Abstract
Hepatitis C virus (HCV) exhibits a high genetic diversity and is classified into 6 genotypes, which are further divided into 66 subtypes. Current sequencing strategies require prior knowledge of the HCV genotype and subtype for efficient amplification, making it difficult to sequence samples with a rare or unknown genotype and/or subtype. Here, we describe a subtype-independent full-genome sequencing assay based on a random amplification strategy coupled with next-generation sequencing. HCV genomes from 17 patient samples with both common subtypes (1a, 1b, 2a, 2b, and 3a) and rare subtypes (2c, 2j, 3i, 4a, 4d, 5a, 6a, 6e, and 6j) were successfully sequenced. On average, 3.7 million reads were generated per sample, with 15% showing HCV specificity. The assembled consensus sequences covered 99.3% to 100% of the HCV coding region, and the average coverage was 6,070 reads/position. The accuracy of the generated consensus sequence was estimated to be >99% based on results from in vitro HCV replicon amplification, with the same extrapolated amount of input RNA molecules as that for the patient samples. Taken together, the HCV genomes from 17 patient samples were successfully sequenced, including samples with subtypes that have limited sequence information. This method has the potential to sequence any HCV patient sample, independent of genotype or subtype. It may be especially useful in confounding cases, like those with rare subtypes, intergenotypic recombination, or multiple genotype infections, and may allow greater insight into HCV evolution, its genetic diversity, and drug resistance development.
INTRODUCTION
Hepatitis C virus (HCV) is characterized by a high genetic diversity and is classified into ≥6 genotypes. Genotypes 1 to 6 have been isolated from multiple patients and are further divided into 66 subtypes (1). At the genotype and subtype levels, HCV exhibits nucleotide sequence divergence rates of approximately 30 and 20%, respectively (2). The pronounced genetic diversity of HCV is a result of both an accumulation of mutations due to a high error rate of its viral RNA-dependent RNA polymerase (RdRp) and the long-term association of the virus with the human population (3). Furthermore, intra- and intergenotypic recombinant HCV strains have been described (4–6). Accurate genotyping and subtyping of HCV are important, since current treatment strategies are genotype dependent (7). The standard method for genotyping and subtyping of HCV is the INNO-LiPA, which targets the highly conserved 5′ untranslated region (UTR) and core region of the virus using genotype- and subtype-specific probes. For rare HCV genotypes and subtypes, INNO-LiPA is sometimes unable to fully resolve the subtype, and refinement by population Sanger sequencing may be needed. During HCV drug development, sequencing strategies are essential to provide insight into viral genetic changes in response to antiviral drug pressure. The most common sequencing methods are population Sanger sequencing and deep sequencing of the drug target gene. Both of these methods require amplification of the gene of interest using genotype- or subtype-specific primers prior to sequencing (8). To increase the understanding of genetic variation and potential drug resistance-associated variants (RAVs) outside drug-targeted genes, approaches to sequencing of the whole HCV genome are warranted. Two different kinds of approaches have been described thus far. The first one involves the amplification of overlapping regions using gene-specific primers coupled with either Sanger sequencing or next-generation sequencing (9–13). This method enables efficient sequencing of known genotypes and subtypes. However, the enormous genetic diversity of HCV renders this method less useful for sequencing rare or unknown HCV subtypes and genotypes. In addition, the amplification of the viral population might be skewed by the use of genotype-specific primers due to primer misalignment to particular viral variants. The second method for sequencing the whole HCV genome involves a metagenomic sequencing approach known as RNA sequencing (RNA-Seq) to sequence the total RNA in a sample. Ninomiya et al. (14) demonstrated successful sequencing of HCV from two clinical samples using RNA-Seq technology. However, the human background was high, with only about 0.01% of the 15 to 94 million reads generated per sample being HCV specific. Moreover, the generated HCV genomes were missing parts of 5′ UTR, core, and E2. In a recent study by Malbeouf et al. (15), a novel sequence-independent RNA amplification method, the NuGEN Ovation RNA-Seq system, coupled with next-generation sequencing was used for capturing complete protein-coding regions of HIV, respiratory syncytial virus, and West Nile virus (15). In this study, we evaluated a similar method described by Malbeouf et al. (15) for sequencing of the full-length HCV genome from HCV replicon transcripts as well as 17 patient samples with both common and rare genotypes and subtypes. This method has the potential to allow an analysis of HCV evolution and variability along the entire genome for any genotype and subtype, providing important information on viral variants that might impact clinical and therapeutic outcomes.
MATERIALS AND METHODS
RNA transcripts from HCV plasmids.
RNA transcripts were generated from two HCV genotype 2a plasmids (2a-RlucNeo) containing the nonstructural 5B protein (NS5B) as either the wild type or with the S282T substitution, using the T7 RiboMAX Express large-scale RNA production system (Promega), according to the manufacturer's instructions. RNA transcripts were purified using the RNeasy minikit (Qiagen), according to the manufacturer's instructions. RNA concentration was measured on a NanoDrop spectrophotometer (NanoDrop Technologies), and a mixture of 90% wild-type RNA transcript and 10% S282T RNA transcript was generated. The RNA mixture was prepared as 10-fold serial dilutions to generate six different RNA input molecules per reaction, 108 (500 pg), 107 (50 pg), 106 (5 pg), 105 (500 fg), 104 (50 fg), and 103 (5 fg), and was amplified and sequenced as described below.
Clinical samples.
All patients included in the Gilead phase II/III clinical trials described in this study provided informed consent in writing, and the study protocol conformed to the ethical guidelines of the 1975 Declaration of Helsinki, as reflected in a priori approval by the appropriate institutional review committee.
Genotyping of clinical HCV samples.
Clinical samples in phase II/III studies for sofosbuvir (GS-US-334-0110, GS-US-334-0108, GS-US-334-0107, P7977-1231, and P7977-0523) were genotyped using the Siemens Versant HCV genotype INNO-LiPA 2.0 (Innogenetics, Ghent, Belgium), and the NS5B gene was amplified and sequenced by DDL (DDL Diagnostic Laboratory, Rijswijk, The Netherlands). Viral load was measured using the Cobas AmpliPrep/Cobas TaqMan HCV test, version 2.0 (Roche Molecular Systems). Seventeen patient samples with HCV genotypes 1 to 6 were selected and subjected to full-length HCV genome sequencing.
RNA isolation from clinical samples.
Plasma samples were thawed and centrifuged at 3,600 rpm for 10 min at 4°C to remove cellular debris. HCV RNA was isolated from 200 μl of plasma using the QIAamp MinElute virus spin kit (Qiagen), as per the manufacturer's instructions. Instead of the carrier RNA provided in the kit, 50 μg of linear acrylamide (Life Technologies) was used as a carrier. HCV RNA was eluted with 20 μl of nuclease-free water. The extracted RNA was treated with Turbo DNase (Life Technologies), using the manufacturer's instructions to remove DNA from the sample. Finally, RNA was aliquoted and stored at −80°C until used.
