate subunit of neuronal calcium channels and alter their membrane trafficking. We consider the structural basis for binding and action of each of these classes of drugs below.
Phenylalkylamines.
The phenylalkylamines have two aromatic moieties connected by a central aliphatic chain containing a tertiary amino group (Sampson and Kass, 2011). Like local anesthetics, they are thought to enter the intracellular mouth of the pore through the open activation gate and bind to a specific receptor site in a protonated positively charged form. They exhibit both frequency dependence and voltage dependence of channel block (Hondeghem and Katzung, 1984). Frequency-dependent block of calcium channels arises when repeated opening of the pore in rapidly firing cells allows more rapid access of the drugs to their receptor site and increases the fraction of calcium channels that are blocked. Verapamil, the only phenylalkylamine widely used in therapy, is effective in the treatment of atrial arrhythmias, particularly those that arise by re-entry of action potentials conducted back from the atrioventricular node. This therapeutic use depends upon rapid frequency-dependent block of calcium channels by verapamil binding to its receptor site in the pore of calcium channels.
Extensive studies by photoaffinity labeling (Striessnig et al., 1990) and site-directed mutagenesis (Hockerman et al., 1995, 1997a) have mapped the amino acid residues that are required for high-affinity binding of verapamil and derivatives to the S6 segments in domains III and IV of the cardiac CaV1.2 calcium channel. Molecular modeling suggested that these amino acid residues face the lumen of the pore and cooperate to form the drug-binding site (Cheng et al., 2009). In agreement with these prior molecular models, the three-dimensional structure of CavAb places these amino acid residues in positions to cooperate in forming a single drug-binding site in the central cavity of the pore (Fig. 4A, green). Drug access to this site would be controlled by opening and closing of the intracellular inactivation gate as envisioned in the modulated receptor hypothesis (Hille, 1977). Moreover, the dramatic conformational change in the central cavity and drug-binding site during slow inactivation (Fig. 2C) could provide a molecular basis for high affinity binding to inactivated calcium channels, which is another hallmark of the actions of phenylalkylamines (Hondeghem and Katzung, 1984).
Fig. 4.
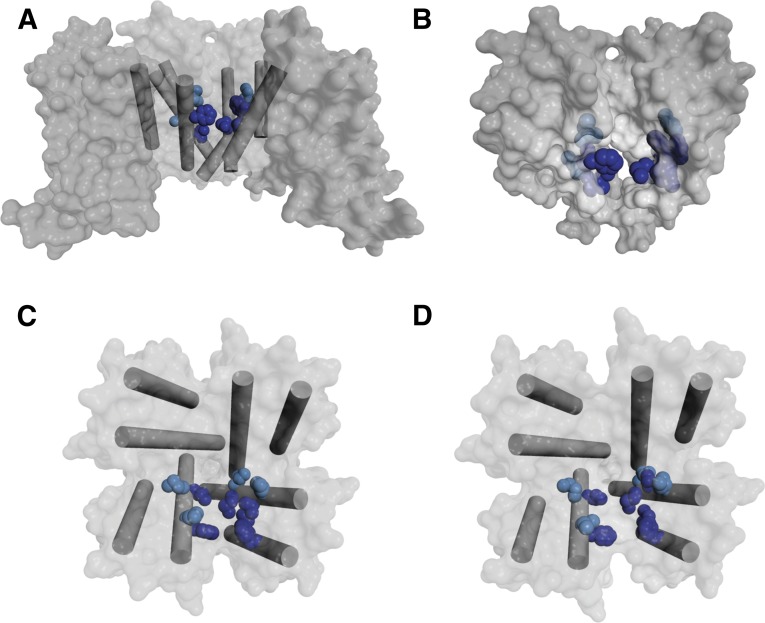
Amino acid side chains in the phenylalkylamine-binding site. (A) Top view of the pore module of CavAb in the preopen state, with amino acid side chains analogous to those implicated in phenylalkylamine binding illustrated in green and amino acid side chains specific for dihydropyridine illustrated in blue. (B) Side view of CavAb in the slow-inactivated state, with amino acid side chains analogous to those implicated in phenylalkylamine binding illustrated in green. (C) Top view of CavAb pore module in the preopen state, with the S5 and S6 segments illustrated as cylinders and amino acid side chains analogous to those implicated in phenylalkylamine binding illustrated in dark green for CaV1.2-specific residues and in light green for CaV-conserved residues. (D) Similar view to (C) of the inactivated state.
