A novel Pat1-dependent mechanism is identified for the protection of kinetochore-associated Cse4 from ubiquitination in order to ensure faithful chromosome segregation and genomic stability.
Abstract
Evolutionarily conserved histone H3 variant Cse4 and its homologues are essential components of specialized centromere (CEN)-specific nucleosomes and serve as an epigenetic mark for CEN identity and propagation. Cse4 is a critical determinant for the structure and function of the kinetochore and is required to ensure faithful chromosome segregation. The kinetochore protein Pat1 regulates the levels and spatial distribution of Cse4 at centromeres. Deletion of PAT1 results in altered structure of CEN chromatin and chromosome segregation errors. In this study, we show that Pat1 protects CEN-associated Cse4 from ubiquitination in order to maintain proper structure and function of the kinetochore in budding yeast. PAT1-deletion strains exhibit increased ubiquitination of Cse4 and faster turnover of Cse4 at kinetochores. Psh1, a Cse4-specific E3-ubiquitin ligase, interacts with Pat1 in vivo and contributes to the increased ubiquitination of Cse4 in pat1∆ strains. Consistent with a role of Psh1 in ubiquitination of Cse4, transient induction of PSH1 in a wild-type strain resulted in phenotypes similar to a pat1∆ strain, including a reduction in CEN-associated Cse4, increased Cse4 ubiquitination, defects in spatial distribution of Cse4 at kinetochores, and altered structure of CEN chromatin. Pat1 interacts with Scm3 and is required for its maintenance at kinetochores. In conclusion, our studies provide novel insights into mechanisms by which Pat1 affects the structure of CEN chromatin and protects Cse4 from Psh1-mediated ubiquitination for faithful chromosome segregation.
INTRODUCTION
The kinetochore is composed of centromeric (CEN) DNA, associated protein complexes, and a distinct chromatin structure (Bloom and Carbon, 1982) and is essential for faithful chromosome segregation in every eukaryotic system (Allshire and Karpen, 2008; Verdaasdonk and Bloom, 2011; Burrack and Berman, 2012; Choy et al., 2012; Maddox et al., 2012). The dissection of kinetochore structure and function of budding yeast has actively been pursued since the discovery of centromeres in 1980 (Clarke and Carbon, 1980). About 71 kinetochore proteins have been identified (Lechner and Carbon, 1991; Pot et al., 2003; Crotti and Basrai, 2004; Cho et al., 2010; Gonen et al., 2012; Mishra et al., 2013); many of them are evolutionarily conserved, with orthologues found in fungi, flies, worms, and humans (Hartzog et al., 1996; Kitagawa and Hieter, 2001; Meraldi et al., 2006; Przewloka and Glover, 2009; Lampert and Westermann, 2011). Although CEN function is evolutionarily conserved, the CEN DNA sequence differs among organisms, ranging from the ∼125 base pairs of unique DNA sequences in budding yeasts to several mega–base pairs of DNA composed of repeated sequences, species-specific satellite DNA arrays, or retrotransposon-derived sequences in other organisms (Clarke and Carbon, 1980; Allshire and Karpen, 2008; Verdaasdonk and Bloom, 2011; Burrack and Berman, 2012; Maddox et al., 2012). Despite such a variation in DNA sequence, CEN DNA in every eukaryotic organism examined so far is marked with specialized nucleosomes carrying an evolutionarily conserved histone H3 variant (Cse4 in budding yeast, Cnp1 in fission yeast, CID in fruit fly, HTR12 in Arabidopsis, and CENP-A in humans; Allshire and Karpen, 2008; Verdaasdonk and Bloom, 2011; Burrack and Berman, 2012; Maddox et al., 2012). In budding yeast, ubiquitin-mediated proteolysis of Cse4 is important for kinetochore function and faithful chromosome segregation (Collins et al., 2004; Au et al., 2008, 2013; Deyter and Biggins, 2014). For example, an E3-ubiquitin ligase, Psh1, targets excess Cse4 for ubiquitination and proteolysis in order to prevent Cse4 mislocalization to non-CEN regions (Hewawasam et al., 2010, 2014; Ranjitkar et al., 2010). Because Psh1 also associates with CEN DNA, it is proposed that either the association of Cse4 with Scm3 and/or assembly of CEN chromatin exclude Psh1-mediated ubiquitination of CEN-associated Cse4 (Hewawasam et al., 2010).
We recently identified an evolutionarily conserved protein Pat1 (protein associated with topoisomerase II; PATL1 in humans) as a structural component of budding yeast kinetochores (Mishra et al., 2013). Pat1 associates with CEN chromatin and regulates kinetochore structure to promote faithful chromosome segregation (Haase et al., 2013; Mishra et al., 2013). It is proposed that there are two pools of Cse4 molecules at the budding yeast kinetochores (Lawrimore et al., 2011; Haase et al., 2013). The core Cse4 molecules mediate attachment of kinetochores to microtubules, whereas the accessory (or peri-CEN) molecules may be required for the rapid assembly of Cse4 in case of its removal from the centromeres (Haase et al., 2013). Deletion of PAT1 results in depletion of the peri-CEN Cse4 molecules (Haase et al., 2013; Scott and Bloom, 2014). However, the molecular mechanism by which Pat1 regulates Cse4 molecules at kinetochores remains unknown.
In this study, we report that Pat1 protects Cse4 from Psh1-mediated ubiquitination. In the absence of Pat1, we observed increased Cse4 ubiquitination regulated by the cell cycle and faster turnover of Cse4 at kinetochores. Transient induction of PSH1 in a wild-type strain exhibits phenotypes similar to a pat1Δ strain, such as increased Cse4 ubiquitination, reduced Cse4 at kinetochores, and alterations in structural integrity of CEN chromatin. Pat1 interacts with Scm3 and regulates the centromeric levels of Scm3. Together our data uncover a novel Pat1-dependent mechanism for the maintenance of peri-CEN Cse4 molecules at kinetochores, which includes the protection of Cse4 from Psh1-mediated ubiquitination.
RESULTS
pat1Δ strains exhibit increased ubiquitination of Cse4
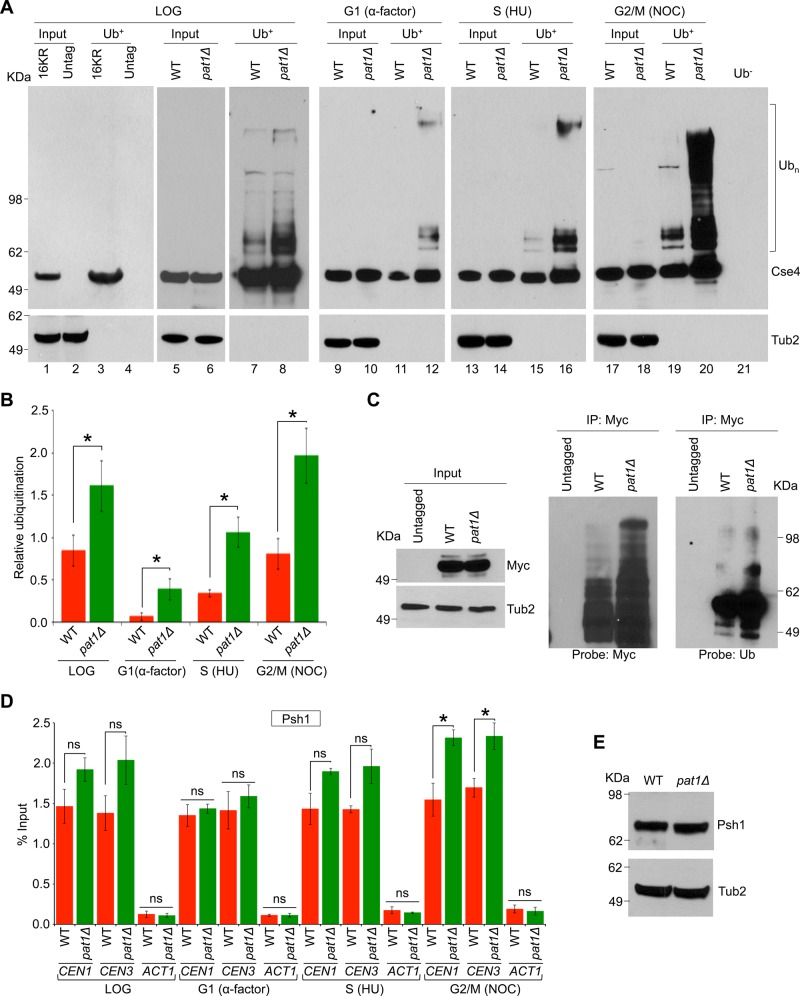
Pat1 is a structural component of the budding yeast kinetochore (Mishra et al., 2013), and deletion of PAT1 exhibits reduction in CEN-associated Cse4 and depletion of peri-CEN Cse4 molecules (Haase et al., 2013). Ubiquitination of Cse4 by an E3-ubiquitin ligase, Psh1, regulates cellular levels of Cse4 (Hewawasam et al., 2010; Ranjitkar et al., 2010; Au et al., 2013). Because Psh1 associates with CEN DNA (Hewawasam et al., 2010), we reasoned that reduced levels of Cse4 at CEN DNA in pat1∆ strains (Haase et al., 2013) may be due to increased ubiquitination of Cse4. Hence we examined the ubiquitination state of endogenously expressed Cse4 in wild-type and pat1Δ strains. Controls included a strain expressing cse416KR, a mutant allele of Cse4 that cannot be ubiquitinated because all lysine residues are replaced with arginine (Ranjitkar et al., 2010; Au et al., 2013), and an untagged wild-type strain. Agarose beads with tandem ubiquitin-binding entities (Ub+) were used in an affinity assay (Hjerpe et al., 2009; Au et al., 2013) to pull down ubiquitin-associated proteins, and ubiquitination status of Cse4 was examined by Western blot analysis. Untagged wild-type and cse416KR strains do not show a laddering pattern for Cse4 (Figure 1A, lanes 3 and 4). A faster-migrating species is observed in the Ub+ pull down from all strains that were similar in size to the input band. Because cse416KR cannot be ubiquitinated (Ranjitkar et al., 2010), this faster-migrating species likely represents unmodified forms of Cse4, which interact with ubiquitinated proteins bound to Ub+ agarose. Similar observations were made previously in Ub+ affinity agarose experiments with wild-type Cse4 and cse416KR (Au et al., 2013; Hewawasam et al., 2014). In a logarithmically growing culture, higher levels of ubiquitinated Cse4 were observed in a pat1Δ strain, as evident from the laddering pattern when compared with the wild-type strain (Figure 1A, lanes 7 and 8 and input control lanes 5 and 6). To examine whether Cse4 ubiquitination is affected by the cell cycle stage, we assayed ubiquitination of Cse4 in wild-type and pat1Δ strains that were synchronized in G1 (α-factor treatment), S (hydroxyurea treatment), and G2/M (nocodazole treatment) stages of the cell cycle. Hydroxyurea blocks the synthesis of deoxyribonucleotides, inducing a DNA replication–dependent checkpoint in S phase (Weinert et al., 1994), whereas nocodazole inhibits polymerization of microtubules, leading to a cell cycle arrest in mitosis (Jacobs et al., 1988). The cell cycle synchronization was verified by fluorescence-activated cell sorting (FACS) analysis (Supplemental Figure S1A). In wild-type cells, the ubiquitination of Cse4 was much higher in mitotic (G2/M) cells (Figure 1A, lane 19 and input control lane 17), and was barely detectable in G1 and S phase–arrested cells (Figure 1A, lanes 11 and 15 and input control lanes 9 and 13). In a pat1Δ strain, maximum Cse4 ubiquitination was also observed in mitotic (G2/M) cells (Figure 1A, lane 20 and input control lane 18), however, it was consistently higher than the wild-type strain in all cell cycle stages examined (Figure 1A, lanes 12, 16, and 20, and input control lanes 10, 14, and 18). Control experiments performed with a wild-type strain with agarose beads without Ub-binding activity (Ub−) do not exhibit a laddering pattern for Cse4 (Figure 1A, lane 21). Next we quantified the fraction of ubiquitinated Cse4 and normalized this to the total Cse4 levels (input) in each stage of the cell cycle as described previously (Au et al., 2013). The enrichment of ubiquitinated Cse4 was significantly higher (twofold to threefold) in mitotic (G2/M) cells than in the G1 or S phase cells (Figure 1B). The increased ubiquitination of Cse4 in mitosis was verified by assaying cells synchronized in G1 (α-factor treatment) and released into pheromone-free medium (Supplemental Figure S2, A–D). Consistent with the increased ubiquitination of Cse4 in nocodazole-treated cells (Figure 1A), higher levels of ubiquitinated Cse4 were observed in cells undergoing mitosis (Supplemental Figure S2, A–D). To determine whether only ubiquitination of Cse4 is increased in pat1Δ strains or the overall ubiquitination activity is up-regulated, we examined the ubiquitination of histone H2B in wild-type and pat1Δ strains. The levels of histone H2B ubiquitination were largely similar between wild-type and pat1Δ strains, suggesting that the increased ubiquitination of Cse4 in a pat1Δ strain is specific to Cse4 (Supplemental Figure S2E).
