Abstract
Various mutations in the rpoB gene, which encodes the RNA polymerase β subunit, are associated with increased vancomycin (VAN) resistance in vancomycin-intermediate Staphylococcus aureus (VISA) and heterogeneously VISA (hVISA) strains. We reported that rpoB mutations are also linked to the expression of the recently found “slow VISA” (sVISA) phenotype (M. Saito, Y. Katayama, T. Hishinuma, A. Iwamoto, Y. Aiba, K Kuwahara-Arai, L. Cui, M. Matsuo, N. Aritaka, and K. Hiramatsu, Antimicrob Agents Chemother 58:5024–5035, 2014, http://dx.doi.org/10.1128/AAC.02470-13). Because RpoC and RpoB are components of RNA polymerase, we examined the effect of the rpoC(P440L) mutation on the expression of the sVISA phenotype in the Mu3fdh2*V6-5 strain (V6-5), which was derived from a previously reported hVISA strain with the VISA phenotype. V6-5 had an extremely prolonged doubling time (DT) (72 min) and high vancomycin MIC (16 mg/liter). However, the phenotype of V6-5 was unstable, and the strain frequently reverted to hVISA with concomitant loss of low growth rate, cell wall thickness, and reduced autolysis. Whole-genome sequencing of phenotypic revertant strain V6-5-L1 and comparison with V6-5 revealed a second mutation, F562L, in rpoC. Introduction of the wild-type (WT) rpoC gene using a multicopy plasmid resolved the sVISA phenotype of V6-5, indicating that the rpoC(P440L) mutant expressed the sVISA phenotype in hVISA. To investigate the mechanisms of resistance in the sVISA strain, we independently isolated an additional 10 revertants to hVISA and VISA. In subsequent whole-genome analysis, we identified compensatory mutations in the genes of three distinct functional categories: the rpoC gene itself as regulatory mutations, peptidoglycan biosynthesis genes, and relQ, which is involved in the stringent response. It appears that the rpoC(P440L) mutation causes the sVISA phenotype by augmenting cell wall peptidoglycan synthesis and through the control of the stringent response.
INTRODUCTION
Methicillin-resistant Staphylococcus aureus (MRSA) remains one of the major causes of both health care- and community-associated infections. Although vancomycin (VAN) is the first-line antibiotic for the treatment of serious MRSA infections, high rates of VAN treatment failure for MRSA strains with susceptibility to an acceptable MIC (i.e., 2 mg/liter) have led to questions regarding its efficacy (1–6).
Although heterogeneously vancomycin-intermediate S. aureus (hVISA) is classified as a “susceptible” strain according to the Clinical and Laboratory Standards Institute (CLSI) criteria (7), it generates VISA at high frequency. Recently, we isolated a new type of VISA phenotype (designated “slow VISA” [sVISA]) from the hVISA strain Mu3 using one-step VAN selection (8). The sVISA strain showed an extremely-slow-growth phenotype and higher levels of VAN resistance than extant VISA strains. Moreover, sVISA phenotypes rapidly reverted to the hVISA phenotype upon removal of VAN from culture media. This transient high-resistance acquisition by hVISA may be the reason for the frequent VAN-refractory MRSA infections (9).
Various mutations in the rpoB gene, which encodes the RNA polymerase β subunit, are associated with an increased VAN MIC in vancomycin-intermediate S. aureus (VISA) and heterogeneously VISA (hVISA) strains (10–12). We recently reported associations of rpoB mutations with expression of the sVISA phenotype (8). Although its resistance mechanism or mechanisms are unknown, we previously identified a single mutation in rpoC encoding another RNA polymerase subunit in the VISA strain Mu3fdh2*V6-5 (V6-5), which was derived from hVISA clinical strain Mu3 (13). Thus, in the present study, we reconfirmed that V6-5 expressed a typical sVISA phenotype and investigated the genetic mechanisms that lead to transient high resistance of the sVISA strain. A total of 11 revertants of the rpoC mutant sVISA strain were isolated, and whole-genome analyses were used to identify mutations that were associated with the transient sVISA phenotype. Presumable compensatory (or complementary) rpoC mutations were found in 3 out of 10 revertant strains. The other revertants were found to possess mutations in peptidoglycan biosynthesis genes (4 strains) and the stringent response gene relQ (3 strains).
MATERIALS AND METHODS
Bacterial strains and growth conditions.
All bacterial strains, plasmids, and primers used in this study are listed in Table 1. The rpoC mutant VISA strain Mu3fdh2*V6-5 was obtained by selecting Mu3fdh2* hVISA cells using a brain heart infusion (BHI) agar plate containing 6 mg/liter VAN (13). The clinically isolated VISA and hVISA strains Mu50 and Mu3 and the sVISA strain 6R-P (8) were used as reference strains. All S. aureus strains were cultivated in BHI broth or agar (Difco Laboratories, Detroit, MI) with aeration at 37°C, unless indicated otherwise.
TABLE 1.
List of bacterial strains and plasmids used in this study
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| Strains | ||
| S. aureus | ||
| Mu3 | hVISA clinical isolate from JUHa in 1996 | 36 |
| Mu50 | VISA clinical isolate from JUH in 1996 | 36 |
| 6R-P | sVISA strain; mutant isolated from Mu3 by selection with 6 mg/liter VAN | 8 |
| Mu3fdh2* | Mu3-derived construct with its fdh2 (SAVH_2293) replaced by fdh2*b | 13 |
| Mu3fdh2*V6-5 | VISA mutant strain obtained by selection of Mu3fdh2* with 6 mg/liter VAN; spontaneously generates LC, SC, and PC | 13 |
| V6-5 | Mu3fdh2*V6-5 with PC morphology | This study |
| V6-5-L1 to -L11 | Derivative of Mu3fdh2*V6-5 with LC morphology | This study |
| E. coli | ||
| JM109 | General purpose host strain for molecular cloning | TaKaRa Bio |
| Plasmids | ||
| pKE515 | E. coli-S. aureus shuttle vector for complementation of rpoC and relQ genes | 14 |
| pSR_rpoC | pKE515 carrying 3.8-kb DNA fragment containing WT rpoC | This study |
| pSR_relQ | pKE515 carrying 1.0-kb DNA fragment containing WT relQ | This study |
JUH, Juntendo University Hospital.
fdh2* indicates the fdh2(A297V) mutation, which is found in clinical VISA strain Mu50, although the introduction of the mutation did not confer the VISA phenotype on Mu3.
Recombinant DNA techniques.
Extraction and purification of plasmid DNA from Escherichia coli, DNA isolation from S. aureus, restriction endonuclease digestion, ligation reactions, and DNA cloning were performed as previously described (13). S. aureus electroporation was performed using a Gene Pulser system (Bio-Rad, Hercules, CA) as previously described (13). The plasmid pSR_rpoC was constructed with Mu3 chromosomal DNA and was amplified using the BamHI-rpoC-CP1 and KpnI-rpoC-CP2 primers (see Table S1 in the supplemental material) and then was inserted into the BamHI and KpnI sites in the pKE515 plasmid vector (14). The integrity of the cloned rpoC gene was determined by sequencing the recombinant plasmid using the primers listed in Table S1. The plasmid pSR_relQ containing the relQ gene region was amplified using the rshB-SP1 and rshB-SP2 primers, and the resulting fragment was ligated into the SmaI site of the pKE515 vector.
Antimicrobial susceptibility testing.
MICs were determined using the Etest (AB Biodisk, Solna, Sweden) in accordance with the manufacturer's recommendations. In these experiments, BHI agar was used instead of Mueller-Hinton (MH) agar because BHI agar is more supportive of the VISA phenotype than other tested agars (15).
Phenotype assays.
