Abstract
WWOX, the WW domain-containing oxidoreductase gene at chromosome region 16q23.3–q24.1, spanning chromosomal fragile site FRA16D, encodes the 46 kDa Wwox protein, a tumor suppressor that is lost or reduced in expression in a wide variety of cancers, including breast, prostate, ovarian, and lung. The function of Wwox as a tumor suppressor implies that it serves a function in the prevention of carcinogenesis. Indeed, in vitro studies show that Wwox protein interacts with many binding partners to regulate cellular apoptosis, proliferation, and/or maturation. It has been reported that newborn Wwox knockout mice exhibit nascent osteosarcomas while Wwox+/− mice exhibit increased incidence of spontaneous and induced tumors. Furthermore, absence or reduction of Wwox expression in mouse xenograft models results in increased tumorigenesis, which can be rescued by Wwox re-expression, though there is not universal agreement among investigators regarding the role of Wwox loss in these experimental models. Despite this proposed tumor suppressor function, the overlap of the human WWOX locus with FRA16D sensitizes the gene to protein-inactivating deletions caused by replication stress. The high frequency of deletions within the WWOX locus in cancers of various types, without the hallmark protein inactivation-associated mutations of “classical” tumor suppressors, has led to the proposal that WWOX deletions in cancers are passenger events that occur in early cancer progenitor cells due to fragility of the genetic locus, rather than driver events which provide the cancer cell a selective advantage. Recently, a proposed epigenetic cause of chromosomal fragility has suggested a novel mechanism for early fragile site instability and has implications regarding the involvement of tumor suppressor genes at chromosomal fragile sites in cancer. In this review, we provide an overview of the evidence for WWOX as a tumor suppressor gene and put this into the context of fragility associated with the FRA16D locus.
Keywords: FRA16D, fragile sites, FOR gene, WOX1, WWOX-human gene, Wwox- mouse gene, Wwox- human/mouse protein
Introduction
WWOX, WW domain containing oxidoreductase, is a large gene (1.2 Mb open reading frame) with a relatively small, 2.2 kb transcript.1 A schematic of WWOX is denoted in Figure 1(b) showing nine exons and a particularly large eighth intron spanning 779,639 bp.2 The Wwox protein contains two WW binding domains at its N terminal region and a short-chain dehydrogenase/reductase (SDR) domain in its central region.1 Characterization of these domains has been an important part of describing the cellular and physiological functions of Wwox.
Figure 1.
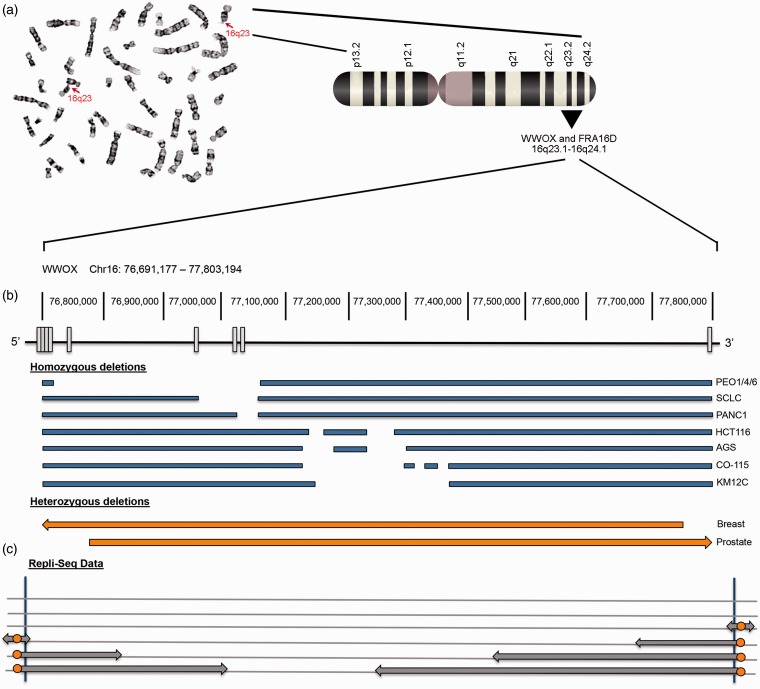
Chromosome fragility and genetic alterations at the WWOX/FRA16D locus. (a) Metaphase spread showing G-banded metaphase chromosomes and homozygous breaks at 16q23, with corresponding chromosome 16 ideogram, identifying the colocalization of WWOX and FRA16D. (b) Schematic of the WWOX gene showing exons as white boxes below their corresponding genomic location. Genetic alterations are listed in blue (homozygous) or orange (heterozygous) with deletions denoted as gaps. Samples shown for homozygous deletions are epithelial cancer cell lines: PEO1/4/6 cell lines derived from ovarian adenocarcinoma, SCLC derived from small cell lung carcinoma including the cell lines WX330 and NCI-H69, PANC1 cell line derived from pancreatic ductal adenocarcinoma, HCT116 cell line derived from colorectal carcinoma, AGS cell line derived from stomach adenocarcinoma, CO-115 and KM12C cell lines were derived from colon carcinomas. Heterozygous deletions were detected in breast and prostate cancers via PCR microsatellite analysis. (c) Replication-sequencing data generated from ENCODE for the WWOX locus in epithelial cells. Cell cycle phases are indicated on the left with S phase subdivided into four fractions. The WWOX locus relies on long-traveling forks (gray arrows) emanating from replication origins (orange circles) located in the flanking regions to converge in G2 phase, resulting in late completion of replication for the center of the gene. (A color version of this figure is available in the online journal.)
WW domains are small protein modules named for their unique structure: two conserved tryptophan (W) residues spaced approximately 20 amino acids apart.3 Functionally, WW domains are recognized for their involvement in protein–protein interactions and grouped according to their binding preference to proline-rich ligands. Initially, Wwox was thought to belong to Group I after demonstrating binding of proteins harboring PPxY motifs (where P is proline, Y is tyrosine, and x is any amino acid) through its first WW domain.4–6 However, a recent study by Abu-Odeh et al. employed an MS-based screen which confirmed previous PPxY protein interactions, but also demonstrated that the majority of Wwox interacting proteins did not contain PY motifs.7 Rather, these proteins exhibited PPxF or LPxF motifs (where F is phenylalanine and L is leucine), suggesting that perhaps WW1 domain of WWOX binds non-canonical proline-rich motifs. Many studies have shown that WW1 domain interacting proteins do not interact with the WW2 domain.5–7 After showing that the second WW domain, WW2, contains two distinct amino acid residues within the WW binding pocket, compared to WW1, McDonald et al. proposed that the WW2 domain serves as a chaperone for augmenting physiological binding of WW1.4
When the WWOX gene was cloned in 2000, Bednarek et al. predicted that the SDR domain enabled Wwox to play a role in steroid metabolism.1 Seven years later, a Wwox knockout mouse was generated by Aqeilan et al. and found to succumb postnatally to a severe metabolic syndrome.8 The phenotype of this mouse was carefully characterized through immunohistochemistry and affymetrix gene expression analysis to demonstrate that Wwox plays critical functions in both gonadal development and the steroidogenesis pathway.9 Subsequent lipoprotein profiles in liver-specific Wwox knockout mice have suggested a role for the protein in cholesterol homeostasis and fatty acid biosynthesis/triglyceride metabolism. In support of this proposed role of Wwox in lipoprotein and steroid metabolism, Iatan et al. have characterized variants within the WWOX gene which segregate with dyslipidemia in two French Canadian families.10 Interestingly, WWOX expression levels are highest in hormonally regulated tissues such as the ovary, prostate, and testes.1 Thus, Wwox protein appears to play critical roles in lipoprotein, high-density lipoprotein, and sex steroid metabolism, although the contribution of the SDR domain to this phenotype remains to be confirmed.
