Abstract
The understanding of the complex role of the bile acid-gut microbiome axis in health and disease processes is evolving rapidly. Our focus revolves around the interaction of the gut microbiota with liver diseases, especially cirrhosis. The bile acid pool size has recently been shown to be a function of microbial metabolism of bile acid and regulation of the microbiota by bile acids is important in the development and progression of several liver diseases. Humans produce a large, conjugated hydrophilic bile acid pool, maintained through positive-feedback antagonism of FXR in intestine and liver. Microbes use bile acids, and via FXR signaling this results in a smaller, unconjugated hydrophobic bile acid pool. This equilibrium is critical to maintain health. The challenge is to examine the manifold functions of gut bile acids as modulators of antibiotic, probiotic and disease progression in cirrhosis, metabolic syndrome and alcohol use. Recent studies have shown potential mechanisms explaining how perturbations in the microbiome affect bile acid pool size and composition. With advancing liver disease and cirrhosis, there is dysbiosis in the fecal, ileal and colonic mucosa, in addition to a decrease in bile acid concentration in the intestine due to the liver problems. This results in a dramatic shift toward the Firmicutes, particularly Clostridium cluster XIVa and increasing production of deoxycholic acid (DCA). Alcohol intake speeds up these processes in the subjects with and without cirrhosis without significant FXR feedback. Taken together, these pathways can impact intestinal and systemic inflammation while worsening dysbiosis. The interaction between bile acids, alcohol, cirrhosis and dysbiosis is an important relationship that influences intestinal and systemic inflammation, which in turn determines progression of the overall disease process. These interactions and the impact of commonly used therapies for liver disease can provide insight into the pathogenesis of inflammation in humans.
Introduction
A new concept has emerged in recent years regarding the human body. It is a complex ecosystem constituted on a cellular basis primarily of prokaryotes and archaea whose gene content is estimated to encode 99% of functional genes (1). Colonized at birth, or perhaps prior to (2), a series of stochastic events and selection pressures culminate in what is recognized as an “adult” microbiota by the first year of age (3). Microbial density in the large bowel reaches an impressive 1011 cells cm−3. Of the 55 bacterial known phyla, selection pressures winnow diversity down to two predominant phyla, the Firmicutes and the Bacteroidetes, in addition to minor representation by Actinobacteria and Proteobacteria (4). The Bacteroidetes are gram-negative, non-sporeforming, anaerobic, rod-shaped bacteria. The Firmicutes inhabiting the gut are gram-positive, anaerobic, low G+C bacteria. As an example of the taxonomic hierarchy, the secondary bile acid producing species, Clostridium scindens, is found in the genus Lachnoclostridium, within the family Lachnospiraceae, within the order Clostridiales, within the class Clostridia, within the phylum Firmicutes, within the kingdom Bacteria and the domain Prokarya. Bile acids represent a major positive or negative selective pressure on the gut microbome. Bile acids accomplish negative selection pressures directly through antimicrobial properties and indirectly through activation of FXR-induced anti-microbial peptide synthesis in the small bowel (5). Conversely, bile acids elicit positive selection pressures directly through growth stimulation by biotransformations of the steroid nucleus and hydroxy groups that provide a source of or sink for electrons during fermentative metabolism (Figure 1). Bile salt deconjugation is a source of amino acids taurine and glycine that are known to cause blooms of certain microbial taxa of medical importance (6). In addition, the production of secondary bile acids by certain gut microbes may represent an important means whereby competitors are excluded given that secondary bile acids are significantly more toxic to certain bacteria than primary bile acids. We will explore here the human experience of the bile acid-gut microbiome-liver axis in health and cirrhosis.
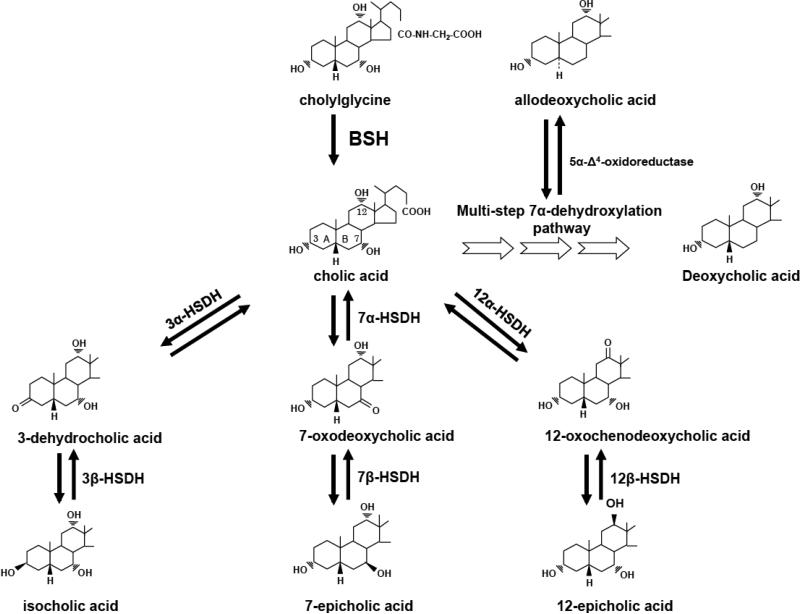
Figure 1. Bacterial bile salt-biotransforming reactions in the human intestinal tract.
