Abstract
Rapidly growing mycobacteria are rarely found in central nervous system infections. We describe a case of polymicrobial infection in a brain abscess including two rapidly growing Mycobacterium species, M. immunogenum and M. llatzerense. The Mycobacterium isolates were distinguishable by molecular methods, and whole-genome sequencing showed <60% pairwise nucleotide identity.
CASE REPORT
A 40-year-old woman with a past medical history notable for migraines and frequent childhood sinopulmonary infections presented with 7 days of unremitting right frontal headache and 2 days of night sweats, chills, vomiting, left-sided vision loss, and right leg numbness. Unlike her prior migraines, this headache was more intense and not relieved by rest in a dark room; it was also associated with visual impairments and sensation abnormalities. Throughout childhood, the patient had suffered from recurrent upper and lower respiratory tract infections, for which she had received frequent antibiotics, chest percussion, and sinus irrigation using a neti pot with unfiltered, unsterilized tap water. As an adult, the frequency of her respiratory infections decreased to approximately annually, with her last one being 4 months prior to admission. The patient was born and raised in rural Pennsylvania and had regularly consumed unpasteurized milk and drunk well water. In the preceding months, the patient had swum in a river in northern California and traveled to France, where she had consumed various soft cheeses.
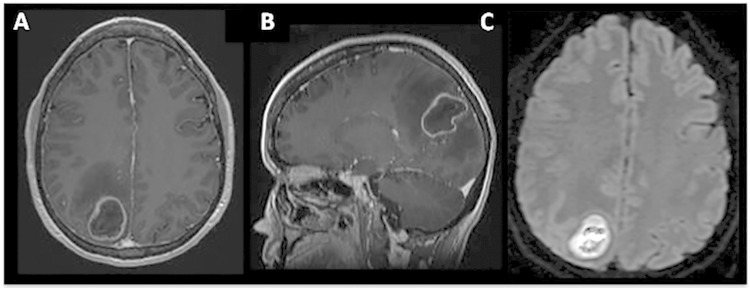
Upon presentation, the patient was moderately distressed but alert and oriented to person, place, and time. She had a temperature of 37°C, a heart rate of 51 beats per minute, a blood pressure of 122/66 mm Hg, a respiratory rate of 12 breaths per minute, and an oxygen saturation of 100% on room air. Her physical exam was remarkable for a left lower homonymous quandrantanopia and reduced sensation to light touch on her right lower leg. She had symmetric facial musculature, a clear oropharynx, no paranasal or frontal sinus tenderness, and no neck stiffness. Her lungs were clear to auscultation bilaterally, cardiac exam demonstrated sinus rhythm with a soft systolic murmur, musculoskeletal strength and reflexes were within normal limits, and there were no skin lesions. A complete blood count was notable for a white blood cell count of 11.3 × 109/liter (90% neutrophils), hemoglobin at 11.2 g/dl, and a platelet count of 287 × 109/liter. The C-reactive protein level was 7.4 mg/liter, and an erythrocyte sedimentation rate was 30 mm/h. T1-weighted magnetic resonance imaging (MRI) of the brain demonstrated a 3.7-cm by 2.8-cm by 3.0-cm rim-enhancing lesion in the right parietal/occipital lobe, with restricted diffusion upon diffusion-weighted MRI (Fig. 1). The patient underwent emergent craniotomy and drainage of pressurized purulent fluid contained within the cavity.
FIG 1.
(A and B) A T1-weighted MRI demonstrates a 3.0-cm by 3.7-cm by 2.8-cm rim-enhancing lesion within the right parietal lobe. (C) A diffusion-weighted MRI demonstrates a bright signal within the abscess, indicating restricted diffusion.
Gram staining demonstrated Gram-positive cocci and Gram-positive rods, and the patient was started on empirical antibiotic therapy with vancomycin, metronidazole, and ceftriaxone. No acid-fast bacilli or fungal organisms were visible on direct stains. Bacterial cultures were positive for Streptococcus anginosus group and Peptostreptococcus. A transthoracic echocardiogram was negative for valvular disease or a patent foramen ovale. Serum HIV antibody (Ab) and antigen (Ag), cryptococcal Ag, Toxoplasma IgG and IgM Ab, and Coccidioides IgG and IgM Ab assays were all negative. Blood and urine cultures were negative, and the results of two QuantiFERON-TB Gold assays were indeterminate. As all identified bacteria were susceptible to beta-lactam antibiotics, the patient's antimicrobial regimen was narrowed to ceftriaxone (2 g once daily). The patient was continued on ceftriaxone therapy, with a stable visual field defect, resolved headache and sensory changes, and a collapsed abscess cavity on imaging at 5 weeks postprocedure.
