Abstract
The Drosophila brain is comprised of neurons formed by approximately 100 lineages, each of which is derived from a stereotyped, asymmetrically dividing neuroblast. Lineages serve as structural and developmental units of Drosophila brain anatomy and reconstruction of lineage projection patterns represents a suitable map of Drosophila brain circuitry at the level of neuron populations (“macro-circuitry”). Two phases of neuroblast proliferation, the first in the embryo and the second during the larval phase (following a period of mitotic quiescence), produce primary and secondary lineages, respectively. Using temporally controlled pulses of hydroxyurea (HU) to ablate neuroblasts and their corresponding secondary lineages during the larval phase, we analyzed the effect on development of primary and secondary lineages in the late larval and adult brain. Our findings indicate that timing of neuroblast re-activation is highly stereotyped, allowing us to establish “birth dates” for all secondary lineages. Furthermore, our results demonstrate that, whereas the trajectory and projection pattern of primary and secondary lineages is established in a largely independent manner, the final branching pattern of secondary neurons is dependent upon the presence of appropriate neuronal targets. Taken together, our data provide new insights into the degree of neuronal plasticity during Drosophila brain development.
Keywords: Ablation, Brain, Development, Drosophila, Lineage
Introduction
The Drosophila brain develops from a stereotyped set of embryonically-born stem cells, called neuroblasts. Each neuroblast is defined by its expression of a unique combination of transcriptional regulators (Skeath and Thor, 2003; Urbach and Technau, 2003b). Neuroblasts divide asymmetrically, each mitotic division resulting in a self-renewing neuroblast and a “ganglion mother cell,” which divides once more giving rise to two postmitotic neurons. In holometabolous insects, such as Drosophila, neuroblasts undergo two phases of proliferation. The first phase occurs during the embryonic period; the second one takes place in the larva. In the embryo, a neuroblast divides five to eight times, producing groups (“lineages”) of 10–20 embryonic (“primary”) neurons each (Larsen et al., 2009). Neurons belonging to the same lineage share a number of fundamental morphological characteristics: cell bodies remain clustered together in the outer layer (cortex) of the brain and their axons fasciculate into a common tract (primary axon tract; PAT). In cases where clones of differentiated primary neurons have been labeled it became apparent that neurons of one lineage also share one or a few specific brain compartments in which they form synaptic contacts. For example, four lineages (MB1–4) are restricted to the calyx and lobes of the mushroom body (Ito et al., 1997) and one lineage (BAmv3) forms the projection neurons of the larval antennal lobe (Das et al., 2013; Python and Stocker, 2002; Ramaekers et al., 2005).
At the end of embryogenesis, most neuroblasts enter a period of quiescence. Only five neuroblasts (MB1–4, BAlc/LNb) continuously divide between embryogenesis and early metamorphosis (Ito and Hotta, 1992; Ito et al., 1997; Stocker et al., 1997). All other neuroblasts exit the quiescent phase and re-enter the cell cycle between approximately 20 and 48h after hatching (Ito and Hotta, 1992). During this secondary phase of proliferation, which lasts to the end of the larval stage, most neuroblasts generate an average of 150 postembryonic (“secondary”) neurons (Bello et al., 2008). Similar to primary neurons, secondary neurons of a given lineage form coherent clusters of neuronal cell bodies and project axons which bundle together as the secondary axon tract (SAT). Secondary axon tracts form a stereotyped, conspicuous pattern that is visible from the larva through metamorphosis into the adult stage (Lovick et al., 2013; Wong et al., 2013). Differentiation of secondary neurons (i.e. sprouting of branches and formation of synapses) occurs during metamorphosis, along with remodeling of primary neurons; both secondary neurons and remodeled primary neurons form the adult brain circuitry.
The mechanism triggering the larval (”secondary”) phase of proliferation involves signals derived from the surface glia surrounding the neuroblasts (Ebens et al., 1993). The insulin pathway, which links larval growth in general to the nutritional state, plays an important role in secondary neuroblast proliferation as well (Chell and Brand, 2010). Many aspects of how secondary neuroblast proliferation is initiated remain unknown. In particular, it is not clear whether and how the identity of a neuroblast influences the time point at which it enters mitosis. The time period over which neuroblasts start to divide lasts for more than 24 hours, though the order in which neuroblasts resume proliferation and produce their respective secondary lineages has not been documented. In other words, in any given larva, some neuroblasts enter mitosis considerably earlier than others. Given the high degree of stereotypy of neuroblasts in the embryo (Urbach and Technau, 2003a; Younossi-Hartenstein et al., 1996), and of lineages and their SATs in the late larva (Pereanu and Hartenstein, 2006), we assumed that the birth order of secondary lineages is also highly invariant: a neuroblast of a given identity will always re-enter mitosis at the same time point. To test this hypothesis we used the drug hydroxyurea (HU), a compound known to arrest actively dividing cells, to ablate proliferating neuroblasts and therefore secondary neurons (lineages) they give rise to (de Belle and Heisenberg, 1994; Prokop and Technau, 1994). If our assumption is correct, applying HU at a specific time point should always affect the same set of lineages. We systematically administered short HU pulses during and after the 20–48h period when neuroblasts enter their larval phase of proliferation and analyzed the effect on the development of secondary lineages in the late larval and adult brain using global markers for SATs (anti-Neurotactin/BP106, anti-Neuroglian/BP104), as well as several lineage-specific Gal4 lines.
Our data demonstrate that the time points at which secondary neuroblasts start to divide are indeed fairly stereotyped, which allowed us to reconstruct a “birth calendar” for all lineages. Knowing the birth date of a lineage is of importance for future experiments targeting that particular lineage for ablation or lineage-specific manipulation by mosaic analysis. Aside from establishing lineage birth dates, our results also provide new insights into the degree of plasticity in Drosophila brain development. Trajectories of secondary axon tracts appear to be established largely independently of each other. Similarly, the structure of primary neurons in the larval and adult brain is mostly unaffected by the loss of secondary lineages. In contrast to the apparent rigid nature in which axonal trajectories are established, the final patterning of terminal arbors by secondary lineages appears to depend upon the presence of corresponding neuronal targets (loss of target tissue leads to the absence of terminal arbors by surviving secondary lineages in that region).
Materials and Methods
Genetics
Flies were grown at 25°C using standard fly media unless otherwise noted. per-Gal4 (Kaneko and Hall, 2000), en-Gal4 (Tabata et al., 1995), ple-Gal4 (TH-Gal4; Friggi-Grelin et al., 2002; #8848, Bloomington Drosophila Stock Center, University of Indiana, IN, USA), GH146-Gal4 (a gift from R.F. Stocker, University of Fribourg, Switzerland; Stocker et al., 1997), UAS-mcd8::GFP (Lee and Luo, 1999; #5137, BDSC).
Immunohistochemistry
Samples were fixed in 4% formaldehyde or 4% methanol-free formaldehyde in phosphate buffer saline (PBS, Fisher-Scientific, pH = 7.4; Cat No. #BP399-4). Tissues were permeabilized in PBT (PBS with 0.1–0.3% Triton X-100, pH = 7.4) and immunohistochemistry was performed using standard procedures (Ashburner, 1989). The following antibodies were provided by the Developmental Studies Hybridoma Bank (Iowa City, IA): mouse anti-Bruchpilot (Brp, 1:20), mouse anti-Neurotactin (BP106, 1:10), rat anti-DN-Cadherin (DN-EX #8, 1:20), and mouse anti-Neuroglian (BP104, 1:30). Secondary antibodies, IgG1 (Jackson ImmunoResearch; Molecular Probes) were used at the following dilutions: Cy5-conjugated anti-rat Ig (1:100), Cy3-conjugated anti-mouse Ig (1:200), Cy5-conjugated anti-mouse Ig (1:250); Alexa 546-conjugated anti-mouse (1:500), DynaLight 649-conjugated anti-rat (1:400), Alexa 568-conjugated anti-mouse (1:500).
Hydroxyurea (HU) Ablation Experiments
Hydroxyurea (HU, Sigma) acts as a DNA-synthesis inhibitor which blocks the normal function of nucleotide reductase (Timson, 1975) and is lethal to S-phase cells (Furst and Mahowald, 1985). HU has been used in Drosophila to ablate adult muscle precursors (Broadie and Bate, 1991) as well as central brain neuroblasts (de Belle and Heisenberg, 1994; Stocker et al., 1997). Procedure for preparation of HU was adapted from Broadie and Bate (1991). HU was administered to fly larvae through the diet. Briefly, HU was dissolved in distilled water at a concentration of 50mg/ml. The dissolved HU was then added to partially cool melted fly media to achieve a final concentration of 5mg/ml. After thorough mixing, the HU media was poured onto 60 × 15mm petri dishes to cool. Food plates were made fresh (<1 day beforehand) for each experiment.
To ablate neuroblasts, staged larvae were allowed to grow on standard media at 25°C in petri dishes until time of ablation. Larvae were quickly transferred via blunted forceps to food plates containing 5mg/ml of HU for four hours. This is sufficient time for the HU to accumulate to doses high enough to kill actively dividing neuroblasts (Broadie and Bate, 1991; Truman and Bate, 1988; White and Kankel, 1978). After four hours, larvae were transferred to petri dishes containing standard media and grown until dissected as either wandering L3 or adults. Fly stocks and larvae for experiments were grown at 25°C.
Confocal Microscopy
Staged Drosophila larval and adult brains labeled with suitable markers were viewed as whole-mounts by confocal microscopy [LSM 700 Imager M2 using Zen 2009 (Carl Zeiss Inc.); lenses: 40× oil (numerical aperture 1.3)]. Complete series of optical sections were taken at 2-µm intervals. Captured images were processed by ImageJ or FIJI (National Institutes of Health, http://rsbweb.nih.gov/ij/ and http://fiji.sc/) and Adobe Photoshop.
Generation of three-dimensional models
Digitized images of confocal sections were imported into FIJI (Schindelin et al., 2012; http://fiji.sc/). Complete series of optical sections were taken at 2-µm intervals. Since sections were taken from focal planes of one and the same preparation, there was no need for alignment of different sections. Models were generated using the 3-dimensional viewer as part of the FIJI software package. Digitized images of confocal sections were imported using TrakEM2 plugin in FIJI software (Cardona et al., 2012). Surface renderings of larval brains stained with anti-Bruchpilot were generated as volumes in the 3-dimensional viewer in FIJI. Cell body clusters were indicated on surface renderings using TrakEM2. Digital atlas models of cell body clusters and SATs were created by manually labeling each lineage and its approximate cell body cluster location in TrakEM2.
