ABSTRACT
Human cytomegalovirus (HCMV) tegument protein pUL47 is an interaction partner of pUL48 and highly conserved among herpesviruses. It is closely associated with the capsid and has an important function early in infection. Here, we report a specific role of pUL47 in the tegumentation of capsids in the cytoplasm. A newly generated mutant virus (TB-47stop), in which expression of pUL47 is blocked, exhibited a severe impairment in cell-to-cell spread and release of infectivity from infected cells. Ultrastructural analysis of TB-47stop-infected cells clearly showed cytoplasmic accumulations of nonenveloped capsids that were only partially tegumented, indicating that these capsids failed to complete tegumentation. Nevertheless, these accumulations were positive for HCMV inner tegument proteins pp150 and pUL48, suggesting that their attachment to capsids occurs independently of pUL47. Despite these morphological alterations, fully enveloped virus particles were found in the extracellular space and at the viral assembly complex (vAC) of TB-47stop-infected cells, indicating that pUL47 is not essential for the generation of virions. We confirmed findings that incorporation of pUL48 into virions is impaired in the absence of pUL47. Interestingly, pUL47 exhibited a strong nuclear localization in transfected cells, whereas it was found exclusively at the vAC in the context of virus infection. Colocalization of pUL47 and pUL48 at the vAC is consistent with their interaction. We also found a shift to a more nuclear localization of pUL47 when the expression of pUL48 was reduced. Summarizing our results, we hypothesize that pUL48 directs pUL47 to the vAC to promote tegumentation and secondary envelopment of capsids.
IMPORTANCE Generation of infectious HCMV particles requires an organized and multistep process involving the action of several viral and cellular proteins as well as protein-protein interactions. A better understanding of these processes is important for understanding the biology of HCMV and may help to identify targets for antiviral intervention. Here, we identified tegument protein pUL47 to function in tegumentation and proper trafficking of capsids during late phases of infection. Although pUL47 is not essential for the generation and release of infectious virions, its absence led to massive accumulations of partially tegumented capsids at the cell periphery. Detection of pUL48 at these accumulations indicated a pUL47-independent attachment of pUL48 to the capsid. On the other hand, localization of pUL47 to the vAC during infection appeared to be dependent on tegument protein pUL48, which suggests an intricate interplay of these proteins for normal generation of infectious virus progeny.
INTRODUCTION
Virions of human cytomegalovirus (HCMV) consist of the four major components that are characteristic for herpesvirus particles: the double-stranded DNA (dsDNA) genome that is packaged in an icosahedral capsid, the tegument, and the viral envelope. The tegument is a layer of viral and cellular proteins that has a critical role for the generation of virions by connecting the envelope with the capsid. It is a very complex structure that in the case of HCMV consists of more than 38 different viral proteins, a number of cellular proteins, and mRNAs (1–3). HCMV capsids acquire most of their tegument and their envelope at the cytoplasmic viral assembly complex (vAC). The vAC is characteristic of HCMV-infected cells and the result of a virally induced reorganization of Golgi and endosomal membranes (4–13). Many tegument proteins have been found to accumulate at the vAC during infection (10, 14–23), which emphasizes their role for virion assembly. In addition to their function as structural virion components, tegument proteins act at several steps of the viral infection cycle. These include initiation and regulation of viral replication, release of viral DNA into the nucleus, immune evasion, intracellular trafficking of virus particles, and assembly of infectious progeny (24–32). Formation of the tegument during virion assembly is a poorly understood process. There is increasing evidence of an underlying intricate network of protein-protein interactions that regulates tegument formation, which starts with the attachment of viral proteins to the capsid. The tegument is divided into a capsid-proximal, or inner, and a capsid-distal, or outer, tegument layer. HCMV tegument proteins of the inner tegument are tightly associated with the capsid and therefore likely candidates to regulate the first steps of tegumentation (33). These proteins include the HCMV proteins pp150, pUL48, and pUL47. Whereas pp150 is an HCMV-specific tegument protein, for pUL48, known as the large tegument protein, and pUL47, homologous proteins have been identified in all herpesvirus subfamilies (34). The UL32-encoded pp150 has been shown to function in stabilizing capsids and in their intracellular trafficking (14, 16, 35, 36). While pUL48 is essential for viral replication, pUL47 belongs to the class of HCMV proteins that augments viral growth (37, 38). A mutant virus with a large substitution of the UL47 open reading frame (ORF) has been shown to accumulate 100-fold-reduced extracellular virus titers (29). pUL47, with a molecular weight of 110 kDa, is a component of the inner tegument of mature virions but is also found in dense bodies (DB) and noninfectious enveloped particles (NIEPS) (39). It is part of a larger protein complex in infection consisting of at least pUL47, pUL48, pUL69, and the UL86-encoded major capsid protein (MCP) (29). pUL47 is self-interacting to form oligomers and is also directly interacting with the large tegument protein pUL48 (40). Incorporation of pUL48 into virions is impaired in the absence of pUL47, which results in virions defective for both pUL47 and pUL48. These virions exhibit a defect after virus entry and prior to immediate early (IE) gene transcription, which may occur at the level of genome release into the nucleus (29).
The homologs of pUL47 and pUL48 in the alphaherpesviruses herpes simplex virus 1 (HSV-1) and pseudorabiesvirus (PrV) are encoded by the UL37 and UL36 ORFs and are known as pUL37 and pUL36. Several functions have been ascribed to pUL37 during early and late phases of infection (41–47). Upon entry, pUL37 remains associated with intracytoplasmic capsids to facilitate together with pUL36 their transport along microtubules to the nucleus (48–54). Although pUL37 is not required, its presence accelerates capsid transport to the nucleus (53, 54). In addition, pUL37 is involved in modulating cellular responses by triggering NF-κB activation through tumor necrosis factor (TNF) receptor-associated factor 6 (55). During the late phase of infection, pUL37 regulates virion maturation, capsid trafficking, and egress (51, 52, 56–60). In infections with mutant viruses lacking pUL37, nonenveloped capsids accumulate in the cytoplasm as a result of either a complete block of secondary envelopment, as seen in HSV-1, or a drastic but not lethal defect with only little envelopment, as observed in PrV. Consequently, egress of infectious virions depends highly on pUL37 in HSV-1 and PrV (51, 60–62). Attachment of pUL37 to capsids is required for directing them to the trans-Golgi network (TGN), where secondary envelopment occurs. Capsids are targeted to the TGN by an interaction of pUL37 with cellular protein PBAG1 (57, 63). Binding of pUL37 to capsids is regulated by the large tegument protein pUL36 and requires their interaction (59, 64). Attachment of pUL37 seems to start already in the nucleus (64–67). The interaction of pUL36 and pUL37 is highly conserved in alphaherpesviruses and an essential requirement for virion morphogenesis (41–47). Functions of the individual complex partners are strongly intertwined with their complex formation. The localization of pUL37 at the TGN is independent of capsids and requires the presence pUL36 (58, 60). In turn, the interaction with pUL37 stabilizes pUL36 (47).
The exact role of HCMV pUL47 during the late phase of infection is not clear, which is surprising in the light of the strong conservation of this protein and its importance for virion morphogenesis in other herpesviruses. To gain insight into the role of HCMV pUL47 during the late phase of infection, we generated a mutant virus, TB-47stop, that is unable to express pUL47. This mutant virus was used to evaluate viral growth in the absence of pUL47 and to determine its role in tegumentation and secondary envelopment of HCMV by electron microscopy after high-pressure freezing and freeze substitution. Growth properties of our TB-47stop virus were similar to those of a previous UL47 mutant (29) with the addition of a severe defect in cell-to-cell spread and an impaired egress of virus particles. Ultrastructural examination of viral morphogenesis and immunofluorescence revealed that pUL47 is involved in the processes of tegumentation, proper capsid trafficking, and their trapping at the vAC, resulting in the accumulation of partially tegumented and nonenveloped capsids in the cytoplasm when pUL47 is missing. Furthermore, generation of mutant viruses with Flag-tagged pUL47 enabled the detection of pUL47 in immunofluorescence analysis. The localization of pUL47 at the vAC was independent of the presence of capsids and was regulated by pUL48. Collectively, these results offer new insights into the function of pUL47 during the late phase of HCMV infection and into the regulation of the intracellular localization of pUL47. We propose a mechanism by which localization of pUL47 at the vAC is regulated by its complex partner pUL48 and promotes tegumentation, secondary envelopment, and correct trafficking of capsids to generate mature infectious virus progeny.
MATERIALS AND METHODS
Cells.
Human foreskin fibroblasts (HFFs) and human embryonic lung fibroblasts (MRC-5; European Collection of Cell Cultures) were maintained in minimal essential medium (MEM; Gibco-BRL) supplemented with 10% fetal calf serum (FCS; Gibco-BRL) and 1% nonessential amino acids (Biochrom AG). HFFs were used until passage 29 for infection studies. MRC-5 cells were used between passages 24 and 30 for the reconstitution of virus mutants. Human epithelial carcinoma cells (HeLa) were maintained in Dulbecco's modified Eagle medium (DMEM; Gibco-BRL) supplemented with 10% FCS and 1% nonessential amino acids. HeLa cells were used in transfection experiments and indirect immunofluorescence analyses. All media were additionally supplemented with 1% penicillin-streptomycin (100-fold; PAA Laboratories) and 1% l-glutamine (200 mM; PAA Laboratories).
BAC mutagenesis and virus reconstitution.
Recombinant bacterial artificial chromosomes (BACs) were generated using a markerless two-step RED-GAM recombination protocol (68, 69). RED recombination was used to modify the TB40-BAC4 BAC that is derived from the endotheliotropic HCMV strain TB40E (70).
The BAC of mutant virus TB-47stop was generated with primers ep-UL47stop-for (5-GGGTCGCGCGGCCGTTTGGCCACCGCGCGCGCGTCCATGtagCAAGGCGCACGGTAGATTAGGATGACGACGATAAGTAGGG) and ep-UL47stop-rev (5-CTGCTCTATCAACTTTTTGAAATCTACCGTGCGCCTTGctaCATGGACGCGCGCGCGGTGCAACCAATTAACCAATTCTGATTAG). For the BAC of the revertant virus, TB-47rev, the stop codon and the frameshift in the TB-47stop BAC were repaired by RED recombination with primers ep-UL47resc-for (5-GGGTCGCGCGGCCGTTTGGCCACCGCGCGCGCGTCCATGatggctAGGCGCACGGTAGATTAGGATGACGACGATAAGTAGGG) and ep-UL47resc-rev (5-CTGCTCTATCAACTTTTTGAAATCTACCGTGCGCCTagccatCATGGACGCGCGCGCGGTGCAACCAATTAACCAATTCTGATTAG). For insertions of the Flag tag sequence into the TB40-BAC4, we used primers ep-Nflag47-for (5-GTCGCGCGGCCGTTTGGCCACCGCGCGCGCGTCCATGATGgattacaaggacgatgatgacaagGCAAGGCGAGGATGACGACGATAAGTAGGG), ep-Nflag47-rev (5-GCTGCTCTATCAACTTTTTGAAATCTACCGTGCGCCTTGCcttgtcatcatcgtccttgtaatcCATCATGGCAACCAATTAACCAATTCTGATTAG) for the mutant virus TB-Nflag47 and primers ep-Cflag47-for (5-GCCCGCCGGCTCGACATCGGTGTCCCTGCCGCCGGCCTCGgattacaaggacgatgatgacaagCCATGAAAAGGATGACGACGATAAGTAGGG) and ep-Cflag47-rev (5-ATGTCGCCCTGGTGGCAGCTGGCCTGCGTGACTTTCATGGcttgtcatcatcgtccttgtaatcCGAGGCCGCAACCAATTAACCAATTCTGATTAG) to generate the TB-Cflag47 virus. Primers contained sequences of homology upstream and downstream of the sites to be mutated, the mutation (lowercase), and sequences homologous to the pEPkan-S template plasmid (underlined). All generated recombinant BAC DNAs were controlled for integrity and correctness by restriction length polymorphism and sequencing of the mutated region.
