Abstract
Glioma is the most aggressive brain tumor with high invasiveness and poor prognosis. More reliable, sensitive and practical biomarkers to reveal glioma high invasiveness remain to be explored for the guidance of therapy. We herein evaluated the diagnostic and prognostic value of aldehyde dehydrogenase 1A1 (ALDH1A1) in the glioma specimens from 237 patients, and found that ADLH1A1 was frequently overexpressed in the high-grade glioma (WHO grade III-IV) as compared to the low-grade glioma (WHO grade I-II) patients. The tumor cells with ALDH1A1 expression were more abundant in the region between tumor and the borderline of adjacent tissue as compared to the central part of the tumor. ALDH1A1 overexpression was associated with poor differentiation and dismal prognosis. Notably, the overall and disease-free survivals of the patients who had ALDH1A1+ tumor cells sparsely located in the adjacent tissue were much worse. Furthermore, ALDH1A1 expression was correlated with the “classical-like” (CL) subtype as we examined GBM specimens from 72 patients. Multivariate Cox regression analysis revealed that ALDH1A1 was an independent marker for glioma patients’ outcome. Mechanistically, both in vitro and in vivo studies revealed that ALDH1A1+ cells isolated from either a glioblastoma cell line U251 or primary glioblastoma cells displayed significant invasiveness, clonogenicity, and proliferation as compared to ALDH1A1- cells, due to increased levels of mRNA and protein for matrix metalloproteinase 2, 7 and 9 (MMP2, MMP7 and MMP9). These results indicate that ALDH1A1+ cells contribute to the progression of glioma including invasion, proliferation and poor prognosis, and suggest that targeting ALDH1A1 may have important implications for the treatment of highly invasive glioma.
Keywords: ALDH1A1, glioma, invasion, matrix metalloproteinase, prognosis
Introduction
Glioma is one of the most common solid malignancies in brain with atrocious patient survival. It is a heterogeneous disease with regard to its morphological presentation. The patients with glioma have poor outcomes, the median survival ranging from 12-15 months [1]. Many genetic factors alterations have been correlated with poor or better prognosis in glioma [2]. The immunohistochemistry (IHC) test to aim some specific antigens, such as methyl guanine methyl transferase (MGMT), glycoprotein (transmembrane) nmb (GPNMB), lysyl oxidase-like 3 (LOXL3) and interleukin 13 receptor, alpha 2 (IL13RA2), in tumour cells have significantly improved the diagnosis of glioma and the prediction of prognosis [3,4]. Marie Le Mercier, et al. revealed the existence of several GBM subtypes by IHC analysis of TP53, platelet-derived growth factor receptor alpha (PDGFRA) and epidermal growth factor receptor (EGFR) [5]. Although these and other molecules are demonstrated for their correlation to the malignancy and prognosis of glioma [6-8], more sensitive, reliable and practical indicators to reveal high invasiveness remain explored not only for pathological diagnosis but also for prognostic prediction and targeted therapy.
Aldehyde dehydrogenase 1A1 (ALDH1A1), a member of ALDH1 family of enzymes, is a detoxification enzyme in the metabolism of aldehydes to their corresponding carboxylic acids. In liver, cytosolic ALDH1A1 contributes primarily to the biosynthesis of retinoic acid (RA) from vitamin A [9,10]. ALDH1A1 is also found in human and murine hematopoietic progenitor or stem cells and other cancer stem cells, such as colorectal carcinoma, prostate cancer, lung cancer and breast cancer [11-16]. However, the role of ALDH1A1 in glioma as putative prognostic and significance marker remains nebulous [17,18].
The aim of our study was to investigate ALDH1A1 expression in different grade gliomas, and to reveal its correlation with clinicopathological features and prognosis of glioma as well as molecular classification of GBM. We performed IHC staining of ALDH1A1 on the glioma specimens obtained from 237 patients with different WHO grades. Both in vitro and in vivo studies by using one glioma cell line and primary cells from a glioma patient were executed to explore the corresponding mechanism underneath the role of ALDH1A1 on glioma progression. Our study indicates that ALDH1A1+ cells are highly invasive and ALDH1A1 can be applied as a biomarker to predict patients’ outcome. Thus, targeting ALDH1A1+ cells will be propitious for effective therapy against glioma.
Materials and methods
Tissue specimens, patient characteristics and immunohistochemistry (IHC)
Glioma tissues were surgically obtained from 166 patients from Southwest Hospital, Third Military Medical University between 2006 and 2009, and 71 patients from Tiantan Hospital, Capital Medical University between 2006 and 2010. Patients were informed for the procedures that were conducted according to the guidelines of the Research Ethics Committees of both institutions. Table 1 shows the main clinicopathological information of the glioma patients. Classification of the glioma was determined according to the criteria of World Health Organization (WHO) 2007. Follow-up data from 114 of 237 patients were collected by the intervals of 4-6 months at periodic visits. Follow-up time was defined as the time from the date of surgical pathology diagnosis to the date of death or the date of the last visit. The median follow-up time was 28 months (ranging from 4-118). The disease-free survival was defined as the time between the date of surgical pathology diagnosis and the date of the last follow-up examination when the patient was diagnosed as disease-free, or the date of the first recurrence regardless of local or regional occurrence.
Table 1.
Clinical Characteristics of Study Specimen
| Grade (WHO) | Case | Mean age (years) | Gender | Resection range | Radio-therapy | Chemo-therapy | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||||
| ≤ 40.8 | > 40.8 | Male | Female | Part | All | Yes | No | Yes | No | ||
| I | 18 | 15 | 3 | 9 | 9 | 1 | 17 | 15 | 3 | 0 | 18 |
| II | 99 | 64 | 35 | 55 | 44 | 25 | 74 | 63 | 36 | 22 | 77 |
| III | 48 | 16 | 32 | 35 | 13 | 15 | 33 | 40 | 8 | 26 | 22 |
| IV | 72 | 19 | 53 | 42 | 30 | 27 | 45 | 53 | 19 | 16 | 56 |
| Total | 237 | 114 | 123 | 141 | 96 | 68 | 169 | 171 | 66 | 64 | 173 |
Categorical data are presented as numbers.
