Abstract
Vibrio parahaemolyticus is an important causative agent of gastroenteritis, with the consumption of contaminated seafood being the major transmission route. Resistance to penicillin is common among V. parahaemolyticus strains, whereas cephalosporin resistance remains rare. In an attempt to assess the current prevalence and characteristics of antibiotic resistance of this pathogen in common food samples, a total of 54 (17% of the total samples) V. parahaemolyticus strains were isolated from 318 meat and seafood samples purchased from supermarkets and wet markets in Shenzhen, China, in 2013. These isolates exhibited high-level resistance to ampicillin, yet they were mostly susceptible to other antimicrobials, except for two that were resistant to extended-spectrum cephalosporins. The β-lactamase gene blaPER-1 was detectable in one strain, V. parahaemolyticus V43, which was resistant to both third- and fourth-generation cephalosporins. Compared to other blaPER-1-positive V. parahaemolyticus strains reported in our previous studies, strain V43 was found to harbor an ∼200-kb conjugative plasmid carrying genes that were different from the antimicrobial resistance genes reported from the previous studies. The β-lactamase gene blaCMY-2 was detectable for the first time in another V. parahaemolyticus isolate, V4, which was resistant to third-generation cephalosporins. This blaCMY-2 gene was shown to be located in an ∼150-kb IncA/C-type conjugative plasmid with a genetic structure consisting of traB-traV-traA-ISEcp1-blaCMY-2-blc-sugE-encR-orf1-orf2-orf3-orf4-dsbC-traC, which is identical to that of other IncA/C conjugative plasmids in Enterobacteriaceae, albeit with a different size. These findings indicate that the transmission of extended-spectrum-β-lactamase (ESBL) and AmpC β-lactamase genes via conjugative plasmids can mediate the development of extended-spectrum cephalosporin resistance in V. parahaemolyticus, thereby posing a potential threat to public health.
INTRODUCTION
Vibrio parahaemolyticus is a halophilic Gram-negative bacterium and one of the most important seafood-borne pathogens worldwide. It is widely distributed in estuarine-marine environments and seafood products, often causing infections via the consumption of raw or undercooked seafood (1, 2). The most common symptoms of V. parahaemolyticus infections include gastroenteritis, diarrhea, headache, nausea, and vomiting. For children, the elderly, and people with suppressed immune systems, V. parahaemolyticus intestinal infections can be fatal (3). The pathogenicity of V. parahaemolyticus is always associated with the presence of the toxin genes tdh (encoding the thermostable direct hemolysin [TDH]) and its homolog trh (encoding the thermostable direct hemolysin [TDH]-related hemolysin) (4). The U.S. Food and Drug Administration estimates that approximately 4,500 infections occur per year in the United States, and over the past 15 years, outbreaks have been increasing in terms of scale and frequency (3). In recent years, the number of V. parahaemolyticus infections in China has increased significantly (1).
For life-threatening infections caused by V. parahaemolyticus, antibiotic therapy (mainly including tetracycline, quinolones, and third-generation cephalosporins) is necessary (5). The increasing prevalence of antimicrobial resistance in V. parahaemolyticus, presumably due to the extensive use of antimicrobials in clinical treatment and aquaculture systems, was recently reported (6–9). To date, resistance to ampicillin, streptomycin, kanamycin, tetracycline, chloramphenicol, and ciprofloxacin has been reported (9). Recent studies also reported resistance to third-generation cephalosporins in V. parahaemolyticus strains due to carriage of an extended-spectrum-β-lactamase (ESBL) gene, blaPER-1 (10). However, other ESBL and AmpC-type genes are not detectable in V. parahaemolyticus.
With the rapid development of the aquaculture industry, infections associated with V. parahaemolyticus in seafood have become increasingly common in China and other Asian countries. In China, research on the epidemiology and antimicrobial resistance of V. parahaemolyticus from food products, and meat products in particular, is very rare. This study reports the surveillance of the prevalence and antimicrobial resistance of V. parahaemolyticus collected from different food products, including shrimps, pork, beef, and chicken, in Shenzhen, China. The data will provide valuable information for the future control of V. parahaemolyticus infections in China and neighboring countries.
MATERIALS AND METHODS
Isolation and identification of V. parahaemolyticus.
A total of 318 food samples, including those from shrimps, chicken, pork, and beef, were collected from supermarkets and wet markets in Shenzhen, China, from June to September 2013. The isolation of V. parahaemolyticus was conducted as previously described, with some modifications (11). Briefly, food samples were purchased and transferred to a laboratory within 3 h under low-temperature conditions (stored in insulated bags containing ice cubes). Five grams of food samples was homogenized in 45 ml of alkaline peptone water (APW) and incubated overnight at 37°C. A loopful of culture was then streaked on thiosulfate citrate-bile salts-sucrose (TCBS) agar. After 16 h of incubation at 37°C, suspicious colonies (blue or green) were picked and streaked on CHROMagar plates for identification. The mauve colonies on CHROMagar plates, presumptively identified as V. parahaemolyticus, were transferred into brain heart infusion (BHI) broth with 3% NaCl and incubated overnight at 37°C. The bacterial culture was stored in 15% glycerol at −80°C for further characterization.
