Abstract
Purpose
While many Mycosis Fungoides (MF) patients presenting with stage I disease enjoy an indolent disease course and normal life expectancy, about 15-20% of them progress to higher stages, and most ultimately succumb to their disease. Currently, it's not possible to predict which patients will progress and which patients will have a stable disease. Previously, we conducted microarray analyses with RT-PCR validation of gene expression in biopsy specimens from 60 stage I-IV CTCL patients, identified three distinct clusters based upon transcription profile and correlated our molecular findings with 6 years of clinical follow up.
Experimental Design
We test by RT-PCR within our prediction model the expression of ∼240 genes that were previously reported to play an important role in CTCL carcinogenesis. We further extend the clinical follow up of our patients to 11 years. We compare the expression of selected genes between MF/Sézary Sydrome and benign inflammatory dermatoses that often mimic this cancer.
Results
Our findings demonstrate that 52 out of the ∼240 genes can be classified into cluster 1-3 expression patterns and such expression is consistent with their suggested biological roles. Moreover, we determined that 17 genes (CCL18, CCL26, FYB, T3JAM, MMP12, LEF1, LCK, ITK, GNLY, IL2RA, IL-26, IL-22, CCR4, GTSF1, SYCP1, STAT5A, TOX) are able to both identify patients who are at risk of progression and also distinguish MF/SS from benign mimickers.
Conclusions
This study, combined with other gene expression analyses, prepares the foundation for the development of personalized molecular approach towards diagnosis and treatment of CTCL.
Keywords: Cutaneous T-Cell Lymphoma (CTCL), Mycosis Fungoides (MF), Sézary Syndrome (SS), prognostic markers, diagnostic markers, expression profiling
Introduction
Cutaneous T-Cell Lymphomas (CTCL) are rare and sometimes lethal malignancies. CTCL's are a heterogeneous group of Non-Hodgkin's lymphoproliferative disorders characterized by localization of neoplastic T lymphocytes to the skin(1). Previous epidemiologic studies based on the Surveillance, Epidemiology and End Results (SEER) databases documented that this disease is on the rise in the United States and around the world(2). However, recent findings in the United States suggest that the overall national incidence of this cancer has stabilized at a rate of ∼10 cases per million per year in the last decade(3). Different regional variations in CTCL incidence have been reported, where ∼14-16 cases per million individuals per year were diagnosed in San Francisco, California, while only 6-7 cases per million individuals per year were diagnosed in Iowa during 2000-2009(3). Recent studies also revealed that CTCL may occur in married couples(4) and cluster in families(5). Recent investigations demonstrated geographic clustering of CTCL cases, therefore implying possible existence of an environmental trigger for this malignancy (6) though this remains to be confirmed.
Mycosis Fungoides (MF) and Sézary Syndrome (SS) together represent the most common forms of CTCL and account for >50% of all CTCL cases(1). In Caucasians MF/SS affect individuals over 55 years of age, while in African-Americans, Hispanics and Arabic individuals this disease can present at a significantly younger age (i.e. 20s and 30s)(7).
In early disease stages that can last for many years, classic MF presents as flat erythematous skin patches and thin plaques with sharply defined borders resembling benign inflammatory dermatoses, while at later stages, MF cells can form thicker plaques or tumors over larger areas of skin and may disseminate to lymph nodes and internal organs(1). Early diagnosis of CTCL is often challenging since this cancer can masquerade clinically as other entities such as chronic eczematous dermatitis, psoriasis or fungal infections(8). Even histopathology and current PCR studies for T cell receptor clonality are sometimes not sufficient for definitive diagnosis(8). This, unfortunately, results in a delay, often by many years, in the diagnosis of this cancer.
In advanced disease, malignant cells can sometimes spread to involve lymph nodes and peripheral blood, leading to the leukemic form of MF. SS is now believed to be a distinct entity, different from leukemic MF since it often presents de-novo without any evidence of preceding MF(8), arises from different cancer initiating cells (i.e., central memory T cells)(9) and carries a more uniformly poor prognosis(8). This leukemic form of CTCL is characterized by a triad of erythroderma, lymphadenopathy and detection of malignant T cells with cerebriform nuclei on a peripheral blood smear(1).
Clinical disease stage at the time of diagnosis remains the best predictor of survival and progression for MF. Early stages (i.e., stage IA and IB) often exhibit an indolent disease course, with normal or near normal life expectancy(10-12). In contrast, advanced stages and/or SS are associated with recalcitrant disease and poor 5-year survival rate(1). The majority of MF patients present with an early stage (i.e., IA or IB) disease(10-12). However, ∼15-20% of these patients will progress to higher stages and may ultimately succumb to their cancer(10-12). At the same time, while many patients with advanced CTCL stages will experience an aggressive course of disease progression, this is highly variable and a small minority of these patients survive for much longer than 5 years. Improving our ability to effectively diagnose MF/SS(i.e. being able to distinguish it molecularly from benign mimickers) and, most importantly, developing molecular tools to identify patients at risk of progression at early disease stages will enable us to personalize our management approach towards diagnosis and treatment of this cancer.
To discover novel prognostic molecular markers and to gain additional insight in disease etiology we had previously performed a microarray and subsequent RT-PCR analyses of gene expression in biopsy specimens from 60 stage I-IV MF/SS patients(13, 14). These patients were initially followed for 6 years. The original gene expression analyses revealed three distinct transcription profile clusters (i.e. clusters 1, 2 and 3), where clusters 1 and 3 contained a mix of stage I–IV disease patients, while cluster 2 contained mostly stage I and only a few cases of advanced disease patients(13, 14). All stage IV MF/SS patients fell into clusters 1 and 3(13, 14). The described three distinct transcription profile clusters were associated with different clinical courses. Cluster 2 genes corresponded to the best clinical outcome and good response to therapy while cluster 1 and 3 molecular signature patterns were associated with the worst and intermediate clinical outcomes, respectively, and poor response to therapy(13, 14). Due to relatively short initial clinical follow up period (6 years) these trends did not reach statistical significance at that time(13, 14).
In the current work we performed a literature review that highlighted ∼240 genes whose expression and function is believed to be important in CTCL pathogenesis diagnosis and/or treatment. We subsequently tested the expression of these genes by RT-PCR in our patient population to identify additional genes that fit into the above described three cluster prediction model. A number of genes that fit this model were then selected for further testing and their expression patterns were compared between CTCL lesional skin vs. normal skin from healthy volunteers vs. lesional skin from patients affected by benign dermatoses that often masquerade as CTCL (e.g. chronic eczema, psoriasis and pityriasis rubra pilaris). This was done to identify which prognostic markers may also have a diagnostic value in this cancer.