FIGURE 1:
Cse4 ubiquitination is cell cycle dependent and is increased in pat1Δ strains. (A) Cell cycle–dependent ubiquitination of Cse4, which is increased in a pat1Δ strain. Western blotting showing the levels of Cse4 ubiquitination (Ubn) in wild-type (WT, YMB6398) and pat1Δ (YMB8422) strains in cells grown to different stages of the cell cycle: logarithmic phase (LOG), G1 cells synchronized with α-factor, S phase cells synchronized with (HU), and G2/M cells synchronized with nocodazole (NOC). Wild-type strain with mutant cse416KR in which all lysine residues are changed to arginine (16KR; Au et al., 2013) and an untagged strain (untag; YPH499) were used as control in ubiquitin pull-down experiments. Eluted proteins were analyzed by Western blotting with α-Myc (Cse4) or α-Tub2 (served as a loading control) antibodies. Molecular weight markers for protein size in kilodaltons. (B) Quantification of relative ubiquitination of Cse4 in different cell cycle stages. Ratio of ubiquitinated Cse4 (bracket in A) to the total Cse4 levels (input) in WT and pat1Δ strains was calculated. The histogram represents the average of three biological replicates with SE. *p < 0.05, Student's t test. Statistically significant differences were observed between G1 and S or G2/M phases of the cell cycle in WT (p values: G1 vs. S, 0.004; G1 vs. G2/M, 0.008; S vs. G2/M, 0.034) and pat1Δ (p values: G1 vs. S, 0.019; G1 vs. G2/M, 0.005; S vs. G2/M, 0.037) strains. (C) Ubiquitination of Cse4 is increased in a pat1Δ strain. Western blotting showing the levels of Cse4 ubiquitination in wild-type (WT, YMB6398), pat1Δ (YMB8422), and untagged control (YPH499) strains grown to logarithmic phase of growth. Endogenously expressed Cse4-Myc was immunoprecipitated. Eluted proteins were analyzed by Western blotting with α-Myc (Cse4), α-Ub, or α-Tub2 (served as a loading control) antibodies. Molecular weight markers for protein size in kilodaltons. (D) Association of Psh1 with CEN chromatin through the cell cycle. ChIP-qPCR showing the levels of Psh1 in WT (YMB8126) and pat1Δ (YMB9096) strains at CEN (CEN1, CEN3) and non-CEN (ACT1) regions. Enrichment was determined by qPCR and is shown as percentage input. Average from three biological experiments ± SE . *p < 0.05; ns, statistically not significant, Student's t test. (E) Western blotting using α-TAP antibodies revealed that protein levels of Psh1 are not different between WT (YMB8126) and pat1Δ (YMB9096) strains. Molecular weight markers for protein size in kilodaltons.
To confirm further and validate the use of Ub+ agarose for ubiquitination status of Cse4, we immunoprecipitated endogenously expressed Myc-tagged Cse4 and examined the ubiquitination status of Cse4 by Western blot analysis using anti-ubiquitin and anti-Myc antibodies. Consistent with results from Ub+ experiments (Figure 1A), higher levels of Cse4 ubiquitination were observed in a pat1Δ strain than with the wild-type strain (Figure 1C). Control experiments with an untagged strain do not exhibit a laddering pattern for Cse4 (Figure 1C).
Chromatin immunoprecipitation (ChIP) experiments followed by quantitative PCR (qPCR) were done to examine whether the increased ubiquitination of Cse4 in pat1∆ strains may be due to higher levels of Psh1 at CEN. CEN levels of Psh1 in a logarithmically growing culture were not significantly different between the wild-type (1.46% of input at CEN1 and 1.38% at CEN3) and pat1Δ (1.92% at CEN1 and 2.03% at CEN3) strains, although slightly higher levels were observed in pat1Δ strains (Figure 1D). To examine whether CEN association of Psh1 is cell cycle regulated, we did ChIP experiments using cells synchronized in G1 with α-factor, in S phase with hydroxyurea, or in G2/M with nocodazole treatment. Cell cycle synchronization was confirmed by FACS analysis (Supplemental Figure S1B). No significant enrichment of Psh1 was detected at either CEN or non-CEN ACT1 regions in ChIP-qPCR experiments with an untagged strain (Supplemental Figure S3). CEN levels of Psh1 were not significantly different between wild-type and pat1Δ strains in G1 and S phase of the cell cycle (Figure 1D). However, in G2/M cells, CEN levels of Psh1 were significantly higher in the pat1Δ strain (2.31% at CEN1 and 2.33% at CEN3) than the wild type (1.54% at CEN1 and 1.69% at CEN3; p < 0.05; Figure 1D). No significant enrichment of Psh1 was observed at ACT1 locus used as a control (Figure 1D). Protein expression level of Psh1 was similar between the wild-type and pat1Δ strains (Figure 1E).
Centromeric Cse4 exchanges rapidly in pat1Δ strains
Increased ubiquitination of Cse4 (Figure 1A) and depletion of peri-CEN Cse4 molecules in a pat1Δ strain (Haase et al., 2013) suggest that Pat1 may protect CEN-bound Cse4 from ubiquitination and make it more stable at the kinetochore. Hence we examined whether a deletion of PAT1 results in a faster exchange of Cse4-GFP at metaphase kinetochores by using a fluorescence recovery after photobleaching (FRAP) assay (Pearson et al., 2004). The Cse4-GFP strain does not exhibit growth defects at 25 and 38°C, suggesting that GFP tagging of Cse4 does not affect its function (Figure 2A). Cse4-GFP–labeled centromeres were photobleached, and FRAP (pre and post) was followed at 1-min intervals for 10 min. The average fluorescence recovery of Cse4-GFP after photobleaching in the wild-type strain was low at ∼13% ± 3 (mean ± SE, n = 6, after 5 min), indicating that Cse4 is a stable component of the kinetochore and that centromeres remained positioned within their respective spindle half after metaphase congression (Figure 2B). The low level of Cse4-GFP fluorescence recovery as observed here for the wild-type strain has been reported previously (Pearson et al., 2004; Lawrimore et al., 2011). Consistent with our hypothesis, the average Cse4-GFP fluorescence recovery in the pat1Δ strain was 23% ± 5 (mean ± SE, n = 13, after 5 min; Figure 2, B–D, and Supplemental Figure S4), which is significantly faster than that for the wild-type strain (p < 0.05). Taken together, these observations support the conclusion that Pat1 is required for stable maintenance of Cse4 at the kinetochores and protects it from ubiquitination.
FIGURE 2:
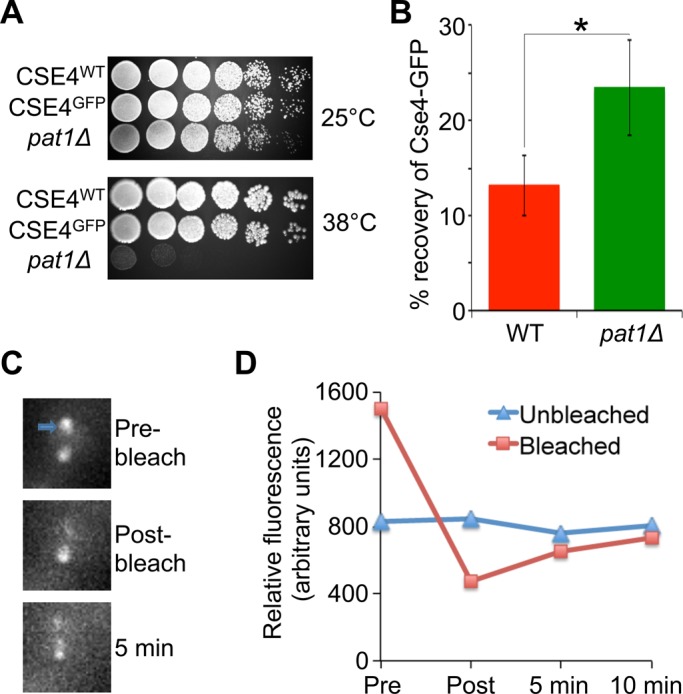
Cse4 exchange at kinetochores is faster in pat1Δ strains. (A) Cse4-GFP strains do not exhibit growth defects or temperature sensitivity to growth phenotypes. Serial dilutions (fivefold) of WT (KBY8116), Cse4-GFP (KBY2012), and pat1Δ (YMB8936) strains grown on YPD at 25 and 38°C. (B) Average Cse4-GFP fluorescence recovery for WT (KBY2012) and pat1Δ (KBY8166) strains 5 min postbleaching ± SE. *p < 0.05, Student's t test. (C) Representative images showing photobleaching and recovery of Cse4-GFP fluorescence in a pat1Δ strain. Cse4-GFP-labeled centromeres were photobleached during metaphase. One of the two kinetochore clusters was bleached (blue arrow). Fluorescence recovery observed after 5 min of photobleaching. (D) Fluorescence recovery of the bleached spot in a pat1Δ strain from representative image shown in C.