IHC staining was performed on the paraffin sections of glioma tissues. Briefly, the sections were immersed in a 10 mM citrate buffer (pH 6.0), and incubated in a microwave oven at 100°C for 5 mins and then 45°C for 10 mins. Endogenous peroxidase activity was blocked with 3% H2O2 at 37°C for 30 mins. The protein abundances of ALDH1A1, p53, PDGFRA, EGFR and Ki67 were detected through incubation with the primary antibodies of anti-human ALDH1A1 (1:600) (clone 44/ALDH, mouse monoclonal IgG1; BD Pharmingen, San Diego, CA), p53 (1:500) (ZSGB-BIO ORIGENE, Beijing, China), PDGFRA (1:100) (Thermo Scientific, China), EGFR (1:100) (ZSGB-BIO ORIGENE, Beijing, China), and Ki67 (1:200) (ZSGB-BIO ORIGENE, Beijing, China) overnight at 4°C. Then, the corresponding poly-clonal goat anti-mouse or anti-rabbit immunoglobulin G secondary antibodies (Dako Cytomation, Glostrup, Denmark) were incubated at 37°C for 30 min. The tissue sections were stained with diaminobenzidine (DAB) (Dako Cytomation) as a substrate for color development and counterstained with hematoxylin. Positive and negative controls were included in each immunohistochemical reaction.
Scores were determined according to the percentages of positive tumor cells and intensities of staining. We defined the observation that was less than 5% as score 0 (negative), between 5% to 25% positive cells as score 1 (weak expression), between 26% to 50% score 2 (moderate expression); and between 51% to 75% as score 3 (strong expression), and more than 75% as scores 4 (strongest expression). A “total staining score” of 0-12 was gained from multiplying the scores of the percentages of positive tumor cells by the scores of the intensities of positive tumor cells [19]. The “low score group or negative one” was defined if the “total staining score” of ALDH1A1, p53, PDGFRA, EGFR, or Ki67 expression in tumor cells was less than 6, while the “high score group or positive one” was defined if the “total staining score” of ALDH1A1, p53, PDGFRA, EGFR, or Ki67 expression in tumor cells was equal or larger than 6.
For the analysis of the relationship between glioma invasion and distribution of ALDH1A1 positive tumor cells, we defined the invasion frontier as the areas within 200 µm of boundary between tumor and adjacent normal brain tissue. At least ten fields in each area of the section were randomly selected for calculating average percentage of positive cells over total cancer cells. Secondly, for the analysis of the relationship between ALDH1A1 expression in the normal brain tissue adjacent to invasion frontier and outcome of the patients and if the number of ALDH1A1+ cells in these area was ≥ 10 under five high power fields, we defined the observation as positive.
ALDEFLUOR assay and flow cytometry
The ALDEFLUOR kit (Stem Cell Technologies, Canada) was used to detect tumor cells with high or low ALDH enzymatic activity. Briefly, glioblastoma cells were suspended in ALDEFLUOR assay buffer containing ALDH substrate, and BODIPY-aminoacetaldeh-tyde (BAAA) was added and then incubated at 37°C for 45 mins. Diethylaminobenzal-dehyde (DEAB), an inhibitor of ALDH, served as negative controls. Application of 7-Amino-actinomycin D (7-AAD) staining solution (BD Pharmingen, USA) was used to exclude dead cells. Cells were assayed for experiments after the sort was completed.
RNA extraction and real-time PCR
Total RNA was extracted with RNAiso reagent (TakaRa, Japan). Reverse-transcription and qu-antitative real-time PCR were performed by using an One Step SYBR Prime Script RT-PCR kit (TakaRa). The primers designed for the genes were shown in Table S1. The 2-ΔΔCT method was applied to determine the expression of each gene by ALDH1A1+ relative to ALDH1A1- glioblastoma cells. GAPDH expression was used for normalization. All experiments were performed at least in quadruplicate, and results were plotted as the mean ± SD.
Western blotting
The tumor cells of ALDH1A1+ and ALDH1A1- from both U251 cell line and the primary glioblastoma sample were analyzed by western blotting by using NE-PER Cytosol Extraction Kit (Thermo Scientific, PA). Primary antibodies included mouse monoclonal anti-human ALDH1A1 (1:1000) (BD Pharmingen, San Diego, CA), rabbit polyclonal anti-human MMP2 (1:500) (ZSGB-BIO ORIGENE, Beijing, China), mouse monoclonal anti-human MMP7 (1:500) (ZSGB-BIO ORIGENE, Beijing, China), and rabbit polyclonal anti-human MMP9 (1:500) (ZSGB-BIO ORIGENE, Beijing, China). After the application of horseradish peroxidase-labeled corresponding secondary antibodies, chemiluminescence was detected by SuperSignal West Pico (Pierce, PA) and Image Quant 5.0 was applied for quantification.
Cell culture
The human glioma cell line U251 was purchased from Cell Bank of Shanghai Institute of Cell Biology, Chinese Academy of Sciences. The primary glioma cell line was made from the glioblastoma sample surgical removed from a female patient, 48 years old. Both cell lines were cultured in Dulbecco’s modified Eagle’s medium (Gibco, USA) containing 10% fetal bovine serum at 37°C in humidified air with 5% CO2.
Colony formation assay
Four-hundred viable glioblastoma cells from U251 cell line or primary tumor cells per well were seeded in each well of 6-well plates and cultured in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal bovine serum. After incubation for ten days, colonies that contained more than 50 cells were counted and stained by crystal violet.
CCK8 assay
FACS-sorted ALDH1A1+ and ALDH1A1- tumor cells from U251 or primary glioblastoma cells were plated in 96-well plates with 100 μl DMEM containing 10% FBS at 1,000 cells per well. Cell Counting Kit-8 (CCK8) assays was employed for quantitation of cell viability. Briefly, one day after culture, 10 μl of CCK8 solution was added to each well and incubated at 37°C for 2 h. OD value at 450 nm for each well was then recorded by spectrophotometer for continuous eight days.
Invasion assay
Invasion transwell with aperture 8 µm (Corning Millipore BD) was coated with 10 µl of Matrigel™/DMEM (1:1, v/v) at 37°C for 30 min. ALDH1A1+ and ALDH1A1- glioma cells were both seeded in the upper chambers of transwell at the density of 1.5×104 in 200 µl of serum-free DMEM. The lower chambers were filled with 500 µl of DMEM with 10% fetal bovine serum. After 48 h incubation, the filter membranes were fixed with 4% formaldehyde for 15 min. Next, the cells on the upper chamber were scraped by cotton swab, and the high invasiveness cells on the lower chambers were stained with crystal violet for 15 min. Finally, the high invasiveness cells were counted in ten different microscopic visual areas at 400 magnification.
Animal survival time assay
The tumor cells of ALDH1A1+ and ALDH1A1- from U251 cell line were orthotopically transplanted into the brains of 6-week-old female NOD-SCID mice with 5×104 cells per mouse (n = 6). The survival time was recorded.
