Lin et al. identified a mutant that selectively affects dendrite growth in neurons with large dendrite arbors without affecting dendrite growth in neurons with small dendrite arbors or the animal overall. This mutant disrupts a putative amino acid transporter, Pathetic (Path), that localizes to the cell surface and endolysosomal compartments in neurons.
Keywords: Drosophila, dendrite, growth control, transporter
Abstract
Dendrites exhibit enormous diversity in form and can differ in size by several orders of magnitude even in a single animal. However, whether neurons with large dendrite arbors have specialized mechanisms to support their growth demands is unknown. To address this question, we conducted a genetic screen for mutations that differentially affected growth in neurons with different-sized dendrite arbors. From this screen, we identified a mutant that selectively affects dendrite growth in neurons with large dendrite arbors without affecting dendrite growth in neurons with small dendrite arbors or the animal overall. This mutant disrupts a putative amino acid transporter, Pathetic (Path), that localizes to the cell surface and endolysosomal compartments in neurons. Although Path is broadly expressed in neurons and nonneuronal cells, mutation of path impinges on nutrient responses and protein homeostasis specifically in neurons with large dendrite arbors but not in other cells. Altogether, our results demonstrate that specialized molecular mechanisms exist to support growth demands in neurons with large dendrite arbors and define Path as a founding member of this growth program.
How cells achieve their correct size and how this relates to organ size control are fundamental, unresolved questions in biology. As animals grow, most tissues scale proportional to animal growth by the addition of new cells. In contrast, nervous system growth largely entails the growth of existing cells. For example, many types of sensory neurons are born early and must grow continuously during animal development to maintain coverage of a growing receptive field (Bloomfield and Hitchcock 1991; Parrish et al. 2009). Within the nervous system, different types of neurons have different growth requirements, depending on the size and complexity of their dendrite and axon arbors. For example, mature cerebellar Purkinje neurons have dendrites that are more than two orders of magnitude longer than dendrites of nearby granule cells (Fiala et al. 2008). Likewise, dendrite arbor size scales with increased body size for many types of neurons across phylogeny (Purves and Lichtman 1985); thus, neurons in larger animals must support greater growth demands. Given this enormous range in cell size, it seems likely that distinct mechanisms must exist to support growth in small and large neurons; however, whether neurons with extreme growth requirements, such as Purkinje neurons, have specialized machinery to support their growth demands remains unknown.
Size control in neurons depends in part on specification of neuronal type. Some neurons have an intrinsic growth program that operates largely independently of external influences. For example, the size and shape of some isolated retinal ganglion cells grown in culture are comparable with corresponding cells in vivo, suggesting that growth properties in these neurons are intrinsically encoded (Montague and Friedlander 1991). Likewise, following widespread genetic ablation of ganglion cells, spared ganglion cells adopt a normal size independently of contacts with other ganglion cells, further suggesting that size is intrinsically determined in ganglion cells (Lin et al. 2004). In some scenarios, the expression of particular transcription factors dictates dendrite arbor size and complexity. For example, levels of the transcription factor cut dictate dendrite arbor size in Drosophila dendrite arborization (da) neurons; loss of cut reduces arbor size in da neurons with large dendrite arbors, whereas ectopic cut expression drives overgrowth of da neurons with small dendrite arbors (Grueber et al. 2003a). Similarly, levels of the MEC-3 transcription factor specify elaborate (low) or simple (high) dendrite arbors in Caenorhabditis elegans sensory neurons (Smith et al. 2013). However, the downstream factors that facilitate growth and whether they are materially different in neurons with small and large dendrite arbors remain unknown.
Drosophila peripheral nervous system (PNS) neurons have type-specific dendrite arbors that vary in size by several orders of magnitude in total dendrite length (Grueber et al. 2002), providing a tractable system to study dendrite growth control. Here we report our identification and characterization of path, which encodes a putative amino acid transporter that is broadly expressed in neurons and nonneuronal cells but is preferentially required for growth in neurons with large dendrite arbors. Dendrite growth in different types of neurons with large dendrite arbors arrests at the same value of total dendrite length in path mutants despite the fact that dendrites in these neurons normally grow to very different sizes, suggesting that path defines a program required for extreme growth in neurons. Consistent with this notion, mutation of path impinges on nutrient responses and protein homeostasis in neurons with large arbors but not in other cells. Altogether, our studies suggest that Path functions as part of a nutrient sensor in neurons and define a novel form of growth control required for extreme growth demands in neurons.
Results
path is required for late stage dendrite growth
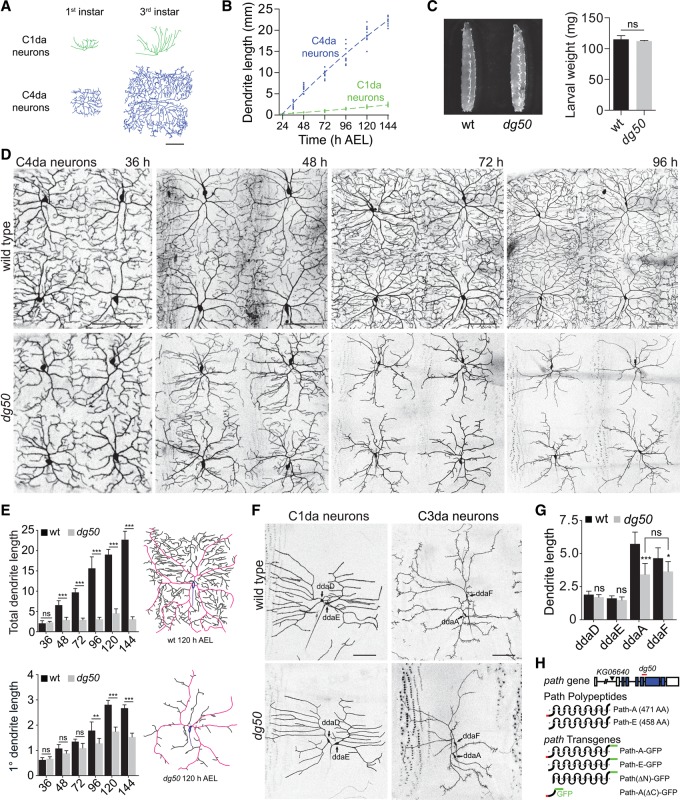
Drosophila da neurons are born embryonically, and dendrites of these neurons grow continuously during larval development to maintain proportional body wall coverage (Parrish et al. 2009; Jiang et al. 2014). Different classes of da neurons cover different territories and, as a result, support different levels of growth (Fig. 1A,B; Grueber et al. 2002; Parrish et al. 2009). Among these neurons, class I da (C1da) neurons have the simplest dendrite arbors and occupy the smallest territory, whereas class IV da (C4da) neurons have the most complex arbors and occupy the largest territory; C4da dendrite length is nearly 10-fold greater than C1da dendrite length by the end of larval development (Fig. 1B).
Figure 1.
Identification of a novel regulator of dendrite growth. (A,B) Growth control in da neurons. (A) Traces of representative C1da and C4da neurons in first instar and third instar larvae. (B) Plots depicting C1da (green) and C4da (blue) dendrite growth during larval development. Points mark measurements from individual neurons. n = 10 for each time point. (C–E) path regulates dendrite growth in C4da neurons. (C) dg50 is dispensable for larval growth. (Left) Wild-type and dg50 mutant larvae at 120 h after egg laying (AEL). (Right) Weight of wild-type and dg50 mutant larvae. Mean and standard deviation for five independent measurements (80 larvae each) are shown. (D) Time course of dendrite growth in C4da neurons (ppk-CD4-tdTomato) at 36, 48, 72, and 96 h AEL in wild-type control and dg50 mutants. (E) Mean total dendrite length and primary dendrite length (in millimeters) of individual ddaC C4da neurons of the indicated genotype at the indicated times are shown. n = 10 neurons for each time point. Traces of representative C4da neurons from 120 h AEL wild-type and dg50 mutant larvae are shown with primary dendrites shaded in magenta. (F,G) Effects of dg50 on dendrite growth in different classes of da neurons. (F) Dendrites of C1da neurons (Gal498b, UAS-CD4-tdGFP) and C3da neurons (ss-myrGFP) in wild-type control and dg50 mutant larvae are shown. (G) Mean total dendrite length of C1da and C3da neurons of the indicated genotypes. n = 10 neurons for each. (*) P < 0.05; (**) P < 0.01; (***) P < 0.001; (ns) not significant compared with wild-type controls; one-way ANOVA with a post-hoc Dunnett's test. In this and all subsequent figures, dorsal is up, and anterior is left. Bars, 50 μm. (H) Schematic depicting the path locus, including alleles used in this study, Path polypeptides that differ by 13 amino acids at the N terminus (red), and path transgenes used in this study. Boxes indicate transmembrane domains, and the gray dashed line represents a membrane.
We reasoned that if neurons with large dendrite arbors have specialized machinery to support increased growth demands, then mutations in such machinery would preferentially affect C4da dendrite growth. We therefore screened for mutations that affected growth of C4da dendrites but not C1da dendrites. Specifically, we EMS-mutagenized a Drosophila line bearing ppk-CD4-tdTomato, which labels C4da neurons, and screened mutants for defects in C4da dendrite arbor size. From this screen, we identified a homozygous viable mutant (dendrite growth defective 50 [dg50]) with severe C4da dendrite growth defects but no obvious effect on animal growth (Fig. 1C,D). In dg50 mutants, early growth and patterning of C4da neurons proceed normally (Supplemental Fig. S1), but, beginning in late first larval instar larvae, two growth defects are manifest. First, C4da dendrite length plateaus at ∼3.5 mm, which dg50 mutants reach by ∼36–40 h after egg laying (AEL), whereas control C4da dendrites continue growing throughout larval development, reaching ∼20 mm (Fig. 1D,E). Second, C4da dendrite arbors were remodeled after reaching their maximum length in dg50 mutants: Terminal dendrites were progressively lost, whereas primary dendrites continued to grow, maintaining a constant value of total dendrite length (Fig. 1D,E; Supplemental Fig. S2).
