Summary
Human intestinal organoids (HIOs) are a tissue culture model in which small intestine-like tissue is generated from pluripotent stem cells. By carrying out unsupervised hierarchical clustering of RNA-sequencing data, we demonstrate that HIOs most closely resemble human fetal intestine. We observed that genes involved in digestive tract development are enriched in both fetal intestine and HIOs compared to adult tissue, whereas genes related to digestive function and Paneth cell host defense are expressed at higher levels in adult intestine. Our study also revealed that the intestinal stem cell marker OLFM4 is expressed at very low levels in fetal intestine and in HIOs, but is robust in adult crypts. We validated our findings using in vivo transplantation to show that HIOs become more adult-like after transplantation. Our study emphasizes important maturation events that occur in the intestine during human development and demonstrates that HIOs can be used to model fetal-to-adult maturation.
Highlights
-
•
HIOs derived from hPSCs remain fetal in vitro
-
•
HIOs become adult-like when transplanted into mice
-
•
Transcriptional profiling across time reveals hallmarks of human gut maturation
-
•
The intestinal stem cell protein OLFM4 is a marker of human crypt maturation
Human pluripotent stem cell-derived intestinal organoids (HIOs) are an in vitro model of the small intestine. Spence and colleagues used transcriptional profiling to demonstrate that HIOs remain fetal in vitro and show that they undergo maturation into adult-like tissue when transplanted in vivo. Their results demonstrate that HIOs are a valuable in vitro model to study the fetal intestine.
Introduction
Three-dimensional in vitro models of the human intestine, such as human intestinal organoids (HIOs), offer immense promise for gastrointestinal (GI) research. HIOs offer the unique ability to understand physiologic interactions of the intestine in an in vitro setting and have potential for future applications in tissue engineering and regenerative medicine. Reports include the use of HIOs for studying viral infections (Finkbeiner et al., 2012), bacterial infections (Leslie et al., 2015), inflammatory bowel disease (Xue et al., 2013), and development of enteroendocrine and β cells (Chen et al., 2014; Du et al., 2012). HIOs are generated using directed differentiation of human pluripotent stem cells (hPSCs) in a stepwise differentiation process that mimics embryonic development (Finkbeiner and Spence, 2013; McCracken et al., 2011; Spence et al., 2011; Wells and Spence, 2014).
We previously showed that HIOs contain both mesenchymal and epithelial cell populations, including the four major epithelial cell types of the small intestine (enterocytes, goblet, Paneth, and enteroendocrine cells) as well as LGR5+ epithelial cells. Moreover, we demonstrated that some cell types were functional, absorbing peptides (enterocytes) and secreting mucin (goblet cells). These findings suggested that HIOs represent mature, functional intestinal tissue (Spence et al., 2011). This conclusion was in contrast to findings in other hPSC-derived lineages, such as pancreatic β-like cells and in lung organoids, which were shown to be more similar to fetal rather than adult tissue (Dye et al., 2015; Hrvatin et al., 2014).
Interestingly, follow-up studies by other groups and ours using HIOs have suggested that HIOs are less mature than adult tissue (Fordham et al., 2013), and HIOs transplanted into the mouse kidney capsule exhibit enhanced cellular function and morphology (Watson et al., 2014). However, the maturity of HIO tissues in vitro remained uncertain, as it was unclear if HIOs represented tissue that was fetal in nature or, alternatively, if expression levels of some genes simply remained low in vitro. Therefore, in this study we sought to do the following: (1) investigate the developmental state of maturity of in-vitro-derived HIOs, (2) determine if HIOs are more similar to human fetal or adult intestinal tissue, and (3) identify cellular and molecular hallmarks of human intestinal maturation. Using RNA-sequencing (RNA-seq) and transcriptome-wide comparisons of HIOs, human fetal, and human adult intestinal tissue, our data show that while HIOs possess markers for functional cell types as originally reported, they are globally more similar to fetal human intestinal tissue than to adult intestine. We also show that the enhanced cellular differentiation and enhanced cellular function after in vivo engraftment of HIOs under the mouse kidney capsule correspond to mature adult-like intestinal tissue. Hallmarks of this maturation process include strong induction of antimicrobial peptide genes produced by Paneth cells and enhanced expression of genes required for digestion. We also found that expression of the Notch-dependent intestinal stem cell (ISC) marker OLFM4 (van der Flier et al., 2009b; VanDussen et al., 2012) is very weakly expressed in HIOs and human fetal tissue, and that acquisition of high levels of OLFM4 expression in the crypt is a hallmark of intestinal maturation. This finding also was confirmed in the developing mouse intestine. Taken together, our results identify major cellular changes that take place during human fetal-to-adult intestinal maturation, demonstrate that acquisition of OLMF4 expression in the crypt is a hallmark of maturation, and highlight that HIOs are a tractable model system that represents human fetal intestinal tissue.
Results
Transcriptome-wide Comparisons Reveal that HIOs More Closely Resemble Fetal Rather Than Adult Intestine
To understand the maturation status of HIOs based on global gene expression patterns, we conducted RNA-seq and compared global gene expression data obtained for undifferentiated H9 stem cells, definitive endoderm, and whole HIOs to existing RNA-seq datasets for fetal and adult human mucosa (epithelium and sub-epithelial mesenchyme) (Table S1). We employed principal component analysis (PCA) to examine variation in transcript abundance among the 23,615 genes contained in each of the RNA-seq datasets. The reduction of this highly multi-dimensional dataset into two dimensions, or principle components, enables the unbiased comparison and visualization of total transcriptional activity between samples. The results suggest that the HIO, human fetal, and adult transcriptomes differ substantially from embryonic stem cell (ESC) and definitive endoderm precursors (Figure 1A). Furthermore, the variation captured in the first two principal components (PC1 and PC2) demonstrates broad similarity between human GI tissues (fetal and adult) and the HIO tissue generated in vitro, suggesting a high degree of similarity in global transcriptional activity among these samples. In the PCA, HIOs clustered more closely with other fetal GI tissue, whereas adult tissues tended to cluster separately. This clustering was more apparent when PCA was conducted with HIO, fetal intestine, and adult intestine only (Figure S1). Since the variation in the larger dataset (Figure 1A) is largely driven by differences between more undifferentiated cells (ESCs and endoderm) and differentiated GI tissue, we conducted additional analyses to examine similarities between different tissues.
Figure 1.
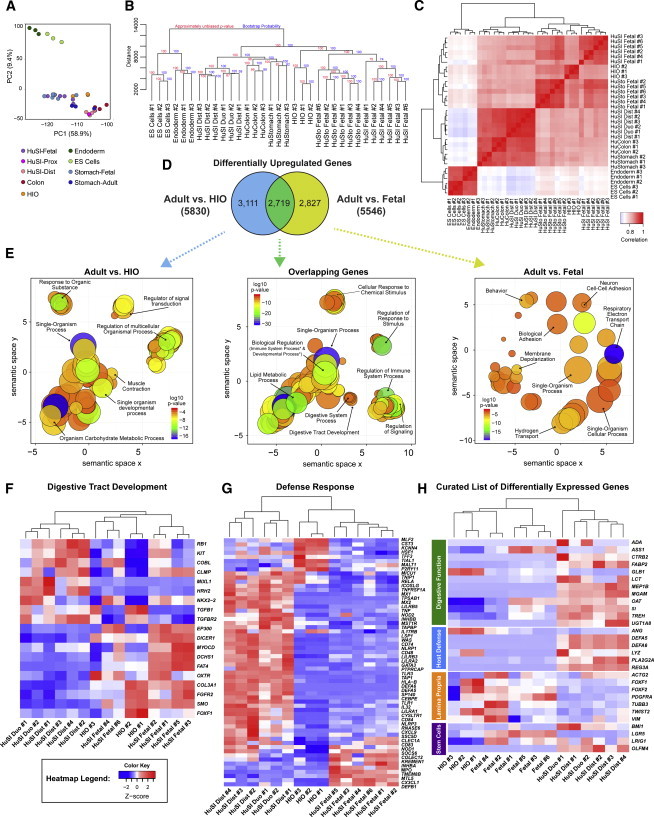
HIOs Resemble Fetal Intestine
(A) PCA was conducted on undifferentiated H9 ESCs (n = 3 independent experiments), definitive endoderm (n = 3 independently differentiated from H9s), HIOs (n = 3 independently differentiated from H9s), adult distal small intestine (n = 4 independent biological specimens), adult duodenum (n = 2 independent biological specimens), adult colon (n = 3 independent biological specimens), adult stomach (n = 3 independent biological specimens), fetal stomach (n = 6 independent biological specimens), and fetal small intestine (n = 6 independent biological specimens) (see also Table S1). The top two principal components account for 68.3% of the variation in the data and show that HIOs group with adult and fetal tissues of the intestine, stomach, and colon, all of which are clearly distinct from endoderm and ESCs.