Results
HU pulses applied at a defined time interval ablate distinct secondary lineages without altering the projection of other lineages
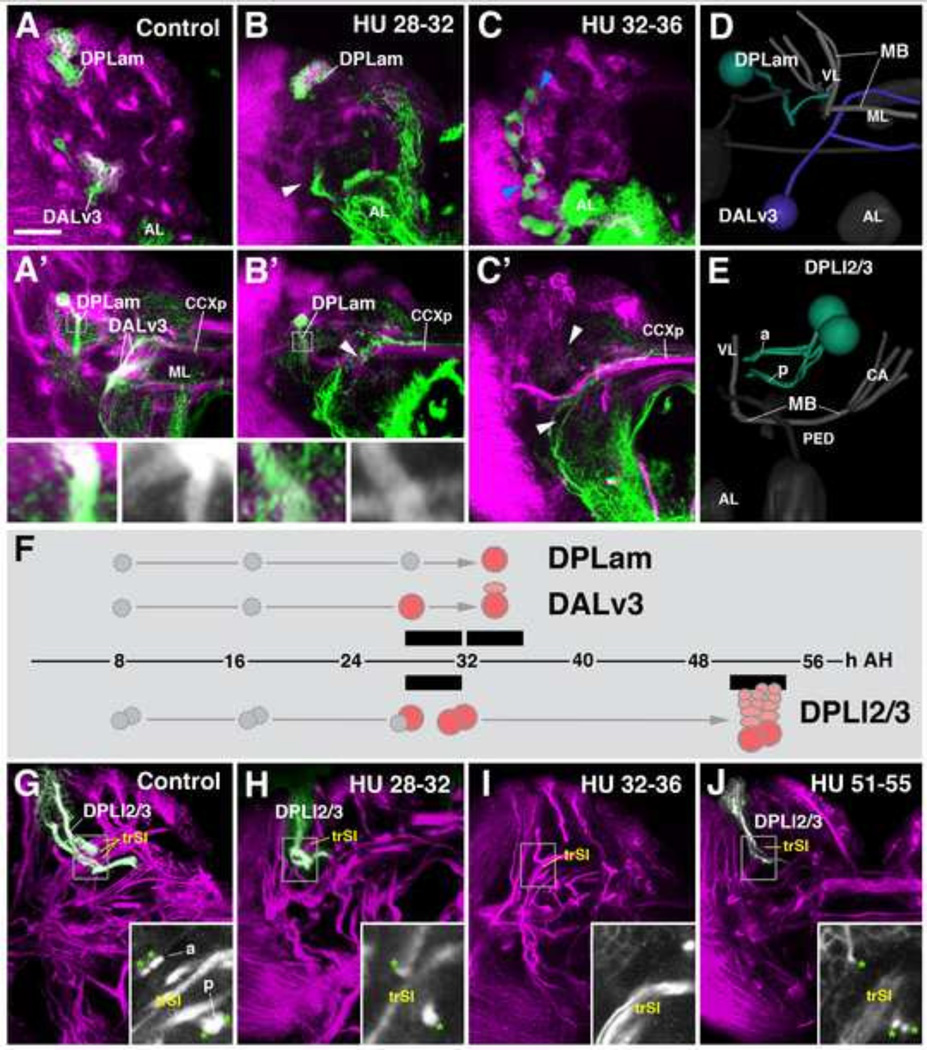
Engrailed (en)-Gal4 is expressed, among others, in two brain lineages, DPLam and DALv3 (Kumar et al., 2009; Fig. 1A–D). HU application prior to 28h after-hatching (AH) has no effect on either of these lineages. Pulses from 28–32h AH ablated DALv3 in many specimens, leading to the absence of the cluster of cell bodies in the cortex and the secondary axon tract in the neuropil (arrowheads in Fig. 1B, B’). The second en-Gal4-positive lineage, DPLam, is never affected by 28–32h HU pulses; its cell body cluster is present at its normal location (Fig. 1B) and its secondary axon tract follows its normal trajectory (Fig. 1B’, see inset). Both DALv3 and DPLam are consistently ablated when applying HU at 32–36h AH (Fig. 1C, C’, F). These results indicate that secondary lineages have fairly invariant birth dates, defined by the time at which the secondary neuroblast enters its larval phase of proliferation. The results further demonstrate that the ablation of subsets of lineages leaves the development of other lineages unaffected, making it possible to identify these lineages based on their location and axonal trajectory. Some lineages, like DALv3, seem to have a more sharply defined birth date, in that HU prior to a certain time point (e.g., 28h AH) leaves the lineage intact in all cases, whereas it always ablates that lineage in the subsequent interval (e.g., 28–32h AH). However, most lineages, like DPLam, show more variability, where HU at one interval ablates a lineage only in a certain fraction of cases; applying HU at the subsequent interval would enhance the fraction, or move it to 100% (see also below).
Figure 1.
Hydroxyurea (HU) ablates neural lineages in a time-dependent manner. Panels A–C’ and G–J show z-projections of confocal sections of late larval brain, labeled with anti-Neurotactin (BP106, magenta). In A–C’, two lineages, DPLam and DALv3, are labeled by GFP driven by engrailed-Gal4 (green). Panels of the upper row (A–C) show a section of the anterior brain cortex; A’–C’ depict a brain section at the level of the mushroom body medial lobe (ML) and the primordium of the central complex (CCXp). In G–J, the lineage pair DPLl2/3 is highlighted (green). 3D digital models in panels D and E illustrate location and tracts of lineages DPLam, DALv3, and DPLl2/3 within a single brain hemisphere (D; anterior view; E: lateral view). The mushroom body (MB) and antennal lobe (AL) are shown for reference. Panel F represents a time line (hours after hatching, AH) where intervals at which HU was administered are shown as black bars. Symbols flanking the time line above and below represent the onset of proliferation (red circles) of the neuroblasts forming lineages DPLam, DALv3, and DPLl2/3, deduced from HU effects shown in this figure. Thus, pulses of HU at 28–32 h AH (B, B’) results in ablation of DALv3; note cluster of en-positive neurons present in brain cortex of control (A), absent in HU-treated animal (arrowhead in B). Likewise, the branched axon tract of DALv3 (A’) is absent in HU-treated animal (arrowhead in B’). By contrast, lineage DPLam is unaffected by HU administration between 28 and 32h. Insets in A’ and B’ show the characteristic, vertically oriented axon tract of DPLam in control (A’) and experimental animal (B’). Application of HU at 32–36h ablates both DALv3 and DPLam secondary neurons (white arrowheads in C’). Note that primary neurons of both lineages, which also express en-Gal4-driven GFP, are not affected by the HU pulse (blue arrowheads in C). DPLl2 and 3 form a pair of neighboring lineages in the dorso-lateral brain cortex (E, G). Each has two hemilineages, one (a) projects along the dorsal brain surface before entering the neuropil, the other (p) invades the neuropil after a short distance. Note presence of all four hemilineage tracts, marked by green asterisks, in inset of panel G. The intermediate transverse superior fascicle (trSI) is shown as reference. HU pulses from 28–32h eliminated one of the DPLl lineages (note a single a and p hemilineage tract in inset of panel H). HU application from 32–36 generally ablated both DPLl lineages (inset of panel I). Later HU pulses (e.g., from 51–55h, as shown in panel J) resulted in truncated lineages, since the neuroblasts were able to generate part of their progeny before being blocked by HU. For abbreviations of compartments and fascicles see Tab. 1. Scale bar: 25µm.
Whereas ablating a neuroblast at the time before it enters its first mitosis should result in the absence of the entire lineage, later HU pulses should give the neuroblast time to start proliferating and produce a certain number of neurons before arresting it, which would result in the formation of small (‘truncated’) lineages. This hypothesis could be confirmed for most lineages and is illustrated in Fig. 1F–J. DPLl2 and DPLl3 form a pair of secondary lineages whose tracts extend close to each other; they are easily recognized because of their bifurcated axon tracts which pass the trSI fascicle at its dorsal and ventral side, respectively (Cardona et al., 2010; Lovick et al., 2013; Fig. 1E, G). HU pulses from 36–40h AH consistently ablated both of these lineages (not shown); pulses from 28–32h or 32–36h ablated one or both lineages (Fig. 1H, I), indicating there is a degree of variability to the time of birth of DPLl2/3 and other lineages (see also below). If HU pulses were applied after 50h AH, truncated versions of DPLl2/3 and most other lineages can be observed at their normal position and with normal axon trajectory (Fig. 1J).
Larval HU pulses do not hinder the development of primary neurons or glial cells during the larval period
Primary neurons and glia are born and differentiate during the embryonic phase. In the late larva, primary neurons can be distinguished from secondary neurons by their large cell bodies located deep in the cortex and by the fact that they form branched neurites in the neuropil. The en-Gal4 driver is expressed in both primary and secondary components of DPLam and DALv3. HU pulses at 32–36h AH ablated secondary neurons, but left primary neurons intact (Fig. 1C). To confirm that larval HU pulses do not prevent the proper projection of primary neurons we used the TH-Gal4 driver line which is expressed in a small number of dopaminergic (D) neurons belonging to seven primary lineages whose projections in the larval brain are known (Blanco et al., 2011; Cruz et al., 2015). Supplementary Figure S1 shows DA clusters DL1 (lineages CP2/3), DL2a (lineages BLVa1/2), and DM1b (lineage DPMl1) in the late larval brain of a control animal (Fig. S1A–C) and a HU treated animal (Fig. S1D–F). Location and number of DA neurons as well as their axonal projection and arborization occurs normally in the HU treated animal. Note, for example, profuse arborization of the DL1 cluster in the anterior compartments (SMP, IPa, LAL) surrounding the lobes of the mushroom body in the control and experiment (Fig. S1A, D). Note also the characteristic trajectory of DL1 axons which form part of the obP fascicle. In the control, these axons are sandwiched between the secondary tracts of CP2/3 and CP1/4 (Fig. S1B, inset); in the experiment (Fig. S1E, inset), the secondary CP2/3 tract is ablated (arrowhead), but primary DL1 axons appear at their normal position dorsal of CP1/4, whose secondary neurons are born after 32h AH and are not affected by the 28–32h HU pulse applied in this experiment.
Neurons of the larval brain are invested by several types of glial cells, including two types of neuropil glia: cortex glia, and surface glia (for review, see Hartenstein, 2012). These cells are born as primary glia in the embryo. Additional, secondary glia are produced by a few select lineages, notably some of the dorsomedial type II lineages (Izegina et al., 2009; Viktorin et al., 2012; Omoto et al., 2015). However, these additional glial cells, recognizable by the specific marker Repo, do not begin to differentiate until late larval stages, thus primary glia are solely responsible for forming a stable scaffold around neurons and proliferating neuroblasts. Similar to primary neurons, these primary glia were not affected by the early larval pulses of HU (data not shown), suggesting that the time-dependent ablation of lineages described in this work is most likely due to a direct effect of HU on neuroblasts as they re-enter mitosis.