The WWOX gene also spans the common chromosomal fragile site (CFS) FRA16D. The term “fragile site” was first used to describe gaps that appeared on chromosomes in culture following replication stress.11 Initially, replication stress was induced by treatments of folate deprivation or dihydrofolate reductase inhibition,11 but the mild polymerase inhibitor, aphidicolin, is now traditionally used to precipitate CFSs.12 Fragile sites gained attention when the rare fragile site FRAXA, located at Xq27, was associated with X-linked mental retardation, also known as fragile X syndrome.13 Common fragile sites were identified when a group of different, recurring fragile loci were observed in control samples when studying FRAXA.12 Fragile sites are classified as rare or common depending on their frequency within the population.14 CFSs are identifiable in all individuals, while rare fragile sites are seen in less than 5% of the population. Rare fragile sites segregate in a Mendelian manner and exhibit nucleotide expansions, such as the CCG trinucleotide repeat that characterizes FRAXA.15 In general, CFSs harbor large transcriptionally active genes, usually greater than 1 Mb in length.16 The two most frequently activated and well-studied CFSs in lymphocytes are FRA3B at 3p14.2, encoding FHIT and FRA16D encoding WWOX.17
In addition to being located at fragile sites, FHIT and WWOX have also both been reported to be tumor suppressor genes,8,18 whose loss is associated with numerous human cancers.19,20 In 1979, the first chromosomal translocation associated with familial cancer, a renal cell carcinoma, was mapped to a breakpoint in 3p21.21 Following the identification of CFSs in 1984, scientists quickly noticed that the non-random alterations associated with cancer frequently coincided with the location of CFSs.22 Accordingly, the 3p21 breakpoint was later hypothesized to be near FRA3B at 3p14.223 and the FHIT gene was cloned at 3p14.2 in 1996,24 showing the overlap of FHIT and FRA3B loci. In parallel to the search for CFSs, cancer researchers were attempting to map tumor suppressor genes by homozygosity mapping. In the 1990s loss of heterozygosity (LOH) and allelic imbalances in breast cancers were mapped to the long arm of chromosome 16,25 the location of FRA16D and WWOX.1
Two opposing views dominate the discussion regarding the role of CFSs in cancer. One school of thought is that genomic instability created by cancer progression causes collateral damage to FHIT and WWOX. In this way, they are thought to be unselected “passenger” mutations in cancers.26 The counterargument is that deletions and other genomic alterations at FHIT and WWOX loci at CFSs occur early in cancer initiation or progression, and cancer cells with clonally unique FHIT or WWOX gene deletions are selectively expanded due to loss of tumor suppressor functions such as protection of genome stability27 or programmed apoptosis.28,29 In this review, we discuss the arguments for WWOX as a tumor suppressor and put this into context of recent discoveries regarding CFSs.
Chromosomal fragility
Understanding the mechanism underlying chromosomal fragility is an important precursor to determining the biological effects of gene deletions at fragile sites. Until three years ago, the prevailing explanation for chromosomal fragility was the presence of sequences affecting replication fork dynamics. The replication fork is generated when the helicase/topoisomerase complex travels just upstream of polymerase to uncoil and separate the DNA strands. With aphidicolin-mediated replication stress, the helicase proceeds uncoupled from DNA polymerase, generating long stretches of single-stranded DNA. Nucleotide repeats, associated with rare fragile sites, were believed to form secondary structures within these stretches of single-stranded DNA and impair movement of the replication fork, causing its collapse and the DNA breaks seen cytogenetically as chromosomal breaks.16 Since rare, heritable fragile sites were discovered to involve expansion of nucleotide repeats, it was logical to suppose that common fragile sites also owed their fragility to the actual nucleotide sequence. As these sequences were discovered and reported, efforts were made to discover sequence elements within them that could account for their fragility. A theory that was widely investigated was put forward by the Kerem laboratory, a proposal that flexible regions of DNA within CFS loci could cause secondary structures that could interfere with replication fork progression under conditions of mild replication stress.30 However, it was difficult to identify sequences that would generate predictable secondary structures in single-stranded DNA for common fragile sites that would be true only of fragile sites.17,31 For instance, the FRA3B and FRA16D loci are AT rich compared to the rest of the genome, but this in itself cannot explain fragility, as comparable AT enriched genomic regions are not fragile. Other theories proposed that differences in chromatin and replication-associated proteins at the fork were responsible for differences in susceptibility to DNA breaks in fragile versus non-fragile regions.32
In 2011, the laboratory of Michelle Debatisse investigated epigenetic mechanisms associated with FRA3B.33 Letessier et al. used DNA combing experiments, where stretched-out DNA strands are pulse labeled with fluorescent tags in vivo during replication, to evaluate replication fork speed and symmetry. Surprisingly, the authors found that replication forks were not stalling along the FHIT locus. They employed replication-sequencing (repli-seq) data to evaluate the distribution of replication initiation and termination events at FRA3B and showed that in lymphoblasts (the white blood cell type previously used to identify and describe the frequency of CFS) the FRA3B locus has no replication initiation origin within a central 700 kb region of the 1.2 Mb gene. This is a significant scarcity of replication origins since other genomic regions of similar size have approximately 10 replication origins. With no central replication origins, replication forks from flanking regions of the locus must cover very long distances (10 times longer than normal) to converge and complete replication. If a replication stressor such as aphidicolin is applied to lymphoblasts, replication at loci like FRA3B may not reach completion before entering G2 and mitosis phases, causing chromosomal gaps in mitosis, the defining feature of CFSs. Letessier et al. also examined repli-seq data for FRA3B in fibroblasts, cells derived from connective tissue. Fibroblasts exhibited a different replication origin pattern that corresponded to the lack of fragility at 3p14.2 in fibroblasts compared to lymphoblasts. In this way, Letessier et al. showed that different cell types exhibit different common fragile sites based on an epigenetically determined feature, the placement of replication origins across genomic loci in cells of specific tissue origins.33
Genetic alterations and fragility of Wwox
If fragility of FRA16D is indeed due to unfinished replication caused by a scarcity of replication origins, a review of the published deletion locations of WWOX should correspond to those regions prone to breakage. Genetic alterations at the WWOX locus occur through LOH, homozygous deletions, and translocations.20 LOH, or loss of one allele or a portion of one allele of the WWOX gene, has been reported for 52% of hepatocellular carcinomas, 53% of prostate carcinomas, 67% of breast carcinomas as well as for lung and gastric cancer.34–36 Homozygous deletions between exons 4 and 9 have been characterized in colon, ovarian, small cell lung, and pancreatic cancer cell lines.34,37 Translocations have been reported in multiple myeloma38 and the colon cancer cell line, HCT116.39 Likewise, aberrant transcripts have been reported for Wwox,40–42 also showing loss of exons 4 and 8 in the majority of cases, indicating a similar pattern for these exon deletions. Figure 1 shows examples of homozygous deletions and LOH that have been documented for WWOX in several cancer types and cell lines. Most losses occur between exons 4 and 8, the region that corresponds to the late-replicating region by repli-seq data. Altogether, the Debatisse theory of fragility supports the genetic alteration patterns thus far reported for WWOX.
