Highlight
Transcriptomic analysis of the responses of Physcomitrella patens gametophytes to differential CO2 to O2 concentrations reveals extensive transcriptional reprogramming, photosynthetic acclimation, and altered oxidative signalling and defence responses.
Key words: Elevated carbon dioxide, microarray, Physcomitrella patens, sub-ambient oxygen.
Abstract
It is widely accepted that atmospheric O2 has played a key role in the development of life on Earth, as evident from the coincidence between the rise of atmospheric O2 concentrations in the Precambrian and biological evolution. Additionally, it has also been suggested that low atmospheric O2 is one of the major drivers for at least two of the five mass-extinction events in the Phanerozoic. At the molecular level, our understanding of the responses of plants to sub-ambient O2 concentrations is largely confined to studies of the responses of underground organs, e.g. roots to hypoxic conditions. Oxygen deprivation often results in elevated CO2 levels, particularly under waterlogged conditions, due to slower gas diffusion in water compared to air. In this study, changes in the transcriptome of gametophytes of the moss Physcomitrella patens arising from exposure to sub-ambient O2 of 13% (oxygen deprivation) and elevated CO2 (1500 ppmV) were examined to further our understanding of the responses of lower plants to changes in atmospheric gaseous composition. Microarray analyses revealed that the expression of a large number of genes was affected under elevated CO2 (814 genes) and sub-ambient O2 conditions (576 genes). Intriguingly, the expression of comparatively fewer numbers of genes (411 genes) was affected under a combination of both sub-ambient O2 and elevated CO2 condition (low O2–high CO2). Overall, the results point towards the effects of atmospheric changes in CO2 and O2 on transcriptional reprogramming, photosynthetic regulation, carbon metabolism, and stress responses.
Introduction
The moss Physcomitrella patens is a non-vascular, multicellular land plant believed to have diverged from the land plant lineage more than 400 million years ago (Nishiyama et al., 2003). P. patens has a relatively simple morphology and single-celled layer anatomy, thereby requiring constant co-equilibration of tissue water content with the environment (Reski, 1999; Quatrano et al., 2007; Charron and Quatrano, 2009; Cho et al., 2009). Such simple anatomical features imply the evolution of considerable intrinsic cellular and molecular mechanisms in response to abiotic stresses, and there is evidence to suggest that P. patens is highly tolerant of abiotic stresses (Frank et al., 2005; Cho et al., 2009; Mishler and Oliver, 2009; Koster et al., 2010).
It is widely accepted that atmospheric O2 has played a key role in the development of life on Earth, as evident from the coincidence between the rise of atmospheric O2 concentrations in the Precambrian and biological evolution (Lenton, 2003, Taylor and McElwain, 2010). Our understanding of the variations in atmospheric O2 concentrations throughout the Phanerozoic is largely derived from models based on geochemical cycling of carbon and sulphur, with predictions of atmospheric O2 as low as 13–20% at times in the Mesozoic (Belcher and McElwain, 2008; Berner, 2009). It has been suggested that low atmospheric O2 is one of the major drivers for at least two of the five mass-extinction events in the Phanerozoic (Lenton, 2003; Huey and Ward, 2005).
Variations exist in the responses of plants to oxygen deprivation. For example, marine angiosperms such as Zostera marina (eelgrass) exhibit a high tolerance to oxygen deprivation (Pulido and Borum, 2010), and some species that inhabit marshes and wetlands can tolerate low oxygen availability for short periods (Luo et al., 2012). At the molecular level, our understanding of the responses of plants to sub-ambient O2 concentrations is largely confined to studies of the responses of underground organs, e.g. roots, to hypoxic conditions (Bailey-Serres et al., 2012), although developing embryos in seeds (particularly in large fruits) can also experience periods of oxygen deprivation (Crawford, 2012). Oxygen deprivation often results in elevated CO2 levels, particularly under waterlogged conditions, due to slower gas diffusion in water compared to air (Greenway et al., 2006). When subjected to high concentrations of CO2, germinating chickpeas (Cicer arietinum) are less tolerant of oxygen deprivation (Crawford, 2012), suggesting an interaction between low O2 levels and elevated CO2 concentrations.
In this study, changes in the transcriptome of the P. patens arising from exposure to sub-ambient O2 (oxygen deprivation) and elevated CO2 were examined to further our understanding of the responses of non-vascular plants to changes in atmospheric composition. The results revealed previously unknown regulatory events associated with major physiological and biochemical responses to elevated CO2 or sub-ambient O2 alone, or in combination.
Material and methods
Plant growth
P. patens ecotype ‘Gransden 2004’ were propagated on cellophane overlay BCDAT agar plates under controlled conditions: photosynthetic photon flux density of 50 μmol s-1 m-2, 16h light/8h darkness, relative humidity of 75%, and 23°C in a Versatile Environmental Test Chamber, MLR-351 H (Sanyo, Japan). A mortar and pestle was used to homogenize 10–12-day-old moss protonemata in sterile distilled water. About 1–2mL of homogenized protonemal tissue was inoculated on sterile muslin cloth placed on top of sterile water-soaked Jiffy-7 peat pellets (Jiffy Products International AS, Kristansand, Norway) in GA7 Magenta boxes (Magenta Corporation, Chicago, IL, USA) containing 50–60mL sterile water. Adhesive microfiltration discs (18.6mm Ø, with inner filter area diameter of 10mm, and 2 μm pore size) (TQPL, Hampshire, UK) were pasted over a 5mm hole drilled through the lids used to close the G7 Magenta boxes. The containers were then sealed with Leukopor tape and incubated for 4–6 weeks under controlled conditions.
The 4–6-week-old P. patens gametophytes were transferred to Conviron BDW40 walk-in plant growth rooms (Conviron Europe Ltd., Isleham, UK) in University College Dublin Péac (Program for Experimental Atmospheres and Climate) and grown under the various conditions listed in Table 1. P. patens were acclimatized for 1 week under ambient CO2 and O2 conditions before being transferred to conditions of 1500 ppmV CO2 and ambient O2 (21%) (subsequently referred to as the ‘elevated CO2 treatment’), ambient CO2 (400 ppmV) and 13% O2 (subsequently referred to as the ‘sub-ambient O2 treatment’), and a combination of 1500 ppmV CO2 and 13% O2 (subsequently referred to as the ‘low O2–high CO2 treatment’), at a photosynthetic photon flux density of 50–70 μmol s-1 m-2, and 16h light/8h darkness. For control, gametophytes were kept on Jiffy-7 peat pellets in G7 Magenta boxes with microfiltration discs under ambient CO2 and O2 condition. Relative humidity within the chambers was maintained at 80% and chambers were set for midday peak temperature of 28°C and a night-time temperature of 18°C. P. patens gametophytes were grown under ambient and modified atmospheres for 7 days after which they were harvested at the same time and snap frozen in liquid N2 and stored at −80°C before RNA isolation. Special care was taken to avoid peat contamination.
Table 1.
Summary of experimental conditions used in this study
| Experimental condition | CO2 levels (ppm) | O2 levels (ppm) | CO2 to O2 ratio | CO2 to O2 ratio relative to ambient CO2 and O2 levels |
|---|---|---|---|---|
| Ambient | 400 | 210 000 | 0.0019:1 | 1:1 |
| Elevated CO2 | 1500 | 210 000 | 0.0071:1 | 3.75:1 |
| Sub-ambient O2 | 400 | 130 000 | 0.0030:1 | 1:0.619 |
| Low O2–high CO2 | 1500 | 130 000 | 0.0115:1 | 3.75:0.619 |
CO2 measurement using gas chromatography
Gas chromatography analyses were conducted for gas samples from within the GA7 Magenta containers where the P. patens were grown to determine the efficiency of gas exchange via the adhesive microfiltration discs fixed on the container lids. CO2 analysis was carried out on a gas chromatograph (Shimadzu GC-2014; Shimadzu Europa GmbH, Duisburg, Germany) fitted with an electron capture detector (carrier gas N2 at a flow rate of 20mL min-1) with an automated injection system (Loftfield et al., 1997). The gas peak area was recorded with the Peaksimple software (SRI Inc., Menlo Park, CA, USA) and used for determination of CO2 concentrations. The concentration of CO2 within the Magenta GA7 containers corresponded to the CO2 concentration within the growth chamber, indicating efficient gas exchange of gases via the adhesive microfiltration disc. CO2 concentration is used here as a proxy for efficiency of gas exchange via the adhesive microfiltration discs.
Total RNA isolation
Total RNA was extracted from P. patens gametophytes using the RNeasy Mini Kit (Qiagen, Manchester, UK) according to the protocol recommended by the manufacturer. Total RNA was eluted with 30 to 50 μL of RNAse-free water. Qualities and quantities of total RNA were determined using a Nanodrop ND 1000 Spectrophotometer (Nanodrop Technologies, Hemel Hempstead, UK). Total RNA integrity was determined with an Agilent 2100 BioAnalyzer (Agilent Technologies, Inc., Santa Clara, CA, USA).