Full-length HCV genome amplification and sequencing.
RNA was reverse transcribed and amplified using the Ovation RNA-Seq V2 system (NuGEN, San Carlos, CA). Briefly, cDNA was generated by reverse transcriptase that extended random hexamers hybridizing across the genome. Double-stranded DNA (dsDNA) was generated and amplified using single-primer isothermal linear amplification (SPIA) (16). The manufacturer's protocol was modified in the following ways: to remove RNA secondary structures present in the HCV genome, cDNA synthesis was performed at 52°C for 20 min. For amplification cleanup, dsDNA was purified using s 1:1.4 volume ratio of dsDNA and AMPure RNAclean beads. The final amplified product was purified using a 1:0.8 volume ratio of product and AMPure XP beads (Beckman Coulter Genomics, Danvers, MA), as described previously (15). The purified products were eluted in 30 μl of Tris-EDTA (TE) buffer and stored at −20°C. The quality and concentration of amplified products were assessed by Bioanalyzer (Agilent Technologies) and NanoDrop (Thermo Scientific) measurements. The amplified products were fragmented using the Covaris system (Woburn, MA), and paired-end indexed libraries were created for each sample using Ovation Ultralow DR multiplex systems (NuGEN), according to the manufacturer's instructions. The indexed libraries were subjected to Illumina MiSeq deep sequencing using the MiSeq reagent 300-cycle kit and bidirectional sequencing of 150 bp. The libraries for the RNA transcripts and the patient samples were sequenced in pools of 24 and 8 samples, respectively. Library preparation, multiplexing, and deep sequencing were performed at Centrillion Biosciences (Palo Alto, CA).
Full-length HCV genome sequencing data analysis.
Contigs were generated de novo for each sample using Vicuna (Broad Institute, Inc.) (17). The generated contigs were aligned to a subtype-specific reference using MOSAIK version 1.1.0017 (18) and merged to generate a draft full-genome assembly. If the aligned contigs disagreed at a particular locus, the most frequent nucleotide was used. If all contigs had a deletion at a locus or if no contigs covered a locus, the corresponding reference base was used. The subtype-specific reference was selected based on the results of INNO-LiPA and/or NS5B genotyping for each sample.
In order to iteratively refine the draft assembly, reads were aligned to this draft assembly, and a consensus sequence was generated based on the alignments. If more than one nucleotide occurred at >15%, the appropriate mixture was used. Regions that lacked coverage were replaced with the corresponding reference region. This was repeated a total of three times to produce the final full-genome assembly.
Raw reads from the FASTQ files were trimmed and filtered based on quality score and length. Trimming was done on reads when the quality score decreased to <15, and reads <50 nucleotides after trimming were removed. The trimmed reads were aligned to the final assembly sequence to generate a final consensus sequence. A cutoff of 10 reads per position was used to generate the consensus sequence. Insertions and deletions (indels) were reported from reads for which the indel(s) did not result in a frameshift and when present in >50% of the viral population. Nucleotide mixtures were reported in the consensus when present at >15% of the viral population. For the noncoding regions, the 5′ UTR and the 3′ UTR, possible indels were manually investigated by studying the alignments of the raw reads for these regions.
To obtain a relative sequence length of the generated consensus sequences, they were compared to the HCV 1a strain H77 reference genome (GenBank accession no. AF009606).
Genetic variability across the HCV genome.
The background variation introduced during amplification and sequencing was estimated from the in vitro-transcribed RNA samples. We defined variation as the percent composition of all but the most prevalent nucleotide or amino acid at each position using the trimmed and filtered reads. The average variation and a 95% confidence interval were calculated for each of the in vitro-transcribed RNA samples.
Intrahost nucleotide and amino acid variation across the HCV genome for the patient samples were evaluated the same way. For both the in vitro-transcribed RNA samples and the patient samples, the percent variation per position was plotted for each sample.
Resistance-associated variants.
The presence of potential resistance-associated variants (RAVs) in NS3, NS5A, and NS5B were evaluated in the produced full-genome sequences from the samples from these 17 patients. The following changes from the genotype 1a reference (H77) were investigated: NS3 position 36any, 43any, 54any, 55any, 80any, 122R, 155any, 156any, 168any, 170A/L/T, and 175L; NS5A positions 24G/N/R, 28A/G/T, 30any, 31any, 32L, 38F, 58D, 92K/T, and 93any; and NS5B positions 96any, 142T, 159any, 282any, 289any, 316any, 320any, 321any, 390I, 414I/T/V, 415Y, 419any, 422K, 423any, 445F, 448any, 452any, 482any, 486any, 494A, 495any, 496S, 499A, 554S, 556G, and 559G.
Determination of taxonomic group of unaligned reads.
To determine the taxonomic group of the unaligned reads and possible presence of unexpected HCV genotypes, analysis was conducted using the NCBI BLAST 2.2.28 (19). A random subset of 50,000 of reads that did not align to HCV from each sample was aligned to the NCBI nucleotide (nt) database. The BLAST output was sorted into taxonomic groups using a script.
Phylogenetic analysis of full-length HCV genome sequences.
The generated full-genome sequences were aligned using Clustal W (20). In addition, to study the genetic similarity of these sequences to previously described full-length sequences, the 5 most genetically similar sequences for each the full-length sequence of each sample were included in the alignment, as determined by nucleotide BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi). The alignment was cut into E1/E2, NS3/4A, NS5A, and NS5B genes. Neighbor-joining phylogenetic trees were inferred in MEGA 5.2 (21) using the Tajima-Nei model and gamma-distributed rates among sites (α = 0.5). The confidence of the branches was assessed by the bootstrap test using 500 replicates. The phylogenetic trees were visualized using FigTree version 1.3.1 (http://tree.bio.ed.ac.uk/).
Nucleotide sequence accession numbers.
All consensus genomes were submitted to the NCBI GenBank database (accession no. KM587614 to KM587630).
RESULTS
Full-genome sequencing of in vitro-transcribed HCV RNA.