Since fragility studies were initially conducted in lymphoblasts,12 identifying the cell-type specific mechanism of fragility has prompted a re-evaluation of CFSs in different cell types. As such, Hosseini et al. evaluated cells from epithelial tissues, the most common tissue origin for human cancers, for location and frequency of common fragile sites.43 The authors used epithelial cell lines from breast, colorectal, and bronchial epithelial tissue to evaluate common fragile sites from metaphase spreads in order to appropriately provide a new CFS ranking for epithelial cells. They also evaluated repli-seq data provided by publicly available databases (ENCODE) in order to put their results into context with the epigenetic theory of fragility proposed by the Debatisse laboratory. Cytogenetic analyses demonstrated that the most fragile locus in all epithelial cells examined, with the exception of the bronchial epithelial cell line, was FRA16D rather than FRA3B. This implies that FRA16D is the most fragile locus in most epithelial-derived human cancers. In a similar study, Le Tallec et al. analyzed epithelial (colorectal and breast) and erythroid-derived cancer cell lines to identify CFSs and quantify fragility in these cell types.44 FRA16D was the only fragile site identified in all four cell types compared (lymphoblasts, fibroblasts, epithelial, and erythroid cells) and was consistently one of the top three CFSs for epithelial cells, showing more fragility than FRA3B in all cell lines except one.44 This is a significant finding as FRA3B has overshadowed FRA16D for nearly three decades as the “most fragile site.” It would then follow that fragility of different tumor suppressor genes should be put into context of the cell type from which the tumor originates.
Putative Wwox biological functions
In order to determine the relevance of FRA16D fragility and resulting WWOX deficiency in cancers, a review of recent functional studies is required to determine putative roles of Wwox in tumor suppression. The function of Wwox can be defined, in part, by the company of the partners its binds. A growing list of Wwox interacting proteins have been described and include transcription factors p73,45 Ap2γ and γ,46 Jun,47 Runx2,48 and PPxY independent interactions with p53 and Mdm2.49 These transcription factors regulate pathways involving cellular apoptosis, proliferation, and development.50 Other Wwox interacting proteins of notable physiological relevance include: ErbB4,5 a receptor tyrosine kinase involved in mitogenesis and differentiation, and Dvl-2,51 a stabilizing protein involved in the non-canonical Wnt/β-catenin pathway which elicits cell growth and proliferation. Not only does Wwox serve a variety of functions in distinct intracellular pathways contingent on the identity of its binding partner, Gourley et al. also show that Wwox modulates interactions between tumor cells and the extracellular matrix (ECM).28 Through in vitro and in vivo studies involving the transfection of WWOX into ovarian cancer cell lines, the authors showed that adhesion of tumor cells to fibronectin in the ECM is dependent on WWOX expression via membrane integrin α3.28 This suggests that Wwox serves important, yet diverse intracellular and extracellular functions.
Wwox cellular localization also provides some clues to its function. Most studies report Wwox protein localized to the cytoplasm where it sequesters transcription factors (Ap2γ and p73)46 or signaling proteins such as Dvl-2, to prevent nuclear import and transactivation or Wnt pathway activity.5,51 In this way, Wwox deficiency could result in the deregulation of numerous physiologically relevant pathways through binding and repression of a variety of transcription factors and signaling proteins in the cytoplasm. Other reports have suggested that Wwox localizes to the golgi apparatus,2 as well as the nucleus and mitochondria.49,52 With the large array of binding partners identified, it is possible that Wwox participates in pathways located in distinct compartments within the cell. The case for definition of Wwox function becomes more complex in some instances, as with ErbB4 and Wwox binding in breast cancer cells.5 Here, Wwox competes for ErbB4 intracellular domain binding, with other WW domain containing proteins, Yap and Itch, involved in the same pathway but producing opposing downstream effects.5 Although studies are beginning to shed light on the myriad functions of Wwox, it is clear that much work is to be done in order to characterize the full extent and particular mechanisms in which Wwox deficiency may potentiate tumor cells. It will be necessary to learn much more about which WW domain containing proteins are expressed in specific tissues or niches where Wwox is expressed, and how these expression patterns change with development of cancer and progressive stages of cancer, in order to follow the signal pathways directed by Wwox within the complex WW domain network of signaling proteins.
Wwox deficiency in cancers
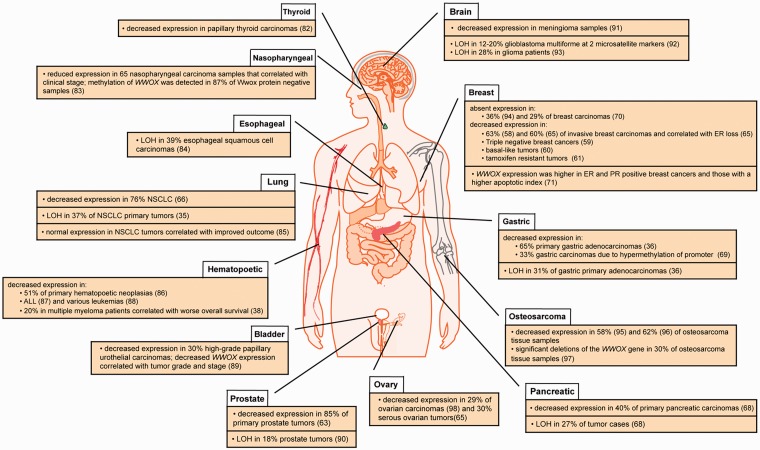
Evidence for WWOX as a tumor suppressor gene began with reports that a variety of epithelial cancers, including breast carcinomas, preinvasive breast lesions, prostate and hepatic carcinomas exhibited LOH at 16q.53,54 Several efforts then culminated in cloning of the WWOX gene and identifying its overlap with FRA16D on the long arm of chromosome 16.1,40 Since then, studies have confirmed that WWOX expression is decreased or absent in many human cancers, reviewed in Gardenswartz and Aqeilan20 and highlighted in Figure 2. Interestingly, Wwox deficiency is often correlated with breast, prostate, and ovarian cancers,55,56 tissues for which WWOX expression is highest. Studies have demonstrated the important physiological role that Wwox plays in mammary gland ductal development as knockout mice exhibit impaired ductal growth and increased fibronectin levels.57 There is also evidence of significant Wwox deficiency in human invasive breast carcinoma tissue samples58 and breast cancer cell lines.42 Furthermore, reduced WWOX expression is highly correlated with various clinicopathologic factors such as triple negative breast cancers,59 basal phenotypes,60 and tamoxifen resistance.61
Figure 2.
WWOX/Wwox expression alterations in human cancers. Summary of reports showing correlations between reduced/absent WWOX/Wwox expression, LOH (loss of heterozygosity), or normal WWOX expression in various common human cancers. (A color version of this figure is available in the online journal.)
In terms of prostate cancer, WWOX has been identified as an area of genomic loss in tumors based on aCGH62 and its expression has been found to be decreased in 84% of tumors analyzed via immunohistochemistry.63 Furthermore, Qin et al. found that WWOX overexpression in vitro induced apoptosis and stalled cell growth, suggesting a mechanism for WWOX tumor suppressor function in prostate cancer cells. As mentioned earlier, a similar mechanism for the involvement of WWOX in ovarian cancers has also been proposed.28 Accordingly, decreased WWOX expression has been significantly correlated with advanced clinical stage (FIGO Stage IV), decreased overall survival, and lymph node metastasis in ovarian cancers.64,65
WWOX deregulation has also been associated with a number of non-hormonally regulated cancers such as lung, pancreatic, gastric, bone, and skin cancers, as well as others highlighted in Figure 2. Non-small cell lung tumor samples were found to exhibit decreased WWOX expression,35,66 which correlated with certain histotypes as well as increased tumor aggressiveness.67 Pancreatic and gastric cancers also demonstrated WWOX LOH as well as decreased expression in primary tumors and cell lines.36,68 Furthermore, WWOX transfection inhibited pancreatic cell colony formation via apoptosis.68 In summary, WWOX expression is frequently decreased in a wide variety of cancers. In some cases, WWOX plays a key developmental function (mammary ducts) but more often it is involved in promoting apoptosis, as in the case of detached ovarian and prostate cancer cells, revealing its tumor suppressive function.