MRC-5 cells were used for reconstitution of recombinant viruses and virus stock production. The reconstitution from BAC DNA was performed exactly as previously described (71). Reconstitution of the TB-47stop virus from TB-47stop BAC DNA was possible but required an extended period of cultivation until cells were completely infected and resulting virus stocks exhibited virus yields lower than 104 infectious particles per ml. pUL47-expressing MRC-5 cells, designated t-MRC47, were generated by lentiviral transduction to generate virus stocks of TB-47stop virus with higher titers. Electroporation of these cells with TB-47stop BAC DNA and subsequent propagation of TB-47stop virus resulted in phenotypically complemented virus, TB-47stop+UL47.
Generation of t-MRC47 lentivirus-transduced MRC-5 expressing pUL47.
A lentivirus expressing a C-terminally Myc-tagged pUL47 was generated by using the Gateway cloning system (Invitrogen) and the lentiviral vector pLenti6.2/V5-DEST (Invitrogen). The sequence of the myc tag was added to the UL47 sequence by PCR using primers ex-47-for (5-TCGGATCCATGATGGCAAGGCGCACGG) and ex-47myc-rev (5-ACGAATTCTCACAGATCTTCTTCAGAAATAAGTTTTTGTTCTGGCGAGGCCGGCGG) and cloned into the Gateway vector pENTR1A (Invitrogen) by using BamHI and EcoRI restriction sites. The UL47myc sequence was transferred from the pENTR1A vector into the pLenti6.2/V5-DEST vector by the LR reaction according to the manufacturer's protocol, resulting in pLenti-UL47myc. Purified DNA of pLenti-UL47myc was used to generate lentivirus stock with the ViraPower lentiviral expression system (Invitrogen). This lentivirus was used for transduction of MRC-5 in the presence of 10 μg/ml Polybrene (Sigma). Lentivirus-transduced cells were selected by a subsequent incubation with 5 μg/ml blasticidin (Invitrogen) for at least 5 days and their subsequent cultivation with 1 μg/ml blasticidin for further expansion. Aliquots of t-MRC47 cells were stored in liquid nitrogen and freshly thawed for reconstitution and propagation of TB-47stop virus.
Antibodies.
HCMV-encoded and cellular proteins were detected by use of monoclonal antibodies (MAbs) in focus expansion assays, titration, Western blot analyses, and indirect immunofluorescence studies. The MAbs used in the present study included those directed against pp28 (clone 5C3; Santa Cruz Biotech), pp65 (MAb 28-77), pp150 (UL32, Xp-1) (72), MCP (MAb 28-4), IE1/2 (MAb 63-27), actin (Sigma-Aldrich), and FlagM2 (Sigma-Aldrich). HCMV tegument proteins pUL71 and pUL48 were detected with previously described polyclonal antibodies (71, 73). The anti-pp71 polyclonal antibody was received from T. Stamminger, University of Erlangen (74). The UL47-specific polyclonal sera used in Western blot analyses was generated in rabbits injected with a pUL47 C-terminal fragment comprising amino acid residues 433 to 983 of the HCMV strain TB40-BAC4. For antibody generation, we followed the same strategy as previously described for that of a polyclonal antibody against pUL48 (71). Goat anti-mouse and goat anti-rabbit secondary antibodies conjugated with Alexa Fluor 488 or 555 (Invitrogen) were used for focus expansion assay and immunofluorescence studies; goat anti-mouse or goat anti-rabbit secondary antibodies conjugated with horseradish peroxidase (HRP; Pierce) were used for Western blot analyses, and an HRP-conjugated rabbit anti-mouse antibody (Dako) was used for titration.
Indirect immunofluorescence.
For indirect immunofluorescence studies of HCMV-infected cells, HFFs were seeded in μ-Slide eight-well plates (Ibidi GmbH) and synchronized for 48 h by serum starvation. Cells were then infected at a multiplicity of infection (MOI) of 0.5 to 3. At the required time postinfection, cells were washed twice with phosphate-buffered saline (PBS) and fixed with 4% paraformaldehyde (PFA) in PBS for 10 min at 4°C and stained as described previously (71). Ibidi slides allowed direct imaging without prior mounting by use of an inverted fluorescence microscope. If not stated otherwise, confocal images were taken with a 63× objective lens of an Axio-Observer.Z1 fluorescence microscope equipped with an Apotome (Zeiss) and Axiovision 4.8 software.
For indirect immunofluorescence of transfected cells, HeLa cells were grown on glass coverslips in 24-well plates and transfected using Lipofectamine LTX and fixed with 4% PFA after 24 h, as described previously (71).
Growth analysis.
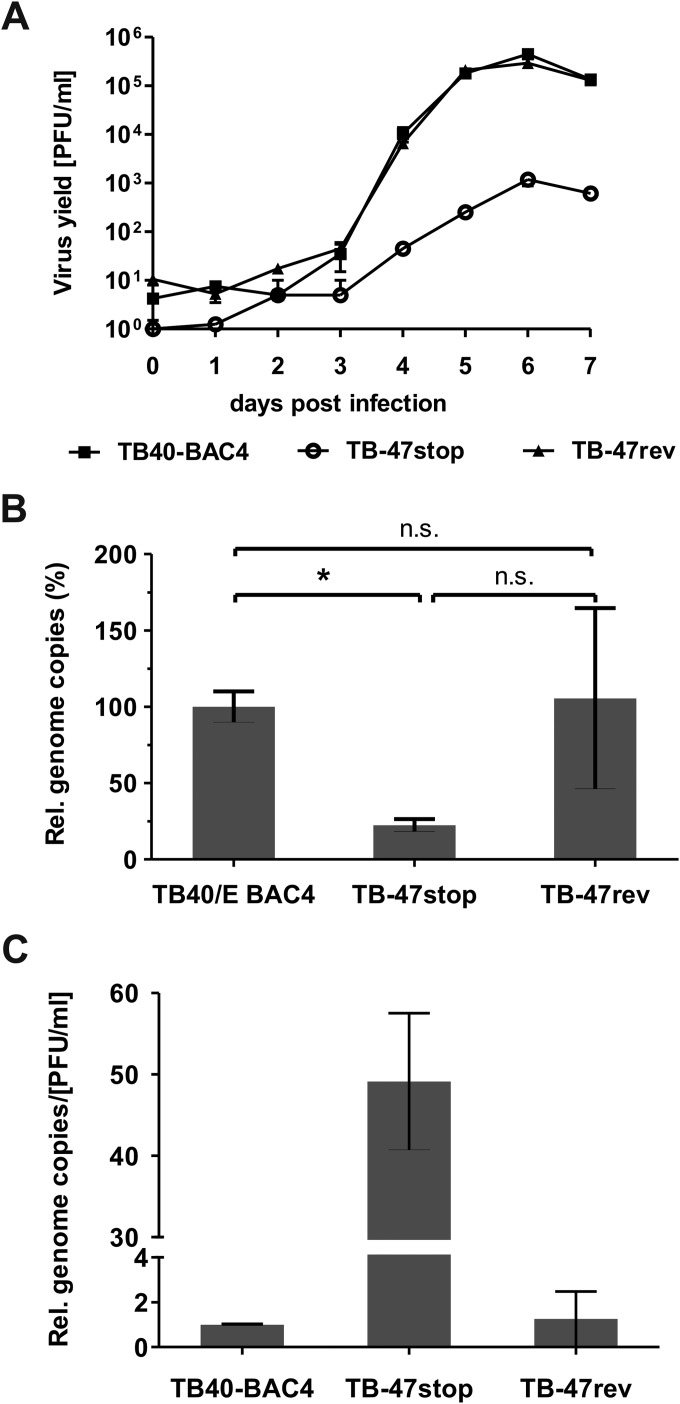
For single-step growth kinetics analysis, HFFs were infected with the respective viruses (TB40-BAC4, TB-47stop+UL47, and TB-47rev) at an MOI of 3 for 24 h at 37°C. The inocula were removed the next day and replaced with fresh medium after washing with PBS. Equal infection rates were controlled by titration of the inocula. Supernatants from virus-infected cells were collected every day until day 7 and stored at −80°C. Virus yields from at least four individual supernatants for each virus were determined on HFFs by titration as described previously (73).
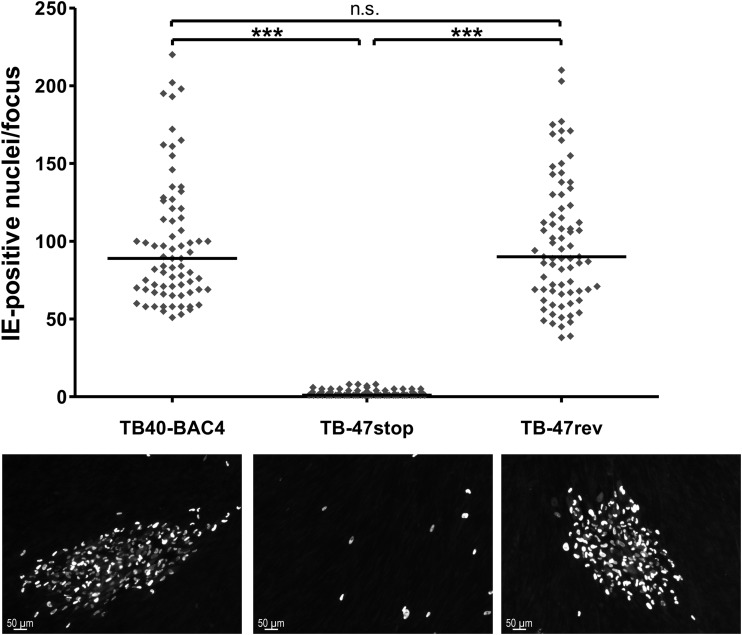
Focal virus expansion was determined by infecting confluent HFFs with 50, 100, and 150 PFU of the respective viruses, followed by incubation for 9 days at 37°C under a 0.6 to 0.7% methylcellulose overlay. The overlay was renewed once at day 5 postinfection. For this experiment, the not-complemented TB-47stop virus was used. At day 9 postinfection, cells were fixed with methanol and HCMV-infected cells were visualized by indirect immunofluorescence staining for the HCMV IE1/2 antigen. Images of virus-infected cell foci were acquired by using the Axio-Observer.Z1 confocal microscope with a 10× objective lens and the Axiovision software. The number of immediate-early (IE) antigen-positive nuclei for each focus of at least 75 foci for each virus from three independent experiments were counted by using the manual counting tool of the software Adobe Photoshop. Statistical analysis was performed by applying the Student t test.
Western blotting.
For Western blot analysis, HFFs were synchronized for 48 h by serum starvation and subsequently infected with the respective viruses at an MOI of 1. At day 5 postinfection, cells were washed twice with PBS and lysed in radioimmunoprecipitation assay (RIPA) buffer (75). After the addition of 4× loading buffer, cell lysates were sonicated for 1 min and protein denaturation was performed at 95°C for 10 min. The lysates were then subjected to 7.5% SDS-PAGE and transferred to polyvinylidene difluoride (PVDF) membranes for Western blot analyses.
Virion gradient purification.
Virion purification of TB40-BAC4, TB-UL47stop, and TB-UL47rev viruses was performed essentially as previously described (75, 76). Supernatants of at least five fully infected 75-cm2 flasks were collected, and cellular debris was removed by low-speed centrifugation (6,000 rpm, 4°C, 15 min). Virus particles were pelleted by ultracentrifugation for 1 h at 4°C at 23,000 rpm in an SW-28 rotor (Beckman). Pelleted virus particles were resuspended in 2 ml of 0.04 M sodium phosphate buffer (pH 7.4) and layered onto glycerol-tartrate gradients formed in 0.04 M sodium phosphate. Virus particles were then separated by centrifugation for 90 min at 6°C and 23,000 rpm in an SW-41 rotor. The virion layer was visualized with incandescent light and collected from the gradient. Virions extracted from the gradient were diluted with the sodium phosphate buffer and again centrifuged at 23,000 rpm for 1 h using an SW-41 rotor. The virions were processed for Western blot analysis by adding RIPA buffer and 4× loading buffer to the pellet.