On postoperative day 11, the growth of a few colonies was noted on the Middlebrook 7H11 agar inoculated with abscess material, and on postoperative day 13, the BacT/Alert MP mycobacterial broth culture bottle turned positive. Kinyoun stains from both culture medium isolates were positive for acid-fast bacilli, and within 3 days subcultures showed visible colonies with identical morphologies on Lowenstein-Jensen (LJ) and 7H11 media. Mycobacterial DNA probes performed on the isolates were negative for Mycobacterium tuberculosis. Sequencing of the 7H11 isolate rpoB gene at the National Jewish health laboratory in Denver, CO, showed identification as Mycobacterium llatzerense, but in-house 16S rRNA gene sequencing analysis from the broth culture isolate was consistent with Mycobacterium immunogenum. To resolve this discrepancy, repeat isolation was performed from the two media, with partial sequencing of the 16S rRNA gene using specific V1-V3 PCR primers (1). This showed 100% identity to Mycobacterium immunogenum DSM 45595T with the broth medium isolate. The 7H11 isolate 16S rRNA PCR product had 100% identity to a number of uncultured bacterial sequences from treated drinking water biofilms present in GenBank (2) as well as to a previously described clinical M. llatzerense isolate (3). It also showed identity in 420/421 bp (99.8%) to Mycobacterium llatzerense MG13T and in 437/439 bp (99.5%) to Mycobacterium aubagnense U8T. Because mycobacterial 16S rRNA sequences from different species can have very high sequence similarity, additional analysis was needed to confirm species identification (4).
As no reference genomes were available for either of these species and to ensure correct identification of the putative M. llatzerense isolate, we performed shotgun next-generation sequencing (NGS) to assemble and recover draft genome sequences. Genomic DNA was isolated from LJ slants swabbed with subcloned isolates using the Qiagen EZ1 DNA tissue kit. One nanogram of genomic DNA was used as input into the Nextera DNA sample preparation kit (Illumina, San Diego, CA) according to the manufacturer's protocol, except that reagent volumes were cut in half. NGS libraries were sequenced using an Illumina MiSeq 2- by 80-bp sequencer (M. immunogenum) or a HiSeq2500 2- by 250-bp sequencer (M. llatzerense). Raw reads were filtered for a quality score of >30, and adapters were trimmed using cutadapt (5). Paired-end reads were then assembled using SPAdes v3.1.1 at default parameters (6). Genome annotation was performed with Prokka v1.10 (7).
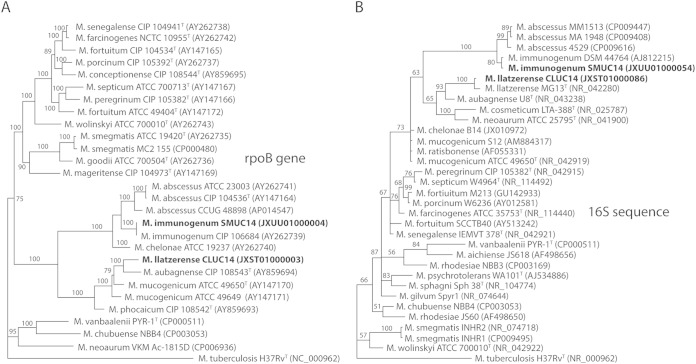
The full-length rpoB and 16S rRNA sequences from the two genomes confirmed the prior identification of the M. immunogenum isolate based on partial rpoB and 16S rRNA sequencing and indicated that the second isolate was indeed M. llatzerense (Fig. 2). Since there were no full-length rpoB sequences available for M. llatzerense, we performed phylogenetic analysis using partial sequences from 4 conserved loci (recA, the 16S to 23S rRNA gene internal transcribed spacer [ITS], hsp65, and rpoB) and found our isolate to cluster with other M. llatzerense strains, confirming its identification (see Fig. S1 in the supplemental material). The minor sequence variations seen in these loci between different M. llatzerense strains are likely due to geographical variation of this species. The Mycobacterium llatzerense genome assembly yielded 6,091,591 bp in 103 contiguous sequences (contigs) for an N50 contig length of 114,066 bp with 5,939 genes and 5,879 coding DNA sequences (CDS) present. The Mycobacterium immunogenum genome assembly yielded 5,566,719 bp in 102 contigs for an N50 contig length of 161,477 bp with 5,562 genes and 5,497 CDS present. Across aligned regions with >5 kb of synteny, the two genomes demonstrated an average nucleotide identity of approximately 60%. An all-by-all blastp search of annotated proteins with matching best hits (two-way method) revealed an average two-way amino acid identity of 65.3% across 2,769 proteins shared between the two new Mycobacterium proteomes (8).