Illustration of the phenylalkylamine-binding site by highlighting the side chains of amino acid residues in CavAb that are homologous to those that form this receptor site on CaV1.2 channels shows that this set of amino acid residues are located in the pore-lining transmembrane segments (Fig. 4B, side view). These residues are organized to form a coherent drug-binding site, as illustrated in the top view in Fig. 4C (dark green, CaV1.2-specific residues; light green, CaV-conserved residues). The detailed conformation of this site is changed in the slow-inactivated state of CavAb (Fig. 4D), which is likely to be analogous to the voltage-dependent inactivated state of calcium channels. As two of the S6 segments move away from the axis of the pore and two move toward it, the side chains of the amino acid residues that interact with phenylalkylamines in two adjacent S6 segments move farther apart. Thus, these models provide a preliminary view of possible state-dependent binding interactions underlying frequency- and voltage-dependent block of calcium channels by phenylalkylamines.
Molecular modeling and ligand-docking methods have been used to predict the binding pose of the phenylalkylamines in their receptor site (Lipkind and Fozzard, 2003; Cheng et al., 2009). In the initial study, based on the structure of the KcsA potassium channel, the bound phenylakylamine was fit in a half-folded, L-shaped conformation, with the positively charged ammonium group projecting toward the negatively charged ion selectivity filter, aromatic ring A in the central cavity, and aromatic ring B projecting toward the interface between domains III and IV (Lipkind and Fozzard, 2003). In the second model, based on the KvAP structure, the elongated phenylalkylamine molecule is predicted to lie in a more extended conformation, with the conserved nitrile group projecting toward the ion selectivity filter with Ca2+ bound in it, the positively charged ammonium group placed at the axis of the pore in the focus of the helical dipole of the four P-helices, and the aromatic rings and methoxy groups making hydrophobic and hydrogen-binding interactions with amino acid side chains and the backbone of the S6 segments (Cheng et al., 2009). These models both satisfy many of the requirements derived from structure-function studies; therefore, studies of bound phenylalkylamines in crystal structures of calcium channels or their homologs will be required to precisely define the position of the bound drug and thereby determine the structural basis for state-dependent binding.
Benzothiazepines.
Like verapamil, the benzothiazepine diltiazem is thought to bind in the pore of calcium channels and derive frequency and voltage dependence of its blocking activity according to the modulated receptor hypothesis. The amino acid residues that form the diltiazem-binding site are partially overlapping with the amino acid residues in the verapamil/phenylalkylamine-binding site (Kraus et al., 1996; Hockerman et al., 2000), but the binding of these two classes of drugs is not competitive. The conformation of the side chains of these amino acid residues would also be changed in the slow-inactivated state.
Dihydropyridines.
The dihydropyridines can function as either activators or inhibitors of calcium channels; therefore, they must act as gating modifiers rather than pore blockers and they must bind to a site outside of the ion conduction pathway to avoid blocking it (Catterall and Striessnig, 1992; Catterall et al., 2005b). In some cases, enantiomeric pairs are activators or inhibitors, respectively, indicating that very subtle changes in the drug-receptor interaction are sufficient to convert from agonist to antagonist action (Kokubun et al., 1986). The dihydropyridine-binding site was initially probed by photoaffinity labeling (Striessnig et al., 1991), which revealed interaction with the S6 segments in domains III and IV and led to the proposal of a domain-interface model for drug binding and action (Catterall and Striessnig, 1992; Catterall et al., 2005b). Site-directed mutagenesis studies of these channel segments (Mitterdorfer et al., 1996; Peterson et al., 1996, 1997; Schuster et al., 1996) identified both conserved amino acid residues that are required for drug binding and Cav1.2-specific residues that create the selective high-affinity binding site for dihydropyridines in cardiac calcium channels. Moreover, nine amino acid residues in the IIIS5, IIIS6, and IVS6 segments were shown to be sufficient to confer high-affinity binding and selective channel activation and inhibition by dihydropyridines, including enantiomeric pairs of activators and inhibitors (Hockerman et al., 1997b; Ito et al., 1997; Sinnegger et al., 1997). Overall, these reconstitution studies gave a remarkably complete transfer of dihydropyridine pharmacology to the completely dihydropyridine-insensitive CaV2.1 and CaV2.3 channels.