The 3, 7, and 12 hydroxy group carbons of cholic acid are numbered and the AB rings are identified. BSH, bilesalt hydrolase; HSDH, hydroxysteroid dehydrogenase. Reprinted from Ref. 16.
Modification of bile acids by microbiota in healthy individuals
Exogenous and endogenous compounds that enter the gut rapidly affect the structure and function of the gut microbiome. Dietary components such as fats can increase the levels of bile acids entering the gut. Work done during the 1970's and 1980's firmly demonstrated that diets rich in animal protein and fat significantly increased total fecal bile acids (7, 8). A diet high in animal protein and saturated fat stimulates bile secretion through enhanced signaling by cholecystokinin. Consumption of a Western-style diet (high saturated-fat/high simple sugar) shifts the microbiome structure toward bile-resistant members, capable of producing toxic and pro-inflammatory products from bile salts, namely secondary bile acids (9) and hydrogen sulfide from taurine (6). Studies coupling gnotobiotic mice with metatranscriptomics and metabolomics demonstrate that simple-sugar diets promote degradation of protective host mucin providing greater access of toxic microbial metabolites to the gut epithelium (10, 11). Rapid alteration of the gut microbiome through animal-based diet has been observed to significantly increase fecal bile acid levels, the activity of microbial bile acid modifying enzymes in feces (9). Plant-based diets have been shown to result in lower fecal bile acid levels compared to animal-based diets (9).
Indeed, bile acid feeding largely reproduces the effects observed in high-animal protein and fat diets (12, 13). Expansion of the Firmicutes at the expense of Bacteroidetes, reduced bacterial diversity and density was observed during cholic acid feeding (12). Rats fed a “medium cholic acid diet” (est. 0.98 mM) or “high cholic acid diet” (est. 2.55 mM) resulted in shifts in a balanced Firmicutes/Bacteroides ratio of 54.1%/30.7% to 98.6% of the microbiome represented by Firmicutes (12). Bile acids, particularly DCA, are thus a major factor shaping the mammalian gut microbiome.
While the mammalian liver is responsible for producing bile acids in the body, germ-free animals retain a global pool of only primary bile acids (14). Thus, early studies in germ-free animals demonstrated that gut microbes have evolved biochemical pathways to modify host bile acids. Indeed, host-microbe coevolution is evident as the secondary bile acids, lithocholic acid, is a high affinity ligand for the vitamin D receptor, while deoxycholic acid is a high affinity activator of TGR-5 (15). As bile salts transit through the small bowel, populations of bacteria increase from 103 per ml in the duodenum to approaching 1011 per ml as bile salts enter the large bowel (16). Species of Lactobacillus, and members of the family Enterobacteriaceae express bile salt hydrolase (BSH) enzymes capable of hydrolyzing bile salts to free taurine and glycine and bile acids (16). As expected, overexpression of recombinant BSH by E. coli in the small bowel of mice resulted in malabsorption of lipids due presumably to reduced capacity to form mixed micelles (17). Significant weight loss, reduction in serum cholesterol and triglycerides, and increased BA synthesis ensues through expression of BSH relative to BSH- E. coli control (17). Disruption of mixed micelle formation would lead to significant loss of nutrients into the colon, and would be expected to alter the microbiome composition. It may be speculated that sub-therapeutic levels of antibiotics fed to agricultural animals may result in weight gain through reduced BSH activity in the small bowel.