WWOX: a non-canonical tumor suppressor
Watanabe et al. challenged the notion of WWOX as a tumor suppressor due to their finding that tissues from 10 of out of 16 gastric tumors and five out of five breast tumors exhibited normal or increased WWOX expression.52 Subsequent studies evaluating many more tumor samples and in many more cancers have demonstrated a significant correlation with decreased WWOX expression in gastric cancers36,69 and breast cancers,58,65,70 sometimes also correlating with more aggressive disease and worse prognosis.59,61,71 It is also possible that losses or changes in the balance of expression of specific Wwox-binding or competing WW-domain proteins could lead in some cases to gain of WWOX expression.
Initial studies of WWOX also described a lack of point mutations along the gene length, a classic sign of a tumor suppressor.1 A recent examination of breast cancer tissue samples identified 13 point mutations in 81 breast cancer tissue samples.72 One mutation was considered a previously reported polymorphism,64 however of the remaining 12, four were categorized as non-sense mutations and eight as missense mutations in the coding sequence. Still, numerous reports failed to find non-polymorphic point mutations within Wwox1,64,66,68,73 suggesting a need for confirmation of these findings. We propose that due to its location at a CFS, the WWOX locus is so sensitive to damage-induced chromosomal gaps or breaks at late-replicating sites within this large gene, that LOH and homozygous deletions occur at a much higher frequency than might be expected for occurrence of point mutations. In addition, WWOX silencing occurs via other classical tumor suppressor mechanisms such as hypermethylation of promoter regions and ubiquitination in breast, lung, bladder, and prostate cancers.74,75
Interestingly, the overlap of putative tumor suppressor genes and CFSs is highly conserved over many species, including mice.76,77 Mouse models have confirmed the tumor suppressive function of the WWOX ortholog. Wwox+/− mice have a significantly higher incidence of spontaneous lung and mammary tumors and are more sensitive to DNA damaging agents compared to wild-type mice.45,46 Significantly, some of these tumors from Wwox+/− mice retained one intact Wwox allele, suggesting haploinsufficiency as a mechanism for loss of tumor suppressive function. Complete knockout mice exhibit osteosarcomas by 3–4 weeks of age8 and Wwox hypomorphic mice display spontaneous B-cell lymphomas.78 On the other hand, although the Wwox+/− mouse on a C3H mammary tumor-susceptible genetic background exhibited enhanced mammary tumorigenesis,79 mice with targeted deletions of Wwox in mammary tissue did not show a higher incidence of mammary tumors.57,80 These conflicting results could be explained by the varying impact of ECM interactions with cancers—as Wwox was decreased in all tissues in the C3H mouse, but only in epithelial mammary cells in the conditional knockouts. In this way, the interactions with the ECM, such as that proposed by Gourley et al. in ovarian caner cells,28 could contribute to its tumor suppressor function in mammary cancers. It is also possible that loss of Wwox contributes to the progression of mammary carcinogenesis rather than the initiation.
Despite much evidence suggesting an association of WWOX deficiency in various cancers and the selective advantage it affords cancer cells, speculation continues regarding the role of tumor suppressor genes located at CFSs, such as WWOX and FHIT, in carcinogenesis. Proponents of the passenger theory, regard homozygous deletions, a hallmark of tumor suppressor genes in cancer, as bystander effects of WWOX and FHIT fragility.26 Taken together with the replication origin pattern of fragility, one would predict that reduced WWOX expression would be correlated with nearly all epithelial cancers, since it is the most fragile locus in epithelial cells. However, McAvoy et al. determined the frequency of fragile sites for various cancers and then compared the expression of corresponding genes located at their CFS loci.81 Interestingly, there was no correlation between fragility and the frequency with which the corresponding genes were deleted. Furthermore, some cancers did not have inactivation of any of the large genes located at CFSs, such as WWOX, suggesting specific selection criteria for CFS gene inactivation in different cancers. Recently, Le Tallec et al. demonstrated through elegant and detailed erythroid and epithelial cell CFS profiling that over 50% of recurrent cancer deletions originate from CFSs associated with large genes, demonstrating new significance for the contribution of genes located at CFSs in cancer.44
Conclusion
Due to its location at a CFS, its lack of point mutations, and conflicting mouse mammary cancer model data, the function of WWOX as a tumor suppressor gene has been challenged. We have reviewed data showing significantly decreased expression of WWOX in various human cancers, in some cases correlating with histotype and prognosis. In addition, there are numerous studies that imply involvement of WWOX in apoptosis, cell growth, and/or proliferation, all functions that contribute to a tumor suppressor role.
In light of the recent findings highlighting the exquisite fragility of FRA16D, we propose that replication stress caused by the underlying genomic instability of cancer cells results in deletions at WWOX due to its overlap with the FRA16D. This mechanism of alteration to expression could explain or contribute to the scarcity of point mutations found within the WWOX gene. The presence of WWOX in clonal populations of heterogenous cancer cells suggests that following FRA16D-associated chromosome rearrangements, WWOX deficiency may provide a selective advantage for these cells—possibly through tissue- and cell-specific interactions with intracellular and extracellular proteins expressed in the particular cancer tissues. For example, in prostate tissues, the binding of WWOX with the transcription factor Ap2γ prevents Ap2γ activity in the nucleus and subsequent activation of ERBB2, a mediator of androgen receptor activity and prostate cancer cell growth.63 In a Wwox-deficient state, Ap2γ is free to activate ERBB2 and promote cancer cell growth. However, in other tissues, which lack ERBB2 and androgen receptors, loss of Wwox might not provide a growth advantage and so not promote the clonal expansion of those cells; therefore, Wwox deficiency would not be associated with cancers in those tissue types. This idea is in accord with the findings by McAvoy et al. that WWOX deficiency was not found in all cancer cells where WWOX/FRA16D was the most fragile CFS—perhaps it is only found at high frequency in tissues in which its myriad protein–protein interactions enable it to provide a selective advantage. One could speculate that with the many putative binding partners and pathways associated with Wwox, and the array of complex inter-relationships among them, such as those involving Wwox, ErbB4, Yap, and Itch, that many possible scenarios complicate elucidation of a Wwox tumor suppressor role in mouse mammary tumor models.
The proposal by Le Tallec et al. that over 50% of deletions in human cancers occur at CFSs emphasizes the importance of defining specific sites of chromosome fragility in different cancer types and understanding the biological context and function of genes located at those sites. The discoveries regarding CFSs have highlighted the importance of Wwox owing to its location at FRA16D, the most frequent fragile site in epithelial cells, the most common origin of human cancers. More research is necessary to understand the biological and tumor suppressor functions of WWOX. It is necessary to understand when WWOX loss occurs in cancer progression but also to continue illuminating the various functions of Wwox protein in distinct tissue types to more fully characterize its tumor suppressor function.
Acknowledgements
We thank Teresa Druck for figure preparation and Huebner lab members for constructive feedback. This research has been supported by 9T32 OD010429, U01 CA154200, and R01 CA120516.
Authors’ Contributions
MSS wrote the manuscript. KH edited and provided helpful discussions.