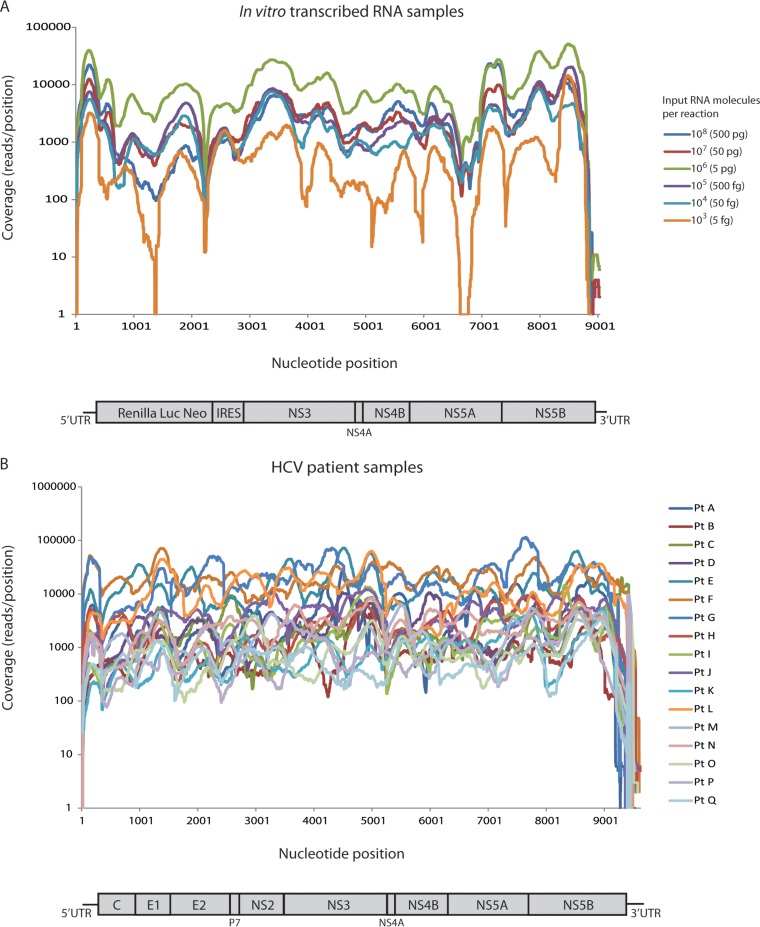
To assess the accuracy and detection of 10% S282T of the full-length HCV genome sequencing assay, amplification and sequencing were performed on in vitro-transcribed RNA samples. RNA transcripts from 2a replicons, wild-type, and the NS5B S282T mutant were used with six different RNA input molecules per reaction: 108 (500 pg), 107 (50 pg), 106 (5 pg), 105 (500 fg), 104 (50 fg), and 103 (5 fg). Each sample was spiked with the 10% S282T variant. In parallel, a no-template control (NTC) sample containing water instead of RNA template was amplified and sequenced. The samples were amplified using the NuGEN Ovation RNA-Seq system, and all amplification reactions generated sufficient final product to make Illumina libraries and subsequent sequencing on the MiSeq platform. On average, 653,935 reads were generated per sample, of which 63.5% of the reads were aligned to the 2aRlucNeo reference sequence (Table 1). The reads were assembled de novo, and a consensus sequence was generated for each sample. A complete consensus sequence of the 2aRlucNeo coding region (8,474 nucleotides) was generated for 5 of 6 samples. In the sample with the lowest number of RNA input molecules per reaction (103), the generated consensus sequence spanned 96% of the coding region. The average coverage for the six samples ranged from 2,091 to 27,462 reads per position (Table 1 and Fig. 1A). Of the >1 million reads generated for the NTC sample, 0.03% were specific to the 2aRlucNeo. These reads originated from cross talk between samples and were most likely due to errors introduced in the sample-specific barcodes. The non-HCV sequences of the no-template control were primer and adaptor sequences originating from the amplification and sequencing process.
TABLE 1.
Full-genome sequencing of in vitro-transcribed RNA (2aRlucNeo)
| No. of input RNA molecules per reaction | Input RNA amt per reaction | Total no. of MiSeq reads | % reads aligned to HCV (2aRlucNeo reference sequence) | Avg coverage (no. of reads per position) | % (no. total no) for: |
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| Assembled 5′ UTR length | Sequence identity of assembled 5′ UTR to reference | Sequence identity CDSa | Assembled 3′ UTR length | Sequence identity of assembled 3′ UTR to reference | S282T 10%b | |||||
| 108 | 500 pg | 351,921 | 88.7 | 9,404 | 91 (328/361) | 100 (328/328) | 100 (8,474/8,474) | 58 (136/234) | 100 (136/136) | 10.6 (688/6,464) |
| 107 | 50 pg | 292,867 | 93.8 | 7,880 | 90 (326/361) | 100 (326/326) | 100 (8,474/8,474) | 58 (136/234) | 100 (136/136) | 9.7 (874/9,036) |
| 106 | 5 pg | 1,006,481 | 93.1 | 27,462 | 91 (328/361) | 100 (328/328) | 100 (8,474/8,474) | 79 (186/234) | 100 (186/186) | 9.7 (1,024/10,552) |
| 105 | 500 fg | 283,822 | 71.8 | 6,844 | 91 (328/361) | 100 (328/328) | 100 (8,474/8,474) | 22 (52/234) | 100 (52/52) | 11 (1,144/10,364) |
| 104 | 50 fg | 457,311 | 29.2 | 4,419 | 90 (326/361) | 100 (326/326) | 100 (8,474/8,474) | 21 (48/234) | 100 (48/48) | 14.6 (786/5,392) |
| 103 | 5 fg | 1,531,207 | 4.2 | 2,091 | 84 (302/361) | 100 (302/302) | 99.98 (8,146/8,148) | 19 (44/234) | 100 (44/44) | 5.9 (16/272) |
| NTCc | 0 | 1,811,726 | 0.03 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
Identity was calculated as the number of exact matches to the 8,474-nucleotide-long coding sequences (CDS) of 2aRlucNeo, excluding the 5′ UTR and 3′ UTR.
Percent hits and hits per total number of reads of the drug-resistant NS5B mutation S282T, which was added at 10% in all dilutions.
NTC, no-template control.
FIG 1.
Read coverage throughout the HCV genome. (A) Comparison of read coverages for in vitro-transcribed RNA samples with different number of input RNA molecules per reaction. (B) Comparison of read coverage throughout the HCV genome for the patient samples. Coverage was computed as total number of reads spanning a given position. Pt, patient.
The generated consensus sequence was compared with the 2aRlucNeo reference, and a 100% match was found in all samples, except for the 5-fg sample, in which 2 nucleotide differences were found in the assembled coding region. In all samples, partial 5′ UTR and 3′ UTR sequences were obtained, and these sequences had 100% identity to 2aRlucNeo (Table 1). It was not possible to generate a consensus sequence from the NTC sample.
The frequency of the S282T substitution was consistent with the 10% S282T addition for the samples with 108 (500 pg) to 105 (500 fg) input RNA molecules per reaction (range, 9.7 to 11%), whereas the lower-RNA-input samples of 104 (50 fg) and 103 (5 fg) molecules per reaction had S282T detected at 14.6% and 5.9%, respectively.
Full-length HCV genome sequencing of HCV patient samples.