To determine whether dg50 specifically affects growth in neurons with large dendrite arbors, we assayed dg50 mutants for effects on growth of C1da and C3da neurons, which normally have small and medium-sized dendrite arbors, respectively (Fig. 1F,G). In wild-type larvae, dendrites of dorsal C1da neurons (ddaD and ddaE) grew to a total length of 1.8 mm, on average, at 120 h AEL. C1da dendrites in dg50 mutants grew to a comparable size and exhibited no measurable patterning defects. We also noted that dg50 had no effect on dendrite growth in PNS neurons with even smaller dendrite arbors, including chordotonal (ch) neurons and bipolar dendrite (bd) neurons (Supplemental Fig. S3). Thus, dg50 has no effect on dendrite growth in PNS neurons with small dendrite arbors.
Next, we monitored the effects of dg50 on growth of the C3da neurons ddaA and ddaF, which have medium-sized dendrite arbors (Fig. 1F,G). In control larvae, C3da dendrites reach a total length of 5.3 mm (ddaA) and 4.8 mm (ddaF), on average, at 120 h AEL. In contrast, C3da dendrites in dg50 mutants grew to only 3.5 mm, comparable with the upper limit for C4da total dendrite length in dg50 mutants. dg50 therefore appears to impose an upper limit on dendrite growth in PNS neurons; in dg50 mutants, neurons with small dendrite arbors (bd, ch, and C1da) are unaffected, whereas dendrite growth in neurons with larger arbors (ddaA, ddaF, and ddaC) arrests at a fixed dendrite length despite the fact that these neurons normally grow to very different sizes.
To identify the molecular basis for these growth defects, we screened for chromosome deficiencies that failed to complement dg50. Heterozygosity for dg50 had no effect on dendrite growth, but placing dg50 in trans to Df(3L)BSC773 phenocopied dg50 homozygotes (Supplemental Fig. S1), localizing dg50 to a small interval and demonstrating that dg50 is a recessive, loss-of-function allele. Df(3L)BSC773 deletes 64 genes, including path, which has been implicated in growth control (Goberdhan et al. 2005); thus, we tested whether dg50 is allelic to path. Indeed, homozygosity for a P-element insertion allele of path (pathKG06640) or heteroallelic combination of pathKG06640 with dg50 or Df(3L)BSC773 similarly affected C4da dendrite growth (Supplemental Fig. S1). Furthermore, dg50 carries a 172-nucleotide deletion in path that causes a frameshift and premature stop, truncating the transmembrane domains. Thus, dg50 should render Path nonfunctional (Fig. 1H). Finally, ubiquitous expression of GFP-tagged Path (Actin-Gal4, UAS-pathA-GFP) rescued dg50 dendrite phenotypes (Supplemental Fig. S1), demonstrating that dendrite defects in dg50 mutants were caused by loss of path function and that UAS-pathA-GFP is a functional transgene. We conclude that dg50 is a loss-of-function allele of path.
path encodes an amino acid transporter of the solute carrier 36 (SLC36) family (Fig. 1H), many members of which are highly expressed in vertebrate nervous systems, including the two putative mammalian orthologs SLC36A1 and SLC36A4 (Sagné et al. 2001; Bermingham and Pennington 2004; Roshanbin et al. 2014). SLC36 transporters appear to be involved in transport of amino acids and their derivatives; thus, they may directly modulate cellular nutrient availability. However, requirements for SLC36 transporters in nervous system development have not been examined in vivo.
path is required for extreme dendrite growth in neurons
Our initial characterization of path suggested that neurons grow to a fixed limit in the absence of path. Since path transcript is maternally deposited (Tomancak et al. 2002), we examined whether maternal path supported dendrite growth in path zygotic-null mutants. To this end, we used the dominant female sterile technique (Perrimon 1984) to generate homozygous mutant pathdg50 germline clones and assayed for maternal path contribution to dendrite growth. Notably, C4da dendrites in larvae lacking both maternal and zygotic path function arrested at the same value of dendrite growth as zygotic path mutants (Supplemental Fig. S1), demonstrating that path function is indeed dispensable for dendrite growth up to this fixed limit. Taken together, these results suggest that da neurons can support dendrite growth up to a maximum of ∼3.5 mm without path, but growth beyond this limit requires path.
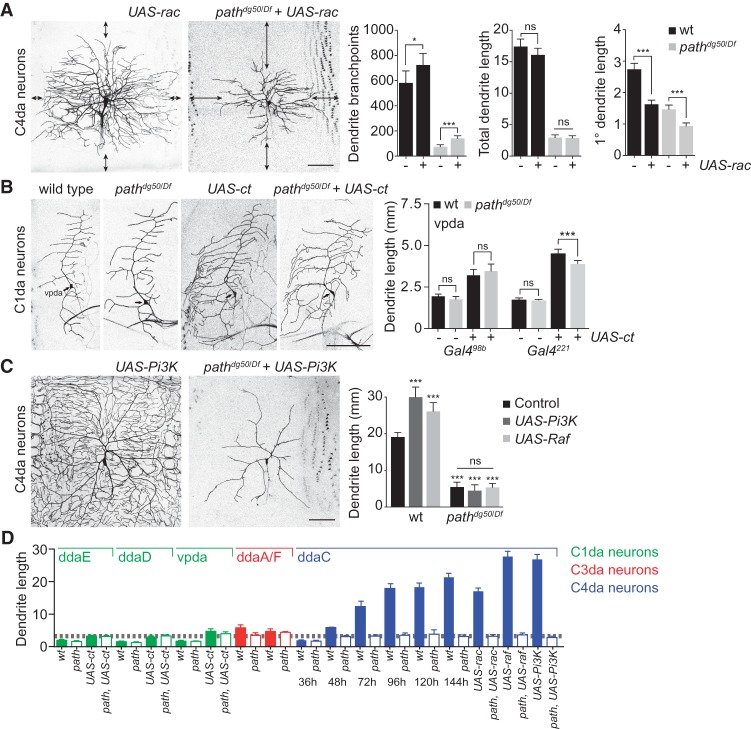
In path mutants, dendrite patterning is altered to facilitate maximal extension of major dendrites. Whereas major dendrite branches continue to elongate throughout larval development in path mutants, overall dendrite length is unchanged after 36 h AEL, and terminal dendrites are lost by retraction (Fig. 1D; Supplemental Fig. S2). This phenotype might reflect a deficit in branching or an intrinsic hierarchy in which primary dendrite growth predominates. We therefore investigated the relationship between dendrite patterning and path-dependent dendrite growth. To this end, we overexpressed the small GTPase Rac, which drives ectopic dendrite branching in C4da neurons (Lee et al. 2003), and monitored the effects on dendrite growth in wild-type and path mutant larvae. Although total dendrite length was unaffected, rac overexpression drove ectopic dendrite branching in both wild-type and path mutant larvae (Fig. 2A), demonstrating that path mutant C4da neurons can support branching. Strikingly, this increase in terminal dendrite branching was accompanied by a significant decrease in primary dendrite growth in both control and path mutant larvae. As a result, distal portions of the territory normally covered by C4da neurons were completely devoid of dendrites. Thus, although path mutant C4da neurons can support dendrite branching, path affects dendrite growth independently of dendrite patterning, and, in path mutants, primary dendrite growth normally appears to be prioritized at the expense of terminal branching.
Figure 2.
path is required for dendrite growth above a fixed limit. (A) path affects dendrite growth independently of dendrite patterning. (Left) Representative images of wild-type or path mutant C4da neurons expressing UAS-rac (ppk-Gal4, UAS-rac). (Right) Quantification of dendrite branching and length in the indicated genotypes. Arrows mark uncovered territory. (*) P < 0.05; (***) P < 0.001; (ns) not significant compared with controls that do not express UAS-Rac; unpaired t-tests with Welch's correction. (B) Forced overgrowth in C1da dendrites requires path. C1da neurons (vpda) are shown for the indicated genotypes. UAS-ct expression induces dendrite overgrowth in C1da neurons of wild-type and path mutant larvae, but this overgrowth is limited in path mutants. Quantification shows mean dendrite length and standard deviation in vpda C1da neurons of the indicated genotype. (C) path is epistatic to treatments that increase C4da dendrite growth. (Left) Representative images of wild-type or path mutant C4da neurons expressing UAS-Pi3K92E. (Right) Quantification of dendrite growth in wild-type or path mutant larvae expressing UAS-Pi3K92E or UAS-Raf in C4da neurons. (***) P < 0.001; (ns) not significant compared with wild-type controls unless otherwise indicated in B and C; one-way ANOVA with a post-hoc Dunnett's test. n = 10 neurons for each genotype. Bars, 50 μm. (D) Summary of dendrite measurements highlighting the upper limit for path-independent dendrite growth (gray dashed line).