Statistical analyses
Unpaired t test was applied to compare the expression of ALDH1A1 with clinicopathological characteristics of glioma by using GraphPad Prism 5 (San Diego, CA, USA). The SPSS statistical software package (standard version 17.0, Chicago, IL, USA) was used to analyze the relationship between the expression of ALDH1A1 and overall survival (OS) and disease-free survival (DFS) of the patients. Cox proportional hazards regression model was employed for multivariate survival analysis. A significance of 5% was adopted. P-value < 0.05 was considered statistically significant.
Results
ALDH1A1 expression is correlated with glioma histological grade and predicts worse prognosis
We applied IHC staining to evaluate the expression of ALDH1A1 in the primary glioma specimens from 237 patients. As shown in Figure 1A, ALDH1A1 staining of tumor cells showed a punctuated intracellular morphology. A significantly heterogeneity in staining patterns was observed in the tumor cells. The staining intensity and positive area were fairly variable in different grade glioma. We quantitatively evaluated the surgical specimens of 117 low grade glioma patients (LGG) (WHO I-II) and 120 high grade glioma (HGG) patients (WHO III-IV). ALDH1A1 overexpression with IHC staining score over 5 was found respectively in 10 (8.5%) of 117 LGG patients and 41 (34.2%) of 120 (HGG) patients (WHO III-IV) (Figure 1A). The overall expression scores were 3.92 ± 3.80 in 120 HGG patients as compared with 2.00 ± 2.57 in 117 LGG ones (WHO I-II) (Figure 1B), which is statistically significant. Furthermore, the specimens with high expression of ALDH1A1 also exhibited high level of proliferation protein Ki67. ALDH1A1 level was statistically correlated with Ki67 (Figure 1C).
Figure 1.
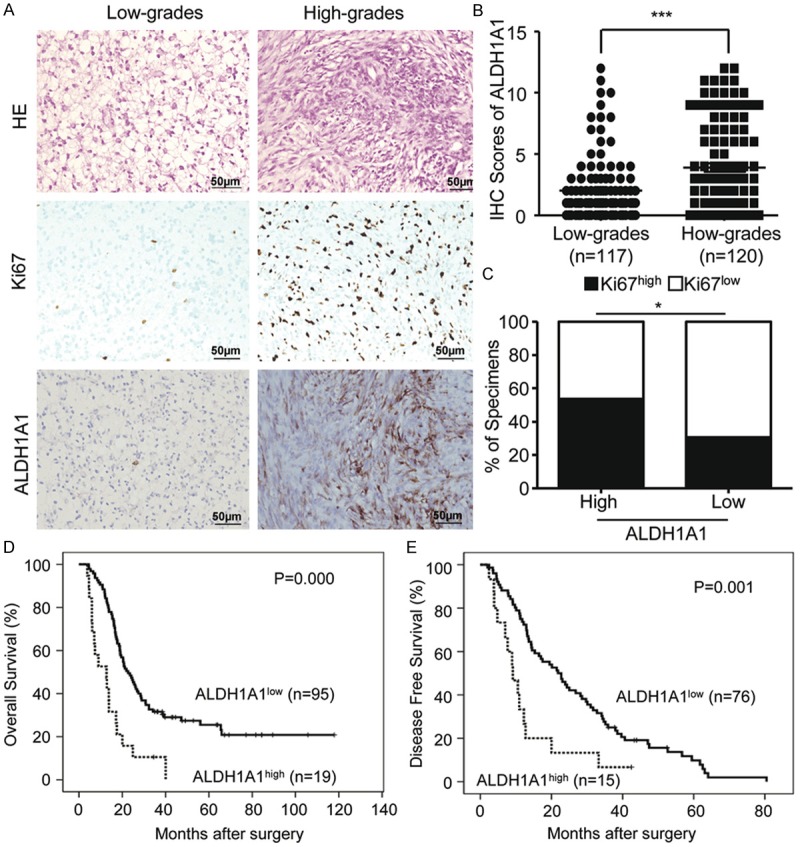
Correlations between clinicopathology characteristics and ALDH1A1 expression on the specimens from 237 glioma patients by immunohistochemical and Kaplan-Meier estimation. A. No or less ALDH1A1+ and Ki67+ cells are seen in low-grade gliomas (LGGs) (WHO I-II), but more ALDH1A1+ and Ki67+ cells are found in high-grade gliomas (HGGs) (WHO III-IV). B. Quantitative analysis of ALDH1A1+ cells in LGGs and HGGs shows more ALDH1A1+ cells in HGGs than those in LGGs. C. ALDH1A1 levels are correlated with Ki67 expression in human glioma specimens. D. Kaplan-Meier estimation performed on all of the glioma patients indicates that the patients with ALDH1A1+ expression have shorter overall survival time than the patients with ALDH1A1- expression (ALDH1A1+, n = 19 and ALDH1A1-, n = 95). (P < 0.001, log-rank test). E. Kaplan-Meier estimation performed on all of the glioma patients indicates that the patients with ALDH1A1+ expression have shorter disease-free survival time than those with ALDH1A1- expression (ALDH1A1+, n = 15 and ALDH1A1-, n = 76) (P < 0.001, log-rank test). *indicates significant difference (P < 0.05), ***P < 0.001.
The correlation of ALDH1A1 expression with patient survival was then analyzed in the glioma patients who had follow-up data. We found that median survival time of ALDH1A1 low expression patients was 31 months (ranging from 4 to more than 118 months), whereas the median survival of ALDH1A1 high expression patients was 14 months (ranging from 4 to more than 40 months). The estimated overall survival rate of low score group of glioma patients (n = 95) was significantly better than that of high score group of glioma patients (n = 19) (P = 0.000) (Figure 1D). Disease-free survival rate was remarkably different between the patients with low and ones with high expression of ALDH1A1 (P = 0.001) (Figure 1E). By using Cox proportional hazards model, we performed multivariate analysis to evaluate the independent predictive value of ALDH1A1 expression for OS as shown in Table 2. We found that ALDH1A1 expression was the independent negative prognostic factor for both overall survival (HR = 1.917; 95% CI = 1.131-3.436; P = 0.017) and histological grade (HR = 5.394; 95% CI = 2.790-10.431; P = 0.000). Therefore, ALDH1A1 can be used as a biomarker for patients’ outcome, and its detection in the glioma specimens can predict patients prognosis.
Table 2.