(B) A dendrogram was generated based on hierarchical clustering of the gene sets using Canberra distance. Fetal tissues are more similar to each other than to their respective adult tissues, and HIOs cluster within the same clade as the fetal tissues. Branch lengths indicate the level of dissimilarity between samples, with longer branch lengths indicating a greater level of dissimilarity. Red labels at branch point correspond to the approximately unbiased (AU) p value. AU > 95 indicates that a given branch assignment is strongly supported by the data. Green labels at each branch point correspond to the bootstrap probability (BP) of a cluster, defined as the frequency of a given relationship among the bootstrap replicates. ES, ESCs; HuSI, human small intestine; HuSI Duo, human small intestine duodenum; HuSI Dist, human distal small intestine; HuSto, human stomach.
(C) Spearman ranking was used to cluster samples and generate a heatmap of similarity, with dark red indicating the highest level of similarity between samples. The dendrogram indicates that HIOs are most similar to human small intestine.
(D) Differential gene expression analysis was carried out to identify genes that are upregulated in adult intestine compared to either HIOs (blue circle) or fetal intestine (yellow circle). The gene lists were then compared to determine how many genes overlap between those two comparisons (green), and the results are shown as a Venn diagram.
(E) The list of genes from each of the three categories of the Venn diagram were subjected to GO analysis to identify enriched GO terms. The results are shown as REVIGO (Supek et al., 2011) scatterplots in which similar GO terms are grouped in arbitrary two-dimensional space based on semantic similarity. Each circle indicates a specific GO term and circle sizes are indicative of how many genes are included in each term, where larger circles indicate a greater number of genes that are included in that GO term. Colors indicate the level of significance of enrichment of the GO term. Asterisks indicate terms that are hidden under other circles.
(F and G) Heatmaps were generated for gene sets belonging to the specific GO terms (F) Digestive Tract Development (GO:0048565) and (G) Defense Response (GO:0006952), and they further demonstrate that HIOs are more similar to fetal intestine with respect to these biological functions of the intestine.
(H) A curated heatmap was generated to examine particular genes that were chosen as being some of the most highly differentially expressed genes and were also of biological interest.
See also Figure S1.
We conducted unsupervised hierarchical clustering of the global gene expression data, based on the Canberra distance (Figure 1B) and Spearman rank correlation (Figure 1C), to assess the relationship of HIOs to the other tissues. In both analyses, stem cells and definitive endoderm formed a distinct out-group relative to the GI tissues, consistent with PCA. Interestingly, human tissues segregate according to their maturation status (fetal or adult) rather than regional identity, consistent with previously published data in mice showing that gene expression in early embryonic stomach and intestine is remarkably similar, and that regional gene expression patterns do not occur until late fetal life (Li et al., 2009). Based on Canberra distance, HIOs shared a third-tier clade with the fetal stomach and fetal small intestine, supporting the high degree of similarity between HIOs and fetal GI tissue relative to adult GI tissue. Despite clustering with fetal stomach, it is important to note that HIOs expressed the intestine-specific marker CDX2 (Figure S3A; Spence et al., 2011). In the Spearman rank correlation (Figure 1C), HIOs also broadly grouped with fetal stomach and fetal small intestine; however, in this analysis, HIOs were most closely related to fetal small intestine. Additional analyses conducted on a targeted subset of data including only HIOs and fetal and adult small intestinal samples consistently demonstrated that HIOs were most closely related to fetal intestine (Figure S1).
To further assess the differences between adult intestine and HIOs and to examine these differences relative to those that exist when comparing adult and fetal human intestines, we carried out differential gene expression analysis (adult versus HIO and adult versus fetal). We identified genes significantly upregulated in adult tissue relative to HIOs (Table S2), and we identified the proportion of these genes that also were upregulated in adult tissue relative to fetal intestine (Table S3). We found that 2,719 genes were commonly upregulated in adult intestine relative to HIOs and fetal tissue, representing 49% of upregulated genes in the adult versus fetal comparison and 46.6% of upregulated genes in the adult versus HIO comparison (Figure 1D). In addition, this comparison allowed us to identify genes uniquely upregulated in the adult versus HIO and in adult versus fetal comparisons. To further explore unique and common genes, we used the Gene Ontology Enrichment Analysis and Visualization (GOrilla) and Reduce and Visualize Gene Ontology (REVIGO) (Eden et al., 2007, 2009; Supek et al., 2011) discovery tools to examine biological processes that are enriched in each gene list (Figure 1E). GOrilla identifies significantly enriched gene ontology (GO) terms from a given gene list, and REVIGO then reduces redundancies by clustering semantically related terms and outputting them as a scatter plot for visualization. There were 240 significant GO terms identified by GOrilla from the list of 3,111 genes that were uniquely upregulated in adult versus HIO tissue. REVIGO reduced these to 72 unique GO terms (Figure 1E, left; Table S4). For the 2,287 genes uniquely upregulated in adult versus fetal intestine, there were 68 significant GO terms identified by GOrilla, which were reduced to 24 unique GO terms in REVIGO (Figure 1E, right; Table S5). Impressively, there were 609 significant GO terms identified from the list of 2,719 overlapping genes (Figure 1E, middle; Table S6). The list of GO terms enriched in the overlapping gene set was then reduced to 103 unique GO terms in REVIGO.
We chose to more closely examine biologically relevant GO terms identified by our analysis that were associated with the intestine (Table S6). To do so, we used fragments per kilobase of exon per million fragments mapped (FPKM) values generated from the RNA-seq dataset, and generated heatmaps for differentially expressed genes that fall under the GO terms “digestive tract development” (GO:0048565; Figure 1F) and “defense response” (GO:0006952; Figure 1G). Interestingly, the hierarchical clustering of the human adult and fetal samples with the HIO samples based solely on the genes contained within the GO category digestive tract development resulted in a dendrogram in which HIOs and fetal samples are intermingled, demonstrating the close relationship that HIOs share with fetal intestine with respect to this set of genes (Figure 1F). Some of the genes within the digestive tract development GO category that were highly expressed in both HIOs and fetal intestine included developmental signaling pathways important in fetal mouse intestine development, such as FGF and HH signaling (Nyeng et al., 2011; Walton et al., 2012).
A large number of genes involved in the defense response are highly expressed in adult intestine but expressed at low levels in both fetal tissue and HIOs (Figure 1G). These include genes involved in host defense against bacteria such as Paneth cell defensins (DEFA5 and DEFA6) and genes for the intracellular bacterial-sensing NOD proteins. Upon examination of gene lists for differential expression (Tables S2 and S3), we also observed that expression of genes involved in digestion, Paneth cell function, and, most interestingly, the ISC marker OLFM4 (van der Flier et al., 2009b; VanDussen et al., 2012) was markedly increased in the adult. We curated a heatmap to highlight a subset of genes involved in the following: (1) digestive function, (2) Paneth cell function, (3) ISC regulation, and (4) expression in the lamina propria. Cumulatively, these data also demonstrated a strong correlation between HIO and fetal tissue compared to the adult (Figure 1H).
HIOs Develop Mature Architecture after In Vivo Transplantation
Previous work demonstrated that in vivo engraftment of HIOs into the kidney capsule of immunocompromised NSG (NOD-scid IL2Rgammanull) mice (Ito et al., 2002) resulted in tissue with enhanced cellular differentiation and morphology (Watson et al., 2014). We sought to examine this further and determine if in vivo engraftment of HIOs would follow maturation into adult-like tissue based on predicted fetal-adult differences observed in the RNA-seq analysis (Figures 1F–1H). HIOs were grown in vitro for ∼3 weeks and divided into two groups. One group was implanted under the kidney capsule of NSG mice; a matched control group was cultured in vitro only. Transplanted and control HIOs were compared after 16 weeks. Control HIOs that remained in vitro possessed epithelial thickenings that resembled villus-like domains, but generally lacked the crypt-villus architecture and underlying core of mesenchymal tissue (lamina propria) that typifies the human adult small intestine. However, transplanted HIOs (tHIOs) developed an architecture that more closely resembled the adult intestine with true villi containing a lamina propria (Figure 2A; Figure 6).