References
- 1.Bednarek AJ, Laflin KJ, Daniel RL, Liao Q, Hawkins KA, Aldaz CM. WWOX, a novel WW domain-containing protein mapping to human chromosome 16q23.3–16q24.1, a region frequently affected in breast cancer. Cancer Res 2000; 60: 2140–5. [PubMed] [Google Scholar]
- 2.Ludes-Meyers JH, Bednarek AK, Popescu NC, Bedford M, Aldaz CM. WWOX, the common chromosomal fragile site, FRA16D, cancer gene. Cytogenet Genome Res 2003; 100: 101–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sudol M, Recinos CC, Abraczinskas J, Humbert J, Farooq A. WW or WoW: the WW domains in a union of bliss. IUBMB 2005; 57: 773–8. [DOI] [PubMed] [Google Scholar]
- 4.McDonald CB, Buffa L, Bar-Mag T, Salah Z, Bhat V, Mikles DC, Deegan BJ, Seldeen KL, Malhotra A, Sudol M, Aqeilan RI, Nawaz Z, Farooq A. Biophysical basis of the binding WWOX tumor suppressor to WBP1 and WBP2 adaptors. J Mol Biol 2012; 422: 58–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aqeilan RI, Donati V, Palamarchuk A, Trapasso F, Kaou M, Pekarsky Y, Sudol M, Croce CM. WW domain-containing proteins, WWOX and YAP, compete for interaction with ErbB-4 and modulate its transcriptional function. Cancer Res 2005; 15: 6764–71. [DOI] [PubMed] [Google Scholar]
- 6.Ludes-Meyers JH, Kil H, Bednarek AK, Drake J, Bedford MT, Aldaz CM. WWOX binds the specific proline-rich ligand PPXY: identification of candidate interacting proteins. Oncogene 2004; 23: 5049–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Abu-Odeh M, Bar-Mag T, Huang H, Kim T, Salah Z, Abdeen SK, Sudol M, Reichmann D, Sidhu S, Kim PM, Aqeilan RI. Characterizing WW domain interactions of tumor suppressor WWOX reveals its association with multiprotein networks. J Biol Chem 2014; 289: 8865–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aqeilan RI, Trapasso F, Hussain S, Costinean S, Marshall D, Pekarsky Y, Hagan JP, Zanesi N, Kaou M, Stein GS, Lian JB, Croce CM. Targeted deletion of Wwox reveals a tumor suppressor function. PNAS 2007; 104: 3949–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aqeilan RI, Hagan JP, de Bruin A, Rawahneh M, Salah Z, Gaudio E, Siddiqui H, Volinia S, Alder H, Lian JB, Stein GS, Croce CM. Targeted ablation of the WW domain-containing oxidoreductase tumor suppressor leads to impaired steroidogenesis. Endocrinology 2009; 150: 1530–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iatan I, Choi HY, Ruel I, Reddy MV, Kil H, Lee J, Abu Odeh M, Salah Z, Abu-Remaileh M, Weissglas-Volkov D, Nikkola E, Civelek M, Awan Z, Croce CM, Aqeilan RI, Pajukanta P, Aldaz CM, Genest J. The WWOX gene modulates HDL and lipid metabolism. Circ Cardiovasc Genet 2014; epub ahead of print PMID: 24871327. [DOI] [PMC free article] [PubMed]
- 11.Sutherland GR. Heritable fragile sites on human chromosomes I. Am J Hum Genet 1979; 31: 125–35. [PMC free article] [PubMed] [Google Scholar]
- 12.Glover TW, Berger C, Coyle J, Echo B. DNA polymerase α inhibition by aphidicolin induces gaps and breaks at common fragile sites in human chromosomes. Hum Genet 1984; 67: 136–42. [DOI] [PubMed] [Google Scholar]
- 13.Sutherland GR. Heritable fragile sites on human chromosomes. Am J Hum Genet 1982; 34: 452–8. [PMC free article] [PubMed] [Google Scholar]
- 14.Schwartz M, Zlotorynski E, Kerme B. The molecular basis of common and rare fragile sites. Cancer Lett 2006; 232: 13–26. [DOI] [PubMed] [Google Scholar]
- 15.Kremer EJ, Pritchard M, Lynch M, Yu S, Holman K, Baker E, Warren ST, Schlessinger D, Sutherland GR, Richards RI. Mapping of DNA instability at the fragile X to a trinucleotide repeat sequence p(CCG)n. Science 1991; 252: 1711–14. [DOI] [PubMed] [Google Scholar]
- 16.Durkin SG, Glover TW. Chromosome fragile sites. Annu Rev Genet 2007; 41: 169–92. [DOI] [PubMed] [Google Scholar]
- 17.Helmrich A, Stout-Weider K, Herman K, Schrock E, Heiden T. Common fragile sties are conserved features of human and mouse chromosomes and relate to large active genes. Genome Res 2006; 16: 1222–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fong LY, Fidanza V, Zanesi N, Lock LF, Siracusa LD, Mancini R, Siprashvili Z, Ottey M, Martin SE, Druck T, McCue PA, Croce CM, Huebner K. Muir-Torre-like syndrome in Fhit deficient mice. Proc Natl Acad Sci USA 2000; 97: 4742–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huebner K, Croce CM. Cancer and the FRA3B/FHIT fragile locus: it’s a HIT. British J Cancer 2008; 88: 1501–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gardenswartz A, Aqeilan RI. WW domain-containing oxidoreductase’s role in myriad cancers: clinical significance and future implications. Exp Biol Med 2014; 3: 253–63. DOI: 10.1177/1535370213519213. [DOI] [PubMed] [Google Scholar]
- 21.Cohen AJ, Frederick PL, Berg S, Marchetto DJ, Tsai S, Jacobs SC, Brown RS. Hereditary renal-cell carcinoma associated with a chromosomal translocation. N Engl J Med 1979; 301: 592–5. [DOI] [PubMed] [Google Scholar]
- 22.Yunis J, Soreng AL. Constitutive fragile sites and cancer. Science 1984; 226: 1199–204. [DOI] [PubMed] [Google Scholar]
- 23.Glover TW, Coyle-Morris JF, Li FP, Brown RS, Berger CS, Gemmill RM, Hecht F. Translocation t(3;8)(p14.2;q24.1) in renal cell carcinoma affects expression of the common fragile site at 3p14(FRA3B) in lymphocytes. Cancer Genet Cytogenet 1988; 31: 69–73. [DOI] [PubMed] [Google Scholar]
- 24.Ohta M, Inoue H, Cotticelli MG, Kastury K, Baffa R, Palazzo J, Siprashvii Z, Mori M, McCue P, Druck T, Croce CM, Huebner K. The FHIT gene, spanning the chromosome 3p14.2 fragile site and renal carcinoma-associated t(3;8) breakpoint, is abnormal in digestive tract cancers. Cell 1996; 84: 587–97. [DOI] [PubMed] [Google Scholar]
- 25.Aldaz CM, Chen T, Sahin A, Cunningham J, Bondy M. Comparative allelotype of in situ and invasive human breast cancer: high frequency of microsatellite instability in lobular breast carcinomas. Cancer Res 1995; 55: 3976–81. [PubMed] [Google Scholar]