Univariate and multivariate analysis of different prognostic parameters
| Variable | All cases | Univariate analysisa | Multivariate analysisb | ||
|---|---|---|---|---|---|
|
| |||||
| Mean survival (months) | p Value | HR (95% CI) | p Value | ||
| Sex | 0.344 | 0.072 | |||
| Male | 66 | 27 | 1.0 | ||
| Female | 48 | 30 | 0.661 (0.421 to 1.038) | ||
| Age at surgery | 0.000 | 0.194 | |||
| ≤ 42.6 yc | 52 | 50 | 1.0 | ||
| > 42.6 y | 62 | 19 | 1.396 (0.844 to 2.309) | ||
| Histological grade (WHO) | 0.000 | 0.000 | |||
| I-II | 39 | 48 | 1.0 | ||
| III-IV | 75 | 18 | 5.394 (2.790 to 10.431) | ||
| ALDH1A1 Expression | 0.000 | 0.017 | |||
| Low Score (0-5) | 95 | 31 | 1.0 | ||
| High Score (6-12) | 19 | 14 | 1.917 (1.131 to 3.436) | ||
Log-rank test;
Cox regression model;
Mean age.
HR, hazard ratio; CI, confidence interval.
High expression of ALDH1A1 is correlated with molecular classification of GBM
Molecular classification of glioblastoma has been suggested in the pathological examination of GBM and the individualized treatment of the patients [5,20]. To address whether ALDH1A1 expression is correlated with molecular classification of GBM, we performed IHC staining to analyze the individual expression of TP53, PDGFRA and EGFR in our series of specimens from 72 GBM patients (Figure 2A). Overall, 36 from 72 samples (50%) were defined as “Pronural-Like” (PNL) subtype as characterized by TP53- and/or PDGFRA-positive staining, which included 4 (12.5%) samples with ALDH1A1 overexpression. Other 13 (18.1%) samples belonged to “Classical-Like” (CL) subtype with EGFR-positive but TP53- and PDGFRA-negative staining, and most of them (11 samples) (84.6%) were ALDH1A1 overexpression. The remaining 23 (31.9%) samples that did not fit any of these criteria were classified as “Other”, and only 5 from them (21.7%) exhibited ALDH1A1 overexpression. Statistical analysis revealed that the expression of ALDH1A1 was significantly different among theses subtypes (Figure 2B, P = 0.0001). These results indicate that ALDH1A1 expression is correlated with the malignance and molecular classification of GBM.
Figure 2.
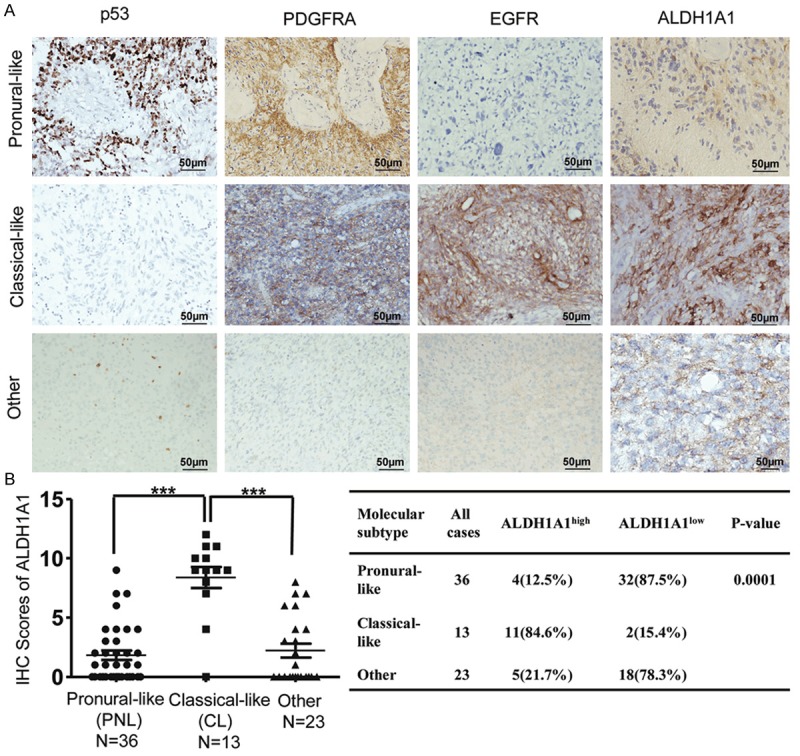
Correlation between molecular classification of GBM and ALDH1A1 expression in the specimens from 72 GBM patients. A. IHC detection of ALDH1A1 expression in each molecular type of GBM reveals that ALDH1A1 is higher in “classical-like” GBM as compare to the other types of “pronural-like” and “other” ones. B. Quantitative analysis of correlation of molecular classification of GBM with ALDH1A1 expression. ***indicates significant difference (P < 0.001).
Detection of ALDH1A1+ invasive cells is informative for prediction of glioma patients’ prognosis
Invasion is one of important features of glioma [21]. A correlation of ALDH1A1 expression with invasiveness of glioma cells was indicated by our immunohistochemical analysis of glioma patients’ specimens with clear boundary where the number of ALDH1A1+ cells was increased in the invasive frontier area as compared with the non-invasive frontier area (Figure 3A). Quantitatively, the percentage of ALDH1A1+ glioma cells in the invasive frontier area was significantly higher than that in the non-invasive frontier area (49% ± 6% vs. 14% ± 3%, p < 0.0001). Importantly, by further examination of the specimens with clear boundary from 20 glioma patients, we noticed that there were some ALDH1A1+ cells located in the “normal” brain tissue adjacent to the invasive frontier (Figure 3B). Furthermore, Kaplan-Meier analysis indicated that the patients with more numbers of these located ALDH1A1+ cells had worse prognosis than those with less or negative ones (Figure 3C). Thus, our data indicate that detection of ALDH1A1+ cells located in the “normal” brain tissue adjacent to the invasive frontier is not only informative for prognosis prediction but also instructive for designing more effective regimens to treat glioma patients.
Figure 3.