Figure 2.
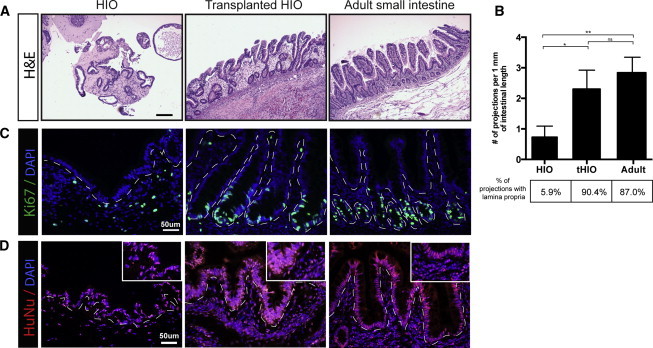
HIOs Develop Mature Architecture after In Vivo Transplantation
(A) H&E sections of an HIO, tHIO, and human intestine demonstrate that HIOs lack the characteristic crypt-villus units that are found in the adult small intestine and that transplanted HIOs (tHIOs) have a morphology that resembles the adult intestine.
(B) Villi morphology was quantified by counting the number of projections with a vimentin-positive core. tHIOs contain a similar number of villi per millimeter of intestinal length as adult intestine, whereas HIOs contain less projections and even fewer true villi. Three independently derived biological replicates for each tissue type with five fields of view for each sample were analyzed. Averages were compared using unpaired t tests. ∗p value 0.01–0.05, ∗∗p value 0.001–0.01.
(C) Ki67 staining demonstrates that the crypt-like domains are also the site of localized proliferation in both tHIOs and adult tissue.
(D) Staining for human nuclear antigen confirms that the tissue of intestinal morphology found in tHIOs is of human origin and not derived from the mouse host.
Figure 6.
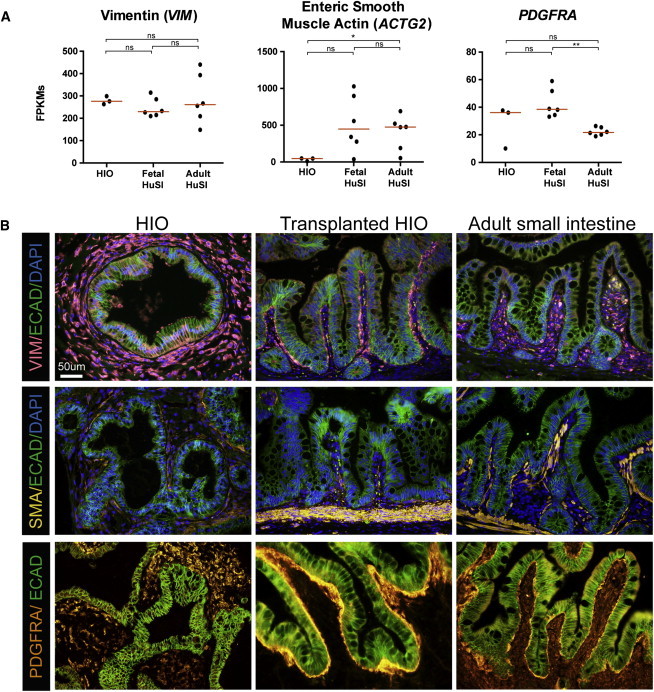
HIOs Express Mesenchymal Cell Markers but Become Organized Like Adult Intestine after Transplantation
(A) Normalized FPKMs were plotted for HIOs (n = 3 independently differentiated from H9s), fetal small intestine (n = 6 independent biological specimens), and adult small intestine (n = 6 independent biological specimens), with each sample represented by a black dot. Red bars indicate the average FPKM value for each group. VIM is expressed to a similar level across HIOs and fetal and adult human intestines. HIOs express lower levels of ACTG2 than adult tissue, with fetal tissue expressing a range of levels. Most of the HIO and fetal intestine samples express a higher level of PDGFRa than adult tissue.
(B) The expression trends are confirmed by immunofluorescence staining with most notable changes in the spatial arrangement of positive cells for all three markers. All samples are biological replicates with an n. ∗p value 0.01–0.05, ∗∗p value 0.001–0.01, ∗∗∗p value 0.0001–0.001.
Projections resembling villi were counted and normalized per unit of length for HIO, tHIO, and adult intestine. Projections were categorized as villus-like, if they did not possess a lamina propria, or as true villi, if they possessed a lamina propria determined by the presence of vimentin staining within the center of the projection (Figure 3). The percentage of projections that were true villi for each condition was determined (Figure 2B). HIOs contained less than one projection per millimeter of epithelial length and only 5.6% of them could be classified as villi. In comparison, tHIO and adult intestine had two to three projections per millimeter of epithelium, of which 90.4% and 87% were true villi with a lamina propria, respectively (Figure 2B). Furthermore, proliferation was localized to the crypt-like domains in tHIOs as indicated by Ki67 staining (Figure 2C). Staining for human nuclear antigen confirmed that the transplanted intestinal tissues were of human origin and therefore derived from transplanted HIOs (Figure 2D).
Figure 3.
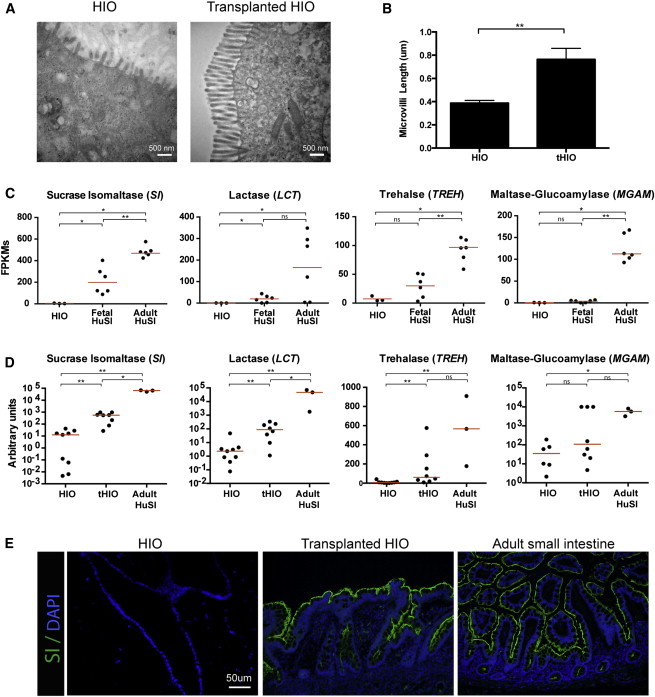
HIOs Express Brush Border Enzymes after In Vivo Transplantation
(A) HIOs have sparse and short microvilli compared to the microvilli present in tHIO cells, as determined by electron microscopy.
(B) Quantification of microvilli length indicates tHIOs have microvilli that are approximately two times longer than microvilli on HIO cells. Six independently derived HIOs and six tHIOs were analyzed with five measurements taken per sample. Averages were compared using unpaired t tests. ∗∗p value 0.001–0.01.
(C) Normalized FPKMs were plotted for HIOs (n = 3 independently differentiated from H9s), fetal small intestine (n = 6 independent biological specimens), and adult small intestine (n = 6 independent biological specimens), with each sample represented by a black dot. Red bars indicate the average FPKM value for each group. Brush border enzymes involved in carbohydrate digestion are expressed at low levels in both HIOs and fetal intestine, whereas they are highly expressed in adult intestine. ∗p value 0.01–0.05, ∗∗p value 0.001–0.01, ∗∗∗p value 0.0001–0.001. All samples are biological replicates.
(D) Transplantation of HIOs into the kidney capsule results in an increase in the expression of brush border enzymes. qRT-PCR analysis was carried out by normalizing gene expression to the housekeeping gene GAPDH and plotting values as a.u. (n = at least 3 biological replicates for each tissue).