Methods
Patients and Samples
All patients were enrolled in the IRB-approved study protocol with informed consent in accordance with the Declaration of Helsinki(13, 14). CTCL patients were recruited from the Cutaneous Lymphoma Clinic at the Dana Farber Cancer Institute (DFCI)/Brigham and Women's Hospital (BWH). All tissue samples were obtained and processed as previously described(13). Briefly, six-mm punch biopsies from involved skin were collected from patients between January 26, 2003 and June 1, 2005. The obtained 6 mm biopsies were immediately snap-frozen in liquid nitrogen. Tissue was powdered in liquid nitrogen (Cryo-Press; Microtec Co, Chiba, Japan), and total RNA was extracted using Trizol (Invitrogen, Carlsbad, CA) and converted to cDNA using the iScript RT-PCR kit (Bio-Rad, Hercules CA) according to the manufacturer's instructions.
The historic cohort of patients from Boston (n=60), which was initially reported in 2007(13), was at the heart of extensive research that led to multiple publications in the field(13-20). For these patients, 11 years of clinical follow-up data was analyzed in the same way as in previous reports(13-20). The biopsy samples analyzed in this study are the same samples that were analyzed in our previous papers(13-20). The diagnosis and clinical staging were established according to the diagnostic criteria of CTCL(21).
Similarly, volunteers with normal healthy skin (n=6) and benign inflammatory dermatoses (n=17) were recruited from the outpatient dermatology clinic of the University of British Columbia (Vancouver, Canada) with informed consent(17). Full-thickness lesional skin punch biopsies were obtained under local anesthesia as previously described(13, 14, 17).
Quantitative Real-Time Reverse Transcription-PCR Gene Expression Analysis
While microarray analysis provides an unprecedented capacity for whole genome expression profiling, it has a number of inherent pitfalls that have been described elsewhere (22-24). Quantitative Real-Time PCR (RT-PCR) serves as the “gold standard” method for evaluation of gene expression(23), which was the main reason for using this approach in the current study. Before initiating this study, we searched PubMed, Medline and Web of Science databases using terms “genetic”, “gene expression”, “gene expression profiling” and “CTCL”, “Mycosis Fungoides” or “Sézary Syndrome” to identify studies published in English. Based on this search >400 studies were reviewed and 241 genes of interest were selected. Gene expression was tested via RT-PCR in CTCL patients' lesional skin, normal skin form healthy volunteers and lesional skin from patients with benign inflammatory dermatoses as previously described(14, 17, 19). Primer pair sequences for tested genes and control housekeeping genes are listed in supplementary table 1. RT-PCR was performed utilizing the obtained cDNA from patients and iScript RT-PCR mix (Bio-Rad, Mississauga, Ontario) on Bio-Rad iCycler as previously described(14-16). The expression was standardized using genorm method(25) utilizing ACTB, SDHA, YWHAZ and HMBS housekeeping genes. For every gene analyzed the highest expression value in our samples was set as 1 fold of expression similarly to the protocol in our previous studies(14, 19).
Statistical analyses
Disease progression and disease-specific survival was analyzed using XLSTAT software (Addinsoft, New York, NY) to obtain Kaplan-Meier curves as previously described(14). p values were calculated using the logrank test(26). Patient multivariate analysis was performed using the Cox proportional hazards regression method taking into account multiple progression events for each patient.
Results
The three signature gene expression model identifies novel prognostic markers for CTCL
One of the important criticisms of our previous microarray and RT-PCR gene expression studies(13, 14) was that they did not include numerous genes that were reported in literature by other authors to be important in the pathogenesis, diagnosis and treatment of CTCL. To address this concern, in the current study we conducted a literature search and identified ∼240 genes (Supplemental Table 1) that were previously reported to play an important role in CTCL. We tested their expression in our cohort of patients for which extensive clinical follow up is available. This analysis demonstrated that a number of previously reported genes can be classified into cluster 1, cluster 2 or cluster 3 expression patterns (as shown in Figures 1-3) and such expression is congruent with their suggested biological role in CTCL pathogenesis.
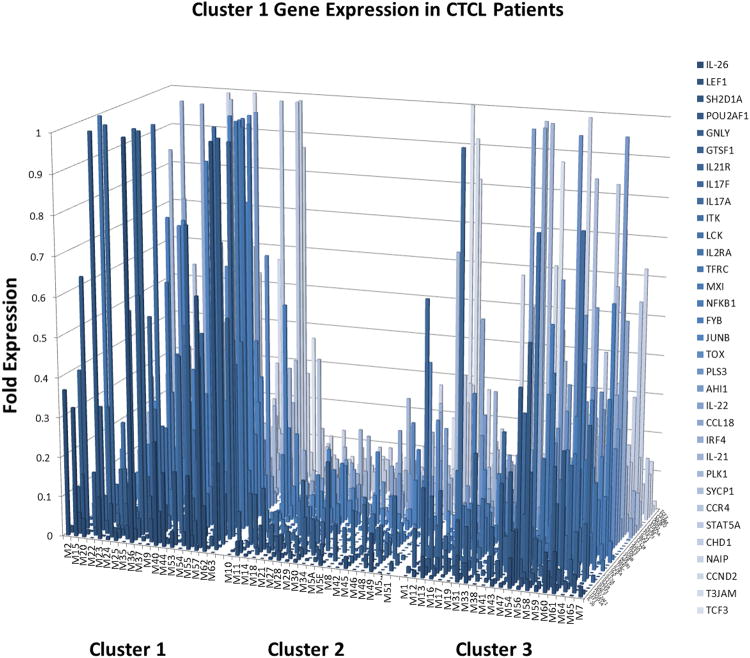
Figure 1.
Expanded RT-PCR analysis of gene expression reveals 33 genes that are expressed in poor and intermediate prognosis cluster 1 and 3 patients, but not in favorable prognosis cluster 2 patients.
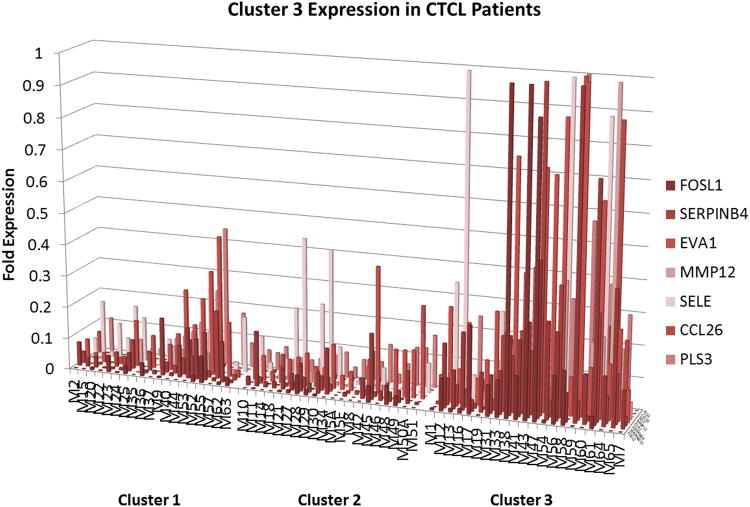
Figure 3.