The HCV genomes of 17 patient samples with genotype 1a, 1b, 2a, 2b, 2c, 2j, 3a, 3i, 4a, 4d, 5a, 6a, 6e, or 6j were successfully sequenced using the full-length HCV genome sequencing assay. The viral load ranged from 1 million to 23 million IU/ml, which corresponds to the in vitro-transcribed RNA samples with 106 and 105 RNA molecules per reaction after taking dilution factors from RNA extraction and amplification into account. In 14 of the 17 patient samples, the consensus sequence spanned the complete coding region of 9,036 nucleotides relative to the H77 reference genome. A few bases at the end of NS5B lacked sufficient coverage in 3 of 17 samples, generating 99.3 to 100% of the coding sequence. The insufficient coverage was likely due to the difficulty of primer annealing in the 3′ region due to RNA secondary structures. This was compounded by the difficulty associated with assembly through the poly(U/C) region in 3′ UTR affecting the proximal NS5B 3′ end. On average, 3.7 million reads were generated per sample, of which 14.6% of the reads aligned to HCV (Table 2). The average coverage throughout the genome ranged from 684 to 20,734 reads per position and sample (Table 2 and Fig. 1B). A drop in coverage was consistently seen in the end of the genome; in the 5′UTR, a drop of 50% was detected in most samples, whereas in the 3′ UTR, a drop to zero reads was detected either before or after the poly(U) region for all samples (Fig. 1B).
TABLE 2.
Full-HCV-genome sequencing of patient samples
| Patient IDa | HCV viral load (IU/ml) | Estimated RNA input copies per reaction | GTb | Total no. of MiSeq reads | % reads aligned to HCVc | Avg coveraged | Length of assembled 5′ UTR (%)e | Length of assembled HCV coding region (%)f | Length of assembled 3′ UTR (%)g |
|---|---|---|---|---|---|---|---|---|---|
| A | 5,660,000 | ∼105 | 1a | 3,259,050 | 21.6 | 4,675 | 1–341 (100) | 342–9312 (99.28) | 0 |
| B | 23,700,000 | ∼106 | 1b | 1,795,928 | 47.7 | 971 | 1–341 (100) | 342–9377 (100) | 9378–9420 (15.6) |
| C | 9,100,000 | ∼105 | 2a | 3,999,578 | 7.6 | 2,540 | 1–341 (100) | 342–9377 (100) | 9378–9469 (34.2) |
| D | 9,980,000 | ∼105 | 2b | 4,575,692 | 9.7 | 2,815 | 1–341 (100) | 342–9377 (100) | 9378–9400 (8.6) |
| E | 1,952,468 | ∼105 | 2b | 3,637,670 | 42.0 | 16,283 | 1–341 (100) | 342–9377 (100) | 9378–9425 (17.8) |
| F | 18,700,000 | ∼106 | 2c | 5,101,944 | 39.9 | 20,734 | 1–341 (100) | 342–9377 (100) | 9378–9583 (76.5) |
| G | 22,600,000 | ∼106 | 2c | 2,780,471 | 47.3 | 24,000 | 1–341 (100) | 342–9377 (100) | 9378–9583 (76.5) |
| H | 5,620,000 | ∼105 | 2j | 3,002,747 | 6.92 | 2,736 | 1–341 (100) | 342–9377 (100) | 9378–9486 (40.3) |
| I | 2,300,000 | ∼105 | 3a | 4,030,933 | 10.1 | 2,150 | 15–341 (95.6) | 342–9377 (100) | 9378–9489 (41.4) |
| J | 1,820,000 | ∼105 | 3i | 2,690,695 | 11.6 | 4,163 | 1–341 (100) | 342–9377 (100) | 9378–9466 (32.8) |
| K | 8,400,000 | ∼105 | 4a | 2,550,342 | 3.0 | 992 | 13–341 (96.2) | 342–9377 (100) | 9378–9450 (26.9) |
| L | 5,820,000 | ∼105 | 4d | 2,770,483 | 37.8 | 14,020 | 1–341 (100) | 342–9377 (100) | 9378–9583 (76.5) |
| M | 5,820,000 | ∼105 | 5a | 3,165,617 | 4.3 | 1,822 | 20–341 (94.1) | 342–9377 (100) | 9378–9395 (6.3) |
| N | 32,300,000 | ∼106 | 6a | 3,106,860 | 6.4 | 2,785 | 1–341 (100) | 342–9371 (99.93) | 0 |
| O | 13,300,000 | ∼106 | 6a | 2,999,607 | 2.5 | 943 | 26–341 (92.4) | 342–9374 (99.97) | 0 |
| P | 5,500,000 | ∼105 | 6e | 2,997,274 | 2.3 | 878 | 1–341 (100) | 342–9377 (100) | 9378–9387 (3.7) |
| Q | 21,300,000 | ∼106 | 6j | 3,260,145 | 1.5 | 684 | 1–341 (100) | 342–9377 (100) | 9378–9395 (6.3) |
ID, identification.
GT, genotype.
Percentage of reads aligning to the subtype-specific reference sequence.
Average coverage was calculated as the average number of reads per positions in the HCV genome.
Assembled 5′UTR relative to positions 1 to 341 in H77.
Length of assembled HCV coding region (core to NS5B) relative to positions 342 to 9377 in H77 (GenBank accession no. AF009606).
Assembled 3′UTR relative to positions 9378 to 9646 in H77.
Background variation in the in vitro-transcribed RNA samples.
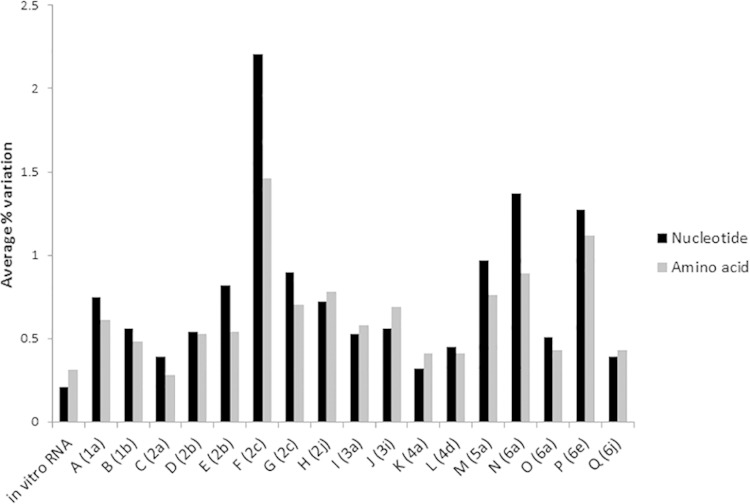
The amount of background variation in the in vitro RNA transcript samples was investigated to calculate the error rate of the full-length HCV genome sequencing assay. Due to the clonal origin of the in vitro-transcribed RNA, the genetic variations in the generated sequence reads were likely due to amplification or sequencing errors, except the 10% addition of S282T in each sample. The average percent nucleotide variation per position was calculated for each sample (Table 3). For the samples with 106 and 105 RNA input molecules per reaction, which corresponds to the RNA input from the patient samples, the average background variation was 0.21% (95% confidence interval [CI] 0.207, 0.213%). The nucleotide variation was plotted per position for each sample (see Fig. S1 in the supplemental material). The average nucleotide variation was similar in all samples; however, the maximum nucleotide variation detected for each sample was highest in the sample that had 103 input RNA molecules per reaction (Table 3; see also Fig. S1). For the in vitro-transcribed RNA samples, an increased level of nucleotide variation was detected in the end of NS5A, which was associated with a 10-fold decrease in coverage seen in all samples at this location (see Fig. S1). However, no statistical correlation was detected between coverage and level of variation (data not shown). Moreover, the specific decrease in coverage in the end of NS5A was not detected in the patient samples (Fig. 1B). On the amino acid level, the samples with 106 and 105 RNA input molecules per reaction had an average variation of 0.31% (Fig. 2).