Doubling time (DT) and autolytic assays were performed as previously described (8). Cell wall thicknesses were measured in electron microscope images (model H-7700; Hitachi, Tokyo, Japan) at a final magnification of 20,000 as previously described (13).
Appearance rates of large colonies of sVISA.
The appearance rates of large colonies (LC variants) of the sVISA strains V6-5 and 6R-P were determined as previously described (8).
Isolation of LC variants from the sVISA strain V6-5.
A total of 105 sVISA cells were inoculated into 10 tubes containing 4 ml of BHI broth. After 20 h of cultivation at 37°C, portions of the 10 cultures containing about 103 CFU were spread onto BHI agar plates. Single large colonies were then picked from plates after 24-h incubation. A total of 10 colonies were isolated and established as large colony (LC) strains and were used in further studies.
Whole-genome sequencing and identification of mutations.
DNA extraction was performed using a Qiagen genome-tip system (Qiagen), and library preparations were performed using Nextera XT DNA sample preparation kits (Illumina, Inc., San Diego, CA). Pools of six samples were sequenced on the Illumina MiSeq platform, and 250-bp end reads were generated. Reads from bacterial strains were then mapped to the reference whole-genome sequence of the Mu3 strain (accession no. NC_009782.1), and mutations were identified using Genome Traveler software (version 2.1.21; In Silico Biology, Inc., Yokohama, Japan).
RNA preparation and microarray analysis.
RNA extraction, cDNA labeling, hybridization, and data analyses of microarray data were performed according to previously described protocols (16). In brief, cells were grown mid-exponentially at 37°C to an optical density at 660 nm (OD660) of approximately 0.6. A customized high-density synthetic oligonucleotide array of pre-methicillin-resistant S. aureus (pre-MRSA) (clinical strain N315) was designed using the 2,814,816-bp chromosome genome sequence, which included 2,628 predicted open reading frames (ORFs) (GenBank accession no. NC_002745) (NimbleGen Systems, Inc., Madison, WI). Subsequently, 1,128 ORFs that were absent in N315 cells but present in the 13 clinical S. aureus strains COL, JH1, JH9, MRSA252, MSSA476, Mu3, Mu50, MW2, NCTC 8325, Newman, RF122, USA300 FPR, and USA300 TCF and plasmid pLW043 were added to the array (a total of 3,846 ORFs per array). Each microarray probe (60-mer) was replicated three times to facilitate intraarray reproducibility. Arrays were then scanned using a NimbleGen MS200 microarray scanner (NimbleGen Systems, Inc.), and data were extracted using NimbleScan software (NimbleGen Systems, Inc.).
Statistical analyses of the microarray data were performed after normalization of signal intensity using a robust multichip analysis (RMA) algorithm (17) and global normalization. Mean normalized signal intensities were calculated, and differences between strains were identified using Student's t test. Subsequently, n-fold change ratios were calculated from mean normalized signal intensities.
Microarray data accession number.
The sequences from gene expression of the Mu3fdh2*, V6-5, V6-5-L1, -L2, -L3, -L5, -L7, -L8, and -L9 strains have been deposited in the NCBI under GEO accession no. GSE65338 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE65338).
RESULTS
The rpoC(P440L) mutation leads to expression of the sVISA phenotype. (i) Mu3fdh2*V6-5 cells express the sVISA phenotype.
Instability of growth rates and VAN resistance were significant phenotypes of sVISA cells. Colony-size morphologies were assessed in overnight cultures of Mu3fdh2*V6-5 that were established by selection with VAN as a VISA strain from the hVISA strain Mu3fdh2* (13). Colony sizes of Mu3fdh2*V6-5 on BHI agar plates after 24 h of incubation were heterogeneous and included large (LC), small (SC), and pinpoint (PC) colonies, as observed in the rpoB(R512P) mutant sVISA strain 6R-P (8). Mu3fdh2*V6-5 cells generated large colonies at a frequency of 0.3%, which was more frequent than in 6R-P cells (3 × 10−7).
Doubling times (DTs) and VAN MICs of the pinpoint (V6-5) and large (V6-5-L1) strains were measured after establishment from the Mu3fdh2*V6-5 strain and colony purification. As shown in Table 2, the DT of V6-5 (72.9 min) was much longer than in the sVISA strain 6R-P (62.2 min) and the large variant V6-5-L1 (41 min). The VAN MIC of V6-5 was 16 mg/liter, as determined by Etest at 72 h, and was greater than that for the VISA strain Mu50, which is similar to that of the sVISA strain 6R-P. In contrast, the MIC of V6-5-L1 decreased to 4 mg/liter, which is similar to that of the hVISA strain Mu3fdh2*. These data demonstrate the sVISA phenotype in V6-5 cells.
TABLE 2.
Identification and characterization of sVISA with the rpoC(P440L) mutation
| Strain | VAN resistance phenotypea | DT (min) | VAN MIC (mg/liter) at: |
Appearance rate (%) of large colonies | Cell wall thickness (mean ± SD nm) | Mutated gene | Amino acid substitution(s) | Reference | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 24 h | 48 h | 72 h | 96 h | 120 h | ||||||||
| V6-5 | sVISA | 72.9 | 6 | 12 | 16 | 16 | 16 | 10/2,910 (0.3) | 33.4 ± 2.5 | rpoC | P440L | This study |
| V6-5-L1 | hVISA | 41.0 | 3 | 4 | 4 | 4 | 4 | NDb | 29.5 ± 2.2 | rpoC | P440L, F652L | This study |
| Mu3fdh2* | hVISA | 37.7 | 3 | 3 | 3 | 3 | 4 | ND | 27.8 ± 1.9 | 13 | ||
| Mu50 | VISA | 40.3 | 8 | 12 | 12 | 12 | 12 | ND | 33.9 ± 3.7 | 36 | ||
| 6R-P | sVISA | 62.2 | 8 | 12 | 16 | 16 | 16 | 3/1.0 × 107 | 33.4 ± 2.2 | rpoB | R512P | 8 |
sVISA, slow VISA; hVISA, heterogeneously VISA.
ND, not determined.
(ii) Cell wall thickening and reduced autolytic activity in the rpoC(P440L) mutant sVISA strain V6-5.
Increased cell wall thickness and reduced autolytic activity were observed in the sVISA strain 6R-P, which carried an rpoB gene mutation (8). Subsequently, cell wall thickness and Triton X-100-induced autolytic activity were evaluated in the rpoC(P440L) mutant sVISA strain and in the LC variant strain. Electron microscope observations of the V6-5 strain showed comparable cell wall thicknesses (33.4 ± 2.5 nm) to those of the 6R-P (33.4 ± 2.2 nm) and Mu50 VISA (33.9 ± 3.7 nm) strains but greater thicknesses than in the parental strain, Mu3fdh2* (27.8 ± 1.9 nm). However, the V6-5-L1 strain showed partial loss of cell wall thickness (29.5 ± 2.2 nm) compared with V6-5 cells.