Dendrite growth in neurons with small arbors (e.g., C1da neurons) was unaffected by mutation of path, possibly because dendrite length in these neurons was below the threshold for sensitivity to path function. If this was indeed the case, increasing dendrite length beyond this threshold in C1da neurons should require path function. Ectopic expression of the homeodomain transcription factor cut drives exuberant dendrite growth in C1da neurons (Grueber et al. 2003a); therefore, we monitored the effects of path mutation on the ability of C1da neurons to support cut-induced exuberant dendrite growth. Consistent with prior reports, UAS-cut expression induced dendrite overgrowth in each of the C1da neurons (vpda, ddaD, and ddaE) (Fig. 2B; Supplemental Fig. S4); we focused our analysis on vpda because our Gal4 drivers expressed most stably in that neuron. We found that two different C1da Gal4 drivers (Gal498b and Gal4221) became active at different time points (first instar larva and stage 17 embryo, respectively) and therefore supported different levels of UAS-cut-induced dendrite growth (3.0 mm and 4.8 mm, respectively) (Fig. 2B). We reasoned that if path is required for dendrite growth beyond ∼3.5 mm in da neurons, path mutation should affect UAS-cut-induced dendrite growth in C1da neurons when driven by Gal4221 but not Gal498b. This is precisely what we observed. Whereas UAS-cut expression induced dendrite overgrowth in all C1da neurons in path mutants, the extent of growth was constrained only when UAS-cut was driven by Gal4221. The increased growth demands imposed by ectopic cut expression therefore require path only when dendrites grow beyond the empirically determined threshold (∼3.5 mm), further demonstrating a role for path in supporting extreme dendrite growth. Different classes of da neurons develop at different rates (Parrish et al. 2009) and therefore reach this growth limit at different times (C4da neurons, 48 h AEL; C3da neurons, 72 h AEL; cut-expressing C1da neurons, 96 h AEL). Thus, the growth limit for path-independent dendrite growth in C4da neurons appears to be independent of larval size and developmental progression.
Next, we investigated whether treatments that stimulate exuberant dendrite growth can modify the growth limit in path mutant larvae. Expression of UAS-Pi3K92E or UAS-Raf in C4da neurons drives excessive dendrite branch elongation while causing little effect on patterning of C4da dendrites (Fig. 2C), resulting in dendrite arbors with a 50% or 37% increase, respectively, in total dendrite length. However, mutation of path was epistatic to expression of either Pi3K92E or Raf in growth control of these neurons, with dendrite growth arresting at the same value in the absence or presence of these transgenes, further underscoring the notion that path mutation imparts a fixed growth limit on neurons (Fig. 2D).
path is a permissive factor for dendrite growth
Path is required to support growth in neurons with large dendritic arbors, so we wondered whether ectopic expression of path could promote exuberant growth in neurons with small dendrite arbors. To test this possibility, we overexpressed path (UAS-pathA-GFP) in C1da neurons of otherwise wild-type larvae and monitored the effects on dendrite growth. (Supplemental Fig. S5). Indeed, we found that overexpression of path could promote a modest but significant increase in C1da dendrite growth. In contrast, path overexpression had no effect on the total dendrite length of C4da neurons. Given the relatively modest effect of path overexpression on C1da dendrites and the absence of any effect on C4da neurons, we conclude that the final size of dendritic arbors in these neurons is likely regulated by other factors, with path required to support growth beyond a fixed limit.
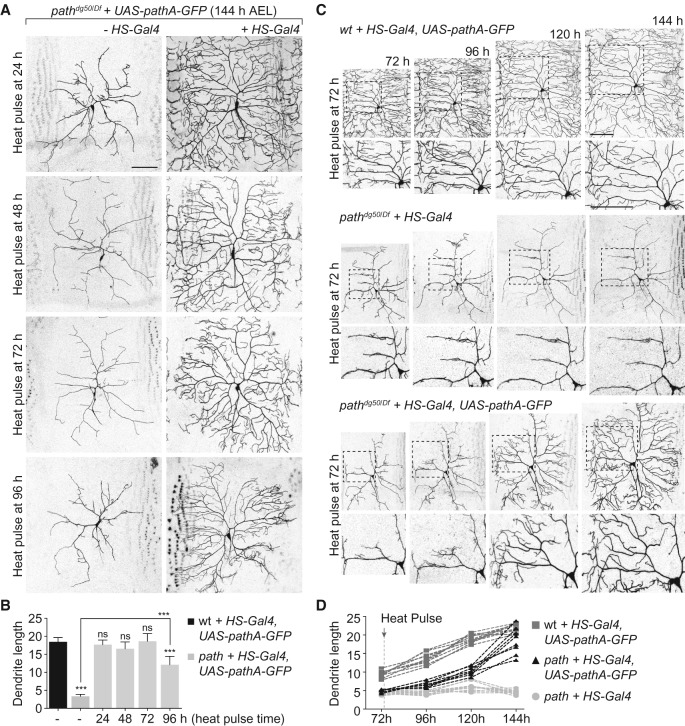
We next investigated the relationship between path-dependent and path-independent growth. To this end, we tested whether supplying path to path-deficient neurons after they reached the limit of path-independent growth was sufficient to support extreme dendrite growth. For these assays, we used Heat Shock Gal4 (HS-Gal4) to inducibly and ubiquitously express UAS-pathA-GFP at different stages during larval development and monitored effects on C4da dendrite growth using live time-lapse microscopy. As described above, ubiquitous expression of UAS-pathA-GFP was sufficient to mitigate the dendrite growth defects of path mutants but did not induce dendrite overgrowth, suggesting that path is a permissive factor for dendrite growth.
Dendrite defects in path mutants first appear after 36 h AEL (Fig. 1D); therefore, we tested whether an early pulse of UAS-pathA-GFP expression (1 h of heat shock at 24 h AEL) would suffice for dendrite growth. Indeed, transient early UAS-pathA-GFP expression supported wild-type growth in C4da neurons (Fig. 3A,B). Next, we assayed the effects of resupplying UAS-pathA-GFP at different time points during larval development: 48 h AEL, shortly after C4da neurons cross the threshold for path-dependent growth and growth defects first appear; 72 h AEL, when growth and patterning defects are manifest in path mutants; and 96 h AEL, when wild-type C4da dendrites have grown 400% more than path mutants. To our surprise, resupplying UAS-pathA-GFP at each time point was sufficient to significantly rescue dendrite growth defects of path mutants (Fig. 3A,B). Thus, growth defects of path mutants are reversible. We note that resupplying path expression at 96 h AEL led to incomplete rescue at 144 h AEL (Fig. 3B). Prior studies have shown that structural plasticity in C4da neurons is progressively limited during larval development as a result of dendrite–substrate interactions (Grueber et al. 2003b; Sugimura et al. 2003; Parrish et al. 2009; Jiang et al. 2014); our findings suggest that this restriction in plasticity depends on proper dendrite growth during early larval stages. Remarkably, the rescued neurons were comparable with wild-type controls in size and pattern even when path was resupplied after growth defects were manifest, suggesting that factors other than path determine the final size of C4da neurons.
Figure 3.
path is a permissive factor for dendrite growth. (A,B) Growth defects of path mutants are reversible. path mutant [pathdg50/Df(3L)BSC773] larvae carrying a path rescue transgene (UAS-pathA-GFP) without (left) or with (right) a heat-inducible Gal4 driver (HS-Gal4) were heat-shocked for 1 h at 37°C at the indicated time, and effects on dendrite growth were assessed at 144 h AEL using ppk-CD4-tdTomato to label C4da dendrites. (B) Quantification of C4da dendrite length (in millimeters) in path mutants at 144 h AEL after resupplying UAS-pathA-GFP at the indicated times. n = 8 neurons for each genotype. (C,D) Resupplying path to path mutants induces heterochronic growth. (C) Time-lapse imaging of dendrite growth prior to a 1-h heat shock (72 h AEL; left), 24 h after heat shock (96 h AEL), 48 h after heat shock (120 h AEL), and 72 h after heat shock (144 h AEL) in larvae of the indicated genotype. (D) Plot depicting growth profile of C4da dendrites in the indicated genotypes following induction of path expression. Note that path expression induces rapid late stage growth in path mutants. n = 8 neurons for each genotype. (**) P < 0.01; (***) P < 0.001; (ns) not significant compared with wild-type controls unless otherwise indicated; one-way ANOVA with a post-hoc Dunnett's test. Bars, 50 μm.
We next used time-lapse microscopy to characterize dendrite growth in path mutant neurons before and after resupplying path expression. Prior to induction of path expression at 72 h, path mutant C4da dendrites exhibited severe growth deficits (Fig. 3C). However, 24 h after transient pathA-GFP expression, path mutant C4da neurons exhibited sprouting of new dendrites that grew extensively over the next 48 h, eventually regenerating wild-type dendrite coverage in these neurons. Whereas path mutant C4da neurons exhibited no net growth over this time lapse, the “rescued” neurons exhibited a significant increase in growth rate and overall growth compared with wild-type controls during this time lapse (Fig. 3C,D). Thus, altering the timing of path expression alters growth rate and timing but not the final size of dendrite arbors, suggesting that additional mechanisms exist that sense and/or constrain size in these neurons.