(E) Immunofluorescence shows that sucrase isomaltase protein expression is not detectable in HIOs, but is evident in tHIOs and resembles the expression seen in adult intestine.
See also Figures S2 and S3.
HIOs Have Enhanced Brush Border Enzyme Expression after In Vivo Transplantation
HIOs possess the predominant epithelial cell types of the small intestine that are present in both fetal and adult human intestine (Figure 3; Figure S2; Spence et al., 2011). Enterocytes of the small intestine are apically lined with microvilli, which are referred to as the brush border of the intestine. Transmission electron microscopy analysis and comparison of HIOs to tHIOs revealed that the brush border in HIOs is not as well developed (Figure 3A); tHIOs have microvilli that are nearly twice as long as the microvilli present in HIOs (Figure 3B). These findings are consistent with previous reports in which the microvilli in the fetal intestine were reported to be an average of 1 μm in length by 8 weeks of gestation and grow in length from 9–10 weeks of gestation through 15 weeks of gestation (Kelley, 1973; Moxey and Trier, 1979). Studies in the adult have shown that jejunal microvilli range from 0.67 to 1.3 μm in length (Brown and Badylak, 2014), with duodenal microvilli being slightly shorter than jejunal microvilli (Helander and Fändriks, 2014). Consistent with microvilli function, our transcriptome-wide analysis suggested that many critical functional enzymes are expressed at low levels in HIOs and fetal tissue compared to adult (Figures 1G and 3C). For example, FPKM values for the brush border enzymes sucrase isomaltase (SI), trehalase (TREH), lactase (LCT), and maltase glucoamylase (MGAM) demonstrated that expression of these genes was lowest in HIOs and highest in adult intestine (Figure 3C).
To demonstrate that the changes we saw in expression of brush border enzymes were not due to variations in the tissue collected for RNA-seq, we examined expression of multiple epithelial genes (Figure S3A). Through this analysis, we found that there were no consistent trends in epithelial-specific gene expression between any of the samples, suggesting that there were no underlying differences in the samples that could skew our interpretations. For example, there was no consistent evidence to suggest that certain samples had an over-representation of epithelial gene expression compared to the others. This data provided confidence that gene expression differences are reflective of true biological differences.
To determine if brush-border-related gene expression in tHIOs resembles adult tissue, we examined RNA expression of a number of genes by qRT-PCR. Expression in HIOs, tHIOs, and adult intestine was determined and is shown as arbitrary units of expression. We confirmed by qRT-PCR that tHIOs had enhanced expression of many genes, including SI, LCT, and FABP2, relative to HIOs (Figure 3D; Figure S1B). This adult-like pattern of gene expression in the tHIOs was not universally applicable, as some genes, including TREH, MGAM, and MEP1B (Figure 3D; Figure S3B), did not show robust induction after transplantation. Immunofluorescence staining confirmed that tHIOs appeared indistinguishable from adult small intestine in the expression of the brush border enzyme SI, whereas SI was undetectable in control HIOs (Figure 3E).
Paneth Cell Differentiation Is a Hallmark of Intestinal Maturation
Paneth cells play many important roles in the small intestine, acting as a niche for ISCs (Sato et al., 2011b) and secreting a variety of host-defense molecules including antimicrobial peptides (Clevers and Bevins, 2013). Lysozyme (LYZ), an enzyme that destroys the cell wall of bacteria, was the first host-defense product identified in Paneth cells (Deckx et al., 1967; Peeters and Vantrappen, 1975). RNA-seq data confirmed our previous findings (Spence et al., 2011) that LYZ is expressed in HIOs and also demonstrated that the human fetal intestine expresses low levels of LYZ compared to adult intestine (Figure 4A). LYZ expression was variable in HIOs and therefore was not significantly different from either fetal or adult intestine (Figure 4A). Our analysis suggested that HIOs do not possess fully differentiated Paneth cells since other Paneth cell genes/protein had low expression, including DEFA5 (Mallow et al., 1996) and REG3A (Clevers and Bevins, 2013), similar to fetal tissue (Figures 1F and 4A). These results were consistent with previous data showing that DEFA5 expression is low in the fetal intestine and increases as the intestine matures (Mallow et al., 1996).
Figure 4.
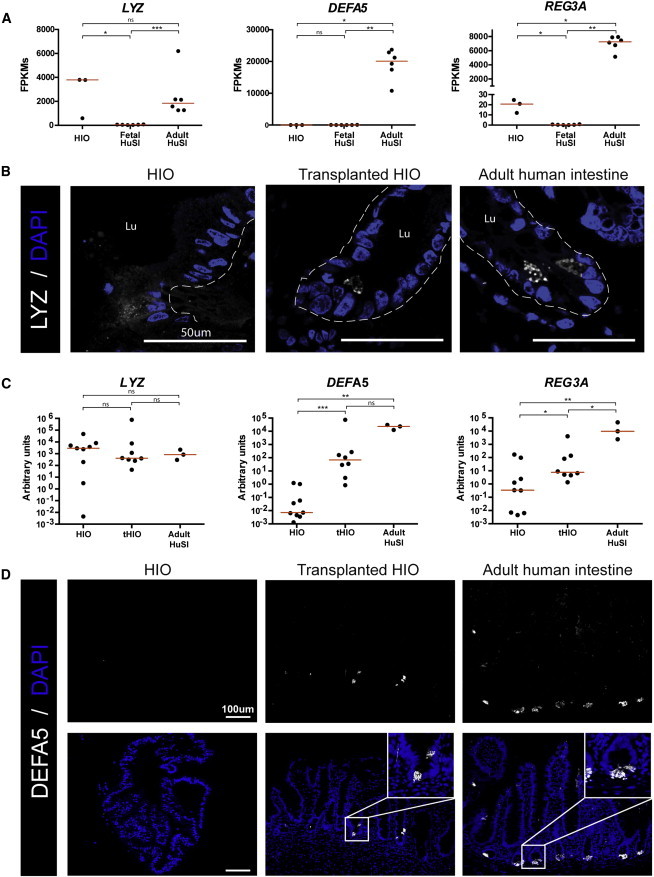
HIOs Form Crypts In Vivo Containing Mature Paneth Cells
(A) Normalized FPKMs were plotted for HIOs (n = 3 independently derived H9-HIOs), fetal small intestine (n = 6 independent biological specimens), and adult small intestine (n = 6 independent biological specimens), with each sample represented by a black dot. Red bars indicate the average FPKM value for each group. Lysozyme (LYZ) is expressed in HIOs at a similar level to adult intestine; however, mature Paneth cell markers DEFA5 and REG3A are both expressed at high levels in adult intestine and expressed at very low levels in both HIO and fetal intestine. ∗p value 0.01–0.05, ∗∗p value 0.001–0.01, ∗∗∗p value 0.0001–0.001. All samples are biological replicates.
(B) Immunofluorescence staining shows that lysozyme is diffusely expressed in HIOs, but is localized to punctate granules in transplanted HIOs and adult intestine.
(C) Transplantation of HIOs (tHIOs) into the kidney capsule results in an increase in expression of Paneth cell genes. qRT-PCR analysis was carried out by normalizing gene expression to the housekeeping gene GAPDH and plotting values as a.u. (n = at least 3 biologically independently differentiated HIOs and tHIOs, or independent biological intestinal specimens).
(D) Immunofluorescence staining shows that α-defensin 5 is not detectable in HIOs, but shows a similar staining pattern in both tHIOs and adult intestine.
Given that Paneth cells reside in the intestinal crypts and that Paneth cell genes were different between HIOs/fetal and adult intestine, we wanted to determine if tHIOs exhibited enhanced Paneth cell gene expression and localization to crypt-like domains. One of the hallmarks of mature Paneth cells is the packaging of antimicrobial peptides and LYZ into secretory granules. Consistent with our previous findings and with reports of LYZ expression in the fetal mouse intestine (Spence et al., 2011), LYZ protein expression was found as diffuse cytoplasmic staining in HIOs; the appearance of distinct secretory granules was not obvious (Figure 4B). This is in contrast to the strong granular lysozyme staining observed in Paneth cells of the adult intestine and tHIOs (Figure 4B). We also examined mRNA expression for DEFA5 and REG3A in HIOs, tHIOs, and adult tissue. High expression of DEFA5 and REG3A was observed in tHIOs, similar to adult intestine and distinct from HIOs (Figure 4C). Consistent with qRT-PCR data, protein expression of α-defensin 5 was not detected by immunofluorescence in HIOs, but was present in granular structures in the crypt-like domains of tHIOs and the crypts of the adult intestine (Figure 4D). Collectively, these results suggest that HIOs contained immature cells that diffusely expressed lysozyme but lacked other antimicrobial peptides. Furthermore, transplantation of HIOs into the murine kidney capsule resulted in Paneth cells that resembled those found in the adult human intestine.