RT-PCR analysis of gene expression reveals 7 genes that are preferentially expressed in the intermediate prognosis cluster 3 patients.
Proto-oncogenes, inflammatory cytokines, cell cycle, novel cancer testis genes were expressed in aggressive disease clusters 1 and 3 (Figure 1, Figure 3 and Supplementary Table 2). In total, 33 out of ∼240 genes tested fit into cluster 1 (poor prognosis cluster) expression pattern with partial overlap with cluster 3 (intermediate prognosis cluster) patients (Figure 1), while 7 gene were preferentially expressed in cluster 3 patients (Figure 3). In these clusters we observed the expression of cell survival and cell cycle genes CCND2, NFKB1, PLK1, NAIP; putative oncogenes JUNB, TOX, AHI1; novel cancer testis genes GTSF1, SYCP1 as well as embryonic stem cell genes TCF3, EVA1, CHD1; genes promoting inflammatory T cell signaling ITK, LCK, FYB, GNLY, CCL18, CCL26, E-Selectin; skin homing chemokine receptor CCR4;cytokines (and their cognate receptors) that were reported to be secreted by the Th17 cells IL-26, IL-17A, IL-17F, IL-21 and IL-21R;the IL-22 cytokine; actin binding protein PLS3; matrix metalloproteinase MMP12; downstream positive regulator of WNT/β-catenin signalling LEF1; transcription factors STAT5A, MXI and POU2AF; markers of T cell activation TFRC, IRF4; and other signaling genes including T3JAM, FOSL1, SHD1A, SERPINB4 (Figure 1, Figure 3 and Supplementary Table 2). As described in detail in supplementary table 2 many of these genes were reported to play cancer promoting roles in CTCL and other cancers. Moreover, several of them (e.g. CCR4, IL2RA and TOX) are recognized or proposed as therapeutic targets in CTCL.
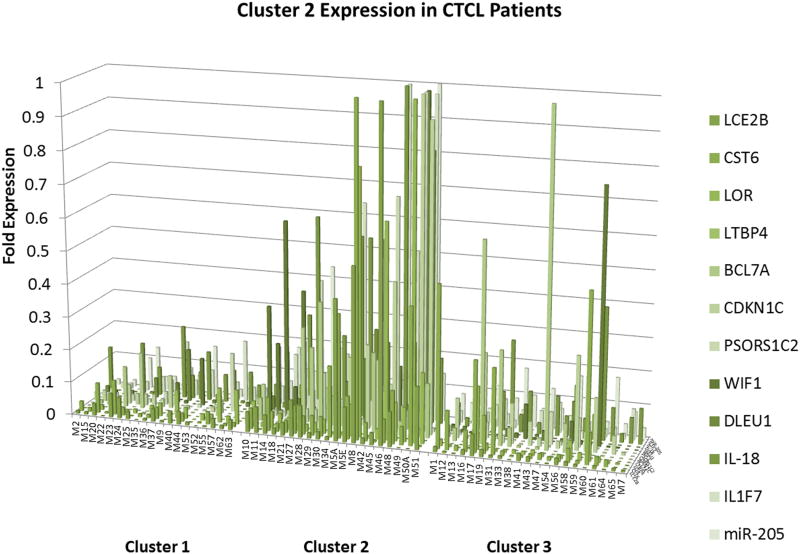
On the other hand, putative tumor suppressor genes CDKN1C, BCL7A DLEU1, miR-205 and CST6; epidermal differentiation genes LCE2B, LOR; TGF-βsignaling gene LTBP4; WNT/β-catenin pathway antagonist WIF1 (WNT Inhibitory Factor 1) and psoriasis susceptibility gene PSORS1C2 were expressed in a favorable prognosis cluster 2 (Figure 2). Also, IL-18 cytokine, which is known to induce interferon-γ response (possibly targeting malignant infiltrating T cells), and its downstream target IL1F7 (also known as IL-37) were also upregulated in this cluster of patients (Figure 2). As discussed in supplementary table 2 many of these genes were reported to act as tumor suppressors in CTCL and other cancers and were shown to be downregulated in neoplasia.
Figure 2.
RT-PCR analysis of gene expression reveals 12 genes that are expressed in favorable prognosis cluster 2 patients and partially overlapping with patients in Cluster 3.
We previously demonstrated based on 6 years of clinical follow up that these three signature gene expression patterns were associated with different clinical outcomes in CTCL patients(14). However, at that time these trends did not reach statistical significance(14). In the current work we extended the clinical follow up of our patients until 2014 (11 years of clinical follow up).
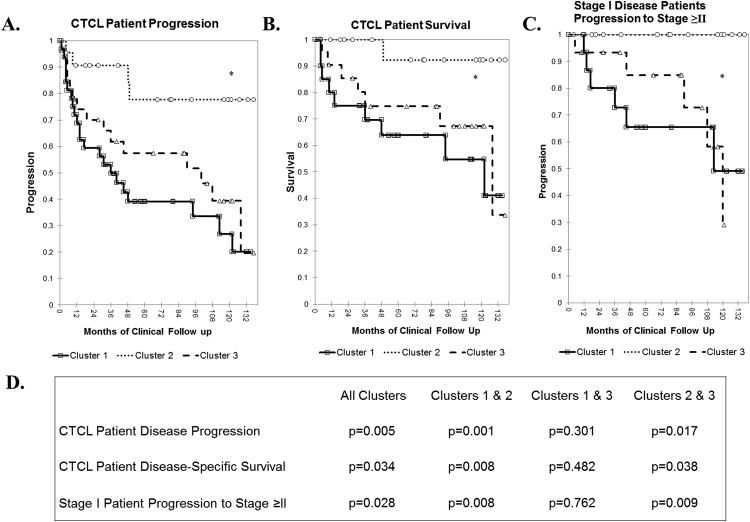
The new extended 11-year clinical analysis of CTCL progression confirms our previous observations and documents that cluster 2 had many fewer number of progression events (i.e., advancement to a higher CTCL stage and/or death) than clusters 1 and 3 (Figure 4A). Logrank test of the presented Kaplan-Meier analysis documents that these three clusters are statistically different (p=0.005). Similarly, with respect to survival, cluster 2 patients enjoyed a favorable 11-year survival, while clusters 1 and 3 patients experienced an overall poor survival (Figure 4B). Statistical significance was observed for survival differences between the three clusters (p=0.034). All 60 patients were analyzed in the above described analyses.
Figure 4.
Kaplan-Meier analysis of CTCL patient disease outcomes A. CTCL patient overall disease progression (defined as progression to a higher clinical stage and/or death, p=0.005) B. CTCL patient disease-specific survival, p=0.034 C. Progression of patients with stage I disease to more advance stages (i.e., stage ≥II). D. p-Values for Kaplan-Meier comparisons for each individual cluster pair.