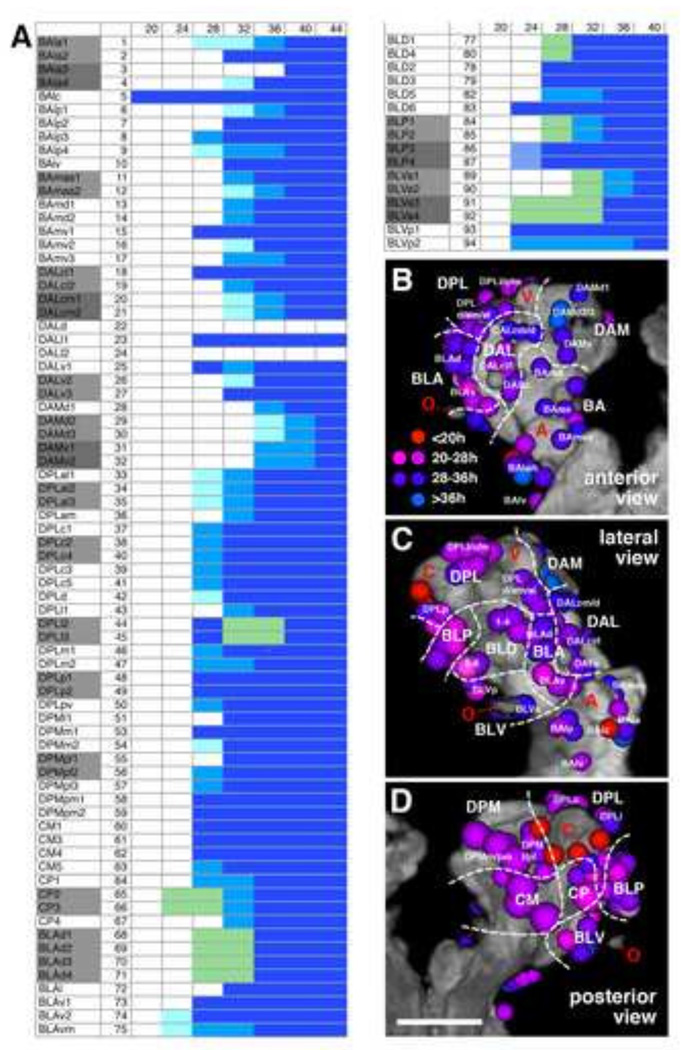
Calendar of birthdates of secondary lineages
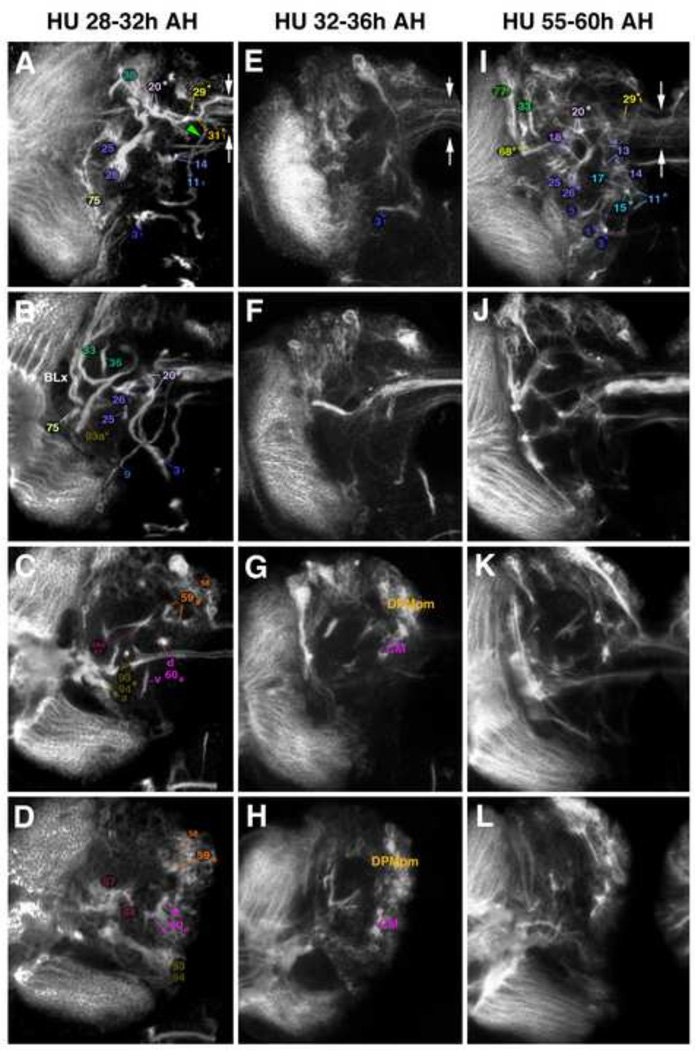
Following treatments with HU at defined intervals, brains dissected at the late larval stage and labeled with anti-neurotactin (BP106) to visualize secondary axon tracts were assayed for the presence or absence of specific lineages. Given their characteristic shape and position (see Fig. 1G–J), tracts remaining in HU treated animals could be assigned to specific lineages in most cases (Fig. 2, 3). Taking the earliest time interval at which application of HU ablates a lineage as a rough birth date of that lineage we established a temporal chart of birth dates for all secondary lineages (Fig. 4A). As explained above, most lineages show a certain degree of variability. The variability could in part be artifactual, reflecting merely that the level of HU (which depends on the feeding of the larva) reached a critical threshold somewhat later in one case versus another. This idea is supported by the observation that by slightly shifting the interval of HU application (e.g., 33–37h AH vs 32–36h AH) one obtained, for selected lineages, different ratios of ablated vs non-ablated. For example, BAla3, not affected by application at 32–36h, was affected in about half of the cases at 33–37h; likewise, DALcm1/2 and DPLl2/3, ablated in a fraction of cases with HU pulses between 32–36h, were always gone with 33–37h pulses (data not shown).
Figure 2.
Ablation of secondary lineages by timed 4h pulses of hydroxyurea (HU). All panels of this and the following Figure, Fig. 3, show z-projections of contiguous confocal sections of a late third instar larval left brain hemisphere labeled with BP106, representing brain slices of 15–20 µm thickness. Panels of both Figures 2 and 3 are arranged in four rows and three columns. Pairs of vertical arrows indicate the midline. Z-projections of the first row (A, E, I) correspond to an anterior level (mushroom body lobes; MB). Panels of the second row (B, F, J) represent a “subanterior” level (FBpr primordium of fan-shaped body; EBpr primordium of ellipsoid body). The third row (C, G, K) corresponds to the level of the great commissure (GC) and dorsal commissural tracts forming the posterior plexus of the fan-shaped body (FBppl). The fourth row (D, H, L) represents a posterior level (CA calyx of mushroom body). All panels of one column show brain of larva subjected to a specific HU regimen (Figure 2A–D: control; Figure 2E–H: HU pulse at 20–24h after hatching (AH); Figure 2I–L: HU pulse at 24–28h AH; Figure 3A–D: HU at 28–32h AH; Figure 3E–H: HU at 32–36h AH; Figure 3I–L: HU at 55–60h AH). Secondary axon tracts (SATs) of individual lineages are annotated with a unique numerical identifier (for tabulated listing of lineages see Fig. 4A). Numbers followed by an asterisk indicate tracts formed by more than one SAT (typically two SATs) which cannot be followed separately. For example, “20*” stands for “20 and 21”. Lower case letters ‘d’ and ‘v’ indicate dorsal or ventral hemi-/sublineage tracts formed by the CP lineages and CM lineages. Subscripted ‘1’ or ‘2’ indicate cases where the SAT of a closely spaced lineage pair or group of lineages is still identifiable in experimental animals, but is reduced in size. Compare, for example the SAT pair formed by BAmas1/2 (‘11*’ in Fig. 2A, E, I; point where the two tracts split is indicated by red arrowhead in E and I) with the thin tract (’111’; lack of split indicated by green arrowhead) resulting from HU pulse at 28–32h AH (Fig. 3A). For abbreviations of compartments and fascicles see Tab. 1. Scale bar: 50µm.
Other abbreviations: BLx BL lineage group.
Figure 3.
Ablation of secondary lineages by timed 4h pulses of hydroxyurea (HU) (continued). For explanation of panels, see legend to Figure 2.
Figure 4.
(A) Secondary lineage birthdates. All lineages are listed at the left; for lineage abbreviations in this panel and in panels (B–D), see Table 1 and Lovick et al. (2013). The horizontal axis represents the time axis, subdivided into 4h intervals. Numbers at the top indicate hours after hatching. Onset of blue shading indicates time interval at which the corresponding secondary lineage is ablated by HU pulse (dark blue = ablation in >90% of cases; medium blue = ablation in 50%–90%; light blue = ablation in 10–50%). Grey shading in left column points out lineage pairs with highly similar or identical axonal trajectory in larval brain. Green shading is applied in cases where one member of a lineage pair was consistently ablated in a high fraction of cases, whereas the other member was spared. For example, during a 20–24 or 24–28h interval, one lineage of the CP2/CP3 pair is ablated; due to the identical axonal trajectory of CP2 and CP3, it is not possible to determine which of the two was affected. (B–D): Correlation of birth date and location of a lineage. Digital three-dimensional models of larval brain hemispheres, showing position of neuropil entrypoints of lineages (colored spheres) in relationship to neuropil topography (gray). The neuropil surface model was generated by volume-rendering of a series of confocal sections of a brain hemisphere labeled with the synaptic marker nc82 (Brp; see Lovick et al., 2013). Four prominent elements of the neuropil surface are indicated in red lettering (A antennal lobe; C calyx; V tip of vertical lobe; O optic lobe). The three panels represent different view points (B: anterior; C: lateral; D: posterior). White hatched lines demarcate territories occupied by the different lineage groups that are annotated in white lettering (eg. BA, BLA). Coloring of a lineage indicates its birth date, based on time point when it was ablated by a 4h HU pulse. Color key (see panel B): red = birth date before 20h; magenta = birth date 20–28h; violet = birth date 28–36h; blue = birth date 36–44h. Scale bar: 50µm.
With the exception of the four MB lineages and the BAlc/lAL lineage (which reportedly never cease their proliferative activity; Ito and Hotta, 1992), all lineages have a birth date between 20 and 40h AH. Lineages born early or late during this interval are generally intermingled and show no clear topological pattern (Fig. 4B). Possible exceptions are lineages located dorso-medially in the anterior brain, including the five DAM lineages (DAMd1–3, 28–30; DAMv1–2, 31–32) and the medial DAL lineages (DALcm1–2, 20–21; DALd, 22), which are among the latest born lineages (32–36h AH; Figs. 3E, 4B), and the postero-medially located type II lineages (DPMm1, 53; DPMpm1–2, 58–59; CM1–4, 60–62), which form a coherent group born relatively early (24–28h AH; Tab. 1; Figs. 2L, 4B). The very first born lineages (birth date 20–24h AH) are clustered laterally around the optic lobe and comprise one or two representatives each of the four BL groups (BLA: BLAv2-vm, 74–75; BLD: BLD6, 83; BLP: BLP3–4, 86–87; BLV: BLVa3–4, p1–2, 91–94; Figs. 2F–H, 4B). Also one lineage of the CP2/3 pair is consistently affected with HU pulses as early as 20–24h AH [compare thick tract “65d*”, formed by the two lineages CP2/3, in control (Fig. 2C) with thin corresponding tract, “65*1 in Fig. 2G]. Aside from CP2/3, several other lineages form pairs or small groups, whereby the cell body clusters are neighbors in the brain cortex and the axon tracts extend very close to each other, or even merge, so that they cannot be distinguished in the neuropil of the larval brain. These paired/clustered lineages include BAla1/2, BAla3/4, BAlp2/3, BAmas1/2, BAmv1/2, DALcl1/2, DALcm1/2, DALv2/3, DAMd2/3, DAMv1/2, DPLal1–3, DPLc2/4, DPLl2/3, DPLp1/2, DPMpl1/2, CP2/3, BLAd1–4, BLP1/2, BLP3/4, BLVa1/2, and BLVa3/4. It is noteworthy that, almost without exception, individual members of these pairs/clusters have different birth dates. For example, following HU treatment from 20–24h AH, one out of the two CP2/3 lineages or BLP3/4 lineages was ablated (Fig. 4A). HU pulses at 24–28h AH consistently ablated two out of the four BLAd1–4 lineages (68–71; Figs. 2I, 4A). Pulses from 28–32h ablated one lineage of the BAmas1/2 and BAla3/4 pair, and two of the DPLal1–3 triplet (11–12, 3–4, and 33–35, respectively; Figs. 3A–B; 4A).
Table 1.
List of abbreviations of neuropil fascicles and larval neuropil compartments (left) and adult neuropil compartments (center, right).
| Fascicles | Abbr. | Compartments (adult) |
Abbr. | Compartments (adult) cont’d |
Abbr. |
|---|---|---|---|---|---|
| Commissure of the lateral accessory lobe | LALC | Antennal lobe | AL | Posteriorlateral protocerebrum | PLP |
| Dorsolateral root of the fan-shaped body | dlrFB | Antenno-mechanosensory | AMMC | Anterior periesophageal neuropil | PENPa |
| Dorsomedial root of the fan-shaped body | dmrFB | and motor center | Protocerebral bridge | PB | |
| Great commissure | GC | Anterior optic tubercle | AOTU | Subesophageal ganglion | SEG |
| Intermediate superior transverse fascicle | trSI | Bulb | BU | Superior intermediate | SIP |
| Longitudinal superior medial fascicle | loSM | Ellipsoid body | EB | protocerebrum | |
| Anterior loSM | loSMa | Fan-shaped body | FB | Superior lateral protocerebrum | SLP |
| Posterior loSM | loSMp | Inferior protocerebrum | IP | Anterior SLP | SLPa |
| Medial antennal lobe tract | mALT | Anterior IP | IPa | Posterior SLP | SLPp |
| Medial equatorial fascicle | MEF | Lateral IP | IPl | Superior medial protocerebrum | SMP |
| Oblique posterior fascicle | obP | Medial IP | IPm | Inferior ventrolateral cerebrum | VLCi |
| Posterior lateral fascicle | PLF | Posterior IP | IPp | Ventrolateral protocerebrum | VLP |
| Subellipsoid body commissure | SuEC | Lateral accessory lobe | LAL | Anterior VLP | VLPa |
| Lateral horn | LH | Posterior VLP | VLPp | ||
| Compartments (larval) | Abbr. | Mushroom body | MB | Ventromedial cerebrum | VMC |
| Central complex primordium | CCXp | Calyx | CA | Anterior VMC | VMCa |
| Ellipsoid body primordium | EBpr | Medial lobe | ML | Infracommissural VMC | VMCi |
| Fan-shaped body primordium | FBpr | Pedunculus | PED | Postcommissural VMC | VMCpo |
| Spur | SPU | Precommissural VMC | VMCpr | ||
| Vertical lobe | VL | Supracommissural VMC | VMCs | ||
| Noduli | NO | ||||
Differences in the birth dates of secondary lineages also do not seem to reflect gross differences in projection pattern. According to the recent mapping of the projection of secondary lineages in the adult brain (Ito et al., 2013; Wong et al., 2013; Yu et al., 2013) one can clearly recognize lineages with local projections (one or two neighboring compartments) from other lineages with far flung projections (e.g., the antennal lobe lineages connecting the ventral deuterocerebrum with the dorsal protocerebrum or the lineages with long commissural axons connecting the ventrolateral protocerebrum of both hemispheres). Members of both classes, small local and large projection, are found among the early-born or the late-born lineages. For example, the DAMd2/3 pair which forms widespread connections between the superior medial protocerebrum and the posterior ventromedial cerebrum is born around the same time as the DAMv1/2 pair which develops only local projections in the superior medial protocerebrum (Wong et al., 2013). Born during the earliest interval (20–24h AH), the CP2/3 pair forms large projections between posterior dorso-lateral compartments (lateral horn, superior lateral protocerebrum) and anterior-medial compartments (mushroom body lobes, fan-shaped body; Wong et al., 2013). The BLP3/4 pair, born as early as CP2/3, has restricted arborizations in the lateral horn.