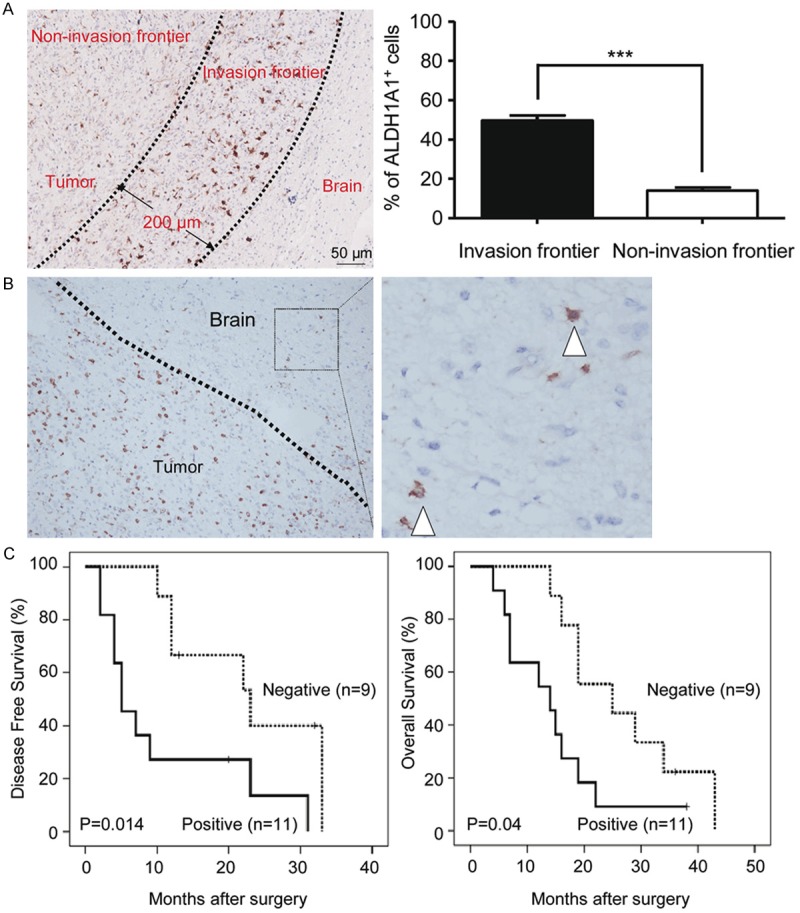
IHC analysis of ALDH1A1+ cells’ distribution in glioma specimens. A. Representative IHC images show the difference in the distribution of ALDH1A1+ cells between the invasive frontier area and the non-invasive frontier area. Two extending lines from the glioma are drawn to distinguish the invasive frontier area and non-invasive frontier area (left); Quantitative analysis reveals that ALDH1A1+ cells are increased in the invasive frontier area as compared to the non-invasive frontier area (right). B. Representative IHC images show ALDH1A1+ cells in the “normal” brain tissue adjacent to the invasive frontier. C. Kaplan-Meier estimation performed on some of the glioma patients indicates that the patients (n = 11) with ALDH1A1+ cells in the “normal” brain tissue adjacent to the invasive frontier have shorter disease-free survival time than those (n = 9) with less or no ALDH1A1+ cells. (P < 0.001, log-rank test). Data in A presented as the mean ± SEM. The Student’s t-test was used for between ALDH1A1high and ALDH1A1low analysis. ***indicates significant difference (P < 0.001).
ALDH1A1+ glioma cells possess higher capabilities of colony formation and proliferation
To reveal the mechanism(s) underneath the correlation of high expression of ALDH1A1 with poorer prognosis of glioma patients, we performed both in vitro and in vivo studies by using U251 glioma cell line and one primary glioma cell line established from a female patient. We first examined the proportion of ALDH1high cells in the glioma cell line U251, and found that ALDH1high cells accounted for 6.9% ± 2.1% of the whole-cell population (n = 14) (Figure S1A). Similar results were also obtained that ALDH1high cells accounted for 4.2% ± 1.9% of the whole-cell population (n = 14) from primary glioma cells (Figure S1A). We then measured expressions of various subtypes of ALDH1 family on the cells sorted through ALDEFLUOR™ kit. Analysis by qRT-PCR revealed that ALDH1high cells highly expressed ALDH1A1 gene as compared with ALDH1low ones, whereas the differences of other subtypes of ALDH family between them were not obvious (Figure S1B). Thus, ALDH1A1 was the main molecular subtype of ALDH family in the sorted ALDH1high glioma cells, and we named this group of cells as ALDH1A1+ glioma cells and the sorted ALDH1low glioma cells as ALDH1A1- glioma cells.
We then examined the bio-behaviors of the sorted ALDH1A1+ cells and found that these cells possessed higher capabilities of colony formation as compared with ALDH1A1- ones (Figure 4A). The percentage of colonies formed by ALDH1A1+ cells were higher than those formed by ALDH1A1- cells in U251 (16.3% ± 3.2% vs. 7.1% ± 1.5%, P < 0.05) and primary glioblastoma cells (8.4% ± 0.9% vs. 4.1% ± 0.6%, P < 0.05). Proliferation assay revealed that ALDH1A1+ cells sorted from either U251 or primary glioma cells were highly proliferated (Figure 4B, P < 0.05). Furthermore, orthotopic xenotransplantation murine model studies demonstrated that SCID mice transplanted with ALDH1A1+ cells were alive for a period from 48 to 78 days, whereas ALDH1A1- cells-injected SCID mice lived for a statistically significant longer period ranging from 90 to 121 days (Figure 4C; P = 0.001). These data suggest that ALDH1A1+ cells play a major role for glioma progression.
Figure 4.
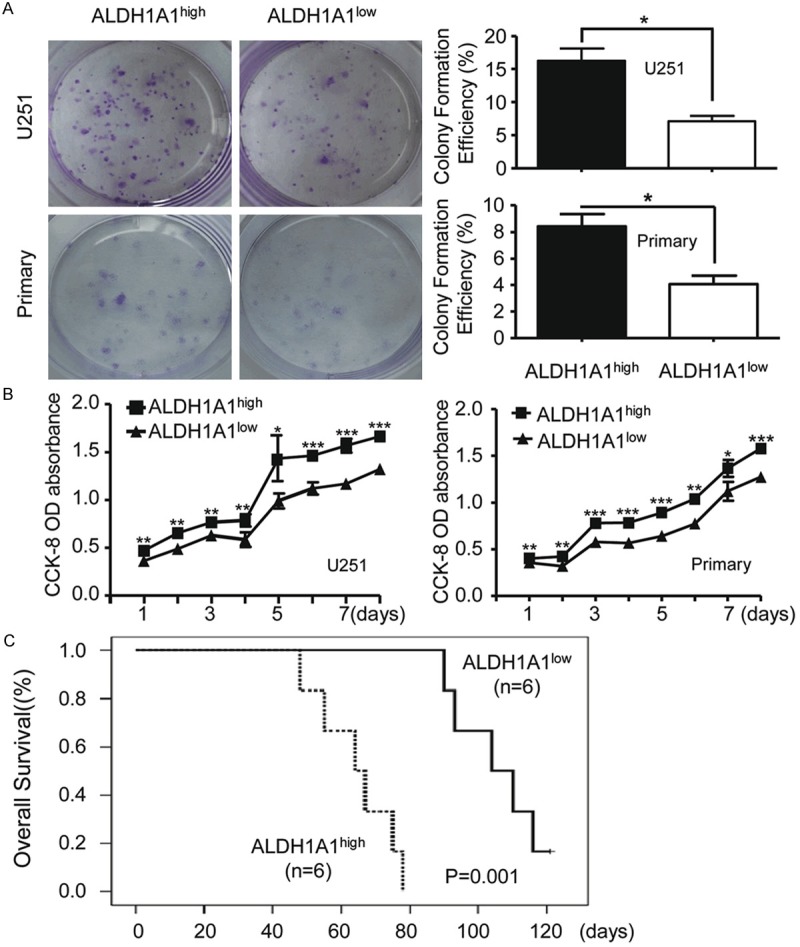
ALDH1A1 glioma cells are capable of colony formation and proliferation. A. Colony formation of FACS-sorted ALDH1A1+ and ALDH1A1- cells yielded from U251 and primary glioblastoma cells (left). Quantitative analysis shows significant difference between colony formation by ALDH1A1+ and that by ALDH1A1- cells (right). B. Quantitative analysis by cell counting kit-8 (CCK8) assay shows significant difference between proliferation capability by ALDH1high and that by ALDH1low cells isolated from U251 and primary glioma cells. C. Kaplan Meier survival curve of overall survival (OS) indicates the association between FACS-sorted ALDH1A1+ and ALDH1A1- cells of U251 cells in SCID. Animals xenografted with ALDH1A1+ cells had a mean OS of 64 days as compared to 89 days for those with ALDH1A1- ones. (P = 0.001, log-rank test). Each value is the mean ± SEM. The Student’s t-test was used for between ALDH1A1high and ALDH1A1low analysis. *indicates significant difference (P < 0.05), **P < 0.01, ***P < 0.001.