Psh1 contributes to the increased ubiquitination of Cse4 in a pat1∆ strain
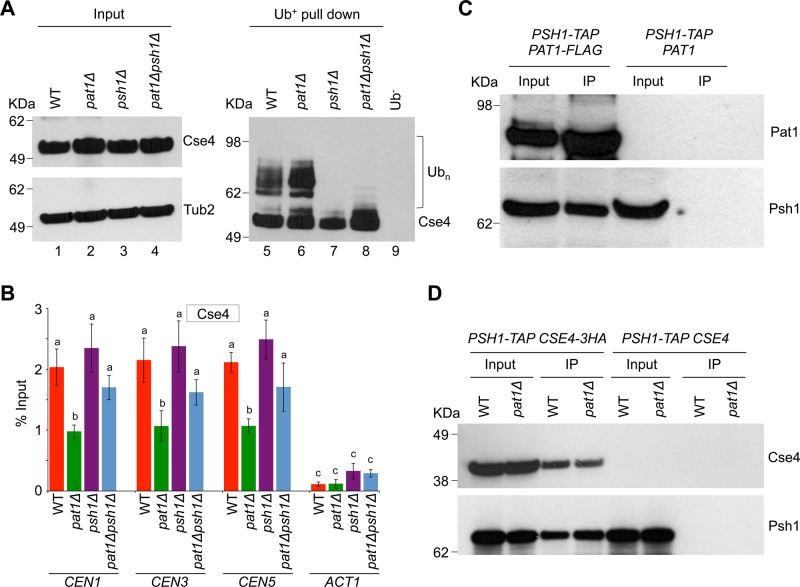
The increased ubiquitination of Cse4 and higher levels of CEN-bound Psh1 in pat1∆ strains (Figure 1, A and C) during mitosis prompted us to investigate whether Psh1 contributes to the increased ubiquitination of Cse4 in the pat1∆ strain. We created a pat1∆ psh1∆ strain and assayed the ubiquitination of Cse4 in this strain and compared it to that of wild-type, pat1∆, and psh1∆ strains. No laddering pattern for Cse4 was observed in control experiments performed with a wild-type strain using agarose beads without Ub-binding activity (Ub−; Figure 3A, lane 9). As observed previously (Figure 1A), the ubiquitination of Cse4 was much higher in the pat1Δ strain (Figure 3A, lane 6 and input control lane 2) than the wild-type strain (Figure 3A, lane 5 and input control lane 1). Consistent with previous reports, Cse4 ubiquitination was greatly diminished in the psh1Δ strain (Figure 3A, lane 7 and input control lane 3). The increased ubiquitination of Cse4 observed in pat1∆ strains was also diminished in the pat1Δ psh1Δ strain (Figure 3A, lane 8 and input control lane 4).
FIGURE 3:
Psh1 contributes to the ubiquitination of Cse4 in a pat1Δ strain. (A) Deletion of PSH1 reduces the levels of Cse4 ubiquitination in a pat1Δ strain. Western blotting showing the levels of Cse4 ubiquitination (Ubn) in logarithmic growing cultures of WT (YMB6398), pat1Δ (YMB8422), psh1Δ (YMB9311), and pat1Δpsh1Δ (YMB9312) strains. Antibodies used were α-Myc (Cse4) or α-Tub2 (served as a loading control). Molecular weight markers for protein size in kilodaltons. (B) Deletion of PSH1 results in higher levels of Cse4 at kinetochores in the pat1Δ psh1Δ strain. WT (YMB6398), pat1Δ (YMB8422), psh1Δ (YMB9311), and pat1Δpsh1Δ (YMB9312) strains were grown to logarithmic phase of growth in YPD at 30°C, and ChIP for Cse4 was performed. The enrichment levels of Cse4 at CEN (CEN1, CEN3, CEN5) and non-CEN (ACT1) regions are presented. Enrichment was determined by qPCR and is shown as percentage input. Average from three biological experiments ± SE. Values sharing the same letter are not significantly different at a 5% level based on analysis of variance (p > 0.05). (C) Pat1 interacts in vivo with Psh1. Immunoprecipitation experiments were done using agarose beads conjugated with α-Flag antibodies using cell extracts from strain YMB9280 expressing Flag-tagged Pat1 and TAP-tagged Psh1 (PSH1-TAP PAT1-FLAG) and a control strain YMB8126 expressing TAP-tagged Psh1 (PSH1-TAP PAT1) from their native promoters. Strains were grown in YPD at 30°C. Eluted proteins were analyzed by Western blotting with α-Flag (Pat1) and α-TAP (Psh1) antibodies. Molecular weight markers for protein size in kilodaltons. (D) Deletion of PAT1 does not significantly affect the in vivo interaction between Psh1 and Cse4. Immunoprecipitation experiments were performed using agarose beads conjugated with α-HA antibodies using cell extracts from WT (YMB9281) and pat1Δ (YMB9282) strains expressing TAP-tagged Psh1 and HA-tagged Cse4 (PSH1-TAP CSE4-3HA) from their native promoters. For control experiments, WT (YMB8126) and pat1Δ (YMB9096) strains expressing TAP-tagged Psh1 (PSH1-TAP CSE4) from their native promoters were used. Strains were grown in YPD at 30°C. Eluted proteins were analyzed by Western blotting with α-HA (Cse4) and α-TAP (Psh1) antibodies. Protein signals were quantified using ImageJ software (Schneider et al., 2012). Molecular weight markers for protein size in kilodaltons.
Next we performed ChIP-qPCR to examine the levels of Cse4 at kinetochores in these strains. In agreement with previous observations (Haase et al., 2013), the levels of Cse4 at CENs (CEN1, CEN3, and CEN5) were reduced in pat1Δ strains (Figure 3B). Consistent with the ubiquitination state of Cse4 (Figure 3A), the levels of Cse4 at kinetochores were higher in the pat1Δ psh1Δ strain than in the pat1Δ strain (Figure 3B). To examine whether Pat1 interacts with Psh1 in vivo, we performed immunoprecipitation experiments using a strain that expressed both Flag-tagged Pat1 and TAP-tagged Psh1 from their endogenous promoters. Our results showed an interaction between Pat1 and Psh1 (Figure 3C). No interactions were observed in control experiments using a wild-type strain carrying only TAP-tagged Psh1 (Figure 3C). Taken together, these observations show that Psh1 contributes to increased ubiquitination of Cse4 in a pat1Δ strain.
Previous studies showed an in vivo interaction between Psh1 and Cse4 (Hewawasam et al., 2010; Ranjitkar et al., 2010). We did immunoprecipitation experiments to determine whether the increased ubiquitination of Cse4 in pat1∆ strains is due to altered interaction of Psh1 and Cse4 in these strains. Experiments were done with wild-type and pat1Δ strains expressing TAP-tagged Psh1 and HA-tagged Cse4 from their endogenous promoters. The interaction of Psh1 and Cse4 is not significantly different in the pat1∆ strains compared to the wild-type strain (Figure 3D). No signals were observed in control experiments using a wild-type strain carrying only TAP-tagged Psh1 (Figure 3D).
Induction of PSH1 results in increased Cse4 ubiquitination, reduction in centromere-associated Cse4, and altered spatial distribution of Cse4 at kinetochores
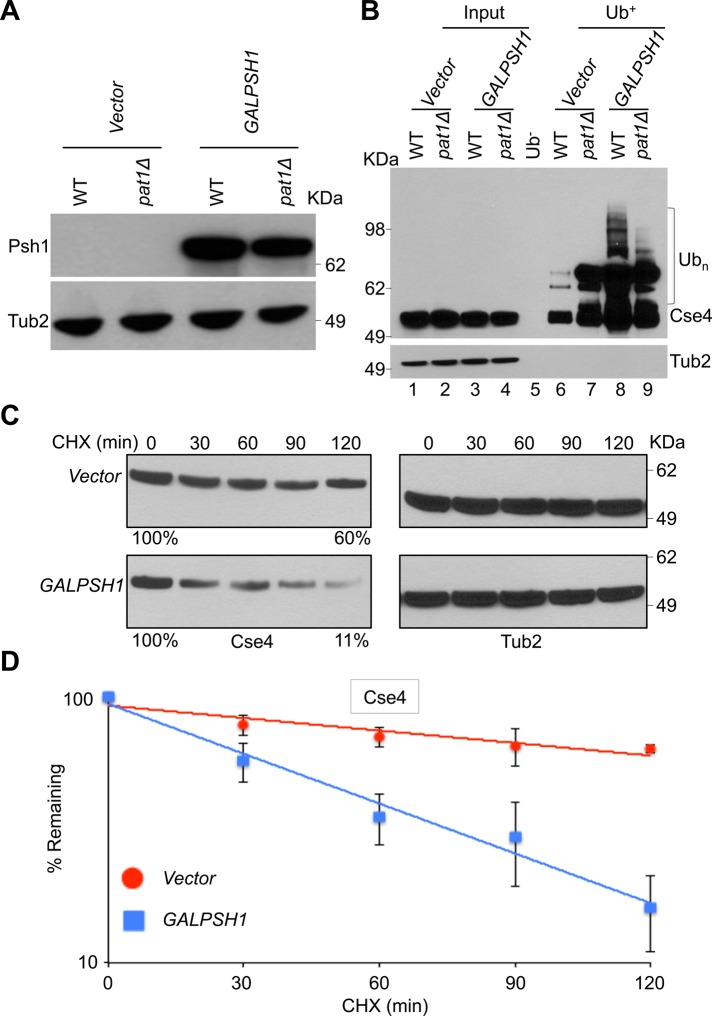
Our results show that Psh1 contributes to increased ubiquitination of Cse4 in a pat1∆ strain. Hence we examined whether transient induction of PSH1 exhibits phenotypes similar to those observed in a pat1∆ strain. Because constitutive overexpression of PSH1 causes loss of viability in wild-type and pat1Δ strains (Supplemental Figure S5), all experiments were done with a GALPSH1 strain grown for 6 h in galactose medium (Figure 4, A and B). In agreement with our hypothesis, transient induction of PSH1 showed increased Cse4 ubiquitination in a wild-type strain compared with the vector control (Figure 4B, lanes 6 and 8). Higher levels of ubiquitinated Cse4 were observed in a pat1∆ strain; however, no major differences in Cse4 ubiquitination with and without induction of PSH1 were observed in a pat1∆ strain (Figure 4B, lanes 7 and 9). Control experiments performed using chromatin extracts from the wild-type strain with agarose beads without Ub-binding activity (Ub−) exhibited no laddering pattern for Cse4 (Figure 4B, lane 5). Consistent with the role of Psh1 in Cse4 proteolysis (Hewawasam et al., 2010; Ranjitkar et al., 2010), transient induction of PSH1 in a wild-type strain caused faster degradation of Cse4 (approximately sixfold to sevenfold) than the vector control (Figure 4, C and D).