Microarray analysis
Microarray analyses were conducted using a two-colour loop design (Churchill, 2002) to compensate for dye bias as the amount of label per amount of RNA is different for Cy3 and Cy5, by making duplicate hybridizations with the same sample using both Cy3 and Cy5 labelling and averaging the ratios from dye-swapped hybridizations. Three biological replicates from each experimental condition were analysed in duplicate by MOgene LC (St. Louis, MO, USA), a Agilent-certified service provider specializing in P. patens microarray analysis. The chip used was a 60-base oligomer cDNA array comprising a total of 22 895 ESTs generated by Leeds University, UK, and the National Institute for Basic Biology, Japan (Cuming et al., 2007; Rensing et al., 2008).
Total RNA (2.5 μg) was labelled using the ULS-Cy 3/5 ULS RNA Labelling Kit (Product #EA-006; Kreatech Biotechnology, San Diego, CA, USA). Following labelling of RNA with Cy3 and Cy5 fluorescent dyes, equal amounts of RNA (1 μg) were mixed in nuclease-free water and processed using the Gene Expression Hybridization Kit (Product #5188–5242; Agilent Technologies Inc.). For hybridization, samples were placed between the Agilent backing slide and microarray chip, sealed in the hybridization chamber, and hybridized for 17h in a 60°C rotating hybridization oven. After hybridization, the slides were washed sequentially in 6× SSC buffer (0.15M NaCl, 15nM Na-acetate, pH 7.0) at room temperature, then 0.1× SSC on ice. Nitrogen gas was used to dry the slides before scanning using a DNA Microarray Scanner (#G2565BA; Agilent Technologies Inc.) with the Agilent Scan Control software. The fluorescent intensities of each feature were extracted using Feature Extraction Software with default parameters (Ver. 9.1, Agilent Technologies Inc.). The raw intensity data were then log (base e = 2.718) transformed for normalization before ANOVA (mixed model) analysis. When the raw intensities of both Cy3 and Cy5 channels were below 150 (typical background intensity is 40) and the signal-to-background ratio below 2, genes were removed from further analysis. Log ratios among different samples and the P-values were calculated using a mixed model (Wolfinger et al., 2001). A significant change in gene expression was based on the 2-fold cut-off with P-values <0.05 (Cuming et al., 2007).
Microarray data were normalized against ambient condition (ambient CO2 and O2) to examine the effect of different concentrations of CO2 and O2 on gene expression in P. patens gametophytes. Recent annotations of the Physcomitrella gene models were retrieved from the Department of Energy Joint Genome Institute (http://genome.jgi-psf.org/Phypa1_1/Phypa1_1.home.html) (Zimmer et al., 2013). Functional annotations based on BLAST2GO analysis were used for a detailed analysis of the response of P. patens gametophytes to the different experimental atmospheres in this study.
Semi-quantitative reverse transcriptase PCR
Reverse transcriptase (RT)-PCR was conducted as previously described (Xiong et al., 2009; O’Donoghue et al., 2013). Briefly, total RNA was isolated using the Qiagen RNeasy kit according to the manufacturer’s recommendations. The quality and quantity of the total RNA was determined using a NanoDrop 1000 spectrophotometer. One microgram of total RNA was treated with 1U of DNase I (Invitrogen, Hemel Hempstead, UK) before being used for cDNA synthesis using 200U of M-MLV Reverse Transcriptase (Invitrogen). The resultant cDNA was used for PCR amplification using 0.5U of Go-Taq DNA polymerase (Promega, Southampton, UK). The sequences of the primers used for amplification are listed in Supplementary Table S1.
Results and discussion
In this study, the effects of a 7-day exposure to elevated CO2 (1500 ppmV), sub-ambient O2 (13%), and low O2–high CO2 [combination of elevated CO2 (1500 ppmV) and sub-ambient O2 (13%)] on transcriptomic changes in P. patens gametophytes were examined. No observable morphological changes were observed in P. patens gametophytes under these modified atmospheric conditions (data not shown).
Elevated CO2 evoked large-scale transcriptome response in P. patens gametophytes
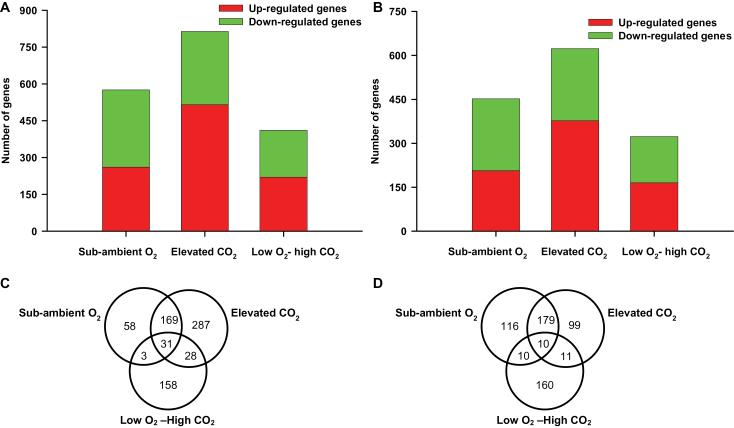
Microarray analysis revealed that the expression of a large number of genes (n = 814, fold change ≥ 2; P < 0.05), relative to ambient conditions (control), were significantly affected when P. patens gametophytes were grown under elevated CO2 conditions. The expression of a relatively smaller number of genes was observed under sub-ambient O2 and low O2–high CO2 treatment, in which a total of 576 and 411 genes were significantly affected, respectively (Fig. 1A). Of the 814 genes significantly affected under elevated CO2, 63% were up-regulated, whereas only 45% and 54% of the transcripts were up-regulated in response to sub-ambient O2, and low O2–high CO2 treatment, respectively (Fig. 1A). Interestingly, the capacity of elevated CO2 to up-regulate gene expression in P. patens gametophytes was reduced by half when atmospheric O2 content was reduced from 21% to 13% (Fig. 1A), suggesting a possible interaction between atmospheric concentrations of CO2 and O2 on plant responses. Of the 814 genes expressed in response to elevated CO2, a total of 623 (377 up-regulated and 246 down-regulated) genes showed homology to annotated genes from Arabidopsis thaliana (Fig. 1B). Out of 576 expressed genes in response to sub-ambient O2, 206 up-regulated and 246 down-regulated genes showed homology with A. thaliana genes. In low O2–high CO2 treatment, out of 411 expressed genes, 165 up-regulated and 158 down-regulated Physcomitrella genes showed homology with A. thaliana genes (Fig. 1B).
Fig. 1.
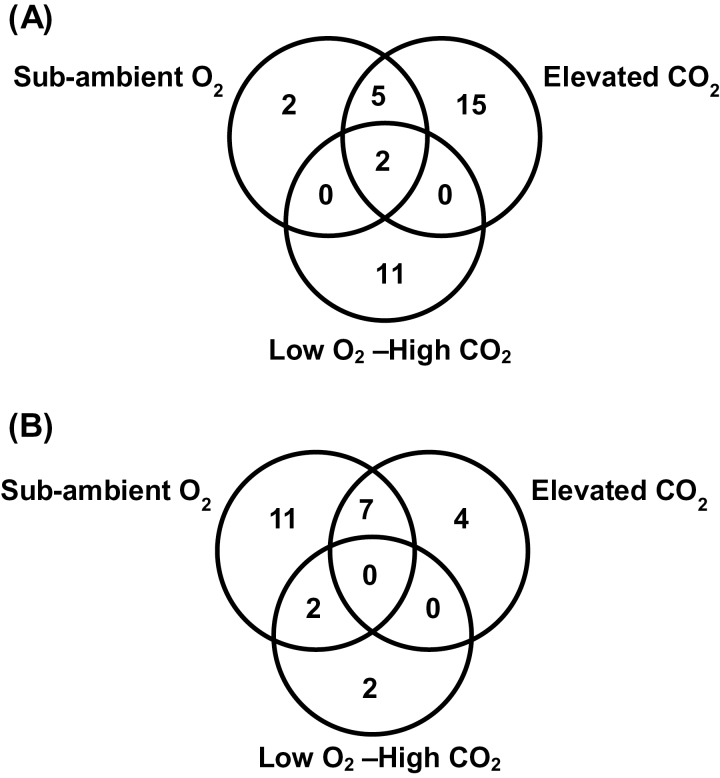
Microarray analysis of the number of genes whose expression are affected in P. patens gametophytes following a 7-day exposure to sub-ambient O2 (13%), elevated CO2 (1500 ppmV), and low O2–high CO2 compared to ambient CO2 and O2 level. (A) The number of transcripts exhibiting significant changes, relative to control ambient conditions (fold change ≥ 2; P ≤ 0.05, red and green indicates number of up- or down-regulated genes respectively) are indicated. (B) Numbers of P. patens transcripts homologous to Arabidopsis genes are indicated. (C) Venn diagram showing the number of P. patens genes commonly or exclusively up-regulated and (D) down-regulated in response to elevated CO2 (1500 ppmV), sub-ambient O2 (13%), and low O2–high CO2 compared to control ambient environment.