In the large bowel, BSH is a redundant feature of the microbiome, and one of significant importance to individual microbes, and presumably for the overall microbiome. Functional metagenomic screening of 101 BSH-positive clones from a human fecal sample found that of those with taxonomic information, BSH were located in the three major bacterial divisions Firmicutes (30%), Bacteroidetes (14.4%) and Actinobacteria (8.9%) (18). Indeed, the only two archaeal species known to inhabit the human gut, Methanobrevibacter smithii and Methanospera stadmanae possess BSH (18). To date, no BSH inhibitors have been reported, though the effect of inhibiting BSH on microbiome structure and function would be of significant interest. It is well known for instance, that many pathogens (19) and indeed probiotic bacteria require BSH to colonize (20). The antimicrobial nature of bile salts, particularly glycine-conjugates, suggests that one role of BSH is detoxification (16,19). Even if a particular microbial species, or several lack BSH, they are in a sense “covered” due to the presence other species that rapidly hydrolyze bile salts. It may be that natural selection has favored this redundancy of BSH across diverse taxonomic groups to maintain microbiome functional homeostasis in the face of perturbations that may affect one taxonomic group more severely than others (3). If an important function, such as BSH is limited to a narrow taxonomy, the loss of BSH function may become a perturbation with cascading effects altering microbiome structure/function and thus the host.
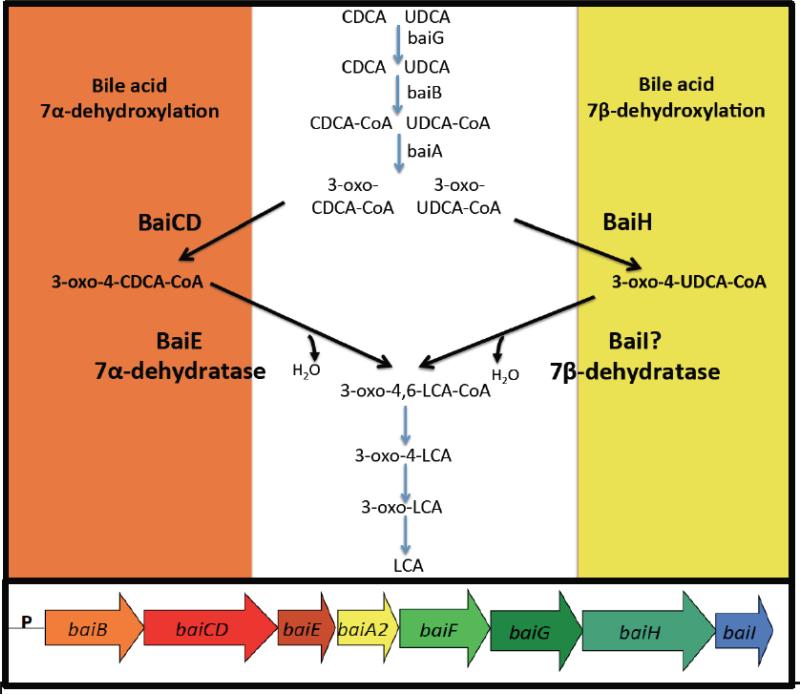
In contrast to BSH, the ability to produce secondary bile acids is limited to a few species in the genus Clostridium, gram positive members of the Firmicutes (16). Their numbers average in healthy individuals 103 to 105 per gram wet weight. Representing less than 0.025% of the microbiome by cell density, they none-the-less are capable of converting several hundred milligrams of primary bile acids to secondary bile acids daily. Bile acid 7α-dehydroxylation of human primary bile acids cholic acid and chenodeoxycholic acid proceeds by a complex, multi-step pathway culminating in more hydrophobic bile acids deoxycholic acid, and lithocholic acid, respectively (16). Lithocholic acid can also be generated from the 7β-dehydroxylation of ursodeoxycholic acid (UDCA). UDCA can be introduced exogenously via therapies (Ursodiol), or endogenously produced by epimerization of the 7α-hydroxy by 7α-hydroxysteroid dehydrogenases (HSDH) and 7β-dehydrogenases found in single species (Clostridium absonum) or two separate species expressing one or the other HSDH (21). The evolution of the 7β-dehydroxylation pathway appears to have arisen by gene-duplication events. There is significant genetic and enzymatic overlap in the bile acid 7α-dehydroxylation and the bile acid 7β-dehydroxylation pathways (16). Transport (baiG), ligation to Coenzyme A (baiB, baiF), oxidation of the 3α-hydroxy (baiA) and the reductive arm of the pathway are common to both 7α-hydroxy and 7β-hydroxy bile acids (Figure 2). The two unique steps, oxidation of C4-C5 and bile acid 7-dehydration require separate, but homologous enzymes. The baiE and baiI encode a bile acid 7α-dehydratase, and putative bile acid 7β-dehydratase, respectively (Figure 2), are found on the same multi-gene operon, and encode homologous enzymes in the SnoEL family of proteins. The baiCD and baiH are also encoded on the same operon, and are clearly homologous sharing high amino acid sequence identity and are both members of the “Old Yellow” family of proteins (22). It is suggested that epimerization of CDCA to UDCA functions to form a less toxic bile acid, and equilibrium constants favor generation of UDCA (23). Thus, duplication and subsequent divergence of the baiE and baiCD toward recognition of 7β-hydroxy-bile acids would open up an additional niche. Bile acid 7α-dehydroxylating bacteria express a 7α-HSDH, possibly to regulate flux of primary bile acids through the pathway (7-oxo-bile acids cannot be 7α/β-dehydroxylated) and would be necessary to reduce 7-oxo-bile acids generated from microbial species expressing 7α/β-HSDH. Removal of the 7-hydroxy group yields a net 2 reduction; bile acids are electron sinks for these fermenting bacteria. However, this small population of microbes has a disproportionate effect on the structure of the microbiome as DCA is ten times more antimicrobial than CA.