FIG 2.
Phylogenetic trees of Mycobacterium species based on the full-length rpoB gene (A) and 16S rRNA (B) sequences. Isolates sequenced in this study are shown in bold.
The whole genome that matched M. llatzerense most closely in the NCBI database was M. chubuense NBB4 (GenBank accession number CP003053), with an average nucleotide identity of 81.8% and average amino acid identity of 71.6% across a core 2,932 proteins. The whole genome that matched M. immunogenum most closely was Mycobacterium abscessus subsp. bolletii strain 103 (GenBank accession number JAOK01000000), with an average nucleotide identity of 87.9% and average amino acid identity of 91.0% across 3,987 shared proteins. Both strains contained beta-glucosidase but not alpha-glucosidase genes and a katG catalase gene without associated isoniazid resistance changes (9). M. llatzerense contained a blaF beta-lactamase gene, while M. immunogenum had a carbapenem-hydrolyzing Klebsiella pneumoniae carbapenemase (KPC) beta-lactamase precursor protein that is common to the M. abscessus group (10).
Rapidly growing mycobacteria are very rarely reported from neurological infections and brain abscesses (11). A review from 2008 found 19 cases of central nervous system (CNS) infections by these organisms, of which only two cases had predominantly brain abscesses (12). Most patients presented with chronic meningitis, and the majority had a primary source with secondary dissemination to the CNS. Mycobacterial brain abscesses are commonly seen in patients with underlying immunodeficiency syndromes. We considered this possibility in the current case, especially given the patient's history of recurrent childhood respiratory infections. However, a preliminary evaluation for immunodeficiency has thus far been negative, as the patient has normal T lymphocyte levels and activation and normal levels of IgA, IgM, IgE, and IgG subclasses. The patient's subacute course was characterized by headache and visual changes, her history of sinopulmonary infections, and the fact that the coisolated bacterial species are common constituents of both upper airway and oropharyngeal flora, suggesting an initial respiratory source of inoculation. Several rapidly growing mycobacterial species, including M. llatzerense and M. immunogenum, have been isolated from water sources, suggesting potential water exposure to the mycobacterium species ultimately isolated from the abscess (13–15). However, environmental cultures were not available to investigate this possibility.
The majority of rapidly growing mycobacterial infections in the CNS have been attributed to Mycobacterium fortuitum, M. abscessus, and Mycobacterium mucogenicum (12). Besides being associated with the Streptococcus anginosus group and Peptostreptococcus, this case was associated with M. immunogenum and M. llatzerense, two species which have not previously been associated with CNS infections. M. immunogenum was first described in 2001 as a member of the M. abscessus-Mycobacterium chelonae group, with unique susceptibility and genotypic characteristics (15). It has most commonly been associated with cutaneous infections, but respiratory, eye, and disseminated diseases have also been described (16). Mycobacterium llatzerense was first described in 2008 as a contaminant of hemodialysis water and is most closely related to M. aubagnense and M. mucogenicum (17). Subsequent case reports have isolated M. llatzerense from an abdominal abscess and infected lung in the setting of pneumonia (3, 18). It is unlikely that our findings represent contamination, as M. llatzerense has not been previously isolated in our laboratory and no other rapidly growing mycobacteria were isolated in our laboratory during this time frame.
As the isolates were phenotypically indistinguishable in our laboratory, identification of mixed infection with two rapidly growing mycobacterial species required molecular methods. Initial results from rpoB and 16S rRNA gene sequencing of separate isolates from two different media were discordant, leading to further investigation, which revealed that there were in fact two distinct RGM isolated from the brain abscess culture. The different initial growth characteristics seen here may be related to medium formulation and/or temperature requirements. For example, some strains of M. llatzerense are reported to grow only at 22°C and 30°C, while other strains can grow at 37°C (3, 17); in our laboratory, all mycobacterial media are incubated at 36°C. This case underscores the possibility, as shown here, that cultures containing rapidly growing mycobacteria may consist of more than one species, potentially impacting the response to therapy, and suggests one application of whole-genome sequencing in discriminating between mixed bacterial populations in polymicrobial infections. Susceptibility testing of the M. immunogenum isolate showed susceptibility to amikacin (MIC ≤ 8 μg/ml) and clarithromycin (MIC ≤ 0.25 μg/ml) and resistance to cefoxitin (MIC > 128 μg/ml), ciprofloxacin (MIC = 4 μg/ml), doxycycline (MIC > 16 μg/ml), and trimethoprim-sulfamethoxazole (MIC > 4/76 μg/ml), interpreted using CLSI guidelines for rapidly growing mycobacteria (19). Testing of the M. llatzerense isolate was not possible due to the absence of growth in susceptibility medium, which may be related to the media used or the temperature requirements of this isolate.