Since each cluster had a large number of stage I disease patients (i.e., 11/19 stage I patients in cluster 1, 18/20 patients in cluster 2 and 14/21 patients in cluster 3), we specifically analyzed the progression of these patients towards more advanced disease (i.e., stage≥II) with respect to their genetic clusters. As presented in Figure 4C, Cluster 1 and 3 stage I patients had the highest 11-year progression rates. Strikingly, none of the cluster 2 stage I patients have progressed towards advanced disease (i.e., progression rate of 0%) during the period of 11 years. In addition, we conducted Kaplan-Meier comparisons for each individual cluster pair. p values for these analyses are presented in Figure 4D.
Finally, for this patient cohort we conducted a multivariate analysis of disease progression based on gender, age and clinical disease stage at the time of diagnosis. As we expected, the clinical stage at the time of diagnosis was a strong predictor of cancer progression in our patients (Supplementary Table 3). Specifically, stage ≥III patients had a ∼12 fold risk of progressing to higher stages and/or dying from their disease, when compared to stage I disease patients. Stage II disease patients had a 4.7 fold risk of progression and/or death. Also, consistent with the trends reported in the literature, a weak association was documented between male sex and disease progression (Supplementary Table 3). Based on our analysis, age alone was not an independent risk factor for disease progression (Supplementary Table 3).
Comparison of gene expression between CTCL, normal skin and lesional skin from benign inflammatory dermatoses patients
The early stages of MF are often difficult to distinguish clinically from other benign entities including chronic eczema, psoriasis and pityriasis rubra pilaris(8). Furthermore, detection of T cell clonality in itself is also not diagnostic of CTCL since a number of benign dermatoses (e.g. lichen planus, pityriasis lichenoides, lichen sclerosus and pigmented purpura) too can have a dominant T cell clone as measured by diagnostic PCR-based techniques(8). Histological diagnosis is often difficult since in early patch disease stages malignant lymphocytes represent only 5-10% of the total inflammatory infiltrate. Even in advanced stages, using all available clinical and laboratory tools, it is often difficult to distinguish patients with advanced erythrodermic MF and SS disease from patients presenting with non-malignant erythrodermas secondary to psoriasis, pityriasis rubra pilaris and atopic dermatitis(8). Hence, new genetic markers are urgently needed to distinguish CTCL from various benign inflammatory dermatoses.
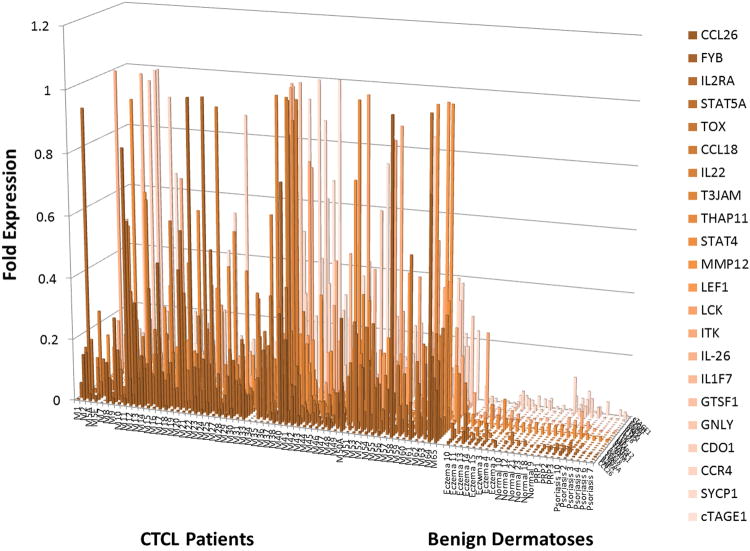
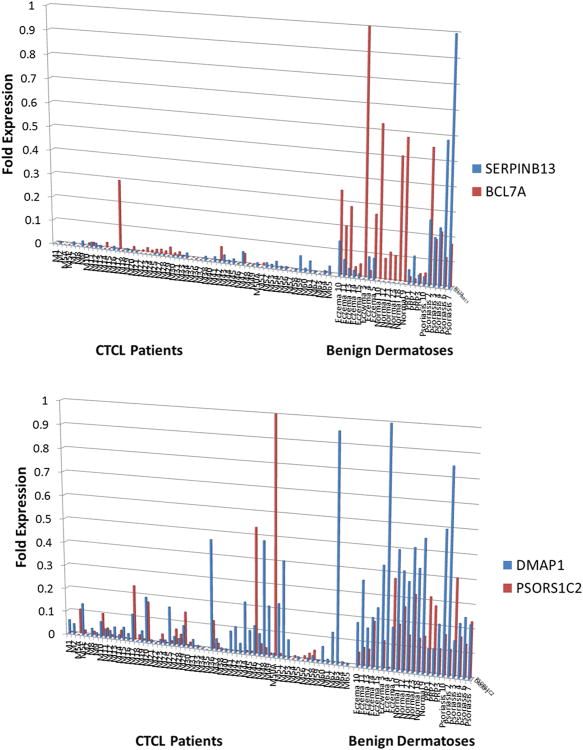
To address this we compared the expression of the above described candidate genes between CTCL lesional skin (n=60), skin biopsies from healthy volunteers and patients with chronic eczema, psoriasis, pityriasis rubra pilaris (PRP). Strikingly numerous genes that were upregulated in clusters 1 and 3 CTCL patients (Figures 1 and 3) were preferentially expressed in CTCL, but not in benign skin samples (Figure 5, Supplementary Table 4). Specifically, this analysis demonstrated that CCL18, CCL26, FYB, T3JAM, MMP12, LEF1, LCK, ITK, GNLY, IL2RA, IL-26, IL-22, CCR4, GTSF1, SYCP1, STAT5A and TOX can jointly be used as diagnostic and poor prognostic markers in CTCL patients. This comparative expression analysis further revealed that select genes (SERPINB13 and BCL7A) were preferentially upregulated in benign skin conditions, but not in CTCL (Figure 6A). PSORS1C2 and WIF1 cluster 2 genes were expressed in normal skin and in indolent CTCL cases (Figure 6B and Supplementary Figure 1).
Figure 5.
RT-PCR analysis of gene expression identified 22 genes that are preferentially expressed in CTCL, but not in normal skin from healthy volunteers or benign inflammatory dermatoses that often masquerade as CTCL (e.g., psoriasis, chronic eczema and pityriasis rubra pilaris or PRP). Notably, 17 of the above genes are upregulated in cluster 1 and 3 patients and potentially can serve as diagnostic and prognostic markers at the same time.
Figure 6.
RT-PCR analysis of gene expression identified several genes that are (A) upregulated in normal skin and benign dermatoses, but not in CTCL or (B) upregulated in benign skin samples and in indolent/stable CTCL.