Real-time PCR.
Viral DNA was isolated and purified from cell culture supernatants at day 7 of single-step growth kinetics analysis by using the High Pure viral nucleic acid kit (Roche). Genome copies were determined by real-time PCR with the Brilliant QPCR master mix (Stratagene), including Sybr green and primers HCMV Pol-CP1-For (5-AGGCCGTTACTGTCTGCAGG) and HCMV-Pol-CP1-Rev (5-CGGCCTCGTAGTGAAAATTAATGG.) Relative genome copy numbers were calculated by using a standard curve derived from defined dilutions of BAC DNA of TB-47rev and the threshold cycle (CT) values of the tested viral DNAs. Relative genomic copies of TB40-BAC4 were set at 100%. Real-time PCR was performed for at least four individual supernatants for each virus.
Transmission electron microscopy.
The methods of high-pressure freezing (HPF), freeze substitution, and Epon embedding were performed as previously described, with slight modifications (77, 78). Briefly, HFFs (2.3 × 104/well) were seeded in μ-Slide eight-well plates containing carbon-coated sapphire discs (3 mm in diameter, 50-μm thickness; Engineering Office, M. Wohlwend GmbH). At a confluence of at least 90% (usually after 1 day of cultivation), cells were infected with the respective viruses at an MOI between 1 and 2. Five days postinfection, cells on sapphire discs were fixed by HPF using the Compact 01 high-pressure freezer (Engineering Office, M. Wohlwend GmbH). The remaining cells in the μ-Slide eight-well plate were fixed with 4% PFA for 10 min at 4°C and subjected to indirect immunofluorescence staining to control for infection rates and if possible for typical mutant virus alterations. After HPF, cells on the sapphire discs were subjected to freeze substitution (77, 79) and subsequently embedded in Epon (Fluka). For transmission electron microscopy (TEM), ultrathin sections were cut with an ultramicrotome (Ultracut UCT; Leica), placed on Formvar-coated single-slot grids (slot dimensions, 2 mm by 1 mm; Plano GmbH), and examined in a Jeol JEM-1400 transmission electron microscope equipped with a charge-coupled device (CCD) camera at an acceleration voltage of 120 kV.
Quantitative analyses by TEM.
Morphogenesis stages of virus particles in the area of the vAC were quantified as previously described (73) in two-dimensional electron micrographs that were taken from randomly selected infected cells from at least three independent experiments for each virus. Only cells that had more than 10 and less than 100 particles at the area of the vAC were quantified. For each analyzed cell, the numbers of the various virus particles were determined from at least four micrographs which together covered the entire area of the vAC. Micrographs were taken at a magnification of ×25,000, which was sufficient to distinguish the various morphogenesis stages of the virus particles.
RESULTS
Growth characteristics of a newly generated UL47 stop mutant virus.
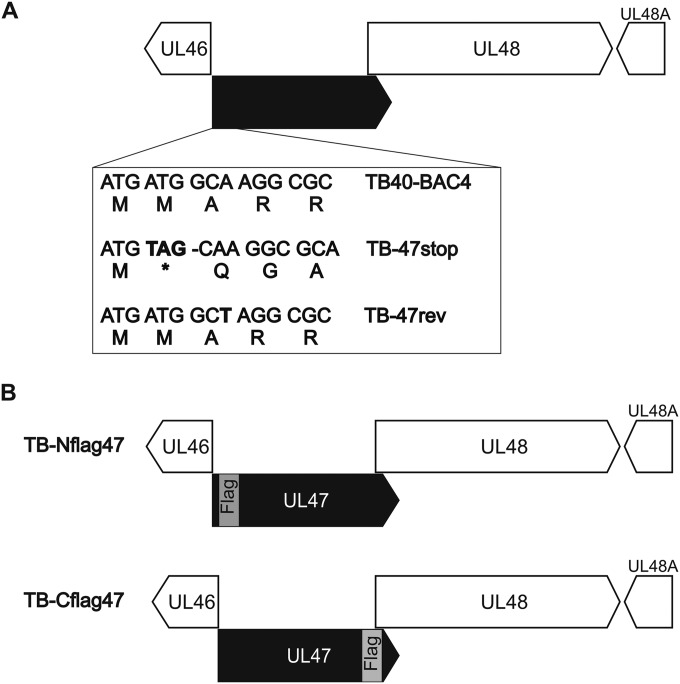
A role of pUL47 for the generation of infectious virus particles and during viral entry prior to immediate early (IE) gene expression has been shown previously for a mutant of AD169, lacking most of the UL47 sequence (29). To elucidate the exact function of pUL47 during HCMV morphogenesis especially in the late stages of virion assembly, a new mutant virus, TB-47stop, was generated in the backbone of an infectious clone (TB40-BAC4) of the endotheliotropic HCMV strain TB40/E (70). To minimize unwanted effects on the expression of neighboring genes, a stop codon was generated at amino acid position 2 by introducing nucleotide substitutions A4T and T5A of the UL47 open reading frame (ORF) (Fig. 1A). The stop codon is followed by a deletion of nucleotide G7, causing a frameshift to completely prevent expression of pUL47. These mutations were introduced downstream of the UL46 ORF. A revertant virus, TB-47rev, was generated by mutating the UL47 sequence of TB-47stop back to the original sequence. The additional introduction of the A9T mutation in UL47 resulted in a silent mutation in pUL47 of the TB-47rev virus which allows distinction of the revertant virus from parental TB40-BAC4. During reconstitution of the TB-47stop virus, we noticed severely impaired growth in cell culture accompanied by insufficient extracellular virus yields for high-MOI infections. To complement for the growth impairment, TB-47stop virus was propagated in pUL47-expressing MRC-5 cells, designated t-MRC47. Mutant virus propagated on t-MRC47 (TB-47stop+UL47) is phenotypically complemented but unable to express pUL47 from the viral genome (data not shown). We first determined the focal spread of our TB-47stop virus by performing a focus expansion assay (Fig. 2). Remarkably, focal spread of the TB-47stop virus in HFFs was almost abrogated since many single immediate early (IE) antigen-positive cells and only very small foci with a maximum of eight IE antigen-positive cells were counted at day 9 postinfection. In contrast, wild-type TB40-BAC4 and revertant viruses exhibited foci with a median number of 80 IE antigen-positive cells. This implies that the focal spread of HCMV requires pUL47. We then compared viral growth of TB-47stop virus to that of TB-47rev and wild-type TB40-BAC4 viruses in single-step growth kinetics analysis by infecting fibroblasts at an MOI of 3 (Fig. 3A). Infection with the TB-47stop virus resulted in up to 1,000-fold-lower virus yields in supernatants of infected fibroblasts than in those infected with TB40-BAC4 and revertant viruses. Comparable growth levels of TB40-BAC4 and TB-47rev viruses indicated to us that the mutation of TB-47stop was successfully restored in the revertant virus. Given the previously reported impaired infectivity of virus particles in the absence of pUL47 (29), we tested to what extent this contributed to the lower virus yields by single-step growth kinetics analysis by determining the particle-to-PFU ratios for all three viruses. We used the culture supernatants of a single-step growth kinetics assay at day 7 postinfection and analyzed them in two different ways. Infectious virus yields were determined by titration, and viral genome copies were quantified from equal volumes of these supernatants by real-time PCR. The relative genome copy numbers were calculated from a standard curve, and the number of genomic copies of TB40-BAC4 virus was set at 100%. We found less viral DNA in supernatants from TB-47stop-infected cells than in TB40-BAC4 virus-infected cells, whereas TB-47rev and TB40-BAC4 viruses released comparable DNA levels (Fig. 3B). Relative infectivity is expressed as the ratio between the relative genome copy number and the virus yield in the supernatant (Fig. 3C). The comparison of the three viruses revealed a 49-fold-lower relative infectivity of virus particles released from TB-47stop virus-infected cells, indicating an important role of pUL47 in the production of infectious virus progeny. In contrast to the AD169-based mutant virus, we found fewer genomes released from TB-47stop virus infections, which suggests that either formation or egress of virus particles requires pUL47 (29).
FIG 1.
Generation of UL47 mutants. (A) Introduction of nucleotide substitutions A4T and T5A and deletion of nucleotide G7 of the UL47 ORF in mutant virus TB-47stop resulted in a stop codon (bold) that is followed by a frameshift. The stop codon and the frameshift mutation were repaired in mutant virus TB-47rev. Introduction of the A9T substitution (bold) in that mutant virus led to mutation, allowing differentiation of the wild-type virus TB40-BAC4 and the TB-47rev virus. (B) Generation of two mutant viruses that express pUL47 with either an N-terminal Flag tag (TB-Nflag47; insertion after the second methionine) or a C-terminal Flag tag (TB-Cflag47; insertion before the terminal proline).
FIG 2.
Focal virus expansion. Each data point (rhombus) of the graph represents the number of immediate early (IE) antigen-positive nuclei of one focus at day 9 postinfection. The bold horizontal lines in the graph depict the median number of IE antigen-positive cells/focus for each virus. For the statistical analysis, the two-tailed Student t test was used (***, P < 0.0001; n.s., not significant). Representative images of foci of TB40-BAC4, TB-47stop, and TB-47rev viruses are shown (lower panels). HCMV-infected cell nuclei (white) were stained with an anti-IE1/2 MAb and a goat anti-mouse secondary antibody conjugated with Alexa Fluor 555 (Invitrogen).
FIG 3.
Viral growth in the absence of pUL47. (A) Single-step growth kinetics assays of wild-type TB40-BAC4, revertant TB-47rev, and mutant TB-47stop viruses were performed by infecting HFFs at an MOI of 3. Virus yields of the supernatants were determined at the indicated times. Virus yields at day 0 represent the input virus remaining after infection. Shown are the mean and standard deviation of results for four experiments. (B) Relative genome copies in the supernatants of TB40-BAC4, TB-47stop, and TB-47rev viruses at day 7 postinfection. Relative genomic copies were determined by real-time PCR and calculated from a standard curve. Relative genome copies of TB40-BAC4 were set as 100%. For the statistical analysis, the two-tailed Student t test was used (*, P = 0.0189; n.s., not significant. (C) The graph depicts the ratio between the relative genome copies and the virus yields in the supernatants of TB40-BAC4, TB-47stop, and TB-47rev viruses at day 7 postinfection (standard deviations are shown).
TB-47stop exhibits a defect in virion morphogenesis.