OLFM4 Is a Marker of Mature Intestine
Our results demonstrate that the significant differences in Paneth cell maturity between HIO/fetal intestine and tHIO/adult intestine also were correlated with robust developmental changes in the well-characterized stem cell gene OLFM4 (van der Flier et al., 2009b; VanDussen et al., 2012; Figure 1). We observed low OLFM4 transcript abundance in the HIOs and fetal intestine RNA-seq datasets relative to the adult intestine, which exhibited ∼69- and 108-fold higher expression, respectively (Figure 5A). In contrast, LGR5 was expressed in all tissues examined and showed the most robust expression in the human fetal intestine (Figure 5A), which is consistent with other reports of high LGR5 expression in human fetal small intestine (Fordham et al., 2013). We recently showed robust LGR5 expression in LGR5:eGFP HIOs and tHIOs (Watson et al., 2014) and confirmed this by in situ hybridization (Figure 5D; Figure S5), suggesting that the observed changes in OLFM4 are a unique developmental event. Thus, we further investigated OLFM4 gene and protein expression within HIOs, tHIOs, and adult intestine (Figures 5B and 5C). qRT-PCR analysis demonstrated an increase in OLFM4 expression after HIO transplantation that was comparable to expression in the adult intestine (Figure 5B). At the protein level, OLFM4 was expressed in HIOs in small patches of epithelial cells that were sporadically distributed throughout the epithelium of the organoid. In serial section analysis, OLFM4+ patches were not common and were not localized to any obvious morphological structures (Figure 5C; data not shown). In contrast, OLFM4 expression was regularly observed and spatially restricted to crypt-like domains in tHIOs in the same manner as observed in the adult intestine.
Figure 5.
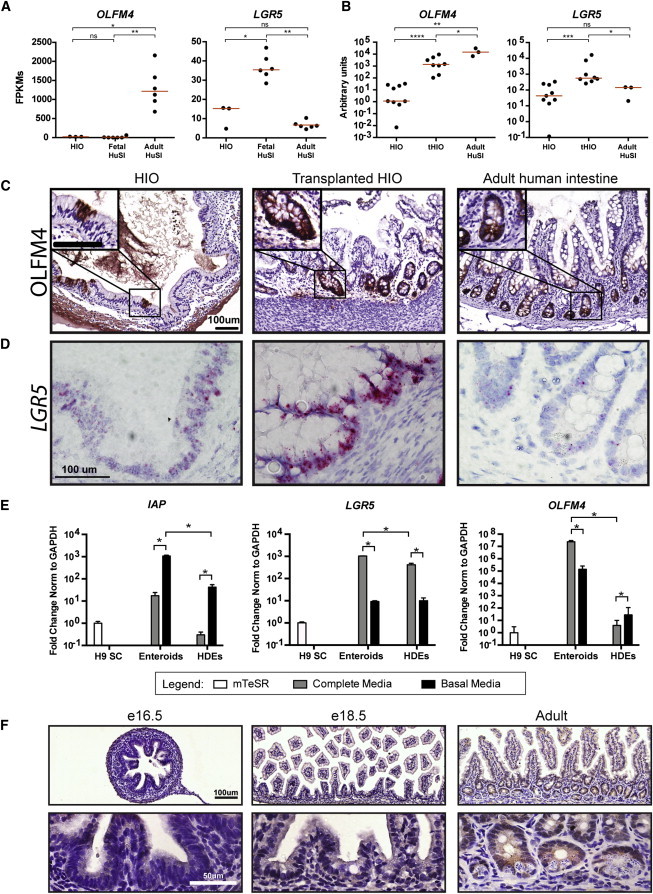
OLFM4 Is a Marker of Mature Intestine
(A) Normalized FPKMs were plotted for HIOs (n = 3 independently differentiated from H9s), fetal small intestine (n = 6 independent biological specimens), and adult small intestine (n = 6 independent biological specimens), with each sample represented by a black dot. Red bars indicate the average FPKM value for each group. LGR5 is expressed to a similar level in both HIOs and adult human intestine; however, there is higher expression detected in human fetal intestine. In contrast, OLFM4 is expressed at a very low level in both HIOs and fetal intestine, but is dramatically higher in adult intestine. All samples are biological replicates.
(B) OLFM4 expression is enhanced in tHIOs and is similar to the level of expression in adult intestine, whereas the level of expression in HIOs is much lower. qRT-PCR analysis was carried out by normalizing gene expression to the housekeeping gene GAPDH and plotting values as a.u. (n = at least 3 biological replicates for each tissue).
(C) Immunohistochemistry demonstrates that HIOs do express OLFM4 protein; however, OLFM4 is sporadically expressed throughout HIOs and cannot be detected in all histological sections. After transplantation, OLFM4 is readily detectable in all histological sections, and it is localized to crypt-like domains in a pattern similar to adult human intestine.
(D) In situ hybridization confirms that tHIOs express a higher level of LGR5 and that HIOs express a level similar to adult intestine.
(E) Epithelium-only cultures were used to functionally analyze the responsiveness of enterocyte and stem cell gene expression to changes in culture conditions. Mature human enteroids express a higher level of IAP than immature HDEs when grown in complete media, which favors stem cells (gray bars, left). An increase in IAP expression in basal media for both enteroids and HDEs confirms that removal of pro-stem cell factors results in increased differentiation as expected (black bars, left). LGR5 is expressed to similar levels in both enteroids and HDEs grown in complete media (gray bars, middle). In contrast, OLFM4 is highly expressed in enteroids, but is barely detected in HDEs (gray bars, right). Growth of both enteroids and HDEs in basal media results in a reduction of LGR5 expression (black bars, middle); however, OLFM4 expression is only reduced in enteroids, but not HDEs grown in basal media (black bars, right). n = 3 biological replicates for all tissues. ∗p value 0.01–0.05.
(F) To confirm the developmental pattern of OLFM4 seen in human tissues, we also examined histological sections of mouse intestine at different developmental ages. OLFM4 was only detected in the adult intestine and not at any of the embryonic ages examined.
See also Figures S4 and S5.
Many reports have demonstrated OLFM4 to be a reliable marker of LGR5+ crypt base columnar (CBC) stem cells in the adult small intestine (Schuijers et al., 2014; van der Flier et al., 2009a). To further probe the stem cell populations in HIOs, we compared HIOs to adult human crypt-derived enteroids (Sato et al., 2011a) in functional experiments. Importantly, enteroids are epithelium-only structures. Therefore, to carry out fair comparisons between the two systems, we used gentle enzymatic digestion and mechanical disruption to remove the mesenchymal cell population from HIOs, and we grew the epithelium from HIOs in the same human complete media used to grow enteroids (Figure S4; Table S7). The resulting epithelium contained structures that morphologically resembled human enteroids and are referred to as HIO-derived epithelium (HDE).
To examine the functional response of stem cells in enteroids and HDEs, we carried out a straightforward media-swapping experiment. Enteroids or HDEs were grown in human complete media for 7 days and subsequently either maintained in complete media, which promotes stem cell maintenance, or switched to basal media lacking small molecules and growth factors for an additional 7 days (Table S7). We predicted that complete media would promote maintenance of stem cells and expression of LGR5 and OLFM4 in human enteroids, whereas basal media would result in a reduction of stem cell gene expression and an increase in differentiated cell markers. qRT-PCR expression of intestinal alkaline phosphatase (IAP), which is expressed by mature enterocytes (Hinnebusch et al., 2004), was below the level of detection in HDEs (based on undifferentiated H9 human ESCs as a reference), though it could be detected in enteroids grown in complete media (Figure 5E, gray bars). As predicted, upon switching to basal media, a significant induction of IAP occurred in both HDE and enteroids (Figure 5E, gray versus black bars). Under standard culture conditions in complete media, LGR5 was statistically higher in enteroids than HDEs albeit only modestly so (Figure 5E, gray bars). As predicted, in both enteroids and HDEs, basal media conditions led to reduced expression of LGR5. In contrast to LGR5, OLFM4 was robustly expressed in enteroids and was barely detectable in HDEs. Moreover, basal media conditions led to a significant reduction of OLFM4 only in adult enteroids (Figure 5E, gray versus black bars).