Figure 2. Common and unique biochemical steps in the bile acid 7α-dehydroxylating and 7β-dehydroxylating pathways in Clostridium scindens.
Steps unique to bile acid 7α-DeOH encoded by baiCD and baiE genes (orange region). Step unique to 7β-DeOH of UDCA encoded by baiH gene (yellowr region). baiI gene hypothesized to encode bile acid 7β-dehydratase. Below the pathway is the organization of bai genes in C.scindens.
It is currently unclear whether these organisms are capable of persisting without bile acids, i.e. if we could specifically inhibit the bile acid 7α-dehydroxylation pathway, would these bacteria be outcompeted? What would the structure/function of the microbiome look like if the bile acid 7α-dehydroxylation pathway were inhibited? Such inhibitors may be very useful in preventing and/or treating diseases of the GI tract such as colon cancer, gallstone disease in certain patients. These questions await development of specific inhibitors.
Cirrhosis, bile acids and microbiome
High concentrations of bile salts (mM), and rapid transit time reduces significant bacterial growth in the small bowel as well as competition with the host for nutrients (16, 19). Bile salts also induce the expression of anti-microbial peptides through activation of the FXR demonstrating an indirect mechanism by which bile salts exert control over the location and community structure of the microbiome in the gut (5). As a consequence of end-stage liver disease, cirrhosis, there is a significant reduction in bile flow in the intestines. Since bile acid feeding results in a significant shift in the microbiome toward the Clostridiales within the Firmicutes (12), it may be predicted that decreasing bile acid concentrations in the gut might lead to expansion of taxa at the expense of several taxonomic groups within the Firmicutes.
Recent studies have defined changes in the gut microbiome that occur during cirrhosis (24-27). Beneficial taxa within the Firmicutes such as Lachnospiraceae, Roseburia, Rumminococcaceae and Blautia are underrepresented in cirrhosis (24-27). Members of these taxa are part of the normal microbiota and are key producers of the beneficial short-chain fatty acid, butyrate. These taxa also include members with bile acid 7α-dehydroxylation activity (28). Indeed, conversion of primary bile acids to secondary bile acids is significantly reduced in cirrhotic patients (29).
Several studies have suggested that members of the oral microbiome occurs in the context of cirrhosis, suggesting that bile salts and gastric acid normally exclude oral microbes from colonizing the lower GI tract (25, 27). Gram negative members of the Alcaligenaceae, Enterobacteriaceae, Porphyromonadaceae, and gram positives in the Streptococcaceae are overrepresented in stool of cirrhotic patients (24-27). We have previously shown a direct relationship between cognitive impairment (hepatic encephalopathy) in cirrhotic patients and levels of Alcaligeneaceae and Porphyromonadaceae (24). A recent comparison of gut microbial genome content (metagenome) between cirrhotics and controls suggests enrichment for genes involved in ammonia production, γ-aminobutyric acid (GABA) production, and manganese transport systems, each of which is suggested to play a mechanistic role in cognitive problems associated with liver cirrhosis (27). Further functional genomic approaches (metatranscriptomics) and metabolomics will be necessary in order to determine the role of these products in disease. Reduced colonic levels of bile acids is hypothesized to play an important role in the dysbiosis characterized by a pro-inflammatory, toxic gut microbiome (Figure 3) (27-29).