The likely structure of the dihydropyridine receptor site is illustrated in Fig. 5A by highlighting the amino acid side chains in CavAb that are analogous to those thought to form this receptor site in CaV1.2 channels, as viewed from the side. Darker shading represents the two voltage-sensing domains that flank the receptor site, and lighter shading represents the membrane-exposed surface of the pore domain containing the binding site. Dihydropyridines bind to a site that is formed by amino acid residues in two adjacent S6 segments plus the S5 segment between them. The same amino acid residues are illustrated in the inactivated state of CavAb in Fig. 5B. The binding site is formed on the back side of the S6 segments, oriented away from the pore, plus neighboring amino acid residues in the adjacent S5 segment, as seen in top views of the pore domain (Fig. 5, C and D). Thus, S6 segments from two adjacent subunits or domains cooperate to form the receptor site, as initially proposed in the original domain-interface model of dihydropyridine binding (Catterall and Striessnig, 1992). This site is located very close to the receptor site for phenylalkylamines (Fig. 4A, blue), with some amino acid side chains in position to bind both drugs on the opposite sides of phenyl rings or aliphatic side chains. As expected from the state-dependence of binding of dihydropyridines, the detailed orientation of these amino acid side chains changes significantly in the slow-inactivated state (Fig. 5D). Compared with the preopen state (Fig. 5C), the amino acid side chains from the two S6 segments that form the binding site move apart in the inactivated state (Fig. 5D). This substantial movement of the drug-binding residues should contribute in an important way to the 100- to 1000-fold differences in binding affinity of dihydropyridines for CaV channels.
Fig. 5.
Amino acid side chains in the dihydropyridine-binding site. (A) Side view of intact CavAb in the preopen state with the two voltage-sensing domains illustrated in dark gray and the intervening pore domain illustrated in light gray. Amino acid side chains analogous to those implicated in dihydropyridine binding are illustrated in dark blue for CaV1.2-specific residues and in light blue for CaV-conserved residues. (B) Side view of CavAb in the slow-inactivated state, with amino acid side chains analogous to those implicated in dihydropyridine binding illustrated in blue. (C) Top view of the pore domain of CavAb in the preopen state, with amino acid side chains analogous to those implicated in dihydropyridine binding illustrated in blue. (D) Top view of the pore domain of CavAb in the slow inactivated state, with amino acid side chains analogous to those implicated in dihydropyridine binding illustrated in blue.
Molecular models of dihydropyridine binding have been developed based on the structures of the KcsA and KvAP potassium channels (Lipkind and Fozzard, 2003). These models place the bound drug at the interface between domains III and IV in a position to interact with amino acid residues in the IIIS5, IIIS6, and IVS6 segments, as observed in structure-function studies (Tikhonov and Zhorov, 2009). Specific interactions of the hydrophobic groups on the portside of the boat-like structure of the dihydropyridine ring are predicted to govern the agonist/antagonist properties of the dihydropyridines (Tikhonov and Zhorov, 2009). Although the position of the bound dihydropyridine is similar in the available molecular models, the details of these interactions will differ in models based on the NaVAb structure, which likely resembles calcium channels more closely, and crystal structures of dihydropyridines bound to calcium channels or their homologs are needed to probe structural models for the activation and inhibition of calcium channels by these important drugs.
Conotoxins.
The ω-conotoxins from Conus geographus and other species of cone snails are potent and selective blockers of CaV2.2 channels, which conduct N-type calcium currents that initiate neurotransmitter release at the nerve terminals of primary afferent sensory neurons (Olivera et al., 1994; Catterall et al., 2005b). These toxins are potent analgesics in animal models of neuropathic pain (Miljanich and Ramachandran, 1995). The synthetic version of one of these toxins (ziconitide, Prialt) is effective in patients with intractable pain from cancer and other causes (Lewis et al., 2012). ω-Conotoxins bind in the outer vestibule of calcium channels, and amino acid residues that are required for high-affinity binding have been identified by site-directed mutagenesis (Ellinor et al., 1994; Feng et al., 2001, 2003). Comparison with the structure of CavAb shows that these amino acid residues in mammalian CaV2.2 channels would form a binding site in the vestibule on the extracellular side of the amino acid residues in the ion selectivity filter that determines calcium selectivity. In this position, they would be expected to block ion conductance.
Gabapentinoid Drugs.