There was considerable overlap between the sets of genes up-regulated in sub-ambient O2 and elevated CO2 of 169 genes. Only 3 up-regulated genes showed overlap between sub-ambient O2 and low O2–high CO2 treatment. Elevated CO2 and low O2–high CO2 treatment shared a set of 28 up-regulated genes (Fig. 1C). Of the genes analysed, 58 genes were exclusively up-regulated under sub-ambient O2, whereas the expression of larger numbers of genes were specifically up-regulated under elevated CO2 (287 genes), and low O2–high CO2 treatment (158 genes) (Fig. 1C). Thirty-one genes were up-regulated in all three conditions. There was also a significant overlap between the set of genes down-regulated in the various atmospheres (Fig. 1D). The response to elevated CO2 and sub-ambient O2 was strikingly similar in moss gametophytes, with 179 genes commonly down-regulated. Expression was down-regulated in a greater number of genes under low O2–high CO2 treatment (160 genes) than under elevated CO2 (99 genes) and sub-ambient O2 (116 genes) (Fig. 1D).
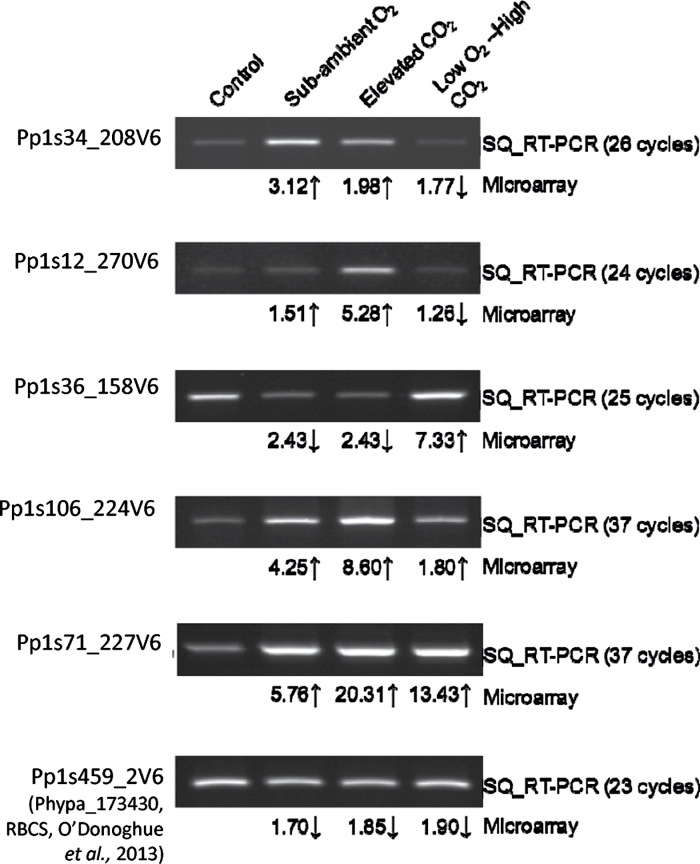
To validate the microarray data, a selection of genes identified as being differentially expressed in microarray experiments under conditions of sub-ambient O2, elevated CO2, and low O2–high CO2 treatment were chosen for semi-quantitative RT-PCR analysis. The results of semi-quantitative RT-PCR showed expression patterns consistent with data from the microarray dataset (Fig. 2). In agreement with O’Donoghue et al. (2013), who successfully used semi-quantitative RT-PCR to validate their microarray dataset from P. patens, the semi-quantitative RT-PCR data presented here clearly show that the microarray data can be used to indicate changes in gene expression levels in P. patens gametophytes under the various atmospheric conditions examined (Fig. 2).
Fig. 2.
RT-PCR validation of microarray data. RNA was extracted from gametophytes and semi-quantitative RT-PCR was performed using primers designed to amplify a selection of genes identified as being differentially expressed in microarray experiments under conditions of sub-ambient O2, elevated CO2, and low O2–high CO2. The corresponding fold-change as identified from microarray experiments is indicated below each gel band. Amplified DNA was run on a 1% (w/v) agarose gel and stained with ethidium bromide before visualization under UV illumination. Images shown are representative of three independent biological replicates.
MapMan molecular functional classification analysis was performed to understand the overall transcriptomic response in P. patens to the different atmospheres using the corresponding A. thaliana locus IDs (AGIs) as a reference to categorize differentially expressed P. patens genes using a web-based interface, The Bio-Array Resource (http://bar.utoronto.ca/welcome.htm) (Provart and Zhu, 2003). Approximately half of the P. patens genes possess homologues in A. thaliana (data not shown). Significantly expressed genes were classified into 31 different functional categories. The data suggest that the expression of genes involved in metabolic processes, stress, photosynthesis and transport functions are significantly altered in response to all 3 atmospheric conditions (Supplementary Fig. S1).
Genes implicated in transcriptional regulation
Microarray results showed that the expression of large numbers of transcription factors was significantly regulated in response to elevated CO2. A total of 16 and 12 transcripts encoding transcription factors were up- and down-regulated, respectively (Supplementary Table S2). MYB transcription factor family proteins, zinc-binding family proteins, and Integrase-type DNA-binding superfamily protein were among the highly expressed transcription-related genes. Interestingly, there was a significant overlap between up- and down-regulated transcription factor genes in response to sub-ambient O2 and elevated CO2. Of the 12 up-regulated and 8 down-regulated genes under sub-ambient O2 (13%), a set of 10 up-regulated and 3 down-regulated genes encoding transcription factors showed common expression patterns with genes expressed under elevated CO2 (Supplementary Table S2 and Supplementary Fig. S2). The effects of a low O2–high CO2 condition were small, with only one and five genes encoding transcription factors in P. patens exclusively up- and down-regulated, respectively (Supplementary Table S2). Together, these data suggest transcriptional reprogramming in response to a 7-day exposure to modified atmospheres, with elevated CO2 and sub-ambient O2 conditions eliciting a greater extent of transcriptional reprogramming compared to a 7-day exposure to a low O2–high CO2 treatment (Supplementary Table S2 and Supplementary Fig. S2). It is unclear why exposure to a low O2–high CO2 treatment resulted in a lesser extent of transcriptional reprogramming. However, it is possible that plants have not, throughout their evolutionary history, experienced this unique atmospheric condition (low O2–high CO2) and, as such, have not evolved the necessary regulatory transcriptional mechanism(s) to respond effectively.
Differential expression of photosynthetic and carbon metabolism genes
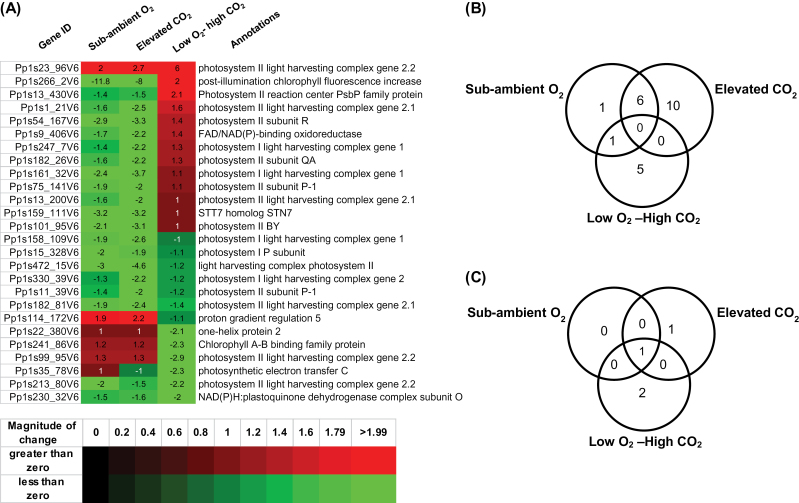
Differential expression of genes encoding proteins involved in photosynthesis and carbohydrate metabolism-related functions was also observed, suggesting the existence of a metabolic response strategy to elevated CO2 or sub-ambient O2 individually, or low O2–high CO2 treatments (Fig. 3) in P. patens gametophytes. Elevated atmospheric CO2 can exert profound effects on photosynthesis in many plants (Stitt and Krapp, 1999; Long et al., 2004; Dermody et al., 2008; Leakey et al., 2009). Bryophytes generally obtain CO2 by diffusion and are not limited by opening and closing of stomata. Atmospheric CO2 diffuses through the cell wall into the cytosol and dissolves in cell wall or apoplast water to form bicarbonate (HCO3 -). It is made available to the site of CO2 consumption by the primary carboxylating enzyme, ribulose-1,5-carboxylase/oxygenase (RuBP) in the chloroplast stroma (Price et al., 2011). Under elevated CO2, an initial increase in the rate of carbon fixation was observed in many C3 plants, resulting in the accumulation of soluble sugars such as glucose, fructose, and sucrose, which are the main products of photosynthetic carbon assimilation (Moore et al., 1997; Ainsworth, 2008). Reduced photosynthetic capacity under long-term exposure to high CO2 is mainly attributed to increased accumulation of storage carbohydrates (Chen et al., 2005; Ainsworth and Long, 2005). This down-regulation or inhibition of photosynthetic activity is generally referred to as the acclimation responses of plants to elevated levels of atmospheric CO2 (Stitt and Krapp, 1999). There is evidence indicating that increased levels of CO2 can modulate expression of several photosynthesis- and Calvin-cycle–related genes (Rogers et al., 1996; Gesch et al., 1998; Moore et al., 1999; Fukayama et al., 2009). Here, P. patens orthologues were identified for many annotated Arabidopsis transcripts functionally associated with the photochemical reactions of photosynthesis, e.g. the light-harvesting complex LHCII. In photosynthetic organisms, LHCII absorbs and transfers excitation energy to the photosystem antenna (Galka et al., 2012). A total of 16 LHCII-related genes were significantly down-regulated under elevated CO2 whereas only eight LHCII genes were down-regulated under sub-ambient O2, of which six were commonly repressed in both conditions. Expression of three transcripts was repressed in response to low O2–high CO2 treatment (Fig. 3A,B; Supplementary Table S3). Only three light reaction-related genes were up-regulated in P. patens gametophytes exposed to a low O2–high CO2 treatment. One gene, Pp1s23_96V6, encoding a chlorophyll a-b binding protein of LHCII type I protein showed induced expression in all three experimental conditions. Moreover, expression of only one and two light reaction-related genes were induced independently in elevated CO2 and low O2-–high CO2 treatment, respectively (Supplementary Table S3). The data suggest a reduction in photochemical reactions of photosynthesis in P. patens gametophytes under elevated CO2 conditions. However, the expression of genes encoding proteins involved in light reactions was not significantly affected in P. patens gametophytes under low O2–high CO2 treatment, indicating that photochemical reactions of photosynthesis are likely to be functional in this environmental regime in moss gametophytes.