- 26.Bignell GR, Greenman CD, Davies H, Butler AP, Edkins S, Andrews JM, Buck G, Chen L, Beare D, Latimer C, Widaa S, Hinton J, Fahey C, Fu B, Futreal PA, Stratton M. Signatures of mutation and selection in the cancer genome. Nature 2010; 463: 893–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saldivar JC, Miuma S, Bene J, Hosseini SA, Shibata H, Sun J, Wheeler LJ, Mathews CK, Huebner K. Initiation of genome instability and preneoplastic processes through loss of Fhit expression. PLoS Genet 2012; 8: 1–16. DOI: 10.1371/journal.pgen.1003077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gourley C, Paige AJW, Taylor KJ, Ward C, Kuske B, Zhang J, Sun M, Janczar S, Harrison DJ, Muir M, Smyth JF, Gabra H. WWOX gene expression abolishes ovarian cancer tumorigenicity in vivo and decreases attachment to fibronectin via integrin α3. Cancer Res 2009; 69: 4835–42. [DOI] [PubMed] [Google Scholar]
- 29.Miuma S, Saldivar JC, Karras JR, Waters CE, Paisie CA, Wang Y, Jin V, Sun J, Druck T, Zhang J, Huebner K. Fhit deficiency-induced global genome instability promotes mutation and clonal expansion. PLoS One 2013; 14: 1–19. DOI: 10.1371/journal.pone.0080730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zlotorynski E, Rahat A, Skaug J, Ben-Porat N, Ozeri E, Hershberg R, Levi A, Scherer SW, Margalit H, Kerem B. Molecular basis for expression of common and rare fragile sites. Mol Cell Biol 2003; 23: 7143–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tsantoulis PK, Kostinas A, Sfikakis PP, Evangelou K, Sideridou M, Levy B, Mo L, Kittas C, Wu X, Papavassiliou AG, Gorgoulis VG. Oncogene-induced replication stress preferentially targets common fragile sites in preneoplastic lesions. A genome-wide study. Oncogene 2008; 27: 3256–64. [DOI] [PubMed] [Google Scholar]
- 32.Jiang Y, Lucas I, Young DJ, Davis EM, Karrison T, Rest JS, Le Beau MM. Common fragile sites are characterized by histone hypoacetylation. Hum Mol Genet 2009; 18: 4501–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Letessier A, Millot GA, Koundrioukoff S, Lachages A, Vogt N, Hansen RS, Malfoy B, Brison O, Debatisse M. Cell-type-specific replication initiation programs set fragility of the FRA3B fragile site. Nature 2011; 470: 120–3. [DOI] [PubMed] [Google Scholar]
- 34.Paige AJW, Taylor KJ, Taylor C, Hillier SG, Farrington S, Scott D, Porteous DJ, Smyth JF, Gabra H, Watson JEV. WWOX: a candidate tumor suppressor gene involved in multiple tumor types. PNAS 2001; 98: 11417–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yendumari S, Kuroki T, Trapasso F, Henry AC, Dumon KR, Huebner K, Williams NN, Kaiser LR, Croce CM. WW domain containing oxidoreductase gene expression is altered in non-small cell lung cancer. Cancer Res 2003; 63: 878–91. [PubMed] [Google Scholar]
- 36.Aqeilan RI, Kuroki T, Pekarsky Y, Albagha O, Trapasso F, Baffa R, Huebner K, Edmonds P, Croce CM. Loss of WWOX expression in gastric carcinoma. Clin Cancer Res 2004; 10: 3053–8. [DOI] [PubMed] [Google Scholar]
- 37.Finnis M, Dayan S, Hobson L, Chenevix-Trench G, Friend K, Ried K, Venter D, Woollatt E, Baker E, Richards R. Common chromosomal fragile site FRA16D mutation in cancer cells. Hum Mol Genet 2005; 14: 1341–9. [DOI] [PubMed] [Google Scholar]
- 38.Jenner MW, Leone PE, Walker BA, Ross FM, Johnson DC, Gonzalez D, Chiecchio L, Dachs Cabanas E, Dagrada GP, Nightingale M, Protheroe RK, Stockley D, Else M, Dickens NJ, Cross NC, Davies FE, Morgan GJ. Gene mapping and expression analysis of 16q loss of heterozygosity identifies WWOX and CYLD as being important in determining clinical outcome in multiple myeloma. Blood 2007; 9: 3291–300. [DOI] [PubMed] [Google Scholar]
- 39.Alsop AE, Taylor K, Zhang J, Gabra H, Paige AJ, Edwards PAW. Homozygous deletions may be markers of nearby heterozygous mutations: the complex deletion at FRA16D in the HCT116 colon cancer cell line removes exons of WWOX. Genes Chromosomes Cancer 2008; 47: 437–47. [DOI] [PubMed] [Google Scholar]
- 40.Ried K, Finnis M, Hobson L, Mangelsdorf M, Dayan S, Nancarrow JK, Woollatt E, Kremmidotis G, Gardner A, Venter D, Baker E, Richards RI. Common chromosomal fragile site FRA16D sequence: identification of the FOR gene spanning FRA16D and homozygous deletions and translocation breakpoints in cancer cells. Hum Mol Genet 2000; 9: 1651–63. [DOI] [PubMed] [Google Scholar]
- 41.Bleau A, Freire J, Pajares MJ, Zudaire I, Anton I, Nistal-Villan E, Redrado M, Zandueta C, Garmendia I, Ajona D, Blanco D, Pio R, Lecanda F, Calvo A, Montuenga LM. New syngenic inflammatory related lung cancer metastatic model harboring double KRAS/WWOX alterations. Int J Cancer 2013; 11: 2516–27. DOI: 10.1002/jic.28574. [DOI] [PubMed] [Google Scholar]
- 42.Driouch K, Prydz H, Monese R, Johansen H, Lidereau R, Frengen E. Alternative transcripts of the candidate tumor suppressor gene, WWOX, are expressed at high levels in human breast tumors. Oncogene 2002; 21: 1832–40. [DOI] [PubMed] [Google Scholar]
- 43.Hosseini SA, Horton S, Saldivar JC, Miuma S, Stampfer MR, Heerema NA, Huebner K. Common chromosome fragile sites in human and murine epithelial cells and FHIT/FRA3B loss-induced global genome instability. Genes Chromosomes Cancer 2013; 52: 1017–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Le Tallec B, Millot GA, Blin ME, Brison O, Dutrillaux B, Debatisse M. Common fragile site profiling in epithelial erythroid cells reveals that most recurrent cancer deletions lie in fragile sites hosting large genes. Cell Report 2013; 4: 420–8. [DOI] [PubMed] [Google Scholar]
- 45.Aqeilan RI, Pekarsky Y, Herrero JJ, Palamarchuk A, Letofsky J, Druck T, Trapasso F, Han S, Huebner K, Croce CM. Functional association between Wwox tumor suppressor protein and p73, a p53 homolog. PNAS 2004; 101: 4401–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Aqeilan RI, Palamarchuk A, Weigel RJ, Herrero JJ, Pekarsky Y, Croce CM. Physical and functional interactions between the Wwox tumor suppressor protein and the Ap-2γ transcription factor. Cancer Res 2004; 64: 8256–61. [DOI] [PubMed] [Google Scholar]
- 47.Gaudio E, Palamarchuk A, Palumbo T, Trapasso F, Pekarsky Y, Croce CM, Aqeilan RI. Physical association with WWOX suppresses c-Jun trancriptional activity. Cancer Res 2006; 24: 11585–9. [DOI] [PubMed] [Google Scholar]
- 48.Del Mare S, Kurek KC, Stein GS, Lian JB, Aqeilan RI. Role of the WWOX tumor suppressor gene in bone homeostasis and the pathogenesis of osteosarcoma. Am J Cancer Res 2011; 5: 585–94. [PMC free article] [PubMed] [Google Scholar]
- 49.Chang NS, Hsu LJ, Lin YS, Lai FJ, Sheu HM. WW domain-containing oxidoreductase: a candidate tumor suppressor. Trends Mol Med 2007; 13: 12–22. [DOI] [PubMed] [Google Scholar]