Path is ubiquitously expressed and localizes to endolysosomal compartments
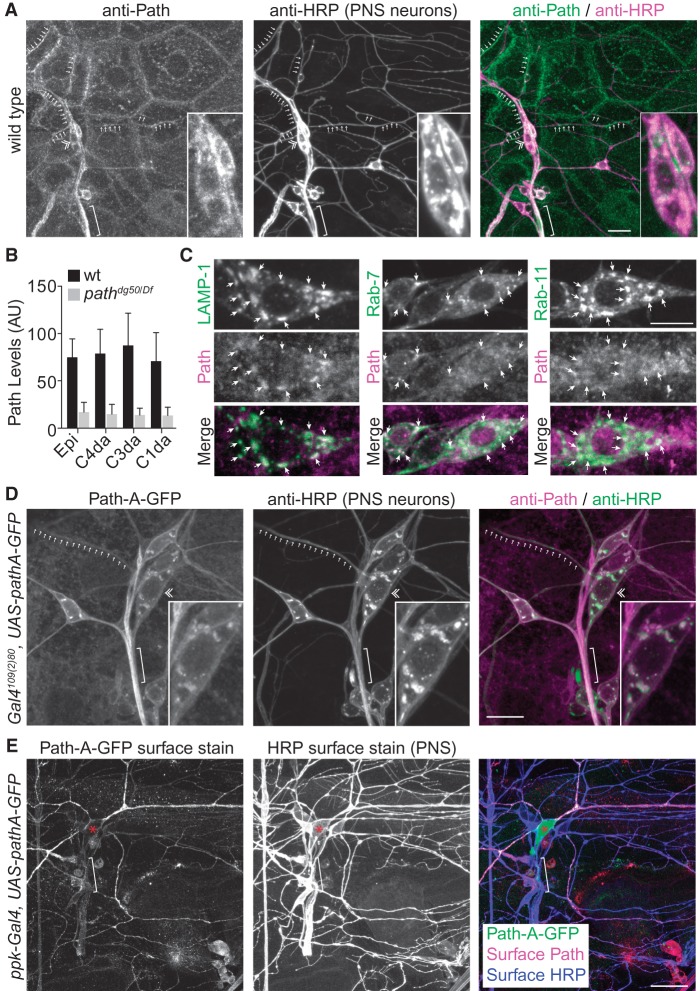
To gain insight into Path's site of action and the basis for the specificity of path mutant growth defects, we monitored distribution of Path in vivo. Since path mutants selectively affect growth in neurons with large dendrite arbors, we anticipated that Path would be selectively expressed in these neurons. However, in third instar larvae, punctate Path immunoreactivity was present in all cells of the body wall (Fig. 4A); this immunoreactivity was absent in pathdg50/Df larvae (Fig. 4B; Supplemental Fig. S6A), demonstrating the specificity of the antibody. In C4da neurons, Path was prominent in the soma and also present in axons and dendrites (Fig. 4A).
Figure 4.
Path is broadly expressed and localizes to the cell surface and endolysosomal compartments. (A) Anti-Path immunostaining of a wild-type third instar fillet reveals expression throughout the body wall. Path localizes to punctate structures within cells and accumulates at junctional domains and in PNS dendrites (arrows) and axons (brackets). A double chevron marks C4da cell body. (B) Quantification of Path levels in different classes of da neurons. Mean pixel intensity and standard deviation for Path immunoreactivity in body wall epithelial cells (epi) and PNS neurons are shown. n = 20 cells each. (C) Path colocalizes with endolysosomal markers. Images depict colocalization of endogenous Path with GFP-LAMP, Rab-7-GFP, or Rab-11-GFP. (D) Path-GFP localizes to axons and dendrites. Representative images of Path-A-GFP and HRP (to label sensory neurons) staining are shown. The inset shows a magnified view of the C4da soma. (E) Path-A-GFP localizes to the plasma membrane in dendrites. Surface-exposed GFP as revealed by immunostaining under nonpermeabilizing conditions (Path-A-GFP surface stain) is shown for a pathdg50/Df mutant expressing UAS-pathA-GFP in C4da neurons. Bars: C, 10 μm; all other panels, 50 μm.
Next, we examined whether Path was present at different levels in neurons with small dendrite arbors and neurons with large dendrite arbors. This was not the case; somatic Path levels were comparable in all da neurons (Fig. 4B). Although the signal in neurites was obscured by epithelial Path expression, we observed no overt difference in Path levels in C1da and C4da dendrites. Likewise, Path levels were comparable in control and UAS-cut-expressing C1da neurons (data not shown). Thus, Path levels do not appear to correlate with dendrite arbor size, consistent with Path functioning as a permissive factor for dendrite growth.
In the course of our analysis, we noted that ∼0.5% of C4da neurons in path mutant larvae exhibited normal growth properties. These “escaper” C4da neurons retained detectable Path immunoreactivity (Supplemental Fig. S6B), likely representing persistent maternally derived protein, indicating that low levels of neuronal Path are sufficient for extreme dendrite growth and suggesting that Path functions cell-autonomously to support neuron growth. We noted an “all or none” response to loss of path: In neurons where detectable Path persisted, dendrite growth was comparable with wild-type controls, whereas neurons without detectable Path did not grow beyond the empirically determined threshold for path-supported growth.
The epithelial expression of Path precluded high-resolution analysis of Path distribution in dendrites; therefore, we focused on characterizing the distribution of endogenous Path in C4da cell bodies. Mammalian SLC36A1 and SLC36A4 transporters localize to endolysosomal compartments (Wreden et al. 2003; Roshanbin et al. 2014), and we likewise found that Path localized to structures labeled by endolysosomal markers in C4da neurons (Fig. 4C). To monitor Path distribution in neurites, we examined intracellular distribution of Path-A-GFP in C4da neurons of path mutants. Consistent with our antibody staining, Path-A-GFP was also distributed along axons and dendrites (Fig. 4D).
One model for Path function in dendrite growth control is that Path localizes to the plasma membrane, where it senses or transports nutrients into the neuron. We therefore examined Path distribution and topology using surface staining of C4da neurons expressing Path-A-GFP, reasoning that GFP should be surface-exposed if Path is targeted to the plasma membrane. As a negative control, we first assayed for surface-exposed GFP in C4da neurons expressing α-tubulin-GFP and observed no surface staining, demonstrating that the plasma membrane was not permeabilized in our staining protocol (Supplemental Fig. S6C). In contrast, surface-exposed Path-A-GFP was detectable throughout the C4da dendrite arbors (Fig. 4E). Path-A-GFP surface staining in dendrites was markedly higher than in axons or cell bodies, but surface staining of other antigens was likewise reduced in C4da axons and cell bodies (for example, HRP immunoreactivity) (Fig. 4E), likely a result of reduced accessibility due to glial wrapping, so we cannot conclusively say whether surface-exposed Path is enriched in dendrites. Overall, these results demonstrate that Path localizes to the cell surface in dendrites of C4da neurons and in endolysosomal compartments, where it likely senses or transports amino acids to support dendrite growth.
path is required cell-autonomously for dendrite growth
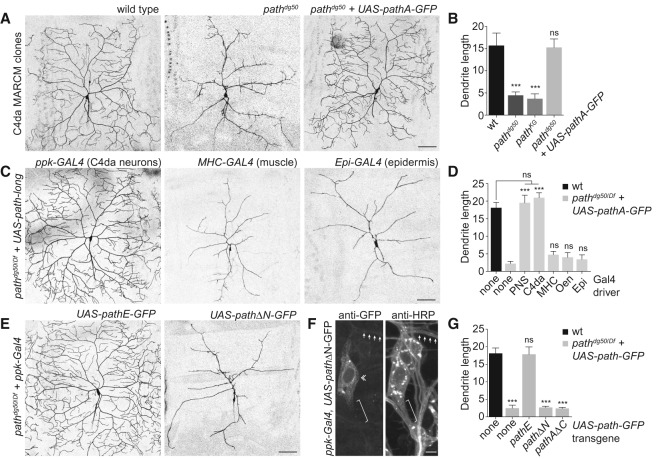
Dendrite growth in PNS neurons depends on both cell-autonomous and cell-nonautonomous cues; therefore, we investigated the site of path action by generating single-cell homozygous path mutant neuron clones in a heterozygous background using the MARCM approach (Lee and Luo 1999) and monitored the effects on dendrite growth. As shown in Figure 5, path mutant C4da MARCM clones exhibited growth defects identical to C4da neurons in path mutant larvae, suggesting that path functions cell-autonomously in C4da neurons to control dendrite growth (Fig. 5A,B). Consistent with this notion, resupplying UAS-pathA-GFP to path mutant C4da MARCM clones rescued the growth defects (Fig. 5A,B). Thus, path functions cell-autonomously to support growth in neurons with large dendrite arbors.
Figure 5.
path is required cell-autonomously for extreme dendrite growth. (A,B) path is required in neurons for dendrite growth. (A) Representative wild-type control, pathdg50, and pathdg50 + UAS-path-GFP ddaC C4da MARCM clones are shown. (B) Quantification of total dendrite length (in millimeters) for ddaC MARCM clones of the indicated genotypes. n ≧ 8 ddaC MARCM clones for each. (C,D) Neuronal path expression is sufficient for extreme dendrite growth. (C) The ability of UAS-path-GFP expression directed by a variety of Gal4 drivers to rescue dendrite growth defects of path mutants was assayed at 120 h AEL. C4da dendrites were visualized by a ppk-CD4-tdTomato reporter in control (UAS-path-GFP/+), path mutant (gal4/+; pathdg50/Df), or UAS-pathA-GFP-expressing path mutant larvae (gal4/UAS-path-A-GFP; pathdg50/Df). Representative images of rescue with the ppk-gal4, MHC-gal4, and A58-gal4 drivers are shown. (D) Quantification of rescue activity of Gal4 drivers at 120 h AEL. Gal4 drivers used to drive UAS-path-A-GFP in the larval body wall were as follows: (none) no Gal4 driver; (PNS [md neurons]) 21-7-Gal4; (C4da [C4da neurons]) ppk-Gal4; (MHC [muscle]) MHC-Gal4; (Oen [oenocytes]) OK72-Gal4; (Epi) A58-Gal4. n = 5 neurons for each genotype. (E–G) The N-terminal domain is required for Path function. (E) Representative images of C4da neurons from path mutant larvae expressing UAS-pathE-GFP (the short isoform of path) (left) or UAS-pathΔN-GFP (a version of path lacking the N-terminal intracellular domain) (right). (F) The N-terminal domain influences Path localization. Antibody staining shows distribution of PathΔN-GFP in C4da neurons (da neurons labeled by anti-HRP immunoreactivity). Arrows mark dendrites, a double chevron marks C4da soma, and a bracket marks the axon. (G) Quantification of rescue activity of neuronal expression of the indicated transgenes in path mutant larvae. n = 8 neurons for each genotype. Error bars represent standard deviation. (***) P < 0.001; (ns) not significant compared with wild-type controls, unless otherwise indicated in B and G, and compared with path mutants in D; one-way ANOVA with a post-hoc Dunnett's test. Bars: A–E, 50 μm; F, 10 μm.