Discussion
This study summarizes many years of research and follow up and describes in detail the findings for the Boston/DFCI/BWHMF/SS cohort of patients. In this work we have completed a comprehensive gene expression analysis for ∼240 genes that were identified in our prior studies and/or were suggested by previous literature reports to play an important role in CTCL lymphomagenesis. Expression of these genes was analyzed in the context of the three signature pattern prediction model that we previously described(13, 14). Based on 11 years of clinical follow up, we document that 52 of these genes are preferentially expressed in various genetic clusters that correlate with different disease outcomes (i.e., overall progression, disease-specific survival and progression of stage I patients to more advanced stages). We further compare the expression of these genes between CTCL and benign inflammatory dermatoses that often mimic CTCL and identify 22 of these genes that are specific for CTCL and 5 genes that are preferentially expressed in benign dermatoses or in indolent CTCL and benign dermatoses.
These results, combined with other expression profiling and meta-analysis studies(27-29) lay the groundwork for the development of personalized molecular approach towards diagnosis and management of CTCL in the future. As highlighted in our findings, a panel of 17 genes: CCL18, CCL26, FYB, T3JAM, MMP12, LEF1, LCK, ITK, GNLY, IL2RA, IL-26, IL-22, CCR4, SYCP1, GTSF1, STAT5A and TOX, can serve a dual role to diagnose CTCL and potentially predict poor clinical disease course. This hypothesis will have to be validated in future prospective studies. As suggested by our Kaplan-Meier analysis, these genes may also prove useful in the prognosis of early stage MF patients. For some time, in other diseases a single diagnostic/prognostic marker (e.g., PSA, CEA, CA-125, LDH or HER2/neu) has influenced medical decision making. However, new molecular genetic approaches may soon enable us to follow a panels of multiple cancer-related genes in our patients(30). Considering that CTCL is a heterogeneous malignancy, following a panel of markers may prove more reliable than analyzing the expression for a single gene(31).
From the above gene list, it is notable that TOX expression was also independently found by two separate laboratories to be a robust diagnostic and prognostic marker for this cancer(17). STAT5A was implicated in carcinogenesis in the early stages of CTCL by activating a oncogenic microRNA miR-155(32). IL-22, a Th22/Th17 cytokine, was proposed to be a dominant cytokine in CTCL tumor microenvironment(33). The chemokine receptor CCR4 has long been reported to be highly expressed in SS and MF(34)and is currentlyan investigational CTCL therapeutic target of Mogamulizumab, a humanized anti-CCR4 antibody(34). IL2RA (IL-2Rα) is expressed in up to 50% of MF/SS cases. Interleukin-2 diphtheria toxin fusion protein (denileukin diftitox) was designed to target this receptor in patients(35). CCL26 was shown to correlate with the clinical itch burden in CTCL patients(36), while another potent T cell chemoatractant CCL18 has been consistently shown to be upregulated in MF and correlate with the types of skin lesions (i.e., patch vs. plaque vs. tumors)(37). As evident from this brief overview, molecular markers identified in our study have a direct clinical correlation to disease symptoms and treatment as reported in the literature. By combining this knowledge with similar studies that identified critical molecular diagnostic and prognostic markers(27-29) we hope to improve our ability to effectively manage this cancer.
This study further highlighted ectopic expression of novel cancer testis genes and embryonic genes GTSF1, SYCP1, TCF3 and CHD1 that were expressed in poor prognosis cluster 1 patients, while cTAGE1, GTSF1 and THAP11 were preferentially expressed in CTCL, but not in benign skin samples. EVA1 (also known as MPZL2), another poor prognosis marker, is expressed early on in the thymus, but then is strongly downregulated during thymocyte developmental progression(38). Also, previous work suggested that a B cell specific gene, B-lymphoid kinase or BLK, is constitutively active in malignant T cells and appears to be a bona fide oncogene which drives malignant T cell proliferation in vitro and tumor formation in vivo(39). In this study we confirm the expression of the aforementioned genes in CTCL and demonstrate that another B cell-specific transcriptional factor POU2AF1 is expressed in poor prognosis cluster 1 patients. Other important putative CTCL oncogenes confirmed by this study include JUNB, PLS3, AHI and PLK1.
For favorable prognosis genes, this study highlights putative tumor suppressor genes BCL7A, CKDN1C, miR-205, DLEU1, IL-18 and WIF1. BCL7A and CDKN1C were previously proposed by our laboratory and others to play important roles in CTCL pathogenesis(15, 16, 40, 41). The miR-205 microRNA was documented to act as a tumor suppressor in melanoma and other cancers, and has the ability to discriminate CTCL from other benign entities(42). DLEU1 (Deleted in Lymphocytic Leukemia 1) long non-coding RNA putative tumor suppressor gene is frequently deleted in B-cell chronic lymphocytic leukemia (B-CLL)(43) and is reported for the first time in this study to play a role in CTCL. The inflammatory cytokine IL-18 is known to induce expression of interferon-γ and promote a Th1 immune response, both of which are associated with disease clearance(44). WNT/β-catenin signaling inhibitor, WIF1 (WNT Inhibitor Factor 1), was previously shown to be downregulated in salivary gland carcinomas, acute lymphoblastic leukemias (ALL)(45) and acute myeloid leukemias (AML)(46). Our study suggests that loss of this gene may also be important for CTCL carcinogenesis. Finally, STAT4 transcription factor appears to play a dual role in CTCL. When compared to benign dermatoses, this gene is upregulated in lesional CTCL skin(18, 47), but its expression appears to be lost in aggressive/recalcitrant disease(18, 31, 47). Previous reports documented that loss of STAT4 expression is associated with a switch from Th1 towards Th2 phenotype in CTCL(18). Loss of STAT4 expression was shown to be a robust and reliable diagnostic marker for SS(31).
While our study shows that several Th17 markers are upre gulated in CTCL, the role of IL-17 (IL-17A, IL-17C and IL-17F) expression and signaling in this cancer remains uncertain. Some studies observed upregulation in expression of these cytokines and, in some cases, their corresponding receptors in CTCL lesional skin by RT-PCR and immunohistochemistry(20, 48-50), in PBMCs from leukemic CTCL patients(49) as well as in immortalized CTCL cell lines(20, 48, 51). At the same time, other studies failed to confirm the expression of IL-17 signaling in CTCL. Careful examination of published literature suggests that while IL-17 may be expressed in CTCL lesional skin in some cases, it is not clear whether this cytokine is produced by malignant T cells, reactive T cells or other cell types.
It was previously demonstrated that fungal (dermatophytes and candida)(52) and/or Staphylococcus aureus(53) infections are potent inducers of IL-17 signaling. Also, many CTCL treatment modalities including interferon(54), imiquimod(55) and phototherapy(56) were shown to induce IL-17 expression in the skin. Hence, it is possible that MF lesions superinfected with bacteria/fungus, recalcitrant treated lesions or lesions that were exposed to UVB may have a higher expression of IL-17 as a result. Hence, future studies will need to clarify whether IL-17 signaling is importantin certain cases of CTCL or an epiphenomenon sporadically observed in a subset of MF lesions.