TABLE 3.
Background variation in the in vitro-transcribed RNA samples
| No. of in vitro-transcribed RNA molecules per reaction | Avg % nt variation ± 95% CI | Maximum nt variation (%)a |
|---|---|---|
| 108 (500 pg) | 0.28 ± 0.0049 | 4.5 |
| 107 (50 pg) | 0.34 ± 0.0078 | 5.3 |
| 106 (5 pg) | 0.28 ± 0.0039 | 2.8 |
| 105 (500 fg) | 0.16 ± 0.0037 | 4.0 |
| 104 (50 fg) | 0.22 ± 0.0067 | 8.0 |
| 103 (5 fg) | 0.19 ± 0.0107 | 21.3 |
This calculation excludes the S282 codon, which was expected to have 10% variant mixture.
FIG 2.
Average nucleotide and amino acid variation. Average nucleotide and amino acid variations were calculated across the genome for each patient sample. Nucleotide variations are shown in black, and amino acid variations are shown in gray. For comparison, the background variation for the in vitro-transcribed RNA sample with 105 to 106 RNA molecules per reaction is shown to the left.
Intrahost HCV nucleotide and amino acid variation.
The high coverage across the HCV genome generated by the full-genome sequencing assay enabled an investigation of the genetic diversity within the viral population on both nucleotide and amino acid levels for each patient sample (Fig. 2; see also Fig. S2 in the supplemental material). The average nucleotide variation in the patient samples was significantly higher (P = 0.01) than the calculated background variation in the in vitro-transcribed RNA samples. Interestingly, the frequency and distribution of variation were substantially different between the patients, with patients F (genotype [GT]2c), N (GT6a), and P (GT6e) having the largest amount of nucleotide and amino acid variation and patients C (GT2a), K (GT4a), and Q (GT6j) having the smallest amount of variation (Fig. 2; see also Fig. S2). Importantly, the level of nucleotide variation detected in these patients was not significantly correlated with the viral load of the sample (P > 0.3). Moreover, nucleotide variation was not correlated with coverage, and no specific hot spots for nucleotide variation were detected in the patient samples. In contrast, amino acid variations were predominantly found in E1/E2 and the end of NS5A for all patients (see also Fig. S2). The regions with the least amino acid variation were the core and NS3.
Presence of potential resistance-associated variants.
The generation of full-length HCV genome sequences enabled an investigation of the presence of potential resistance-associated variations (RAVs) across tested common and rare subtypes. Eleven sites in the NS3 region, 9 sites in the NS5A region, and 26 sites in NS5B described in Materials and Methods were evaluated. No RAVs were detected for any of the samples at positions 43, 54, 55, 155, and 156 in NS3, positions 24, 28, 32, 38, 58, and 92 in NS5A, and positions 96, 142, 159, 282, 316, 320, 321, 390, 448, 452, 495, 496, and 559 in NS5B. Several RAVs were present at baseline and are listed in Tables 4 and 5. The impact of these RAVs on novel NS3, NS5A, and NS5B inhibitors needs to be evaluated further. Importantly, the NS5B S282T substitution associated with resistance to sofosbuvir and mericitabine was not detected in any of the subtypes.
TABLE 4.
Variants with potential NS3 and NS5A resistance-associated amino acid positions
| Patient IDa | GTb | RAV(s) for each position in proteinc: |
|||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NS3 |
NS5A |
||||||||||||||||||||
| V36any | F43any | T54any | V55any | Q80any | S122R | R155any | A156any | D168any | I170A/T/L | L175L | K24G/N/R | M28A/G/T | Q30any | L31any | P32L | S38F | H58D | A92K/L | Y93any | ||
| A | 1a | L | |||||||||||||||||||
| B | 1b | R | L/M | ||||||||||||||||||
| C | 2a | L | G | L | K | M | |||||||||||||||
| D | 2b | L | G | R | L | K | |||||||||||||||
| E | 2b | L | G | R | L | K | M | ||||||||||||||
| F | 2c | L | G | R | L | K | F | ||||||||||||||
| G | 2c | L | G | R | L | K | M | ||||||||||||||
| H | 2j | L | G | R | L | K | M | ||||||||||||||
| I | 3a | L | Q | L | A | ||||||||||||||||
| J | 3i | L | Q | L | K | ||||||||||||||||
| K | 4a | L | L | S | M | ||||||||||||||||
| L | 4d | L | L | R | M | ||||||||||||||||
| M | 5a | L | K | L | T | ||||||||||||||||
| N | 6a | K | R | T | |||||||||||||||||
| O | 6a | K | R | T | |||||||||||||||||
| P | 6e | S | S | ||||||||||||||||||
| Q | 6j | A | T | ||||||||||||||||||
ID, identification.
GT, genotype.
Substitutions show positions in NS3 and NS5A gene with corresponding wild-type amino acid first (from 1a H77 reference genome) and the amino acid(s) associated with second. For each patient, the presence of amino acids associated with resistance is shown in the table. Blank cells indicate identical amino acid as that of 1a wild type, which has not been associated with resistance.
TABLE 5.
Variants with potential NS5B resistance-associated amino acid positions
| Patient IDa | GTb | RAVc |
|||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NI |
NNI |
RBV |
|||||||||||||||||||||||||
| L159any | S282any | C289any | L320any | V321any | S96any | N142T | C316any | M414I/T/V | L419any | R422K | M423any | C445F | Y448any | Y452any | I482any | A486any | V494A | P495any | P496S | A499A | G554S | S556G | D559G | T390I | F415Y | ||
| A | 1a | A | |||||||||||||||||||||||||
| B | 1b | Y | |||||||||||||||||||||||||
| C | 2a | M | I | F | L | A | A | G | Y | ||||||||||||||||||
| D | 2b | M | I | F | L | A | A | G | Y | ||||||||||||||||||
| E | 2b | M | I | F | L | A | A | G | Y | ||||||||||||||||||
| F | 2c | M | I | F | L | A | A | S | G | Y | |||||||||||||||||
| G | 2c | M | I | F | L | A | A | S | G | Y | |||||||||||||||||
| H | 2j | M | V | F | L | A | A | G | Y | ||||||||||||||||||
| I | 3a | F | I | F | L | A | G | Y | |||||||||||||||||||
| J | 3i | F | I | F | L | A | G | Y | |||||||||||||||||||
| K | 4a | F | V | I | F | L | A | G | Y | ||||||||||||||||||
| L | 4d | F | I | I | F | L | A | G | Y | ||||||||||||||||||
| M | 5a | M | I | F | A | G | Y | ||||||||||||||||||||
| N | 6a | M | I | F | L | G | A | A | |||||||||||||||||||
| O | 6a | M | I | F | L | G | A | A | |||||||||||||||||||
| P | 6e | L | I | F | L | A | A | Y | |||||||||||||||||||
| Q | 6j | M | I | F | L | A | A | Y | |||||||||||||||||||
ID, identification.