We next turned to a genetic rescue assay to define the minimal functional site for path to support dendrite growth, testing the ability of transgenic path expression in different tissues to rescue dendrite growth defects of path mutant larvae. Neuronal expression of path using pan-neuronal, PNS-specific, or C4da-specific Gal4 drivers fully rescued the path mutant phenotype (Fig. 5C,D). In contrast, path expression in other body wall tissues had no effect on C4da dendrite growth. Thus, path expression in C4da neurons is both necessary and sufficient for extreme dendrite growth.
The path locus encodes two predicted polypeptides: a short isoform and a long isoform with 13 additional amino acids on the N-terminal intracellular domain (Fig. 1H). Having shown that the long isoform (path-A) was sufficient for extreme dendrite growth, we next tested whether the short isoform (path-E) could likewise support extreme dendrite growth. Indeed, resupplying UAS-pathE-GFP to C4da neurons in path mutant larvae restored wild-type dendrite growth (Fig. 5E,G). In contrast, removing the entire N-terminal intracellular domain (UAS-pathΔN-GFP) abrogated Path function in dendrite growth and prevented Path localization to axons and dendrites (Fig. 5E–G). Finally, the N-terminal domain alone (UAS-pathAΔC-GFP) had no activity in supporting dendrite growth, suggesting that the transmembrane transporter domain is critical for path function. Thus, the N-terminal domain is necessary for Path function, perhaps by modulating Path localization and/or interaction with accessory factors, but Path-long and Path-short are functionally equivalent for dendrite growth.
path regulates protein homeostasis to support dendrite growth
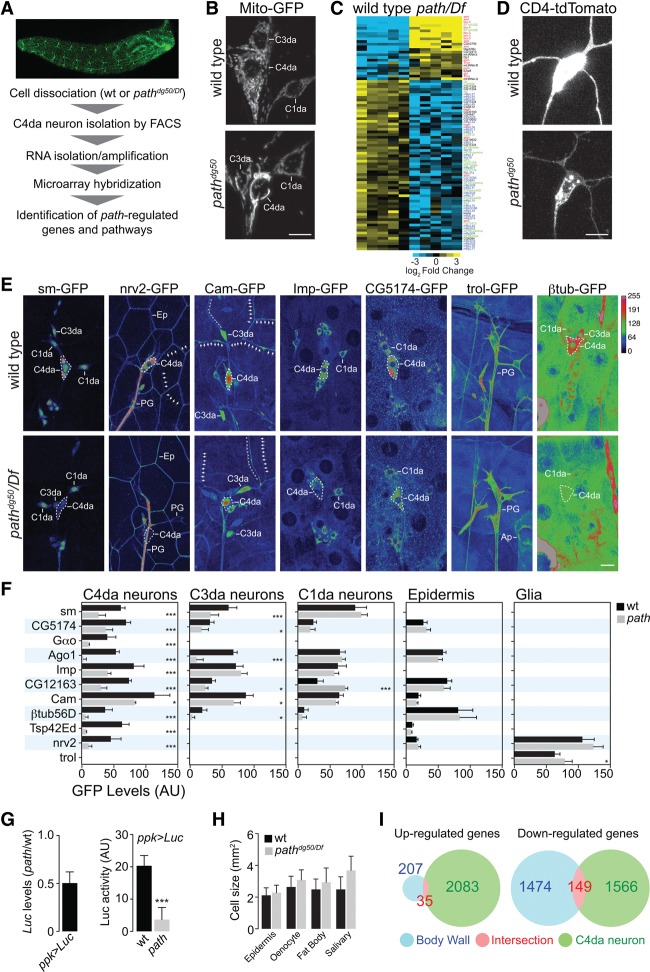
To identify molecular mechanisms of path-dependent growth in C4da neurons, we conducted microarray expression profiling of C4da neurons from wild-type and path mutant larvae (Fig. 6A). We identified >2000 transcripts that were significantly deregulated in path mutant C4da neurons, indicating that path exerts a strong influence on gene expression in these neurons (Supplemental Table S1). Pathway analysis indicated that genes associated with fatty acid metabolism and branched chain amino acid degradation were significantly overrepresented in the set of genes with decreased abundance in path mutant C4da neurons (Supplemental Table S2), consistent with path mutation eliciting a starvation response in C4da neurons and directly or indirectly regulating these pathways. Indeed, path mutant C4da neurons exhibited a striking increase in expression of Drosophila 4E-BP (Thor) (Supplemental Table S1), which is elevated in response to starvation (Teleman et al. 2005; Tettweiler et al. 2005). Additionally, path mutant C4da and, to a lesser degree, C3da neurons exhibited mitochondrial fusion, which occurs in many cell types after nutrient depletion (Rambold et al. 2011), whereas C1da neurons and epithelial cells did not (Fig. 6B; Supplemental Fig. S7). Thus, mutation of path induces an apparent starvation response in neurons with large dendrite arbors but not other cells in the body wall.
Figure 6.
Path affects nutrient signaling and protein homeostasis in C4da neurons. (A) Workflow for microarray expression profiling of path mutant C4da neurons. (B) path affects mitochondria morphology in C4da neurons. PNS neurons from wild-type control (top) and path mutant (bottom) larvae expressing mitochondrial targeted GFP (UAS-mito-GFP) are shown. Levels of apparent mitochondrial fusion ([high] C4da neurons; [low] C3da neurons; [none] C1da neurons) correlate with severity of dendrite growth defects in path mutants. (C) Heat map depicting path-regulated transcripts involved in translation. Significantly deregulated transcripts were identified using SAM analysis with a false discovery rate of 0.01 (Tusher et al. 2001). Transcripts are color-coded according to gene function as follows: (red) translation inhibitor; (green) translation initiation/elongation; (blue) core translational machinery; (black) unknown. (D–F) path affects protein homeostasis in C4da neurons. (D) Levels of the ppk-CD4-tdTomato reporter are reduced in path mutants, although ppk transcription is not affected (Supplemental Table S1). C4da neurons in age-matched wild-type (top) and path mutant (bottom) larvae expressing ppk-CD4-tdTomato were imaged under identical conditions. (E) Representative images depicting expression levels of seven GFP exon trap lines in wild-type or path mutant larvae at 120 h AEL. Images are pseudocolored according to a lookup table (key; top right), C4da cell bodies are outlined with a white dashed line, and cells of interest labeled by each exon trap are indicated: C1da, C3da, and C4da neurons; epithelial cells (Ep); peripheral glia (PG); and apodemes (Ap). Arrows mark C4da dendrites, and double chevrons mark C3da dendrites. Exon traps were imaged under identical conditions in wild-type and path mutant larvae. (F) Mean expression intensity of exon traps in the indicated cell types. n ≧ 8 cells for each bar. (*) P < 0.05; (***) P < 0.001; compared with wild-type controls; one-way ANOVA with a post-hoc Dunnett's test. Error bars indicate standard deviation. Bars, 50 μm. (G) Luciferase mRNA levels and Luciferase activity were assayed in extracts of wild-type and path mutant larvae expressing a UAS-Rluc-Fluc bicistronic reporter transgene in C4da neurons under the control of ppk-Gal4. mRNA levels reflect ΔΔCt values (path mutant/wild-type control; normalized to β-tubulin expression) from three independent biological samples. For Luciferase activity, mean and standard deviation from three independent biological samples of each genotype are shown. (***) P < 0.001; unpaired t-test with Welch's correction. (H) Cell size measurements for the indicated cell types in wild-type or path mutant larvae. Cells were labeled with a membrane reporter (UAS-mCD8-GFP), and cell size (two-dimensional area) was measured by tracing plasma membranes and measuring the area of the resulting polygons. Mean and standard deviation for 100 cells of each genotype are shown. (I) Venn diagram depicting transcriptional changes manifest in the entire body wall (blue) and C4da neurons (green) of path mutant larvae; the region of intersection is shown in red. Significantly deregulated transcripts are identified as in C.
We noted that a large number of transcripts associated with translation were deregulated in C4da neurons of path mutants, and this was of particular interest for several reasons. First, among deregulated translation factors, translational repressors were, in general, increased in abundance in path mutants, whereas positive regulators of translation were reduced in abundance (Fig. 6C), suggestive of a coherent response to reduce translational activity in C4da neurons. Second, transcription of translational machinery is coupled to nutrient availability (Mayer and Grummt 2006); thus, reduced abundance of factors that promote translation was a further indication of a starvation response. Third, we noted that reporter expression (ppk-CD4-tdTomato) was attenuated in C4da neurons of path mutants despite the fact that ppk mRNA levels were apparently not affected (Fig. 6D; Supplemental Table S1). Thus, we hypothesized that path mutation altered protein homeostasis in C4da neurons via effects on protein synthesis, stability, or both, contributing to growth defects.
To monitor the effects of path mutation on protein homeostasis in vivo, we used a collection of GFP protein traps that allows for visualization of proteins expressed from their endogenous loci in Drosophila (Morin et al. 2001). By comparing levels of these protein traps in wild-type and path mutant backgrounds, we aimed to gain insight into the apparent cellular specificity of growth defects in path mutants and explore the generality of protein accumulation defects in path mutant C4da neurons. For these assays, we chose 11 protein traps that displayed a range of expression patterns encompassing the major cell types of the larval body wall (Fig. 6E,F). Additionally, we predominantly chose gene products whose mRNA expression in C4da neurons was unaffected by path (Supplemental Table S1); three traps of gene products with increased mRNA accumulation (sm, Gαo, and Imp,) and one trap of a gene product with reduced mRNA accumulation (CG5174) in path mutant C4da neurons were exceptions to this criteria.