Isolates with typical phenotypes on the TCBS and CHROMagar plates were confirmed by PCR detection of the thermolabile hemolysin (tlh) gene and DNA sequencing. Detection of virulence genes (tdh and trh genes) was also performed using the PCR approach, as previously described (12).
Antimicrobial susceptibility testing.
Antimicrobial susceptibility testing was performed on V. parahaemolyticus isolates using the standard agar dilution method, as described by the Clinical and Laboratory Standards Institute (13). Fourteen antimicrobials were tested, including nalidixic acid, ciprofloxacin, ofloxacin, amikacin, gentamicin, tetracycline, ampicillin, amoxicillin-clavulanic acid, cefoxitin, ceftazidime, meropenem, imipenem, trimethoprim-sulfamethoxazole, and chloramphenicol. The resistance breakpoints published by the Clinical and Laboratory Standards Institute were used (14). Escherichia coli strain ATCC 25922 was included as the quality control strain.
PFGE subtyping.
Pulsed-field gel electrophoresis (PFGE) was conducted to investigate the genetic relatedness of the identified isolates according to the PulseNet protocol (http://www.pulsenetinternational.org/protocols/), with minor modifications. In brief, the cell suspensions (at an optical density at 610 nm [OD610] of 0.9) with proteinase K (1 mg/ml) were immobilized into 2% low-melting-point agarose (Amerson, United Kingdom) at a ratio of 1:1 before incubation in a lysis buffer with proteinase K (0.1 mg/ml). After five consecutive steps of washing with Tris-EDTA buffer (pH 8.0) at 50°C in a water bath shaker, the total DNA was digested with 40 U of NotI or SfiI (New England BioLabs) for 4 h at 37°C or 50°C, respectively. Chromosomal DNA of Salmonella enterica serotype Braenderup (strain H9812) digested with XbaI was used as a reference marker. The DNA bands were separated using a Chef Mapper XA pulsed-field gel electrophoresis system (Bio-Rad Laboratories, Hercules, CA, USA) for 19 h, with an initial switch time of 10 s and final switch time of 35 s at 6 V/cm, at 14°C in 0.5× Tris-boric acid-EDTA buffer. The gels were stained with GoldView dye and photographed using a Bio-Rad Gel Doc 1000 system. A PFGE dendrogram was generated using the BioNumerics 7.1 software.
β-Lactamase gene screening and analysis of genetic environment.
The prevalence of β-lactamase genes among the test strains was determined by PCR assays that covered most of the known β-lactamase genes, as previously described (15). Screening of class I integrons was also performed as described previously (5). The entire sequence of the blaCMY-2 gene was amplified using primers CMY2_F and CMY2_R (Table 1). The amplified product was cloned into a TOPO pCR2.1 vector (Invitrogen) and sequenced with universal primers. Primer walking and PCR mapping were performed to investigate the blaCMY-2 gene environment. A set of primers (Table 1) was designed based on the published flanking sequence of the blaCMY-2 gene (16). The PCR products were sequenced and subjected to sequence alignment analysis using the BioEdit software.
TABLE 1.
PCR primers used to assess the genetic context of β-lactamase genes
| Primer name | Sequence | Length (bp) | Target | Reference or source |
|---|---|---|---|---|
| CMY2_F | GCTGAGAGCTCATGATGAAAAAATCG | 1,146 | blaCMY-2 | This study |
| CMY2_R | GGTACGGATCCTTATTGCAGC | |||
| Primers for blaCMY-2 genetic environment determination | ||||
| TraA-F | ATCAGTTGGCGAATGCCTCA | 1,417 | traA-ISEcp1 | This study |
| ISEcp1-R | AACACGGCTTCATTCGCCCAA | This study | ||
| ISEcp1-F | ATAAAGACCATGCTCTGCGG | 2,537 | ISEcp1- blaCMY-2 | This study |
| CMY-2R | ACGGACAGGGTTAGGATA | 15 | ||
| ISEcp1-F | ATAAAGACCATGCTCTGCGG | 2,800 | ISEcp1- blaCMY-2-blc | This study |
| blc-R | TACCAGGTTCCCAGATAGCG | This study | ||
| ISEcp1-F(B) | ATAAAGACCATGCTCTGCGG | 3,465 | ISEcp1- blaCMY-2-blc-sugE | This study |
| sugE-R(B) | TTGGCCTGAAATACACCCAC | This study | ||
| A-F | CGTAGAGGATCTCAGTTCAG | 3,700 | traB-traV-traA- ISEcp1- blaCMY-2 | 22 |
| A-R | CCTGCCACTGTTTGCCTGTC | |||
| G-F | CGCCTGCTAAAAGCAAGAAT | 2,700 | sugE-ecnR-orf1 | 22 |
| G-R | TGCCATCTGCATGAACAAAT | |||
| H-F | AAGGCTGTTGGCTTCGAGTA | 2,500 | orf1-orf2-orf3 | 22 |
| H-R | TCAGCAGTATCACCCTGCAC | |||
| I-F | ATGTGTTCAAAGCCGAAACC | 2,500 | orf3-orf4-dsbC-traC | 22 |
| I-R | GGTGCCGTAATCGAAGATTT | |||
| Primers for blaPER-1-positive plasmid characterization | ||||
| PER1-F | GCTCCGATAATGAAAGCGT | 520 | blaPER-1 | 15 |
| PER1-R | TTCGGCTTGACTCGGCTGA | |||
| TEM1-F | CATTTCCGTGTCGCCCTTATTC | 800 | blaTEM-1 | 15 |
| TEM1-R | CGTTCATCCATAGTTGCCTGAC | |||
| ISCR1-F | AGACGCCGTGGAAGCGTGTG | 509 | ISCR1 | This study |
| ISCR1-R | GCTCGCTCACACCCTCAGCC | |||
| QnrVC6-F | ATGGAAAAATCAAAGCAATT | 657 | qnrVC6 | 5 |
| QnrVC6-R | TTAGTCAGGAACAATGAT | |||
| Int1-F | GGCATCCAAGCAGCAAGC | Various | Gene cassette (class I integron) | 25 |
| Int1-R | AAGCAGACTTGACCTGAT |
Conjugation and plasmid characterization.