Extensive use of gabapentinoid drugs for the treatment of epilepsy, and especially for the treatment of chronic pain, has evolved over the past decade. Initially, the molecular target and receptor site at which they exert their therapeutic actions were unknown. However, extensive studies now indicate that they bind to the auxiliary α2δ subunit of presynaptic calcium channels. In particular, binding of gabapentinoid drugs to the CaV2.2 channels that conduct the N-type Ca2+ current and initiate neurotransmitter release at the central terminals of nociceptive dorsal root ganglion neurons downregulates channel activity by inhibiting the normal cycle of endocytosis and reinsertion of these channels at the cell surface membrane (Davies et al., 2007). Hopefully, structural studies of α2δ subunits will give novel insights that will help to guide the design of future generations of these surprisingly effective drugs.
Structure and Ion Channel Pharmacology
In drug discovery, high-resolution structures of angiotensin-converting enzymes, human immunodeficiency virus proteases, protein kinases, and many other enzyme targets have led to major advances in the development of compounds with high selectivity for specific target subtypes and much improved safety and efficacy. The revolution in structures of seven-transmembrane G protein–coupled receptors in the past several years is driving a new wave of drug discovery of therapeutic agents that target these signaling molecules with exquisite selectivity and fine-tuned efficacy. The 24-transmembrane voltage-gated ion channels are much more complex, but we look forward to a new era of structure-based drug design for these important therapeutic targets, driven by the rapidly emerging structural information on bacterial homologs, chimeras of bacterial and mammalian channels, and eventually the mammalian sodium and calcium channels themselves.
Authorship Contributions
Participated in research design: Catterall, Swanson.
Performed data analysis: Swanson.
Wrote or contributed to the writing of the manuscript: Catterall, Swanson.
Footnotes
>Research in the authors’ laboratory that is reviewed in this article was supported by research grants from the National Institutes of Health [Grants R01-NS15751, R01-HL112808, and R01-HL117896].
References
- Armstrong CM, Bezanilla F. (1973) Currents related to movement of the gating particles of the sodium channels. Nature 242:459–461. [DOI] [PubMed] [Google Scholar]
- Bagnéris C, DeCaen PG, Naylor CE, Pryde DC, Nobeli I, Clapham DE, Wallace BA. (2014) Prokaryotic NavMs channel as a structural and functional model for eukaryotic sodium channel antagonism. Proc Natl Acad Sci USA 111:8428–8433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bean BP. (1984) Nitrendipine block of cardiac calcium channels: high-affinity binding to the inactivated state. Proc Natl Acad Sci USA 81:6388–6392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruhova I, Tikhonov DB, Zhorov BS. (2008) Access and binding of local anesthetics in the closed sodium channel. Mol Pharmacol 74:1033–1045. [DOI] [PubMed] [Google Scholar]
- Catterall WA. (1986) Molecular properties of voltage-sensitive sodium channels. Annu Rev Biochem 55:953–985. [DOI] [PubMed] [Google Scholar]
- Catterall WA. (2000) From ionic currents to molecular mechanisms: the structure and function of voltage-gated sodium channels. Neuron 26:13–25. [DOI] [PubMed] [Google Scholar]
- Catterall WA. (2010) Ion channel voltage sensors: structure, function, and pathophysiology. Neuron 67:915–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catterall WA, Goldin AL, Waxman SG. (2005a) International Union of Pharmacology. XLVII. Nomenclature and structure-function relationships of voltage-gated sodium channels. Pharmacol Rev 57:397–409. [DOI] [PubMed] [Google Scholar]
- Catterall WA, Mackie K. (2011) Local anesthetics, in Goodman & Gilman’s The Pharmacological Basis of Therapeutics (Brunton L. ed) pp 565–582, McGraw-Hill Co., New York. [Google Scholar]
- Catterall WA, Perez-Reyes E, Snutch TP, Striessnig J. (2005b) International Union of Pharmacology. XLVIII. Nomenclature and structure-function relationships of voltage-gated calcium channels. Pharmacol Rev 57:411–425. [DOI] [PubMed] [Google Scholar]