To corroborate our findings from HIOs and human tissue, we also examined OLFM4 protein expression at different stages of mouse development using immunohistochemistry. OLFM4 protein could not be detected at any of the fetal time points that we examined, but was present in the crypts in histological sections of adult mouse intestine (Figure 5F). These results are supported by microarray studies by others showing a robust increase in OLFM4 gene expression from postnatal day (p)4 to p30 (Schjoldager et al., 2008; GEO profile 45876166). Collectively, our results suggest that acquisition of strong expression of OLFM4 in adult ISCs is a hallmark of the maturation process in humans and mice.
HIO Mesenchymal Cell Populations Gain Adult-like Organization after Transplantation
Given the emergence of villi in tHIOs (Figure 2B), we also wanted to examine cell types found in the lamina propria of the intestine to see how they compare between fetal and adult tissues, and in HIOs, tHIOs, and adult tissue. Based on our RNA-seq analysis, expression of mesenchymal markers vimentin (VIM) and smooth muscle actin (ACTG2) were not statistically different between fetal and adult intestines (Figure 6A). PDGFR-α-positive mesenchymal clusters have been demonstrated to direct the emergence of villi in mice (Walton et al., 2012). Consistent with this notion, PDGFRA expression was statistically higher in fetal intestine compared to adult intestine. All three markers were detectable at the protein level and were expressed throughout the mesenchyme of HIOs (Figure 6B). After transplantation, however, the organization of the mesenchymal cells resembled that seen in adult intestine, most notably for smooth muscle actin (SMA) and PDGFRA. A prominent band of organized SMA+ smooth muscle was present beneath the epithelium of both tHIOs and adult intestine. Likewise, PDGFRA+ cells lined the basal side of the epithelial cells of the villi in both tHIOs and adult intestine, whereas PDGFRA+ cells were spread throughout HIOs.
Discussion
We examined gene expression in human intestinal tissue across the developmental continuum in order to determine maturity of in-vitro-derived HIOs. Using global RNA expression data obtained by RNA-seq, several different analyses demonstrated that the HIO transcriptome closely resembles global gene expression in the human fetal intestine, strongly suggesting that HIOs more accurately represent fetal intestinal tissue than adult intestinal tissue. The adult intestine is a complex organ that performs many important metabolic and immunologic functions and hosts a tremendous population of indigenous bacteria. Our RNA-seq analysis supports the existing body of literature demonstrating that the human fetal intestine is immature with respect to advanced metabolic and host-defense functions, and that acquisition of these features is a hallmark of maturation to adult function. Similar to the human fetal gut, we have demonstrated that HIOs lack strong expression of many digestion-related genes and Paneth cells. We have further demonstrated that HIOs are competent to acquire these attributes of the adult intestine. This process requires transplantation into an in vivo context, suggesting that our in vitro conditions lack important cues for maturation. We suspect that cues from multiple sources are required to induce full maturation. For example, animal studies have highlighted the importance of intestinal microbial colonization in enhancing intestinal function, development of the intestinal immune system, and protection against pathogenic infection (Cash et al., 2006; Hooper, 2004; Hooper et al., 1999, 2001, 2012; Hudault et al., 2001). Moreover, it was recently demonstrated that HIOs transplanted into the kidney capsule could respond to circulating factors (Watson et al., 2014).
Our finding that OLFM4 is not highly expressed in human or mouse fetal intestine but marks ISCs in the adult intestine is intriguing and suggests that there may be significant differences between fetal and adult ISCs. Importantly, the two predominant pathways that regulate adult ISCs, Wnt and Notch, are both thought to be important in late fetal development in mice (Korinek et al., 1998; Tsai et al., 2014; VanDussen et al., 2012; Zhong et al., 2012). Thus, it is possible that acquisition of an adult ISC state occurs gradually across the developmental continuum, such that there are differences in ISC regulation in early fetal development, late fetal development, postnatally, and in adult life. At this time, careful studies across different developmental ages have not been carried out. It is also currently unclear if the gene expression differences between fetal and adult life are functionally significant because, for example, Olfm4 knockout animals do not show a phenotype (Schuijers et al., 2014).
In addition to HIOs, other groups have established primary cell culture models of the human and mouse fetal intestine, called fetal enterospheres (Fordham et al., 2013; Mustata et al., 2013). Mouse fetal enterospheres only retain fetal properties for a limited time because they continue to develop into adult-like organoids. Therefore, it appears as though the developmental program may be hardwired into the epithelium that is removed from the fetal gut, and may pose limitations to the use of primary fetal tissue. While fetal enterospheres are an invaluable tool, our results show that HIOs offer a complementary system that may be more accessible to a wider portion of the research community. Given the difficulty or inability to obtain human fetal tissue for many researchers, in addition to the ethical and legal concerns of using this tissue in biomedical research, our data suggest that HIOs provide an appropriate alternative model.
Taken together, our results show that HIOs represent the fetal/immature intestine, and we have identified several hallmarks of fetal-to-adult intestine development that can be modeled in HIOs transplanted into mice. Given the limitations in acquiring human fetal tissue and the even greater limitations of performing functional experiments on this tissue, HIOs provide unique opportunities to explore human fetal intestinal biology. Importantly, we also have demonstrated that discoveries made using HIOs are recapitulated during fetal development and adult maturation in mice, highlighting the power of this system to make important discoveries.
Experimental Procedures
Generation of HIOs
HIOs were generated as previously described (McCracken et al., 2011; Spence et al., 2011; Xue et al., 2013). For details, see the Supplemental Experimental Procedures.
Human Tissue
Human adult small intestinal tissue was collected from deceased donors through the Gift of Life, Michigan. Institutional review board (IRB) approval was obtained for use of human tissues (University of Michigan IRB HUM00024263). Segments of the small intestine were collected, and mucosal scrapings were used to generate enteroids or collected and snap frozen for RNA analysis. Whole-thickness tissue was fixed and embedded in paraffin or cryomold protectant (O.C.T., Fisherbrand) for histological purposes. HIOs were collected in the same manner.
Growth of Adult Human Enteroids
Human enteroids were isolated as previously described (Miyoshi et al., 2012). Enteroids were propagated in complete media as described in Table S7. Conditioned media were generated from Wnt3a-expressing L cells (ATCC CRL-2647) and R-spondin 2-expressing cells (Bell et al., 2008). Biological replicates of enteroids consisted of ∼15–20 enteroids per replicate.
RNA-Seq and Analysis
RNA was extracted using the Trizol extraction protocol from the manufacturer (Life Technologies). Library preparation and sequencing of HIOs was performed in the University of Michigan DNA Sequencing Core on the Illumina Hi-Seq 2000 platform using TruSeq library preparations. The Bioinformatics Core at the University of Michigan processed all RNA-seq data. See the Supplemental Experimental Procedures for a detailed description of methods and for details regarding informatics analysis. All data and analyses are available online (https://github.com/hilldr/Finkbeiner_StemCellReports2015).
Kidney Capsule Implants
As previously described (Watson et al., 2014), HIOs that had been in culture for ∼3 weeks were removed from matrigel and embedded into purified type I collagen (BD Biosciences) and allowed to form a solid plug before being placed back into standard growth media overnight. NSG mice (NOD-scid IL2Rgammanull) (Ito et al., 2002) were used for the implants. All animal work was reviewed and approved by institutional animal care and use committees.
qRT-PCR, Histology, and Microscopy
The qRT-PCR, histology, immunostaining, and microscopy were essentially carried out as previously described (Rockich et al., 2013; Spence et al., 2009, 2011). For additional details, see the Supplemental Experimental Procedures.