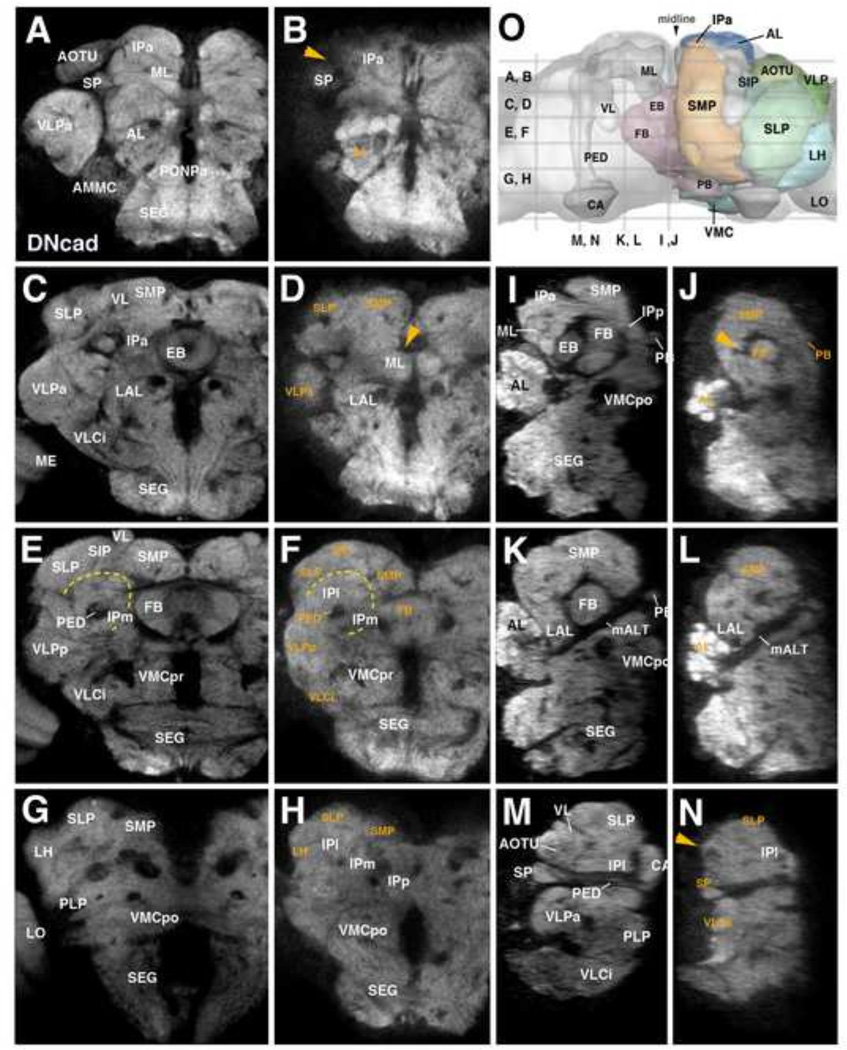
Ablation of secondary lineages causes strong effects on adult neuropil organization
Given the size of primary versus secondary lineages (Bello et al., 2008; Larsen et al., 2009), the secondary neurons account for 80 – 90% of the neurons of the adult central brain. Hence it stands to reason that the volume of the neuropil compartments, formed by neuronal arborizations and synapses, also depends largely on the secondary neurons, and would be decreased if secondary neurons were absent. The analysis of adult brains of animals treated with HU during the larval stage confirms this notion: the loss of neuropil volume is correlated with the time of HU application, and time points between 32–36h and 36–40h AH, causing the virtual absence of secondary lineages in the larva (see above), lead to the strongest effects in the adult brain (Fig. 5). Despite this, many animals underwent metamorphosis and were able to eclose (approximately 86% 32–36h and 30% 36–40h HU treated animals were sub-viable and were dissected out of the pupal case (data not shown)). Eclosing adults were essentially immobile, exhibiting little or no spontaneous movement or reflex action (J.L., unpublished observation).
Figure 5.
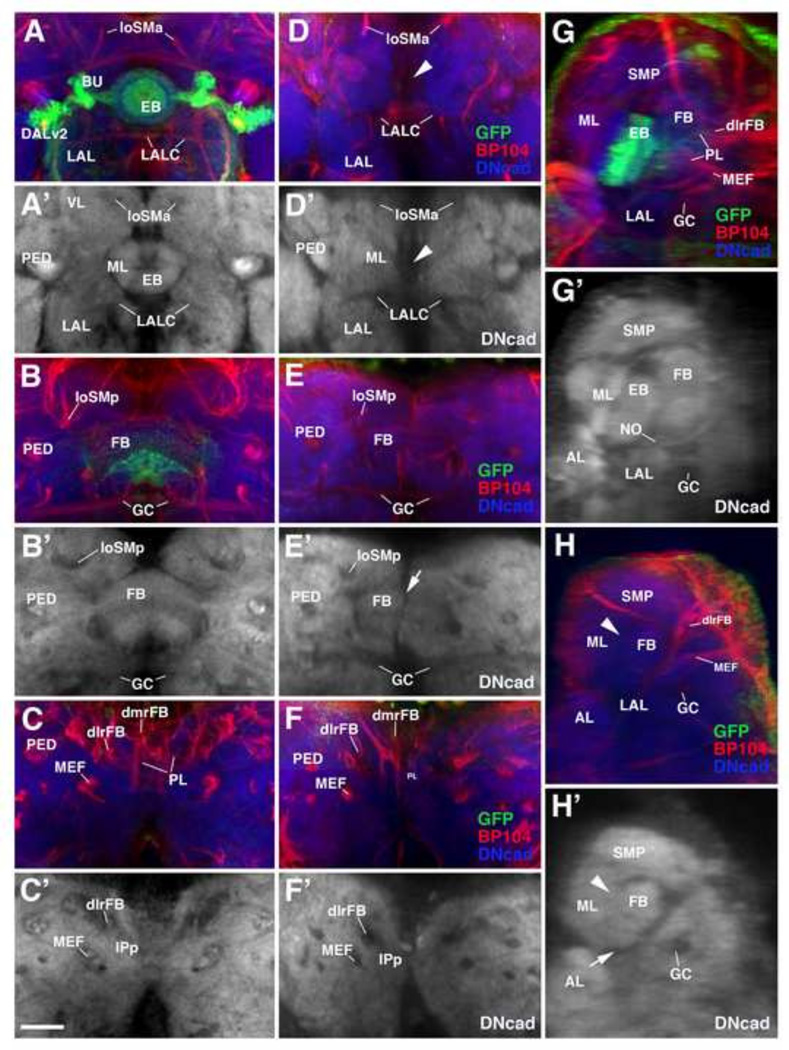
Effects of HU-mediated ablation of secondary lineages on adult brain neuropils. Z-projections of frontal (A–H) and sagittal (I–N) confocal sections of brains of control (left column: A, C, E, G; third column: I, K, M) and HU treated animals (second column: B, D, F, H; right column: J, L, N). HU was applied during 32–36h interval after hatching. Brain neuropil is labeled with anti-DNcad. Compartments are annotated in white lettering; orange lettering highlights compartments most strongly affected by HU treatment. Level of sections are indicated in panel O, which shows central brain neuropil in a dorsal view. Compartments of mushroom body and central complex are outlined and annotated for left hemisphere; right hemisphere shows outlines and names of other compartments that are visible in dorsal view. Orange arrowheads throughout the Figure point at locations where compartments were completely ablated by HU pulse (AOTU in panels (B) and (N), EB in panels (D) and (J)). Dashed yellow line in panels (E) and (F) outlines the inferior protcerebrum (IP), a region largely unaffected by HU treatment. For abbreviations of compartments and fascicles see Tab. 1. Scale bar: 50µm.
Other abbreviations: LO lobula of the optic lobe.
Compartments affected most strongly following neuroblast ablation are those known to be formed mostly of secondary neurons (e.g. the mushroom body, whose α/β and α’/β’ neurons are all born post-embryonically) and compartments that are newly formed during metamorphosis and therefore are likely comprised preferentially of secondary neurons, including the central complex (ellipsoid body/EB, fan-shaped body/FB, noduli; arrowhead in Fig. 5D, J) and anterior optic tubercle (AOTU; arrowhead in Fig. 5B, N). Aside from the AOTU, other compartments closely associated with the input from the optic lobe, whose neurons differentiate during metamorphosis, are also strongly affected by larval HU treatment; these compartments include the ventrolateral protocerebrum (VLPa, VLPp; Fig. 5D, F, N) and the lateral horn (LH; Fig. 5H). Least affected are compartments whose volume normally does not increase significantly during metamorphosis, including the inferior protocerebrum (IPa, IPl, IPm, IPp; Fig. 5B, F, H, N) and ventromedial cerebrum (VMCpo, VMCpr; Fig. 5F, H; Pereanu et al., 2010).
HU-induced defects of the central complex, illustrated in more detail in Fig. 6, are most severe. The central complex, and particularly the ellipsoid body, is formed by a small number of lineages. DALv2 includes large field (ring) neurons of the ellipsoid body (EB) and the dorsal Type II lineages DPm1/DM1, DPMpm1/DM2, DPMpm2/DM3, and CM4/DM4 form the small field (columnar) neurons connecting the EB with the fan-shaped body (FB) and protocerebral bridge (Wong et al., 2013; Yang et al., 2013). The ablation of DALv2, labeled by per-Gal4 (Fig. 6A, arrowhead in D, G, arrowhead in H), is always associated with the elimination of the EB as defined by synaptic markers such as DNcad or Nc82 (Fig. 6A’, arrowhead in D’; G’, arrowhead in H’). The same holds true for the noduli, formed by the columnar neurons of the dorsal Type II lineages, which are not detectable in adult brains where these lineages were ablated (not shown). By contrast, a strongly reduced FB is always present in HU treated animals (Fig. 6E, E’, H, H’). Interestingly, the FB in experimental animals is commonly split in the midline (arrowhead in Fig. 6E’). This might be due to the fact that (among all of the Type II lineages) the crossed axons of DPMm1/DM1 and DPMpm1/DM2, both of which innervate the contralateral half of the FB, are missing; as a result, the cohesion between the left and right half of the FB could be compromised.
Figure 6.