FIGURE 4:
Induction of Psh1 causes increased ubiquitination and increased proteolysis of Cse4. (A) Western blot analysis was done using whole-cell protein extracts prepared from WT (YMB6398) and pat1Δ (YMB8948) strains transformed with vector (pRS426 GAL1) or GALPSH1HA (pMB1628). Strains were grown to logarithmic phase of growth in synthetic medium, and gene expression was induced in the presence of galactose plus raffinose (2% each) at 30°C for 6 h. Blots were probed with α-HA for expression of GALPSH1HA and α-Tub2 (served as a loading control) antibodies. Molecular weight markers for protein size in kilodaltons. (B) Transient induction of PSH1 increases Cse4 ubiquitination. Western blotting showing the levels of Cse4 ubiquitination in WT (YMB6398) and pat1Δ (YMB8948) strains transformed with vector (pRS426 GAL1) or GALPSH1HA (pMB1628). Strains were grown to logarithmic phase of growth in synthetic medium, and gene expression was induced in the presence of galactose plus raffinose (2% each) at 30°C for 6 h. Ubiquitin pull down was performed as described in Materials and Methods. Eluted proteins were analyzed by Western blotting with α-Myc (Cse4) or α-Tub2 (served as a loading control) antibodies. Molecular weight markers for protein size in kilodaltons. (C) Transient induction of PSH1 causes increased proteolysis of Cse4. WT (YMB6398) strains transformed with vector (pRS426 GAL1) or GALPSH1HA (pMB1628) were grown to logarithmic phase of growth in synthetic medium, and gene expression was induced in the presence of galactose plus raffinose (2% each) at 30°C for 6 h. Cycloheximide (100 μg/ml) was added to the cultures, and samples were collected at indicated time intervals. Protein extracts were prepared and analyzed by Western blotting with α-Myc (Cse4) or α-Tub2 (served as a loading control) antibody. Molecular weight markers for protein size in kilodaltons. (D) Rate of Cse4 degradation is faster in a WT strain with transient induction of PSH1. Signal intensities from Western blots shown in C were quantified and statistically analyzed as described previously (Au et al., 2008). Average from three biological experiments ± SE.
ChIP experiments were used to determine whether increased ubiquitination of Cse4 contributes to lower levels of Cse4 at CEN in GALPSH1 strains. ChIP-qPCR revealed that the level of CEN-associated Cse4 was reduced significantly in wild-type and pat1Δ strains with GALPSH1 when compared to the levels observed in a wild-type strain with vector alone (Figure 5A). Of note, the transient induction of PSH1 in a wild-type strain exhibits CEN-associated Cse4 levels similar to those observed in a pat1Δ strain (Figure 5A). No significant enrichment of Cse4 was observed at the non-CEN HML locus (Figure 5A).
FIGURE 5:
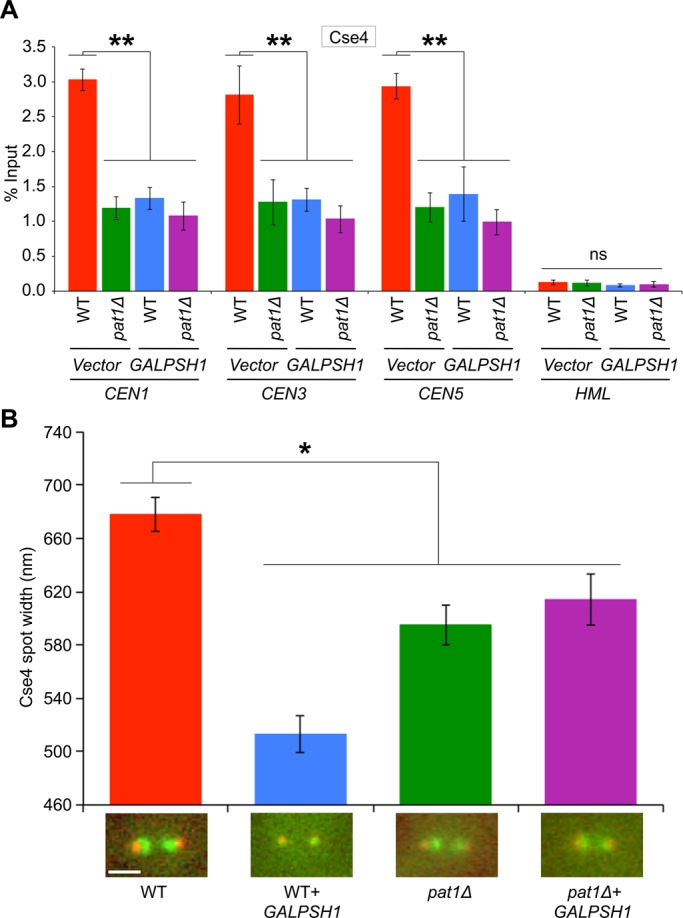
Induction of Psh1 causes reduction in CEN levels of Cse4, and its spatial distribution is altered at the kinetochores. (A) Transient induction of PSH1 causes reduction in CEN levels of Cse4. WT (YMB6398) and pat1Δ (YMB8948) strains transformed with vector (pRS426 GAL1) or GALPSH1HA (pMB1628) were grown to logarithmic phase of growth in synthetic medium, and gene expression was induced in the presence of galactose plus raffinose (2% each) at 30°C for 6 h. ChIP for Cse4 was performed. The enrichment levels of Cse4 at CEN (CEN1, CEN3, CEN5) and non-CEN (HML) regions are presented. Enrichment was determined by qPCR and is shown as percentage input. Average from three biological experiments ± SE. **p < 0.01, ns, statistically not significant, Student's t test. (B) Transient induction of PSH1 causes alterations in the appearance of Cse4 at kinetochores. The height of the cluster of Cse4-GFP at kinetochores was determined from line scans (perpendicular to the spindle axis) through the protein cluster as previously described (Haase et al., 2013). The height was determined from the full-width, full-maximum of the Gaussian distribution. The average spot width of Cse4-GFP foci is shown in WT (KBY2012) and pat1Δ (KBY8166) strains overexpressing PSH1 (GALPSH1HA, pMB1628) for 6 h. Sample sizes range from 11 to 15 cells for each condition, and representative images from each condition are shown. *p < 0.05, Student's t test.
We previously showed that reduced levels of CEN-bound Cse4 contribute to its altered spatial distribution in pat1∆ strains (Haase et al., 2013). Given the similarity of phenotypes of pat1∆ with the transient induction of PSH1, we reasoned that spatial distribution of Cse4 at metaphase kinetochores could be altered in GALPSH1 strains. Our results supported this hypothesis, as the spot width of Cse4, which spans 678 nm, was decreased to 513 nm upon transient induction of PSH1 in a wild-type strain (Figure 5B). Consistent with our previous results (Haase et al., 2013), a decrease of Cse4 spot width to 595 nm was observed in a pat1Δ strain. The spatial distribution of Cse4 remained unaltered upon transient induction of PSH1 in a pat1∆ strain (Figure 5B). Induction of PSH1 specifically affects the distribution of Cse4, as the distribution of another kinetochore protein, Ndc80, was not altered in a GALPSH1 strain (Supplemental Figure S6) or a pat1∆ strain (Haase et al., 2013).
Induction of PSH1 results in structural changes of CEN chromatin and kinetochores
The reduced levels of CEN-associated Cse4 led us to examine whether transient induction of PSH1 causes defects in the structural integrity of CEN chromatin similar to that observed in a pat1∆ strain, by using DraI accessibility assay (Saunders et al., 1990; Meluh et al., 1998; Mythreye and Bloom, 2003; Crotti and Basrai, 2004; Yu et al., 2011; Mishra et al., 2013). Induction of PSH1 increased DraI accessibility of CEN3 chromatin in a wild-type strain (sixfold to eightfold) compared to the strain with vector alone (Figure 6). In agreement with previous results (Mishra et al., 2013), increased DraI accessibility of CEN3 chromatin was observed in the pat1Δ strain with vector alone (Figure 6). There was a slightly higher DraI accessibility of CEN3 in the pat1∆+GALPSH1 strain, although this was not statistically significant. Deletion of PAT1 or transient overexpression of PSH1 did not affect the structure at non-CEN regions such as ADP1 (H3 chromatin; Figure 6).
FIGURE 6:
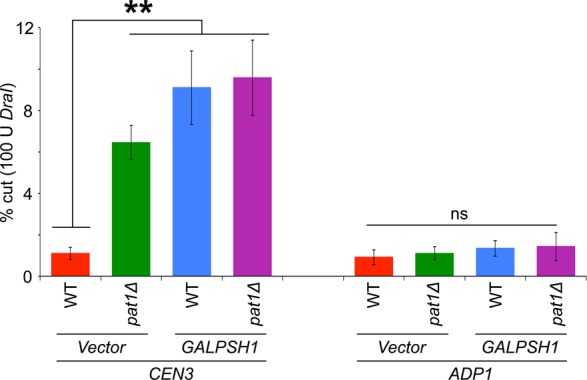
Induction of Psh1 causes structural alterations in CEN chromatin. Transient induction of PSH1 increases the accessibility of DraI to CEN chromatin. WT (YMB6398) and pat1Δ (YMB8948) strains transformed with vector (pRS426 GAL1) or GALPSH1HA (pMB1628) were grown to logarithmic phase of growth in synthetic medium, and gene expression was induced in the presence of galactose plus raffinose (2% each) at 30°C for 6 h. Nuclei were prepared and treated with DraI restriction enzyme to measure enzymatic accessibility as described previously (Mishra et al., 2013). DraI accessibility of CEN chromatin (CEN3) is significantly increased upon overexpression of PSH1 in WT and pat1Δ strains but had no significant affect at H3-chromatin (ADP1). Average from three biological experiments ± SE . **p < 0.01, ns, statistically not significant, Student's t test.
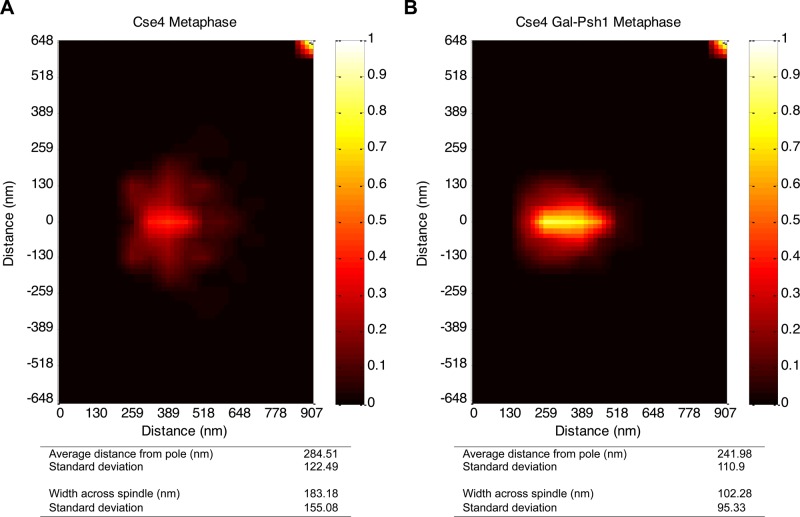
Next we examined whether induction of PSH1 alters the two-dimensional distribution of Cse4 at the kinetochores similar to that observed in a pat1∆ strain (Haase et al., 2013). The distance of Cse4 from the spindle pole body and the spread of Cse4 perpendicular to the spindle pole body were measured. Cse4 distribution in both the wild-type and pat1Δ strains was within 5 nm relative to the spindle pole body (distance from spindle pole: wild type, 286 ± 120 nm; pat1Δ, 281 ± 74 nm). However, the spread of Cse4 perpendicular to the spindle (width across spindle) was reduced about twofold in pat1Δ strains (wild type, 181 ± 155 nm; pat1Δ, 108 ± 102 nm; Haase et al., 2013). Cse4 distribution in the wild-type strain without PSH1 overexpression was similar to previous observations (distance from spindle pole, 285 ± 122 nm; width across spindle, 183 ± 155 nm; n = 208; Figure 7A; Haase et al., 2013). However, the wild-type strain with transiently induced PSH1 levels exhibited a change in the two-dimensional structure of Cse4 similar to that observed for a pat1Δ strain. The distance of Cse4 from the spindle pole was 242 ± 111 nm, whereas the spread of Cse4 perpendicular to the spindle (width across spindle) was reduced to 102 ± 95 nm (n = 204; Figure 7B). Transient induction of PSH1 in a pat1Δ strain did not further exacerbate the two-dimensional structure of Cse4 (Haase et al., 2013).