A filter mating experiment was carried out to study the transferability of resistance phenotypes. Overnight cultures of donors (V. parahaemolyticus) and recipients (sodium-azide-resistant E. coli strain J53) were mixed together at a donor-to-recipient ratio of 2/1, plated on a filter membrane, and selected on MacConkey agar with ceftazidime (4 μg/ml) and sodium azide (100 μg/ml) to select transconjugants.
The major incompatibility (Inc) groups of plasmids from transconjugants and parental strains were identified by a PCR-based replicon typing (PBRT) method (16). S1 nuclease PFGE (S1-PFGE) was used to analyze the plasmids that played a role in conferring resistance to β-lactams, according to the standard PFGE protocol described above, except for the use of S1 nuclease instead in the digestion step. After washing with Tris-EDTA buffer (pH 8.0), total DNA was digested with 50 U of S1 nuclease (Thermal Scientific) for 30 min at 37°C. The linear plasmids were separated from the chromosomal DNA by electrophoresis for 20 h, with an initial switch time of 2.16 s and final switch time of 63.8 s at 6 V/cm at 14°C in 0.5× Tris-boric acid-EDTA buffer, using a Chef Mapper XA pulsed-field electrophoresis system (Bio-Rad Laboratories, Hercules, CA, USA). Southern blot hybridization was carried out in accordance with the manufacturer's instructions for digoxigenin (DIG)-High Prime DNA labeling and detection starter kit II (Roche Diagnostics), using the blaCMY-2 and blaPER-1 digoxigenin-labeled probes.
For blaPER-1-positive strains, a comparison was conducted between a strain from this study and another two blaPER-1-positive V. parahaemolyticus stains identified in our previous studies (5, 10). The primers used to amplify different β-lactamase genes, the qnrVC gene, and integrons are listed in Table 1.
RESULTS AND DISCUSSION
Prevalence of V. parahaemolyticus in different food samples in Shenzhen.
A total of 54 (17% of the total samples) V. parahaemolyticus isolates from different food samples were recovered from 318 food samples in wet markets and supermarkets. Of these isolates, 20/76 (26%), 19/84 (23%), 9/85 (11%), and 6/73 (8%) were isolated from shrimps, chicken, pork, and beef, respectively. V. parahaemolyticus was considered the most important pathogen causing seafood-borne infections (17). Reports about V. parahaemolyticus from other kinds of foods are scarce. In this study, V. parahaemolyticus was detected in all types of meat products, such as chicken, pork, and beef. The prevalence of V. parahaemolyticus in chicken was similar to that in shrimps, whereas its prevalence in pork and beef was slightly lower than that in shrimps. It appears that the chicken, pork, and beef food samples might be cross-contaminated during the process of handling and storage. More in-depth investigations of the origin of V. parahaemolyticus in foods other than seafood should be performed.
In order to determine whether these isolates were virulent, the prevalence of the tdh and trh genes was screened by PCR. Although the tdh and trh genes are known to occur in 99% of clinical isolates (1), none of the 54 food isolates contained these two virulence genes. This finding was consistent with those of previous studies indicating a low prevalence of tdh and trh genes in V. parahaemolyticus from food samples (7, 9). The relationship between clinical and environmental strains should be investigated further in the future.
Antimicrobial susceptibility profile and PFGE typing.
Antimicrobial susceptibility testing of the 54 V. parahaemolyticus isolates showed that relatively high rates of resistance to ampicillin (100%) and tetracycline (11%) were observed. In addition, five isolates (9%) were found to be resistant to sulfamethoxazole-trimethoprim, and one was resistant to chloramphenicol. Importantly, two isolates exhibited resistance to cefoxitin and ceftazidime. Consistent with a previous study (9), multiple antimicrobial resistance V. parahaemolyticus was not common. The detection of V. parahaemolyticus isolates that are resistant to third-generation cephalosporins in mainland China has not been reported. Our susceptibility testing results indicated that front-line drugs, including cefoxitin, ceftazidime, and fluoroquinolones, remained highly effective against V. parahaemolyticus infections. However, our finding of emerging V. parahaemolyticus resistant to third-generation cephalosporins has raised a concern for public health, despite being nonpathogenic, as the transfer of resistance determinants from food isolates to other pathogenic Vibrio strains would have serious consequences.