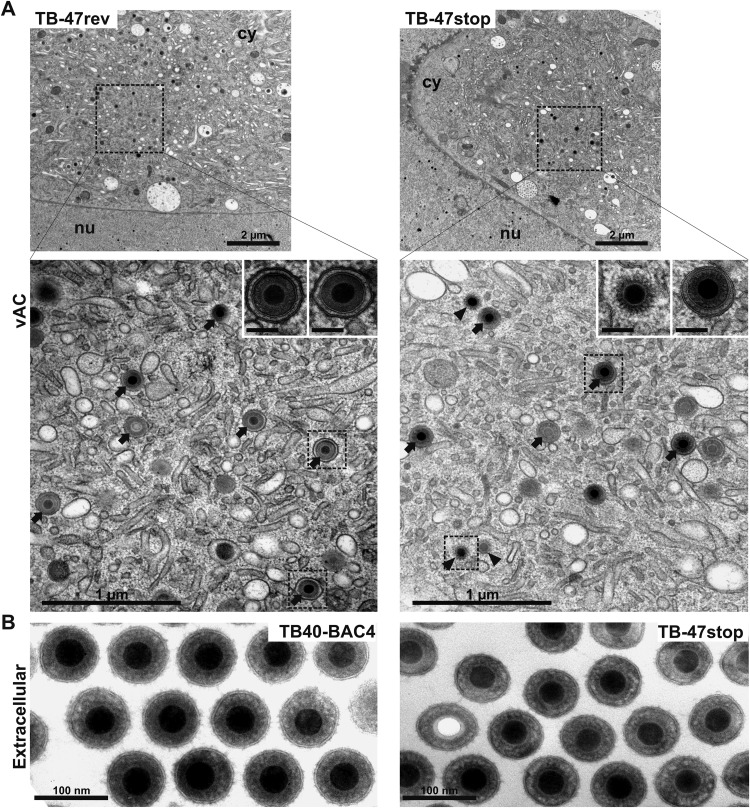
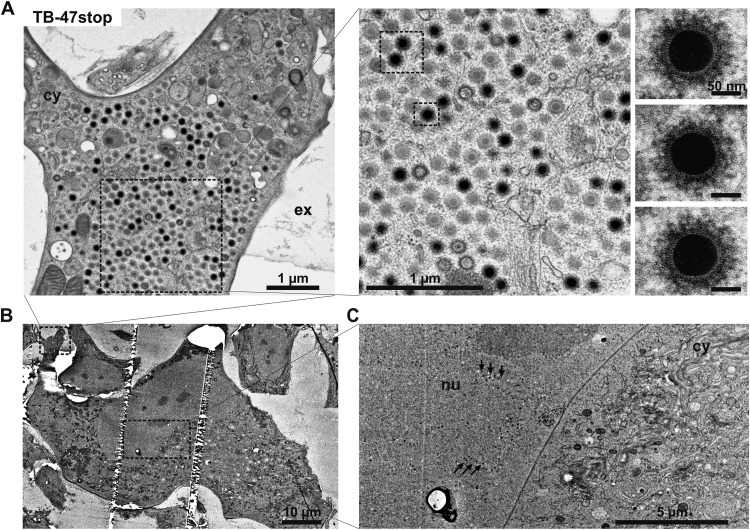
To investigate possible defects in late stages of virus morphogenesis, we performed an electron microscopic investigation of TB-47stop-infected cells at day 5 postinfection. First, we focused on the ultrastructure of intra- and extracellular TB-47stop virus particles in comparison to those of wild-type virus TB40-BAC4 and the revertant virus, since the impaired infectivity of TB-47stop particles could be based on alterations of the tegument. The ultrastructures of intracellular and extracellular virions (Fig. 4), especially the thickness and appearance of the tegument layer, were undistinguishable between all three viruses, arguing against a major role of pUL47 in tegument formation. Next, we looked at nuclear and cytoplasmic stages of virus morphogenesis to determine if there was a defect accounting for the impairment in virus release. Nuclear stages of HCMV morphogenesis appeared normal in TB-47stop virus-infected cells, as the structure and ratios of A, B, and C capsids were undistinguishable for all three viruses (data not shown). To clarify whether the growth defect of the TB-47stop virus was based on defective secondary envelopment, we determined the ratio of fully enveloped capsids, membrane-attached capsids, and capsids with no contact to membranes (naked) in the vACs of infected fibroblasts (Table 1). We included in our quantification only cells that showed similar densities of nuclear capsids to ensure similar stages of infection. Other than cells lacking any particles associated with the vAC (at least seven TB-47stop virus-infected cells), we found an increase of nonenveloped capsids in vACs of the TB-47stop virus (Fig. 4A; Table 1). Numbers of fully enveloped particles were slightly reduced in the absence of pUL47. Nevertheless, the majority of virus particles in the area of the vAC were fully enveloped. Consistent with this, we frequently found virions in the extracellular space of TB-47stop virus-infected cells (Fig. 4B).
FIG 4.
Ultrastructural visualization of a viral assembly complex (vAC) and extracellular particles of the indicated viruses at day 5 postinfection. (A) Overview of vACs in the cytoplasm (cy) together with a part of the nucleus (nu). The lower panels show higher magnification of fully enveloped (arrows) and naked (arrowheads) particles within the vACs and detailed images in which the structures of the tegument in fully enveloped particles (TB-47rev and TB-47stop) and naked particles (TB-47stop) are depicted. Scale bars, 100 nm. (B) Extracellular HCMV particles. DNA-filled capsids are indicated by their electron-dense cores, whereas noninfectious virions exhibit electron-translucent cores.
TABLE 1.
Ultrastructural quantification of HCMV particles in vACs of fibroblasts infected with the respective virus at day 5 postinfection
| Virus | No. of vACs quantified | % of particles (mean ± SD)a |
||
|---|---|---|---|---|
| Enveloped | Membrane attached | Naked | ||
| TB40-BAC4 | 17 | 54.8 ± 23.6 | 29.9 ± 10.8 | 15.3 ± 14.9 |
| TB-47stop | 30 | 47.5 ± 25.1 | 30.3 ± 17.7 | 22.2 ± 23.4 |
| TB-47rev | 36 | 59.1 ± 23.1 | 31.1 ± 18.1 | 9.9 ± 9.6 |
Values are mean percentages ± standard deviation (SD) of enveloped particles, nonenveloped particles attached to membranes, and nonenveloped particles not attached to membranes (naked particles). There were no significant differences between the percentages (Kruskal-Wallis and Dunn's multiple-comparison tests; P < 0.05).
The most obvious phenotypic alteration in cells infected with TB-47stop virus was that in many TB-47stop virus-infected cells, a large quantity of nonenveloped virus capsids accumulated in the cell periphery (Fig. 5A). These accumulations were densely packed with mainly C capsids and a few empty capsids. The electron-dense material that was decorating the outside of these capsids appeared to be different in density and thickness from the classical tegument structure of fully enveloped virions found at the vAC (Fig. 5A, right panel). We interpret this as a result of a defect in acquisition of the normal tegument.
FIG 5.
Ultrastructure of a TB-47stop-infected fibroblast at day 5 postinfection. (A) Accumulation of nonenveloped and partially tegumented HCMV capsids in the cell periphery. The higher-magnification images in the right panel and the insets show fuzzy material decorating the capsid. (B) Overview of the cell depicted in panel A. (C) The nucleus (nu) of the depicted cell contains large numbers of capsids (arrows). No capsids are found in the depicted area of the vAC within the cytoplasm (cy).
In conclusion, our ultrastructural data suggest that tegumentation and secondary envelopment can proceed without pUL47, but cytoplasmic trafficking of capsids seems to be altered in the absence of pUL47. As a result, nonenveloped capsids accumulate at the periphery of the cell. All these alterations likely contribute to the growth impairment of TB-47stop virus.
Intracellular distribution of viral proteins in the absence of pUL47.
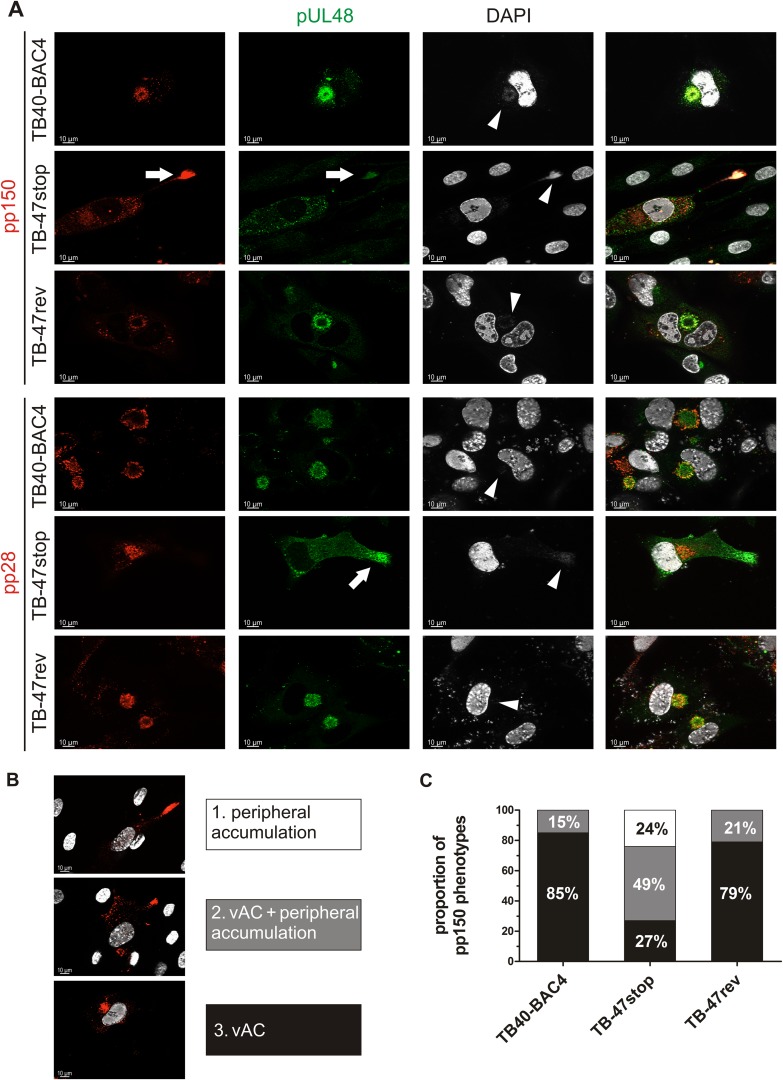
To support the ultrastructural differences in TB-47stop virus-infected cells, we investigated the intracellular distribution of viral proteins in the absence of pUL47. Furthermore, we sought to identify tegument proteins which can bind to capsids independently of pUL47. We hypothesized that, most likely, proteins of the inner tegument will be detected at capsid accumulations of TB-47stop virus-infected cells. Therefore, we performed indirect immunofluorescence stainings for tegument proteins pp150 (UL32) and pUL48. HCMV pp150 is known to directly interact with capsids (16, 76) and to be involved in stabilizing capsids (36). It has also been used as a marker to study intracellular capsid trafficking (76). We hypothesized that pp150 binding to capsids is independent of pUL47 because it is not part of the pUL47/pUL48/MCP/pUL69 complex (29) and likely attaches to capsids in the nucleus (76, 80). By staining for pUL48, we wanted to clarify whether pUL47 regulates the attachment of pUL48 to capsids. Staining for tegument protein pp28 was included because this protein is strongly localized to the vAC during the entire course of infection and most likely incorporated into virions there (15, 81). Indirect immunofluorescence analysis was performed on infected cells that were treated identically to those used for electron microscopic investigations. The majority of TB40-BAC4- and TB-47rev-infected fibroblasts exhibited concentrated signals for pp150, pUL48, and pp28 at the area of the vAC at day 5 postinfection (Fig. 6A). In contrast, in TB-47stop-infected fibroblasts, pp150 signals were found predominantly at the nucleocapsid accumulations in the cell periphery. The presence of viral DNA at these accumulations was indicated by DAPI (4′,6-diamidino-2-phenylindole) signals. These accumulations were also positive for pUL48 but devoid of pp28, which remained localized at the vAC. This suggested to us that attachment of both pp150 and pUL48 does not require pUL47. In agreement with our ultrastructural data, we noted that the intracellular distribution of pp150 in TB-47stop virus-infected cells exhibited various patterns (Fig. 6B). In the absence of pUL47, signals for pp150 signals were either strictly localized at the cell periphery, a phenotype that was restricted to the TB-47stop virus, or at the vAC, or at both sites (Fig. 6). A quantification of these phenotypes of virus-infected cells at day 5 postinfection showed that the concentration of pp150 signals at the vAC was the predominant phenotype in wild-type and revertant virus-infected cells (Fig. 6C). In contrast, the most abundant phenotype in TB-47stop virus-infected cells was characterized by an accumulation of pp150 signals at the cell periphery together with signals at the vAC. In conclusion, the quantification of pp150 phenotypes supports the result from ultrastructural data that pUL47 has an important role in capsid trafficking and tegumentation.
FIG 6.
Indirect immunofluorescence of HFFs infected with TB40-BAC4, TB-47stop, or TB-47rev virus at day 5 postinfection. (A) Intracellular distributions of viral proteins were compared by staining pUL48 (green) with our polyclonal serum and counterstaining with either anti-pp150 MAb or anti-pp28 MAb (red). White arrows indicate peripheral accumulations of pUL48 and pp150 in TB-47stop-infected cells. White arrowheads depict DAPI accumulation at the vAC and at the periphery. (B) Intracellular distribution of pp150 was categorized into three phenotypes: accumulations only at cell extensions (1), accumulations at cell extensions and at the vAC (2), and strict localization at the vAC (3). The nuclei were stained with DAPI and are visible in white. (C) Summary of the relative proportions of the three categories from three independent experiments normalized to 100%. For each individual experiment and virus, the intracellular distribution of pp150 was categorized from 25 infected cells that were positive for both pp150 and pUL48.