- 50.Del Mare S, Salah Z, Aqeilan RI. WWOX: its genomics, partners and functions. J Cell Biochem 2009; 108: 737–45. [DOI] [PubMed] [Google Scholar]
- 51.Bouteille N, Driouch K, Hage PE, Sin S, Formstecher E, Camonis J, Lidereau R, Lallemand F. Inhibition of the Wnt/β-catenin pathway by the WWOX tumor suppressor protein. Oncogene 2009; 28: 2569–80. [DOI] [PubMed] [Google Scholar]
- 52.Watanabe A, Hippo Y, Taniguchi H, Iwanari H, Yashiro M, Hirakawa K, Kodama T, Aburatani H. An opposing view on WWOX protein function as a tumor suppressor. Cancer Res 2003; 63: 8629–33. [PubMed] [Google Scholar]
- 53.Chen T, Sahin A, Aldaz M. Deletion map of chromosome 16q in ductal carcinoma in situ of the breast: refining a putative tumor suppressor gene region. Cancer Res 1996; 56: 5605–9. [PubMed] [Google Scholar]
- 54.Latil A, Cussenot O, Fournier G, Driouch K, Lidereau R. Loss of heterozygosity at chromosome 16q in prostate adenocarcinoma: identification of three independent regions. Cancer Res 1997; 57: 1058–62. [PubMed] [Google Scholar]
- 55.Ramos D, Aldaz CM. WWOX, a chromosomal fragile site gene and its role in cancer. Adv Exp Med Biol 2006; 587: 149–59. [DOI] [PubMed] [Google Scholar]
- 56.Lewandowska U, Zelazowski M, Seta M, Byczewska M, Pluciennik E, Bednarek AK. WWOX, the tumor suppressor gene affected in multiple cancers. J Physiol Pharmacol 2009; 60: 47–56. [PubMed] [Google Scholar]
- 57.Abdeen SK, Salah Z, Khawaled S, Aqeilan RI. Characterization of WWOX inactivation in murine mammary gland development. J Cell Physiol 2013; 228: 1391–6. [DOI] [PubMed] [Google Scholar]
- 58.Guler G, Uner A, Guler N, Iliopoulos D, Hauck WW, McCue P, Huebner K. The fragile genes FHIT and WWOX are inactivated coordinately in invasive breast carcinoma. Cancer 2004; 100: 1605–14. [DOI] [PubMed] [Google Scholar]
- 59.Guler G, Himmetoglu C, Jimenez RE, Geyer SM, Wang WP, Costinean S, Pilarski RT, Morrison C, Suren D, Liu J, Chen J, Kamal J, Shapiro CL, Huebner K. Aberrant expression of DNA damage response proteins is associated with breast cancer subtype and clinical features. Breast Cancer Res 2011; 129: 421–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Guler G, Huebner K, Himmetoglu C, Jimenez RE, Costinean S, Volinia S, Pilarski RT, Hayran M, Shapiro CL. Fhit, Wwox and AP2γ expression levels correlate with basal phenotype in breast cancer. Cancer 2009; 115: 899–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Guler G, Iliopoulos D, Guler N, Himmetoglu C, Hayran M, Huebner K. Wwox and Ap2γ expression levels predict tamoxifen response. Clin Cancer Res 2007; 13: 6115–21. [DOI] [PubMed] [Google Scholar]
- 62.Watson JE, Doggett NA, Albertson DG, Andaya A, Chinnaiyan A, van Dekken H, Ginzinger D, Haqq C, James K, Kamkar S, Kowbel D, Pinkel D, Schmitt L, Simko JP, Volik S, Weinberg VK, Paris PL, Collins C. Integration of high-resolution array comparative genomic hybridization analysis of chromosome 16q with expression array data refines common regions of loss at 16q23-qter and identifies underlying candidate tumor suppressor genes in prostate cancer. Oncogene 2004; 23: 3487–94. [DOI] [PubMed] [Google Scholar]
- 63.Qin HR, Iliopoulos D, Semba S, Fabbri M, Druck T, Volinia S, Croce CM, Morrison CD, Klein RD, Huebner K. A role for the WWOX gene in prostate cancer. Cancer Res 2006; 66: 6477–81. [DOI] [PubMed] [Google Scholar]
- 64.Paige AJW, Zucknick M, Janczar S, Paul J, Mein CA, Taylor KJ, Stewart M, Gourley C, Richardson S, Perren T, Ganesan TS, Smyth JF, Brown R, Gabra H. WWOX tumor suppressor gene polymorphisms and ovarian cancer pathology and prognosis. Eur J Cancer 2010; 46: 818–25. [DOI] [PubMed] [Google Scholar]
- 65.Nunez MI, Rosen DG, Ludes-Meyers JH, Abba MC, Kil H, Page R, Klein-Szanto AJP, Godwin AK, Liu J, Mills GB, Aldaz CM. WWOX protein expression varies among ovarian carcinoma histotypes and correlates with less favorable outcome. BMC Cancer 2005; 5: 1–10. DOI: 10.1186/1471-2407-5-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Baykara O, Demirkaya A, Kaynak K, Tanju S, Toker A, Buyru N. WWOX gene may contribute to progression of non-small-cell lung cancer (NSCLC). Tumour Biol 2010; 4: 315–20. [DOI] [PubMed] [Google Scholar]
- 67.Donati V, Fontanini G, Dell’Omodarme M, Prati MC, Nuti S, Lucchi M, Mussi A, Fabbri M, Basolo F, Croce CM, Aqeilan RI. WWOX expression in different histologic types and subtypes of non-small cell lung cancer. Clin Cancer Res 2007; 13: 884–91. [DOI] [PubMed] [Google Scholar]
- 68.Kuroki T, Yendamuri S, Trapasso F, Matsuyama A, Aqeilan RI, Alder H, Rattan S, Cesari R, Nolli ML, Williams NN, Mori M, Kanematsu T, Croce CM. The tumor suppressor gene WWOX at FRA16D is involved in pancreatic carcinogenesis. Clin Cancer Res 2004; 1: 2459–65. [DOI] [PubMed] [Google Scholar]
- 69.Maeda N, Semba S, Nakayam S, Yanagihara K, Yokozaki K, Yokozaki H. Loss of WW domain-containing oxidoreductase expression in the progression and development of gastric carcinoma: clinical and histopathologic correlations. Virchows Arch 2010; 457: 423–32. [DOI] [PubMed] [Google Scholar]
- 70.Wang X, Chao L, Ma G, Chen L, Zang Y, Sun J. The prognostic significance of WWOX expression in patients with breast cancer and its association with the basal-like phenotype. J Cancer Res Clin Oncol 2011; 137: 271–8. [DOI] [PubMed] [Google Scholar]
- 71.Pluciennik E, Kusinska R, Potemski P, Kubiak R, Kordek R, Bednarek AK. WWOX—the FRA16D cancer gene: expression correlation with breast cancer progression and prognosis. Eur J Surg Oncol 2006; 32: 153–7. [DOI] [PubMed] [Google Scholar]
- 72.Ekizoglu S, Muslumanoglu M, Dalay N, Buyru N. Genetic Alterations of the WWOX gene in breast cancer. Med Oncol 2011; 3: 1529–35. DOI: 10.1007/s12032-011-0080-0. [DOI] [PubMed] [Google Scholar]
- 73.Paige AJW, Taylor KJ, Taylor C, Hillier SG, Farrington S, Scott D, Porteous DJ, Smyth JF, Gabra H, Watson JEV. WWOX: a candidate tumor suppressor gene involved in multiple tumor types. PNAS 2001; 98: 11417–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Iliopoulos D, Guler G, Han SY, Johnston D, Druck T, McCorkell KA, Palazzo J, McCue PA, Baffa R, Huebner K. Fragile genes as biomarkers: epigenetic control of WWOX and FHIT in lung, breast and bladder cancer. Oncogene 2005; 24: 1625–33. [DOI] [PubMed] [Google Scholar]