Among the 54 isolates, a total of 49 PFGE patterns were identified, and 5 strains were untypeable. The PFGE dendrogram (see Fig. S1 in the supplemental material) showed that most isolates did not exhibit significant genetic relatedness, and no extensive clonal dissemination features were observed. However, there were several clusters in which >90% genetic similarity was observed between isolates from different sources. This finding provides evidence of cross-contamination between different types of food in supermarkets.
β-Lactamase gene screening.
β-Lactamase gene screening was performed on two V. parahaemolyticus isolates that were resistant to third-generation cephalosporins. For strain V43, which was resistant to all tested β-lactams, including cefepime, but not imipenem or meropenem, blaTEM-1 and blaPER-1 were detectable. Since the β-lactamase gene blaPER-1 was previously detectable in V. parahaemolyticus isolated from shrimps in Hong Kong in our laboratory (5, 10), this blaPER-1-positive strain was selected for further characterization in this study. On the other hand, the β-lactamase gene blaCMY-2 was detected for the first time in strain V4, which was resistant to ampicillin, amoxicillin-clavulanic acid, cefotaxime, and ceftriaxone, and had reduced susceptibility to ceftazidime but not cefepime or carbapenems (Table 2).
TABLE 2.
Antimicrobial susceptibility profiles of extended-spectrum-cephalosporin-resistant V. parahaemolyticus strains (V4, V43, 2010V36, and 2011V1) and the corresponding transconjugants
| Strain | β-Lactamase gene | MIC (mg/liter) ofa: |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AMP (32) | CTX (4) | CRO (4) | CAZ (16) | CEP (16) | TET (16) | GEN (16) | NAL (32) | CIP (4) | CHL (32) | KAN (64) | TRI (16) | ||
| J53b | 1 | ≤0.125 | ≤0.125 | ≤0.125 | ≤0.125 | 2 | 1 | 2 | ≤0.125 | 2 | 0.25 | ≤0.125 | |
| V4 | blaCMY-2 | 128 | 8 | 32 | 8 | 0.5 | 32 | 1 | 0.5 | ≤0.125 | 2 | 8 | 16 |
| V4-J53 | blaCMY-2 | >128 | 16 | 64 | 64 | 0.5 | 64 | 1 | 2 | ≤0.125 | 64 | 1 | >64 |
| V43 | blaPER-1 | >128 | >128 | >128 | >128 | 32 | 32 | 8 | 1 | ≤0.125 | 2 | 8 | 32 |
| V43-J53 | blaPER-1 | 128 | 128 | 128 | >128 | 16 | 64 | 8 | 2 | ≤0.125 | 32 | 0.5 | >64 |
| 2010V36 | blaPER-1 | >128 | >128 | >128 | >128 | 32 | 1 | 1 | 0.25 | ≤0.125 | 0.25 | 2 | 16 |
| 2010V36-J53 | blaPER-1 | >128 | >128 | >128 | >128 | 16 | 2 | 1 | 2 | ≤0.125 | 2 | 2 | >64 |
| 2011V1 | blaPER-1 | >128 | >128 | >128 | >128 | 16 | 1 | 4 | 0.5 | ≤0.125 | 16 | 64 | >64 |
| 2011V1-J53 | blaPER-1 | >128 | >128 | >128 | >128 | 32 | 2 | 4 | 4 | ≤0.125 | 16 | 64 | >64 |
Numbers in parentheses are the breakpoint for each agent. AMP, ampicillin; CTX, cefotaxime; CRO, ceftriaxone; CAZ, ceftazidime; CEP, cefepime; TET, tetracycline; GEN, gentamicin; NAL, nalidixic acid; CIP, ciprofloxacin; CHL, chloramphenicol; KAN, kanamycin; TRI, trimethoprim.
J53, E. coli recipient.
Characterization of blaPER-1-bearing plasmids in V. parahaemolyticus.