FIGURE 7:
Induction of Psh1 causes structural alterations of Cse4 at kinetochores. Statistical probability map from experimental images representing kinetochore-associated Cse4 upon PSH1 induction in a WT (KBY2012) strain. Statistical analysis of images is shown at the bottom. Distance from the spindle pole is the average distance. The width across the spindle is obtained by adding the average distances below and above the spindle axis. The detailed procedure used to prepare statistical probability maps was described previously (Haase et al., 2013). (A) Experimental statistical probability map for Cse4 in a WT strain without PSH1 induction. Map was generated from 208 images. (B) Experimental statistical probability map for Cse4 in a WT strain with PSH1 induction (GALPSH1). Map was generated from 204 images.
Pat1 interacts with Scm3 and pat1∆ strains showed reduced levels of centromeric Scm3
The kinetochore protein Scm3 is a Cse4-specific assembly factor and protects centromeric Cse4 from Psh1-mediated ubiquitination (Hewawasam et al., 2010). Because reduced levels of peri-CEN Cse4 contribute to its altered spatial distribution in pat1∆ strains (Haase et al., 2013), we examined whether Pat1 interacts with Scm3 in vivo and affects the levels of centromeric Scm3. We constructed a strain that expressed HA-tagged Pat1, Flag-tagged Scm3, and Myc-tagged Cse4 from their endogenous promoters. No signals were observed in a control experiment using an untagged Pat1 strain (Figure 8A). As previously reported, we observed an interaction between Scm3 and Cse4 (Camahort et al., 2007; Mizuguchi et al., 2007; Stoler et al., 2007; Mishra et al., 2011). Immunoprecipitation results showed that Pat1 interacts with Scm3 and Cse4 but not with histone H3 in vivo.
FIGURE 8:
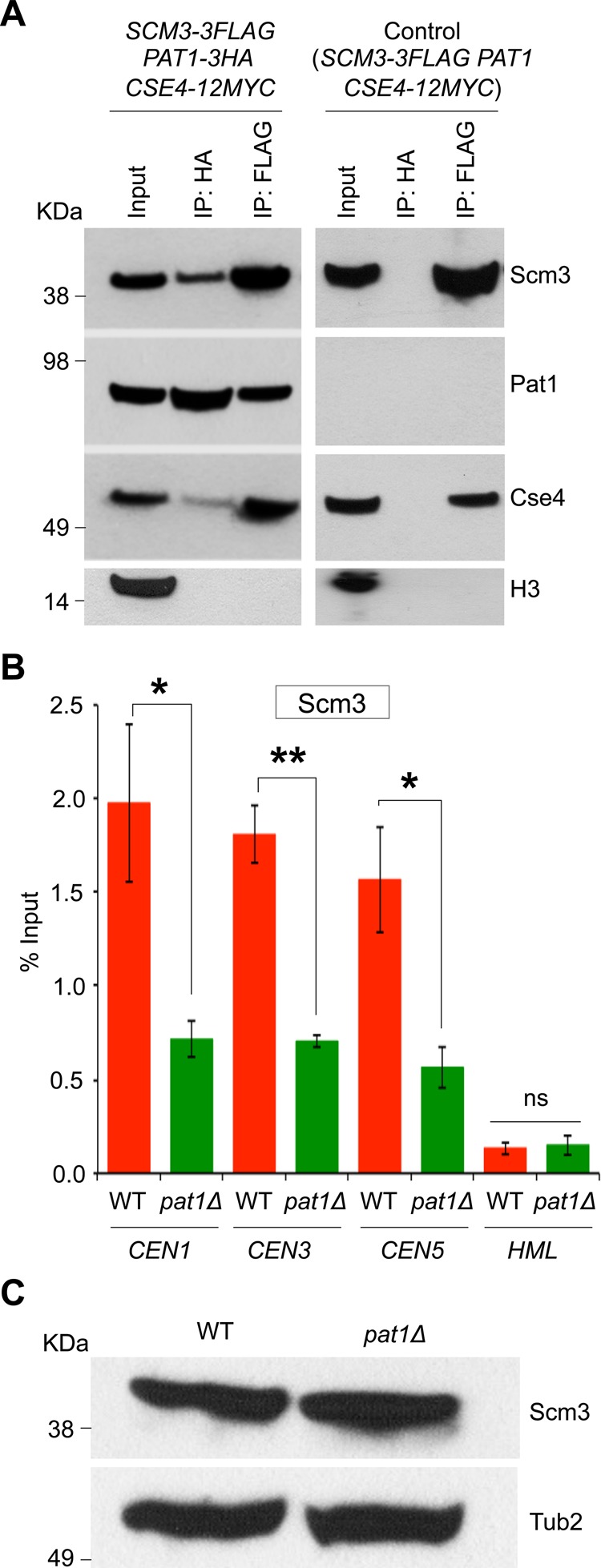
Pat1 interacts with Scm3, and pat1∆ strains showed reduced levels of centromeric Scm3. (A) Western blots of proteins copurifying with Pat1 and Scm3. Coimmunoprecipitation experiments were done using agarose beads conjugated with α-HA or α-Flag antibodies using cell extracts from strain (YMB7963) grown in YPD at 30°C expressing HA-tagged Pat1, Flag-tagged Scm3, and Myc-tagged Cse4 from their native promoters at the endogenous locus. A strain expressing Flag-tagged Scm3, Myc-tagged Cse4, and untagged Pat1 (JG595) from their native promoters at the endogenous locus was used for control experiments. Eluted proteins were analyzed by Western blotting with α-HA (Pat1), α-Flag (Scm3), α-Myc (Cse4), and α-H3 antibodies. (B) CEN association of Scm3 is reduced in pat1∆ strains. CEN levels of Scm3 were assayed by ChIP analysis of Scm3-Flag at CEN (CEN1, CEN3, and CEN5) and non-CEN (HML) DNA in WT (RC154) and pat1Δ (YMB8473) strains grown in YPD at 30°C. Enrichment was determined by qPCR and is shown as percentage input. Average from three biological experiments ± SE. *p < 0.05, **p < 0.01, Student's t test. (C) Expression of Scm3 is not affected in pat1∆ strains. Western blots showing protein levels of Scm3-Flag and Tub2 for strains used in B.
ChIP-qPCR experiments showed that the level of CEN-associated Scm3 was reduced significantly in pat1Δ (0.71% of input at CEN1, 0.70% at CEN3, and 0.56% at CEN5) compared with the levels observed in a wild-type strain (1.97% at CEN1, 1.80% at CEN3, and 1.56% at CEN5; Figure 8B). No significant enrichment of Scm3 was detected at the non-CEN HML locus (Figure 8B). Western blotting revealed that the reduction in CEN-associated Scm3 in the pat1Δ strain was not due to reduction in the protein levels of Scm3 (Figure 8C). The interaction of Pat1 with Scm3 and the reduced levels of centromeric Scm3 suggest that Pat1, together with Scm3, plays a critical role in protecting centromeric Cse4 from Psh1-mediated ubiquitination.
DISCUSSION
Evolutionarily conserved histone H3 variant Cse4 and its homologues are essential for chromosome segregation (Allshire and Karpen, 2008; Verdaasdonk and Bloom, 2011; Burrack and Berman, 2012; Maddox et al., 2012). In budding yeast, two pools of Cse4 molecules have been identified at kinetochores (Lawrimore et al., 2011; Haase et al., 2013). The complex with core Cse4 molecules interacts with kinetochore microtubules, and the accessory Cse4 molecules that reside at the peri-CEN region may be required for rapid assembly of Cse4 upon eviction from centromeres (Haase et al., 2013). Peri-CEN Cse4 pools are reduced in a pat1∆ strain, which exhibits defects in chromosome segregation and altered structure of CEN chromatin (Haase et al., 2013; Mishra et al., 2013; Scott and Bloom, 2014). Here we show that Pat1 interacts with Scm3 and E3-ubiquitin ligase Psh1 to maintain peri-CEN Cse4 at kinetochores by protecting it from Psh1-mediated ubiquitination. These conclusions are based on results showing that pat1∆ strains exhibit 1) an increase in Cse4 ubiquitination, 2) a faster turnover of Cse4 at kinetochores, 3) reduced Cse4 ubiquitination when combined with psh1∆, and 4) reduced levels of centromeric Scm3. Furthermore, transient induction of PSH1 in a wild-type strain exhibits phenotypes similar to a pat1Δ strain, such as increased Cse4 ubiquitination, reduction in CEN-associated Cse4, altered structure of CEN chromatin, and distribution of Cse4 at the kinetochores.
Our results show that Pat1 protects CEN-associated Cse4 pools from Psh1-mediated ubiquitination. A higher enrichment of ubiquitinated Cse4 was observed in a pat1Δ strain, particularly in G2/M cells, which correlated with increased levels of CEN-associated Psh1. Consistent with this hypothesis, Cse4 molecules from peri-CEN chromatin were depleted in a pat1∆ strain (Haase et al., 2013). We propose that the macromolecular kinetochore complex protects the core Cse4 molecules and renders these molecules inaccessible to Psh1 activity; however, the peri-CEN Cse4 pools are vulnerable to ubiquitin-mediated proteolysis. This dynamic property of Cse4 ubiquitination is important for mitosis, as the pat1Δ strain exhibits spindle checkpoint–dependent cell cycle delay and errors in chromosome segregation (Wang et al., 1996; Haase et al., 2013; Mishra et al., 2013). Furthermore, we showed that ubiquitination of Cse4 is cell cycle regulated in wild-type cells. The abundance of ubiquitinated Cse4 increases from S phase to G2/M cells and decreases rapidly in cells synchronized in G1, suggesting a high degree of coordination between Cse4 ubiquitination and cell cycle. Future studies should give us insights into the functional relevance of the cell cycle–regulated ubiquitination of Cse4.