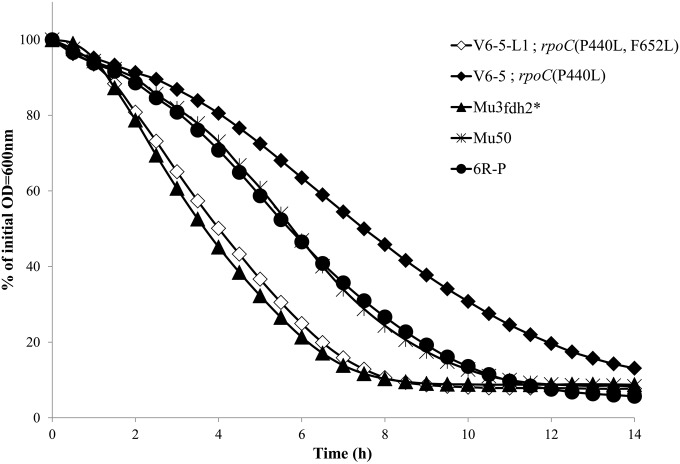
Autolytic activity (Fig. 1) was significantly reduced in the V6-5 strain compared with those in Mu3fdh2* and parental Mu3 strains, and it was greater than those in the Mu50 and 6R-P strains. In contrast, V6-5-L1 autolytic activity was restored to levels that were comparable with those in the Mu3fdh2*strain. These data indicated that V6-5 cells express the sVISA phenotypes and that V6-5-L1 cells are phenotypic hVISA revertants.
FIG 1.
Autolytic activity in rpoC(P440L) mutant sVISA strain V6-5 and its phenotypic revertant strain, V6-5-L1. Shown is Triton X-induced autolysis of V6-5 [rpoC(P440L)], V6-5-L1 [rpoC(P440L F652L)], Mu3fdh2*, Mu3, and VISA strain Mu50. Autolysis was measured as the decline in optical density (OD) versus time and is expressed as the percentage of the initial optical density.
(iii) The rpoC(P440L) mutation is associated with expression of the sVISA phenotype.
Whole-genome sequencing of the sVISA strain V6-5 and the revertant strain V6-5-L1 was performed, and comparisons of sequence data revealed that the V6-5-L1 strain carries a P440L mutation and a second mutation in the rpoC gene. The 652nd residue of V6-5 (Phe) was mutated to leucine in V6-5-L1 strain (Table 2), and the F652L mutation was considered complementary to the dysfunction caused by the P440L mutation. In subsequent complementation experiments (Table 3), introduction of the pSR_rpoC plasmid (wild-type [WT] rpoC) into V6-5 cells significantly decreased the VAN MIC (from 16 mg/liter to 4 mg/liter) to levels that were comparable with those in the parent strain, Mu3fdh2*. Moreover, introduction of the pSR_rpoC plasmid shortened the DT from 71 min to 43 min in V6-5 cells. In contrast, this vector plasmid had little effect on VAN MIC or DT in V6-5 cells, indicating that the rpoC(P440L) mutation is associated with expression of the sVISA phenotype.
TABLE 3.
Introduction of the WT rpoC gene cures V6-5 of the sVISA phenotype
| Strain |
rpoC gene in: |
DT (min)a | VAN MIC (mg/liter) ata: |
|||||
|---|---|---|---|---|---|---|---|---|
| Chromosome | Plasmid | 24 h | 48 h | 72 h | 96 h | 120 h | ||
| V6-5(pSR_rpoC) | P440L | pSR(WT rpoC) | 43.0 | 4 | 4 | 4 | 4 | 4 |
| V6-5(pSR) | P440L | pSR | 72.7 | 4 | 12 | 16 | 16 | 16 |
| V6-5 | P440L | (−)b | 71.0 | 4 | 12 | 16 | 16 | 16 |
| Mu3fdh2* | WT | (−) | 37.1 | 3 | 3 | 3 | 3 | 3 |
When the MICs and doubling times for the strains carrying the pSR plasmids were measured, 10 mg/liter chloramphenicol was added to the BHI agar plates and medium to maintain the plasmids.
(−), no plasmids.
Identification of mutations associated with the phenotypic reversion.
The present V6-5 sVISA strain rapidly reverted to the hVISA phenotype, and the associated genetic mechanisms were investigated in 10 independently isolated LC variants of V6-5 using phenotypic and genetic analyses.
Ten LC variants were designated V6-5-L2 to -11 in order of isolation. After purification, the DTs and MICs of VAN were determined in all strains, and cell wall thicknesses and autolytic activities were determined in 6 of 10 strains. In these experiments, decreases in DT ranged from 41.3 to 54.7 min among sVISA LC strains, and MICs were decreased to 3 to 8 mg/liter (Table 4), indicating decreases in sVISA VAN resistance compared with the levels in VISA and hVISA strains.
TABLE 4.
Nonsynonymous mutations additionally incorporated in the large-colony variants obtained from sVISA strain V6-5 harboring the rpoC(P440L) mutation
| Variant strain | ORF ID no. | Gene name | Gene product | Nucleotide change | Amino acid changea |
|---|---|---|---|---|---|
| V6-5-L2b | SAHV_0541 | rpoC | RNA polymerase β′ subunit | C610318G | Leu 440→Val |
| V6-5-L3 | SAHV_0234 | Acetyl-CoAc transferase | C274549T | Asp 243→Asn | |
| SAHV_0541 | rpoC | RNA polymerase β′ subunit | C610935A | Asp 645→Glu | |
| SAHV_0541 | rpoC | RNA polymerase β′ subunit | G610937T | Arg 646→Leu | |
| SAHV_1004 | Na+/H+ antiporter homologue | A1061220T | Leu 527→Phe | ||
| V6-5-L4 | SAHV_0541 | rpoC | RNA polymerase β′ subunit | T610954A | Phe 652→Ile |
| SAHV_2402 | sbi | IgG-binding protein | C2549778T | Ala 284→Val | |
| V6-5-L5 | SAHV_1000 | relQ | GTP pyrophosphokinase | T1055998A | Tyr 29(*) |
| V6-5-L6 | SAHV_1762 | RNA polymerase sporulation-specific sigma factor | G1913693T | Lys 144→Asn | |
| SAHV_2108 | murZ | UDP-N-acetylglucosamine 1-carboxyvinyltransferase | G2252973C | Thr 240→Arg | |
| V6-5-L7 | SAHV_2145 | glmM | Phosphoglucosamine mutase | A2298569T | Phe 5→Ile |
| V6-5-L8 | SAHV_2108 | murZ | UDP-N-acetylglucosamine 1-carboxyvinyltransferase | C2253663A | Gly 10→Val |
| V6-5-L9 | SAHV_0157 | capJ | Capsular polysaccharide synthesis enzyme Cap5J | T176380A | Phe 3→Ile |
| SAHV_1000 | relQ | GTP pyrophosphokinase | C1056191T | Gln 94(*) | |
| V6-5-L10 | SAHV_0962 | Exonuclease RexA | C1015940T | Silent | |
| SAHV_0997 | Globin family protein | G1054322A | Ala 115→Val | ||
| SAHV_1000 | relQ | GTP pyrophosphokinase | A1056194T | Arg 95(*) | |
| V6-5L-11 | SAHV_2108 | murZ | UDP-N-acetylglucosamine 1-carboxyvinyltransferase | G2252775A | Thr 306→Ile |
(*), stop codon.
This was the only strain in which the original mutation was altered into a new one.
Acetyl-CoA, acetyl coenzyme A.
Subsequent whole-genome sequencing of revertant and parental V6-5 sVISA strains identified mutations in revertants, as listed in Table 4. Although mutations varied between revertants, all of them carried at least one mutation in the genes coding for (i) the RNA polymerase beta′ subunit, RpoC, (ii) RelQ, or (iii) the peptidoglycan biosynthesis molecules GlmM and MurZ (Table 5).
TABLE 5.