- 75.Mahajan NP, Whang YE, Mohler JL, Earp HS. Activated tyrosine kinase Ack1 promotes prostate tumorigenesis: role of Ack1 in polyubiquitination of tumor suppressor Wwox. Cancer Res 2005; 65: 10514–23. [DOI] [PubMed] [Google Scholar]
- 76.Krummel KA, Denison SR, Calhoun E, Phillips LA, Smith DI. The common fragile site FRA16D and its associated gene WWOX are highly conserved in the mouse at Fra8E1. Genes Chromosomes Cancer 2002; 34: 154–67. [DOI] [PubMed] [Google Scholar]
- 77.Pekarsky Y, Druck T, Cotticelli MG, Ohta M, Shou J, Mendrola J, Montgomery JC, Buchberg AM, Siracusa LD, Maneti G, Gong LY, Dragani TA, Croce CM, Huebner K. The murine Fhit locus; isolation, characterization and expression in normal and tumor cells. Cancer Res 1998; 58: 3401–8. [PubMed] [Google Scholar]
- 78.Ludes-Meyers JH, Kil H, Nunez MI, Conti CJ, Parker-Thornburg J, Bedfrod MT, Aldaz CM. Wwox hypomorphic mice display a higher incidence of B-cell lymphomas and develop testicular atrophy. Genes Chromosomes Cancer 2007; 46: 1129–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Abdeen SK, Salah Z, Maly B, Smith Y, Tufail R, Abu-Odeh M, Zanesi N, Croce CM, Nawaz Z, Aqeilan RI. Wwox inactivation enhances mammary tumorigenesis. Oncogene 2011; 30: 3900–6. [DOI] [PubMed] [Google Scholar]
- 80.Ferguson BW, Gao X, Kil H, Lee J, Benavides F, Abba MC, Aldaz CM. Conditional Wwox deletion in mouse mammary gland by means of two cre recombinase approaches. PLoS One 2012; 7: 1–10. DOI: 10.1371/journal.pone.0036618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.McAvoy S, Ganapathiraju SC, Ducharme-Smith AL, Pritchett JR, Kosari F, Perez DS, Zhu Y, James CD, Smith DI. Non-random inactivation of large common fragile site genes in different cancers. Cytogenet Genome Res 2007; 118: 260–9. [DOI] [PubMed] [Google Scholar]
- 82.Dias EP, Pimenta FJ, Sarquis MS, Dias Filho MA, Aldaz CM, Fujii JB, Gomez RS, De Marco L. Association between decreased WWOX protein expression and thyroid cancer. Thyroid 2007; 11: 1055–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yang Z, Lan H, Chen X, Li P, Li S, Li W, Mo W, Tang A. Molecular alterations of the WWOX gene in nasopharyngeal carcinoma. Neoplasma 2014; 61: 170–6. [DOI] [PubMed] [Google Scholar]
- 84.Kuroki T, Trapasso F, Shiraishi T, Alder H, Mimori K, Mori M, Croce CM. Genetic alterations of the tumor suppressor gene WWOX in esophageal squamous cell carcinoma. Cancer Res 2002; 62: 2258–60. [PubMed] [Google Scholar]
- 85.Becker S, Markova B, Wiewrodt R, Hoffarth S, Hahnel PS, Pleiner S, Schmidt LH, Breitenbuecher F, Schuler M. Functional and clinical characterization of the putative tumor suppressor WWOX in non-small cell lung cancer. J Thorac Oncol 2011; 12: 1976–83. [DOI] [PubMed] [Google Scholar]
- 86.Ischii H, Vecchione A, Furukawa Y, Sutheesophon K, Han SY, Druck T, Trapasso F, Nishimura M, Saito Y, Ozawa K, Croce CM, Huebner K, Furukawa Y. Expression of FRA16D/WWOX and FRA3B/FHIT genes in hematopoietic malignancies. Mol Cancer Res 2003; 13: 940–7. [PubMed] [Google Scholar]
- 87.Chen X, Zhang H, Li P, Yang Z, Qin L, Mo W. Gene expression of WWOX, FHIT and p73 in acute lymphoblastic leukemia. Oncol Lett 2013; 4: 963–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Cui Z, Lin D, Cheng F, Luo L, Kong L, Xu J, Hu J, Lan F. The role of the WWOX gene in leukemia and its mechanisms of action. Oncol Rep 2013; 6: 2154–62. [DOI] [PubMed] [Google Scholar]
- 89.Ramos D, Abba M, Lopez-Guerrero JA, Rubio J, Solsona E, Almenar S, Llonbart-Bosch A, Aldaz CM. Low levels of WWOX protein immunoexpression correlate with tumour grade and a less favorable outcome in patients with urinary bladder tumours. Histopathology 2008; 52: 831–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Cheng I, Levin AM, Tai YC, Plummer S, Chen GK, Neslund-Dudas C, Casey G, Rybicki BA, Witte JS. Copy number alterations in prostate tumors and disease aggressiveness. Genes Chromosomes Cancer 2012; 51: 66–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Aarhus M, Bruland O, Bredholt G, Lybaek H, Husebye ES, Krossnes BK, Vedeler C, Wester K, Lund-Johansen M, Knappskogg PM. Microarray analysis reveals down-regulation of the tumour suppressor gene WWOX and up-regulation of the oncogene TYMS in intracranial sporadic cancers. J Neurooncol 2008; 88: 251–9. [DOI] [PubMed] [Google Scholar]
- 92.Kosla K, Pluciennik E, Kurzyk A, Jesionek-Kupnicka D, Kordek R, Potemski P, Bednarek AJ. Molecular analysis of WWOX expression correlation with proliferation and apoptosis in glioblastoma multiforme. J Neurooncol 2011; 101: 207–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yu K, Fan J, Ding X, Li C, Wang J, Xiang Y, Wang QS. Association study of a functional copy number variation in the WWOX gene with risk of gliomas among Chinese people. Int J Cancer 2014; 7: 1687–91. DOI: 10.1002/jic/28815. [DOI] [PubMed] [Google Scholar]
- 94.Aqeilan RI, Hagan JP, Aqeilan A, Pichiorri F, Fong LY, Croce CM. Inactivation of the Wwox gene accelerates forestomach tumor progression in vivo. Cancer Res 2007; 67: 5606–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kurek K, Del Mare S, Salah Z, Abdeen S, Sadiq H, Lee S, Gaudio E, Zanesi N, Jones KB, DeYoung B, Amir G, Gebhardt M, Warman M, Stein GS, Stein JL, Lian LB, Aqeilan RI. Frequent attenuation of the WWOX tumor suppressor in osteosarcoma is associated with increased tumorigenicity and aberrant RUNX2 expression. Cancer Res 2010; 70: 5577–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Yang J, Cogdell D, Yang D, Hu L, Li H, Zheng H, Du X, Pang Y, Trent J, Chen K, Zhang W. Deletion of the WWOX gene and frequent loss of its protein expression in human osteosarcoma. Cancer Lett 2010; 1: 31–8. [DOI] [PubMed] [Google Scholar]
- 97.Yang J, Zhao L, Tian W, Liao Z, Zheng H, Wang G, Chen K. Correlation of WWOX, RUNX2 and VEGFA protein expression in human osteosarcoma. BMC Med Genomics 2013; 6: 1–8. http://www.biomedcentral.com/1755-8794/6/56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lan C, Chenggang W, Yulan B, Xiaohui D, Junhui Z, Xiao W. Aberrant expression of WWOX protein in epithelial ovarian cancer: a clinicopathologic and immunohistochemical study. Int J Gynecol Pathol 2012; 2: 125–32. [DOI] [PubMed] [Google Scholar]