GT, genotype.
Substitutions show positions in NS5B gene and their wild-type amino acid first (from 1a H77 reference genome) and the amino acid(s) associated with resistance second. For each patient, the presence of amino acids associated with resistance is shown in the table. Blank cells indicate identical amino acid as that of 1a wild type, which has not been associated with resistance. NI, nucleoside inhibitor; NNI, non-nucleoside inhibitor; RBV, ribavirin.
To assess the comparability between the full-genome sequencing assay and the standard NS5A and NS5B deep-sequencing assay, the amino acid frequencies in samples in which results from both assays were available were compared. An identical amino acid was detected at each RAV position, and the frequencies of the amino acids were comparable between the two assays.
Phylogenetic analysis.
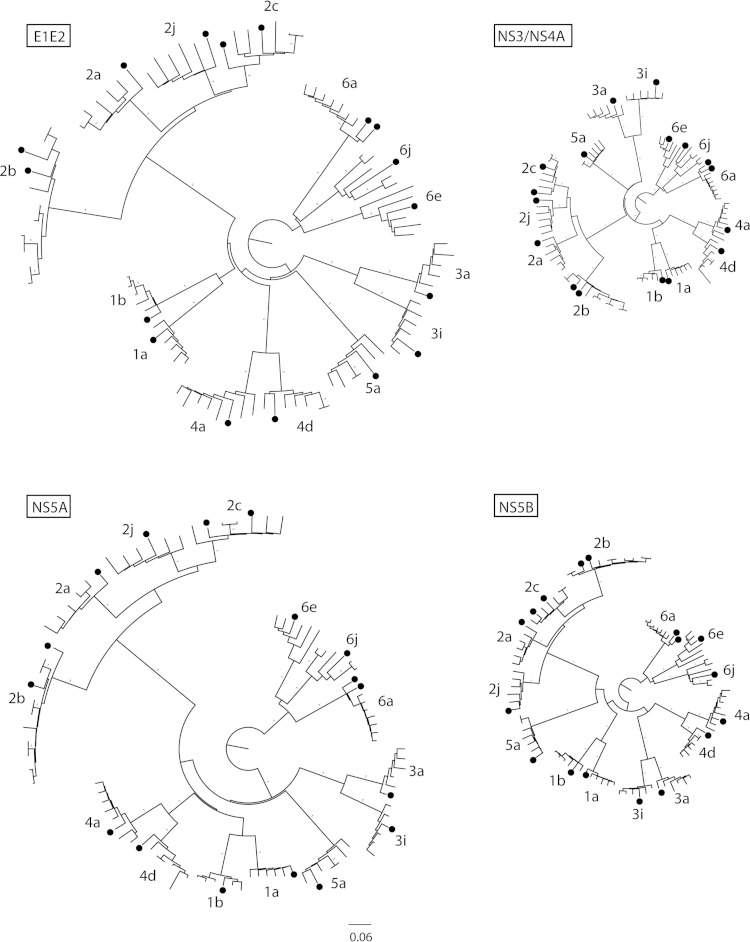
To study the genotype diversity at different regions of the HCV genome and determine the genetic relationships between the 17 nearly full-length HCV sequences in relation to other publically available HCV genome sequences, neighbor-joining phylogenetic trees were inferred for the E1/E2, NS3/4A, NS5A, and NS5B regions (Fig. 3). Analysis of the phylogenetic relationships showed that the sequences determined in this study were genetically similar to publically available sequences with the same subtype; however, the sequences were not genetically identical to each other or previously described sequences. Moreover, the phylogenetic analysis confirmed the HCV subtype classification, as assessed by NS5B sequencing of each patient sample for each genomic region. Furthermore, the genetic distance between the sequences was higher in the E1/E2 and NS5A regions than that in the NS3/4A and NS5B regions.
FIG 3.
Phylogenetic analysis of the generated HCV sequences from the patient samples. Neighbor-joining trees were constructed for E1/E2, NS3/NS4A, NS5A, and NS5B genes from the patient full-genome sequences. Patient sequences are shown by filled circles. Branches with a 100% bootstrap value are denoted by a 1. All trees were rooted to genotype 6 and are shown on the same scale.
HCV-specific reads and unaligned reads.
The number of reads aligning to HCV was lower in the patient samples than that in the in vitro RNA sample with 105 RNA molecules per reaction, despite a similar number of input RNA molecules in the patient samples. The HCV-specific reads in the patient samples ranged between 1.5% and 47% (Table 2), whereas the in vitro-transcribed RNA sample had 71.8% of the reads aligning to the reference (Table 1). To investigate the reason for this difference, we performed a BLAST analysis of 50,000 randomly selected unaligned reads from 6 samples. The taxonomic composition of the unaligned reads in the patient samples was 70 to 98% metazoan, 0 to 18% bacterial, 0.3 to 12% viral, and 2 to 16% unassigned sequences (no hit or low complexity). For the in vitro-transcribed RNA, the composition of the unaligned reads was 1% metazoan, 40% bacterial, 32% viral (HCV), and 27% unassigned sequences (no hit or low complexity) (see Fig. S3 in the supplemental material). The main difference in the composition of the unaligned reads between the patient samples and the in vitro-transcribed RNA was the increased human background in the patient samples. The viral sequences found among the 50,000 read sets were HCV reads with the same subtype as that of the corresponding sample.
DISCUSSION
HCV sequencing is used frequently during drug development to determine the substitutions associated with resistance to direct-acting antivirals (DAAs) in both in vitro selections and clinical trials. Moreover, during the development of pangenotypic HCV compounds, efficient sequencing strategies that are independent of HCV subtype or genotype are essential for resistance testing of rare subtypes. Sequencing of the complete HCV genome can give important information regarding genetic changes and potential drug resistance-associated mutations that might be present outside the target gene of the drug. In this study, we report the successful sequencing of complete HCV genomes from 17 patient samples with genotypes 1 to 6, including samples with subtypes that have limited sequence information, using a subtype-independent full-genome sequencing assay. In comparison to previously described methods for full-genome sequencing of HCV, using either traditional sequence-specific primers (9–12) or RNA-Seq technology (14), the full-genome sequencing method described here has the advantage of being subtype and genotype independent and having a significantly improved sensitivity compared to that of RNA-Seq. Furthermore, the highly variable E2 region, which previously was shown to be difficult to assemble due to sequence variability (14), was successfully sequenced and assembled for all samples using the de novo assembly approach described here.