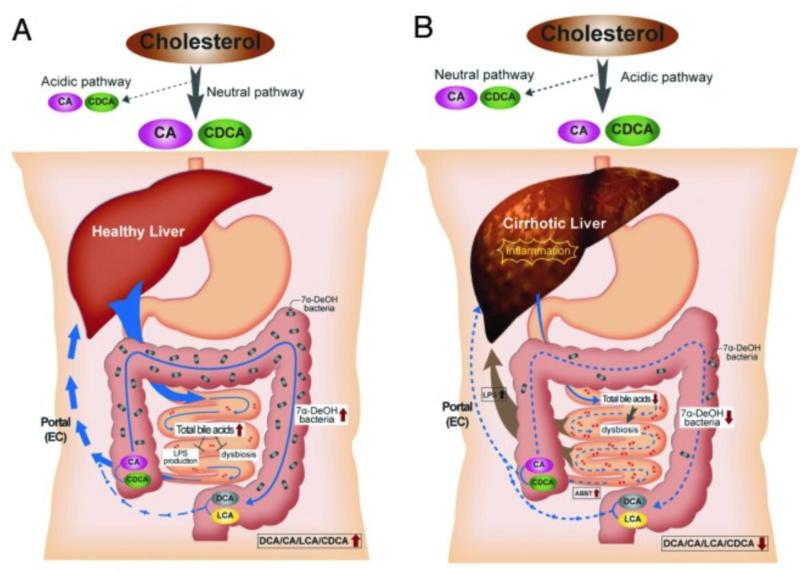
Figure 3. Proposed model for the relationship between bile acids, the microbiome and cirrhosis.
A. In healthy individuals, cholesterol is primarily converted to CA and CDCA by the neutral bile acid biosynthetic pathway. Sufficient quantities of bile salts enter the small intestine to prevent dysbiosis and the release of inflammatory markers (i.e., LPS). Bile acid 7α-dehydroxylating bacteria are found in normal range (103–105 cells per gram wet weight), and the ratio of secondary to primary bile acids in stool is high. B. In cirrhosis, the neutral pathway is repressed due to downregulation of CYP7A1 by proinflammatory cytokines, and the acidic pathway is the primary pathway for bile acid synthesis. Dysbiosis occurs due to lower concentration of bile salts entering the small bowel. This dybiosis is characterized by inflammation due to an increase in organisms with potent LPS such as members of the Enterobacteriaceae. The population of 7α-dehydroxylating bacteria in the colon is hypothesized to decrease due to lower levels of primary bile acids which are thought to serve the as an energy source. Consequently, ratio of secondary/primary bile acids is low in cirrhosis. Reproduced with permission from Gut Microbes (28)
Effects of alcohol on fecal bile acid levels and the microbiome
A link has been found between the secondary bile acid DCA, hepatic stellate cells and hepatocarcinoma in a mouse model of liver cancer (30). Bile acid products of the gut microbiome may induce inflammation in liver leading to cirrhosis, as stellate cells are active in this process (31). Alcohol is known to stimulate bile acid synthesis in humans (31). Recently, it has been reported that alcohol induces bile acid synthesis through activation of hepatic cannabinoid receptor type 1 (Cb1r) and CREBH (32). Kakiyama et al (2014) have shown that cirrhotic patients who are currently drinking have significantly higher levels of fecal secondary bile acids compared to cirrhotic patients abstaining from alcohol (> 6 months), nonalcoholic cirrhotics, drinkers without cirrhosis and even healthy control who abstain from alcohol (33). Cirrhotic patients actively drinking also showed significant increases in mRNA of proinflammatory markers (TNF-α, IL-6, IL-1β, MCP-1) in colonic but not ileal tissue compared with all other groups (33). COX-2 was also significantly higher in both ileum and colon of actively drinking cirrhotic patients relative to other groups; however, mRNA levels were highest in the colon (33). This study by Kakiyama et al. (2014) also suggested that alcohol-induced activation of Cb1r overrides downregulation of Cyp7A1 by FGF-19 (33). This is supported by increased serum levels of conjugated-DCA, and by the finding that mRNA levels of FXR and FGF-19, but not SHP, were increased in the ileum and sigmoid colon of drinkers but not nonalcoholic cirrhotics or control patients (33).