Overall, we observed striking concordance between path-dependent effects on cell growth and protein trap levels. Each of the 10 protein traps normally expressed in C4da neurons exhibited significantly reduced levels at 120 h AEL in path mutants, suggesting that path broadly promotes protein accumulation in these neurons. In contrast, these protein traps were expressed at comparable levels in wild-type and path mutant C4da neurons at 36 h AEL, prior to the onset of growth defects (Supplemental Fig. S8), after which time, protein trap levels were progressively reduced. Therefore, expression of the protein traps closely follows the progression of growth defects in path mutant C4da neurons. In C3da neurons, levels of many protein traps were reduced as well, but the effect was less severe. In contrast, levels of protein traps in neurons with small dendrite arbors (C1da neurons and bd neurons), peripheral glia, epidermal cells, and muscle were unaffected by path mutation, demonstrating remarkable specificity in cell types with path-dependent effects on protein accumulation.
In principle, altered protein levels of path mutants could be manifest at the level of either synthesis (mRNA translation) or decay (protein stability). To more directly examine the relationship between path function and translation in C4da neurons in vivo, we turned to cell type-specific Luciferase reporter assays. Similar in vivo reporter approaches have been used to dissect tissue-specific functions of other translational regulators (Schleich et al. 2014). We expressed a UAS-Rluc-Fluc reporter specifically in C4da neurons of path mutant larvae or wild-type controls and measured both protein and mRNA expression levels by Luciferase activity assays and quantitative RT–PCR (qRT–PCR), respectively. This revealed only a modest decrease in mRNA levels in path mutants (Fig. 6G). In contrast, Luciferase activity decreased to background levels in path mutants. These data are consistent with a strong decrease in translational output per mRNA in path mutant C4da neurons.
To further assess the cellular specificity of the response to path mutation, we monitored the effects of path mutation on growth of other larval cell types. However, larval cell types other than C4da/C3da neurons exhibited little or no dependence on path for growth (Fig. 6H). Finally, we used microarray analysis to survey gene expression responses to loss of path that were manifest across the larval body wall and observed minimal overlap with path-responsive changes in gene expression of C4da neurons (Fig. 6I; Supplemental Tables S3, S4). We conclude that path mutation induces a starvation response and alters protein homeostasis specifically in neurons with extreme growth requirements (C4da and C3da neurons).
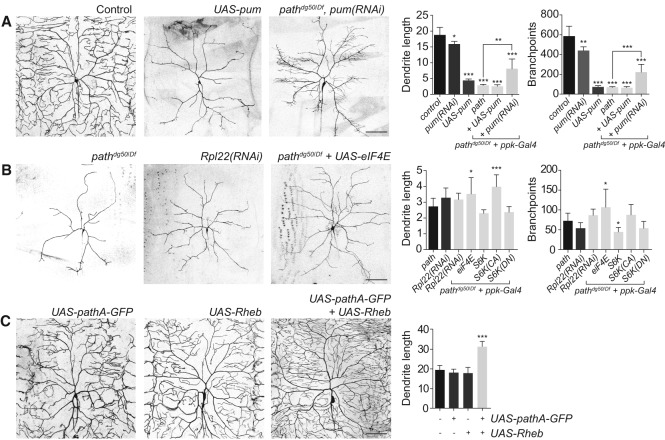
Our microarray analysis indicated that translational repressors, including pum, were up-regulated in path mutant C4da neurons, whereas core translational machinery was down-regulated; thus, we tested whether reducing pum function or increasing translational activity abrogated dendrite growth defects of path mutants. As previously reported (Ye et al. 2004), we found that pum overexpression strongly attenuated terminal dendrite growth in an otherwise wild-type background, but pum overexpression failed to enhance the growth defect of path mutants (Fig. 7A), consistent with pum up-regulation contributing to growth defects of path mutants. Whereas RNAi knockdown of pum in C4da neurons of otherwise wild-type larvae caused a modest growth deficit in C4da neurons, pum(RNAi) significantly increased dendrite growth in path mutant larvae (Fig. 7A), demonstrating that pum up-regulation is a functionally relevant output of path; up-regulation of other translational repressors (aret, bru-3, etc.) (Fig. 6C) may similarly contribute to path mutant dendrite growth defects.
Figure 7.
Path regulates dendrite growth via effects on translation. (A) Increased translational repression contributes to growth defects of path mutants. Representative images of C4da dendrites are shown for a wild-type control, neuronal expression of UAS-pum or UAS-pum(RNAi), and path mutant larvae expressing UAS-pum(RNAi) in neurons. Quantification of dendrite growth phenotypes is shown for the indicated genotypes. (B) Reduced translational capacity likely contributes to dendrite growth defects in path mutants. RNAi of core translational machinery such as Rpl22 causes growth defects similar to path, whereas overexpression of eIF4e partially mitigates path mutant growth defects. Quantification of dendrite growth phenotypes is shown for the indicated genotypes. (C) path can cooperate with TORC1 to promote dendrite growth. (Left) Representative C4da neurons expressing UAS-pathA-GFP, UAS-Rheb, or both are shown. (Right) Quantification of the effects of UAS-pathA-GFP and/or UAS-Rheb expression on C4da dendrite growth. n ≧ 7 cells for each bar. (*) P < 0.05; (**) P < 0.01; (***) P < 0.001; compared with wild-type controls in A and C and path mutants in B; one-way ANOVA with a post-hoc Dunnett's test. Error bars indicate standard deviation. Bars, 50 μm. Dendrite length measurements are in millimeters.
As with pum overexpression, RNAi knockdown of core translational machinery (Rpl22) caused dendrite growth defects comparable with path mutants (Fig. 7B), but the effects of Rpl22(RNAi) and path mutation were not additive, consistent with path affecting dendrite growth by modulating translation. If the dendrite growth defects in path mutants were due to reduced translational output, we reasoned, then treatments that increase translation should mitigate the dendrite growth defect of path mutants. This is what we observed. Overexpression of either eIF4E, which promotes cap-dependent translation (Richter and Sonenberg 2005) and is down-regulated in path mutant C4da neurons (Supplemental Fig. S9), or a constitutively active form of the ribosomal protein S6 kinase (CA-S6k), which promotes cell growth by increasing translational activity, increased C4da dendrite growth in path mutants (Fig. 7B). We note that the efficacy of these treatments in rescuing growth defects of path mutant neurons was limited, and this was likely due in part to the growth inhibitory effect of up-regulated translational repressors (Figs. 6C, 7A). Altogether, these results suggest that Path regulates dendrite growth at least in part via effects on translation.
The best-characterized substrates for SLC36 transporters are amino acids (Thwaites and Anderson 2011), and SLC36 transporters can modulate TORC1 activity in cultured cells (Heublein et al. 2010); therefore, we hypothesized that Path supports dendrite growth by modulating TORC1 activity in response to amino acids. If Path was functioning as a nutrient sensor for TORC1, we reasoned that TORC1 inactivation should mimic loss of path, and activation of TORC1 should bypass requirements for path. This is not what we observed. First, loss of neuronal TORC1 function (Tor mutation, Tor dominant negative, Raptor mutation, and RagA dominant negative) led to a less severe growth defect in C4da neurons than loss of path function (Supplemental Fig. S10), suggesting that path likely engages growth machinery other than just TORC1. Second, overexpression of Rheb, which can bypass the nutrient requirement for cell growth (Garami et al. 2003; Saucedo et al. 2003); Tor; raptor; or a constitutive active version of Akt, which can activate TORC2 in response to amino acid starvation under certain conditions (Tato et al. 2011), had no positive effect on dendrite growth of path mutants (Supplemental Fig. S10). However, coexpression of UAS-pathA-GFP and UAS-Rheb had a synergistic effect on dendrite growth (Fig. 7C), suggesting that path can stimulate dendrite growth together with TORC1. We conclude that Path likely functions through TORC1 as well as additional pathways to support the extreme growth requirements of neurons with large dendrite arbors.
Discussion
Neurons exhibit striking diversity in size and shape, but whether large neurons have specialized machinery to support their growth remains largely unknown. Our identification and characterization of Path demonstrates that, at least in Drosophila, neurons with large dendrite arbors have additional requirements to meet extreme growth demands. In neurons with large dendrite arbors, the response to path mutation is complex; despite manifesting signs of starvation and defects in protein homeostasis, primary dendrites continue to grow. Thus, dendrite patterning appears to be hierarchical and context-dependent: In path mutants, neurons lack the ability to generate material for overall arbor expansion; hence, material is preferentially allocated to facilitate primary dendrite elongation.
Independent of path, dendrites of PNS neurons grow to a maximum of ∼3.5 mm, and growth beyond this limit requires path. How this limit is determined is currently unknown. One possibility is that the limit may be the result of some cellular commodity being exhausted in the absence of path function. If this is the case, path-independent and path-dependent growth programs could use much of the same cellular machinery. Consistent with this scenario, neurons that do not require path for normal dendrite growth do require path for exuberant growth and hence have the machinery necessary for path-dependent growth. By extension, path may be broadly engaged in response to extreme growth demands and/or stress in neurons and nonneuronal cells, although path is dispensable for growth of most larval cell types under laboratory conditions.
Unexpectedly, the timing and pattern of C4da dendrite growth are remarkably labile. In the absence of path activity, dendrites grow to a maximum of ∼3.5 mm and persist at that size even as arbors are remodeled to maximize primary dendrite elongation. Resupplying path to these neurons even after several days of growth arrest completely reversed the growth defects despite the fact that structural plasticity in C4da dendrites, as in many other systems, is progressively limited (Ramoa et al. 1988; Grueber et al. 2003b; Sugimura et al. 2003; Parrish et al. 2009). Our findings suggest that restriction of structural plasticity in C4da neurons depends on the extent of dendrite growth during early developmental stages; it remains to be seen whether a similar relationship is manifest in other systems. Interestingly, resupplying path expression following several days of growth arrest led to more rapid dendrite growth than at any period in wild-type development, suggesting that growth inhibitory cues or other signaling pathways that coordinate dendrite growth and animal growth normally temper dendrite growth at earlier developmental stages.