Doubling time, mutation, antibiogram, and cell wall thickness of rpoC(P440L) mutant sVISA strain V6-5-derived LC variants
| Strain | Mutationa |
Colony size variant | DT (min) | MIC (mg/liter)b |
Cell wall thicknessc |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene or ORF ID no. | Amino acid change | VAN |
BHA |
Mean ± SD (nm) | Ratio | |||||||
| BHA | MHA | TEC | MU | DAP | LZD | |||||||
| Mu50 | LC | 40.3 | 12 | 6 | 12 | 0.19 | 4 | 0.38 | 33.9 ± 3.7 | 1.22 | ||
| Mu3 | LC | 29.9 | 3 | 1.5 | 8 | 0.25 | 2 | 0.75 | NDc | |||
| Mu3fdh2* | LC | 37.7 | 3 | 1.5 | 12 | 0.25 | 1.5 | 0.5 | 27.8 ± 1.9 | 1 (reference) | ||
| V6-5 | PC | 72.9 | 16 | 8 | 24 | 0.094 | 3 | 0.38 | 33.4 ± 2.5 | 1.20 | ||
| RpoC group | ||||||||||||
| V6-5-L1 | rpoC | F652L | LC | 41.0 | 4 | 2 | 16 | 0.19 | 2 | 0.5 | 29.5 ± 2.2 | 1.06 |
| V6-5-L2 | rpoC | L440V | LC | 41.3 | 4 | 2 | 16 | 0.19 | 2 | 0.5 | 28.0 ± 2.3 | 1.01 |
| V6-5-L4 | rpoC | F652I | LC | 42.4 | 6 | 2 | 16 | 0.125 | 3 | 0.5 | ND | |
| sbi | A284V | |||||||||||
| V6-5-L3 | rpoC | D645E, R646L | SC | 50.5 | 8 | 3 | 16 | 0.125 | 2 | 0.5 | 30.7 ± 3.1 | 1.10 |
| SAHV_0234 | D243N | |||||||||||
| SAHV_1004 | L527F | |||||||||||
| RelQ group | ||||||||||||
| V6-5-L5 | relQ | Y29(*) | SC | 54.7 | 8 | 4 | 8 | 0.064 | 2 | 0.38 | 29.3 ± 3.5 | 1.05 |
| V6-5-L9 | relQ | Q94(*) | SC | 51.2 | 8 | 4 | 8 | 0.094 | 3 | 0.38 | 28.7 ± 2.0 | 1.03 |
| capJ | F3I | |||||||||||
| V6-5-L10 | relQ | R95(*) | SC | 52.1 | 8 | 4 | 8 | 0.094 | 2 | 0.38 | ND | |
| SAHV_0997 | A115V | |||||||||||
| PG group | ||||||||||||
| V6-5-L7 | glmM | F5I | SC | 49.6 | 4 | 2 | 6 | 0.064 | 3 | 0.75 | 23.5 ± 2.0 | 0.85 |
| V6-5-L11 | murZ | T306I | SC | 48.4 | 3 | 2 | 6 | 0.094 | 2 | 0.5 | ND | |
| V6-5-L8 | murZ | G10V | SC | 48.7 | 4 | 1.5 | 8 | 0.064 | 2 | 0.5 | 28.1 ± 1.6 | 1.01 |
| V6-5-L6 | murZ | T240R | SC | 50.2 | 4 | 1.5 | 8 | 0.094 | 2 | 0.5 | ND | |
| SAHV_1762 | K144N | |||||||||||
Boldface type indicates the mutation was identified in a single gene. (*), stop codon.
The MIC was determined by Etest after 72 h of incubation on a brain heart infusion agar (BHA) or Mueller-Hinton agar (MHA) plate. VAN, vancomycin; TEC, teicoplanin; MU, mupirocin; DAP, daptomycin; LZD, linezolid.
ND, not determined.
(i) The RpoC group.
Mutations in the rpoC gene were found in V6-5-L2, V6-5-L3, and V6-5-L4 strains (Tables 4 and 5; RpoC group). The V6-5-L3 strain had two additional rpoC mutations that substituted for the 645th (Asp) and 646th (Arg) residues with Glu and Leu, respectively. The 652nd residue (Phe) in V6-5-L4 was mutated to Ile at the same position as for the V6-5-L1 strain. In the crystal structures of Thermus thermophilus and Escherichia coli RNA polymerase (18, 19), amino acid residues corresponding to positions 440 and 646 to 652 in the sVISA strain V6-5 are conformational neighbors. Thus, the rpoC(P440L) mutation in V6-5 is likely affected by conformational changes that are caused by these additional rpoC mutations in LC variants.
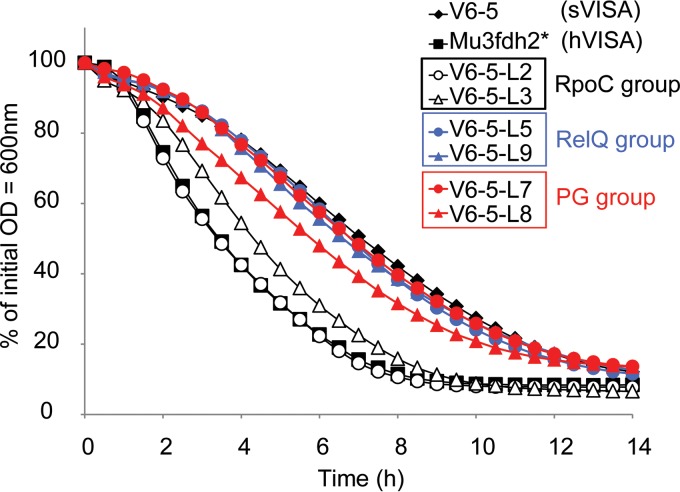
Notably, V6-5-L2 had a second mutation in the 440th codon (CTA→GTA), resulting in substitution of the 440th residue Leu to Val. The VAN MIC decreased to 4 mg/liter in the V6-5-L2 strain, and the DT increased to 41.3 min. Moreover, neither thickening of the cell wall nor reduction in autolytic activity was observed as in the V6-5 strain, and wall thicknesses and autolytic activities were restored to levels similar to those of the parent strain, Mu3fdh2* (Table 5 and Fig. 2). These results strongly indicate an association between the rpoC(P440L) mutation in V6-5 and the sVISA phenotype.
FIG 2.
Autolytic activities in LC variants. Shown is Triton X-induced autolysis of V6-5-L2 and V6-5-L3 (the RpoC group), V6-5-L5 and V6-5-L9 (the RelQ group), V6-5-L7 and V6-5-L8 (the PG group), V6-5, and Mu3fdh2*.
(ii) The RelQ group.
The gene relQ encodes the 211-amino-acid protein RelQ in S. aureus. Mutations in relQ were found in 3 strains (Tables 4 and 5; RelQ group). The V6-5-L5 strain carried a single mutation in the relQ gene that led to an amino acid substitution for the 29th residue Tyr with a stop codon (*). Deletion of amino acids 115 to 143 abolished the synthetic activity of (p)ppGpp (20). Thus, decreased DT and VAN MIC in V6-5-L5 may reflect loss of RelQ function, following mutation of Y29(*) and deletion of the active site. Accordingly, after introduction of wild-type relQ into the V6-5-L5 strain using plasmid pSR, complementation experiments showed restoration of VAN resistance and DT (Table 6). In contrast, introduction of the pSR_relQ into Mu3fdh2* did not affect VAN resistance or DT, indicating that expression of multiple copies of RelQ does not affect VAN resistance and that the loss of RelQ function in the V6-5-L5 strain contributed to the loss of the sVISA phenotype in rpoC(P440L) mutant cells.
TABLE 6.