In summary, this study combined with other gene expression profiling analyses prepares the groundwork for the development of personalized molecular approachtowards diagnosis, prognosis, and treatment of CTCL. In the future, it will be important to optimize a panel of genes and select 5-10 robust markers to diagnose and prognosticate this malignancy.
Supplementary Material
Statement of Translational Relevance.
The majority of Mycosis Fungoides (MF) cases (i.e., ≥70%) present with stage I disease and ∼80% of these patients experience an indolent clinical course. Unfortunately, it is currently not possible to predict which patients will progress and which ones will remain stable based on the available clinical or pathological criteria. Furthermore, at early stages this malignancy often masquerades as chronic eczema, psoriasis or other benign inflammatory dermatoses, which often delays the definitive diagnosis for months or years. The presented three cluster molecular signature model highlights novel prognostic markers, many of which are already linked to disease pathogenesis. Moreover, comparison of gene expression between Mycosis Fungoides/Sézary Sydrome and benign inflammatory dermatoses uncovers a panel of 17 markers that are able not only to identify patients, who are at risk of progression, but also distinguish MF from its benign mimickers. This knowledge will aid future molecular diagnosis and prognosis for these cancers.
Acknowledgments
We thank Mr. Gregory Cormack and RNomique Centre at the University of Sherbrooke for their technical assistance in performing molecular experiments.
Financial Support: This work was supported by the Canadian Dermatology Foundation research grants to Dr. Sasseville, Dr. Litvinov and Dr. Zhou, the Fonds de la recherche en santé du Québec (FRSQ) research grant to Dr. Sasseville (FRQS# 22648), Canadian Institutes of Health Research to Dr. Zhou, and the National Institutes of Health SPORE in Skin Cancer (P50 CA093683) to Dr. Kupper.
Footnotes
Conflict-of-interest disclosure: The authors declare no competing financial interests.
References
- 1.Willemze R, Jaffe ES, Burg G, Cerroni L, Berti E, Swerdlow SH, et al. WHO-EORTC classification for cutaneous lymphomas. Blood. 2005;105:3768–85. doi: 10.1182/blood-2004-09-3502. [DOI] [PubMed] [Google Scholar]
- 2.Criscione VD, Weinstock MA. Incidence of cutaneous T-cell lymphoma in the United States, 1973-2002. Arch Dermatol. 2007;143:854–9. doi: 10.1001/archderm.143.7.854. [DOI] [PubMed] [Google Scholar]
- 3.Korgavkar K, Xiong M, Weinstock M. Changing incidence trends of cutaneous T-cell lymphoma. JAMA dermatology. 2013;149:1295–9. doi: 10.1001/jamadermatol.2013.5526. [DOI] [PubMed] [Google Scholar]
- 4.Hazen PG, Michel B. Hodgkin's disease and mycosis fungoides in a married couple. Dermatologica. 1977;154:257–60. doi: 10.1159/000251078. [DOI] [PubMed] [Google Scholar]
- 5.Hodak E, Klein T, Gabay B, Ben-Amitai D, Bergman R, Gdalevich M, et al. Familial mycosis fungoides: report of 6 kindreds and a study of the HLA system. Journal of the American Academy of Dermatology. 2005;52:393–402. doi: 10.1016/j.jaad.2003.12.052. [DOI] [PubMed] [Google Scholar]
- 6.Litvinov IV, Tetzlaff MT, Rahme E, Habel Y, Risser DR, Gangar P, et al. Identification of geographic clustering and regions spared by cutaneous T-cell lymphoma in Texas using 2 distinct cancer registries. Cancer. 2015:n/a–n/a. doi: 10.1002/cncr.29301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sun G, Berthelot C, Li Y, Glass DA, 2nd, George D, Pandya A, et al. Poor prognosis in non-Caucasian patients with early-onset mycosis fungoides. Journal of the American Academy of Dermatology. 2009;60:231–5. doi: 10.1016/j.jaad.2008.09.063. [DOI] [PubMed] [Google Scholar]
- 8.Jawed SI, Myskowski PL, Horwitz S, Moskowitz A, Querfeld C. Primary cutaneous T-cell lymphoma (mycosis fungoides and Sezary syndrome): part I. Diagnosis: Clinical and histopathologic features and new molecular and biologic markers. Journal of the American Academy of Dermatology. 2014;70:205 e1–16. doi: 10.1016/j.jaad.2013.07.049. quiz 21-2. [DOI] [PubMed] [Google Scholar]
- 9.Campbell JJ, Clark RA, Watanabe R, Kupper TS. Sezary syndrome and mycosis fungoides arise from distinct T-cell subsets: a biologic rationale for their distinct clinical behaviors. Blood. 2010;116:767–71. doi: 10.1182/blood-2009-11-251926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Benton EC, Crichton S, Talpur R, Agar NS, Fields PA, Wedgeworth E, et al. A cutaneous lymphoma international prognostic index (CLIPi) for mycosis fungoides and Sezary syndrome. European journal of cancer. 2013;49:2859–68. doi: 10.1016/j.ejca.2013.04.018. [DOI] [PubMed] [Google Scholar]
- 11.Agar NS, Wedgeworth E, Crichton S, Mitchell TJ, Cox M, Ferreira S, et al. Survival outcomes and prognostic factors in mycosis fungoides/Sezary syndrome: validation of the revised International Society for Cutaneous Lymphomas/European Organisation for Research and Treatment of Cancer staging proposal. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2010;28:4730–9. doi: 10.1200/JCO.2009.27.7665. [DOI] [PubMed] [Google Scholar]
- 12.Talpur R, Singh L, Daulat S, Liu P, Seyfer S, Trynosky T, et al. Long-term outcomes of 1,263 patients with mycosis fungoides and Sezary syndrome from 1982 to 2009. Clinical cancer research: an official journal of the American Association for Cancer Research. 2012;18:5051–60. doi: 10.1158/1078-0432.CCR-12-0604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shin J, Monti S, Aires DJ, Duvic M, Golub T, Jones DA, et al. Lesional gene expression profiling in cutaneous T-cell lymphoma reveals natural clusters associated with disease outcome. Blood. 2007;110:3015–27. doi: 10.1182/blood-2006-12-061507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Litvinov IV, Jones DA, Sasseville D, Kupper TS. Transcriptional profiles predict disease outcome in patients with cutaneous T-cell lymphoma. Clinical cancer research: an official journal of the American Association for Cancer Research. 2010;16:2106–14. doi: 10.1158/1078-0432.CCR-09-2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Litvinov IV, Kupper TS, Sasseville D. The role of AHI1 and CDKN1C in cutaneous T-cell lymphoma progression. Experimental dermatology. 2012;21:964–6. doi: 10.1111/exd.12039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Litvinov IV, Zhou Y, Kupper TS, Sasseville D. Loss of BCL7A expression correlates with poor disease prognosis in patients with early-stage cutaneous T-cell lymphoma. Leukemia & lymphoma. 2012 doi: 10.3109/10428194.2012.717695. [DOI] [PubMed] [Google Scholar]