Statistical Analyses
Data are expressed as the median of each sample set with each data point represented in the plots. Fetal samples from different post-conception days were treated as one group, available adult small intestinal samples were treated as another, and then HIO and tHIOs were treated as their own groups. Unless otherwise noted in the figure legend, pairwise comparisons were carried out as indicated with GraphPad Prism 5.0 software using unpaired Mann-Whitney t tests due to the non-normal distribution of the data. Significant values are denoted as follows: ∗p value 0.01–0.05, ∗∗p value 0.001–0.01, and ∗∗∗p value 0.0001–0.001. At least three biological replicates for each group were used in all experiments.
Author Contributions
S.R.F. and J.R.S. conceived the study. S.R.F., D.R.H., M.J.T., P.H.D., Y.-H.T., A.M.C., C.H.A., M.M.M., C.L.W., and J.J.F. conducted experiments. S.R.F., D.R.H., M.J.T., P.H.D., Y.-H.T., P.J.D., and J.R.S. analyzed results. R.N., M.T., M.M.M., O.D.K., N.F.S., M.A.H., D.H.T., and P.J.D. provided critical material support. S.R.F. and J.R.S. wrote the manuscript. All authors read and edited the manuscript and approved the final contents.
Acknowledgments
We thank Rich McEachin and Manjusha Pande at the University of Michigan Bioinformatics Core Facility. We also thank the Gift of Life, Michigan, for access to human intestinal tissue. This work was supported by grants from the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK, K01DK091415 and U01DK103141) to J.R.S. and by the University of Michigan Center for Gastrointestinal Research (UMCGR) (NIDDK, 5P30DK034933). Research reported in this publication and conducted by S.R.F. was supported by an NIDDK training grant, “Training in Basic and Translational Digestive Sciences” (T32DK094775), a postdoctoral fellowship from The Hartwell Foundation, and the National Center for Advancing Translational Sciences of the NIH (2UL1TR000433). Additionally, this work was supported by a grant from the California Institute for Regenerative Medicine (RN3-06525) to O.D.K. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Published: June 4, 2015
Footnotes
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/4.0/).
Accession Numbers
The accession number for all sequences reported in this paper is EMBL-EBI ArrayExpress: E-MTAB-3158.
Supplemental Information
References
- Bell S.M., Schreiner C.M., Wert S.E., Mucenski M.L., Scott W.J., Whitsett J.A. R-spondin 2 is required for normal laryngeal-tracheal, lung and limb morphogenesis. Development. 2008;135:1049–1058. doi: 10.1242/dev.013359. [DOI] [PubMed] [Google Scholar]
- Brown B.N., Badylak S.F. Extracellular matrix as an inductive scaffold for functional tissue reconstruction. Transl. Res. 2014;163:268–285. doi: 10.1016/j.trsl.2013.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cash H.L., Whitham C.V., Behrendt C.L., Hooper L.V. Symbiotic bacteria direct expression of an intestinal bactericidal lectin. Science. 2006;313:1126–1130. doi: 10.1126/science.1127119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y.-J., Finkbeiner S.R., Weinblatt D., Emmett M.J., Tameire F., Yousefi M., Yang C., Maehr R., Zhou Q., Shemer R. De novo formation of insulin-producing “neo-β cell islets” from intestinal crypts. Cell Rep. 2014;6:1046–1058. doi: 10.1016/j.celrep.2014.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clevers H.C., Bevins C.L. Paneth cells: maestros of the small intestinal crypts. Annu. Rev. Physiol. 2013;75:289–311. doi: 10.1146/annurev-physiol-030212-183744. [DOI] [PubMed] [Google Scholar]
- Deckx R.J., Vantrappen G.R., Parein M.M. Localization of lysozyme activity in a Paneth cell granule fraction. Biochim. Biophys. Acta. 1967;139:204–207. doi: 10.1016/0005-2744(67)90136-2. [DOI] [PubMed] [Google Scholar]
- Du A., McCracken K.W., Walp E.R., Terry N.A., Klein T.J., Han A., Wells J.M., May C.L. Arx is required for normal enteroendocrine cell development in mice and humans. Dev. Biol. 2012;365:175–188. doi: 10.1016/j.ydbio.2012.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dye B.R., Hill D.R., Ferguson M.A., Tsai Y.-H., Nagy M.S., Dyal R., Wells J.M., Mayhew C.N., Nattiv R., Klein O.D. In vitro generation of human pluripotent stem cell derived lung organoids. eLife. 2015;4:e05098. doi: 10.7554/eLife.05098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eden E., Lipson D., Yogev S., Yakhini Z. Discovering motifs in ranked lists of DNA sequences. PLoS Comput. Biol. 2007;3:e39. doi: 10.1371/journal.pcbi.0030039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eden E., Navon R., Steinfeld I., Lipson D., Yakhini Z. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics. 2009;10:48. doi: 10.1186/1471-2105-10-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkbeiner S.R., Spence J.R. A gutsy task: generating intestinal tissue from human pluripotent stem cells. Dig. Dis. Sci. 2013;58:1176–1184. doi: 10.1007/s10620-013-2620-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkbeiner S.R., Zeng X.-L., Utama B., Atmar R.L., Shroyer N.F., Estes M.K. Stem cell-derived human intestinal organoids as an infection model for rotaviruses. MBio. 2012;3:e00159. doi: 10.1128/mBio.00159-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fordham R.P., Yui S., Hannan N.R.F., Soendergaard C., Madgwick A., Schweiger P.J., Nielsen O.H., Vallier L., Pedersen R.A., Nakamura T. Transplantation of expanded fetal intestinal progenitors contributes to colon regeneration after injury. Cell Stem Cell. 2013;13:734–744. doi: 10.1016/j.stem.2013.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helander H.F., Fändriks L. Surface area of the digestive tract - revisited. Scand. J. Gastroenterol. 2014;49:681–689. doi: 10.3109/00365521.2014.898326. [DOI] [PubMed] [Google Scholar]
- Hinnebusch B.F., Siddique A., Henderson J.W., Malo M.S., Zhang W., Athaide C.P., Abedrapo M.A., Chen X., Yang V.W., Hodin R.A. Enterocyte differentiation marker intestinal alkaline phosphatase is a target gene of the gut-enriched Kruppel-like factor. Am. J. Physiol. Gastrointest. Liver Physiol. 2004;286:G23–G30. doi: 10.1152/ajpgi.00203.2003. [DOI] [PubMed] [Google Scholar]
- Hooper L.V. Bacterial contributions to mammalian gut development. Trends Microbiol. 2004;12:129–134. doi: 10.1016/j.tim.2004.01.001. [DOI] [PubMed] [Google Scholar]
- Hooper L.V., Xu J., Falk P.G., Midtvedt T., Gordon J.I. A molecular sensor that allows a gut commensal to control its nutrient foundation in a competitive ecosystem. Proc. Natl. Acad. Sci. USA. 1999;96:9833–9838. doi: 10.1073/pnas.96.17.9833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper L.V., Wong M.H., Thelin A., Hansson L., Falk P.G., Gordon J.I. Molecular analysis of commensal host-microbial relationships in the intestine. Science. 2001;291:881–884. doi: 10.1126/science.291.5505.881. [DOI] [PubMed] [Google Scholar]
- Hooper L.V., Littman D.R., Macpherson A.J. Interactions between the microbiota and the immune system. Science. 2012;336:1268–1273. doi: 10.1126/science.1223490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrvatin S., O’Donnell C.W., Deng F., Millman J.R., Pagliuca F.W., DiIorio P., Rezania A., Gifford D.K., Melton D.A. Differentiated human stem cells resemble fetal, not adult, β cells. Proc. Natl. Acad. Sci. USA. 2014;111:3038–3043. doi: 10.1073/pnas.1400709111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudault S., Guignot J., Servin A.L. Escherichia coli strains colonising the gastrointestinal tract protect germfree mice against Salmonella typhimurium infection. Gut. 2001;49:47–55. doi: 10.1136/gut.49.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito M., Hiramatsu H., Kobayashi K., Suzue K., Kawahata M., Hioki K., Ueyama Y., Koyanagi Y., Sugamura K., Tsuji K. NOD/SCID/gamma(c)(null) mouse: an excellent recipient mouse model for engraftment of human cells. Blood. 2002;100:3175–3182. doi: 10.1182/blood-2001-12-0207. [DOI] [PubMed] [Google Scholar]