Defects of the central complex following HU-mediated ablation of secondary lineages. Z-projections of frontal (A–F’) and sagittal (G–H’) confocal sections of brains of control (left column: A–C’; right column: G, G’) and HU-treated animals (middle column: D–F’; right column: H, H’). Rows represent corresponding planes of sections along the antero-posterior axis or medio-lateral axis (A/A’, D/D’: ellipsoid body; B/B’, E/E’: fan-shaped body; C/C’, F/F’: posterior roots of the central complex; G/G’, H/H’: 5µm lateral of midline). HU was applied during 28–32h interval after hatching. Axon fascicles formed by secondary axon tracts are globally labeled by anti-Neuroglian (BP104; red in A–H). Brain neuropil is labeled with anti-DNcad (blue in A–H; white in A’-H’). The DALv2 lineage, forming wide field (R-) neurons of the ellipsoid body and fan-shaped body is labeled by GFP driven by per-Gal4 (green in A, D, B, E, C, F, G, H). Ablation of secondary lineages, including DALv2, by the HU pulse leads to complete elimination of the ellipsoid body (EB in A/A’, G/G’; arrowhead in D/D’, H/H’). The fan-shaped body and the posterior roots of the central complex (FB in B/B’, E/E’, G/G’, H/H’; dlrFB and dmrFB in C/C’, F/F’, G, H) are strongly reduced. Note vertical cleft in midline of reduced fan-shaped body in experimental animals (arrow in E’). For abbreviations of compartments and fascicles see Tab. 1. Scale bar: 25µm.
Other abbreviations: PL plexus of the fan-shaped body.
Effects of the ablation of secondary lineages on neuronal branching morphogenesis
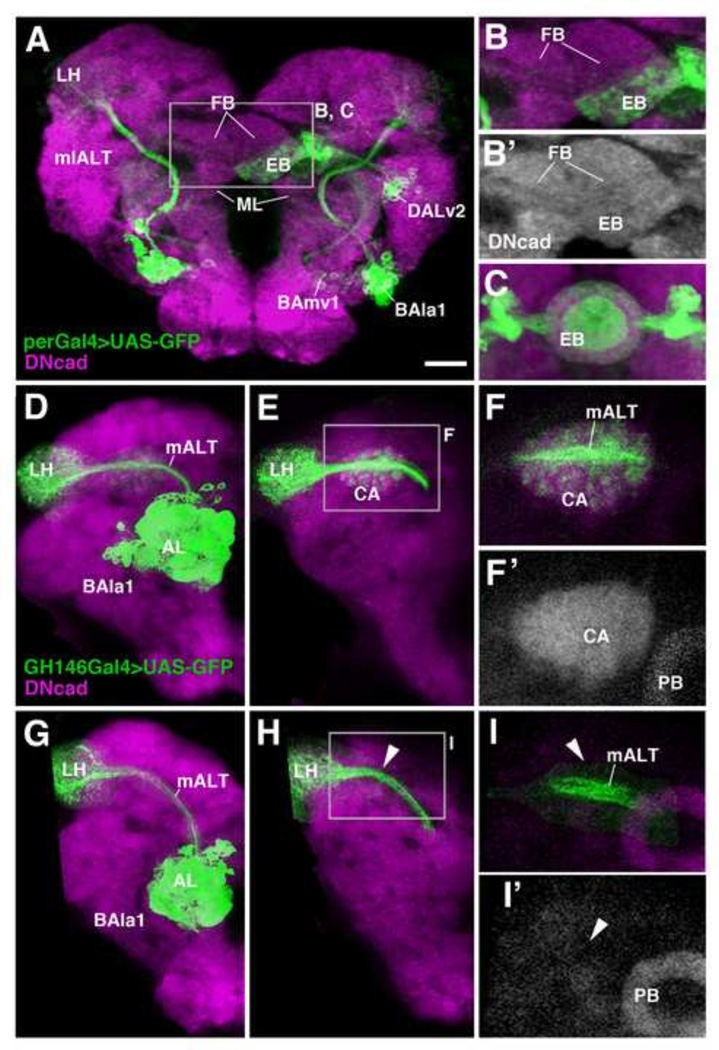
HU pulses between 24 and 32h AH frequently result in brain asymmetries, ablating a given lineage on one side, but not the other. This effect can be explained in light of the more or less pronounced variability in the birth dates of lineages: whereas the neuroblast of a lineage might start to divide at 24h in the left hemisphere, its counterpart in the opposite hemisphere might be delayed by a few hours. An example is shown in Fig. 7A–C, where DALv2 is ablated on the left, but not the right. This unilateral ablation resulted in a rudimentary, misshapen “hemi-ellipsoid body” which is closely attached to the ventral surface of the (rudimentary) right fan-shaped body (Fig. 7B, B’). Furthermore, in this and the three other cases of unilateral DALv2 ablation we were able to observe, the terminal arborizations of the non-ablated DALv2 showed a pattern that significantly deviated from the normal pattern. Thus, terminal fibers formed regularly spaced aggregates, separated by signal-free gaps, as opposed to a normal ellipsoid body (created by the overlap of neuronal arborizations created by neurons on both sides) where the DALv2 terminal arbors fill a smooth and continuous, ring-shaped volume (Fig. 7C). We conclude that interactions between DALv2 axons and their contralateral counterparts determine the pattern and spacing of terminal branches of this lineage.
Figure 7.
HU-mediated ablation of secondary lineages reveals neuronal interactions. (A, B, B’): Unilateral ablation of the DALv2 lineage, labeled by per-Gal4 (green), in left brain hemisphere. All panels show z-projections of frontal confocal sections at level of central complex (EB ellipsoid body; FB fan-shaped body). Neuropil is labeled by anti-DNcad (magenta or white) Note asymmetric axonal arborization of remaining right hemispheric DALv2 (EB in A, B/B’), and coarse-grained texture of terminal fibers (B) compared to control (C). (D–I) Terminal arborization of ventral antennal lobe projection neurons of lineage BAla1 (labeled by GH146-Gal4) in the mushroom body calyx (CA) and lateral horn (LH) of control animal (D–F’) and HU-treated animal (G–I’; pulse between 0–4h after hatching). (D) and (G) show z-projections of confocal sections including both anterior levels (AL antennal lobe), which contain BAla1 cell bodies and dendrites, and posterior levels (LH lateral horn; CA calyx) where BAla1 axons terminate. (E–F’) and (H–I’) focus on the posterior level for better resolution. Note dense, evenly-spaced terminal arbors of BAla1 in both lateral horn and calyx of control (E–F’). In HU-treated animal (G–I’), the calyx is ablated (arrowhead in I’), and terminal branches of BAla1 towards this structure are absent (arrowheads in H, I). By contrast, the projection to the lateral horn (LH) is undisturbed (H). For abbreviations of compartments and fascicles see Tab. 1. Scale bar: 25µm.
A significant branching defect of one secondary lineage in reaction to the lack of another lineage could also be observed for the antennal projection lineages BAmv3/adNB and BAlc/lNB, both of which are labeled by GH146-Gal4 (Das et al., 2013; Lai et al., 2008; Stocker et al., 1997). BAmv3 and BAlc project along the mALT towards the calyx (CA) and lateral horn (LH; Fig. 7D–F). As the mALT passes along the anterior surface of the calyx, BAlc/BAmv3 axons send short branches into the calyx (Fig. 7F). Early HU treatment ablates the secondary MB lineages, resulting in the strong reduction or absence of the CA (Fig. 7I, I’). In these animals, no side branches emerge from the BAlc/BAmv3 axons (Fig. 7I), indicating that signals specific to the CA are required to induce branching off the main BAlc/BAmv3 axons.
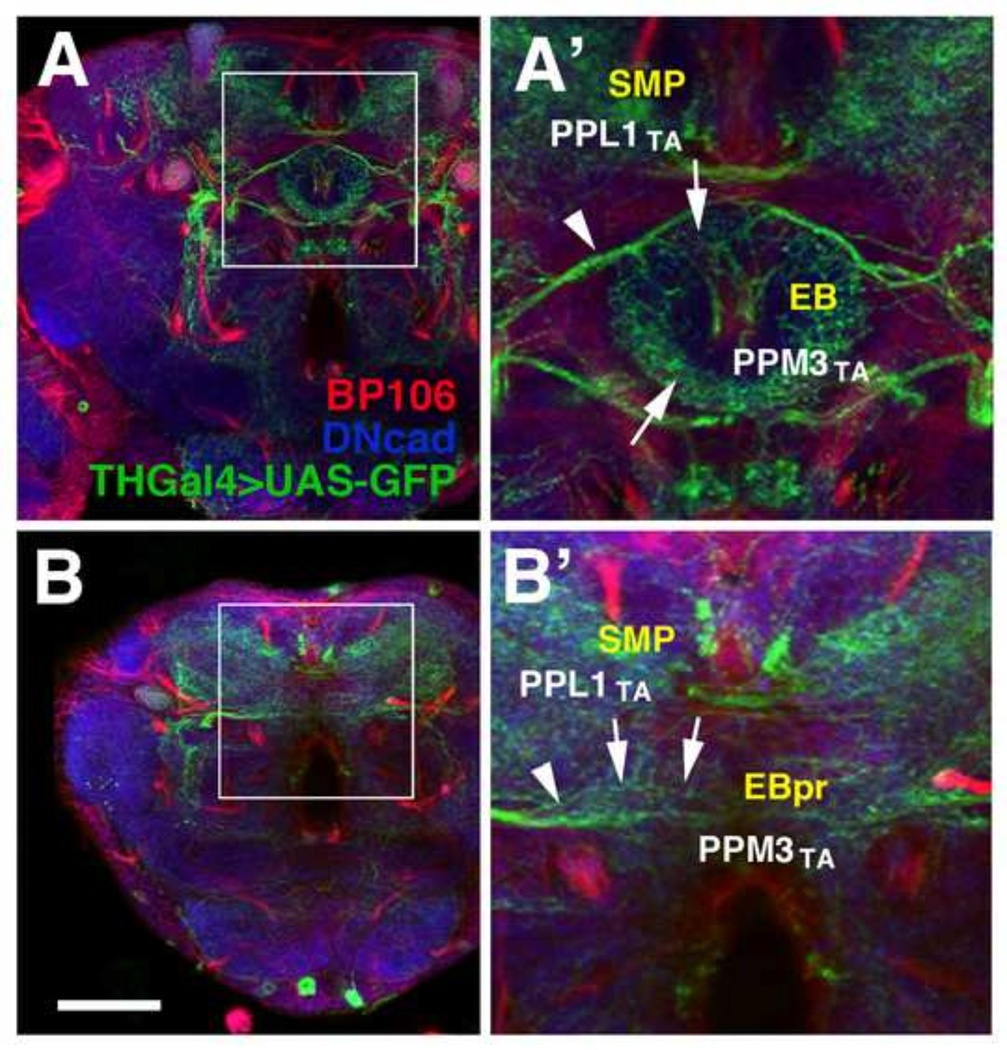
To test the effect of widespread ablation of secondary lineages on the differentiation of primary neurons we analyzed the structure of the TH-Gal4-positive neurons in adult brains of animals treated with HU between 32 and 36h AH. Previous studies had shown that primary neurons prune back their neurite tree at the onset of metamorphosis (Blanco et al., 2011; Consoulas et al., 2000; Marin et al., 2005; Roy et al., 2007; Weeks, 1999). Subsequently, a new neurite tree is reassembled that often resembles the primary tree, but also can show new, adult-specific features. As shown in Fig. 8 and Supplementary Figure S2, TH-Gal4-positive neurons of HU treated animals appear in their normal pattern (compare Fig. S2F–J with Fig. S2A–E) and exhibit a densely branched neurite tree, as shown in Fig. S2F’/G’ for the superior medial protocerebrum (SMP), the (rudimentary) medial lobe (ML), and the anterior inferior protocerebrum (IPa) surrounding the medial lobe. Terminal arborizations of the PPM3 neurons, which predominantly innervate the central complex, are present, but are abnormally shaped. For example, terminal branches of PPM3 neurons innervating the ellipsoid body (EB) follow the circular shape of this compartment (Fig. 8A, A’; Cruz et al., 2015). In the absence of the secondary DALv2 lineage that scaffolds the EB, PPM3 axons follow the trajectory towards the position where the EB would normally appear (Fig. 8B’). However, terminal branches sprouting from these axons are arranged along a horizontal line, rather than a circle (arrows in Fig. 8B’).