How might Path regulate dendrite growth? The most straightforward model is that Path is required for transmembrane transport of essential amino acids, which in turn modulate activity of mTORC1 and other amino acid-responsive signaling pathways. Path localizes to the cell surface and endolysosomal compartments; thus, it could be involved in transporting amino acids into the cell or out of the lysosome. Several lines of evidence suggest that either scenario is unlikely. First, although one vertebrate Path counterpart, SLC36A1, promotes amino acid transport out of lysosomes and, as a result, antagonizes mTORC1 activity (Zoncu et al. 2011), Path is most closely related to SLC36A4 (Schiöth et al. 2013), which has no effect on lysosomal amino acid content. Second, Path is most active at neutral pH (Goberdhan et al. 2005); thus, it seems unlikely that it is involved in amino acid transport out of lysosomes. Third, although Path displays high affinity for a variety of amino acids, it exhibits minimal processivity in transport assays (Goberdhan et al. 2005), so it seems unlikely that it would substantially contribute to amino acid flux.
While it is possible that Path requires an unidentified cofactor for maximal transport activity, an alternative explanation is that Path may function as a transceptor, a transporter-like protein with a receptor function, to regulate signaling in response to metabolites independently of its transport capacity (Thevelein and Voordeckers 2009). Examples of transceptors include the system A amino acid transporter SNAT2, which regulates transcription in response to amino acid binding (Hyde et al. 2007), and GLUT2, a sugar transporter that has transport-independent functions as a receptor for extracellular glucose (Stolarczyk et al. 2010). Thus, Path could function at the cell surface to report on environmental conditions, gating downstream signaling in response to extracellular amino acid availability. In this model, low levels of extracellular amino acids or the absence of Path would signal the cell to marshal its resources and arrest dendrite growth, whereas high levels of amino acids would activate signaling that regulates protein homeostasis, supporting extreme dendrite growth. With this in mind, it will be intriguing to further characterize the N-terminal intracellular domain, as the N-terminal domain is a likely site for protein–protein interactions and is required for Path function.
Many degenerative disorders preferentially affect neurons with large axons/dendrites, including Purkinje neurons, Betz cells, motoneurons, and sensory neurons. Thus, it is plausible that defects in growth machinery that contribute to extreme neuron growth contribute to pathology in these diseases. Path orthologs are present in vertebrates (Schiöth et al. 2013), but their in vivo function remains unknown. Interestingly, two mammalian SLC36 transporters—SLC36A1 and SLC36A4—are highly expressed in the nervous system (Sagné et al. 2001; Bermingham and Pennington 2004; Roshanbin et al. 2014). Thus, it will be of great interest to determine whether these SLC36 transporters are required for extreme dendrite growth similar to Path.
Materials and methods
Fly husbandry
A list of alleles used in this study is available in the Supplemental Material. EMS mutagenesis was conducted as previously described (Lee et al. 2015). Transgenic lines were generated using P-element-mediated integration, except for UAS-Luciferase reporter lines, which were generated using ϕC31-mediated integration.
Live imaging
Embryos were collected on yeasted grape juice agar and aged at 25°C in a moist chamber. At the appropriate time, a single embryo/larva was mounted in 90% glycerol under coverslips sealed with grease and imaged on a Leica SP5 microscope with a 40× 1.25 NA lens. For quantitation of dendrite phenotypes, image stacks of dendrites in segments A3–A4 were captured from eight to 10 larvae. For time-lapse analysis, larvae were imaged at the indicated time, recovered to yeasted agar plates with vented lids, aged at 25°C, and imaged again.
Immunohistochemistry
Larval fillets were dissected/processed as described (Grueber et al. 2002) with the exception of tissue for anti-Path staining, which was fixed in 10% TCA for 15 min, washed five times in PBS-Triton X-100 (0.3%), and then processed as other tissue. Samples were stained with the following antibodies: HRP conjugated with Cy2 or Cy3 (1:200; Jackson ImmunoResearch), mCD8 (1:100; Life Technologies), anti-Path (1:500), 50 ng/mL DAPI (Life Technologies), and secondary antibodies (1:250) from Jackson ImmunoResearch.
Surface staining
Tissue was fixed in 4% formaldehyde/PBS, and subsequent washes and antibody incubations were conducted in the absence of detergent to prevent cell permeabilization.
Path antibody
Anti-Path antibodies were produced in guinea pigs using a KLH-conjugated peptide derived from the N-terminal cytosolic domain (KIQPRKSDTEQALAGN) common to all predicted Path polypeptides (Pierce Antibodies). Antibodies were affinity-purified using the antigenic peptide. Specificity was confirmed by immunohistochemistry of path mutant body wall preparations.
Molecular biology
UAS-path transgenes
The path-RA (path-long) and its derivatives were PCR-amplified from cDNA clone RH24992 (Drosophila Genomics Resource Center); path-RE (path-short) and its derivatives were isolated via RT–PCR from larval body wall RNA.
ss-myr-GFP transgene
The GMR15F10 ss enhancer (Pfeiffer et al. 2008) was PCR-amplified and shuttled into a Gateway destination cassette (Life Technologies) that was cloned into pJFRC19 as a HindIII/SpeI fragment.
UAS-Rluc-Fluc bicistronic reporter transgene
A bicistronic reporter containing the Renilla luciferase coding sequence followed by the CrPV IGRI IRES driving Firefly luciferase (pUASTattB-Rluc-IRES-Fluc) was amplified by PCR from a mammalian expression vector (Landry et al. 2009) using primer AGD 215_bicis-F (ATCGCGGCCGCACTAGTATGACTTCGAAAGTTTATGA) and reverse primer AGD216_bicis-R (ATTGGTACCACCGGTGCTAGCTTACACGGCGATCTTTCCGC). The fragment was cloned into the pUASTattB vector using NotI and KpnI restriction sites, and transgenic lines were generated by targeted insertion into an attP docker line at 22A3 (Bestgene).
Luciferase assays
Larvae (120 h AEL) were filleted in ice-cold PBS (10 per sample), and body wall tissue was lysed with 1× luciferase cell culture lysis reagent (Promega). Total protein concentration of each lysate was measured using a quick-start Bradford protein assay (Bio-Rad), and the concentrations of all samples were normalized by diluting lysates in lysis reagent as needed. Twenty microliters of cell lysate was analyzed for Renilla luciferase using the Luciferase assay system (Promega), and luminescence was measured using a Victor V plate reader (PerkinElmer). All values were corrected to blank wells. For measuring relative Renilla transcript levels, SYBR Green-based qRT–PCRs were used. RNA purification was performed with a RNAqueous-Micro total RNA isolation kit (Thermo Fisher). qRT–PCRs were done as previously described. β-Tubulin was used as a normalization control for experiments analyzing mRNAs. Primer sequences for Renilla and β-tubulin were as follows: Ren F, GGGTGCTTGTTTGGCATTTC; Ren R, GGCCATTCATCCCATGATTC; β-tub F, AGACAAGATGGTTCAGGT; and β-tub R, CGAGGCTCTCTACGATAT.
Measurements
Two-dimensional (2D) projections of Z-stacks were used for computer-assisted dendrite tracing with Neurolucida (MBF Bioscience), and features were measured using the traces. To monitor expression of GFP exon traps, we traced the outline of cells of interest in 2D projections of Z stacks using ImageJ and measured the mean pixel intensity within the cell. For these experiments, we imaged control and path mutant larvae using identical settings, including the same number and thickness of optical sections.
Statistical analysis
Differences between group means were analyzed via ANOVA with a post-hoc Dunnett's test; pairwise comparisons of group means were done with unpaired t-tests with Welch's correction.
Accessions
Microarray data are available at the NCBI Gene Expression Omnibus (accession no. GSE64477).
Supplementary Material
Acknowledgments
We thank the Bloomington Drosophila Stock Center, Peter Soba, Bing Ye, and Tom Neufeld for fly stocks; Vassie Ware, Paul Lasko, and the Developmental Studies Hybridoma Bank for antibodies; the Drosophila Genomics Resource Center and Robert Edwards for cDNA clones; Sunnie Thompson for providing plasmids used to generate UAS-Luciferase reporters; Marvin Nayan for assistance with EMS screening; Jon Ahn for assistance with stereology; Peter Soba and Kazuo Emoto for advice and reagents; and Michael Kim and Dave Parichy for critical reading of the manuscript. This work was supported by National Institute of Mental Health R00-MH084277, National Institute of Neurological Disorders and Stroke R01-NS076614, a March of Dimes Basil O'Connor Starter Scholar Award, and a Klingenstein Fellowship in Neuroscience to J.Z.P.; a Benjamin Hall Graduate Fellowship to W.-Y.L.; a National Science Foundation Graduate Research Fellowship (DGE-1256032) to C.W.; and a National Institutes of Health Institutional Training Grant for Neurobiology (T32-GM007108) to K.L.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.259119.115.