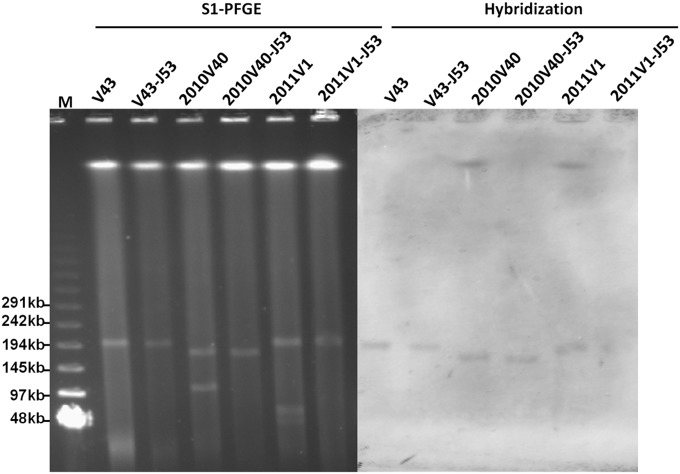
Two genetically unrelated blaPER-1-positive isolates, V. parahaemolyticus 2010V36 and 2011V1 (see Fig. S2 in the supplemental material), which were isolated from our previous studies, were selected for further characterization, along with strain V43 recovered in this study (5, 10). Conjugation experiments showed that plasmids carrying blaPER-1 in these three V. parahaemolyticus strains were transferrable to E. coli J53. The transconjugants V43-J53, 2010V36-J53, and 2011V1-J53 were found to have acquired phenotypic resistance to most of the β-lactam antibiotics tested, including resistance/intermediate resistance to fourth-generation cephalosporins, such as cefepime, but not carbapenems (Table 2). Understanding the molecular mechanisms by which V. parahaemolyticus exhibits resistance or intermediate resistance to fourth-generation cephalosporins, especially those involving acquisition of the ESBL gene blaPER-1, will allow us to define the breakpoint of cefepime for V. parahaemolyticus, since this is the only report of resistance/intermediate resistance to cefepime in V. parahaemolyticus. On the basis of the breakpoints of E. coli, as well as a comparison between the drug susceptibility levels of transconjugants and the test V. parahaemolyticus strains, our data indicate that a breakpoint of 16 mg/liter or 8 mg/liter is more appropriate. Apart from the blaPER-1-associated phenotypes, other phenotypes, such as those showing resistance to tetracycline and trimethoprim in V43, resistance to trimethoprim in 2010V36, and resistance to chloramphenicol, kanamycin, and trimethoprim in 2011V1, could also be transferred to E. coli J53 through conjugation. These resistance phenotypes were consistent with those conferred by the integrons carrying different resistance genes that were detectable in the conjugative plasmids (Table 3). S1 nuclease PFGE showed that V43 contained one plasmid with a size of ∼200 kb that could be transferred to E. coli J53. Southern hybridization confirmed the presence of blaPER-1 on this plasmid (Fig. 1). Strain 2010V36 was shown to harbor two plasmids, with sizes of ∼175 kb and ∼110 kb, with the ∼175-kb plasmid being transferrable to E. coli J53. The plasmid size in 2010V36 here was different from that in the previous study, presumably because the use of the S1-PFGE method offered more accurate plasmid size determination in this study. Southern hybridization also confirmed the presence of a blaPER-1 gene on the ∼175-kb plasmid. Strain 2011V1 was found to contain three plasmids with sizes of ∼200 kb, ∼70kb, and ∼45 kb. The ∼200-kb plasmid was shown to be self-transmissible. Southern hybridization also confirmed the presence of blaPER-1 in the ∼200-kb plasmid (Fig. 1).
TABLE 3.
Comparison of genetic contents of blaPER-1-bearing plasmids recovered from three V. parahaemolyticus strains and the corresponding transconjugants
| Genetic component | Strain |
Transconjugant |
||||
|---|---|---|---|---|---|---|
| V43 | V43-J53 | 2010V36 | 2010V36-J53 | 2011V1 | 2011V1-J53 | |
| blaPER-1 | + | + | + | + | + | + |
| blaTEM-1 | + | + | − | − | − | − |
| ISCR1 | + | + | + | + | + | + |
| qnrVC6 | − | − | − | − | + | + |
| Class I integrons | arr-3, dfrA27 | arr-3, dfrA27 | arr-3, dfrA27 | arr-3, dfrA27 | aacA3, catB2, dfrA1, aadA1 | aacA3, catB2, dfrA1, aadA1 |
| No. of plasmids (size [kb]) | 1 (200) | 1 (200) | 2 (175, 110) | 1 (175) | 3 (200, 70, 45) | 1 (200) |
FIG 1.
S1-PFGE patterns and Southern blot hybridization of blaPER-1-positive strains and the corresponding transconjugants. M, Lambda ladder PFGE marker (New England BioLabs).
In our previous study, the partial genetic environment of blaPER-1 was determined for strain 2011V1 (5). To check whether blaPER-1 in V43 and 2010V36 also contained genetic structures similar to that of strain 2011V1, further characterization of plasmids recovered from V43, 2010V36, and 2011V1 was performed using primers to amplify the flanking region of blaPER-1, with blaPER-1 of 2011V1 as a template for primer design (Table 1). The results, as summarized in Table 3, showed that the genetic structures of these plasmids were different, despite the fact that the plasmids from strains V43 and 2011V1 were both about 200 kb in size. In addition to blaPER-1, a plasmid from V43 also contained a blaTEM-1 gene, ISCR1, and an integron carrying the arr-3 and dfrA27 genes; although the conjugative blaPER-1-containing plasmid from strain 2010V36 contained ISCR1 and an integron carrying the arr-3 and dfrA27 genes, it was slightly smaller in size (∼175 kb) than the other two plasmids. The conjugative plasmid from strain 2011V1 contained ISCR1 and a new integron carrying the aacA3, catB2, dfrA1, and aadA1 genes. In addition, it carried a novel PMQR gene, qnrVC6 (5). These data indicate that all three blaPER-1-bearing plasmids recoverable from three different V. parahaemolyticus strains were genetically different. A further sequencing approach will be applied to understand the evolutionary features of these three plasmids in V. parahaemolyticus.
Characterization of plasmids carrying the blaCMY-2 gene in V. parahaemolyticus.