Figure 8.
HU-mediated ablation of secondary lineages does not affect the adult differentiation of dopaminergic primary neurons expressing TH-Gal4. Axon fascicles formed by secondary lineages are globally labeled by anti-Neuroglian (BP104, red), and neuropil is labeled by anti-DNcad (blue). (A, A’): Z-projection of frontal confocal sections of control animal at level of ellipsoid body (EB). (B, B’): Corresponding z-projection of HU-treated animal (HU pulse at 32–36h AH). White arrows indicate terminal arbors and white arrowheads point to long axons. Shown are terminal arborizations of the dopaminergic neuronal groups PPL1 (PPL1TA), which innervates the superior medial protocerebrum (SMP) and PPM3 (PPM3TA), which normally follows the ring shape of the ellipsoid body (EB). In HU treated animal, the EB is rudimentary (“EBpr” in panel B’) and PPM3 projections follow a straight course across the midline. Scale bar: 50µm.
Discussion
Hydroxyurea-mediated ablation of neural lineages
In this study we used hydroxyurea (HU) to ablate secondary neural lineages, following the previously published regimen of HU application that was established to kill mushroom body neuroblasts (de Belle and Heisenberg, 1994; Prokop and Technau, 1994). HU blocks DNA synthesis by inhibiting ribonucleotide reductase and causes cell death in S-phase. Since HU does not affect gene transcription or protein synthesis and therefore specifically targets dividing cells, it is used as an anti-neoplastic drug in a number of different cancers. As a means to block cell division and/or ablate specific lineages, HU has been applied in several previous studies in both vertebrates (e.g., Xenopus: Harris and Hartenstein, 1992) and invertebrates (e.g., Becker et al., 1998; Broadie and Bate, 1991; Malun, 1998; Pfister et al., 2007). In Drosophila, a short pulse of HU given right after hatching ablates the four mushroom body lineages (as well as one other lineage of antennal lobe projection neurons (BAlc/lNB)), which allows for the study of different aspects of mushroom body function in Drosophila larvae and adults (Sweeney et al., 2012). The reason why HU specifically targets neuroblast lineages, rather than all dividing cell populations in the larva, lies in the peculiar, stem cell-like mode of neuroblast mitosis (generation of neuronal progeny through a series of asymmetric divisions). On the other hand, adult progenitors in other tissues, such as the imaginal discs, the intestine, or the musculature, divide symmetrically and asynchronously or parasynchronously. During any given 4 hour time interval, some of these progenitors are in the S-phase of mitosis (and thereby sensitive to HU), but most will not be, and will continue to proliferate. Since adult progenitors show a great deal of regulative capacity (e.g. the imaginal leg disc: Kiehle and Schubiger, 1985; imaginal wing disc: Milán et al., 1997; Neufeld et al., 1998; Wartlick et al., 2011), organ size will be affected little or not at all following a HU pulse. By contrast, a neural lineage results from a single, asymmetrically dividing neuroblast, and will be missing in its entirety once the neuroblast is ablated.
Applying BrdU, Ito and Hotta (1992) had documented the time course of appearance of dividing neuroblasts. Their data showed a roughly linear increase in the number of BrdU positive clusters at the brain surface from 20 to 50h AH, a finding that is matched by our data presented in this study. Since all neuroblasts re-enter mitosis within a relatively short time period of about 20h it is not possible to delete individual lineages, using 4h pulses. Based on our data (Fig. 4), a HU pulse administered from 20–24h AH resulted in the ablation of an average of six lineages (in addition to the MB lineages and BAlc). A pulse from 24–28h added another 33 lineages to that number, indicating that most secondary lineages are born during this time window. An average of 22 lineages appear in the 28–32h interval, 19 in the 32–36h interval, and only 2 after 36h (Fig. 4). For one lineage, DALl2, we could not establish a birth date. DALl2 has a short tract that only touches the surface of the VLP compartment before terminating, therefore not providing a distinctive enough pattern that would allow us to determine whether this lineage is present in an experimental brain where many surrounding tracts are missing.
As expected from earlier works on the four MB lineages and BAlc/lNB, secondary lineages in general have a fairly invariant birth date. For example, among the lineages ablated by the 20–24h HU pulse are always BLD6, BLVp1, one of the CP2/3 pair, and one of the BLP3/4 pair. Lineages affected last (36–40h) include the DAMd2/3 and the DAMv1/2 pairs. The birth dates established by the HU application, for these and all other secondary lineages, correlate well with the labeling of developing lineages using insc-Gal4 (Lovick et al., 2015b). The HU data further suggest that the time of birth is fixed more precisely for some lineages than for others. Thus, lineages such as BAmv1 or DALcl1 were never affected with 20–24h (or earlier) HU pulses, and were always absent following 24–28h (or later) pulses (Fig. 4), suggesting that the birth date of these lineages invariably falls into the 24–28h interval. In contrast, most lineages were affected with increasing severity over a longer period. For example, lineages DPLal1/2 were infrequently ablated with 24–28h HU pulses; they were absent most of the time following 28–32h pulses, and always gone with pulses 32–36h or later (Fig. 4). This finding points at a certain variability in the timing of reactivation of the DPLal neuroblasts. We can only speculate why the variability in birth date seems higher in some versus other lineages. It is possible that certain neuroblasts are more sensitive to the HU: if applied at a given interval, HU would reach a concentration that, due to variability in feeding or other factors, falls in a range (x1–x2). x1 is sufficient to arrest neuroblast A, whereas another neuroblast, B, requires level x2. In that case, HU application at that time interval will always ablate lineage A, but variably B. To address this question, more detailed studies, focusing on a few, selected lineages, would be required.
Mechanisms involved in the asynchronous reactivation of neuroblasts
Our findings demonstrate that neurons of central brain secondary lineages arise asynchronously during early larval development. Secondary neuroblasts sequentially reactivate (exit quiescence) over a period of approximately twenty hours extending from 20–40h post-hatching. With the exception of five pairs of neuroblasts, which continuously divide (BAlc/lNB and the four MB neuroblasts), central brain neuroblasts resume proliferation beginning with those located most laterally, near to the optic lobe (BLAv2/vm, BLD6, BLP3/4, BLVa3/4, BLVp1/2). This observation matches previous reports in the ventral nerve cord that neuroblasts located most laterally exit quiescence first (Chell and Brand, 2010; Ito and Hotta, 1992; Truman and Bate, 1988).
The fact that neuroblasts reactivate at different time points, rather than simultaneously, is a curious phenomenon, given that extrinsic factors seem to be primarily responsible to drive neuroblast re-entry into the cell cycle. Recent findings showed that amino acids, which activate the Tor pathway in the fat body, and insulin from cortex glia which activates PI3K/Akt signaling within the neuroblasts themselves (Chell and Brand, 2010; Sousa-Nunes et al., 2011), are responsible for neuroblast reactivation. Additional extrinsic factors have also been implicated in regulating exit from quiescence. These include the cell adhesion molecule E-Cadherin expressed by glia/neuroblast lineages (Dumstrei et al., 2003), which promotes neuroblast proliferation, the glia-secreted glycoprotein Anachronism (Ana; Ebens et al., 1993), which prevents cell-cycle re-entry after quiescence and the ECM molecule Terribly Reduced Optic Lobes (Trol; Voigt et al., 2002), which counteracts Ana and enhances exit from quiescence, possibly through positive regulation of FGF and Hedgehog signaling pathways in neuroblasts (Barrett et al., 2008; Park et al., 2003). Given that all neuroblasts should have equal access to any of these hemolymph- and glia-derived stimuli, the question arises why they react to them in different ways, some neuroblasts re-entering mitosis earlier than others.
There are a number of explanations for this phenomenon. First, there might be subtle quantitative differences in the density of glial cells or glia-neuroblast contacts which could account for the fact that signals passed between glia and neuroblasts have a different strength at different locations. This possibility is not likely, given that (with the exception of the DAM/DALcm lineages which, as a cohesive group, reactivate relatively late) in most locations, early and late re-activating neuroblasts are neighbors. Another explanation would be to postulate the existence of intrinsic factors within the neuroblast that modulate the effect of how neuroblasts at different locations react to extrinsic re-entry stimuli. Along this line, it has been shown that precocious VNC neuroblast re-activation through overexpression of PI3K within neuroblasts does not alter the order in which the neuroblasts begin dividing (lateral to medial; Chell and Brand, 2010) or the duration of time in which they divide, suggesting that neuroblasts have a cell intrinsic timer which dictates the timing and duration of neuroblast proliferation. The importance and molecular nature of intrinsically timed factors for controlling cell fate and neuroblast proliferation has been amply documented. A stereotyped sequence of transcription factors, Hb, Kr, Pdm, Cas, and Grh is expressed in many embryonic neuroblasts and acts to control the fate of neurons born at the respective time points when these molecules are present (Brody and Odenwald, 2005; Pearson and Doe, 2004). In many lineages the sequence of transcription factors resumes in the larva. For example, the VNC neuroblast Nb 3-3 ends on Cas in the embryo and becomes quiescent; subsequently, it re-activates in the early larva with the expression of Cas, and then switches to Svp (Tsuji et al., 2008). Abdominal Nb3-3 neuroblasts, as a result of expression of the Hox gene AbdA, do not become quiescent; they switch from Cas to Svp already in the late embryo. These results indicate that neuroblasts have an intrinsic timer (responsible for switching between different transcription factors) that is “remembered” throughout the phase of quiescence. This timer could be responsible to modulate a neuroblast’s response to the “wake-up” stimuli acting on it from the outside during the larval phase.
We would like to point out that one aspect of the mitosis re-entry pattern of neuroblasts could be most easily explained by assuming local cell-cell interactions. This is the finding that in many of the cases where lineages are neighbors and have similar or identical trajectory (e.g., DALcl1/2, CP2/3, BLVa3/4), one member of the pair/small group is born earlier than the other member(s). For example, 24–28h HU pulses consistently ablated one of the DALcl1/2 pair (DALcl1) and two of the BLAd1–4 group. This slight difference in birth date among members of a lineage pair is corroborated by an independent analysis where the early larval development of secondary lineages was imaged directly (Lovick et al., 2015b). It is possible that in these cases, cell-cell interactions between neighboring neuroblasts of a lineage pair plays a role in tuning the exact time point when they re-enter mitosis.
Ablation of secondary lineages reveals extrinsic mechanisms controlling neuronal differentiation
The targeted ablation of specific brain compartments, or populations of neurons, has been an important means to study questions of neuronal development, function, and plasticity. Typically, these ablation studies relied on surgically disrupting cohesive fiber bundles, such as sensory nerves or central brain tracts, and then assaying the effects on other populations of neurons that formed pre- or postsynaptic connections with the ablated fibers. Among many other results, these experiments demonstrated the high degree of plasticity in the nervous system of both vertebrates and invertebrates. For example, dendrites, deprived of one of their normal inputs, reacted by reshaping their branching pattern whereby they gained access to other inputs (Mizrahi and Libersat, 2001; Murphey and Chiba, 1990). Due to its small size, Drosophila has not been used frequently as a model to address neural development by surgical means. However, “genetic ablation” can substitute for surgery, and a number of important insights were gained from genetically removing certain cell types, or tissues, and study the effect on the remaining cells of the nervous system. For example, genetically ablating all sensory neurons in embryos did not prevent the structural and functional differentiation of basic motor circuits underlying peristaltic behavior (Suster and Bate, 2002), indicating that the presence of sensory afferents (or activity) is not required for the proper wiring of motor neurons and many interneurons.