Total fecal bile acids and fecal secondary bile acids in currently drinking alcoholic cirrhotics were three to four times the levels of both nonalcoholic and abstinent alcoholic cirrhotic patients (8.9 μmol/g vs. 2.9 μmol/g vs. 2.2 9 μmol/g, respectively) (33). DCA as been reported to activate cell-signaling pathways (EGFR, AKT, ERK 1/2, PKC, β-catenin, Cox-2), generate stress through reactive oxygen species (NADPH oxidase, PLA2), and inflammation NF-κB and proinflammatory cytokine synthesis (34). Interestingly, the gut microbiome of currently drinking alcoholic cirrhotic patients was largely unchanged with the exception of a significant increase in the Veillonellaceae within the phylum Firmicutes, and a significant decrease in Bacteroidaceae and Porphyromonadaceae within the phylum Bacteroidetes (33). Recall that a major shift occurred in the microbiome of rats fed high levels of CA resulting in members of the Firmicutes dominating at the expense of gram negative taxa (12). However, despite bile acid concentrations in the small and large bowel of human alcoholic cirrhotics surpassing healthy control levels, dysbiosis persisted in cirrhotic patients. Thus, while bile acids are important in shaping the healthy adult microbiome, they are only one factor within a larger context of factors shaping the dysbiosis recently defined in alcoholic cirrhosis.
Probiotics and Secondary Bile Acids
A recent randomized, double-blind placebo-controlled study of cirrhotic patients evaluated the efficacy of Lactobacillus rhamnosus GG ATCC strain 53103(LGG) on cognition, systemic inflammation, fecal bile acids, and gut microbiome composition (35). Bajaj et al. (2014) observed a significant reduction in serum endotoxin, and TNF-α in the LGG group only (35). At baseline, the placebo and LGG treatment group had no significant differences in stool microbiome. However, a significant increase was reported in members of the Firmicutes (Lachnospiraceae and Clostridiales XIV) and a decline in taxa associated with worsening disease and cognitive problems (Enterobacteriaceae and Porphyromonadaceae) in the LGG-group, but not the placebo group (35). A significant increase in DCA was also observed in the placebo group relative to baseline levels (35). A recent clinical trial achieved similar results through use of the antibiotic rifaximin, which targets bacterial function, resulted in improvement in dysbiosis, reduced DCA levels, and reduced endotoxemia (36).
Recent work by Zhang et al. (2014) examined the effect of another probiotic bacterium, Lactobacillus casei Zhang on fecal bile acids, inflammation during high-fat sucrose (HFS) diet in a model of type II diabetes (37). They report that blood lipid-lowering effects of L. casei Zhang are due to fecal bile acid elimination, supported by significant reduction of plasma bile acids, and a significant increase in total fecal bile acids (37). However, despite significant increases in bile acid input, secondary bile acid levels were significantly lower in probiotic + HFS diet relative to HFS diet alone. qPCR quantification revealed significant increases in the levels of Clostridium scindens in HFS diet relative to control chow diet and HFS + probiotic (37). A diminution in the levels of C. scindens or in general bile acid 7α-dehydroxylating activity by C. scindens and other species capable of secondary bile acid formation, may be responsible for decreased secondary bile acid levels. Decades of research have implicated increased fecal, and serum DCA levels with colon cancer, and now liver cancer. Probiotics may thus be an important tool in reducing in reducing the levels and activity of bile acid 7α/β-dehydroxylating bacteria finding applications in prevention of diseases of the GI tract among alcoholics and individuals consuming a high fat, high simple sugar diet.
Conclusions
The ‘omics’ revolution and attention to our ‘microbial co-conspirators’ is leading to new thinking about treating disease in the GI tract and beyond. For instance, cognitive problems associated with liver disease are treated by targeting the gut microbiome (25). This long reach of the human microbiome and the notion that the human body is an ecosystem is underscored by recent applications of systems biology to cirrhosis (25,26,29,33,36). Cholestasis appears to generate system inflammation due largely to the absence of adequate bile salt concentrations in the small bowel allowing for colonization of the small bowel by LPS producing members of the oral microbiota (27,28). Dysbiosis associated with alcohol, and cirrhosis of the liver is currently being defined by high-throughput techniques defining community structure and gene content (16s rDNA sequencing , metagenomics), gene expression (metatranscriptomics) and metabolic phenotype (metabolomics). The bile acid pool size and composition is the result of a complex interaction between the three domains of life, eukarya, prokarya and archaea that make up the human ecosystem. Defining the role of bile acids in complex human diseases, changes in the composition of bile acids as a result of diet, antibiotics, probiotics, and other perturbations will require the ability to measure changes in bile acid modifying microbes, host regulation of synthesis and excretion of bile acids and how these networks of genes and metabolites are predictably altered. This must necessarily be the goal of 21st century bile acid-microbiome research.