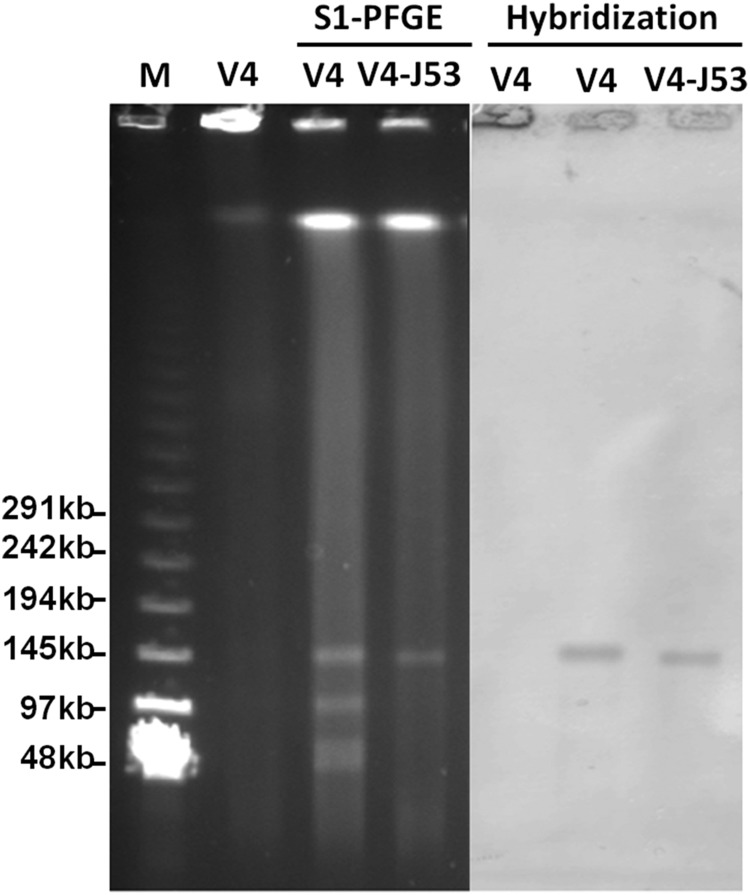
Conjugation experiments were also performed on the V4 strain, with the results showing that the plasmid containing the blaCMY-2 gene in this strain could be transferred to E. coli J53. The transconjugant V4J53 exhibited slightly higher MICs of various β-lactam antimicrobials than those of the original V4 strain (Table 2). Interestingly, V4 did not exhibit resistance to chloramphenicol, whereas V4J53 became resistant to chloramphenicol, presumably due to the fact that some resistance genes might be better expressed in E. coli (Table 2). This phenomenon was also observable in our previous study (10). S1 nuclease PFGE and Southern hybridization analysis showed that the V4 strain harbored 4 plasmids, with sizes ranging from ∼45 kb to ∼150 kb. The conjugative plasmid detected in transconjugant V4J53 was ∼150 kb. Southern hybridization confirmed that the blaCMY-2 gene was located in this transmissible plasmid in both V4 and V4J53 (Fig. 2). Plasmid typing indicated that the transmissible plasmid belonged to the IncA/C type plasmid. Plasmids harboring blaCMY-2 have been found to belong to different plasmid types (IncA/C, IncI1, etc.) detectable in members of Enterobacteriaceae recovered from various sources. The sizes of the plasmids varied from ∼45 kb to ∼200 kb (18–21). IncA/C plasmids carrying blaCMY-2 with a similar size (∼150 kb) were mainly reported in E. coli and S. enterica (20). It is likely that the blaCMY-2-bearing IncA/C plasmid in this strain originated from Enterobacteriaceae; alternatively, the formation of this plasmid may be due to mobilization of the blaCMY-2 gene to different plasmid backbones through ISEcp1-mediated transposition activities (22).
FIG 2.
S1-PFGE patterns and Southern blot hybridization of V4 and transconjugant V4J53. M, lambda ladder PFGE marker (New England BioLabs); V4, V4 genomic DNA without S1 nuclease digestion.
The flanking sequence of blaCMY-2 was amplified by PCR mapping, according to published sequences, and confirmed by sequencing. It was found that the arrangement of the genetic environment of blaCMY-2 was in the order traB-traV-traA-ISEcp1-blaCMY-2-blc-sugE-encR-orf1-orf2-orf3-orf4-dsbC-traC (Fig. 3). The β-lactamase gene blaCMY-2 is the most commonly reported plasmid-carried AmpC β-lactamase gene in Salmonella spp., E. coli, and other species of Enterobacteriaceae worldwide (23). This gene renders the host organism resistant to a variety of β-lactams, including oxyimino-cephalosporins and cephamycins (23). The genetic content in the vicinity of the blaCMY-2 gene usually comprised the ISEcp1 and blc-sugE elements in its upstream and downstream regions, respectively (22). Therefore, the V. parahaemolyticus V4 strain is genetically similar to those strains described in several other reports (22, 24). A comparison with other published sequences in the NCBI database indicated that the genetic segment in which the blaCMY-2 gene is located has been identified in plasmids of Salmonella spp., E. coli, Klebsiella pneumoniae, Providencia stuartii, and Shigella sonnei (22–24). However, the sizes of these previously reported plasmids were different from that of the V4 strain.
FIG 3.