Introduction of the WT relQ gene into strain V6-5-L5 restores the sVISA phenotype
| Strain |
relQ gene |
DT (min)a | MIC (mg/liter) ata: |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Chromosome | Plasmid | VAN |
TECb |
||||||||
| 48 h | 72 h | 96 h | 120 h | 48 h | 72 h | 96 h | 120 h | ||||
| Mu3fdh2* | WT | (−)c | 37.9 | 3 | 3 | 3 | 3 | 12 | 12 | 12 | 12 |
| Mu3fdh2*(pSR_relQ) | WT | pSR(WT relQ) | 41.3 | 3 | 3 | 3 | 3 | 12 | 12 | 12 | 12 |
| Mu3fdh2*(pSR) | WT | pSR | 37.8 | 3 | 3 | 3 | 3 | 12 | 12 | 12 | 12 |
| V6-5 | WT | (−) | 72.3 | 12 | 16 | 16 | 16 | 24 | 24 | 24 | 24 |
| V6-5-L5 | Y29(*)d | (−) | 54.4 | 6 | 6 | 8 | 8 | 8 | 8 | 8 | 12 |
| V6-5-L5(pSR_relQ) | Y29(*) | pSR(WT relQ) | 88.1 | 8 | 12 | 12 | 12 | 16 | 24 | 24 | 24 |
| V6-5-L5(pSR) | Y29(*) | pSR | 56.2 | 4 | 6 | 6 | 6 | 8 | 8 | 8 | 8 |
When the MICs and DTs for the strains carrying the pSR plasmids were measured, 10 mg/liter chloramphenicol was added to the BHI agar plates and medium to maintain the plasmids.
TEC, teicoplanin.
(−), no plasmids.
(*), stop codon.
(iii) The PG group.
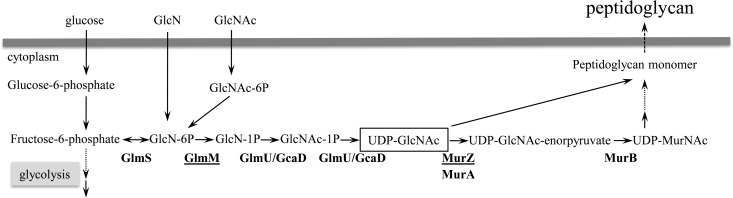
Mutations in the peptidoglycan (PG) synthesis genes murZ and glmM were found in 3 and 1 of the 10 LC revertants, respectively (Tables 4 and 5; PG group). GlmM converts glucosamine-6-phosphate (GlcN-6P) to GlcN-1P and lies in the main pathway for UDP-GlcNAc, which is the cardinal precursor metabolic for cell wall peptidoglycan. MurZ catalyzes the biosynthesis of the first intermediate for the production of peptidoglycan-monomer units from UDP N-acetylglucosamine (UDP-GlcNAc) (Fig. 3). Mutation of a single gene was sufficient to reduce VAN resistance in three of four strains (V6-5-L7, V6-5-L8, and V6-5-L11) (Table 5).
FIG 3.
Metabolic pathway for the synthesis of cell wall peptidoglycan. GlmM converts GlcN-6P to GlcN-1P as part of the UDP-GlcNAc synthetic pathway and produces the cardinal precursor metabolite for cell wall peptidoglycan. MurZ is the first enzyme in the biosynthetic pathway for production of peptidoglycan monomer units from UDP-GlcNAc.
Cell wall thickness and autolytic activity in the LC variants.
The rpoC(P440L) mutation in the V6-5 strain conferred thickening of the cell wall (Tables 2 and 5), whereas the V6-5-L3 (RpoC group) and V6-5-L5 and V6-5-L9 (RelQ group) strains showed partial loss of cell wall thickening, and the VAN MIC decreased to 8 mg/liter (Table 5). The V6-5-L8 strain carried a murZ(G10V) mutation and had a reduced cell wall thickness of 28.1 + 1.6 nm, comparable to that of Mu3fdh2* (27.8 ± 1.9 nm). Moreover, cell wall thickness was significantly decreased to 23.5 ± 2.0 nm in the glmM(F5I) revertant, less than that of the parental strain, Mu3fdh2*. Accordingly, the VAN MICs of these strains were decreased to 3 to 4 mg/liter, indicating that both murZ and glmM mutations led to decreased VAN resistance in revertants by reducing otherwise augmented cell wall peptidoglycan synthesis in sVISA strain V6-5.
With respect to autolytic activity, the V6-5-L7 (PG group) and V6-5-L5 and V6-5-L9 (RelQ group) strains retained decreased autolytic activity, although the V6-5-L8 murZ mutant strain of the PG group showed slightly restored autolysis activity (Fig. 2), whereas, V6-5-L3 of the RpoC group strain had considerably restored autolytic activity. Moreover, the revertant strain V6-5-L2 harboring the rpoC(L440V) mutation instead of the rpoC(P440L) mutation of the parent strain Mu3fdh2* restored the autolytic activity to the level of the parent, indicating that the rpoC(P440L) mutation in sVISA strain V6-5 was a regulatory mutation of decreased autolytic activity.
Susceptibility tests of antibiotics other than VAN.
In experiments with antibiotics other than VAN (Table 5), susceptibility to daptomycin was affected only marginally with rpoC mutation. Linezolid MICs tended to correlate negatively with VAN MICs, in agreement with Cui et al. (21). Susceptibility to teicoplanin and mupirocin was affected by mutations in rpoC, and although the teicoplanin MIC was 16 mg/liter in all strains of the RpoC group (V6-5-L1, V6-5-L2, V6-5-L3, and V6-5-L4), the MICs of strains in the RelQ and PG groups were decreased to 6 to 8 mg/liter, which was similar to or less than that of the hVISA strain Mu3. MICs of mupirocin for RpoC strains were increased compared with that for sVISA strainV6-5, whereas no increase in mupirocin MIC was observed in the other group of strains.
Transcriptional profiles of sVISA strain V6-5 and its phenotypic revertant strains.
Microarray analysis of the V6-5 and Mu3fdh2* strains revealed as many as 876 differentially expressed genes (≥1.5-fold or <1.5-fold; P value < 0.05) (see Table S2 in the supplemental material). Figure 4 shows representative genes that were differentially expressed in the sVISA strain V6-5 and the phenotypic revertant strains V6-5-L1, -L2, -L3, -L5, -L7, -L8, and -L9 in comparison with Mu3fdh2*.
FIG 4.
Microarray analysis of sVISA strain V6-5 and its revertants: Shown are differentially expressed genes in sVISA strain V6-5 and its phenotypic revertants V6-5L, L1, L2, L4, L6, L7, and L8 compared with genes in hVISA strain Mu3fdh2*.
A total of 431 genes were upregulated in V6-5 relative to Mu3fdh2* (≥1.5-fold; P < 0.05) (see Table S2 in the supplemental material), They included capsule genes, urease-encoding genes ureA, -B, -C, -E, -F, -G, and -D, oligopeptide transporter genes oppD and oppF, genes involved in molybdopterin biosynthesis moaB, mobB, modAB, and moeA, and the ESAT-6 type secretion system (Ess) pathway genes esaA, -B, -C, -D, -E, and -F and essA, -B, and -C (22). As we previously reported, these genes were also upregulated in Mu3-derived sVISA strain 6R-P, which harbored the rpoB(R512P) mutation (8).
Some genes of interest involved in vancomycin resistance were specifically upregulated in V6-5 (and not in 6R-P). They included the walKR genes, which are associated with VAN resistance in clinical and laboratory VISA strains (23, 24).
A total of 445 genes were downregulated (<1.5-fold; P < 0.05) (see Table S2 in the supplemental material) in V6-5, including genes of the pyrimidine synthesis pathway (pyrRPBCAA and -F and tdk), the ribosomal protein genes rpsR and rpsO, the translational genes ptpB, sua5, and prfA, and the teichoic acid biosynthesis gene tagG. These genes were also downregulated in 6R-P (8). Although the nitrate/nitrite reductase-associated genes narG, nasD, nirR, and nasE were downregulated in strain 6R-P, they were not downregulated in V6-5. However, the regulator genes lexA, mepR, and tcaR, the transporter genes mepA, glpT, and msmX, the sortase gene SA2316, and the putative hemK gene were downregulated in both sVISA strains. We previously showed that mutation in the hemK gene converts hVISA strain Mu3 to VISA (13). Finally, the spermidine/putrescine ABC transporter homolog genes potA, B, -C, and -D were specifically downregulated in V6-5.