- 17.Huang Y, Litvinov IV, Wang Y, Su MW, Tu P, Jiang X, et al. Thymocyte selection-associated high mobility group box gene (TOX) is aberrantly over-expressed in mycosis fungoides and correlates with poor prognosis. Oncotarget. 2014;5:4418–25. doi: 10.18632/oncotarget.2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Litvinov IV, Cordeiro B, Fredholm S, Odum N, Zargham H, Huang Y, et al. Analysis of STAT4 expression in cutaneous T-cell lymphoma (CTCL) patients and patient-derived cell lines. Cell cycle. 2014;13:2975–82. doi: 10.4161/15384101.2014.947759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Litvinov IV, Cordeiro B, Huang Y, Zargham H, Pehr K, Dore MA, et al. Ectopic expression of cancer testis antigens in Cutaneous T-Cell Lymphoma (CTCL) patients. Clinical cancer research: an official journal of the American Association for Cancer Research. 2014 doi: 10.1158/1078-0432.CCR-14-0307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krejsgaard T, Litvinov IV, Wang Y, Xia L, Willerslev-Olsen A, Koralov SB, et al. Elucidating the role of interleukin-17F in cutaneous T-cell lymphoma. Blood. 2013;122:943–50. doi: 10.1182/blood-2013-01-480889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Olsen E, Vonderheid E, Pimpinelli N, Willemze R, Kim Y, Knobler R, et al. Revisions to the staging and classification of mycosis fungoides and Sezary syndrome: a proposal of the International Society for Cutaneous Lymphomas (ISCL) and the cutaneous lymphoma task force of the European Organization of Research and Treatment of Cancer (EORTC) Blood. 2007;110:1713–22. doi: 10.1182/blood-2007-03-055749. [DOI] [PubMed] [Google Scholar]
- 22.Chuaqui RF, Bonner RF, Best CJ, Gillespie JW, Flaig MJ, Hewitt SM, et al. Post-analysis follow-up and validation of microarray experiments. Nat Genet. 2002;32(Suppl 1):509–14. doi: 10.1038/ng1034. [DOI] [PubMed] [Google Scholar]
- 23.Morey JS, Ryan JC, Van Dolah FM. Microarray validation: factors influencing correlation between oligonucleotide microarrays and real-time PCR. Biol Proced Online. 2006;8:175–93. doi: 10.1251/bpo126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wurmbach E, Yuen T, Sealfon SC. Focused microarray analysis. Methods. 2003;31:306–16. doi: 10.1016/s1046-2023(03)00161-0. [DOI] [PubMed] [Google Scholar]
- 25.Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3:RESEARCH0034. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mantel N. Evaluation of survival data and two new rank order statistics arising in its consideration. Cancer Chemother Rep. 1966;50:163–70. [PubMed] [Google Scholar]
- 27.van Kester MS, Borg MK, Zoutman WH, Out-Luiting JJ, Jansen PM, Dreef EJ, et al. A meta-analysis of gene expression data identifies a molecular signature characteristic for tumor-stage mycosis fungoides. The Journal of investigative dermatology. 2012;132:2050–9. doi: 10.1038/jid.2012.117. [DOI] [PubMed] [Google Scholar]
- 28.Vega F, Luthra R, Medeiros LJ, Dunmire V, Lee SJ, Duvic M, et al. Clonal heterogeneity in mycosis fungoides and its relationship to clinical course. Blood. 2002;100:3369–73. doi: 10.1182/blood.V100.9.3369. [DOI] [PubMed] [Google Scholar]
- 29.Zhang Y, Wang Y, Yu R, Huang Y, Su M, Xiao C, et al. Molecular markers of early-stage mycosis fungoides. The Journal of investigative dermatology. 2012;132:1698–706. doi: 10.1038/jid.2012.13. [DOI] [PubMed] [Google Scholar]
- 30.Patel LR, Nykter M, Chen K, Zhang W. Cancer genome sequencing: understanding malignancy as a disease of the genome, its conformation, and its evolution. Cancer letters. 2013;340:152–60. doi: 10.1016/j.canlet.2012.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nebozhyn M, Loboda A, Kari L, Rook AH, Vonderheid EC, Lessin S, et al. Quantitative PCR on 5 genes reliably identifies CTCL patients with 5% to 99% circulating tumor cells with 90% accuracy. Blood. 2006;107:3189–96. doi: 10.1182/blood-2005-07-2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kopp KL, Ralfkiaer U, Gjerdrum LM, Helvad R, Pedersen IH, Litman T, et al. STAT5-mediated expression of oncogenic miR-155 in cutaneous T-cell lymphoma. Cell cycle. 2013;12:1939–47. doi: 10.4161/cc.24987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miyagaki T, Sugaya M, Suga H, Kamata M, Ohmatsu H, Fujita H, et al. IL-22, but not IL-17, dominant environment in cutaneous T-cell lymphoma. Clinical cancer research: an official journal of the American Association for Cancer Research. 2011;17:7529–38. doi: 10.1158/1078-0432.CCR-11-1192. [DOI] [PubMed] [Google Scholar]
- 34.Han T, Abdel-Motal UM, Chang DK, Sui J, Muvaffak A, Campbell J, et al. Human anti-CCR4 minibody gene transfer for the treatment of cutaneous T-cell lymphoma. PloS one. 2012;7:e44455. doi: 10.1371/journal.pone.0044455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Foss FM, Waldmann TA. Interleukin-2 receptor-directed therapies for cutaneous lymphomas. Hematology/oncology clinics of North America. 2003;17:1449–58. doi: 10.1016/s0889-8588(03)00110-2. [DOI] [PubMed] [Google Scholar]
- 36.Suga H, Sugaya M, Miyagaki T, Ohmatsu H, Fujita H, Kagami S, et al. Association of nerve growth factor, chemokine (C-C motif) ligands and immunoglobulin E with pruritus in cutaneous T-cell lymphoma. Acta dermato-venereologica. 2013;93:144–9. doi: 10.2340/00015555-1428. [DOI] [PubMed] [Google Scholar]