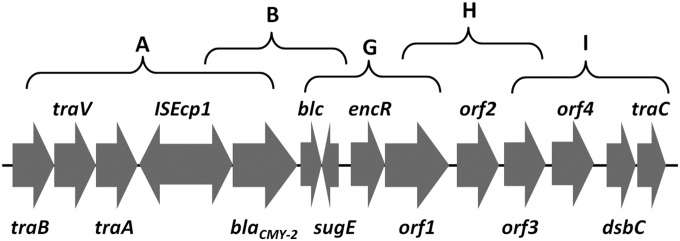
Genetic structure of blaCMY-2-bearing IncA/C conjugative plasmid in V. parahaemolyticus strain V4. A, B, G, H, and I represent the primer targeting regions. The primer sequences are shown in Table 1.
Conclusion.
In this study, we reported the isolation of V. parahaemolyticus from meat products other than seafood. These isolates were susceptible to most of the antimicrobials tested, except that two of them were resistant to extended-spectrum cephalosporins. The β-lactamase gene blaPER-1 was detectable in one V. parahaemolyticus isolate, V43, which was resistant to both third- and fourth-generation cephalosporins. The conjugative blaPER-1-borne plasmid recovered from this strain was structurally different from those reported previously. On the other hand, the β-lactamase gene blaCMY-2 was detected for the first time in another V. parahaemolyticus isolate, V4, which was resistant to the third-generation cephalosporins. The blaCMY-2 gene was shown to be located on an ∼150-kb IncA/C type conjugative plasmid with the genetic structure traB-traV-traA-ISEcp1-blaCMY-2-blc-sugE-encR-orf1-orf2-orf3-orf4-dsbC-traC, which has been reported in other IncA/C conjugative plasmids in Enterobacteriaceae, albeit with a slightly different size. This finding indicated that the transmission of ESBL and AmpC β-lactamase genes via different conjugative plasmids mediated the development of resistance to extended-spectrum cephalosporins and other drugs in V. parahaemolyticus. Such a plasmid is transferrable to Enterobacteriaceae and possibly other pathogenic Vibrio spp., thereby posing a significant threat to public health.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge Edward Chan for critical reading of the manuscript and members of the Sheng lab for fruitful discussion.
This work was supported by the Chinese National Key Basic Research and Development (973) Program (grant 2013CB127200) and the Research Fund for the Control of Infectious Diseases from the Food and Health Bureau, the Government of Hong Kong SAR (grant 13121422 to S.C.).
We declare no conflicts of interest.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.05008-14.
REFERENCES
- 1.Chen W, Xie Y, Xu J, Wang Q, Gu M, Yang J, Zhou M, Wang D, Shi C, Shi X. 2012. Molecular typing of Vibrio parahaemolyticus isolates from the middle-east coastline of China. Int J Food Microbiol 153:402–412. doi: 10.1016/j.ijfoodmicro.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 2.Sani NA, Ariyawansa S, Babji AS, Hashim JK. 2013. The risk assessment of Vibrio parahaemolyticus in cooked black tiger shrimps (Penaeus monodon) in Malaysia. Food Control 31:546–552. doi: 10.1016/j.foodcont.2012.10.018. [DOI] [Google Scholar]
- 3.Gutierrez West CK, Klein SL, Lovell CR. 2013. High frequency of virulence factor genes tdh, trh, and tlh in Vibrio parahaemolyticus strains isolated from a pristine estuary. Appl Environ Microbiol 79:2247–2252. doi: 10.1128/AEM.03792-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tsai SE, Jong KJ, Tey YH, Yu WT, Chiou CS, Lee YS, Wong HC. 2013. Molecular characterization of clinical and environmental Vibrio parahaemolyticus isolates in Taiwan. Int J Food Microbiol 165:18–26. doi: 10.1016/j.ijfoodmicro.2013.04.017. [DOI] [PubMed] [Google Scholar]
- 5.Liu M, Wong MH, Chen S. 2013. Molecular characterisation of a multidrug resistance conjugative plasmid from Vibrio parahaemolyticus. Int J Antimicrob Agents 42:575–579. doi: 10.1016/j.ijantimicag.2013.08.014. [DOI] [PubMed] [Google Scholar]
- 6.de Melo LMR, Almeida D, Hofer E, Dos Reis CMF, Theophilo GND, Santos AFDM, Vieira RHSF. 2011. Antibiotic resistance of Vibrio parahaemolyticus isolated from pond-reared Litopenaeus vannamei marketed in Natal, Brazil. Braz J Microbiol 42:1463–1469. doi: 10.1590/S1517-83822011000400032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Han F, Walker RD, Janes ME, Prinyawiwatkul W, Ge B. 2007. Antimicrobial susceptibilities of Vibrio parahaemolyticus and Vibrio vulnificus isolates from Louisiana Gulf and retail raw oysters. Appl Environ Microbiol 73:7096–7098. doi: 10.1128/AEM.01116-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Laganà P, Caruso G, Minutoli E, Zaccone R, Santi D. 2011. Susceptibility to antibiotics of Vibrio spp. and Photobacterium damsela ssp. piscicida strains isolated from Italian aquaculture farms. New Microbiol 34:53–63. [PubMed] [Google Scholar]