- Catterall WA, Striessnig J. (1992) Receptor sites for Ca2+ channel antagonists. Trends Pharmacol Sci 13:256–262. [DOI] [PubMed] [Google Scholar]
- Cheng RC, Tikhonov DB, Zhorov BS. (2009) Structural model for phenylalkylamine binding to L-type calcium channels. J Biol Chem 284:28332–28342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies A, Hendrich J, Van Minh AT, Wratten J, Douglas L, Dolphin AC. (2007) Functional biology of the α2δ subunits of voltage-gated calcium channels. Trends Pharmacol Sci 28:220–228. [DOI] [PubMed] [Google Scholar]
- Eaholtz G, Scheuer T, Catterall WA. (1994) Restoration of inactivation and block of open sodium channels by an inactivation gate peptide. Neuron 12:1041–1048. [DOI] [PubMed] [Google Scholar]
- Ellinor PT, Zhang JF, Horne WA, Tsien RW. (1994) Structural determinants of the blockade of N-type calcium channels by a peptide neurotoxin. Nature 372:272–275. [DOI] [PubMed] [Google Scholar]
- Feng ZP, Doering CJ, Winkfein RJ, Beedle AM, Spafford JD, Zamponi GW. (2003) Determinants of inhibition of transiently expressed voltage-gated calcium channels by omega-conotoxins GVIA and MVIIA. J Biol Chem 278:20171–20178. [DOI] [PubMed] [Google Scholar]
- Feng ZP, Hamid J, Doering C, Jarvis SE, Bosey GM, Bourinet E, Snutch TP, Zamponi GW. (2001) Amino acid residues outside of the pore region contribute to N-type calcium channel permeation. J Biol Chem 276:5726–5730. [DOI] [PubMed] [Google Scholar]
- Guy HR, Seetharamulu P. (1986) Molecular model of the action potential sodium channel. Proc Natl Acad Sci USA 83:508–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinemann SH, Terlau H, Stühmer W, Imoto K, Numa S. (1992) Calcium channel characteristics conferred on the sodium channel by single mutations. Nature 356:441–443. [DOI] [PubMed] [Google Scholar]
- Hess P, Lansman JB, Tsien RW. (1984) Different modes of Ca channel gating behaviour favoured by dihydropyridine Ca agonists and antagonists. Nature 311:538–544. [DOI] [PubMed] [Google Scholar]
- Hille B. (1977) Local anesthetics: hydrophilic and hydrophobic pathways for the drug-receptor reaction. J Gen Physiol 69:497–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hockerman GH, Dilmac N, Scheuer T, Catterall WA. (2000) Molecular determinants of diltiazem block in domains IIIS6 and IVS6 of L-type Ca(2+) channels. Mol Pharmacol 58:1264–1270. [DOI] [PubMed] [Google Scholar]
- Hockerman GH, Johnson BD, Abbott MR, Scheuer T, Catterall WA. (1997a) Molecular determinants of high affinity phenylalkylamine block of L-type calcium channels in transmembrane segment IIIS6 and the pore region of the α1 subunit. J Biol Chem 272:18759–18765. [DOI] [PubMed] [Google Scholar]
- Hockerman GH, Johnson BD, Scheuer T, Catterall WA. (1995) Molecular determinants of high affinity phenylalkylamine block of L-type calcium channels. J Biol Chem 270:22119–22122. [DOI] [PubMed] [Google Scholar]
- Hockerman GH, Peterson BZ, Sharp E, Tanada TN, Scheuer T, Catterall WA. (1997b) Construction of a high-affinity receptor site for dihydropyridine agonists and antagonists by single amino acid substitutions in a non-L-type Ca2+ channel. Proc Natl Acad Sci USA 94:14906–14911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkin AL, Huxley AF. (1952a) The dual effect of membrane potential on sodium conductance in the giant axon of Loligo. J Physiol 116:497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkin AL, Huxley AF. (1952b) A quantitative description of membrane current and its application to conduction and excitation in nerve. J Physiol 117:500–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hondeghem LM, Katzung BG. (1984) Antiarrhythmic agents: the modulated receptor mechanism of action of sodium and calcium channel-blocking drugs. Annu Rev Pharmacol Toxicol 24:387–423. [DOI] [PubMed] [Google Scholar]
- Ito H, Klugbauer N, Hofmann F. (1997) Transfer of the high affinity dihydropyridine sensitivity from L-type to non-L-type calcium channel. Mol Pharmacol 52:735–740. [DOI] [PubMed] [Google Scholar]