Some characteristic features of gene expression were noticed among the three groups of phenotypic revertants (Fig. 4). For example, the PG mutation group did not revert the enhanced expression of Ess genes, whereas the RelQ group did. Ess is an important factor for colonization and virulence (25, 26). On the other hand, the PG group of revertants had further downregulation of pyrimidine pathway genes beyond the level of rpoC-mediated repression.
Enhanced expression of almost all genes in V6-5 presented in Fig. 4 waned in the phenotypic revertant V6-5-L2 carrying the rpoC(L440V) alternate mutation. This was also the case with V6-5-L1, and the tendency was more pronounced in the revertants with lower VAN MICs. Most notably, the expression of the relA gene encoding GTP pyrophosphokinase was downregulated in V6-5 but reverted to the original level of the parent in the RpoC group of revertant strains (Fig. 4). S. aureus has two other genes encoding GTP pyrophosphatases, relP and relQ. In contrast to the relA gene, the relP gene was upregulated in V6-5, and the expression of relP remained high in the RpoC group of revertants. On the other hand, the expression of relQ in V6-5 did not alter. Also decreased transcription of aminoacyl-tRNA and ribosomal proteins was noticed in V6-5, which was greatly relieved in the RpoC group of revertants. Upregulated genes in V6-5, such as capsule genes, molybdopterin biosynthesis genes, Ess genes, and purine metabolic genes, were reverted or downregulated in relQ mutant strains. On the other hand, the downregulated genes sigA and tcaR were reverted in relQ mutant strains. In contrast, the expression of urease genes and peptidoglycan hydrolase genes, such as lytM and sceD, that may be associated with VAN sensitivity (13, 27) remained enhanced in relQ mutant strains. Moreover, the upregulated expression of the relP gene in V6-5 was further augmented in the RelQ group of revertants.
In PG group mutant strains, the enhanced expression of molybdopterin biosynthesis genes, purine metabolism genes, and ABC transporter genes, such as opuCC and opuCB, reverted to the original level of the parent strain. However, the expression of the Ess genes remained high, and pyrimidine metabolism genes remained downregulated in PG group strains.
DISCUSSION
In our recent study, we defined sVISA as a novel VAN resistance phenotype and identified rpoB mutations as a frequent underlying cause (8). In the present study, we showed that the rpoC(P440L) mutation also conferred the sVISA phenotype on an hVISA strain, Mu3fdh2*. Strain V6-5 with a pinpoint colony morphology was rapidly changed into large or small colony variants with either the VISA or hVISA phenotype. Subsequent whole-genome and phenotypic analyses of the variant strains revealed that additional mutations in such genes as rpoC, encoding an RNA polymerase subunit, glmM and mruZ in the peptidoglycan biosynthesis pathway, and the relQ gene involved in the cell wall stress response were correlated with the release of V6-5 from the unstable sVISA phenotype. This clearly showed that the functional integrity of the enzymes involved in peptidoglycan synthesis and cell wall stress response regulatory system are supporting the sVISA phenotype expression of V6-5. We previously reported isolation of 26 sVISA strains from the hVISA strain Mu3 (8). Recently, we found 3 other rpoC mutations in the strains. Whole-genome analyses revealed that the only mutation detected, in 2 out of 3 sVISA strains, was a single nucleotide polymorphism (SNP) in rpoC of the sVISA strains (37), indicating that rpoC mutation does convert hVISA into sVISA.
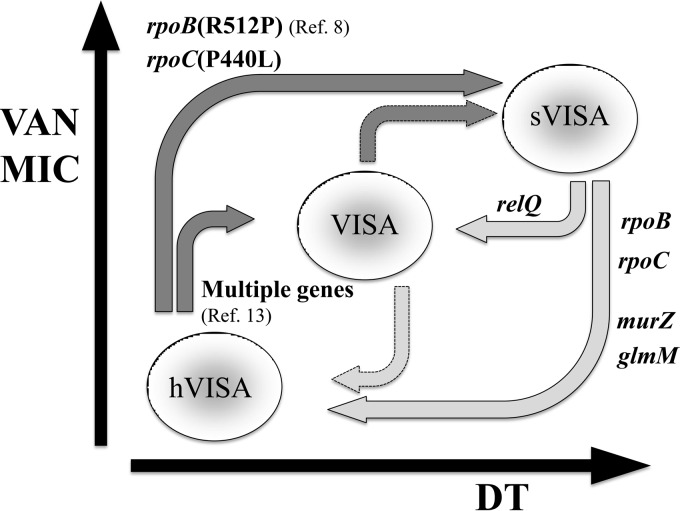
RNA polymerase mutation seems to have the regulatory effect in triggering the entire scheme of sVISA phenotype expression. In the proposed cycle of VAN resistance (Fig. 5), the rpoB(R512P) mutation acts as a functional switch for the reversible phenotype of sVISA. Accordingly, the present data indicate that the rpoC(P440L) mutation is also a regulatory mutation triggering the sVISA phenotype expression. The mutation was specifically replaced by an alternate mutation, L440V, with concomitant loss of the sVISA phenotype in strain V6-5-L2 (Tables 4 and 5 and Fig. 2 and 4). The additional mutations, such as F652L, D645E, R646L, and F652I, found in the rpoC gene of the other LC revertants are considered to be compensatory or complementary mutations that occurred in different codons of the same molecule. It seems likely that the conformational change of RNA polymerase holoenzyme induced by the above mutations in rpoB and rpoC altered the overall gene expression profile toward the mode of survival against cell wall synthesis inhibition and urgent threat of cell lysis.
FIG 5.
Reciprocal relationship between vancomycin (VAN) MICs and doubling times (DTs) of hVISA, VISA, and sVISA strains. The hVISA strain generates VISA and sVISA strains following exposure to VAN. The sVISA strain has a higher VAN MIC and extremely prolonged DT. This step was associated with the mutations rpoB(R512P) and rpoC(P440L). The sVISA phenotype is stable in the presence of VAN but rapidly reverts to VISA and hVISA phenotypes in antibiotic-free media. The ability to revert is associated with mutations in peptidoglycan synthesis genes and the relQ gene and alternative mutations in rpoB and/or rpoC genes. Deep gray and pale gray arrows indicate the presence and absence of VAN, respectively.
The proline of the 440th position in the RNA polymerase β′ subunit is evolutionarily conserved among prokaryotes and is located near the active site of the enzyme (28). P440 and the residues (D645, R646, and F652) found in RpoC group revertants face each other (18, 19). In Escherichia coli, the region corresponding to the 645th to 652nd amino acid residues in S. aureus was identified as a binding site for ppGpp on RNA polymerase (29). ppGpp is a key regulator of the stringent response, which enables bacterial cells to adapt to stressful conditions, such as nutrient starvation and antimicrobial treatment (30). RelQ is a ppGpp synthase, involved in the cell envelope stringent response of S. aureus (20). We found that the relQ mutations reduced the level of vancomycin resistance of sVISA strain V6-5. Microarray analysis revealed some similarities of transcriptional profiles between the RpoC (V6-5-L1, V6-5-L2, and V6-5-L3) and RelQ (V6-5-L5 and V6-5-L9) groups of revertants (Fig. 4). This indicated that physiological changes caused by the rpoC(P440L) mutation covers at least a part of transcriptional changes brought about by the disruption of one of the three rel genes involved in the stringent response (see below). It is also noted that relA, which is the only rel gene that encodes hydrolase activity of ppGpp, is repressed by the rpoC(P440L) mutation, indicating that the ppGpp tends to be accumulated in V6-5 cells.