Fig. 3.
Differentially expressed P. patens genes involved in photochemical reactions of photosynthesis in response to elevated CO2 (1500 ppmV), sub-ambient O2 (13%), and low O2–high CO2. (A) Heat map of the significantly expressed transcripts of photosynthesis based on MapMan functional classification. (B) Venn diagram showing overlapping transcripts that are significantly down-regulated in P. patens gametophytes exposed to elevated CO2, sub-ambient O2, or low O2–high CO2 treatment. (C) Venn diagram showing overlapping transcripts that are significantly up-regulated in P. patens gametophytes exposed to elevated CO2, sub-ambient O2, or low O2–high CO2 treatment.
The effect of different atmospheric conditions on expression of genes involved in CO2 fixation, RuBP regeneration, and starch synthesis in moss gametophytes was also examined. Under elevated CO2 treatment, the expression of a total of 21 genes involved in CO2 fixation, RuBP regeneration, and starch biosynthesis was altered. Expression of a set of 14 and 26 genes involved in CO2 fixation, RuBP regeneration, and starch biosynthesis was altered in response to sub-ambient O2 and low O2–high CO2 treatments, respectively (Table 2, Supplementary Fig. S3). Interestingly, diverse transcriptome responses by moss gametophytes to the low O2–high CO2 treatment condition were observed. Differential expression of a greater number of transcripts (total 13: 4 up-regulated and 9 down-regulated) was specifically detected upon exposure to the low O2–high CO2 treatment condition (Supplementary Fig. S3). These down-regulated P. patens transcripts encode for major components of CO2 fixation: carbonic anhydrase, a protein that catalyses reversible conversion of CO2 to HCO3 -; RuBP small subunit; glyceraldehyde-3-phosphate dehydrogenase; and RuBP activase (Table 2). Overall, 19 genes encoding proteins related to CO2 fixation and RuBP regeneration were down-regulated in P. patens gametophytes under low O2–high CO2 treatment (Table 2). Six transcripts encoding enzymes of starch synthesis, such as ADP-glucose pyrophosphorylase (AGPase) and starch synthase, were highly expressed in P. patens gametophytes under low O2–high CO2 treatment compared to sub-ambient O2 and elevated CO2 (Table 2), suggesting the accumulation of soluble sugars in P. patens gametophytes. For example, P. patens gene Pp1s98_52V6, encoding a glucose-1-phosphate adenylyltransferase large subunit (AGPase), showed greater than 19-fold down-regulation upon exposure to sub-ambient O2, and 13-fold down-regulation to elevated CO2, while expression of the same gene was strongly induced (≥ 4-fold) under low O2–high CO2 treatment (Table 2). Exposure to the low O2–high CO2 condition resulted in elevated expression of genes associated with starch metabolism and starch degradation in moss gametophytes; whereas starch metabolism was partially affected under sub-ambient O2 and elevated CO2 conditions (Table 2). In photosynthetic organisms, photosynthesis is controlled by a sugar-dependent metabolic feedback mechanism. Carbohydrate-dependent feedback inhibition on photosynthetic activity represses expression of RuBP small subunit, RuBP activase, and chlorophyll a/b binding proteins (Gesch et al., 1998; Moore et al., 1999; Stitt and Krapp 1999; Chen et al., 2005). The results suggest that expression of genes functioning in photosynthetic carbon fixation and RuBP regeneration activity may be affected by feedback inhibition mechanisms due to accumulation of soluble sugars. In plants, sugars are important components of the sugar sensing and signalling network that regulate plant growth and development (Rolland et al., 2006). Sugar availability modulates global gene expression patterns via highly complex mechanisms controlling transcription, translation, and protein stability (Rolland et al., 2006). Transcriptome profiling indicates differential responses of P. patens gametophytes at the molecular level to elevated CO2, sub-ambient O2, and low O2–high CO2 treatments. The Hexokinase 1 (HXK1) protein is identified as one of the conserved sugar sensors implicated in controlling expression of genes involved in primary carbon fixation and RuBP regeneration in plants (Jang et al., 1997, Xiao et al., 2000, Rolland et al., 2006).
Table 2.
Altered expression of P. patens genes involved in CO2 fixation, RuBP regeneration and starch synthesis in response to sub-ambient O2, elevated CO2 and low O2-high CO2
| Function | Enzymes | Gene ID | Sub-ambient O2 | Elevated CO2 | Low O2-high CO2 |
|---|---|---|---|---|---|
| CO2 fixation | Carbonic anhydrase | Pp1s264_55V6 | -2.64 | -2.45 | -2.07 |
| Pp1s43_118V6 | - | - | -2.46 | ||
| Pp1s30_234V6 | - | - | 2.34 | ||
| RuBP small subunit | Pp1s204_93V6 | -2.09 | -2.07 | -2.24 | |
| Pp1s188_39V6 | 2.6 | 2.54 | -2.61 | ||
| Pp1s374_50V6 | - | - | -2.4 | ||
| Pp1s66_48V6 | - | - | -2.19 | ||
| Pp1s545_4V6 | - | - | -2.7 | ||
| RuBP activase | Pp1s5_83V6 | - | -2.02 | -2.06 | |
| Pp1s258_44V6 | - | -2.11 | -2.14 | ||
| Pp1s199_130V6 | - | - | -3.09 | ||
| Pp1s199_129V6 | - | - | -2.69 | ||
| GAPDH | Pp1s10_228V6 | 2.42 | 2.01 | 2.52 | |
| Pp1s135_21V6 | -2.81 | -2.42 | - | ||
| Pp1s9_47V6 | -2.29 | - | - | ||
| Pp1s49_34V6 | - | 2.98 | - | ||
| Pp1s414_8V6 | - | -2.22 | -3.36 | ||
| RuBP regeneration | SBPase | Pp1s429_29V6 | - | -2.15 | -2.07 |
| FBPase | Pp1s385_43V6 | -2.07 | - | - | |
| Pp1s242_66V6 | - | -2.12 | -2.24 | ||
| Pp1s163_36V6 | - | - | 3.18 | ||
| Aldolase | Pp1s50_50V6 | - | - | -2.86 | |
| Pp1s475_27V6 | - | - | -2.93 | ||
| Pp1s33_389V6 | - | - | -2.1 | ||
| Starch synthesis | Starch synthase | Pp1s124_155V6 | -4.76 | -4.39 | 13.98 |
| Pp1s234_74V6 | -2.02 | -2.61 | - | ||
| Pp1s93_98V6 | - | 2.86 | - | ||
| Pp1s150_86V6 | - | -2.46 | - | ||
| Pp1s302_51V6 | - | - | 3.3 | ||
| Pp1s104_136V6 | - | - | 2.26 | ||
| GBSS | Pp1s12_341V6 | - | -2.52 | - | |
| ADP-GPP | Pp1s397_21V6 | 4.07 | 2.07 | 11.61 | |
| Pp1s389_5V6 | -2.57 | -2.53 | - | ||
| Pp1s98_52V6 | -19.99 | -13.98 | 4.59 | ||
| Pp1s36_158V6 | -2.43 | -2.43 | 7.33 | ||
| Pp1s2_204V6 | 3.28 | 2.73 | - |
ADP-GPP, ADP-glucose pyrophosphorylase; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; GBSS, granule-bound starch synthase.