References
- Bermingham JR, Pennington J. 2004. Organization and expression of the SLC36 cluster of amino acid transporter genes. Mamm Genome 15: 114–125. [DOI] [PubMed] [Google Scholar]
- Bloomfield SA, Hitchcock PF. 1991. Dendritic arbors of large-field ganglion cells show scaled growth during expansion of the goldfish retina: a study of morphometric and electrotonic properties. J Neurosci 11: 910–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiala JC, Spacek J, Harrish KM. 2008. Dendrite structure. In Dendrites (ed. Stuart G, et al. ), pp. 1–41. Oxford University Press, New York. [Google Scholar]
- Garami A, Zwartkruis FJT, Nobukuni T, Joaquin M, Roccio M, Stocker H, Kozma SC, Hafen E, Bos JL, Thomas G. 2003. Insulin activation of Rheb, a mediator of mTOR/S6K/4E-BP signaling, is inhibited by TSC1 and 2. Mol Cell 11: 1457–1466. [DOI] [PubMed] [Google Scholar]
- Goberdhan DCI, Meredith D, Boyd CAR, Wilson C. 2005. PAT-related amino acid transporters regulate growth via a novel mechanism that does not require bulk transport of amino acids. Development 132: 2365–2375. [DOI] [PubMed] [Google Scholar]
- Grueber WB, Jan LY, Jan YN. 2002. Tiling of the Drosophila epidermis by multidendritic sensory neurons. Development 129: 2867–2878. [DOI] [PubMed] [Google Scholar]
- Grueber WB, Jan LY, Jan YN. 2003a. Different levels of the homeodomain protein cut regulate distinct dendrite branching patterns of Drosophila multidendritic neurons. Cell 112: 805–818. [DOI] [PubMed] [Google Scholar]
- Grueber WB, Ye B, Moore AW, Jan LY, Jan YN. 2003b. Dendrites of distinct classes of Drosophila sensory neurons show different capacities for homotypic repulsion. Curr Biol 13: 618–626. [DOI] [PubMed] [Google Scholar]
- Heublein S, Kazi S, Ogmundsdóttir MH, Attwood EV, Kala S, Boyd CAR, Wilson C, Goberdhan DCI. 2010. Proton-assisted amino-acid transporters are conserved regulators of proliferation and amino-acid-dependent mTORC1 activation. Oncogene 29: 4068–4079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyde R, Cwiklinski EL, MacAulay K, Taylor PM, Hundal HS. 2007. Distinct sensor pathways in the hierarchical control of SNAT2, a putative amino acid transceptor, by amino acid availability. J Biol Chem 282: 19788–19798. [DOI] [PubMed] [Google Scholar]
- Jiang N, Soba P, Parker E, Kim CC, Parrish JZ. 2014. The microRNA bantam regulates a developmental transition in epithelial cells that restricts sensory dendrite growth. Development 141: 2657–2668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry DM, Hertz MI, Thompson SR. 2009. RPS25 is essential for translation initiation by the Dicistroviridae and hepatitis C viral IRESs. Genes Dev 23: 2753–2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T, Luo L. 1999. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron 22: 451–461. [DOI] [PubMed] [Google Scholar]
- Lee A, Li W, Xu K, Bogert BA, Su K, Gao F-B. 2003. Control of dendritic development by the Drosophila fragile X-related gene involves the small GTPase Rac1. Development 130: 5543–5552. [DOI] [PubMed] [Google Scholar]
- Lee J, Peng Y, Lin W-Y, Parrish JZ. 2015. Coordinate control of terminal dendrite patterning and dynamics by the membrane protein Raw. Development 142: 162–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin B, Wang SW, Masland RH. 2004. Retinal ganglion cell type, size, and spacing can be specified independent of homotypic dendritic contacts. Neuron 43: 475–485. [DOI] [PubMed] [Google Scholar]
- Mayer C, Grummt I. 2006. Ribosome biogenesis and cell growth: mTOR coordinates transcription by all three classes of nuclear RNA polymerases. Oncogene 25: 6384–6391. [DOI] [PubMed] [Google Scholar]
- Montague PR, Friedlander MJ. 1991. Morphogenesis and territorial coverage by isolated mammalian retinal ganglion cells. J Neurosci 11: 1440–1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin X, Daneman R, Zavortink M, Chia W. 2001. A protein trap strategy to detect GFP-tagged proteins expressed from their endogenous loci in Drosophila. Proc Natl Acad Sci 98: 15050–15055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrish JZ, Xu P, Kim CC, Jan LY, Jan YN. 2009. The microRNA bantam functions in epithelial cells to regulate scaling growth of dendrite arbors in Drosophila sensory neurons. Neuron 63: 788–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrimon N. 1984. Clonal analysis of dominant female-sterile, germline-dependent mutations in Drosophila melanogaster. Genetics 108: 927–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer BD, Jenett A, Hammonds AS, Ngo T-TB, Misra S, Murphy C, Scully A, Carlson JW, Wan KH, Laverty TR, et al. 2008. Tools for neuroanatomy and neurogenetics in Drosophila. Proc Natl Acad Sci 105: 9715–9720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purves D, Lichtman JW. 1985. Geometrical differences among homologous neurons in mammals. Science 228: 298–302. [DOI] [PubMed] [Google Scholar]
- Rambold AS, Kostelecky B, Elia N, Lippincott-Schwartz J. 2011. Tubular network formation protects mitochondria from autophagosomal degradation during nutrient starvation. Proc Natl Acad Sci 108: 10190–10195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramoa AS, Campbell G, Shatz CJ. 1988. Dendritic growth and remodeling of cat retinal ganglion cells during fetal and postnatal development. J Neurosci 8: 4239–4261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter JD, Sonenberg N. 2005. Regulation of cap-dependent translation by eIF4E inhibitory proteins. Nature 433: 477–480. [DOI] [PubMed] [Google Scholar]
- Roshanbin S, Hellsten SV, Tafreshiha A, Zhu Y, Raine A, Fredriksson R. 2014. PAT4 is abundantly expressed in excitatory and inhibitory neurons as well as epithelial cells. Brain Res 1557: 12–25. [DOI] [PubMed] [Google Scholar]
- Sagné C, Agulhon C, Ravassard P, Darmon M, Hamon M, El Mestikawy S, Gasnier B, Giros B. 2001. Identification and characterization of a lysosomal transporter for small neutral amino acids. Proc Natl Acad Sci 98: 7206–7211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saucedo LJ, Gao X, Chiarelli DA, Li L, Pan D, Edgar BA. 2003. Rheb promotes cell growth as a component of the insulin/TOR signalling network. Nat Cell Biol 5: 566–571. [DOI] [PubMed] [Google Scholar]
- Schiöth HB, Roshanbin S, Hägglund MGA, Fredriksson R. 2013. Evolutionary origin of amino acid transporter families SLC32, SLC36 and SLC38 and physiological, pathological and therapeutic aspects. Mol Aspects Med 34: 571–585. [DOI] [PubMed] [Google Scholar]
- Schleich S, Strassburger K, Janiesch PC, Koledachkina T, Miller KK, Haneke K, Cheng YS, Kuchler K, Stoecklin G, Duncan KE, et al. 2014. DENR-MCT-1 promotes translational re-initiation downstream of uORFs to control tissue growth. Nature 512: 208–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CJ, O'Brien T, Chatzigeorgiou M, Spencer WC, Feingold-Link E, Husson SJ, Hori S, Mitani S, Gottschalk A, Schafer WR, et al. 2013. Sensory neuron fates are distinguished by a transcriptional switch that regulates dendrite branch stabilization. Neuron 79: 266–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolarczyk E, Guissard C, Michau A, Even PC, Grosfeld A, Serradas P, Lorsignol A, Pénicaud L, Brot-Laroche E, Leturque A, et al. 2010. Detection of extracellular glucose by GLUT2 contributes to hypothalamic control of food intake. Am J Physiol Endocrinol Metab 298: E1078–E1087. [DOI] [PubMed] [Google Scholar]
- Sugimura K, Yamamoto M, Niwa R, Satoh D, Goto S, Taniguchi M, Hayashi S, Uemura T. 2003. Distinct developmental modes and lesion-induced reactions of dendrites of two classes of Drosophila sensory neurons. J Neurosci 23: 3752–3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tato I, Bartrons R, Ventura F, Rosa JL. 2011. Amino acids activate mammalian target of rapamycin complex 2 (mTORC2) via PI3K/Akt signaling. J Biol Chem 286: 6128–6142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teleman AA, Chen Y-W, Cohen SM. 2005. 4E-BP functions as a metabolic brake used under stress conditions but not during normal growth. Genes Dev 19: 1844–1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tettweiler G, Miron M, Jenkins M, Sonenberg N, Lasko PF. 2005. Starvation and oxidative stress resistance in Drosophila are mediated through the eIF4E-binding protein, d4E-BP. Genes Dev 19: 1840–1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thevelein JM, Voordeckers K. 2009. Functioning and evolutionary significance of nutrient transceptors. Mol Biol Evol 26: 2407–2414. [DOI] [PubMed] [Google Scholar]
- Thwaites DT, Anderson CMH. 2011. The SLC36 family of proton-coupled amino acid transporters and their potential role in drug transport. Br J Pharmacol 164: 1802–1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomancak P, Beaton A, Weiszmann R, Kwan E, Shu S, Lewis SE, Richards S, Ashburner M, Hartenstein V, Celniker SE, et al. 2002. Systematic determination of patterns of gene expression during Drosophila embryogenesis. Genome Biol 3: RESEARCH0088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G. 2001. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci 98: 5116–5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wreden CC, Johnson J, Tran C, Seal RP, Copenhagen DR, Reimer RJ, Edwards RH. 2003. The H+-coupled electrogenic lysosomal amino acid transporter LYAAT1 localizes to the axon and plasma membrane of hippocampal neurons. J Neurosci 23: 1265–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye B, Petritsch C, Clark IE, Gavis ER, Jan LY, Jan YN. 2004. Nanos and Pumilio are essential for dendrite morphogenesis in Drosophila peripheral neurons. Curr Biol 14: 314–321. [DOI] [PubMed] [Google Scholar]
- Zoncu R, Bar-Peled L, Efeyan A, Wang S, Sancak Y, Sabatini DM. 2011. mTORC1 senses lysosomal amino acids through an inside-out mechanism that requires the vacuolar H+-ATPase. Science 334: 678–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.