Revertant strain V6-5-L5 was unique in that it possessed an early stop codon introduced in the relQ gene (Table 4). The early truncation of the relQ gene cancelled the sVISA phenotype of V6-5 caused by rpoC(P440L) mutation (Table 6). The sVISA phenotype was restored by introduction of the wild-type relQ gene on a multicopy plasmid complemented into V6-5-L5 (Table 6). However, multicopy expression of the relQ gene in the hVISA strain Mu3fdh2*, which possesses the wild-type rpoC gene, did not affect VAN resistance, indicating that mere increase in relQ gene expression does not contribute to sVISA phenotype expression (Table 6). S. aureus possesses two additional small (p)ppGpp synthases, RelP and RelQ, besides RelA, which are involved in the stringent response (20, 31). RelP and RelQ in S. aureus are reported to be induced under the cell envelope stress condition and make the cell tolerate these hazardous conditions (20). Although detailed functional differences between relP and relQ remain unclear, deletion of the relP gene in the V6-5 strain did not affect the sVISA phenotype (data not shown), indicating the essential role of RelQ in the sVISA phenotype. In any case, disruption of relQ seems to have mitigated the sVISA phenotype of V6-5, decreasing the vancomycin MIC and increasing the growth rate (Table 6).
The revertants in the PG group were associated with more significant loss of the sVISA phenotype than those of the RelQ group (Table 5). Activated synthesis and thickening of peptidoglycan layers are the mechanism of the VISA phenotype (32–35). Therefore, it is reasonable that mutations in peptidoglycan biosynthesis pathway genes such as murZ and glmM are likely to lessen VAN resistance (Table 5). It also seems reasonable to assume that RpoC(P440L) mutation not only upregulates some of the critical PG synthesis enzymes, such as glmS, glmM, and murF, but also provides materials and (saved) energies to the PG synthesis pathway by repressing the other metabolic pathways, such as pyrimidine and protein synthesis. In addition to these functions, rpoC regulatory mutation causes reduced autolysis (Fig. 2), and this is not resolved in either one of the RelQ or PG group revertants. The expression of peptidoglycan hydrolase and other genes reported to be involved in autolytic activity of S. aureus was not affected significantly by these mutations (Fig. 4). Since reduced autolysis is another important feature of the sVISA phenotype, elucidation of the mechanism of how the reduced autolysis is brought about by rpoC(P440L) remains one of the most important themes of study.
Supplementary Material
ACKNOWLEDGMENTS
We thank Mitutaka Yoshida (Division of Ultrastructural Research, Juntendo University) for invaluable help in sample preparation and technical support of transmission electron microscopy.
This work was supported by Grant-in-Aid for Scientific Research 24590093 to M.M. from MEXT (Ministry of Education, Culture, Sports and Technology) and in part by Grant-in-Aid S1201013 from the MEXT-Supported Program for the Strategic Research Foundation at Private Universities, 2012 to 2017.
The authors report there are no conflicts of interest for this study.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.00135-15.
REFERENCES
- 1.Hidayat LK, Hsu DI, Quist R, Shriner KA, Wong-Beringer A. 2006. High-dose vancomycin therapy for methicillin-resistant Staphylococcus aureus infections: efficacy and toxicity. Arch Intern Med 166:2138–2144. doi: 10.1001/archinte.166.19.2138. [DOI] [PubMed] [Google Scholar]
- 2.Lodise TP, Graves J, Evans A, Graffunder E, Helmecke M, Lomaestro BM, Stellrecht K. 2008. Relationship between vancomycin MIC and failure among patients with methicillin-resistant Staphylococcus aureus bacteremia treated with vancomycin. Antimicrob Agents Chemother 52:3315–3320. doi: 10.1128/AAC.00113-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Small PM, Chambers HF. 1990. Vancomycin for Staphylococcus aureus endocarditis in intravenous drug users. Antimicrob Agents Chemother 34:1227–1231. doi: 10.1128/AAC.34.6.1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kollef MH. 2007. Limitations of vancomycin in the management of resistant staphylococcal infections. Clin Infect Dis 45:S191–S195. doi: 10.1086/519470. [DOI] [PubMed] [Google Scholar]
- 5.Hsu DI, Hidayat LK, Quist R, Hindler J, Karlsson A, Yusof A, Wong-Beringer A. 2008. Comparison of method-specific vancomycin minimum inhibitory concentration values and their predictability for treatment outcome of meticillin-resistant Staphylococcus aureus (MRSA) infections. Int J Antimicrob Agents 32:378–385. doi: 10.1016/j.ijantimicag.2008.05.007. [DOI] [PubMed] [Google Scholar]
- 6.Gould IM. 2013. Treatment of bacteraemia: methicillin-resistant Staphylococcus aureus (MRSA) to vancomycin-resistant S. aureus (VRSA). Int J Antimicrob Agents 42(Suppl):S17–S21. doi: 10.1016/j.ijantimicag.2013.04.006. [DOI] [PubMed] [Google Scholar]
- 7.CLSI. 2010. Performance standards for antimicrobial susceptibility testing. Approved standard. CLSI document M100-S20. CLSI, Wayne, PA. [Google Scholar]
- 8.Saito M, Katayama Y, Hishinuma T, Iwamoto A, Aiba Y, Kuwahara-Arai K, Cui L, Matsuo M, Aritaka N, Hiramatsu K. 2014. “Slow VISA,” a novel phenotype of vancomycin resistance, found in vitro in heterogeneous vancomycin-intermediate Staphylococcus aureus strain Mu3. Antimicrob Agents Chemother 58:5024–5035. doi: 10.1128/AAC.02470-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hiramatsu K, Katayama Y, Matsuo M, Sasaki T, Morimoto Y, Sekiguchi A, Baba T. 2014. Multi-drug-resistant Staphylococcus aureus and future chemotherapy. J Infect Chemother 20:593–601. doi: 10.1016/j.jiac.2014.08.001. [DOI] [PubMed] [Google Scholar]
- 10.Matsuo M, Hishinuma T, Katayama Y, Cui L, Kapi M, Hiramatsu K. 2011. Mutation of RNA polymerase beta subunit (rpoB) promotes hVISA-to-VISA phenotypic conversion of strain Mu3. Antimicrob Agents Chemother 55:4188–4195. doi: 10.1128/AAC.00398-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Watanabe Y, Cui L, Katayama Y, Kozue K, Hiramatsu K. 2011. Impact of rpoB mutations on reduced vancomycin susceptibility in Staphylococcus aureus. J Clin Microbiol 49:2680–2684. doi: 10.1128/JCM.02144-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gao W, Cameron DR, Davies JK, Kostoulias X, Stepnell J, Tuck KL, Yeaman MR, Peleg AY, Stinear TP, Howden BP. 2013. The RpoB H444Y rifampicin resistance mutation and an active stringent response reduce virulence and increase resistance to innate immune responses in Staphylococcus aureus. J Infect Dis 207:929–939. doi: 10.1093/infdis/jis772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matsuo M, Cui L, Kim J, Hiramatsu K. 2013. Comprehensive identification of mutations responsible for heterogeneous vancomycin-intermediate Staphylococcus aureus (hVISA)-to-VISA conversion in laboratory-generated VISA strains from hVISA clinical strain Mu3. Antimicrob Agents Chemother 57:5843–5853. doi: 10.1128/AAC.00425-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matsuo M, Kurokawa K, Lee BL, Sekimizu K. 2010. Shuttle vectors derived from pN315 for study of essential genes in Staphylococcus aureus. Biol Pharm Bull 33:198–203. doi: 10.1248/bpb.33.198. [DOI] [PubMed] [Google Scholar]