Differentially altered expression of two P. patens transcripts encoding for HXK1 protein was observed. Transcript abundance of P. patens gene Pp1s12_19V6 encoding HXK1 protein was significantly increased in response to low O2–high CO2 treatments (Supplementary Table S4A). Another transcript, Pp1s414_10V6, encoding HXK1 protein was highly repressed under sub-ambient O2 and elevated CO2 conditions. In higher plants, sugar-induced elevation of calcium-dependent protein kinases, SNF1-related kinases, and Ca2+-fluxes are associated with sugar signalling where Ca2+ acts as a second messenger (Rolland et al., 2006). The data presented here revealed down-regulation (≥ 6-fold) of SNF1 kinase homolog 11 gene (Pp1s107_182V6) in response to both sub-ambient O2 and elevated CO2 treatments (Supplementary Table S4A). One calcium-dependent protein kinase gene (Pp1s309_91V6) was induced under elevated CO2. Accumulation of two CBL-interacting serine/threonine-protein kinase genes (Pp1s58_13V6 and Pp1s79_209V6) was detected in gametophytes subjected to sub-ambient O2 and elevated CO2 treatments (Supplementary Table S4A).
Elevations in transcript abundance of the five genes encoding enzymes of glycolysis were observed only in the low O2–high CO2 condition (Supplementary Table S4B). Significant overlap between the sets of genes expressed (one up-regulated and two down-regulated) in sub-ambient O2 and elevated CO2 was observed, suggesting that moss gametophytes responded similarly to the sub-ambient O2 or elevated CO2 treatments at the molecular level. This was supported by another identical expression pattern of genes involved in glycolysis (Supplementary Table S4B). More elevated transcript abundance of the genes encoding enzymes of glycolysis was observed only in the low O2–high CO2 condition (Supplementary Table S4B), indicating that long-term exposure to a higher CO2 to O2 ratio may have triggered foliar respiration in moss gametophytes due to higher substrate availability (Davey et al., 2004; Ainsworth et al., 2006).
Membrane transporters
Large-scale changes in the P. patens transcriptome occurred in response to elevated CO2. A large number of transcripts (n = 31) involved in membrane transport functions was significantly up-regulated in response to the elevated CO2 condition compared to the sub-ambient O2 and low O2–high CO2 conditions, where 12 and 8 genes were up-regulated respectively (Supplementary Table S5A). These membrane transport–related P. patens genes were homologues of Arabidopsis transporter genes such as NRAMP metal ion transporter 4, ABC transporter family protein, glutathione S-conjugate transporter, and calcium-transporting ATPase. A total of 17, 15, and 13 genes were down-regulated under the sub-ambient O2, elevated CO2, and low O2–high CO2 treatments, respectively (Supplementary Table S5B). Of the 31 up-regulated genes in response to elevated CO2 treatment, 22 genes were uniquely expressed in this condition (Supplementary Fig. S4 and Supplementary Table S5A). Interestingly, all eight genes up-regulated in the low O2–high CO2 treatment were exclusively expressed (Supplementary Fig. S4). In the sub-ambient O2 treatment, of the 12 up-regulated membrane transporter genes, nine genes showed overlap with genes up-regulated in the elevated CO2 condition (Supplementary Fig. S4 and Supplementary Table S5A). These data suggest that elevated CO2 intensified expression of membrane transport function. By contrast, exposure to the low O2–high CO2 condition had significantly less effect on the expression of transporter genes in gametophytes.
Effects on hormone metabolism and signal transduction
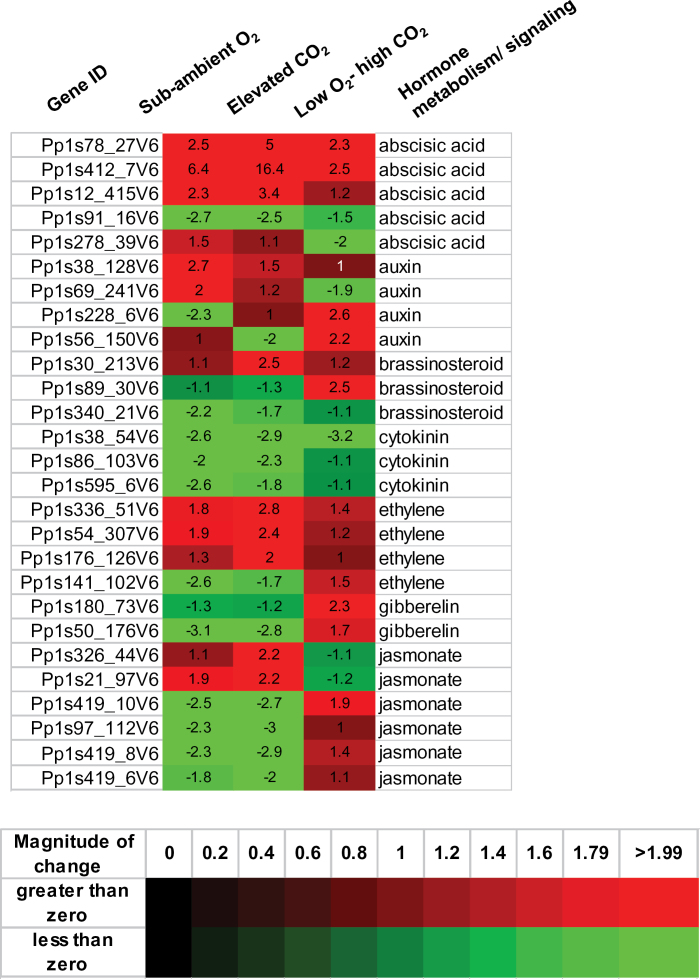
The impact of changing atmospheric CO2 levels on plant hormone metabolism is not well understood (Ribeiro et al., 2012). Plant hormones play vital roles in metabolic adjustment and modulation of gene expression under various environmental conditions to maintain plant growth and development (Bohnert et al., 1995; Kempa et al., 2008). Abscisic acid (ABA) plays a central role in adaptive responses to various abiotic stresses in plants. The ABA biosynthetic pathway in plants is regulated by 9-cis-epoxycarotenoid dioxygenase (NCED) genes. Markedly induced transcript levels of NCED9 (Pp1s412_7V6) were observed in microarray analysis in response to elevated CO2 (fold change ≥ 16) and sub-ambient O2 (fold change ≥ 6) compared to the low O2–high CO2 condition (fold change ≥ 2) (Supplementary Table S6), suggesting that the ABA pool may have increased in moss gametophytes grown in elevated CO2 and sub-ambient O2 conditions. Notably, expression of key genes involved in metabolism of hormones such as auxin, brassinosteroid, cytokinin, ethylene, gibberellin, and jasmonate was altered significantly in moss gametophytes exposed to these different atmospheric conditions (Supplementary Table S6). Heat-map analysis of expressed P. patens genes of hormone metabolism demonstrated differential molecular responses to elevated CO2 and sub-ambient O2 individually and in combination (Fig. 4). MapMan ontology analysis revealed that exposure to elevated CO2 intensified up-regulation of ABA, brassinosteroid, and ethylene metabolism-related genes (Fig. 4 and Supplementary Table S6). In plants, jasmonic acid, a lipid-based hormone that regulates anti-herbivore defences, is a product of the octadecanoid pathway (Wasternack, 2007). Expression of two genes encoding for 12-oxophytodienoate reductase activity (OPR1 and OPR2) was strongly up-regulated and 4 lipoxygenase 3 (LOX3) genes the in octadecanoid pathway were down-regulated, indicating that jasmonate metabolism was also altered by elevated CO2 treatment (Supplementary Table S6). However, elevated CO2 significantly dampened expression of genes involved in gibberellin, cytokinin, and auxin metabolism (Fig. 4 and Supplementary Table S6). Upon exposure to sub-ambient O2 conditions, methylesterase (associated with auxin biosynthesis) and an O-fucosyltransferase-like protein gene involved in cytokinin metabolism were highly expressed in moss gametophytes (Supplementary Table S6), whereas expression of ethylene-, brassinosteroid-, cytokinin-, gibberellin-, and jasmonate-related genes was down-regulated. Interestingly, ABA-, auxin-, and gibberellin-related genes were partially induced or repressed in moss gametophytes grown under the low O2–high CO2 condition (Supplementary Table S6). Additionally, in contrast to elevated CO2 and sub-ambient O2 treatments, exposure to low O2–high CO2 treatment had no effect on expression of ethylene metabolism– or ethylene signalling–related genes (Fig. 4 and Supplementary Table S6). These results suggest that elevated CO2 induced reprogramming of hormone metabolism in moss gametophytes and highlight the differential molecular acclimation potential of P. patens gametophytes to changing environmental conditions.
Fig. 4.
Effects of elevated CO2, sub-ambient O2 and low O2-high CO2 treatments on expression of genes involved in hormone metabolism in P. patens gametophytes. Heat map of the significantly up and down-regulated genes based on MapMan functional classification.