The accuracy of the generated full-genome consensus sequences was estimated to be >99%, based on results from the in vitro-transcribed HCV RNA samples. In addition, the ability to detect 10% S282T using the full-genome sequencing method was evaluated in the in vitro RNA transcript samples, as well the nucleotide background variation introduced during amplification and sequencing. The frequency of S282T detection was close to 10% in all samples, and the average background nucleotide variation was 0.21% in an in vitro-transcribed RNA sample with similar amount of input RNA molecules as that of a patient sample. These results suggest an increased ability to detect minor variants by this method compared to detection with standard population Sanger sequencing. A higher background nucleotide variation was seen in the in vitro-transcribed RNA sample, with the lowest number of input RNA molecules being 103 RNA molecules per reaction. This was expected, since amplification and sequencing errors are more pronounced in samples with a lower number of input molecules due to artifacts generated during amplification (22). It is important to note that for viral infections, such as HCV, in which the plasma viral load is high (usual range, 105 to 107 IU/ml), this sequencing method is highly suitable due to the low background detected for the corresponding amount of input RNA molecules. However, for plasma samples with a lower viral load, the extraction might need to be modified to concentrate the RNA to obtain ≥10,000 RNA input molecules.
HCV is characterized by high genetic variability; a higher evolutionary rate has been described for E1/E2 and the NS5A gene compared to that of the rest of the HCV genome (3, 23). In agreement with previous reports, the intergenotype genetic distance was greater in the E1/E2 and NS5A regions than that in the NS3/4A and NS5B regions, as shown by the phylogenetic analysis. In addition, intrahost genetic variability of the HCV quasispecies showed that amino acid changes were predominantly found in the E1/E2 and NS5A regions in most patients, independent of genotype. The increased variability at these regions may arise through specific selection mechanisms associated with immune escape, where the hypervariable region of the E2 envelope glycoprotein is targeted by neutralizing antibodies, and NS5A contains a high concentration of T- and B-cell epitopes (2, 24). Due to selection pressures, regions targeted by the immune system or by antiviral drugs may require sequence changes for the virus to survive. Interestingly, the overall intrahost HCV nucleotide diversity varied greatly within the samples from the studied patients, and no specific patterns were observed in a comparison of common and rare subtypes. It is possible that high genetic variability in specific regions of the HCV genome are associated with lower treatment response due to an increased likelihood of the presence of preexisting resistance mutations at these locations. Further studies are needed to investigate if sequence diversity is linked to treatment outcome and which regions in the HCV genome might be involved.
Specific amino acids at positions in the NS3, NS5A, and NS5B regions have been associated with drug resistance to DAAs. Investigation of such RAVs in different HCV subtypes and genotypes is of importance, since the presence of RAVs at baseline may affect treatment outcome. Extensive studies have been done in genotype 1 to investigate RAVs in response to treatment with DAAs; however, naturally occurring RAVs in rare subtypes have been studied in only a few reports (9, 25). In this study, we investigated the amino acid composition at RAV positions in NS3, NS5A, and NS5B for the common and rare subtypes described here. We show that the composition of naturally occurring RAVs differs among subtypes, which is of interest, since their presence may impact resistance profiles. Further evaluations are needed to determine the susceptibilities of such RAVs to specific DAAs for each subtype.
The full-genome sequencing assay described in this report is based on random amplification, which can potentially remove the bias associated with sequence-specific primers used in standard sequencing methods to generate a more reliable view of the HCV quasispecies. Moreover, due to random amplification, potential infections by other agents or HCV coinfections could be studied by this method. This method is clinically relevant, since it can facilitate the sequencing of genes targeted by antiviral drugs for all HCV genotypes, including rare subtypes and intergenotypic HCV strains. It is especially relevant for sequencing HCV strains for which sequence information availability is limited for gene-specific primer design. The sequencing information generated can be used for selecting effective antiviral treatment and determining possible drug resistance development. However, it might not be feasible to use this method in a clinical setting because of the limited availability of tools for sequence data processing. Moreover, the higher cost associated with sequencing assays and the nonspecific amplification are limitations of the assay. Further evaluations are needed to determine the sensitivity of the assay.
Due to random amplification, the majority of the generated reads from the patient plasma samples originated from human RNA, which likely originated from human cells present in the plasma and/or human RNA colocalized in HCV virions isolated along with the HCV RNA during extraction. The metazoan sequences were assigned as human or primate sequences in which the primate sequences were most likely misclassified human sequences. The bacterial sequences were classified into different species of proteobacteria, and the origin of these bacterial sequences in the patient samples are unknown. Most likely, they originate from bacterial RNA/DNA present in reagents after production or contamination from the environment. We did not detect any evidence of other RNA virus coinfections in any of the tested patient samples, and the few HCV reads that were not aligning to the selected reference for each patient were composed of reads spanning both sense and antisense directions. Since the positioning of these reads in the alignment was ambiguous, they were removed from the analysis. This artifact was previously described by Malbeouf et al. (15) and likely arose in the NuGEN SPIA amplification step.
Taken together, 17 clinical HCV samples with genotypes 1 to 6 were successfully sequenced using a sequence-independent full-length HCV genome sequencing assay. This method has the potential of facilitating the sequencing of any HCV clinical samples independent of genotype, subtype, or intergenotypic recombination and allowing greater insight into HCV evolution, genetic diversity, and drug resistance development.
Supplementary Material
ACKNOWLEDGMENTS
We thank all the patients who participated in the phase II/III clinical studies and the research staff of the clinical virology department at Gilead. We also thank Diana Brainard of Gilead Sciences for critical review of the manuscript.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.02624-14.