- 15.Hiramatsu K, Hanaki H. 1998. Glycopeptide resistance in staphylococci. Curr Opin Infect Dis 11:653–658. doi: 10.1097/00001432-199812000-00002. [DOI] [PubMed] [Google Scholar]
- 16.Cui L, Lian JQ, Neoh HM, Reyes E, Hiramatsu K. 2005. DNA microarray-based identification of genes associated with glycopeptide resistance in Staphylococcus aureus. Antimicrob Agents Chemother 49:3404–3413. doi: 10.1128/AAC.49.8.3404-3413.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. 2003. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 18.Artsimovitch I, Patlan V, Sekine S, Vassylyeva MN, Hosaka T, Ochi K, Yokoyama S, Vassylyev DG. 2004. Structural basis for transcription regulation by alarmone ppGpp. Cell 117:299–310. doi: 10.1016/S0092-8674(04)00401-5. [DOI] [PubMed] [Google Scholar]
- 19.Zuo Y, Wang Y, Steitz TA. 2013. The mechanism of E. coli RNA polymerase regulation by ppGpp is suggested by the structure of their complex. Mol Cell 50:430–436. doi: 10.1016/j.molcel.2013.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Geiger T, Kästle B, Gratani FL, Goerke C, Wolz C. 2014. Two small (p)ppGpp synthases in Staphylococcus aureus mediate tolerance against cell envelope stress conditions. J Bacteriol 196:894–902. doi: 10.1128/JB.01201-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cui L, Tominaga E, Neoh HM, Hiramatsu K. 2006. Correlation between reduced daptomycin susceptibility and vancomycin resistance in vancomycin-intermediate Staphylococcus aureus. Antimicrob Agents Chemother 50:1079–1082. doi: 10.1128/AAC.50.3.1079-1082.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Anderson M, Aly KA, Chen YH, Missiakas D. 2013. Secretion of atypical protein substrates by the ESAT-6 secretion system of Staphylococcus aureus. Mol Microbiol 90:734–743. doi: 10.1111/mmi.12395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Howden BP, McEvoy CR, Allen DL, Chua K, Gao W, Harrison PF, Bell J, Coombs G, Bennett-Wood V, Porter JL, Robins-Browne R, Davies JK, Seemann T, Stinear TP. 2011. Evolution of multidrug resistance during Staphylococcus aureus infection involves mutation of the essential two component regulator WalKR. PLoS Pathog 7:e1002359. doi: 10.1371/journal.ppat.1002359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shoji M, Cui L, Iizuka R, Komoto A, Neoh HM, Watanabe Y, Hishinuma T, Hiramatsu K. 2011. walK and clpP mutations confer reduced vancomycin susceptibility in Staphylococcus aureus. Antimicrob Agents Chemother 55:3870–3881. doi: 10.1128/AAC.01563-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Korea CG, Balsamo G, Pezzicoli A, Merakou C, Tavarini S, Bagnoli F, Serruto D, Unnikrishnan M. 2014. Staphylococcal Esx proteins modulate apoptosis and release of intracellular Staphylococcus aureus during infection in epithelial cells. Infect Immun 82:4144–4153. doi: 10.1128/IAI.01576-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kneuper H, Cao ZP, Twomey KB, Zoltner M, Jäger F, Cargill JS, Chalmers J, van der Kooi-Pol MM, van Dijl JM, Ryan RP, Hunter WN, Palmer T. 2014. Heterogeneity in ess transcriptional organization and variable contribution of the Ess/type VII protein secretion system to virulence across closely related Staphylocccus aureus strains. Mol Microbiol 93:928–943. doi: 10.1111/mmi.12707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pieper R, Gatlin-Bunai CL, Mongodin EF, Parmar PP, Huang ST, Clark DJ, Fleischmann RD, Gill SR, Peterson SN. 2006. Comparative proteomic analysis of Staphylococcus aureus strains with differences in resistance to the cell wall-targeting antibiotic vancomycin. Proteomics 6:4246–4258. doi: 10.1002/pmic.200500764. [DOI] [PubMed] [Google Scholar]
- 28.Darst SA. 2001. Bacterial RNA polymerase. Curr Opin Struct Biol 11:155–162. doi: 10.1016/S0959-440X(00)00185-8. [DOI] [PubMed] [Google Scholar]
- 29.Ross W, Vrentas CE, Sanchez-Vazquez P, Gaal T, Gourse RL. 2013. The magic spot: a ppGpp binding site on E. coli RNA polymerase responsible for regulation of transcription initiation. Mol Cell 50:420–429. doi: 10.1016/j.molcel.2013.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dalebroux ZD, Swanson MS. 2012. ppGpp: magic beyond RNA polymerase. Nat Rev Microbiol 10:203–212. doi: 10.1038/nrmicro2720. [DOI] [PubMed] [Google Scholar]
- 31.Geiger T, Goerke C, Fritz M, Schäfer T, Ohlsen K, Liebeke M, Lalk M, Wolz C. 2010. Role of the (p)ppGpp synthase RSH, a RelA/SpoT homolog, in stringent response and virulence of Staphylococcus aureus. Infect Immun 78:1873–1883. doi: 10.1128/IAI.01439-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cui L, Ma X, Sato K, Okuma K, Tenover FC, Mamizuka EM, Gemmell CG, Kim MN, Ploy MC, El-Solh N, Ferraz V, Hiramatsu K. 2003. Cell wall thickening is a common feature of vancomycin resistance in Staphylococcus aureus. J Clin Microbiol 41:5–14. doi: 10.1128/JCM.41.1.5-14.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gardete S, Aires-De-Sousa M, Faustino A, Ludovice AM, de Lencastre H. 2008. Identification of the first vancomycin intermediate-resistant Staphylococcus aureus (VISA) isolate from a hospital in Portugal. Microb Drug Resist 14:1–6. doi: 10.1089/mdr.2008.0816. [DOI] [PubMed] [Google Scholar]
- 34.Sieradzki K, Leski T, Dick J, Borio L, Tomasz A. 2003. Evolution of a vancomycin-intermediate Staphylococcus aureus strain in vivo: multiple changes in the antibiotic resistance phenotypes of a single lineage of methicillin-resistant S. aureus under the impact of antibiotics administered for chemotherapy. J Clin Microbiol 41:1687–1693. doi: 10.1128/JCM.41.4.1687-1693.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pereira PM, Filipe SR, Tomasz A, Pinho MG. 2007. Fluorescence ratio imaging microscopy shows decreased access of vancomycin to cell wall synthetic sites in vancomycin-resistant Staphylococcus aureus. Antimicrob Agents Chemother 51:3627–3633. doi: 10.1128/AAC.00431-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hiramatsu K, Aritaka N, Hanaki H, Kawasaki S, Hosoda Y, Hori S, Fukuchi Y, Kobayashi I. 1997. Dissemination in Japanese hospitals of strains of Staphylococcus aureus heterogeneously resistant to vancomycin. Lancet 350:1670–1673. doi: 10.1016/S0140-6736(97)07324-8. [DOI] [PubMed] [Google Scholar]
- 37.Katayama Y, Hishinuma T, Iwamoto A, Saito M, Aiba Y, Sasaki T, Hiramatsu K. 2014. Characterization of slow VISA: a novel phenotype of vancomycin resistance in Staphylococcus aureus, poster C-1383 Abstr 54th Intersci Conf Antimicrob Agents Chemotherapy. American Society for Microbiology, Washington, DC. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.