Footnotes
Disclosure Statement xxxxxxxxxxx
References
- 1.Qin J, Li R, Raes J, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2009;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aagaard K, Ma J, Antony KM, et al. The placenta harbors a unique microbiome. Sci. Transl. Med. 2014;6(237):237ra65. doi: 10.1126/scitranslmed.3008599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ley RE, Peterson DA, Gordon JI. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell. 2006;124:837–848. doi: 10.1016/j.cell.2006.02.017. [DOI] [PubMed] [Google Scholar]
- 4.Eckburg PB, Bik EM, Berstein CN, et al. Diversity of the human intestinal microbical flora. Science. 2005;308(5728):1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Inagaki T, Moschetta A, Lee Y, et al. Regulation of antibacterial defense in the small intestine by the nuclear bile acid receptor. Proc Nat Acad Sci USA. 2006;103(10):3920–3925. doi: 10.1073/pnas.0509592103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Devkota S, Wang Y, Musch MW, et al. Dietary-fat-induced taurocholic acid promotes pathobiont expansion and colitis in Il10−/− mice. Nature. 2012;487(7405):104–108. doi: 10.1038/nature11225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reddy BS. Diet and excretion of bile acids. Cancer Res. 1981;41:3766–3768. [PubMed] [Google Scholar]
- 8.Reddy BS, Wynder EL. Metabolic epidemiology of colon cancer: fecal bile acids and neutral sterols in colon cancer patients with adenomatous polyps. Cancer. 1977;39:2533–2539. doi: 10.1002/1097-0142(197706)39:6<2533::aid-cncr2820390634>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 9.David LA, Maurice CF, Carmody RN, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2013;505:559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sonnenburg JL, Xu J, Leip DD, et al. Glycan foraging in vivo by an intestine-adapted bacterial symbiont. Science. 2005;307:1955–1959. doi: 10.1126/science.1109051. [DOI] [PubMed] [Google Scholar]
- 11.Martens EC, Chiang HC, Gordon JI. Mucosal glycan foraging enhances fitness and transmission of a saccharolytic human gut bacterial symbiont. Cell Host Microbe. 2008;4(5):447–457. doi: 10.1016/j.chom.2008.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Islam KBMS, Fukiya S, Hagio M, et al. Bile acid is a host factor that regulates the composition of the cecal microbiota in rats. Gastroenterology. 2011;141:1773–1781. doi: 10.1053/j.gastro.2011.07.046. [DOI] [PubMed] [Google Scholar]
- 13.Yokota A, Fukiya S, Islam KBMS, et al. Is bile acid a determinant of the gut microbiota on a high-fat diet? Gut Microbes. 2012;3(5):455–459. doi: 10.4161/gmic.21216. [DOI] [PubMed] [Google Scholar]
- 14.Swann JR, Want EJ, Geier FM, et al. Systemic gut microbial modulation of bile acid metabolism in host tissue compartments. Proc Nat Acad Sci USA. 2011;108:4523–4530. doi: 10.1073/pnas.1006734107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Makishima M, Lu TT, Xie W, et al. Vitamin D receptor as an intestinal bile acid sensor. Science. 2002;296(5571):1313–1316. doi: 10.1126/science.1070477. [DOI] [PubMed] [Google Scholar]
- 16.Ridlon JM, Kang D, Hylemon PB. Bile salt biotransformations by human intestinal bacteria. J Lipid Res. 2006;47(2):241–259. doi: 10.1194/jlr.R500013-JLR200. [DOI] [PubMed] [Google Scholar]
- 17.Joyce SA, MacSharry J, Casey PG, et al. Regulation of host weight gain and lipid metabolism by bacterial bile acid modification in the gut. Proc Nat Acad Sci USA. 2014;111(20):7421–7426. doi: 10.1073/pnas.1323599111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jones BV, Begley M, Hill C, et al. Functional and comparative metagenomic analysis of bile salt hydrolase activity in the human gut microbiome. Proc Nat Acad Sci USA. 2008;105(36):13580–13585. doi: 10.1073/pnas.0804437105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Begley M, Gahan CGM, Hill C. The interaction between bacteria and bile. FEMS Micro Rev. 2005;29:625–651. doi: 10.1016/j.femsre.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 20.Begley M, Hill C, Gahan CGM. Bile salt hydrolase activity in probiotics. Appl Environ Microbiol. 2006;72(3):1729–1738. doi: 10.1128/AEM.72.3.1729-1738.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.White BA, Fricke RJ, Hylemon PB. 7β-Dehydroxylation of ursodeoxycholic acid by whole cells and cell extracts of Eubacterium sp. V.P.I. 12708. J Lipid Research. 1982;22:145–153. [PubMed] [Google Scholar]