REFERENCES
- 1.Smith DB, Bukh J, Kuiken C, Muerhoff AS, Rice CM, Stapleton JT, Simmonds P. 2014. Expanded classification of hepatitis C virus into 7 genotypes and 67 subtypes: updated criteria and genotype assignment web resource. Hepatology 59:318–327. doi: 10.1002/hep.26744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Simmonds P. 2004. Genetic diversity and evolution of hepatitis C virus–15 years on. J Gen Virol 85:3173–3188. doi: 10.1099/vir.0.80401-0. [DOI] [PubMed] [Google Scholar]
- 3.Smith DB, Pathirana S, Davidson F, Lawlor E, Power J, Yap PL, Simmonds P. 1997. The origin of hepatitis C virus genotypes. J Gen Virol 78(Pt 2):321–328. [DOI] [PubMed] [Google Scholar]
- 4.Morel V, Fournier C, François C, Brochot E, Helle F, Duverlie G, Castelain S. 2011. Genetic recombination of the hepatitis C virus: clinical implications. J Viral Hepat 18:77–83. doi: 10.1111/j.1365-2893.2010.01367.x. [DOI] [PubMed] [Google Scholar]
- 5.Kalinina O, Norder H, Mukomolov S, Magnius LO. 2002. A natural intergenotypic recombinant of hepatitis C virus identified in St. Petersburg. J Virol 76:4034–4043. doi: 10.1128/JVI.76.8.4034-4043.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hedskog C, Doehle B, Chodavarapu K, Gontcharova V, Crespo Garcia J, De Knegt R, Drenth JP, McHutchison JG, Brainard D, Stamm LM, Miller MD, Svarovskaia E, Mo H. 2014. Characterization of hepatitis C virus intergenotypic recombinant strains and associated virologic response to sofosbuvir/ribavirin. Hepatology 61:471–480. doi: 10.1002/hep.27361. [DOI] [PubMed] [Google Scholar]
- 7.Gane EJ, Stedman CA, Hyland RH, Ding X, Svarovskaia E, Symonds WT, Hindes RG, Berrey MM. 2013. Nucleotide polymerase inhibitor sofosbuvir plus ribavirin for hepatitis C. N Engl J Med 368:34–44. doi: 10.1056/NEJMoa1208953. [DOI] [PubMed] [Google Scholar]
- 8.Murphy DG, Willems B, Deschenes M, Hilzenrat N, Mousseau R, Sabbah S. 2007. Use of sequence analysis of the NS5B region for routine genotyping of hepatitis C virus with reference to C/E1 and 5′ untranslated region sequences. J Clin Microbiol 45:1102–1112. doi: 10.1128/JCM.02366-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Newman RM, Kuntzen T, Weiner B, Berical A, Charlebois P, Kuiken C, Murphy DG, Simmonds P, Bennett P, Lennon NJ, Birren BW, Zody MC, Allen TM, Henn MR. 2013. Whole genome pyrosequencing of rare hepatitis C virus genotypes enhances subtype classification and identification of naturally occurring drug resistance variants. J Infect Dis 208:17–31. doi: 10.1093/infdis/jis679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Okamoto H, Kurai K, Okada S, Yamamoto K, Lizuka H, Tanaka T, Fukuda S, Tsuda F, Mishiro S. 1992. Full-length sequence of a hepatitis C virus genome having poor homology to reported isolates: comparative study of four distinct genotypes. Virology 188:331–341. doi: 10.1016/0042-6822(92)90762-E. [DOI] [PubMed] [Google Scholar]
- 11.Hmaied F, Legrand-Abravanel F, Nicot F, Garrigues N, Chapuy-Regaud S, Dubois M, Njouom R, Izopet J, Pasquier C. 2007. Full-length genome sequences of hepatitis C virus subtype 4f. J Gen Virol 88:2985–2990. doi: 10.1099/vir.0.83151-0. [DOI] [PubMed] [Google Scholar]
- 12.Aizaki H, Aoki Y, Harada T, Ishii K, Suzuki T, Nagamori S, Toda G, Matsuura Y, Miyamura T. 1998. Full-length complementary DNA of hepatitis C virus genome from an infectious blood sample. Hepatology 27:621–627. doi: 10.1002/hep.510270242. [DOI] [PubMed] [Google Scholar]
- 13.Lauck M, Alvarado-Mora MV, Becker EA, Bhattacharya D, Striker R, Hughes AL, Carrilho FJ, O'Connor DH, Rebello Pinho JR. 2012. Analysis of hepatitis C virus intrahost diversity across the coding region by ultradeep pyrosequencing. J Virol 86:3952–3960. doi: 10.1128/JVI.06627-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ninomiya M, Ueno Y, Funayama R, Nagashima T, Nishida Y, Kondo Y, Inoue J, Kakazu E, Kimura O, Nakayama K, Shimosegawa T. 2012. Use of illumina deep sequencing technology to differentiate hepatitis C virus variants. J Clin Microbiol 50:857–866. doi: 10.1128/JCM.05715-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Malboeuf CM, Yang X, Charlebois P, Qu J, Berlin AM, Casali M, Pesko KN, Boutwell CL, DeVincenzo JP, Ebel GD, Allen TM, Zody MC, Henn MR, Levin JZ. 2013. Complete viral RNA genome sequencing of ultra-low copy samples by sequence-independent amplification. Nucleic Acids Res 41:e13. doi: 10.1093/nar/gks794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kurn N, Chen P, Heath JD, Kopf-Sill A, Stephens KM, Wang S. 2005. Novel isothermal, linear nucleic acid amplification systems for highly multiplexed applications. Clin Chem 51:1973–1981. doi: 10.1373/clinchem.2005.053694. [DOI] [PubMed] [Google Scholar]
- 17.Yang X, Charlebois P, Gnerre S, Coole MG, Lennon NJ, Levin JZ, Qu J, Ryan EM, Zody MC, Henn MR. 2012. De novo assembly of highly diverse viral populations. BMC Genomics 13:475. doi: 10.1186/1471-2164-13-475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee WP, Stromberg MP, Ward A, Stewart C, Garrison EP, Marth GT. 2014. MOSAIK: a hash-based algorithm for accurate next-generation sequencing short-read mapping. PLoS One 9:e90581. doi: 10.1371/journal.pone.0090581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Z, Schwartz S, Wagner L, Miller W. 2000. A greedy algorithm for aligning DNA sequences. J Comput Biol 7:203–214. doi: 10.1089/10665270050081478. [DOI] [PubMed] [Google Scholar]
- 20.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 21.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Odeberg J, Ahmadian A, Williams C, Uhlen M, Ponten F, Lundeberg J. 1999. Context-dependent Taq-polymerase-mediated nucleotide alterations, as revealed by direct sequencing of the ZNF189 gene: implications for mutation detection. Gene 235:103–109. doi: 10.1016/S0378-1119(99)00205-X. [DOI] [PubMed] [Google Scholar]
- 23.Gray RR, Parker J, Lemey P, Salemi M, Katzourakis A, Pybus OG. 2011. The mode and tempo of hepatitis C virus evolution within and among hosts. BMC Evol Biol 11:131. doi: 10.1186/1471-2148-11-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dou XG, Talekar G, Chang J, Dai X, Li L, Bonafonte MT, Holloway B, Fields HA, Khudyakov YE. 2002. Antigenic heterogeneity of the hepatitis C virus NS5A protein. J Clin Microbiol 40:61–67. doi: 10.1128/JCM.40.1.61-67.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vallet S, Viron F, Henquell C, Le Guillou-Guillemette H, Lagathu G, Abravanel F, Trimoulet P, Soussan P, Schvoerer E, Rosenberg A, Gouriou S, Colson P, Izopet J, Payan C, ANRS AC11 HCV Group. 2011. NS3 protease polymorphism and natural resistance to protease inhibitors in French patients infected with HCV genotypes 1–5. Antiviral therapy 16:1093–1102. doi: 10.3851/IMP1900. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.