ALDH1A1+ glioma cells possess high invasion capability
Pathological studies of orthotopically transplanted tumors demonstrated that ALDH1A1+ tumor cells exhibited stronger invasiveness and the boundary of transplanted tumor formed from ALDH1A1+ tumor cells was less clear than that formed from ALDH1A1- ones (Figure 5A). To explore the corresponding mechanism, we next examined the invasive capabilities of FCAS-sorted ALDH1A1+ cells. As compared to ALDH1A1- ones, ALDH1A1+ cells respectively sorted from U251 and primary glioma cells had a greater invasiveness (Figure 5B). Gene expression analysis further revealed that ALDH1A1+ tumor cells sorted from either U251 or primary glioma expressed higher levels of MMP2, MMP7 and MMP9 (Figure 5C). Meanwhile, ALDH1A1+ cells had more abundance of MMP2, MMP7 and MMP9 as compared to ALDH1A1- cells (Figure 5D). These results demonstrate that the highly invasive capabilities of ALDH1A1+ cells are associated with the increased levels of MMPs expression.
Figure 5.
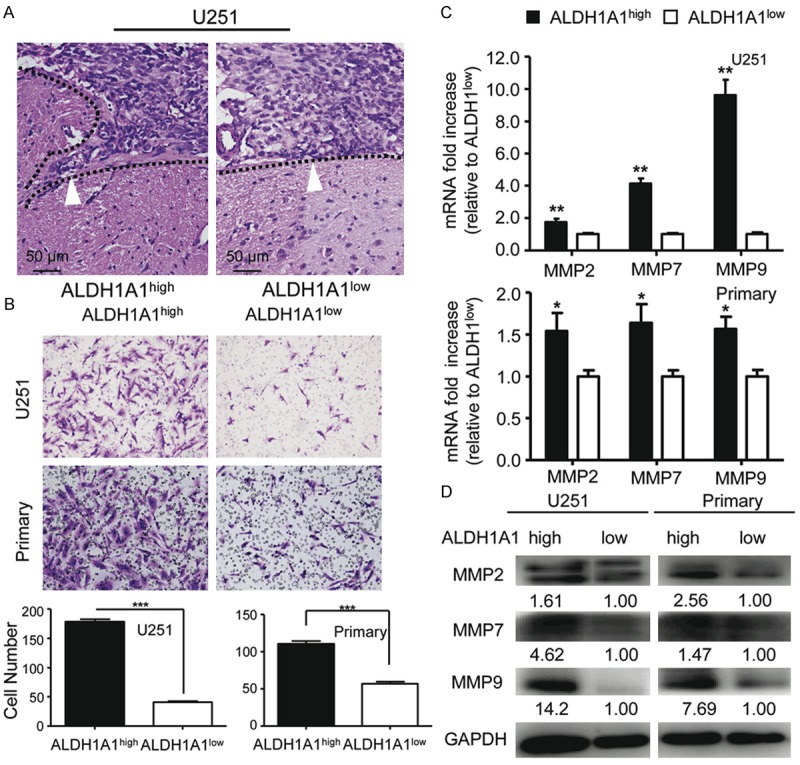
Analysis of invasion of ALDH1A1+ and ALDH1A1- cells and the expression of invasion-associated genes. A. Pathological comparison of the xenografted tumor derived from ALDH1A1+ cells with that from ALDH1A1- cells. B. ALDH1A1+ cells possess higher invasive capability as compared to ALDH1A1- ones. Representative in vitro invasion images of transwell assays (up). Quantitative analysis shows significant difference between invasion by ALDH1A1+ and that by ALDH1A1- cells (low). C. Analysis by qRT-PCR showed high levels of MMP2, MMP7 and MMP9 expression in ALDH1A1+ cells as compared to those in ALDH1A1- cells. D. Western blot analysis indicates more abundance of MMP2, MMP7 and MMP9 expression in ALDH1A1+ as compared to ALDH1A1- cells. Results are expressed as the mean ± SEM. Values shown were normalized to GAPDH. Data were evaluated by Student’s t-test between ALDH1A1high and ALDH1A1low (*p < 0.05, **p < 0.01, ***p < 0.001).
Discussion
Here, we evaluated the clinicopathological characteristics and the prognostic implications of ALDH1A1 expression on the specimens from 237 glioma cases. Our study demonstrate that (1) ALDH1A1 is differently expressed in the specimens of LGG and HGG, (2) ALDH1A1 expression is correlated with the patients’ clinicopathological characteristics including histopathology grade of glioma, (3) ALDH1A1 expression is associate with the molecular classification of GBM, (4) ALDH1A1 is an independent prognostic indicator for the outcome and disease-free survival of glioma patients, (5) the patients with more numbers of ALDH1A1+ cells located in the “normal” brain tissue adjacent to the invasive frontier have worse prognosis than those with less or negative ones, and (6) ALDH1A1+ glioma cells are highly invasive with strong capabilities of proliferation and colony formation. Thus, ALDH1A1+ cells may play an important role in the progression of glioma.