Cse4 is recruited to kinetochores in early S phase, and it remains stably associated with it throughout the cell cycle (Pearson et al., 2004; Boeckmann et al., 2013; Wisniewski et al., 2014). These observations are supported by a low recovery of Cse4-GFP after photobleaching at metaphase kinetochores in a wild-type strain. ChIP assays and cell biology experiments have shown reduced levels of CEN-associated Cse4 in a pat1Δ strain, which were primarily attributed to the loss of peri-CEN Cse4 molecules from kinetochores (Haase et al., 2013). It was unclear whether Pat1 is required for the recruitment of Cse4 molecules or its maintenance at kinetochores. Faster recovery of Cse4-GFP in pat1Δ strains indicates that Cse4 is exchanged rapidly at metaphase kinetochores. These results establish that Pat1 is not required for recruitment of Cse4 molecules to the CEN but is required for the stable maintenance of peri-CEN Cse4 molecules at kinetochores.
We propose that increased levels of ubiquitination of Cse4 in pat1Δ strains are contributed by an E3-ubiquitin ligase, Psh1. Support for this hypothesis is derived from results that showed 1) absence of Cse4 ubiquitination upon deletion of PSH1 in a pat1Δ strain, 2) in vivo interaction of Pat1 with Psh1 and Cse4, and 3) increased levels of CEN-associated Psh1 in a pat1Δ strain during G2/M (mitotic cells), when maximum levels of Cse4 ubiquitination were observed. Further support for a role of Pat1 in regulating peri-CEN Cse4 is derived from our results with transient induction of PSH1, which resulted in phenotypes similar to a pat1Δ strain, such as increased ubiquitination and altered spatial distribution of Cse4.
In addition to regulating the levels of peri-CEN Cse4 (Haase et al., 2013), we now provide evidence that Pat1 interacts with Scm3 and regulates its levels at the CEN. Scm3, a Cse4-specific chaperone, was shown to be involved in the protection of Cse4 from Psh1-mediated ubiquitination (Camahort et al., 2007; Mizuguchi et al., 2007; Stoler et al., 2007; Hewawasam et al., 2010; Mishra et al., 2011; Shivaraju et al., 2011). The in vivo interaction between Pat1 and Scm3 and the reduced levels of CEN-associated Scm3 in pat1∆ strains further support the role of Pat1 in the maintenance of Cse4 at the kinetochores. Unlike Cse4, which shows discrete localization to kinetochore clusters, localization of Scm3 is diffuse throughout the nucleus, and its association with centromeres is cell cycle dependent and dynamic (Luconi et al., 2011; Mishra et al., 2011; Wisniewski et al., 2014). Hence it is not possible to use cell biology approaches similar to the ones used for Cse4 to discern the level of Scm3 in the peri-CEN and determine the relative contributions of Pat1 and Scm3 in Cse4 ubiquitination.
In conclusion, we showed that Pat1 plays a major role for peri-CEN Cse4 molecules to remain stably associated with the kinetochores. CEN chromatin–containing Pat1 shields Cse4 from Psh1-mediated ubiquitination to maintain kinetochore structure and function. In the absence of Pat1, these peri-CEN Cse4 molecules are vulnerable to Psh1-mediated ubiquitination. Our studies provide novel insights into how Pat1 helps to maintain peri-CEN Cse4 molecules at the budding yeast kinetochores.
MATERIALS AND METHODS
Strains, plasmids, and growth conditions
Yeast strains and plasmids used are listed in Table 1. Strains were grown in 1% yeast extract, 2% Bacto-Peptone, and 2% glucose (YPD) or in synthetic yeast medium containing either glucose (2%) or galactose plus raffinose (2% each) with supplements to allow for the selection of plasmids being examined. Wild-type strain with cse416KR (Au et al., 2013) was grown in galactose plus raffinose (2% each) for 4 h to induce the expression of mutant allele of Cse4. Viability assays for the wild-type and pat1Δ strains containing GALPSH1HA (pMB1628) or vector (pRS426 GAL1) were carried out by spotting serial dilutions of equal numbers of cells from three independent transformants for each strain on synthetic medium at 30°C.
TABLE 1:
Strains and plasmids used in this study.
| Saccharomyces cerevisiae strain | Genotype | Source or reference |
|---|---|---|
| BY4741 | MATa ura3Δ0 leu2Δ0 his3Δ1 met15Δ0 | Open Biosystems |
| YMB8126 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 PSH1-TAP::HIS3 | Open Biosystems |
| YMB9096 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 PSH1-TAP::HIS3 pat1Δ::URA3 | This study |
| YMB6398 | MATa ura3-52 lys2-801 ade2-101 trp1Δ63 his3Δ200 leu2Δ1 CSE4-13Myc::LEU2 | Au et al. ([0-9][0-9][0-9][0-9]) |
| YMB8948 | MATa ura3-52 lys2-801 ade2-101 trp1Δ63 his3Δ200 leu2Δ1 CSE4-13Myc::LEU2 pat1Δ::TRP1 | This study |
| YMB8936 | MATa ura3-1 leu2,3-112 his3-1 trp1-1 ade2-1 can1-100 Δbar1 CSE4-12Myc::URA3 SCM3-3Flag::kanMX pat1Δ::TRP1 | This study |
| YMB8422 | MATa ura3-52 lys2-801 ade2-101 trp1Δ63 his3Δ200 leu2Δ1 CSE4-13Myc::LEU2 pat1Δ::URA3 | This study |
| KBY2012 | MATa trp1Δ63 leu2Δ ura3-52 his3 Δ 200 lys2-8Δ1 CSE4GFP::TRP1 (pKK1) SPC29CFP::KAN | Haase et al. ([0-9][0-9][0-9][0-9]) |
| KBY8166 | MATa trp1Δ63 leu2Δ ura3-52 his3Δ200 lys2-8Δ1 CSE4GFP::TRP1 (pKK1) SPC29CFP::KAN pat1Δ:NAT | Haase et al. (2013) |
| KBY8116 | MATa trp1Δ63 leu2Δ ura3-52 his3Δ200 lys2-8Δ1 NDC80GFP::KAN SPC29RFP::Hg | Haase et al. (2013) |
| YMB9280 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 PSH1-TAP::HIS3 PAT1-FLAG::URA3 | This study |
| YMB9281 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 PSH1-TAP::HIS3 CSE4HA::NAT | This study |
| YMB9282 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 PSH1-TAP::HIS3 CSE4HA::NAT pat1Δ::URA3 | This study |
| YMB9311 | MATa ura3-52 lys2-801 ade2-101 trp1Δ63 his3Δ200 leu2Δ1 CSE4-13Myc::LEU2 psh1Δ::KAN | This study |
| YMB9312 | MATa ura3-52 lys2-801 ade2-101 trp1Δ63 his3Δ200 leu2Δ1 CSE4-13Myc::LEU2 psh1Δ::KAN pat1Δ::URA3 | This study |
| RC154 | MATa ura3-1 leu2,3-112 his3-1 trp1-1 ade2-1 can1-100 Δbar1 SCM3-3Flag::KAN NDC10-13Myc::TRP1 | Camahort et al. (2007) |
| YMB8473 | MATa ura3-1 leu2,3-112 his3-1 trp1-1 ade2-1 can1-100 Δbar1 SCM3-3Flag::KAN NDC10-13Myc::TRP1 pat1Δ::URA3 | This study |
| YMB7963 | MATa ura3-1 leu2,3-112 his3-1 trp1-1 ade2-1 can1-100 Δbar1 CSE4-12Myc::URA3 SCM3-3Flag::KAN PAT1-3HA::TRP1 | This study |
| JG595 | MATa ura3-1 leu2,3-112 his3-1 trp1-1 ade2-1 can1-100 Δbar1 PAT1 CSE4-12Myc::URA3 SCM3-3Flag::kanMX | Camahort et al. (2007) |
| YPH499 | MATa ura3-52 lys2-801 ade2-101 trp1Δ63 his3Δ200 leu2Δ1 | Sikorski and Hieter (1989) |
| Plasmid | Description | Reference |
| pRS426-GAL1 | 2μm URA3 GAL1 | Sikorski and Hieter (1989), Mumberg et al. (1994) |
| pMB1628 | 2μm URA3 GAL1/10-PSH1-HA | Gelperin et al. (2005) |
Cell cycle arrest and FACS analysis
Cells were grown in YPD to logarithmic phase at 30ºC, treated for 2 h with 3 μM α-factor (RP01002; GenScript, Piscataway, NJ) to arrest cells in G1, 0.2 M hydroxyurea (HU; H8627; Sigma-Aldrich, St. Louis, MO) to arrest cells in S phase, and 20 μg/ml nocodazole (M1404; Sigma-Aldrich) to arrest cells in G2/M. Samples were analyzed by FACS to confirm the cell cycle arrest using a Becton-Dickinson FACSort flow cytometer and Cell Quest software (BD Biosciences, Boston, MA). Cell cycle stages were determined based on nuclear position and cell morphology in propidium iodide–stained cells using the Zeiss Axioskop 2 microscope (Carl Zeiss, Peabody, MA) as described previously (Calvert and Lannigan, 2010).
Ubiquitin affinity pull-down assay
Ubiquitin affinity pull-down experiments were performed in three biological replicates as described previously (Hjerpe et al., 2009; Au et al., 2013). Briefly, yeast strains were grown to the logarithmic phase in YPD or synthetic medium with either 2% glucose or galactose plus raffinose (2% each) containing supplements to allow selection of plasmids being examined. Cultures were treated with sodium azide (0.1%) for 5 min, and cells were collected. Cell pellets were dissolved in lysis buffer (20 mM Na2HPO4, 20 mM NaH2PO4, 5 mM tetra-sodium pyrophosphate, 50 mM NaF, 1 mM dithiothreitol, 10 mM β-glycerophosphate, 2 mM EDTA, 1% NP-40, 5 mM N-ethylmaleimide, 1 mM phenylmethylsulfonyl fluoride) and protease inhibitor cocktail (P8215; Sigma-Aldrich), and an equal volume of glass beads (425–600 μM) was added. Cells were lysed by bead beating (three times, 2 min each time), followed by vortexing for 60 min at 4°C. Cell lysates were normalized for protein concentration and used for affinity purification with 25 μl of tandem ubiquitin-binding entities Agarose-TUBE1 (UM401; Life Sensors, Malvern, PA; exhibits high affinity to monoubiquitinated and polyubiquitin chain of K48 linkage) or control agarose (UM400; Life Sensors) at 4°C for 16 h. Beads were washed with 1× Tris-buffered saline (TBS) with Tween-20 (0.1%) at room temperature (three times, 5 min each time). The bound proteins were eluted in 2× Laemmli buffer by incubating at 100°C for 10 min and analyzed by Western blotting. Protein intensity signals were quantified using Gene Tools version 3.08 software (SynGene, Cambridge, United Kingdom). Relative ubiquitination of Cse4 in different cell cycle stages was measured as a ratio of ubiquitinated to input Cse4, and statistical significance was determined by Student's t test. We used an independent approach to confirm the results of ubiquitin affinity pull-down assays by performing immunoprecipitation of Cse4, followed by Western blotting as described previously (Ranjitkar et al., 2010; Deyter and Biggins, 2014).