Analysis of intracellular proteins and virion composition of TB-47stop-infected cells.
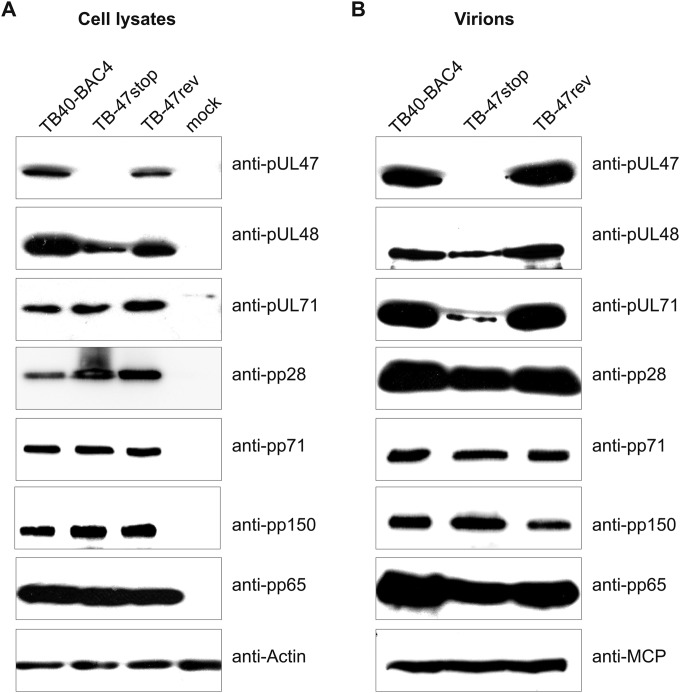
Incorporation of many viral proteins into virions has been shown to be regulated by their interaction with other viral proteins (75, 82, 83). Bechtel and Schenk showed an overall decrease of pUL48 levels in infected cells and an impaired virion incorporation in the absence of pUL47 (29). Therefore, we examined the composition of extracellular virions released from TB-47stop virus-infected cells. As shown in Fig. 7, detection of pUL47 with our polyclonal antibody, directed against a region comprising amino acids 433 to 983, verified that the stop codon in TB-47stop virus indeed prevented expression of pUL47. Consistently, pUL47 was not detected in purified TB-47stop virions. Detection of pUL48 revealed reduced levels in cell lysates as well as in purified virions. In contrast, levels of other tested viral proteins (pUL86 [MCP], pUL32 [pp150], pUL83 [pp65], pUL99 [pp28] and pUL82 [pp71]) in virions and cell lysates were similar in TB-47stop, TB40-BAC4, and TB-47rev viruses. The only exception was tegument protein pUL71, whose presence in virions was clearly reduced whereas intracellular levels of this protein appeared to be unaffected in the absence of pUL47. Thus, TB-47stop virions were deficient for pUL47 and contained reduced amounts of pUL48 and pUL71.
FIG 7.
Analysis of TB-47stop extracellular virion composition. (A) Cell lysates of TB40-BAC4, TB-7stop, and TB-47rev virus-infected HFFs (day 5 postinfection) were processed for Western blot analysis. The blot was probed with individual antibodies against pUL47, pUL48, pUL71, pp71, pp150, and pp28. pp65 and actin served as loading controls. (B) The solubilized proteins of purified extracellular virions were subjected to Western blotting using antibodies against the following viral proteins: pUL47, pUL48, pUL71, pp71, pp150, and pp28. MCP and pp65 served as loading controls.
Localization of pUL47 during infection.
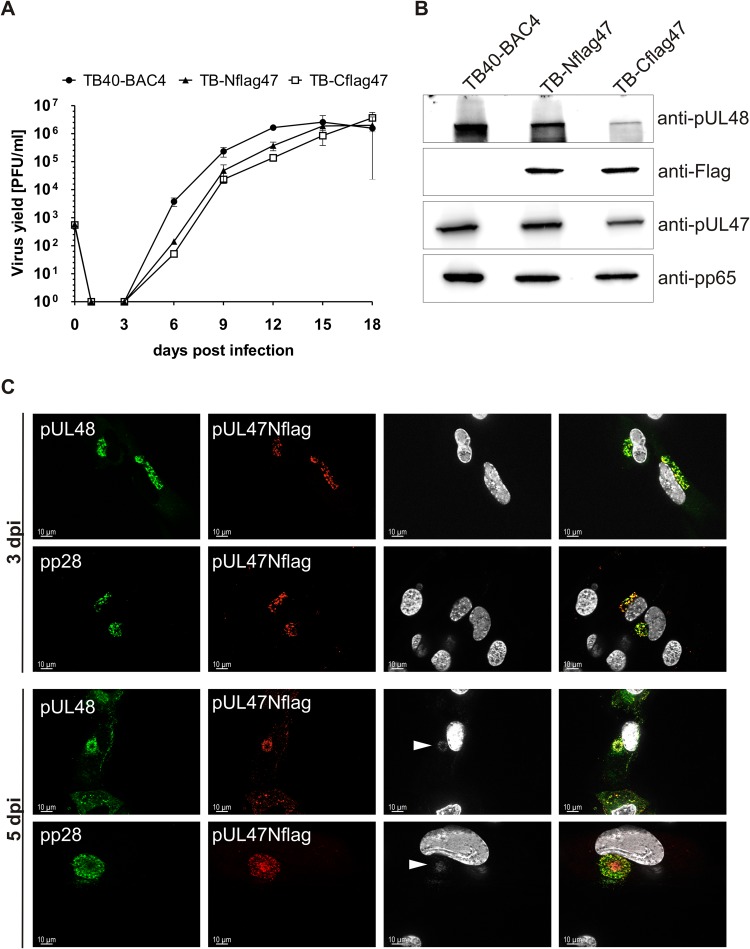
Having shown an important role of pUL47 in tegumentation and particle trafficking during infection, we sought to determine the intracellular localization of this protein in infection. Due to the lack of an antibody detecting pUL47 in indirect immunofluorescence, we generated two mutant viruses that express pUL47 with either an N-terminal or a C-terminal Flag tag, designated TB-Nflag47 and TB-Cflag47, respectively (Fig. 1B). Growth of these mutants was analyzed in a multistep growth kinetics assay to verify that the Flag tag insertions in UL47 did not affect viral growth. Both mutant viruses reached extracellular virus yields that were comparable to those of parental virus (Fig. 8A). Subsequent Western blot analysis of infected cell lysates at day 5 postinfection showed similar protein levels for both Flag tag versions of pUL47 (Fig. 8B). We also checked pUL48 levels to exclude effects on its expression caused by Flag tag insertion directly in front of the start of pUL48 in TB-Cflag47. As shown in Fig. 8B, lysates of TB-Cflag47-infected cells exhibited repeatedly reduced levels of pUL48 compared to levels in cell lysates after TB-Nflag47 and parental virus infection. In the light of these results, we primarily used the TB-Nflag47 mutant for the determination of pUL47 localization during infection. Given the interaction of pUL47 with pUL48 in transfection (40) and in infection (29), we hypothesized that the intracellular distribution of pUL47 is similar to that of pUL48; the latter has been shown to accumulate at the vAC (71). To address this, intracellular distribution of both pUL47 and pUL48 was analyzed at early (day 3 postinfection) and late (day 5 postinfection) times of infection. Since infected cells at day 3 postinfection exhibited different stages of infection, we selected for cells with the least advanced stage. These cells were chosen for a premature vAC and the lack of DAPI signals at this site. Cells of this stage also lacked pp150 signals at the site of the premature vAC, indicating an absence of virus particles (data not shown). As shown in Fig. 8C, pUL47Nflag exhibited a localization pattern similar to those of pUL48 and tegument protein pp28. Their signals were found to be predominantly perinuclear at the site of the premature vAC. The lack of signals for capsid-associated pp150 in the cytoplasm of cells at this stage of infection (data not shown) indicated that the observed pUL47 localization is independent of capsids. At day 5 postinfection, pUL47 signals strongly overlapped those of pUL48 and DAPI signals at the vAC but showed only a partial overlap of pp28 signals. These data are consistent with the formation of a pUL47/pUL48 complex and the accumulation of virus particles at the vAC.
FIG 8.
Characterization of TB-Nflag47 and TB-Cflag47 viruses. (A) Multistep growth kinetics analyses of TB40-BAC4, TB-Nflag47, und TB-Cflag47 viruses were performed by infecting HFFs at an MOI of 0.02. Virus yields in the supernatants of infected cells were determined at indicated times by titration on HFFs. Growth curves show the mean virus yields and standard deviations for three experiments. Virus yields at time zero represent the inoculum. (B) Cell lysates of wild-type TB40-BAC4, TB-Nflag47, and TB-Cflag47 virus-infected HFFs (day 5 postinfection) were processed for Western blot analysis. The blot was incubated with polyclonal anti-pUL48 and anti-pUL47 sera and with an anti-Flag M2 MAb, respectively. pp65 was used as a loading control. (C) Indirect immunofluorescence of HFFs infected with TB-Nflag47 at days 3 and 5 postinfection. The anti-FlagM2 MAb (red) was used to detect pUL47Nflag together with either a polyclonal anti-pUL48 serum (green) or an anti-pp28 MAb (green). The right panels show the merged results; cell nuclei were stained with DAPI and are visible in white. White arrowheads mark the DAPI signals at the vAC at day 5 postinfection.
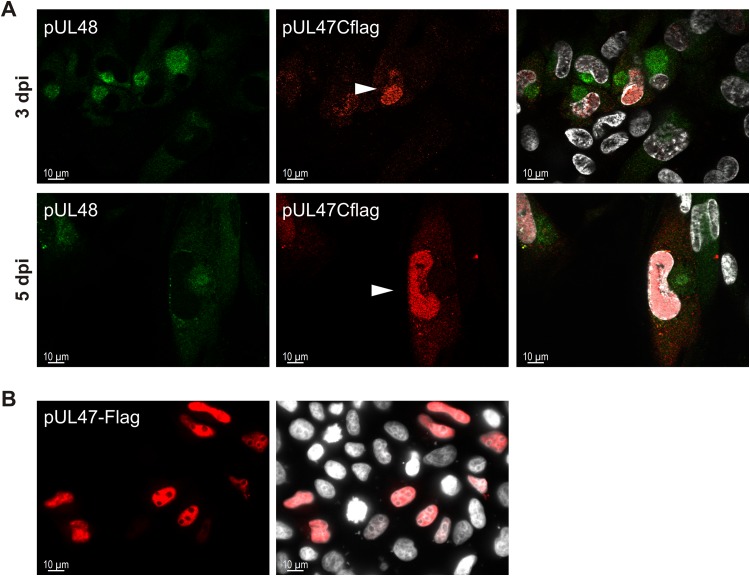
Localization of pUL47 is determined by pUL48.
Because of the interaction of pUL47 with pUL48 and their colocalization during infection, we sought to determine the role of pUL48 in intracellular localization of pUL47. Therefore, we used in indirect immunofluorescence analysis the TB-Cflag47 virus, which showed reduced levels of pUL48 in Western blot analysis and which allows detection of pUL47 by a C-terminal Flag tag. As shown in Fig. 9A, the majority of pUL47Cflag was detected in the nucleus and only very little at the site of the vAC in infections with the TB-Cflag47 virus, whereas pUL48 was still found at the site of the vAC. These changes in intracellular distribution of pUL47 were observed in the majority of infected cells at early and late times of infection. Finally, we determined the localization of pUL47 in transfected cells. Transient expression in HeLa cells showed a strong nuclear localization of pUL47 in the absence of other viral proteins (Fig. 9B). Considering its molecular size of approximately 110 kDa that excludes free diffusion through nuclear pores, pUL47 appears to be actively transported into the nucleus. These data suggest that during infection, pUL47 is translocated from the nucleus to the vAC, as found for TB-Nflag47 infection, and that it fails translocation in cells infected with the TB-Cflag47 virus.