Glioma is the commonest neuroepithelial brain tumor and the prognoses of patients are poor with only 4 to 6 months of population-based survival [22]. ALDH is a family of enzymes assisted by either NAD or NADP as a coenzyme for the metabolism of aldehydes to their corresponding carboxylic acids. The ALDH genes can be subdivided into several families. ALDH1A1, a member of ALDH family, contributes primarily to the biosynthesis of retinoic acid in liver. It has been reported that ALDH1A1 overexpression in some malignant epithelial carcinomas is associated with poor outcomes [23]. S. Alexandra Adam et al. reported that ALDH1A1 expressed and functioned in the early stages of brain development and strong expression of ALDH1A1 was correlated with better survival of glioblastoma patients in a study of 93 GBM cases [17]. Contradictorily, Andrea Schäfer et al. found that among 70 GBM patients, the prognosis of patients with high expression of ALDH1A1 was poor as compared to that of patients with low expression [18]. We speculate that the reasons for the inconsistency may be due to the difference in the number of cases and the race of patients among the studies. Nevertheless, our current study and others indicate that ALDH1A1 expression is inversely correlated with glioma patients’ survival.
Notably, our study demonstrates that the higher the expression of ALDH1A1 on the tumor cells in the boundary region of glioma tumor tissue, the worse the outcome of patients. Furthermore, if more numbers of ALDH1A1+ cells are found located in the “normal” brain tissue adjacent to the invasive frontier on the specimen of a glioma patient, the prognosis of this patient will be much worse so that the dose and the frequency of concurrent chemo-radiation should be increased to strengthen the treatment. Mechanistically, our study reveal that ALDH1A1+ glioma cells are highly invasive due to high levels of MMP2, MMP7 and MMP9. MMPs are well known to be capable of degrading extracellular matrix proteins to promote tumor cell invasion [24]. Thus, ALDH1A1+ glioma cells, by expressing high levels of MMPs, contribute to the invasion of glioma.
The molecular classification of GBM has been suggested for its application in the molecular diagnosis and guidance for treatment of patients as well as prediction of their prognoses. Based on the functional annotation of signature genes generated from high-scale genome-wide profiling studies by gene expression microarrays, three subclasses, i.e., proneural, mesenchymal and proliferative, or four subclasses termed as proneural, mesenchymal, neural, and classical, have been identified for GBM subgroups [20]. An alternative simplified IHC-based approach to quantitatively examine p53, PDGFRA, and EGFR on the surgical specimens reveals two GBM subtypes: the “Classical-like” (CL) subtype which is characterized by EGFR-positive but p53- and PDGFRA-negative staining, and the “Proneural-like” (PNL) subtype which is characterized by p53- and/or PDGFRA-positive staining. The prognosis of “CL” GBM patients is much worse than that of “PNL” and “other” GBM patients. By this alternative IHC method and detection of ALDH1A1, we found that “CL” GBM group had more cases of high expression of ALDH1A1, suggesting that IHC detection of ALDH1A1 may be useful as an additional simple, convenient and reliable way to predict the prognosis of GBM patients.
Currently, several clinical trials targeting some special tumor markers by using various approaches, ranging from immunotherapy to an-tagonists and molecular manipulation of tumor marker expression, are under way in the treatment of some malignancies and might also be broadly applicable to glioma treatment. We speculate that ALDH1A1 will be a potential target for glioma therapy. Meanwhile, our study suggests that ALDH1A1 may be useful as a molecular marker for prognosis prediction of glioma patients.
Acknowledgements
This study was supported by grants from National Basic Research Program of China (973 Program, No. 2010CB529400) and the Outstanding Youth Science Foundation of Chongqing (No. CSTC2013JCYJJQ1003).
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Stupp R, Hegi ME, Mason WP, van den Bent MJ, Taphoorn MJ, Janzer RC, Ludwin SK, Allgeier A, Fisher B, Belanger K, Hau P, Brandes AA, Gijtenbeek J, Marosi C, Vecht CJ, Mokhtari K, Wesseling P, Villa S, Eisenhauer E, Gorlia T, Weller M, Lacombe D, Cairncross JG, Mirimanoff RO. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 2009;10:459–466. doi: 10.1016/S1470-2045(09)70025-7. [DOI] [PubMed] [Google Scholar]
- 2.Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Siu IM, Gallia GL, Olivi A, McLendon R, Rasheed BA, Keir S, Nikolskaya T, Nikolsky Y, Busam DA, Tekleab H, Diaz LA Jr, Hartigan J, Smith DR, Strausberg RL, Marie SK, Shinjo SM, Yan H, Riggins GJ, Bigner DD, Karchin R, Papadopoulos N, Parmigiani G, Vogelstein B, Velculescu VE, Kinzler KW. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Noushmehr H, Weisenberger DJ, Diefes K, Phillips HS, Pujara K, Berman BP, Pan F, Pelloski CE, Sulman EP, Bhat KP, Verhaak RG, Hoadley KA, Hayes DN, Perou CM, Schmidt HK, Ding L, Wilson RK, Van Den Berg D, Shen H, Bengtsson H, Neuvial P, Cope LM, Buckley J, Herman JG, Baylin SB, Laird PW, Aldape K. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell. 2010;17:510–522. doi: 10.1016/j.ccr.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kawaguchi A, Yajima N, Tsuchiya N, Homma J, Sano M, Natsumeda M, Takahashi H, Fujii Y, Kakuma T, Yamanaka R. Gene expression signature-based prognostic risk score in patients with glioblastoma. Cancer Sci. 2013;104:1205–1210. doi: 10.1111/cas.12214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Le Mercier M, Hastir D, Moles Lopez X, De Neve N, Maris C, Trepant AL, Rorive S, Decaestecker C, Salmon I. A simplified approach for the molecular classification of glioblastomas. PLoS One. 2012;7:e45475. doi: 10.1371/journal.pone.0045475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vladimirova V, Waha A, Luckerath K, Pesheva P, Probstmeier R. Runx2 is expressed in human glioma cells and mediates the expression of galectin-3. J Neurosci Res. 2008;86:2450–2461. doi: 10.1002/jnr.21686. [DOI] [PubMed] [Google Scholar]