Immunoprecipitation, protein stability, and Western blotting assays
Immunoprecipitation experiments were performed as described previously (Mishra et al., 2011). Protein samples were prepared using the trichloroacetic acid procedure (Cox et al., 1997), protein concentrations were determined using the DC protein assay (Bio-Rad, Hercules, CA), and equal amounts of protein for each sample were assayed by Western blotting. Primary antibodies used were α-Myc (Z-5, sc-789; Santa Cruz Biotechnology, Dallas, TX), α-TAP (CAB1001; Thermo Scientific, Carlsbad, CA), α-HA (clone 12CA5; Roche, Pleasanton, CA), α-histone H3 (ab1791-100; Abcam, Cambridge, MA), α-histone H2B (ab1790; Abcam), α-Flag (F-3165; Sigma-Aldrich), α-ubiquitin (ab6309-1; Abcam), and α-tubulin 2 (Tub2; Mishra et al., 2011). Secondary antibodies used were sheep α-mouse immunoglobulin (IgG; NA931V; Amersham Biosciences, Amersham, United Kingdom) and donkey α-rabbit IgG (NA934V, Amersham). Protein stability assays were performed as described previously (Au et al., 2008; Au et al., 2013).
Chromatin immunoprecipitation and quantitative PCR
ChIP experiments were carried out in three biological replicates as described previously (Mishra et al., 2007, 2011), with minor modifications. Briefly, cultures were cross-linked with 1% formaldehyde (final concentration) for 15 min and quenched with 125 mM glycine for 5 min at room temperature. Cell pellets were collected by centrifugation, washed with TBS (20 mM Tris-HCl, pH 7.6; 150 mM NaCl) and then used to prepare spheroplasts using Zymolyase 100T. Spheroplasts were washed once with postspheroplasting buffer (1.2 M sorbitol, 1 mM MgCl2, 20 mM Na–1,4-piperazinediethanesulfonic acid, pH 6.8), followed by three washes with FA buffer (50 mM Na-HEPES, pH 7.6, 1 mM EDTA, 1% Triton X-100, 150 mM NaCl, 0.1% Na-deoxycholate) with protease inhibitor cocktail (P8215; Sigma-Aldrich). Spheroplasts were then resuspended in FA buffer with protease inhibitors and sonicated on ice at 30% output cycle for five 12-s bursts applied 2 min apart to obtain an average DNA fragment size of 400 base pairs. The resulting soluble chromatin fraction was used for immunoprecipitation experiments as described previously (Mishra et al., 2007, 2011), using the following antibodies: α-TAP (CAB1001; Thermo Scientific), α-Myc (A7470; Sigma-Aldrich), α-Flag (A2220; Sigma-Aldrich), and α-glutathione-S-transferase (Z-5, sc-459; Santa Cruz Biotechnology). Quantitative-PCR using ChIP DNA (ChIP-qPCR) was performed in the 7500 Fast Real Time PCR System using Fast SYBR Green Master Mix (Applied Biosystems, Foster City, CA) following conditions described in Mishra et al. (2011). The enrichment values were calculated as percentage input using the ΔΔCt method (Livak and Schmittgen, 2001). The primer sequences used in this study are listed in Table 2.
TABLE 2:
Primers used in this study.
| Locus | Forward (5′-3′) | Reverse (5′-3′) | Reference |
|---|---|---|---|
| CEN1 | CTCGATTTGCATAAGTGTGCC | GTGCTTAAGAGTTCTGTACCAC | Choy et al. (2011) |
| CEN3 | GATCAGCGCCAAACAATATGG | AACTTCCACCAGTAAACGTTTC | Choy et al. (2011) |
| CEN5 | AAGAACTATGAATCTGTAAATGACTGATTCAAT | CTTGCACTAAACAAGACTTTATACTACGTTTAG | Choy et al. (2011) |
| ACT1 | AATGGCGTGAGGTAGAGAGAAACC | ACAACGAATTGAGAGTTGCCCCAG | Au et al. (2008) |
| HML | CACAGCGGTTTCAAAAAAGCTG | GGATTTTATTTAAAAATCGAGAGG | Choy et al. (2011) |
| CEN3-DraI | TTGATGAACTTTTCAAAGATGAC | GTCAACGAGTCCTCTCTGGCTA | Mishra et al. (2013) |
| ADP1-DraI | ATCCAAATGTGCTCAAGATAGTAGC | CACCAAACAACATTTACTAGCAGTG | Mishra et al. (2013) |
Preparation of yeast nuclei and DraI accessibility assay
Wild-type and pat1Δ strains carrying vector (pRS426 GAL1) or GALPSH1HA (pMB1628) were grown in synthetic yeast medium with galactose plus raffinose (2% each) for 6 h at 30°C to mid log phase. Nuclei were prepared from the cells, digested with DraI (0 and 100 U/ml of nuclei) for 30 min at 37°C, and DNA was extracted following the procedure described in previous studies (Saunders et al., 1990; Mishra et al., 2013). The susceptibility of chromatin to DraI digestion (100 U/ml) was determined by quantitative real-time PCR as described previously (Mishra et al., 2013) using primers flanking DraI sites of CEN3 (CEN-chromatin) and ADP1 (non-CEN chromatin used as a control). The fraction of DNA digested by the DraI (100 U/ml) enzyme was determined by normalizing the Ct values to an untreated control (no DraI).
Imaging techniques and microscopy
Cultures grown to logarithmic phase at room temperature were used for all imaging experiments. Metaphase cells were selected based on the position of kinetochores (Cse4-GFP) and spindle pole bodies (Spc29-CFP) as live-cell markers (Chen et al., 2000; Maddox et al., 2000). Cells were imaged at room temperature on a Nikon TE-2000E inverted microscope equipped with a 1.4 numerical aperture, 100× Plan-Apo objective (Nikon Instruments, Melville, NY) as described previously (Salmon et al., 2013). Thirteen images stepped through a 200-nm z-axis were obtained for each cell using imaging and acquisition software from MetaMorph (Molecular Devices, Sunnyvale, CA). Kinetochore foci were measured by fitting to a Gaussian distribution to the fluorescent spot. The width of the fluorescent spot was determined from the full-width, full-maximum of the Gaussian distribution. The detailed experimental procedure for the preparation of statistical probability maps was described previously (Haase et al., 2013).
Cse4-GFP FRAP experiments were done using cells grown to logarithmic phase of growth at room temperature and imaging of metaphase cells as described previously (Chen et al., 2000; Maddox et al., 2000). The positions of kinetochores (Cse4-GFP) and spindle pole bodies (Spc29-CFP) were used as live-cell markers for cell cycle stage to identify cells in metaphase. One cluster of Cse4-GFP fluorescence was photobleached with a short, 35-ms exposure of focused 488-nm laser light in metaphase cells. A five-plane fluorescence z-series (0.5-μm steps) was obtained immediately after laser exposure to measure the fluorescence photobleaching and recovery as previously described (Pearson et al., 2004). Cse4-GFP fluorescence intensities were determined by collecting the integrated intensity by placing a 5 × 5–pixel region around the localized fluorescence. The total Cse4-GFP fluorescence intensity was then determined by subtracting the intracellular background levels.
Supplementary Material
Acknowledgments
We thank Jennifer Gerton, Sue Biggins, Roy Parker, Michael Lichten, and Richard Baker for reagents, Kathy McKinnon of the National Cancer Institute Vaccine Branch FACS Core for assistance with FACS, Wei-Chun Au for assistance with the ubiquitin pull-down assay, and members of the Basrai laboratory for discussions. Support for P.K.M., L.E.D., and M.A.B. was provided by the Intramural Research Program of the National Cancer Institute, National Institutes of Health. Support for J.G., J.H., E.Y., and K.B. was derived from National Institute of Health Grant R37 GM32238 to K.B.
Abbreviations used:
- CEN
centromere
- CFP
cyan fluorescent protein
- ChIP
chromatin immunoprecipitation
- FACS
fluorescence-activated cell sorting
- FRAP
fluorescence recovery after photobleaching
- GFP
green fluorescent protein
- qPCR
quantitative PCR.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E14-08-1335) on April 1, 2015.
REFERENCES
- Allshire RC, Karpen GH. Epigenetic regulation of centromeric chromatin: old dogs, new tricks. Nat Rev Genet. 2008;9:923–937. doi: 10.1038/nrg2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Au WC, Crisp MJ, DeLuca SZ, Rando OJ, Basrai MA. Altered dosage and mislocalization of histone H3 and Cse4p lead to chromosome loss in Saccharomyces cerevisiae. Genetics. 2008;179:263–275. doi: 10.1534/genetics.108.088518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Au WC, Dawson AR, Rawson DW, Taylor SB, Baker RE, Basrai MA. A novel role of the N-terminus of budding yeast histone H3 variant Cse4 in ubiquitin-mediated proteolysis. Genetics. 2013;194:513–518. doi: 10.1534/genetics.113.149898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom KS, Carbon J. Yeast centromere DNA is in a unique and highly ordered structure in chromosomes and small circular minichromosomes. Cell. 1982;29:305–317. doi: 10.1016/0092-8674(82)90147-7. [DOI] [PubMed] [Google Scholar]
- Boeckmann L, Takahashi Y, Au WC, Mishra PK, Choy JS, Dawson AR, Szeto MY, Waybright TJ, Heger C, McAndrew C, et al. Phosphorylation of centromeric histone H3 variant regulates chromosome segregation in S. cerevisiae. Mol Biol Cell. 2013;24:2034–2044. doi: 10.1091/mbc.E12-12-0893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrack LS, Berman J. Flexibility of centromere and kinetochore structures. Trends Genet. 2012;28:204–212. doi: 10.1016/j.tig.2012.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvert ME, Lannigan J. Yeast cell cycle analysis: combining DNA staining with cell and nuclear morphology. Curr Protoc Cytom. 2010;32:1–16. doi: 10.1002/0471142956.cy0932s52. Chapter 9: Unit 9. [DOI] [PubMed] [Google Scholar]
- Camahort R, Li B, Florens L, Swanson SK, Washburn MP, Gerton JL. Scm3 is essential to recruit the histone h3 variant cse4 to centromeres and to maintain a functional kinetochore. Mol Cell. 2007;26:853–865. doi: 10.1016/j.molcel.2007.05.013. [DOI] [PubMed] [Google Scholar]
- Chen Y, Baker RE, Keith KC, Harris K, Stoler S, Fitzgerald-Hayes M. The N terminus of the centromere H3-like protein Cse4p performs an essential function distinct from that of the histone fold domain. Mol Cell Biol. 2000;20:7037–7048. doi: 10.1128/mcb.20.18.7037-7048.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho US, Corbett KD, Al-Bassam J, Bellizzi JJ, 3rd, De Wulf P, Espelin CW, Miranda JJ, Simons K, Wei RR, Sorger PK, Harrison SC. Molecular structures and interactions in the yeast kinetochore. Cold Spring Harb Symp Quant Biol. 2010;75:395–401. doi: 10.1101/sqb.2010.75.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choy JS, Acuna R, Au WC, Basrai MA. A role for histone H4K16 hypoacetylation in Saccharomyces cerevisiae kinetochore function. Genetics. 2011;189:11–21. doi: 10.1534/genetics.111.130781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choy JS, Mishra PK, Au WC, Basrai MA. Insights into assembly and regulation of centromeric chromatin in Saccharomyces cerevisiae. Biochim Biophys Acta. 2012;1819:776–783. doi: 10.1016/j.bbagrm.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke L, Carbon J. Isolation of a yeast centromere and construction of functional small circular chromosomes. Nature. 1980;287:504–509. doi: 10.1038/287504a0. [DOI] [PubMed] [Google Scholar]