- Jarvis MF, Honore P, Shieh CC, Chapman M, Joshi S, Zhang XF, Kort M, Carroll W, Marron B, Atkinson R, et al. (2007) A-803467, a potent and selective Nav1.8 sodium channel blocker, attenuates neuropathic and inflammatory pain in the rat. Proc Natl Acad Sci USA 104:8520–8525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kokubun S, Prod’hom B, Becker C, Porzig H, Reuter H. (1986) Studies on Ca channels in intact cardiac cells: voltage-dependent effects and cooperative interactions of dihydropyridine enantiomers. Mol Pharmacol 30:571–584. [PubMed] [Google Scholar]
- Kokubun S, Reuter H. (1984) Dihydropyridine derivatives prolong the open state of Ca channels in cultured cardiac cells. Proc Natl Acad Sci USA 81:4824–4827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krafte DS, Bannon AW. (2008) Sodium channels and nociception: recent concepts and therapeutic opportunities. Curr Opin Pharmacol 8:50–56. [DOI] [PubMed] [Google Scholar]
- Kraus R, Reichl B, Kimball SD, Grabner M, Murphy BJ, Catterall WA, Striessnig J. (1996) Identification of benz(othi)azepine-binding regions within L-type calcium channel α1 subunits. J Biol Chem 271:20113–20118. [DOI] [PubMed] [Google Scholar]
- Lee S, Goodchild SJ, Ahern CA. (2012) Local anesthetic inhibition of a bacterial sodium channel. J Gen Physiol 139:507–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis RJ, Dutertre S, Vetter I, Christie MJ. (2012) Conus venom peptide pharmacology. Pharmacol Rev 64:259–298. [DOI] [PubMed] [Google Scholar]
- Lipkind GM, Fozzard HA. (2003) Molecular modeling of interactions of dihydropyridines and phenylalkylamines with the inner pore of the L-type Ca2+ channel. Mol Pharmacol 63:499–511. [DOI] [PubMed] [Google Scholar]
- McCormack K, Santos S, Chapman ML, Krafte DS, Marron BE, West CW, Krambis MJ, Antonio BM, Zellmer SG, Printzenhoff D, et al. (2013) Voltage sensor interaction site for selective small molecule inhibitors of voltage-gated sodium channels. Proc Natl Acad Sci USA 110:E2724–E2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCusker EC, Bagnéris C, Naylor CE, Cole AR, D’Avanzo N, Nichols CG, Wallace BA. (2012) Structure of a bacterial voltage-gated sodium channel pore reveals mechanisms of opening and closing. Nat Commun 3:1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNamara J. (2011) Pharmacotherapy of the epilepsies, in Goodman & Gilman’s The Pharmacological Basis of Therapeutics (Brunton L. ed) pp 583–608, McGraw-Hill Co., New York. [Google Scholar]
- Michel T, Hoffmann BB. (2011) Treatment of myocardial ischemia and hypertension, in Goodman & Gilman’s The Pharmacological Basis of Therapeutics (Bruntin L. ed) pp 745–788, McGraw-Hill Co., New York. [Google Scholar]
- Miljanich GP, Ramachandran J. (1995) Antagonists of neuronal calcium channels: structure, function, and therapeutic implications. Annu Rev Pharmacol Toxicol 35:707–734. [DOI] [PubMed] [Google Scholar]
- Mitterdorfer J, Wang Z, Sinnegger MJ, Hering S, Striessnig J, Grabner M, Glossmann H. (1996) Two amino acid residues in the IIIS5 segment of L-type calcium channels differentially contribute to 1,4-dihydropyridine sensitivity. J Biol Chem 271:30330–30335. [DOI] [PubMed] [Google Scholar]
- Noda M, Suzuki H, Numa S, Stühmer W. (1989) A single point mutation confers tetrodotoxin and saxitoxin insensitivity on the sodium channel II. FEBS Lett 259:213–216. [DOI] [PubMed] [Google Scholar]
- Olivera BM, Miljanich GP, Ramachandran J, Adams ME. (1994) Calcium channel diversity and neurotransmitter release: the omega-conotoxins and omega-agatoxins. Annu Rev Biochem 63:823–867. [DOI] [PubMed] [Google Scholar]
- Pavlov E, Bladen C, Winkfein R, Diao C, Dhaliwal P, French RJ. (2005) The pore, not cytoplasmic domains, underlies inactivation in a prokaryotic sodium channel. Biophys J 89:232–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payandeh J, Gamal El-Din TM, Scheuer T, Zheng N, Catterall WA. (2012) Crystal structure of a voltage-gated sodium channel in two potentially inactivated states. Nature 486:135–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payandeh J, Scheuer T, Zheng N, Catterall WA. (2011) The crystal structure of a voltage-gated sodium channel. Nature 475:353–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson BZ, Johnson BD, Hockerman GH, Acheson M, Scheuer T, Catterall WA. (1997) Analysis of the dihydropyridine receptor site of L-type calcium channels by alanine-scanning mutagenesis. J Biol Chem 272:18752–18758. [DOI] [PubMed] [Google Scholar]