- 7.Kawaguchi A, Yajima N, Komohara Y, Aoki H, Tsuchiya N, Homma J, Sano M, Natsumeda M, Uzuka T, Saitoh A, Takahashi H, Sakai Y, Fujii Y, Kakuma T, Yamanaka R. Identification and validation of a gene expression signature that predicts outcome in malignant glioma patients. Int J Oncol. 2012;40:721–730. doi: 10.3892/ijo.2011.1240. [DOI] [PubMed] [Google Scholar]
- 8.Kawaguchi A, Iwadate Y, Komohara Y, Sano M, Kajiwara K, Yajima N, Tsuchiya N, Homma J, Aoki H, Kobayashi T, Sakai Y, Hondoh H, Fujii Y, Kakuma T, Yamanaka R. Gene expression signature-based prognostic risk score in patients with primary central nervous system lymphoma. Clin Cancer Res. 2012;18:5672–5681. doi: 10.1158/1078-0432.CCR-12-0596. [DOI] [PubMed] [Google Scholar]
- 9.Vasiliou V, Thompson DC, Smith C, Fujita M, Chen Y. Aldehyde dehydrogenases: from eye crystallins to metabolic disease and cancer stem cells. Chem Biol Interact. 2013;202:2–10. doi: 10.1016/j.cbi.2012.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheung AM, Wan TS, Leung JC, Chan LY, Huang H, Kwong YL, Liang R, Leung AY. Aldehyde dehydrogenase activity in leukemic blasts defines a subgroup of acute myeloid leukemia with adverse prognosis and superior NOD/SCID engrafting potential. Leukemia. 2007;21:1423–1430. doi: 10.1038/sj.leu.2404721. [DOI] [PubMed] [Google Scholar]
- 11.Storms RW, Green PD, Safford KM, Niedzwiecki D, Cogle CR, Colvin OM, Chao NJ, Rice HE, Smith CA. Distinct hematopoietic progenitor compartments are delineated by the expression of aldehyde dehydrogenase and CD34. Blood. 2005;106:95–102. doi: 10.1182/blood-2004-09-3652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hess DA, Wirthlin L, Craft TP, Herrbrich PE, Hohm SA, Lahey R, Eades WC, Creer MH, Nolta JA. Selection based on CD133 and high aldehyde dehydrogenase activity isolates long-term reconstituting human hematopoietic stem cells. Blood. 2006;107:2162–2169. doi: 10.1182/blood-2005-06-2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang EH, Hynes MJ, Zhang T, Ginestier C, Dontu G, Appelman H, Fields JZ, Wicha MS, Boman BM. Aldehyde dehydrogenase 1 is a marker for normal and malignant human colonic stem cells (SC) and tracks SC overpopulation during colon tumorigenesis. Cancer Res. 2009;69:3382–3389. doi: 10.1158/0008-5472.CAN-08-4418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jiang F, Qiu Q, Khanna A, Todd NW, Deepak J, Xing L, Wang H, Liu Z, Su Y, Stass SA, Katz RL. Aldehyde dehydrogenase 1 is a tumor stem cell-associated marker in lung cancer. Mol Cancer Res. 2009;7:330–338. doi: 10.1158/1541-7786.MCR-08-0393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van den Hoogen C, van der Horst G, Cheung H, Buijs JT, Lippitt JM, Guzman-Ramirez N, Hamdy FC, Eaton CL, Thalmann GN, Cecchini MG, Pelger RC, van der Pluijm G. High aldehyde dehydrogenase activity identifies tumor-initiating and metastasis-initiating cells in human prostate cancer. Cancer Res. 2010;70:5163–5173. doi: 10.1158/0008-5472.CAN-09-3806. [DOI] [PubMed] [Google Scholar]
- 16.Croker AK, Goodale D, Chu J, Postenka C, Hedley BD, Hess DA, Allan AL. High aldehyde dehydrogenase and expression of cancer stem cell markers selects for breast cancer cells with enhanced malignant and metastatic ability. J Cell Mol Med. 2009;13:2236–2252. doi: 10.1111/j.1582-4934.2008.00455.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Adam SA, Schnell O, Poschl J, Eigenbrod S, Kretzschmar HA, Tonn JC, Schuller U. ALDH1A1 is a marker of astrocytic differentiation during brain development and correlates with better survival in glioblastoma patients. Brain Pathol. 2012;22:788–797. doi: 10.1111/j.1750-3639.2012.00592.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schafer A, Teufel J, Ringel F, Bettstetter M, Hoepner I, Rasper M, Gempt J, Koeritzer J, Schmidt-Graf F, Meyer B, Beier CP, Schlegel J. Aldehyde dehydrogenase 1A1--a new mediator of resistance to temozolomide in glioblastoma. Neuro Oncol. 2012;14:1452–1464. doi: 10.1093/neuonc/nos270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shi H, Chen S, Jin H, Xu C, Dong G, Zhao Q, Wang W, Zhang H, Lin W, Zhang J, Davidovic L, Yao L, Fan D. Downregulation of MSP58 inhibits growth of human colorectal cancer cells via regulation of the cyclin D1-cyclin-dependent kinase 4-p21 pathway. Cancer Sci. 2009;100:1585–1590. doi: 10.1111/j.1349-7006.2009.01223.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Z, Wang B, Shi Y, Xu C, Xiao HL, Ma LN, Xu SL, Yang L, Wang QL, Dang WQ, Cui W, Yu SC, Ping YF, Cui YH, Kung HF, Qian C, Zhang X, Bian XW. Oncogenic miR-20a and miR-106a enhance the invasiveness of human glioma stem cells by directly targeting TIMP-2. Oncogene. 2015;34:1407–19. doi: 10.1038/onc.2014.75. [DOI] [PubMed] [Google Scholar]
- 21.Yu SC, Xiao HL, Jiang XF, Wang QL, Li Y, Yang XJ, Ping YF, Duan JJ, Jiang JY, Ye XZ, Xu SL, Xin YH, Yao XH, Chen JH, Chu WH, Sun W, Wang B, Wang JM, Zhang X, Bian XW. Connexin 43 reverses malignant phenotypes of glioma stem cells by modulating E-cadherin. Stem Cells. 2012;30:108–120. doi: 10.1002/stem.1685. [DOI] [PubMed] [Google Scholar]
- 22.Kita D, Ciernik IF, Vaccarella S, Franceschi S, Kleihues P, Lutolf UM, Ohgaki H. Age as a predictive factor in glioblastomas: population-based study. Neuroepidemiology. 2009;33:17–22. doi: 10.1159/000210017. [DOI] [PubMed] [Google Scholar]
- 23.Deng S, Yang X, Lassus H, Liang S, Kaur S, Ye Q, Li C, Wang LP, Roby KF, Orsulic S, Connolly DC, Zhang Y, Montone K, Butzow R, Coukos G, Zhang L. Distinct expression levels and patterns of stem cell marker, aldehyde dehydrogenase isoform 1 (ALDH1), in human epithelial cancers. PLoS One. 2010;5:e10277. doi: 10.1371/journal.pone.0010277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roy R, Yang J, Moses MA. Matrix metalloproteinases as novel biomarkers and potential therapeutic targets in human cancer. J. Clin. Oncol. 2009;27:5287–5297. doi: 10.1200/JCO.2009.23.5556. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