The clinical significance of detecting rapidly growing mycobacteria in brain abscesses is unclear. The patient has improved without receiving targeted therapy for these mycobacterial species, and the presence of beta-lactamase and carbapenemase genes in the M. llatzerense and M. immunogenum genomes, respectively, suggest that the prescribed ceftriaxone was ineffective in treating these organisms. Standard mycobacterial treatment regimens, based primarily on results with M. abscessus, M. chelonae, and M. fortuitum, often include amikacin, clarithromycin, and imipenem (11). However, abscess drainage and treatment of the coinfecting bacteria may have been sufficient to treat this infection despite the presence of additional mycobacterial organisms (20). Recent studies have also indicated that synergistic infectivity in polymicrobial populations may play an important role in the pathogenesis of brain abscesses, and it is possible that the mycobacterial species may have contributed to brain abscess formation in this patient (20, 21). The increasing availability of molecular methods, such as whole-genome and metagenomic sequencing, to elucidate the microbial composition of brain abscesses is likely to drive the future development of optimal treatment strategies for these complex polymicrobial infections (22, 23).
Nucleotide sequence accession numbers.
Sequences have been deposited in GenBank for M. llatzerense strain CLUC14 (JXST00000000) and M. immunogenum strain SMUC14 (JXUU00000000).
Supplementary Material
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.00402-15.
REFERENCES
- 1.Zhu D, Tanabe S-H, Yang C, Zhang W, Sun J. 2013. Bacterial community composition of South China Sea sediments through pyrosequencing-based analysis of 16S rRNA genes. PLoS One 8:e78501. doi: 10.1371/journal.pone.0078501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Revetta RP, Gomez-Alvarex V, Gerke TL, Curioso C, Santo Domingo JW, Ashbolt NJ. 2013. Establishment and early succession of bacterial communities in monochloramine-treated drinking water biofilms. FEMS Microbiol Ecol 86:404–414. doi: 10.1111/1574-6941.12170. [DOI] [PubMed] [Google Scholar]
- 3.Cárdenas AM, Gomila M, Lalucat J, Edelstein PH. 2014. Abdominal abscess caused by Mycobacterium llatzerense. J Clin Microbiol 52:1287–1289. doi: 10.1128/JCM.03525-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Roth A, Reischl U, Streubel A, Naumann L, Kroppenstedt RM, Habicht M, Fischer M, Mauch H. 2000. Novel diagnostics algorithm for identification of mycobacteria using genus-specific amplification of the 16S-23S rRNA gene spacer and restriction endonucleases. J Clin Microbiol 38:1094–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Martin M. 2011. cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17:10–12. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 6.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 8.Konstantinidis KT, Tiedje JM. 2005. Towards a genome-based taxonomy for prokaryotes. J Bacteriol 187:6258–6264. doi: 10.1128/JB.187.18.6258-6264.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Musser JM, Kapur V, Williams DL, Kreiswirth BN, van Soolingen D, van Embden JD. 1996. Characterization of the catalase-peroxidase gene (katG) and inhA locus in isoniazid-resistant and -susceptible strains of Mycobacterium tuberculosis by automated DNA sequencing: restricted array of mutations associated with drug resistance. J Infect Dis 173:196–202. doi: 10.1093/infdis/173.1.196. [DOI] [PubMed] [Google Scholar]
- 10.Nessar R, Cambau E, Reyrat JM, Murray A, Gicquel B. 2012. Mycobacterium abscessus: a new antibiotic nightmare. J Antimicrob Chemother 67:810–818. doi: 10.1093/jac/dkr578. [DOI] [PubMed] [Google Scholar]