Elevated CO2 triggers expression of stress-related genes
MapMan analysis of P. patens genes using Arabidopsis homologues as a reference revealed significant alteration in stress-related transcripts (Supplementary Fig. S1). Thirty-three stress-related genes responded to elevated CO2 (22 up-regulated and 11 down-regulated) (Fig. 5A,B and Supplementary Table S7), while altered expression of 29 stress-related transcripts (9 up-regulated and 20 down-regulated) was observed in moss gametophytes exposed to sub-ambient O2 (Supplementary Table S7). By contrast, only 17 (13 up-regulated and 4 down-regulated) stress-associated genes were expressed in response to the low O2–high CO2 condition (Supplementary Table S7). The expression of a comparatively large number of stress- and defence-related genes was repressed in sub-ambient O2 and elevated CO2 conditions (Fig. 5B). We observed an increase in the transcript abundance of the ABA biosynthesis enzyme, PpNCED9 (≥ 16-fold) upon elevated CO2 treatment, suggesting that ABA levels are likely to have increased in moss gametophytes. ABA accumulation in plants under stress conditions induces transcriptional reprogramming, eliciting many responses at the physiological, biochemical, and molecular levels (Shinozaki and Yamaguchi-Shinozaki, 1996). Application of exogenous ABA induced rapid transcriptional responses in P. patens protonemata (Cuming et al., 2007; Richardt et al., 2010 Shinde et al., 2012), and induces stress responses and stress tolerance in P. patens (Frank et al., 2005; Khandelwal et al., 2010; Shinde et al., 2012). Proteins involved in stress signalling and stress tolerance were highly accumulated in P. patens gametophytes after ABA treatment (Wang et al., 2009, Cui et al., 2012). In this study, it was observed that transcripts involved in stress-related functions were highly induced upon elevated CO2 treatment (Supplementary Table S7). Evidence for a triggered transcriptional response of stress-response genes in moss gametophytes grown under elevated CO2 can potentially be attributed to the increased expression of transcripts encoding a key enzyme in the ABA biosynthetic pathway.
Fig. 5.
Effects of elevated CO2, sub-ambient O2, and low O2–high CO2 treatments on expression of stress-related genes in P. patens gametophytes. Venn diagram represents number of commonly and distinctly (A) up-regulated and (B) down-regulated genes.
Recently, increased oxidative stress in plants grown under elevated CO2 was reported (Qiu et al., 2008, Gillespie et al., 2011). In C3 plants, despite expected reduced levels of reactive oxygen species under elevated CO2, oxidative signalling is emerging as an unexpected component of plant response to elevated CO2. Antioxidant enzymes act as scavengers and are associated with cellular detoxification of reactive oxygen species during oxidative stress (Mittler, 2002; 2006). Significant up-regulation of genes encoding enzymes associated with oxidative stress response and signalling and redox regulation was observed in P. patens gametophytes subjected to elevated CO2 treatment (Table 3). A total of 13 and 11 genes encoding for these enzymes and genes with putative functions in detoxification were up-regulated under elevated CO2 and low O2–high CO2 treatment respectively. These genes included peroxidase, catalase, 12-oxophytodienoate reductase, and glutathione s-transferase (Table 3). Only three genes, Pp1s34_208V6 (L-galactono-1,4-lactone dehydrogenase), Pp1s98_250V6 (GDP-D-mannose 3′,5′-epimerase), and Pp1s396_10V6 (SOUL heme-binding family protein), involved in redox balance were up-regulated in P. patens under sub-ambient O2 treatment (Table 3). Additionally, a set of 11 genes mainly encoding peroxidase superfamily protein, thioredoxin superfamily protein, and catalase 1 were significantly up-regulated in P. patens gametophytes under low O2–high CO2 condition. Antioxidant enzymes such as catalase, dehydroascorbate reductase, glutathione-dependent formaldehyde dehydrogenase, and peroxidase were among the highly expressed P. patens genes under this condition (Table 3). Interestingly, only a single transcript coding for VTC2__mannose-1-phosphate guanylyltransferase protein was commonly expressed under elevated CO2 and low O2–high CO2 treatments, indicating differential sensing, signalling, and stress responses in P. patens (Table 3). Together, microarray data indicate that moss gametophytes are likely to have experienced oxidative stress under elevated CO2 levels, which intensified transcriptional responses associated with the acquisition of abiotic stress tolerance.
Table 3.
Up-regulated P. patens genes involved in oxidative signalling and oxidative stress responses
| Gene ID | Annotation | Arabidopsis Gene Identifier | Sub-ambient O2 | Elevated CO2 | Low O2– high CO2 |
|---|---|---|---|---|---|
| Pp1s34_208V6 | L-galactono-1,4-lactone dehydrogenase | AT3G47930 | 3.12 | - | |
| Pp1s98_250V6 | GDP-D-mannose 3\′,5\′-epimerase | AT5G28840 | 2.45 | 2.20 | - |
| Pp1s396_10V6 | SOUL haem-binding family protein | AT5G20140 | 2.80 | 2.70 | - |
| Pp1s98_9V6 | tetraticopeptide domain-containing thioredoxin | AT3G17880 | - | 2.54 | - |
| Pp1s505_9V6 | Rubredoxin-like superfamily protein | AT5G51010 | - | 2.61 | - |
| Pp1s27_275V6 | inositol monophosphatase family protein | AT3G02870 | - | 2.21 | - |
| Pp1s404_1V6 | microsomal glutathione s-transferase, putative | AT1G65820 | - | 2.54 | - |
| Pp1s66_172V6 | glutathione S-transferase PHI 9 | AT2G30860 | - | 2.35 | - |
| Pp1s182_83V6 | microsomal glutathione s-transferase, putative | AT1G65820 | - | 2.21 | - |
| Pp1s224_120V6 | multidrug resistance-associated protein 2 | AT2G34660 | - | 2.02 | - |
| Pp1s21_97V6 | 12-oxophytodienoate reductase 2 | AT1G76690 | - | 2.23 | - |
| Pp1s326_44V6 | 12-oxophytodienoate reductase 1 | AT1G76680 | - | 2.28 | - |
| Pp1s184_82V6 | peroxidase superfamily protein | AT5G06730 | - | 2.55 | - |
| Pp1s114_207V6 | myoinositol-1-phosphate guanylyltransferase | AT4G26850 | - | 2.25 | 4.22 |
| Pp1s20_77V6 | peroxidase superfamily protein | AT5G14130 | - | - | 2.17 |
| Pp1s306_39V6 | peroxidase superfamily protein | AT5G05340 | - | - | 2.91 |
| Pp1s273_43V6 | thioredoxin superfamily protein | AT1G07700 | - | - | 5.83 |
| Pp1s106_67V6 | thioredoxin superfamily protein | AT4G03520 | - | - | 2.42 |
| Pp1s178_130V6 | myoinositol-1-phosphate guanylyltransferase | AT4G26850 | - | - | 2.29 |
| Pp1s71_207V6 | hemoglobin 1 | AT2G16060 | - | - | 2.51 |
| Pp1s40_134V6 | 1-cysteine peroxiredoxin 1 | AT1G48130 | - | - | 2.93 |
| Pp1s98_113V6 | copper chaperone for SOD1 | AT1G12520 | - | - | 2.03 |
| Pp1s223_74V6 | catalase 1 | AT1G20630 | - | - | 6.68 |
| Pp1s506_15V6 | GroES-like zinc-binding dehydrogenase family protein | AT5G43940 | - | - | 2.66 |
Conclusion
Anthropogenic activities have contributed to accelerate the emission of CO2 into the atmosphere. Elevated CO2 is recognized as a major contributing factor to the effects that global climate change exerts on plants (IPCC, 2013). In the present study, changes in the transcriptome of gametophytes of the moss P. patens to differential CO2 to O2 concentrations have been profiled to understand moss acclimation responses to elevated CO2, sub-ambient O2, and low O2–high CO2 conditions at the molecular level. Microarray analyses showed that expression of P. patens genes related to CO2 fixation, RuBP regeneration, and starch synthesis was significantly altered, indicating photosynthetic acclimation of P. patens exposed to high CO2 to O2 concentrations. It is likely that elevated CO2 caused an accumulation of sugars and starch in P. patens as increased transcript abundance of genes encoding enzymes in starch synthesis were observed. Accumulation of soluble sugars may have also contributed to the decrease in RuBP and RuBP activase transcripts in P. patens. However, quantitative analysis of RuBP, chlorophyll, and sugars such as sucrose, fructose, and glucose under elevated CO2 must be conducted before definitive conclusions can be drawn. The result also indicated that elevated CO2 in the presence of ambient O2 and sub-ambient levels of O2 evoked large-scale transcriptional reprogramming of P. patens gametophytes, and changes in oxidative signalling and defence responses. Based on the transcriptome data, it is hypothesized that transcriptional reprogramming may reflect differences in the CO2 to O2 ratio of the imposed experimental atmospheres (Table 1). It will be interesting in future experiments to vary the concentrations of CO2 and O2 and examine the effects of a similar CO2 to O2 ratio for all three atmospheric conditions on the transcriptome. This will enable us to gain better insights into the transcriptional responses exhibited by P. patens gametophytes subjected to different atmospheric CO2 and O2 concentrations.