- Collins KA, Furuyama S, Biggins S. Proteolysis contributes to the exclusive centromere localization of the yeast Cse4/CENP-A histone H3 variant. Curr Biol. 2004;14:1968–1972. doi: 10.1016/j.cub.2004.10.024. [DOI] [PubMed] [Google Scholar]
- Cox JS, Chapman RE, Walter P. The unfolded protein response coordinates the production of endoplasmic reticulum protein and endoplasmic reticulum membrane. Mol Biol Cell. 1997;8:1805–1814. doi: 10.1091/mbc.8.9.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crotti LB, Basrai MA. Functional roles for evolutionarily conserved Spt4p at centromeres and heterochromatin in Saccharomyces cerevisiae. EMBO J. 2004;23:1804–1814. doi: 10.1038/sj.emboj.7600161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deyter GM, Biggins S. The FACT complex interacts with the E3 ubiquitin ligase Psh1 to prevent ectopic localization of CENP-A. Genes Dev. 2014;28:1815–1826. doi: 10.1101/gad.243113.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelperin DM, White MA, Wilkinson ML, Kon Y, Kung LA, Wise KJ, Lopez-Hoyo N, Jiang L, Piccirillo S, Yu H, et al. Biochemical and genetic analysis of the yeast proteome with a movable ORF collection. Genes Dev. 2005;19:2816–2826. doi: 10.1101/gad.1362105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonen S, Akiyoshi B, Iadanza MG, Shi D, Duggan N, Biggins S, Gonen T. The structure of purified kinetochores reveals multiple microtubule-attachment sites. Nat Struct Mol Biol. 2012;19:925–929. doi: 10.1038/nsmb.2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haase J, Mishra PK, Stephens A, Haggerty R, Quammen C, Taylor RM, 2nd, Yeh E, Basrai MA, Bloom K. A 3D map of the yeast kinetochore reveals the presence of core and accessory centromere-specific histone. Curr Biol. 2013;23:1939–1944. doi: 10.1016/j.cub.2013.07.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartzog GA, Basrai MA, Ricupero-Hovasse SL, Hieter P, Winston F. Identification and analysis of a functional human homolog of the SPT4 gene of Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:2848–2856. doi: 10.1128/mcb.16.6.2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewawasam GS, Mattingly M, Venkatesh S, Zhang Y, Florens L, Workman JL, Gerton JL. Phosphorylation by casein kinase 2 facilitates Psh1 protein-assisted degradation of Cse4 protein. J Biol Chem. 2014;289:29297–29309. doi: 10.1074/jbc.M114.580589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewawasam G, Shivaraju M, Mattingly M, Venkatesh S, Martin-Brown S, Florens L, Workman JL, Gerton JL. Psh1 is an E3 ubiquitin ligase that targets the centromeric histone variant Cse4. Mol Cell. 2010;40:444–454. doi: 10.1016/j.molcel.2010.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hjerpe R, Aillet F, Lopitz-Otsoa F, Lang V, England P, Rodriguez MS. Efficient protection and isolation of ubiquitylated proteins using tandem ubiquitin-binding entities. EMBO Rep. 2009;10:1250–1258. doi: 10.1038/embor.2009.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs CW, Adams AE, Szaniszlo PJ, Pringle JR. Functions of microtubules in the Saccharomyces cerevisiae cell cycle. J Cell Biol. 1988;107:1409–1426. doi: 10.1083/jcb.107.4.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagawa K, Hieter P. Evolutionary conservation between budding yeast and human kinetochores. Nat Rev Mol Cell Biol. 2001;2:678–687. doi: 10.1038/35089568. [DOI] [PubMed] [Google Scholar]
- Lampert F, Westermann S. A blueprint for kinetochores—new insights into the molecular mechanics of cell division. Nat Rev Mol Cell Biol. 2011;12:407–412. doi: 10.1038/nrm3133. [DOI] [PubMed] [Google Scholar]
- Lawrimore J, Bloom KS, Salmon ED. Point centromeres contain more than a single centromere-specific Cse4 (CENP-A) nucleosome. J Cell Biol. 2011;195:573–582. doi: 10.1083/jcb.201106036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechner J, Carbon J. A 240 kd multisubunit protein complex, CBF3, is a major component of the budding yeast centromere. Cell. 1991;64:717–725. doi: 10.1016/0092-8674(91)90501-o. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Luconi L, Araki Y, Erlemann S, Schiebel E. The CENP-A chaperone Scm3 becomes enriched at kinetochores in anaphase independently of CENP-A incorporation. Cell Cycle. 2011;10:3369–3378. doi: 10.4161/cc.10.19.17663. [DOI] [PubMed] [Google Scholar]
- Maddox PS, Bloom KS, Salmon ED. The polarity and dynamics of microtubule assembly in the budding yeast Saccharomyces cerevisiae. Nat Cell Biol. 2000;2:36–41. doi: 10.1038/71357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddox PS, Corbett KD, Desai A. Structure, assembly and reading of centromeric chromatin. Curr Opin Genet Dev. 2012;22:139–147. doi: 10.1016/j.gde.2011.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meluh PB, Yang P, Glowczewski L, Koshland D, Smith MM. Cse4p is a component of the core centromere of Saccharomyces cerevisiae. Cell. 1998;94:607–613. doi: 10.1016/s0092-8674(00)81602-5. [DOI] [PubMed] [Google Scholar]
- Meraldi P, McAinsh AD, Rheinbay E, Sorger PK. Phylogenetic and structural analysis of centromeric DNA and kinetochore proteins. Genome Biol. 2006;7:R23. doi: 10.1186/gb-2006-7-3-r23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra PK, Au WC, Choy JS, Kuich PH, Baker RE, Foltz DR, Basrai MA. Misregulation of Scm3p/HJURP causes chromosome instability in Saccharomyces cerevisiae and human cells. PLoS Genet. 2011;7:e1002303. doi: 10.1371/journal.pgen.1002303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra PK, Baum M, Carbon J. Centromere size and position in Candida albicans are evolutionarily conserved independent of DNA sequence heterogeneity. Mol Genet Genomics. 2007;278:455–465. doi: 10.1007/s00438-007-0263-8. [DOI] [PubMed] [Google Scholar]
- Mishra PK, Ottmann AR, Basrai MA. Structural integrity of centromeric chromatin and faithful chromosome segregation requires Pat1. Genetics. 2013;195:369–379. doi: 10.1534/genetics.113.155291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuguchi G, Xiao H, Wisniewski J, Smith MM, Wu C. Nonhistone Scm3 and histones CenH3-H4 assemble the core of centromere-specific nucleosomes. Cell. 2007;129:1153–1164. doi: 10.1016/j.cell.2007.04.026. [DOI] [PubMed] [Google Scholar]
- Mumberg D, Muller R, Funk M. Regulatable promoters of Saccharomyces cerevisiae: comparison of transcriptional activity and their use for heterologous expression. Nucleic Acids Res. 1994;22:5767–5768. doi: 10.1093/nar/22.25.5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mythreye K, Bloom KS. Differential kinetochore protein requirements for establishment versus propagation of centromere activity in Saccharomyces cerevisiae. J Cell Biol. 2003;160:833–843. doi: 10.1083/jcb.200211116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson CG, Yeh E, Gardner M, Odde D, Salmon ED, Bloom K. Stable kinetochore-microtubule attachment constrains centromere positioning in metaphase. Curr Biol. 2004;14:1962–1967. doi: 10.1016/j.cub.2004.09.086. [DOI] [PubMed] [Google Scholar]
- Pot I, Measday V, Snydsman B, Cagney G, Fields S, Davis TN, Muller EG, Hieter P. Chl4p and iml3p are two new members of the budding yeast outer kinetochore. Mol Biol Cell. 2003;14:460–476. doi: 10.1091/mbc.E02-08-0517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Przewloka MR, Glover DM. The kinetochore and the centromere: a working long distance relationship. Annu Rev Genet. 2009;43:439–465. doi: 10.1146/annurev-genet-102108-134310. [DOI] [PubMed] [Google Scholar]
- Ranjitkar P, Press MO, Yi X, Baker R, MacCoss MJ, Biggins S. An E3 ubiquitin ligase prevents ectopic localization of the centromeric histone H3 variant via the centromere targeting domain. Mol Cell. 2010;40:455–464. doi: 10.1016/j.molcel.2010.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmon ED, Shaw SL, Waters JC, Waterman-Storer CM, Maddox PS, Yeh E, Bloom K. A high-resolution multimode digital microscope system. Methods Cell Biol. 2013;114:179–210. doi: 10.1016/B978-0-12-407761-4.00009-9. [DOI] [PubMed] [Google Scholar]
- Saunders MJ, Yeh E, Grunstein M, Bloom K. Nucleosome depletion alters the chromatin structure of Saccharomyces cerevisiae centromeres. Mol Cell Biol. 1990;10:5721–5727. doi: 10.1128/mcb.10.11.5721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott KC, Bloom KS. Lessons learned from counting molecules: how to lure CENP-A into the kinetochore. Open Biol. 2014;4 doi: 10.1098/rsob.140191. 140191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivaraju M, Camahort R, Mattingly M, Gerton JL. Scm3 is a centromeric nucleosome assembly factor. J Biol Chem. 2011;286:12016–12023. doi: 10.1074/jbc.M110.183640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoler S, Rogers K, Weitze S, Morey L, Fitzgerald-Hayes M, Baker RE. Scm3, an essential Saccharomyces cerevisiae centromere protein required for G2/M progression and Cse4 localization. Proc Natl Acad Sci USA. 2007;104:10571–10576. doi: 10.1073/pnas.0703178104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdaasdonk JS, Bloom K. Centromeres: unique chromatin structures that drive chromosome segregation. Nat Rev Mol Cell Biol. 2011;12:320–332. doi: 10.1038/nrm3107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Watt PM, Louis EJ, Borts RH, Hickson ID. Pat1: a topoisomerase II-associated protein required for faithful chromosome transmission in Saccharomyces cerevisiae. Nucleic Acids Res. 1996;24:4791–4797. doi: 10.1093/nar/24.23.4791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinert TA, Kiser GL, Hartwell LH. Mitotic checkpoint genes in budding yeast and the dependence of mitosis on DNA replication and repair. Genes Dev. 1994;8:652–665. doi: 10.1101/gad.8.6.652. [DOI] [PubMed] [Google Scholar]
- Wisniewski J, Hajj B, Chen J, Mizuguchi G, Xiao H, Wei D, Dahan M, Wu C. Imaging the fate of histone Cse4 reveals de novo replacement in S phase and subsequent stable residence at centromeres. Elife. 2014:e02203. doi: 10.7554/eLife.02203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Srinivasan M, Nakanishi S, Leatherwood J, Shilatifard A, Sternglanz R. A conserved patch near the C terminus of histone H4 is required for genome stability in budding yeast. Mol Cell Biol. 2011;31:2311–2325. doi: 10.1128/MCB.01432-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.