FIG 9.
Intracellular distribution of pUL47 in TB-Cflag47 infection. (A) Indirect immunofluorescence of HFFs infected with the TB-Cflag47 virus at days 3 and 5 postinfection. The anti-FlagM2 MAb (red), detecting pUL47Cflag, was used together with the anti-pUL48 antibody (green). The cell nuclei were stained with DAPI and are visible in white. The right panels display the merged results. White arrowheads mark pUL47Cflag-positive cell nuclei of TB-Cflag47-infected cells. (B) Indirect immunofluorescence of HeLa cells transiently expressing pUL47-Flag. Cells were fixed after 24 h, and intracellular distribution of pUL47-Flag was analyzed by staining with the anti-FlagM2 MAb (red). The nuclei were stained with DAPI and are visible in white. Images were taken using an 63× objective and the Zeiss Observer.Z1 fluorescence microscope.
DISCUSSION
In this study, we have analyzed functions of the HCMV tegument protein pUL47. Little was known about its function during HCMV morphogenesis, even though it is a highly conserved tegument protein among herpesviruses (34) with central to essential roles for the generation of infectious progeny in alphaherpesviruses (61, 62). A previous characterization of an AD169-based mutant virus with a large substitution of the UL47 ORF sequence revealed an important function early in HCMV infection (29). The salient finding of our study is that pUL47 also has an important function during virion morphogenesis and in the process of tegumentation of capsids. Furthermore, we show that pUL47 is accumulating at the vAC together with pUL48 and that this accumulation is regulated by pUL48.
The absence of pUL47 results in a severe growth defect, with lower extracellular virus titers and impaired focal spread. A previously characterized UL47 substitution mutant showed impaired replication in fibroblasts and an increased particle-to-PFU ratio together with a delay in the onset of viral gene expression (29). In this study, we evaluated viral growth in the absence of pUL47 in the genetic background of an infectious clone of the endotheliotropic HCMV strain TB40/E (70). Due to the complex organization of the UL47 region and to minimize the risk of affecting the expression of neighboring genes, a mutant virus (TB-47stop) in which expression of pUL47 was abrogated by insertion of a stop codon was generated. We observed up to 1,000-fold-lower numbers of infectious particles in the supernatant of TB-47stop-infected cells than in TB40-BAC4 cells (Fig. 3A) and confirmed for this virus an increase in the particle-to-PFU ratio (29). In addition, a focus expansion assay (Fig. 2) clearly indicated that the absence of pUL47 causes a severe defect in focal spread. While this defect may also be related to a function of pUL47 early in infection and smaller amounts of pUL48 in virions (Fig. 7), it could also be related to a function late in infection, as reported for the alphaherpesvirus counterpart pUL37 (60, 61, 84). In the case of PrV, the pUL37 N terminus is involved in cell-to-cell spread and shares structural similarity with cellular multisubunit tethering complexes which control vesicular trafficking in eukaryotic cells (85). This shows a specific cell-to-cell spread-associated function of pUL37, which could also apply to HCMV pUL47, given the high conservation of this protein among herpesviruses. However, the exact mechanisms behind direct cell-to-cell spread are not entirely clear and it may differ between herpesviruses, considering that HCMV lacks the alphaherpesvirus spread-related glycoproteins E and I (86).
We also observed a reduced incorporation of pUL71 into virions of the TB-47stop virus, while intracellular levels were unaffected (Fig. 7). We can only speculate about the role of pUL71 for the impaired growth of the TB-47stop virus, but it must be mentioned that a mutant virus unable to express pUL71 is characterized by a small-plaque phenotype and impaired secondary envelopment (73). Therefore, a contribution of pUL71 to the TB-47stop phenotype must at least be considered. The finding that intracellular levels of pUL71 were unaffected by the lack of pUL47 (Fig. 7) hints toward an interaction with pUL47, which is interesting but requires further investigation. So far, two large yeast two-hybrid screenings showed no direct interaction of these proteins (87, 88).
In contrast to previous data, we observed that significantly fewer genomes were released from TB-47stop virus-infected fibroblasts (Fig. 3B). This finding is in line with the observation of accumulations of nonenveloped capsid in the cytoplasm of TB-47stop-infected cells, because a large number of capsids will most likely not be released from the cells. Why this has not been found for the AD169-derived UL47 virus mutant is unclear but may be explained by the better adaptation of AD169 to human fibroblasts.
pUL47 is required for tegumentation and proper capsid trafficking. HCMV inner tegument proteins pp150, pUL48, and pUL47 are tightly associated with the capsid (33, 36). From these, pUL48 and pUL47 form a complex that also includes other viral proteins (29, 40). The order of their attachment onto the capsid and how this is regulated is not clear. Here we confirmed previous data of pUL47-independent virion incorporation of pUL48 (Fig. 7). Yet the presence of pUL47 increased the level of pUL48 in the virion by an unknown mechanism. It is reasonable to assume that the formation of the pUL48/pUL47 complex is crucial for efficient incorporation of pUL48 and pUL47 into the virion. It will be of interest to determine the exact role of the pUL48/pUL47 complex in future work. Consistent with its virion incorporation, we detected pUL48 in the absence of pUL47 at accumulations of nonenveloped capsids at the periphery of infected cells by immunostaining. Capsids at these accumulations were also positive for pp150 (Fig. 6). This is in agreement with a direct capsid interaction of pp150 (14, 16, 35) and its function to stabilize capsids (36). It also suggests that the attachment of pp150 to capsids does not require the pUL48/pUL47 complex. The absence of tegument protein pp28 at these accumulations and the lack of the typical tegument structure and the “fuzzy” material at these capsids visualized by electron microscopy led to the conclusion that these capsids failed to acquire the full tegument. The reasons for this are not clear. The accumulation of nonenveloped capsids with a partial tegument points toward a defect in the process of tegumentation. Possible underlying mechanisms may be a defect in the attachment of tegument proteins to capsids or a delay in tegumentation of capsids in the absence of pUL47. Alternatively, an impairment in secondary envelopment could cause premature disassembly of virus particles and thus result in the observed accumulation of nonenveloped capsids with a partial tegument. Capsid accumulations were found preferentially at the apex of spindle-like fibroblasts. Because these regions are the most distal parts of the cell in relation to the vAC, we assume an active transport of capsids along microtubules toward these sites. Although this observation is no formal proof of an active transport of capsids, it indicates that capsid trafficking is possible in the absence of pUL47. Capsid trafficking may be regulated differently in HCMV than in alphaherpesviruses, where in the absence of pUL37, capsid transport to the TGN is blocked (56, 60) and pUL37 is able to interact with cellular motor proteins (57, 63). Interaction with cellular motor proteins in alphaherpesviruses is not restricted to pUL37 but also includes the large tegument protein pUL36 and other inner tegument proteins (e.g., pUS3, ICP0, pUL14, pUL16, or pUL21) but not capsid proteins (e.g., VP5) or outer tegument proteins (e.g., pUL11) (52). To our knowledge, HCMV proteins involved in capsid trafficking have not yet been identified. Therefore, we can only speculate which HCMV proteins regulate capsid trafficking.
Colocalization of pUL47 and pUL48 at the vAC at a stage of infection where no capsids were found at this site indicated to us that their vAC accumulation appears to be independent of capsids (Fig. 8C). Furthermore, accumulation of capsids at the vAC was still possible in the absence of pUL47. In contrast, HSV-1 pUL37 is critical for directing capsids to the TGN after their release from the nucleus (56, 58, 60). Other data report that TGN localization of pUL37 does not require capsids but does require pUL36 (58), which would be in line with our conclusion. Nevertheless, we propose a slightly different role of HCMV pUL47. Capsids are transported independently from pUL47 from the nucleus to the site of the vAC, which is formed around the microtubule-organizing center (MTOC) (4–8, 10). Binding of pUL47 to these capsids at the vAC keeps them there and causes their further tegumentation and secondary envelopment. Capsids at the vAC can still undergo tegumentation and secondary envelopment in the absence of pUL47, but capsids not entering this process accumulate in the periphery as capsids with an incomplete tegument (Fig. 5 and 6). Our model also includes a defect in secondary envelopment, which we could not find in our quantification of envelopment at the vAC by electron microscopy (Table 1). It must be pointed out that we included only virus particles of the vACs in this quantification despite a large number of nonenveloped particles in accumulations in the periphery of cells. Since those accumulations were often found in thin sections other than those with the vAC, we could not include these particles in our quantification. In addition, we found at least seven cells in the TB-47stop infection without any capsids at the vAC. Together, these findings point to a defect in secondary envelopment that is in agreement with the function of pUL37 in alphaherpesviruses (51, 56, 60, 62, 89).
In the light of the pUL47/pUL48 complex formation (29, 40), we supposed that pUL47 would follow pUL48 distribution during infection. The immunofluorescence analyses, by use of the TB-Nflag47 virus (Fig. 1B) expressing pUL47Nflag, confirmed our assumption (Fig. 8C). In addition, in the TB-Cflag47 mutant virus that carries a Flag tag insertion at the C terminus of pUL47, we observed a change to a more-nuclear localization of pUL47 (Fig. 9) and a lower expression level of pUL48 (Fig. 8B). At this point, we can only speculate whether these two observations are connected to each other. The insertion of the Flag tag at the C terminus can have several consequences, such as influencing expression of pUL48 (Fig. 8B) or directly or indirectly affecting the complex formation of pUL47 and pUL48. Since the genomic organization of UL47 and UL48 is very complex, with overlapping transcripts (90), insertion of the Flag tag sequence at the promoter region of UL48 carries a high risk of affecting the transcription of UL48. A requirement of the C terminus of pUL47 for interaction with pUL48 has been shown recently (40). However, the exact sequence requirements in pUL47 are not known. It is also not known whether the Flag tag insertion affects the interaction of pUL47 with other viral or cellular factors. However, in the light of the functional relationship of pUL47 and pUL48 homologs in alphaherpesviruses (41–47) and their interaction in HCMV, it can be assumed that lower levels of pUL48 would also result in more pUL47 that is not in complex with pUL48. Our findings of nuclear localization of pUL47 in transfected cells and nuclear localization of pUL47 in TB-Cflag47 virus infection support this. Nevertheless, this requires further clarification. Currently, we are investigating the role of the pUL47/pUL48 interaction in localization of pUL47 and testing whether the Flag tag at the C terminus of pUL47 is hindering the interaction with pUL48.
In conclusion, we provide evidence for a mechanism by which pUL48 regulates intracellular localization of pUL47 to the vAC, where it functions to capture capsids and to facilitate their further maturation.
ACKNOWLEDGMENTS
The technical assistance of Anke Lüske, Monika Dürre, and Jutta Hegler is gratefully acknowledged. We thank Paul Walther (Electron Microscopy Facility, Ulm, Germany), Ivonne Brock, and Olga Idt (Institute of Virology, Ulm, Germany) for their experimental help.
Ilaria Cappadona was supported by the Graduate School in Molecular Medicine Ulm.