Supplementary data
Fig. S1. MapMan molecular functional classification of P. patens genes (using Arabidopsis homologues as a reference) where the expression levels were significantly up- and down-regulated following a 7-day exposure to (A) sub-ambient O2 (up-regulated), (B) sub-ambient O2 (down-regulated), (C) elevated CO2 (up-regulated), (D) elevated CO2 (down-regulated), (E) Low O2–high CO2 (up-regulated), and (F) Low O2–high CO2 (down-regulated).
Fig. S2. Genes encoding transcriptional regulators that were (A) up- and (B) down-regulated in P. patens gametophytes subjected to by elevated CO2 (1500 ppmV), sub-ambient O2 (13%), and low O2–high CO2.
Fig. S3. Effects of elevated CO2, sub-ambient O2, and low O2–high CO2 treatment on expression of genes encoding proteins involved in CO2 fixation, RuBP regeneration, and starch synthesis in moss P. patens gametophytes. Venn diagram showing the number of commonly and distinctly expressed (up- and down-regulated) genes.
Fig. S4. Membrane transport-related genes that were (A) up- and (B) down-regulated in P. patens gametophytes subjected to by elevated CO2 (1500 ppmV), sub-ambient O2 (13%), and low O2–high CO2.
Table S1. List of probes and their respective primer sequences used for semi-quantitative RT-PCR.
Table S2. Significantly regulated P. patens genes encoding transcription factors following a 7-day exposure to sub-ambient O2 (13%), elevated CO2 (1500 ppmV), and low O2–high CO2 treatment.
Table S3. Effect of elevated CO2 or sub-ambient O2 individually or in combination (low O2–high CO2) on P. patens transcripts functionally associated with the photochemical reactions of photosynthesis.
Table S4A. Altered expression of highly expressed P. patens transcripts involved in signal transduction regulated by metabolic sugar in response to elevated CO2 or sub-ambient O2 individually or in combination (low O2–high CO2).
Table S4B. Regulation of P. patens transcripts involved in glycolysis under elevated CO2 or sub-ambient O2 individually or in combination (low O2–high CO2).
Table S5A. Highly induced P. patens transcripts functionally associated with membrane transport (based on MapMan analysis).
Table S5B. Significantly down-regulated P. patens transcripts functionally associated with membrane transport.
Table S6. Effect of differential CO2 to O2 levels on P. patens transcripts involved in hormone metabolism (based on MapMan analysis using Arabidopsis homologues).
Table S7. Effect of differential CO2 to O2 levels on expression of P. patens stress-associated transcripts (based on MapMan analysis using Arabidopsis homologues).
Acknowledgements
This study was supported by a Science Foundation Ireland (SFI) Research Frontiers Programme Grant (08/SFI/EOB1087) to CK-YN and by the ERC-2011-StG279962-OXYEVOL grant to J.C.M. AB is supported by an Iranian Government Research Scholarship. We thank Drs Matthew Haworth and Caroline Elliott-Kingston for their assistance with plant growth in modified atmospheres. We also thank Sandip S Shinde and Dr Vasanth Singan for assistance in microarray data analysis.
References
- Ainsworth EA. 2008. Rice production in a changing climate: a meta-analysis of responses to elevated carbon dioxide and elevated ozone concentration. Global Change Biology 14, 1642–1650. [Google Scholar]
- Ainsworth EA, Long SP. 2005. What have we learned from 15 years of free-air CO2 enrichment (FACE)? A meta-analytic review of the responses of photosynthesis, canopy properties and plant production to rising CO2 . New Phytologist 165, 351–372. [DOI] [PubMed] [Google Scholar]
- Ainsworth EA, Rogers A, Vodkin LO, Walter A, Schurr U. 2006. The effects of elevated CO2 concentration on soybean gene expression. An analysis of growing and mature leaves. Plant Physiology 142, 135–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey-Serres J, Fukao T, Gibbs DJ, Holdsworh MJ, Lee SC, Licausi F, Perata P, Voesenek LACJ, van Dongen JT. 2012. Making sense of low oxygen sensing. Trends in Plant Science 17, 129–138. [DOI] [PubMed] [Google Scholar]
- Belcher CM, McElwain JC. 2008. Limits for combustion in low O2 redefine paleoatmospheric predictions for the Mesozoic. Science 321, 1197–1200. [DOI] [PubMed] [Google Scholar]
- Berner RA. 2009. Phanerozoic atmospheric oxygen: New results using the GEOCARBSULF model. American Journal of Science 309, 603–606. [Google Scholar]
- Bohnert HJ, Nelson DE, Jensen RG. 1995. Adaptations to environmental stresses. Plant Cell 7, 1099–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charron AJ, Quatrano RS. 2009. Between a rock and a dry place: the water-stressed moss. Molecular Plant 2, 478–486. [DOI] [PubMed] [Google Scholar]
- Chen GY, Yong ZH, Liao Y, Zhang DY, Chen Y, Zhang HB, Chen J, Zhu JG, Xu DQ. 2005. Photosynthetic acclimation in rice leaves to free-air CO2 enrichment related to both ribulose-1, 5-bisphosphate carboxylation limitation and ribulose-1, 5-bisphosphate regeneration limitation. Plant and Cell Physiology 46, 1036–1045. [DOI] [PubMed] [Google Scholar]
- Cho SH, Schwartzenberg K, Quatrano R. 2009. The role of abscisic acid in stress tolerance. Annual Plant Reviews 36, 282–297. [Google Scholar]
- Churchill GA. 2002. Fundamentals of experimental design for cDNA microarrays. Nature Genetics 32, 490–495. [DOI] [PubMed] [Google Scholar]
- Crawford RMM. 2012. Long-term anoxia tolerance in flowering plants. In: Altenbach AV, Bernhard JM, Seckbach S, eds. Anoxia: Evidence for Eukaryote Survival and Paleontological Strategies, Cellular Origin, Life in Extreme Habitats and Astrobiology, Volume 21 Heibelberg: Springer Science+Business BV, 219–246. [Google Scholar]
- Cui S, Hu J, Guo S, Wang J, Cheng Y, Dang X, Wu L, He Y. 2012. Proteome analysis of Physcomitrella patens exposed to progressive dehydration and rehydration. Journal of Experimental Botany 63, 711–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuming AC, Cho SH, Kamisugi Y, Graham H, Quatrano RS. 2007. Microarray analysis of transcriptional responses to abscisic acid and osmotic, salt, and drought stress in the moss, Physcomitrella patens . New Phytologist 176, 275–287. [DOI] [PubMed] [Google Scholar]
- Davey PA, Hunt S, Hymus GJ, DeLucia EH, Drake BG, Karnosky DF, Long SP. 2004. Respiratory oxygen uptake is not decreased by an instantaneous elevation of [CO2], but is increased with long-term growth in the field at elevated CO2 . Plant Physiology 134, 520–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dermody O, Long SP, McConnaughay K, DeLucia EH. 2008. How do elevated CO2 and O3 affect the interception and utilization of radiation by a soybean canopy? Global Change Biology 14, 556–564. [Google Scholar]
- Frank W, Ratnadewi D, Reski R. 2005. Physcomitrella patens is highly tolerant against drought, salt and osmotic stress. Planta 220, 384–394. [DOI] [PubMed] [Google Scholar]
- Fukayama H, Fukuda T, Masumoto C, Taniguchi Y, Sakai H, Cheng W, Hasegawa T, Miyao M. 2009. Rice plant response to long term CO2 enrichment: gene expression profiling. Plant Science 177, 203–210. [Google Scholar]
- Galka P, Santabarbara S, Khuong TTH, Degand H, Morsomme P, Jennings RC, Boekema EJ, Caffarri S. 2012. Functional analyses of the plant photosystem I-light-harvesting complex II supercomplex reveal that light-harvesting complex II loosely bound to photosystem II is a very efficient antenna for photosystem I in state II. Plant Cell 24, 2963–2978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gesch RW, Boote KJ, Vu JCV, Hartwell Allen L, Bowes G. 1998. Changes in growth CO2 result in rapid adjustments of ribulose-1,5-bisphosphate carboxylase/oxygenase small subunit gene expression in expanding and mature leaves of rice. Plant Physiology 118, 521–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie KM, Xu F, Ritcher KT, McGrath JM, Markelz RJ, Ort DR, Leakey AD, Ainsworth EA. 2011. Greater antioxidant and respiratory metabolism in field-grown soybean exposed to elevated O3 under both ambient and elevated CO2 . Plant, Cell and Environment 35, 169–184. [DOI] [PubMed] [Google Scholar]