The present study allows several conclusions in the context of neuronal interactions taking place during brain metamorphosis. First, targeting and arborization of antennal projection neurons depends on extrinsic signals from the target tissues. Two lineages, BAmv3 and BAlc, form projections towards the lateral horn and give off collateral branches towards the calyx. In the absence of the secondary MB lineages, where the calyx is reduced by 90%, BAmv3 does not emit any axon collaterals towards the region where the calyx would normally reside, implying that this projection depends on target-derived signals. There is abundant evidence for extrinsic, target-derived signals triggering or directing axonal growth during the embryonic stage, where cells at the midline of the nervous system control the pattern of commissural axons, emitting repulsive (e.g., Slit, Ephrins) or attractive signals (e.g., Netrins). These signaling pathways show a high degree of conservation between invertebrate models (Drosophila, C. elegans) and vertebrates (Judas et al., 2003; Killeen and Sybingco, 2008). Well studied cases of axon-target interactions at later stages of brain development are rare, in both vertebrate systems and Drosophila. An example from mammalian brain that bears a certain degree of similarity to the antenno-calycal projection discussed here is the cerebro-spinal tract, which forms collaterals to the pontine nuclei in the brain stem. This collateral projection depends on signals from the pons; removal of the pons results in failure of collaterals to form, and ectopic pontine neurons evoke supernumerary collaterals (O’Leary et al., 1991). The molecular nature of the signaling mechanism underlying the cortico-pontine collateral attraction has not yet been elucidated. The interaction between calyx and antenno-calycal afferents described in this study might present an opportunity to screen for elements of the underlying molecular mechanisms controlling axonal collateral growth.
Partial ablation of secondary lineages also revealed the role of extrinsic mechanisms shaping the branching pattern of primary neurons. A small set of primary dopaminergic neurons, the PPM3 neurons, that form part of the CM4 lineage, establish widespread arborizations in the fan-shaped body and the ellipsoid body (Cruz et al., in prep). Rudimentary PPM3 projections invading the midline neuropil at the position where the central complex normally develops are still recognizable following HU pulses between 28 and 36h AH, which ablated secondary lineages that form the major bulk of the volume of the central complex compartments. However, the highly ordered, ring-shaped or layered trajectory of PPM3 terminal branches is absent in the HU treated brains (Fig. 8), indicating that interactions with secondary neurons of the central complex is crucial in shaping the neurite tree of primary neurons.
In conclusion, the temporally controlled HU-mediated ablation of secondary neuroblasts and their lineages provides a set of data that is important for developmental studies of the Drosophila brain. Knowing the exact birth date of a lineage is one of the essential prerequisites when planning to specifically label or genetically manipulate that lineage in a spatiotemporally restricted manner (e.g. Gal4/Gal80ts or other binary repression systems). Our findings will also stimulate further research into the genetic mechanism controlling neuroblast proliferation and quiescence, as well as axonal pathfinding and branching.
Supplementary Material
Highlights.
The fly brain is formed by lineages consisting of primary and secondary neurons.
Using pulses of hydroxyurea during the larval phase, we established birth dates for all secondary lineages.
We show that the tracts of secondary lineages develop independently of each other.
The final branching pattern of secondary neurons appears to be dependent upon presence of appropriate neuronal targets.
Acknowledgements
We thank the members of the Hartenstein laboratory for critical discussions during the preparation of this manuscript. We thank K.T. Ngo, J.J. Omoto, and T. Shenoy for their assistance in several hydroxyurea ablation experiments. We are grateful to the Bloomington Stock Center and R.F. Stocker for fly strains and the Developmental Studies Hybridoma Bank for antibodies. This work was supported by NIH grant (R01 NS29357-15). J.K. Lovick was supported by the USPHS National Research Service Award GM07104.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ashburner M. A laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. Drosophila; pp. 214–217. [Google Scholar]
- Barrett AL, Krueger S, Datta S. Branchless and Hedgehog operate in a positive feedback loop to regulate the initiation of neuroblast division in the Drosophila larval brain. Dev Biol. 2008;317:234–245. doi: 10.1016/j.ydbio.2008.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker T, Bothe G, Harley AR, Macagno ER. Cell proliferation in a peripheral target is required for the induction of central neurogenesis in the leech. J Neurobiol. 1998;34:295–303. doi: 10.1002/(sici)1097-4695(199803)34:4<295::aid-neu1>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- Bello BC, Izergina N, Caussinus E, Reichert H. Amplification of neural stem cell proliferation by intermediate progenitor cells in Drosophila brain development. Neural Dev. 2008;3:5. doi: 10.1186/1749-8104-3-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieber AJ, Snow PM, Hortsch M, Patel NH, Jacobs JR, Traquina ZR, Schilling J, Goodman CS. Drosophila neuroglian: a member of the immunoglobulin superfamily with extensive homology to the vertebrate neural adhesion molecule L1. Cell. 1989;59:447–460. doi: 10.1016/0092-8674(89)90029-9. [DOI] [PubMed] [Google Scholar]
- Blanco J, Pandey R, Wasser M, Udolph G. Orthodenticle is necessary for survival of a cluster of clonally related dopaminergic neurons in the Drosophila larval and adult brain. Neural Dev. 2011;6:34. doi: 10.1186/1749-8104-6-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broadie KS, Bate M. The development of adult muscles in Drosophila: ablation of identified muscle precursor cells. Development. 1991;113:103–118. doi: 10.1242/dev.113.1.103. [DOI] [PubMed] [Google Scholar]
- Brody T, Odenwald WF. Regulation of temporal identities during Drosophila neuroblast lineage development. Curr Opin Cell Biol. 2005;17:672–675. doi: 10.1016/j.ceb.2005.09.013. [DOI] [PubMed] [Google Scholar]
- Cardona A, Saalfeld S, Arganda I, Pereanu W, Schindelin J, Hartenstein V. Identifying neuronal lineages of Drosophila by sequence analysis of their axon tracts. J Neurosci. 2010;30:7538–7553. doi: 10.1523/JNEUROSCI.0186-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chell JM, Brand AH. Nutrition-responsive glia control exit of neural stem cells from quiescence. Cell. 2010;143:1161–1173. doi: 10.1016/j.cell.2010.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consoulas C, Duch C, Bayline RJ, Levine RB. Behavioral transformations during metamorphosis: remodeling of neural and motor systems. Brain Res Bull. 2000;53:571–583. doi: 10.1016/s0361-9230(00)00391-9. [DOI] [PubMed] [Google Scholar]
- Cruz L, Lovick J, Guo M, Hartenstein V. Lineage-based analysis of the dopaminergic system of the Drosophila brain. 2014 (manuscript in preparation) [Google Scholar]
- Das A, Gupta T, Davla S, Prieto-Godino LL, Diegelmann S, Reddy OV, Raghavan KV, Reichert H, Lovick J, Hartenstein V. Neuroblast lineage-specific origin of the neurons of the Drosophila larval olfactory system. Dev Biol. 2013;373:322–337. doi: 10.1016/j.ydbio.2012.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Belle JS, Heisenberg M. Associative odor learning in Drosophila abolished by chemical ablation of mushroom bodies. Science. 1994;263:692–695. doi: 10.1126/science.8303280. 1994. [DOI] [PubMed] [Google Scholar]
- de la Escalera S, Bockamp EO, Moya F, Piovant M, Jiménez F. Characterization and gene cloning of neurotactin, a Drosophila transmembrane protein related to cholinesterases. EMBO J. 1990;9:3593–3601. doi: 10.1002/j.1460-2075.1990.tb07570.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumstrei K, Wang F, Hartenstein V. Role of DE-cadherin in neuroblast proliferation, neural morphogenesis, and axon tract formation in Drosophila larval brain development. J. Neurosci. 2003;23:3325–3335. doi: 10.1523/JNEUROSCI.23-08-03325.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebens AJ, Garren H, Cheyette BN, Zipursky SL. The Drosophila anachronism locus: a glycoprotein secreted by glia inhibits neuroblast proliferation. Cell. 1993;74:15–27. doi: 10.1016/0092-8674(93)90291-w. [DOI] [PubMed] [Google Scholar]
- Friggi-Grelin F, Coulom H, Meller M, Gomez D, Hirsh J, Birman S. Targeted gene expression in Drosophila dopaminergic cells using regulatory sequences from tyrosine hydroxylase. J Neurobiol. 2003;54:618–627. doi: 10.1002/neu.10185. [DOI] [PubMed] [Google Scholar]
- Furst A, Mahowald AP. Cell division cycle of cultured neural precursor cells from Drosophila. Dev Biol. 1985;112:467–476. doi: 10.1016/0012-1606(85)90419-1. [DOI] [PubMed] [Google Scholar]
- Garbe DS, Bashaw GJ. Independent functions of Slit-Robo repulsion and Netrin-Frazzled attraction regulate axon crossing at the midline in Drosophila. J Neurosci. 2007;27:3584–3592. doi: 10.1523/JNEUROSCI.0301-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris WA, Hartenstein V. Neuronal determination without cell division in Xenopus embryos. Neuron. 1991;6:499–515. doi: 10.1016/0896-6273(91)90053-3. [DOI] [PubMed] [Google Scholar]
- Hortsch M, Patel NH, Bieber AJ, Traquina ZR, Goodman CS. Drosophila neurotactin, a surface glycoprotein with homology to serine esterases is dynamically expressed during embryogenesis. Development. 1990;110:1327–1340. doi: 10.1242/dev.110.4.1327. [DOI] [PubMed] [Google Scholar]
- Ito K, Hotta Y. Proliferation pattern of postembryonic neuroblasts in the brain of Drosophila melanogaster. Dev Biol. 1992;149:134–148. doi: 10.1016/0012-1606(92)90270-q. [DOI] [PubMed] [Google Scholar]
- Ito K, Awano W, Suzuki K, Hiromi Y, Yamamoto D. The Drosophila mushroom body is a quadruple structure of clonal units each of which contains a virtually identical set of neurones and glial cells. Development. 1997;124:761–771. doi: 10.1242/dev.124.4.761. [DOI] [PubMed] [Google Scholar]
- Ito M, Masuda N, Shinomiya K, Endo K, Ito K. Systematic analysis of neural projections reveals clonal composition of the Drosophila brain. Curr Biol. 2013;23:644–655. doi: 10.1016/j.cub.2013.03.015. [DOI] [PubMed] [Google Scholar]
- Iwai Y, Usui T, Hirano S, Steward R, Masatoshi T, Uemura T. Axon patterning requires D N-cadherin, a novel neuronal adhesion receptor, in the Drosophila embryonic CNS. Neuron. 1997;19:77–89. doi: 10.1016/s0896-6273(00)80349-9. [DOI] [PubMed] [Google Scholar]
- Judas M, Milosević NJ, Rasin MR, Heffer-Lauc M, Kostović I. Complex patterns and simple architects: molecular guidance cues for developing axonal pathways in the telencephalon. Prog Mol Subcell Biol. 2003;32:1–32. doi: 10.1007/978-3-642-55557-2_1. [DOI] [PubMed] [Google Scholar]
- Kaneko M, Hall JC. Neuroanatomy of cells expressing clock genes in Drosophila: transgenic manipulation of the period and timeless genes to mark the perikarya of circadian pacemaker neurons and their projections. J Comp Neurol. 2000;422:66–94. doi: 10.1002/(sici)1096-9861(20000619)422:1<66::aid-cne5>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- Kiehle CP, Schubiger G. Cell proliferation changes during pattern regulation in imaginal leg discs of Drosophila melanogaster. Dev Biol. 1985;109:336–346. doi: 10.1016/0012-1606(85)90460-9. [DOI] [PubMed] [Google Scholar]