REFERENCES
- 1.Kalejta RF. 2008. Tegument proteins of human cytomegalovirus. Microbiol Mol Biol Rev 72:249–265. doi: 10.1128/MMBR.00040-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Varnum SM, Streblow DN, Monroe ME, Smith P, Auberry KJ, Pas L, Wang D, Ii DGC, Rodland K, Wiley S, Britt W, Shenk T, Smith RD, Nelson JA. 2004. Identification of proteins in human cytomegalovirus (HCMV) particles: the HCMV proteome. J Virol 78:10960–10966. doi: 10.1128/JVI.78.20.10960-10966.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Terhune SS, Schroer J, Shenk T. 2004. RNAs are packaged into human cytomegalovirus virions in proportion to their intracellular concentration. J Virol 78:10390–10398. doi: 10.1128/JVI.78.19.10390-10398.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Das S, Ortiz DA, Gurczynski SJ, Khan F, Pellett PE. 2014. Identification of human cytomegalovirus genes important for biogenesis of the cytoplasmic virion assembly complex. J Virol 88:9086–9099. doi: 10.1128/JVI.01141-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Das S, Pellett PE. 2011. Spatial relationships between markers for secretory and endosomal machinery in human cytomegalovirus-infected cells versus those in uninfected cells. J Virol 85:5864–5879. doi: 10.1128/JVI.00155-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Das S, Vasanji A, Pellett PE. 2007. Three-dimensional structure of the human cytomegalovirus cytoplasmic virion assembly complex includes a reoriented secretory apparatus. J Virol 81:11861–11869. doi: 10.1128/JVI.01077-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cepeda V, Esteban M, Fraile-Ramos A. 2010. Human cytomegalovirus final envelopment on membranes containing both trans-Golgi network and endosomal markers. Cell Microbiol 12:386–404. doi: 10.1111/j.1462-5822.2009.01405.x. [DOI] [PubMed] [Google Scholar]
- 8.Cepeda V, Fraile-Ramos A. 2011. A role for the SNARE protein syntaxin 3 in human cytomegalovirus morphogenesis. Cell Microbiol 13:846–858. doi: 10.1111/j.1462-5822.2011.01583.x. [DOI] [PubMed] [Google Scholar]
- 9.Xie M, Xuan B, Shan J, Pan D, Sun Y, Shan Z, Zhang J, Yu D, Li B, Qian Z. 2015. Human cytomegalovirus exploits interferon-induced transmembrane proteins to facilitate morphogenesis of the virion assembly compartment. J Virol 89:3049–3061. doi: 10.1128/JVI.03416-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sanchez V, Greis KD, Sztul E, Britt WJ. 2000. Accumulation of virion tegument and envelope proteins in a stable cytoplasmic compartment during human cytomegalovirus replication: characterization of a potential site of virus assembly. J Virol 74:975–986. doi: 10.1128/JVI.74.2.975-986.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tandon R, Mocarski ES. 2012. Viral and host control of cytomegalovirus maturation. Trends Microbiol 20:392–401. doi: 10.1016/j.tim.2012.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alwine JC. 2012. The human cytomegalovirus assembly compartment: a masterpiece of viral manipulation of cellular processes that facilitates assembly and egress. PLoS Pathog 8:e1002878. doi: 10.1371/journal.ppat.1002878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hook LM, Grey F, Grabski R, Tirabassi R, Doyle T, Hancock M, Landais I, Jeng S, McWeeney S, Britt W, Nelson JA. 2014. Cytomegalovirus miRNAs target secretory pathway genes to facilitate formation of the virion assembly compartment and reduce cytokine secretion. Cell Host Microbe 15:363–373. doi: 10.1016/j.chom.2014.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.AuCoin DP, Smith GB, Meiering CD, Mocarski ES. 2006. Betaherpesvirus-conserved cytomegalovirus tegument protein ppUL32 (pp150) controls cytoplasmic events during virion maturation. J Virol 80:8199–8210. doi: 10.1128/JVI.00457-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seo JY, Britt WJ. 2007. Cytoplasmic envelopment of human cytomegalovirus requires the postlocalization function of tegument protein pp28 within the assembly compartment. J Virol 81:6536–6547. doi: 10.1128/JVI.02852-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baxter MK, Gibson W. 2001. Cytomegalovirus basic phosphoprotein (pUL32) binds to capsids in vitro through its amino one-third. J Virol 75:6865–6873. doi: 10.1128/JVI.75.15.6865-6873.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Buchkovich NJ, Maguire TG, Alwine JC. 2010. Role of the endoplasmic reticulum chaperone BiP, SUN domain proteins, and dynein in altering nuclear morphology during human cytomegalovirus infection. J Virol 84:7005–7017. doi: 10.1128/JVI.00719-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Buchkovich NJ, Maguire TG, Paton AW, Paton JC, Alwine JC. 2009. The endoplasmic reticulum chaperone BiP/GRP78 is important in the structure and function of the human cytomegalovirus assembly compartment. J Virol 83:11421–11428. doi: 10.1128/JVI.00762-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Homman-Loudiyi M, Hultenby K, Britt W, Soderberg-Naucler C. 2003. Envelopment of human cytomegalovirus occurs by budding into Golgi-derived vacuole compartments positive for gB, Rab 3, trans-Golgi network 46, and mannosidase II. J Virol 77:3191–3203. doi: 10.1128/JVI.77.5.3191-3203.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moorman NJ, Sharon-Friling R, Shenk T, Cristea IM. 2010. A targeted spatial-temporal proteomics approach implicates multiple cellular trafficking pathways in human cytomegalovirus virion maturation. Mol Cell Proteomics 9:851–860. doi: 10.1074/mcp.M900485-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sanchez V, Sztul E, Britt WJ. 2000. Human cytomegalovirus pp28 (UL99) localizes to a cytoplasmic compartment which overlaps the endoplasmic reticulum-golgi-intermediate compartment. J Virol 74:3842–3851. doi: 10.1128/JVI.74.8.3842-3851.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Theiler RN, Compton T. 2002. Distinct glycoprotein O complexes arise in a post-Golgi compartment of cytomegalovirus-infected cells. J Virol 76:2890–2898. doi: 10.1128/JVI.76.6.2890-2898.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Indran SV, Ballestas ME, Britt WJ. 2010. Bicaudal D1-dependent trafficking of human cytomegalovirus tegument protein pp150 in virus-infected cells. J Virol 84:3162–3177. doi: 10.1128/JVI.01776-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith RM, Kosuri S, Kerry JA. 2014. Role of human cytomegalovirus tegument proteins in virion assembly. Viruses 6:582–605. doi: 10.3390/v6020582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tomtishen JP., III 2012. Human cytomegalovirus tegument proteins (pp65, pp71, pp150, pp28). Virol J 9:22. doi: 10.1186/1743-422X-9-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Scalzo AA, Corbett AJ, Rawlinson WD, Scott GM, Degli-Esposti MA. 2007. The interplay between host and viral factors in shaping the outcome of cytomegalovirus infection. Immunol Cell Biol 85:46–54. doi: 10.1038/sj.icb.7100013. [DOI] [PubMed] [Google Scholar]
- 27.Baldick CJ Jr, Marchini A, Patterson CE, Shenk T. 1997. Human cytomegalovirus tegument protein pp71 (ppUL82) enhances the infectivity of viral DNA and accelerates the infectious cycle. J Virol 71:4400–4408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bresnahan WA, Shenk TE. 2000. UL82 virion protein activates expression of immediate early viral genes in human cytomegalovirus-infected cells. Proc Natl Acad Sci U S A 97:14506–14511. doi: 10.1073/pnas.97.26.14506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bechtel JT, Shenk T. 2002. Human cytomegalovirus UL47 tegument protein functions after entry and before immediate-early gene expression. J Virol 76:1043–1050. doi: 10.1128/JVI.76.3.1043-1050.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hayashi ML, Blankenship C, Shenk T. 2000. Human cytomegalovirus UL69 protein is required for efficient accumulation of infected cells in the G1 phase of the cell cycle. Proc Natl Acad Sci U S A 97:2692–2696. doi: 10.1073/pnas.050587597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schierling K, Buser C, Mertens T, Winkler M. 2005. Human cytomegalovirus tegument protein ppUL35 is important for viral replication and particle formation. J Virol 79:3084–3096. doi: 10.1128/JVI.79.5.3084-3096.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Winkler M, Rice SA, Stamminger T. 1994. UL69 of human cytomegalovirus, an open reading frame with homology to ICP27 of herpes simplex virus, encodes a transactivator of gene expression. J Virol 68:3943–3954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu X, Shah S, Lee M, Dai W, Lo P, Britt W, Zhu H, Liu F, Zhou ZH. 2011. Biochemical and structural characterization of the capsid-bound tegument proteins of human cytomegalovirus. J Struct Biol 174:451–460. doi: 10.1016/j.jsb.2011.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fossum E, Friedel C, Rajagopala S, Titz B, Baiker A, Schmidt T, Kraus T, Stellberger T, Rutenberg C, Suthram S, Bandyopadhyay S, Rose D, von Brunn A, Uhlmann M, Zeretzke C, Dong Y-A, Boulet H, Koegl M, Bailer S, Koszinowski U, Ideker T, Uetz P, Zimmer R, Haas J. 2009. Evolutionarily conserved herpesviral protein interaction networks. PLoS Pathog 5:e1000570. doi: 10.1371/journal.ppat.1000570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tandon R, Mocarski ES. 2008. Control of cytoplasmic maturation events by cytomegalovirus tegument protein pp150. J Virol 82:9433–9444. doi: 10.1128/JVI.00533-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dai X, Yu X, Gong H, Jiang X, Abenes G, Liu H, Shivakoti S, Britt WJ, Zhu H, Liu F, Zhou ZH. 2013. The smallest capsid protein mediates binding of the essential tegument protein pp150 to stabilize DNA-containing capsids in human cytomegalovirus. PLoS Pathog 9:e1003525. doi: 10.1371/journal.ppat.1003525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu D, Silva MC, Shenk T. 2003. Functional map of human cytomegalovirus AD169 defined by global mutational analysis. Proc Natl Acad Sci U S A 100:12396–12401. doi: 10.1073/pnas.1635160100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dunn W, Chou C, Li H, Hai R, Patterson D, Stolc V, Zhu H, Liu F. 2003. Functional profiling of a human cytomegalovirus genome. Proc Natl Acad Sci U S A 100:14223–14231. doi: 10.1073/pnas.2334032100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Baldick CJ, Shenk T. 1996. Proteins associated with purified human cytomegalovirus particles. J Virol 70:6097–6105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tullman JA, Harmon ME, Delannoy M, Gibson W. 2014. Recovery of an HMWP/hmwBP (pUL48/pUL47) complex from virions of human cytomegalovirus: subunit interactions, oligomer composition, and deubiquitylase activity. J Virol 88:8256–8267. doi: 10.1128/JVI.00971-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Klupp BG, Fuchs W, Granzow H, Nixdorf R, Mettenleiter TC. 2002. Pseudorabies virus UL36 tegument protein physically interacts with the UL37 protein. J Virol 76:3065–3071. doi: 10.1128/JVI.76.6.3065-3071.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kelly BJ, Mijatov B, Fraefel C, Cunningham AL, Diefenbach RJ. 2012. Identification of a single amino acid residue which is critical for the interaction between HSV-1 inner tegument proteins pUL36 and pUL37. Virology 422:308–316. doi: 10.1016/j.virol.2011.11.002. [DOI] [PubMed] [Google Scholar]
- 43.Mijatov B, Cunningham AL, Diefenbach RJ. 2007. Residues F593 and E596 of HSV-1 tegument protein pUL36 (VP1/2) mediate binding of tegument protein pUL37. Virology 368:26–31. doi: 10.1016/j.virol.2007.07.005. [DOI] [PubMed] [Google Scholar]
- 44.Lee JH, Vittone V, Diefenbach E, Cunningham AL, Diefenbach RJ. 2008. Identification of structural protein-protein interactions of herpes simplex virus type 1. Virology 378:347–354. doi: 10.1016/j.virol.2008.05.035. [DOI] [PubMed] [Google Scholar]