- 9.Jiang Y, Yao L, Li F, Tan Z, Zhai Y, Wang L. 2014. Characterization of antimicrobial resistance of Vibrio parahaemolyticus from cultured sea cucumbers (Apostichopus japonicas). Lett Appl Microbiol 59:147–154. doi: 10.1111/lam.12258. [DOI] [PubMed] [Google Scholar]
- 10.Wong MH, Liu M, Wan HY, Chen S. 2012. Characterization of extended-spectrum-β-lactamase-producing Vibrio parahaemolyticus. Antimicrob Agents Chemother 56:4026–4028. doi: 10.1128/AAC.00385-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Di Pinto A, Terio V, Novello L, Tantillo G. 2011. Comparison between thiosulphate-citrate-bile salt sucrose (TCBS) agar and CHROMagar Vibrio for isolating Vibrio parahaemolyticus. Food Control 22:124–127. doi: 10.1016/j.foodcont.2010.06.013. [DOI] [Google Scholar]
- 12.Paydar M, Teh CSJ, Thong KL. 2013. Prevalence and characterisation of potentially virulent Vibrio parahaemolyticus in seafood in Malaysia using conventional methods, PCR and REP-PCR. Food Control 32:13–18. doi: 10.1016/j.foodcont.2012.11.034. [DOI] [Google Scholar]
- 13.CLSI. 2005. Methods for antimicrobial dilution and disk susceptibility testing of infrequently isolated or fastidious bacteria; proposed guideline. CLSI document M45-P. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 14.CLSI. 2014. Performance standards for antimicrobial susceptibility testing; 23rd informational supplement CLSI document M100-S23. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 15.Dallenne C, Da Costa A, Decre D, Favier C, Arlet G. 2010. Development of a set of multiplex PCR assays for the detection of genes encoding important beta-lactamases in Enterobacteriaceae. J Antimicrob Chemother 65:490–495. doi: 10.1093/jac/dkp498. [DOI] [PubMed] [Google Scholar]
- 16.Carattoli A, Bertini A, Villa L, Falbo V, Hopkins KL, Threlfall EJ. 2005. Identification of plasmids by PCR-based replicon typing. J Microbiol Methods 63:219–228. doi: 10.1016/j.mimet.2005.03.018. [DOI] [PubMed] [Google Scholar]
- 17.Su YC, Liu C. 2007. Vibrio parahaemolyticus: a concern of seafood safety. Food Microbiol 24:549–558. doi: 10.1016/j.fm.2007.01.005. [DOI] [PubMed] [Google Scholar]
- 18.Bortolaia V, Hansen KH, Nielsen CA, Fritsche TR, Guardabassi L. 2014. High diversity of plasmids harbouring blaCMY-2 among clinical Escherichia coli isolates from humans and companion animals in the upper Midwestern USA. J Antimicrob Chemother 69:1492–1496. doi: 10.1093/jac/dku011. [DOI] [PubMed] [Google Scholar]
- 19.Wiesner M, Calva E, Fernandez-Mora M, Cevallos MA, Campos F, Zaidi MB, Silva C. 2011. Salmonella Typhimurium ST213 is associated with two types of IncA/C plasmids carrying multiple resistance determinants. BMC Microbiol 11:9. doi: 10.1186/1471-2180-11-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Call DR, Singer RS, Meng D, Broschat SL, Orfe LH, Anderson JM, Herndon DR, Kappmeyer LS, Daniels JB, Besser TE. 2010. blaCMY-2-positive IncA/C plasmids from Escherichia coli and Salmonella enterica are a distinct component of a larger lineage of plasmids. Antimicrob Agents Chemother 54:590–596. doi: 10.1128/AAC.00055-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Börjesson S, Jernberg C, Brolund A, Edquist P, Finn M, Landen A, Olsson-Liljequist B, Tegmark Wisell K, Bengtsson B, Englund S. 2013. Characterization of plasmid-mediated AmpC-producing E. coli from Swedish broilers and association with human clinical isolates. Clin Microbiol Infect 19:E309–E311. doi: 10.1111/1469-0691.12192. [DOI] [PubMed] [Google Scholar]
- 22.Verdet C, Gautier V, Chachaty E, Ronco E, Hidri N, Decre D, Arlet G. 2009. Genetic context of plasmid-carried blaCMY-2-like genes in Enterobacteriaceae. Antimicrob Agents Chemother 53:4002–4006. doi: 10.1128/AAC.00753-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jacoby GA. 2009. AmpC beta-lactamases. Clin Microbiol Rev 22:161–182. doi: 10.1128/CMR.00036-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Giles WP, Benson AK, Olson ME, Hutkins RW, Whichard JM, Winokur PL, Fey PD. 2004. DNA sequence analysis of regions surrounding blaCMY-2 from multiple Salmonella plasmid backbones. Antimicrob Agents Chemother 48:2845–2852. doi: 10.1128/AAC.48.8.2845-2852.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Khan AA, Cheng CM, Van KT, West CS, Nawaz MS, Khan SA. 2006. Characterization of class 1 integron resistance gene cassettes in Salmonella enterica serovars Oslo and Bareily from imported seafood. J Antimicrob Chemother 58:1308–1310. doi: 10.1093/jac/dkl416. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.