- 37.Miyagaki T, Sugaya M, Suga H, Ohmatsu H, Fujita H, Asano Y, et al. Increased CCL18 expression in patients with cutaneous T-cell lymphoma: association with disease severity and prognosis. Journal of the European Academy of Dermatology and Venereology: JEADV. 2013;27:e60–7. doi: 10.1111/j.1468-3083.2012.04495.x. [DOI] [PubMed] [Google Scholar]
- 38.Guttinger M, Sutti F, Panigada M, Porcellini S, Merati B, Mariani M, et al. Epithelial V-like antigen (EVA), a novel member of the immunoglobulin superfamily, expressed in embryonic epithelia with a potential role as homotypic adhesion molecule in thymus histogenesis. The Journal of cell biology. 1998;141:1061–71. doi: 10.1083/jcb.141.4.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Petersen DL, Krejsgaard T, Berthelsen J, Fredholm S, Willerslev-Olsen A, Sibbesen NA, et al. B-lymphoid tyrosine kinase (Blk) is an oncogene and a potential target for therapy with Dasatinib in cutaneous T-cell lymphoma (CTCL) Leukemia. 2014 doi: 10.1038/leu.2014.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Carbone A, Bernardini L, Valenzano F, Bottillo I, De Simone C, Capizzi R, et al. Array-based comparative genomic hybridization in early-stage mycosis fungoides: recurrent deletion of tumor suppressor genes BCL7A, SMAC/DIABLO, and RHOF. Genes Chromosomes Cancer. 2008;47:1067–75. doi: 10.1002/gcc.20601. [DOI] [PubMed] [Google Scholar]
- 41.van Doorn R, Zoutman WH, Dijkman R, de Menezes RX, Commandeur S, Mulder AA, et al. Epigenetic profiling of cutaneous T-cell lymphoma: promoter hypermethylation of multiple tumor suppressor genes including BCL7a, PTPRG, and p73. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2005;23:3886–96. doi: 10.1200/JCO.2005.11.353. [DOI] [PubMed] [Google Scholar]
- 42.Mittal A, Chitkara D, Behrman SW, Mahato RI. Efficacy of gemcitabine conjugated and miRNA-205 complexed micelles for treatment of advanced pancreatic cancer. Biomaterials. 2014 doi: 10.1016/j.biomaterials.2014.04.053. [DOI] [PubMed] [Google Scholar]
- 43.Liu Y, Corcoran M, Rasool O, Ivanova G, Ibbotson R, Grander D, et al. Cloning of two candidate tumor suppressor genes within a 10 kb region on chromosome 13q14, frequently deleted in chronic lymphocytic leukemia. Oncogene. 1997;15:2463–73. doi: 10.1038/sj.onc.1201643. [DOI] [PubMed] [Google Scholar]
- 44.Jawed SI, Myskowski PL, Horwitz S, Moskowitz A, Querfeld C. Primary cutaneous T-cell lymphoma (mycosis fungoides and Sezary syndrome): part II. Prognosis, management, and future directions. Journal of the American Academy of Dermatology. 2014;70:223 e1–17. doi: 10.1016/j.jaad.2013.08.033. quiz 40-2. [DOI] [PubMed] [Google Scholar]
- 45.Roman-Gomez J, Cordeu L, Agirre X, Jimenez-Velasco A, San Jose-Eneriz E, Garate L, et al. Epigenetic regulation of Wnt-signaling pathway in acute lymphoblastic leukemia. Blood. 2007;109:3462–9. doi: 10.1182/blood-2006-09-047043. [DOI] [PubMed] [Google Scholar]
- 46.Griffiths EA, Gore SD, Hooker C, McDevitt MA, Karp JE, Smith BD, et al. Acute myeloid leukemia is characterized by Wnt pathway inhibitor promoter hypermethylation. Leukemia & lymphoma. 2010;51:1711–9. doi: 10.3109/10428194.2010.496505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dulmage BO, Geskin LJ. Lessons learned from gene expression profiling of cutaneous T-cell lymphoma. The British journal of dermatology. 2013;169:1188–97. doi: 10.1111/bjd.12578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ciree A, Michel L, Camilleri-Broet S, Jean Louis F, Oster M, Flageul B, et al. Expression and activity of IL-17 in cutaneous T-cell lymphomas (mycosis fungoides and Sezary syndrome) International journal of cancer Journal international du cancer. 2004;112:113–20. doi: 10.1002/ijc.20373. [DOI] [PubMed] [Google Scholar]
- 49.Chong BF, Wilson AJ, Gibson HM, Hafner MS, Luo Y, Hedgcock CJ, et al. Immune function abnormalities in peripheral blood mononuclear cell cytokine expression differentiates stages of cutaneous T-cell lymphoma/mycosis fungoides. Clinical cancer research: an official journal of the American Association for Cancer Research. 2008;14:646–53. doi: 10.1158/1078-0432.CCR-07-0610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wu X, Zhou Y, Hwang S. IL-10 is upregulated in advanced mycosis fungoides and is required for maximal tumor development in a murine model of CTCL. Journal of Investigative Dermatology. 2014;134:S93–S. [Google Scholar]
- 51.Krejsgaard T, Ralfkiaer U, Clasen-Linde E, Eriksen KW, Kopp KL, Bonefeld CM, et al. Malignant cutaneous T-cell lymphoma cells express IL-17 utilizing the Jak3/Stat3 signaling pathway. The Journal of investigative dermatology. 2011;131:1331–8. doi: 10.1038/jid.2011.27. [DOI] [PubMed] [Google Scholar]
- 52.Bar E, Whitney PG, Moor K, Reis e Sousa C, LeibundGut-Landmann S. IL-17 regulates systemic fungal immunity by controlling the functional competence of NK cells. Immunity. 2014;40:117–27. doi: 10.1016/j.immuni.2013.12.002. [DOI] [PubMed] [Google Scholar]
- 53.Zielinski CE, Mele F, Aschenbrenner D, Jarrossay D, Ronchi F, Gattorno M, et al. Pathogen-induced human TH17 cells produce IFN-gamma or IL-10 and are regulated by IL-1beta. Nature. 2012;484:514–8. doi: 10.1038/nature10957. [DOI] [PubMed] [Google Scholar]
- 54.Cruz A, Khader SA, Torrado E, Fraga A, Pearl JE, Pedrosa J, et al. Cutting edge: IFN-gamma regulates the induction and expansion of IL-17-producing CD4 T cells during mycobacterial infection. Journal of immunology. 2006;177:1416–20. doi: 10.4049/jimmunol.177.3.1416. [DOI] [PubMed] [Google Scholar]
- 55.van der Fits L, van Kester MS, Qin Y, Out-Luiting JJ, Smit F, Zoutman WH, et al. MicroRNA-21 expression in CD4+ T cells is regulated by STAT3 and is pathologically involved in Sezary syndrome. The Journal of investigative dermatology. 2011;131:762–8. doi: 10.1038/jid.2010.349. [DOI] [PubMed] [Google Scholar]
- 56.Macleod AS, Rudolph R, Corriden R, Ye I, Garijo O, Havran WL. Skin-Resident T Cells Sense Ultraviolet Radiation-Induced Injury and Contribute to DNA Repair. Journal of immunology. 2014 doi: 10.4049/jimmunol.1303297. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.