- 45.Vittone V, Diefenbach E, Triffett D, Douglas MW, Cunningham AL, Diefenbach RJ. 2005. Determination of interactions between tegument proteins of herpes simplex virus type 1. J Virol 79:9566–9571. doi: 10.1128/JVI.79.15.9566-9571.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bucks MA, Murphy MA, O'Regan KJ, Courtney RJ. 2011. Identification of interaction domains within the UL37 tegument protein of herpes simplex virus type 1. Virology 416:42–53. doi: 10.1016/j.virol.2011.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kelly BJ, Bauerfeind R, Binz A, Sodeik B, Laimbacher AS, Fraefel C, Diefenbach RJ. 2014. The interaction of the HSV-1 tegument proteins pUL36 and pUL37 is essential for secondary envelopment during viral egress. Virology 454-455:67–77. [DOI] [PubMed] [Google Scholar]
- 48.Luxton GWG, Lee JI-H, Haverlock-Moyns S, Schober JM, Smith GA. 2006. The pseudorabies virus VP1/2 tegument protein is required for intracellular capsid transport. J Virol 80:201–209. doi: 10.1128/JVI.80.1.201-209.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wolfstein A, Nagel C-H, Radtke K, Döhner K, Allan VJ, Sodeik B. 2006. The inner tegument promotes herpes simplex virus capsid motility along microtubules in vitro. Traffic 7:227–237. doi: 10.1111/j.1600-0854.2005.00379.x. [DOI] [PubMed] [Google Scholar]
- 50.Shanda SK, Wilson DW. 2008. UL36p is required for efficient transport of membrane-associated herpes simplex virus type 1 along microtubules. J Virol 82:7388–7394. doi: 10.1128/JVI.00225-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Roberts AP, Abaitua F, O'Hare P, McNab D, Rixon FJ, Pasdeloup D. 2009. Differing roles of inner tegument proteins pUL36 and pUL37 during entry of herpes simplex virus type 1. J Virol 83:105–116. doi: 10.1128/JVI.01032-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Radtke K, Kieneke D, Wolfstein A, Michael K, Steffen W, Scholz T, Karger A, Sodeik B. 2010. Plus- and minus-end directed microtubule motors bind simultaneously to herpes simplex virus capsids using different inner tegument structures. PLoS Pathog 6:e1000991. doi: 10.1371/journal.ppat.1000991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Granzow H, Klupp BG, Thomas C, Mettenleiter TC. 2005. Entry of pseudorabies virus: an immunogold-labeling study entry of pseudorabies virus: an immunogold labeling study. J Virol 79:3200–3205. doi: 10.1128/JVI.79.5.3200-3205.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Krautwald M, Fuchs W, Klupp BG, Mettenleiter TC. 2009. Translocation of incoming pseudorabies virus capsids to the cell nucleus is delayed in the absence of tegument protein pUL37. J Virol 83:3389–3396. doi: 10.1128/JVI.02090-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu X, Fitzgerald K, Kurt-Jones E, Finberg R, Knipe DM. 2008. Herpesvirus tegument protein activates NF-kappaB signaling through the TRAF6 adaptor protein. Proc Natl Acad Sci U S A 105:11335–11339. doi: 10.1073/pnas.0801617105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pasdeloup D, Beilstein F, Roberts AP, McElwee M, McNab D, Rixon FJ. 2010. Inner tegument protein pUL37 of herpes simplex virus type 1 is involved in directing capsids to the trans-Golgi network for envelopment. J Gen Virol 91:2145–2151. doi: 10.1099/vir.0.022053-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pasdeloup D, Labetoulle M, Rixon FJ. 2013. Differing effects of herpes simplex virus 1 and pseudorabies virus infections on centrosomal function. J Virol 87:7102–7112. doi: 10.1128/JVI.00764-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Desai P, Sexton GL, Huang E, Person S. 2008. Localization of herpes simplex virus type 1 UL37 in the Golgi complex requires UL36 but not capsid structures. J Virol 82:11354–11361. doi: 10.1128/JVI.00956-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Newcomb WW, Brown JC. 2010. Structure and capsid association of the herpesvirus large tegument protein UL36. J Virol 84:9408–9414. doi: 10.1128/JVI.00361-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sandbaumhuter M, Dohner K, Schipke J, Binz A, Pohlmann A, Sodeik B, Bauerfeind R. 2013. Cytosolic herpes simplex virus capsids not only require binding inner tegument protein pUL36 but also pUL37 for active transport prior to secondary envelopment. Cell Microbiol 15:248–269. doi: 10.1111/cmi.12075. [DOI] [PubMed] [Google Scholar]
- 61.Desai P, Sexton GL, McCaffery JM, Person S. 2001. A null mutation in the gene encoding the herpes simplex virus type 1 UL37 polypeptide abrogates virus maturation. J Virol 75:10259–10271. doi: 10.1128/JVI.75.21.10259-10271.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Klupp BG, Granzow H, Mundt E, Mettenleiter TC. 2001. Pseudorabies virus UL37 gene product is involved in secondary envelopment. J Virol 75:8927–8936. doi: 10.1128/JVI.75.19.8927-8936.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.McElwee M, Beilstein F, Labetoulle M, Rixon FJ, Pasdeloup D. 2013. Dystonin/BPAG1 promotes plus-end-directed transport of herpes simplex virus 1 capsids on microtubules during entry. J Virol 87:11008–11018. doi: 10.1128/JVI.01633-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cardone G, Newcomb WW, Cheng N, Wingfield PT, Trus BL, Brown JC, Steven AC. 2012. The UL36 tegument protein of herpes simplex virus 1 has a composite binding site at the capsid vertices. J Virol 86:4058–4064. doi: 10.1128/JVI.00012-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Henaff D, Remillard-Labrosse G, Loret S, Lippe R. 2013. Analysis of the early steps of herpes simplex virus 1 capsid tegumentation. J Virol 87:4895–4906. doi: 10.1128/JVI.03292-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bucks MA, O'Regan KJ, Murphy MA, Wills JW, Courtney RJ. 2007. Herpes simplex virus type 1 tegument proteins VP1/2 and UL37 are associated with intranuclear capsids. Virology 361:316–324. doi: 10.1016/j.virol.2006.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Leelawong M, Lee JI, Smith Ga. 2012. Nuclear egress of pseudorabies virus capsids is enhanced by a subspecies of the large tegument protein that is lost upon cytoplasmic maturation. J Virol 86:6303–6314. doi: 10.1128/JVI.07051-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tischer B, Smith G, Osterrieder N. 2010. En passant mutagenesis: a two step markerless red recombination system. Methods Mol Biol (Clifton, NJ) 634:421–451. doi: 10.1007/978-1-60761-652-8_30. [DOI] [PubMed] [Google Scholar]
- 69.Tischer B, von Einem J, Kaufer B, Osterrieder N. 2006. Two-step red-mediated recombination for versatile high-efficiency markerless DNA manipulation in Escherichia coli. Biotechniques 40:191–198. doi: 10.2144/000112096. [DOI] [PubMed] [Google Scholar]
- 70.Sinzger C, Hahn G, Digel M, Katona R, Sampaio KL, Messerle M, Hengel H, Koszinowski U, Brune W, Adler B. 2008. Cloning and sequencing of a highly productive, endotheliotropic virus strain derived from human cytomegalovirus TB40/E. J Gen Virol 89:359–368. doi: 10.1099/vir.0.83286-0. [DOI] [PubMed] [Google Scholar]
- 71.Brock I, Kruger M, Mertens T, von Einem J. 2013. Nuclear targeting of human cytomegalovirus large tegument protein pUL48 is essential for viral growth. J Virol 87:6005–6019. doi: 10.1128/JVI.03558-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jahn G, Harthus HP, Broker M, Borisch B, Platzer B, Plachter B. 1990. Generation and application of a monoclonal antibody raised against a recombinant cytomegalovirus-specific polypeptide. Klin Wochenschr 68:1003–1007. doi: 10.1007/BF01646545. [DOI] [PubMed] [Google Scholar]
- 73.Schauflinger M, Fischer D, Schreiber A, Chevillotte M, Walther P, Mertens T, von Einem J. 2011. The tegument protein UL71 of human cytomegalovirus is involved in late envelopment and affects multivesicular bodies. J Virol 85:3821–3853. doi: 10.1128/JVI.01540-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tavalai N, Kraiger M, Kaiser N, Stamminger T. 2008. Insertion of an EYFP-pp71 (UL82) coding sequence into the human cytomegalovirus genome results in a recombinant virus with enhanced viral growth. J Virol 82:10543–10555. doi: 10.1128/JVI.01006-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chevillotte M, Landwehr S, Linta L, Frascaroli G, Lüske A, Buser C, Mertens T, von Einem J. 2009. Major tegument protein pp65 of human cytomegalovirus is required for the incorporation of pUL69 and pUL97 into the virus particle and for viral growth in macrophages. J Virol 83:2480–2570. doi: 10.1128/JVI.01818-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sampaio KL, Cavignac Y, Stierhof Y-D, Sinzger C. 2005. Human cytomegalovirus labeled with green fluorescent protein for live analysis of intracellular particle movements. J Virol 79:2754–2767. doi: 10.1128/JVI.79.5.2754-2767.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Buser C, Walther P, Mertens T, Michel D. 2007. Cytomegalovirus primary envelopment occurs at large infoldings of the inner nuclear membrane. J Virol 81:3042–3048. doi: 10.1128/JVI.01564-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Schauflinger M, Villinger C, Mertens T, Walther P, von Einem J. 2013. Analysis of human cytomegalovirus secondary envelopment by advanced electron microscopy. Cell Microbiol 15:305–314. doi: 10.1111/cmi.12077. [DOI] [PubMed] [Google Scholar]
- 79.Walther P, Ziegler A. 2002. Freeze substitution of high-pressure frozen samples: the visibility of biological membranes is improved when the substitution medium contains water. J Microsc 208:3–10. doi: 10.1046/j.1365-2818.2002.01064.x. [DOI] [PubMed] [Google Scholar]
- 80.Tandon R, Mocarski ES. 2011. Cytomegalovirus pUL96 is critical for the stability of pp150-associated nucleocapsids. J Virol 85:7129–7141. doi: 10.1128/JVI.02549-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Seo JY, Britt WJ. 2006. Sequence requirements for localization of human cytomegalovirus tegument protein pp28 to the virus assembly compartment and for assembly of infectious virus. J Virol 80:5611–5626. doi: 10.1128/JVI.02630-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Michael K, Bottcher S, Klupp BG, Karger A, Mettenleiter TC. 2006. Pseudorabies virus particles lacking tegument proteins pUL11 or pUL16 incorporate less full-length pUL36 than wild-type virus, but specifically accumulate a pUL36 N-terminal fragment. J Gen Virol 87:3503–3507. doi: 10.1099/vir.0.82168-0. [DOI] [PubMed] [Google Scholar]
- 83.Michael K, Klupp BG, Mettenleiter TC, Karger A. 2006. Composition of pseudorabies virus particles lacking tegument protein US3, UL47, or UL49 or envelope glycoprotein E. J Virol 80:1332–1339. doi: 10.1128/JVI.80.3.1332-1339.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Jambunathan N, Chouljenko D, Desai P, Charles AS, Subramanian R, Chouljenko VN, Kousoulas KG. 2014. Herpes simplex virus 1 protein UL37 interacts with viral glycoprotein gK and membrane protein UL20 and functions in cytoplasmic virion envelopment. J Virol 88:5927–5935. doi: 10.1128/JVI.00278-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Pitts JD, Klabis J, Richards AL, Smith GA, Heldwein EE. 2014. Crystal structure of the herpesvirus inner tegument protein UL37 supports its essential role in control of viral trafficking. J Virol 88:5462–5473. doi: 10.1128/JVI.00163-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Dingwell KS, Johnson DC. 1998. The herpes simplex virus gE-gI complex facilitates cell-to-cell spread and binds to components of cell junctions. J Virol 72:8933–8942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.To A, Bai Y, Shen A, Gong H, Umamoto S, Lu S, Liu F. 2011. Yeast two hybrid analyses reveal novel binary interactions between human cytomegalovirus-encoded virion proteins. PLoS One 6:e17796. doi: 10.1371/journal.pone.0017796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Phillips SL, Bresnahan WA. 2011. Identification of binary interactions between human cytomegalovirus virion proteins. J Virol 85:440–447. doi: 10.1128/JVI.01551-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Desai PJ. 2000. A null mutation in the UL36 gene of herpes simplex virus type 1 results in accumulation of unenveloped DNA-filled capsids in the cytoplasm of infected cells. J Virol 74:11608–11618. doi: 10.1128/JVI.74.24.11608-11618.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hyun JJ, Park HS, Kim KH, Kim HJ. 1999. Analysis of transcripts expressed from the UL47 gene of human cytomegalovirus. Arch Pharm Res 22:542–548. doi: 10.1007/BF02975323. [DOI] [PubMed] [Google Scholar]