- Peterson BZ, Tanada TN, Catterall WA. (1996) Molecular determinants of high affinity dihydropyridine binding in L-type calcium channels. J Biol Chem 271:5293–5296. [DOI] [PubMed] [Google Scholar]
- Pless SA, Galpin JD, Frankel A, Ahern CA. (2011) Molecular basis for class Ib anti-arrhythmic inhibition of cardiac sodium channels. Nat Commun 2:351. [DOI] [PubMed] [Google Scholar]
- Priest BT, Kaczorowski GJ. (2007) Subtype-selective sodium channel blockers promise a new era of pain research. Proc Natl Acad Sci USA 104:8205–8206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu Y, Rogers J, Tanada T, Scheuer T, Catterall WA. (1995) Molecular determinants of drug access to the receptor site for antiarrhythmic drugs in the cardiac Na+ channel. Proc Natl Acad Sci USA 92:11839–11843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragsdale DS, McPhee JC, Scheuer T, Catterall WA. (1996) Common molecular determinants of local anesthetic, antiarrhythmic, and anticonvulsant block of voltage-gated Na+ channels. Proc Natl Acad Sci USA 93:9270–9275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragsdale DS, McPhee JC, Scheuer T, Catterall WA. (1994) Molecular determinants of state-dependent block of Na+ channels by local anesthetics. Science 265:1724–1728. [DOI] [PubMed] [Google Scholar]
- Ren D, Navarro B, Xu H, Yue L, Shi Q, Clapham DE. (2001) A prokaryotic voltage-gated sodium channel. Science 294:2372–2375. [DOI] [PubMed] [Google Scholar]
- Rohl CA, Boeckman FA, Baker C, Scheuer T, Catterall WA, Klevit RE. (1999) Solution structure of the sodium channel inactivation gate. Biochemistry 38:855–861. [DOI] [PubMed] [Google Scholar]
- Sampson KJ, Kass RS. (2011) Anti-arrhythmic drugs, in Goodman & Gilman’s The Pharmacological Basis of Therapeutics (Brunton L. ed) pp 815–848, McGraw-Hill Co., New York. [Google Scholar]
- Schuster A, Lacinová L, Klugbauer N, Ito H, Birnbaumer L, Hofmann F. (1996) The IVS6 segment of the L-type calcium channel is critical for the action of dihydropyridines and phenylalkylamines. EMBO J 15:2365–2370. [PMC free article] [PubMed] [Google Scholar]
- Shaya D, Findeisen F, Abderemane-Ali F, Arrigoni C, Wong S, Nurva SR, Loussouarn G, Minor DL., Jr (2014) Structure of a prokaryotic sodium channel pore reveals essential gating elements and an outer ion binding site common to eukaryotic channels. J Mol Biol 426:467–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinnegger MJ, Wang Z, Grabner M, Hering S, Striessnig J, Glossmann H, Mitterdorfer J. (1997) Nine L-type amino acid residues confer full 1,4-dihydropyridine sensitivity to the neuronal calcium channel α1A subunit. Role of L-type Met1188. J Biol Chem 272:27686–27693. [DOI] [PubMed] [Google Scholar]
- Striessnig J, Glossmann H, Catterall WA. (1990) Identification of a phenylalkylamine binding region within the α 1 subunit of skeletal muscle Ca2+ channels. Proc Natl Acad Sci USA 87:9108–9112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Striessnig J, Murphy BJ, Catterall WA. (1991) Dihydropyridine receptor of L-type Ca2+ channels: identification of binding domains for [3H](+)-PN200-110 and [3H]azidopine within the α1 subunit. Proc Natl Acad Sci USA 88:10769–10773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stühmer W, Conti F, Suzuki H, Wang XD, Noda M, Yahagi N, Kubo H, Numa S. (1989) Structural parts involved in activation and inactivation of the sodium channel. Nature 339:597–603. [DOI] [PubMed] [Google Scholar]
- Tang L, Gamal El-Din TM, Payandeh J, Martinez GQ, Heard TM, Scheuer T, Zheng N, Catterall WA. (2014) Structural basis for Ca2+ selectivity of a voltage-gated calcium channel. Nature 505:56–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terlau H, Heinemann SH, Stühmer W, Pusch M, Conti F, Imoto K, Numa S. (1991) Mapping the site of block by tetrodotoxin and saxitoxin of sodium channel II. FEBS Lett 293:93–96. [DOI] [PubMed] [Google Scholar]