- Killeen MT, Sybingco SS. Netrin, Slit and Wnt recepetors allow axons to choose the axis of migration. Dev Biol. 2008;323:143–151. doi: 10.1016/j.ydbio.2008.08.027. [DOI] [PubMed] [Google Scholar]
- Kumar A, Fung S, Lichtneckert R, Reichert H, Hartenstein V. Arborization pattern of engrailed-positive neural lineages reveal neuromere boundaries in the Drosophila brain neuropil. J Comp Neurol. 2009;517:87–104. doi: 10.1002/cne.22112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunz T, Kraft KF, Technau GM, Urbach R. Origin of Drosophila mushroom body neuroblasts and generation of divergent embryonic lineages. Development. 2012;139:2510–2522. doi: 10.1242/dev.077883. [DOI] [PubMed] [Google Scholar]
- Lai SL, Awasaki T, Ito K, Lee T. Clonal analysis of Drosophila antennal lobe neurons: diverse neuronal architectures in the lateral neuroblast lineage. Development. 2008;135:2883–2893. doi: 10.1242/dev.024380. [DOI] [PubMed] [Google Scholar]
- Larsen C, Shy D, Spindler S, Fung S, Younossi-Hartenstein A, Hartenstein V. Patterns of growth, axonal extension and axonal arborization of neuronal lineages in the developing Drosophila brain. Dev Biol. 2009;335:289–304. doi: 10.1016/j.ydbio.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T, Lee A, Luo L. Development of the Drosophila mushroom bodies: sequential generation of three distinct types of neurons from a neuroblast. Development. 1999;126:4065–4076. doi: 10.1242/dev.126.18.4065. [DOI] [PubMed] [Google Scholar]
- Lovick JK, Ngo KT, Omoto JJ, Wong DC, Nguyen JD, Hartenstein V. Postembryonic lineages of the Drosophila brain: I. Development of the lineage-associated fiber tracts. Dev Biol. 2013;384:228–257. doi: 10.1016/j.ydbio.2013.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malun D. Early development of mushroom bodies in the brain of the honeybee Apis mellifera as revealed by BrdU incorporation and ablation experiments. Learn Mem. 1998;5:90–101. [PMC free article] [PubMed] [Google Scholar]
- Marin EC, Watts RJ, Tanaka NK, Ito K, Luo L. Developmentally programmed remodeling of the Drosophila olfactory circuit. Development. 2005;132:725–737. doi: 10.1242/dev.01614. [DOI] [PubMed] [Google Scholar]
- Milán M, Campuzano S, García-Bellido A. Developmental parameters of cell death in the wing disc of Drosophila. Proc Natl Acad Sci U S A. 1997;94:5691–5696. doi: 10.1073/pnas.94.11.5691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizrahi A, Libersat F. Synaptic reorganization induced by selective photoablation of an identified neuron. J Neurosci. 2001;21:9280–9290. doi: 10.1523/JNEUROSCI.21-23-09280.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphey RK, Chiba A. Assembly of the cricket cercal sensory system: genetic and epigenetic control. J Neurobiol. 1990;21:120–137. doi: 10.1002/neu.480210109. [DOI] [PubMed] [Google Scholar]
- Neufeld TP, de la Cruz AF, Johnston LA, Edgar BA. Coordination of growth and cell division in the Drosophila wing. Cell. 1998;93:1183–1993. doi: 10.1016/s0092-8674(00)81462-2. [DOI] [PubMed] [Google Scholar]
- O’Leary DD, Heffner CD, Kutka L, López-Mascaraque L, Missias A, Reinoso BS. A target-derived chemoattractant controls the development of the corticopontine projection by a novel mechanism of axon targeting. Development. 1991;2:123–130. [PubMed] [Google Scholar]
- Park Y, Rangel C, Reynolds MM, Caldwell MC, Johns M, Nayak M, Welsh CJ, McDermott S, Datta S. Drosophila perlecan modulates FGF and hedgehog signals to activate neural stem cell division. Dev Biol. 2003;253:247–257. doi: 10.1016/s0012-1606(02)00019-2. [DOI] [PubMed] [Google Scholar]
- Patel NH, Snow PM, Goodman CS. Characterization and cloning of fasciclin III: a glycoprotein expressed on a subset of neurons and axon pathways in Drosophila. Cell. 1987;48:975–988. doi: 10.1016/0092-8674(87)90706-9. [DOI] [PubMed] [Google Scholar]
- Pearson BJ, Doe CQ. Specification of temporal identity in the developing nervous system. Annu Rev Cell Dev Biol. 2004;20:619–647. doi: 10.1146/annurev.cellbio.19.111301.115142. [DOI] [PubMed] [Google Scholar]
- Pereanu W, Hartenstein V. Neural lineages of the Drosophila brain: a three-dimensional digital atlas of the pattern of lineage location and projection at the late larval stage. J Neurosci. 2006;26:5534–5553. doi: 10.1523/JNEUROSCI.4708-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereanu W, Kumar A, Jenett A, Reichert H, Hartenstein V. Development-based compartmentalization of the Drosophila central brain. J Comp Neur. 2010;518:2996–3023. doi: 10.1002/cne.22376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfister D, De Mulder K, Philipp I, Kuales G, Hrouda M, Eichberger P, Borgonie G, Hartenstein V, Ladumer P. The exceptional stem cell system of Macrostomum lignano: screening for gene expression and studying cell proliferation by hydroxyurea treatment and irradiation. Front Zool. 2007;9:4–9. doi: 10.1186/1742-9994-4-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prokop A, Technau GM. Normal function of the mushroom body defect gene of Drosophila is required for the regulation of the number and proliferation of neuroblasts. Dev Biol. 1994;161:321–337. doi: 10.1006/dbio.1994.1034. [DOI] [PubMed] [Google Scholar]
- Python F, Stocker RF. Adult-like complexity of the larval antennal lobe of D. melanogaster despite markedly low numbers of odorant receptor neurons. J Comp Neurol. 2002;445:374–387. doi: 10.1002/cne.10188. [DOI] [PubMed] [Google Scholar]
- Ramaekers A, Magnenat E, Marin EC, Gendre N, Jefferis GS, Luo L, Stocker RF. Glomerular maps without cellular redundancy at successive levels of the Drosophila larval olfactory circuit. Curr Biol. 2005;15:982–992. doi: 10.1016/j.cub.2005.04.032. [DOI] [PubMed] [Google Scholar]
- Roy B, Singh AP, Shetty C, Chaudhary V, North A, Landgraf M, Vijayraghavan K, Rodrigues V. Metamorphosis of an identified serotonergic neuron in the Drosophila olfactory system. Neural Dev. 2007;24:2–20. doi: 10.1186/1749-8104-2-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skeath JB, Thor S. Genetic control of Drosophila nerve cord development. Curr Opin Neurobiol. 2003;13:8–15. doi: 10.1016/s0959-4388(03)00007-2. [DOI] [PubMed] [Google Scholar]
- Snow PM, Patel NH, Harrelson AL, Goodman CS. Neural-specific carbohydrate moiety shared by many surface glycoproteins in Drosophila and grasshopper embryos. J. Neurosci. 1987;7:4137–4144. doi: 10.1523/JNEUROSCI.07-12-04137.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa-Nunes R, Yee LL, Gould AP. Fat cells reactivate quiescent neuroblasts via TOR and glial insulin relays in Drosophila. Nature. 2011;471:508–512. doi: 10.1038/nature09867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stocker RF, Heimbeck G, Gendre N, de Belle JS. Neuroblast ablation in Drosophila P[GAL4] lines reveals origins of olfactory interneurons. J Neurobiol. 1997;32:443–456. doi: 10.1002/(sici)1097-4695(199705)32:5<443::aid-neu1>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- Suster ML, Bate M. Embryonic assembly of a central pattern generator without sensory input. Nature. 2002;416:174–178. doi: 10.1038/416174a. [DOI] [PubMed] [Google Scholar]
- Sweeney ST, Hidalgo A, de Belle JS, Keshishian H. Hydroxyurea ablation of mushroom bodies in Drosophila. Cold Spring Harb Protoc. 2012;2012:231–234. doi: 10.1101/pdb.prot067777. [DOI] [PubMed] [Google Scholar]
- Tabata T, Schwartz C, Gustavson E, Ali Z, Kornberg TB. Creating a Drosophila wing de novo, the role of engrailed, and the compartment border hypothesis. 1995;121:3359–3369. doi: 10.1242/dev.121.10.3359. [DOI] [PubMed] [Google Scholar]
- Timson J. Hydroxyurea. Mutat Res. 1975;32:115–132. doi: 10.1016/0165-1110(75)90002-0. [DOI] [PubMed] [Google Scholar]
- Truman JW, Bate M. Spatial and temporal patterns of neurogenesis in the central nervous system of Drosophila melanogaster. Dev Biol. 1988;125:145–157. doi: 10.1016/0012-1606(88)90067-x. [DOI] [PubMed] [Google Scholar]
- Tsuji T, Hasegawa E, Isshiki T. Neuroblast entry into quiescence is regulated intrinsically by the combined action of spatial Hox proteins and temporal identity factors. Development. 2008;135:3859–3869. doi: 10.1242/dev.025189. [DOI] [PubMed] [Google Scholar]
- Urbach R, Technau GM. Molecular markers for identified neuroblasts in the developing brain of Drosophila. Development. 2003a;130:3621–3637. doi: 10.1242/dev.00533. [DOI] [PubMed] [Google Scholar]
- Urbach R, Technau GM. Early steps in building the insect brain: neuroblast formation and segmental patterning in the developing brain of different insect species. Arthropod Struct Dev. 2003b;32:103–123. doi: 10.1016/S1467-8039(03)00042-2. [DOI] [PubMed] [Google Scholar]
- Voigt A, Pflanz R, Schäfer U, Jäckle H. Perlecan participates in proliferation activation of quiescent Drosophila neuroblasts. Dev Dyn. 2002;224:403–412. doi: 10.1002/dvdy.10120. [DOI] [PubMed] [Google Scholar]
- Wagh DA, Rasse TM, Asan E, Hofbauer A, Schwenkert I, Dürrbeck H, Buchner S, Dabauvalle MC, Schmidt M, Qin G, Wichmann C, Kittel R, Sigrist SJ, Buchner E. Bruchpilot, a protein with homology to ELKS/CAST, is required for structural integrity and function of synaptic active zones in Drosophila. Neuron. 2006;49:833–844. doi: 10.1016/j.neuron.2006.02.008. [DOI] [PubMed] [Google Scholar]
- Wartlick O, Mumcu P, Kicheva A, Bittig T, Seum C, Jülicher F, González-Gaitán M. Dynamics of Dpp signaling and proliferation control. Science. 2011;331:1154–1159. doi: 10.1126/science.1200037. [DOI] [PubMed] [Google Scholar]
- Weeks JC. Steroid hormones, dendritic remodeling and neuronal death: insights from instect metamorphosis. Brain Behav Evol. 1999;54:51–60. doi: 10.1159/000006611. [DOI] [PubMed] [Google Scholar]
- White K, Kankel DR. Patterns of cell division and cell movement in the formation of the imaginal nervous system in Drosophila melanogaster. Dev Biol. 1978;65:296–321. doi: 10.1016/0012-1606(78)90029-5. [DOI] [PubMed] [Google Scholar]
- Wong DC, Lovick JK, Ngo KT, Borisuthirattana W, Omoto JJ, Hartenstein V. Postembryonic lineages of the Drosophila brain: II. Identification of lineage projection patterns based on MARCM clones. Dev Biol. 2013;384:258–289. doi: 10.1016/j.ydbio.2013.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JS, Awasaki T, Yu HH, He Y, Ding P, Kao JC, Lee T. Diverse neuronal lineages make stereotyped contributions to the Drosophila locomotor control center, the central complex. J Comp Neurol. 2013;521:2645–2662. doi: 10.1002/cne.23339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Younossi-Hartenstein A, Nassif C, Green P, Hartenstein V. Early neurogenesis of the Drosophila brain. J Comp Neurol. 1996;370:313–329. doi: 10.1002/(SICI)1096-9861(19960701)370:3<313::AID-CNE3>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- Yu HH, Awasaki T, Schroeder MD, Long F, Yang JS, He Y, Ding P, Kao JC, Wu GY, Peng H, Myers G, Lee T. Clonal development and organization of the adult Drosophila central brain. Curr Biol. 2013;23:633–643. doi: 10.1016/j.cub.2013.02.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Matsumoto T, Mikami S, Nagasawa T, Murakami F. SDF1/CXCR4 signalling regulates two distinct processes of precerebellar neuronal migration and its depletion leads to abnormal pontine nuclei formation. Development. 2009;136:1919–1928. doi: 10.1242/dev.032276. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.