- 11.De Groote MA, Huitt G. 2006. Infections due to rapidly growing mycobacteria. Clin Infect Dis 42:1756–1763. doi: 10.1086/504381. [DOI] [PubMed] [Google Scholar]
- 12.Talati NJ, Rouphael N, Kuppalli K, Franco-Paredes C. 2008. Spectrum of CNS disease caused by rapidly growing mycobacteria. Lancet Infect Dis 8:390–398. doi: 10.1016/S1473-3099(08)70127-0. [DOI] [PubMed] [Google Scholar]
- 13.van Ingen J, Blaak H, de Beer J, de Roda Husman AM, van Soolingen D. 2010. Rapidly growing nontuberculous mycobacteria cultured from home tap and shower water. Appl Environ Microbiol 76:6017–6019. doi: 10.1128/AEM.00843-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dubrou S, Konjek J, Macheras E, Welté B, Guidicelli L, Chignon E, Joyeux M, Gaillard JL, Heym B, Tully T, Sapriel G. 2013. Diversity, community composition, and dynamics of nonpigmented and late-pigmenting rapidly growing mycobacteria in an urban tap water production and distribution system. Appl Environ Microbiol 79:5498–5508. doi: 10.1128/AEM.00900-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wilson RW, Steingrube VA, Böttger EC, Springer B, Brown-Elliott BA, Vincent V, Jost KC, Zhang Y, Garcia MJ, Chiu SH, Onyi GO, Rossmoore H, Nash DR, Wallace RJ. 2001. Mycobacterium immunogenum sp. nov., a novel species related to Mycobacterium abscessus and associated with clinical disease, pseudo-outbreaks and contaminated metalworking fluids: an international cooperative study on mycobacterial taxonomy. Int J Syst Evol Microbiol 51:1751–1764. doi: 10.1099/00207713-51-5-1751. [DOI] [PubMed] [Google Scholar]
- 16.Biggs HM, Chudgar SM, Pfeiffer CD, Rice KR, Zaas AK, Wolfe CR. 2012. Disseminated Mycobacterium immunogenum infection presenting with septic shock and skin lesions in a renal transplant recipient. Transpl Infect Dis 14:415–421. doi: 10.1111/j.1399-3062.2012.00730.x. [DOI] [PubMed] [Google Scholar]
- 17.Gomila M, Ramirez A, Gascó J, Lalucat J. 2008. Mycobacterium llatzerense sp. nov., a facultatively autotrophic, hydrogen-oxidizing bacterium isolated from haemodialysis water. Int J Syst Evol Microbiol 58:2769–2773. doi: 10.1099/ijs.0.65857-0. [DOI] [PubMed] [Google Scholar]
- 18.Teixeira L, Avery RK, Iseman M, Arrossi AV, Harrington S, Stephens K, Winans CG. 2013. Mycobacterium llatzerense lung infection in a liver transplant recipient: case report and review of the literature. Am J Transplant 13:2198–2200. doi: 10.1111/ajt.12318. [DOI] [PubMed] [Google Scholar]
- 19.Clinical and Laboratory Standards Institute (CLSI). 2011. Susceptibility testing of mycobacteria, nocardiae, and other aerobic actinomycetes. Approved standard, 2nd ed CLSI document M24-A2. Clinical and Laboratory Standards Institute, Wayne, PA. [PubMed] [Google Scholar]
- 20.DiGiulio DB, Relman DA. 2009. Majority rules? Tallying the microbial census in an abscess by means of molecular methods. Clin Infect Dis 48:1179–1181. doi: 10.1086/597579. [DOI] [PubMed] [Google Scholar]
- 21.Al Masalma M, Lonjon M, Richet H, Dufour H, Roche P-H, Drancourt M, Raoult D, Fournier P-E. 2012. Metagenomic analysis of brain abscesses identifies specific bacterial associations. Clin Infect Dis 54:202–210. doi: 10.1093/cid/cir797. [DOI] [PubMed] [Google Scholar]
- 22.Mishra AK, Dufour H, Roche P-H, Lonjon M, Raoult D, Fournier P-E. 2014. Molecular revolution in the diagnosis of microbial brain abscesses. Eur J Clin Microbiol Infect Dis 33:2083–2093. doi: 10.1007/s10096-014-2166-z. [DOI] [PubMed] [Google Scholar]
- 23.Kommedal Ø, Wilhelmsen MT, Skrede S, Meisal R, Jakovljev A, Gaustad P, Hermansen NO, Vik-Mo E, Solheim O, Ambur OH, Sæbø Ø, Høstmælingen CT, Helland C. 2014. Massive parallel sequencing provides new perspectives on bacterial brain abscesses. J Clin Microbiol 52:1990–1997. doi: 10.1128/JCM.00346-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.