- Tikhonov DB, Zhorov BS. (2009) Structural model for dihydropyridine binding to L-type calcium channels. J Biol Chem 284:19006–19017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai CJ, Tani K, Irie K, Hiroaki Y, Shimomura T, McMillan DG, Cook GM, Schertler GF, Fujiyoshi Y, Li XD. (2013) Two alternative conformations of a voltage-gated sodium channel. J Mol Biol 425:4074–4088. [DOI] [PubMed] [Google Scholar]
- Vargas E, Yarov-Yarovoy V, Khalili-Araghi F, Catterall WA, Klein ML, Tarek M, Lindahl E, Schulten K, Perozo E, Bezanilla F, et al. (2012) An emerging consensus on voltage-dependent gating from computational modeling and molecular dynamics simulations. J Gen Physiol 140:587–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassilev PM, Scheuer T, Catterall WA. (1988) Identification of an intracellular peptide segment involved in sodium channel inactivation. Science 241:1658–1661. [DOI] [PubMed] [Google Scholar]
- Vilin YY, Ruben PC. (2001) Slow inactivation in voltage-gated sodium channels: molecular substrates and contributions to channelopathies. Cell Biochem Biophys 35:171–190. [DOI] [PubMed] [Google Scholar]
- Wang GK, Quan C, Wang S. (1998) A common local anesthetic receptor for benzocaine and etidocaine in voltage-gated mu1 Na+ channels. Pflugers Arch 435:293–302. [DOI] [PubMed] [Google Scholar]
- West JW, Patton DE, Scheuer T, Wang Y, Goldin AL, Catterall WA. (1992) A cluster of hydrophobic amino acid residues required for fast Na+-channel inactivation. Proc Natl Acad Sci USA 89:10910–10914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarov-Yarovoy V, Baker D, Catterall WA. (2006) Voltage sensor conformations in the open and closed states in ROSETTA structural models of K+ channels. Proc Natl Acad Sci USA 103:7292–7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarov-Yarovoy V, Brown J, Sharp EM, Clare JJ, Scheuer T, Catterall WA. (2001) Molecular determinants of voltage-dependent gating and binding of pore-blocking drugs in transmembrane segment IIIS6 of the Na+ channel α subunit. J Biol Chem 276:20–27. [DOI] [PubMed] [Google Scholar]
- Yarov-Yarovoy V, DeCaen PG, Westenbroek RE, Pan CY, Scheuer T, Baker D, Catterall WA. (2012) Structural basis for gating charge movement in the voltage sensor of a sodium channel. Proc Natl Acad Sci USA 109:E93–E102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarov-Yarovoy V, McPhee JC, Idsvoog D, Pate C, Scheuer T, Catterall WA. (2002) Role of amino acid residues in transmembrane segments IS6 and IIS6 of the Na+ channel α subunit in voltage-dependent gating and drug block. J Biol Chem 277:35393–35401. [DOI] [PubMed] [Google Scholar]
- Yu FH, Catterall WA. (2004) The VGL-chanome: a protein superfamily specialized for electrical signaling and ionic homeostasis. Sci STKE 2004:re15. [DOI] [PubMed] [Google Scholar]
- Yue L, Navarro B, Ren D, Ramos A, Clapham DE. (2002) The cation selectivity filter of the bacterial sodium channel, NaChBac. J Gen Physiol 120:845–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang MM, Green BR, Catlin P, Fiedler B, Azam L, Chadwick A, Terlau H, McArthur JR, French RJ, Gulyas J, et al. (2007) Structure/function characterization of micro-conotoxin KIIIA, an analgesic, nearly irreversible blocker of mammalian neuronal sodium channels. J Biol Chem 282:30699–30706. [DOI] [PubMed] [Google Scholar]
- Zhang X, Ren W, DeCaen P, Yan C, Tao X, Tang L, Wang J, Hasegawa K, Kumasaka T, He J, et al. (2012) Crystal structure of an orthologue of the NaChBac voltage-gated sodium channel. Nature 486:130–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Scheuer T, Catterall WA. (2004a) Reversed voltage-dependent gating of a bacterial sodium channel with proline substitutions in the S6 transmembrane segment. Proc Natl Acad Sci USA 101:17873–17878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Yarov-Yarovoy V, Scheuer T, Catterall WA. (2004b) A gating hinge in Na+ channels; a molecular switch for electrical signaling. Neuron 41:859–865. [DOI] [PubMed] [Google Scholar]