- 22.Kang D, Ridlon JM, Moore DR, 2nd, et al. Clostridium scindens baiCD and baiH genes encode stereo- specific 7alpha/7beta-hydroxy-3-oxo-delta4-cholenoic acid oxidoreductases. Biochim Biophys Acta. 2008;1781(1-2):16–25. doi: 10.1016/j.bbalip.2007.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Macdonald IA, White BA, Hylemon PB. Separation of 7α- and 7β-hydroxysteroid dehydrogenase activities from Clostridium absonum ATCC# 27555 and cellular response of this organism to bile acid inducers. J Lipid Res. 1983;24:1119–1126. [PubMed] [Google Scholar]
- 24.Chen Y, Yang F, Lu H, et al. Characterization of fecal microbial communities in patients with liver cirrhosis. Hepatology. 2011;54:562–572. doi: 10.1002/hep.24423. [DOI] [PubMed] [Google Scholar]
- 25.Bajaj JS, Ridlon JM, Hylemon PB, et al. Linkage of gut microbiome with cognition in hepatic encephalopathy. Am J Physiol Gastrointest Liver Physiol. 2012;302:G168–G175. doi: 10.1152/ajpgi.00190.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bajaj JS, Heuman DM, Hylemon PB, et al. Altered profile of human gut microbiome is associated with cirrhosis and its complications. J Hepatol. 2013 doi: 10.1016/j.jhep.2013.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qin N, Yang F, Li A, et al. Alteration of the human gut microbiome in liver cirrhosis. Nature. 2014;513:59–64. doi: 10.1038/nature13568. [DOI] [PubMed] [Google Scholar]
- 28.Ridlon JM, Alves JM, Hylemon PB, et al. Cirrhosis, bile acids and gut microbiota: unraveling a complex relationship. Gut Microbes. 2013;4(5):1–6. doi: 10.4161/gmic.25723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kakiyama G, Pandak WM, Gillevet PM, et al. Modulation of the fecal bile acid profile by gut microbiota in cirrhosis. J Hepatol. 2013;58:949–955. doi: 10.1016/j.jhep.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yoshimoto S, Loo TM, Atarashi K, et al. Obesity-induced gut microbial metabolite promotes liver cancer through senescence secretome. Nature. 2013;499:97–101. doi: 10.1038/nature12347. [DOI] [PubMed] [Google Scholar]
- 31.Axelson M, Mork B, Sjovall J. Ethanol has an acute effect on bile acid biosynthesis in man. FEBS Lett. 1991;281:155–159. doi: 10.1016/0014-5793(91)80382-d. [DOI] [PubMed] [Google Scholar]
- 32.Chanda D, Kim YH, Li T, et al. Hepatic cannabinoid receptor type 1 mediates alcohol-induced regulation of bile acid enzyme genes expression bia CREBH. PLoS ONE. 8:e68845. doi: 10.1371/journal.pone.0068845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kakiyama G, Hylemon PB, Zhou H, et al. Colonic inflammation and secondary bile acids in alcoholic cirrhosis. Am J Physiol Gastrointest Liver Physiol. 2014;306:G929–G937. doi: 10.1152/ajpgi.00315.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hylemon PB, Zhou H, Pandak WM, et al. Bile acids as regulatory molecules. J Lipid Res. 2009;50:1509–1520. doi: 10.1194/jlr.R900007-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bajaj JS, Heuman DM, Hylemon PB, et al. Randomized clinical trial: Lactobacillus GG modulates gut microbiome, metabolome and endotoxemia in patients with cirrhosis. Aliment Pharmacol Ther. 2014;39:1113–1125. doi: 10.1111/apt.12695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bajaj JS, Heuman DM, Sanyal AJ, et al. Modulation of the metabiome by rifaximin in patients with cirrhosis and minimal hepatic encephalopathy. PLoS ONE. 2013;8:e60042. doi: 10.1371/journal.pone.0060042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang Y, Guo X, Guo J, et al. Lactobacillus casei reduces susceptibility to type 2 diabetes via microbiota-mediated body chloride ion influx. Sci. Report. 2014;4(5654) doi: 10.1038/srep05654. doi: 10.1038/srep05654. [DOI] [PMC free article] [PubMed] [Google Scholar]