- Kelley R.O. An ultrastructural and cytochemical study of developing small intestine in man. J. Embryol. Exp. Morphol. 1973;29:411–430. [PubMed] [Google Scholar]
- Korinek V., Barker N., Moerer P., van Donselaar E., Huls G., Peters P.J., Clevers H. Depletion of epithelial stem-cell compartments in the small intestine of mice lacking Tcf-4. Nat. Genet. 1998;19:379–383. doi: 10.1038/1270. [DOI] [PubMed] [Google Scholar]
- Leslie J.L., Huang S., Opp J.S., Nagy M.S., Kobayashi M., Young V.B., Spence J.R. Persistence and toxin production by Clostridium difficile within human intestinal organoids result in disruption of epithelial paracellular barrier function. Infect. Immun. 2015;83:138–145. doi: 10.1128/IAI.02561-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Udager A.M., Hu C., Qiao X.T., Richards N., Gumucio D.L. Dynamic patterning at the pylorus: formation of an epithelial intestine-stomach boundary in late fetal life. Dev. Dyn. 2009;238:3205–3217. doi: 10.1002/dvdy.22134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallow E.B., Harris A., Salzman N., Russell J.P., DeBerardinis R.J., Ruchelli E., Bevins C.L. Human enteric defensins. Gene structure and developmental expression. J. Biol. Chem. 1996;271:4038–4045. doi: 10.1074/jbc.271.8.4038. [DOI] [PubMed] [Google Scholar]
- McCracken K.W., Howell J.C., Wells J.M., Spence J.R. Generating human intestinal tissue from pluripotent stem cells in vitro. Nat. Protoc. 2011;6:1920–1928. doi: 10.1038/nprot.2011.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi H., Ajima R., Luo C.T., Yamaguchi T.P., Stappenbeck T.S. Wnt5a potentiates TGF-β signaling to promote colonic crypt regeneration after tissue injury. Science. 2012;338:108–113. doi: 10.1126/science.1223821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moxey P.C., Trier J.S. Development of villus absorptive cells in the human fetal small intestine: a morphological and morphometric study. Anat. Rec. 1979;195:463–482. doi: 10.1002/ar.1091950307. [DOI] [PubMed] [Google Scholar]
- Mustata R.C., Vasile G., Fernandez-Vallone V., Strollo S., Lefort A., Libert F., Monteyne D., Pérez-Morga D., Vassart G., Garcia M.-I. Identification of Lgr5-independent spheroid-generating progenitors of the mouse fetal intestinal epithelium. Cell Rep. 2013;5:421–432. doi: 10.1016/j.celrep.2013.09.005. [DOI] [PubMed] [Google Scholar]
- Nyeng P., Bjerke M.A., Norgaard G.A., Qu X., Kobberup S., Jensen J. Fibroblast growth factor 10 represses premature cell differentiation during establishment of the intestinal progenitor niche. Dev. Biol. 2011;349:20–34. doi: 10.1016/j.ydbio.2010.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peeters T., Vantrappen G. The Paneth cell: a source of intestinal lysozyme. Gut. 1975;16:553–558. doi: 10.1136/gut.16.7.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockich B.E., Hrycaj S.M., Shih H.P., Nagy M.S., Ferguson M.A.H., Kopp J.L., Sander M., Wellik D.M., Spence J.R. Sox9 plays multiple roles in the lung epithelium during branching morphogenesis. Proc. Natl. Acad. Sci. USA. 2013;110:E4456–E4464. doi: 10.1073/pnas.1311847110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T., Stange D.E., Ferrante M., Vries R.G.J., Van Es J.H., Van den Brink S., Van Houdt W.J., Pronk A., Van Gorp J., Siersema P.D., Clevers H. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett’s epithelium. Gastroenterology. 2011;141:1762–1772. doi: 10.1053/j.gastro.2011.07.050. [DOI] [PubMed] [Google Scholar]
- Sato T., van Es J.H., Snippert H.J., Stange D.E., Vries R.G., van den Born M., Barker N., Shroyer N.F., van de Wetering M., Clevers H. Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature. 2011;469:415–418. doi: 10.1038/nature09637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schjoldager K.T.B.G., Maltesen H.R., Balmer S., Lund L.R., Claesson M.H., Sjöström H., Troelsen J.T., Olsen J. Cellular cross talk in the small intestinal mucosa: postnatal lymphocytic immigration elicits a specific epithelial transcriptional response. Am. J. Physiol. Gastrointest. Liver Physiol. 2008;294:G1335–G1343. doi: 10.1152/ajpgi.00265.2007. [DOI] [PubMed] [Google Scholar]
- Schuijers J., van der Flier L.G., van Es J., Clevers H. Robust cre-mediated recombination in small intestinal stem cells utilizing the olfm4 locus. Stem Cell Reports. 2014;3:234–241. doi: 10.1016/j.stemcr.2014.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spence J.R., Lange A.W., Lin S.-C.J., Kaestner K.H., Lowy A.M., Kim I., Whitsett J.A., Wells J.M. Sox17 regulates organ lineage segregation of ventral foregut progenitor cells. Dev. Cell. 2009;17:62–74. doi: 10.1016/j.devcel.2009.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spence J.R., Mayhew C.N., Rankin S.A., Kuhar M.F., Vallance J.E., Tolle K., Hoskins E.E., Kalinichenko V.V., Wells S.I., Zorn A.M. Directed differentiation of human pluripotent stem cells into intestinal tissue in vitro. Nature. 2011;470:105–109. doi: 10.1038/nature09691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supek F., Bošnjak M., Škunca N., Šmuc T. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS ONE. 2011;6:e21800. doi: 10.1371/journal.pone.0021800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai Y.-H., VanDussen K.L., Sawey E.T., Wade A.W., Kasper C., Rakshit S., Bhatt R.G., Stoeck A., Maillard I., Crawford H.C. ADAM10 regulates Notch function in intestinal stem cells of mice. Gastroenterology. 2014;147:822–834. doi: 10.1053/j.gastro.2014.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Flier L.G., Haegebarth A., Stange D.E., van de Wetering M., Clevers H. OLFM4 is a robust marker for stem cells in human intestine and marks a subset of colorectal cancer cells. Gastroenterology. 2009;137:15–17. doi: 10.1053/j.gastro.2009.05.035. [DOI] [PubMed] [Google Scholar]
- van der Flier L.G., van Gijn M.E., Hatzis P., Kujala P., Haegebarth A., Stange D.E., Begthel H., van den Born M., Guryev V., Oving I. Transcription factor achaete scute-like 2 controls intestinal stem cell fate. Cell. 2009;136:903–912. doi: 10.1016/j.cell.2009.01.031. [DOI] [PubMed] [Google Scholar]
- VanDussen K.L., Carulli A.J., Keeley T.M., Patel S.R., Puthoff B.J., Magness S.T., Tran I.T., Maillard I., Siebel C., Kolterud Å. Notch signaling modulates proliferation and differentiation of intestinal crypt base columnar stem cells. Development. 2012;139:488–497. doi: 10.1242/dev.070763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walton K.D., Kolterud A., Czerwinski M.J., Bell M.J., Prakash A., Kushwaha J., Grosse A.S., Schnell S., Gumucio D.L. Hedgehog-responsive mesenchymal clusters direct patterning and emergence of intestinal villi. Proc. Natl. Acad. Sci. USA. 2012;109:15817–15822. doi: 10.1073/pnas.1205669109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson C.L., Mahe M.M., Múnera J., Howell J.C., Sundaram N., Poling H.M., Schweitzer J.I., Vallance J.E., Mayhew C.N., Sun Y. An in vivo model of human small intestine using pluripotent stem cells. Nat. Med. 2014;20:1310–1314. doi: 10.1038/nm.3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells J.M., Spence J.R. How to make an intestine. Development. 2014;141:752–760. doi: 10.1242/dev.097386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue X., Ramakrishnan S., Anderson E., Taylor M., Zimmermann E.M., Spence J.R., Huang S., Greenson J.K., Shah Y.M. Endothelial PAS domain protein 1 activates the inflammatory response in the intestinal epithelium to promote colitis in mice. Gastroenterology. 2013;145:831–841. doi: 10.1053/j.gastro.2013.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Z., Baker J.J., Zylstra-Diegel C.R., Williams B.O. Lrp5 and Lrp6 play compensatory roles in mouse intestinal development. J. Cell. Biochem. 2012;113:31–38. doi: 10.1002/jcb.23324. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