- Greenway H, Armstrong W, Colmer TD. 2006. Conditions leading to high CO2 (>5 kPA) in waterlogged-flooded soils ad possible effects on root growth and metabolism. Annals of Botany 98, 9–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huey RB, Ward PD. 2005. Hypoxia, global warming, and terrestrial late Permian extinctions. Science 308, 398–401. [DOI] [PubMed] [Google Scholar]
- IPCC. 2013. Climate change 2013: the physical science basis. Working Group I contribution to the Intergovernmental Panel on Climate Change. (http://www.ipcc.ch/report/ar5/wg1/#.Ulay4VAqjwF)
- Jang JC, León P, Zhou L, Sheen J. 1997. Hexokinase as a sugar sensor in higher plants. Plant Cell 9, 15–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempa S, Krasensky J, Dal Santo S, Kopka J, Jonak C. 2008. A central role of abscisic acid in stress-regulated carbohydrate metabolism. PLoS One 3, e3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khandelwal A, Cho S, Marella H, Sakata Y, Perroud PF, Pan A, Quatrano R. 2010. Role of ABA and ABI3 in desiccation tolerance. Science 327, 546. [DOI] [PubMed] [Google Scholar]
- Koster KL, Balsamo RA, Espinoza C, Oliver MJ. 2010. Desiccation sensitivity and tolerance in the moss Physcomitrella patens: assessing limits and damage. Plant Growth Regulation 62, 293–302. [Google Scholar]
- Leaky ADB, Ainsworth EA, Bernacchi CJ, Rogers A, Long SP. 2009. Elevated CO2 effects on plant carbon, nitrogen, and water relations: six important lessons from FACE. Journal of Experimental Botany 60, 2859–2876. [DOI] [PubMed] [Google Scholar]
- Lenton TM. 2003. The impact of the physical environment. In: L, Rothschild A, Lister, eds. Evolution of Planet Earth: The Impact of the Physical Environment. London: Academic Press, 33–51. [Google Scholar]
- Loftfield N, Flessa H, Augustin J, Besse F. 1997. Automated gas chromatographic system for rapid analysis of the atmospheric trace gases methane, carbon dioxide, and nitrous oxide. Journal of Environmental Quality 26, 560–564. [Google Scholar]
- Long SP, Ainsworth EA, Rogers A, Ort DR. 2004. Rising atmospheric carbon dioxide: plants FACE the future. Annual Review of Plant Biology 55, 591–628. [DOI] [PubMed] [Google Scholar]
- Luo F, Thiele B, Janzik I, Zeng B, Schur U, Matsubara S. 2012. De-submergence responses of antioxidative defense systems in two wetland plants having escape and quiescence strategies. Journal of Plant Physiology 169, 1680–1689. [DOI] [PubMed] [Google Scholar]
- Mishler BD, Oliver MJ. 2009. Putting Physcomitrella patens on the tree of life: the evolution and ecology of mosses. Annual Plant Reviews 36, 1–15. [Google Scholar]
- Mittler R. 2002. Oxidative stress, antioxidants and stress tolerance. Trends in Plant Science 7, 405–410. [DOI] [PubMed] [Google Scholar]
- Mittler R. 2006. Abiotic stress, the field environment and stress combination. Trends in Plant Science 11, 15–19. [DOI] [PubMed] [Google Scholar]
- Moore B, Cheng SH, Sims D, Seemann J. 1999. The biochemical and molecular basis for photosynthetic acclimation to elevated atmospheric CO2 . Plant, Cell and Environment 22, 567–582. [Google Scholar]
- Moore BD, Palmquist DE, Seemann JR. 1997. Influence of plant growth at high CO2 concentrations on leaf content of ribulose-1,5-bisphosphate carboxylase/oxygenase and intracellular distribution of soluble carbohydrates in tobacco, snapdragon, and parsley. Plant Physiology 115, 241–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama T, Fujita T, Shin-I T, Seki M, Nishide H, Uchiyama I, Kamiya A, Carninci P, Hayashizaki Y, Shinozaki K. 2003. Comparative genomics of Physcomitrella patens gametophytic transcriptome and Arabidopsis thaliana: implication for land plant evolution. Proceedings of the National Academy of Sciences USA 100, 8007–8012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donoghue M-T, Chater C, Wallace S, Gray JE, Beerling DJ, Fleming AJ. 2013. Genome-wide transcriptomic analysis of the sporophyte of the moss Physcomitrella patens . Journal of Experimental Botany 64, 3567–3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price GD, Badger MR, von Caemmerer S. 2011. The prospect of using cyanobacterial bicarbonate transporters to improve leaf photosynthesis in C3 crop plants. Plant Physiologist 155, 20–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provart N, Zhu T. 2003. A Browser-based Functional Classification SuperViewer for Arabidopsis Genomics. Current Topics in Computational Molecular Biology 271–272. [Google Scholar]
- Pulido C, Borum J. 2010. Eelgrass (Zostera marina) tolerance to anoxia. Journal of Experimental Marine Biology and Ecology 385, 8–13. [Google Scholar]
- Qiu QS, Huber JL, Booker FL, Jain V, Leakey ADB, Fiscus EL, Yau PM, Ort DR, Huber SC. 2008. Increased protein carbonylation in leaves of Arabidopsis and soybean in response to elevated [CO2]. Photosynthesis Research 97, 155–166. [DOI] [PubMed] [Google Scholar]
- Quatrano RS, McDaniel SF, Khandelwal A, Perroud PF, Cove DJ. 2007. Physcomitrella patens: mosses enter the genomic age. Current Opinion in Plant Biology 10, 182–189. [DOI] [PubMed] [Google Scholar]
- Rensing SA, Lang D, Zimmer AD, Terry A, Salamov A, Shapiro H, Nishiyama T, Perroud PF, Lindquist EA, Kamisugi Y. 2008. The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science 319, 64–69. [DOI] [PubMed] [Google Scholar]
- Reski R. 1999. Molecular genetics of Physcomitrella . Planta 208, 301–309. [Google Scholar]
- Ribeiro DM, Araújo WL, Fernie AR, Schippers JH, Mueller-Roeber B. 2012. Action of gibberellins on growth and metabolism of Arabidopsis plants associated with high concentration of carbon dioxide. Plant Physiology 160, 1781–1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardt S, Timmerhaus G, Lang D, Qudeimat E, Corrêa LG, Reski R, Rensing SA, Frank W. 2010. Microarray analysis of the moss Physcomitrella patens reveals evolutionarily conserved transcriptional regulation of salt stress and abscisic acid signalling. Plant Molecular Biology 72, 27–45. [DOI] [PubMed] [Google Scholar]
- Rogers G, Milham P, Gillings M, Conroy J. 1996. Sink strength may be the key to growth and nitrogen responses in N-deficient wheat at elevated CO2 . Functional Plant Biology 23, 253–264. [Google Scholar]
- Rolland F, Baena-Gonzalez E, Sheen J. 2006. Sugar sensing and signalling in plants: conserved and novel mechanisms. Annual Review of Plant Biology 57, 675–709. [DOI] [PubMed] [Google Scholar]
- Shinde S, Nurul Islam M, Ng CKY. 2012. Dehydration stress-induced oscillations in LEA protein transcripts involves abscisic acid in the moss, Physcomitrella patens . New Phytologist 195, 321–328. [DOI] [PubMed] [Google Scholar]
- Shinozaki K, Yamaguchi-Shinozaki K. 1996. Molecular responses to drought and cold stress. Current Opinion in Biotechnology 7, 161–67. [DOI] [PubMed] [Google Scholar]
- Stitt M, Krapp A. 1999. The interaction between elevated carbon dioxide and nitrogen nutrition: the physiological and molecular background. Plant, Cell and Environment 22, 583–621. [Google Scholar]
- Taylor CT, McElwain JC. 2010. Ancient atmospheres and the evolution of oxygen sensing via the hypoxia-inducible factor in metazoans. Physiology 25, 272–279. [DOI] [PubMed] [Google Scholar]
- Wang X, Yang P, Zhang X, Xu Y, Kuang T, Shen S, He Y. 2009. Proteomic analysis of the cold stress response in the moss, Physcomitrella patens . Proteomics 9, 4529–4538. [DOI] [PubMed] [Google Scholar]
- Wasternack C. 2007. Jasmonates: an update on biosynthesis, signal transduction and action in plant stress response, growth and development. Annals of Botany 100, 681–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfinger RD, Gibson G, Wolfinger ED, Bennett L, Hamadeh H, Bushel P, Afshari C, Paules R. 2001. Assessing gene significance from cDNA microarray expression data via mixed models. Journal of Computational Biology 8, 625–637. [DOI] [PubMed] [Google Scholar]
- Xiao W, Sheen J, Jang JC. 2000. The role of hexokinase in plant sugar signal transduction and growth and development. Plant Molecular Biology 44, 451–461. [DOI] [PubMed] [Google Scholar]
- Xiong TC, Hann CM, Chambers JP, Surget M, Ng CKY. 2009. An inducible, modular system for spatio-temporal control of gene expression in stomatal guard cells. Journal of Experimental Botany 60, 4129–4136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmer AD, Lang D, Buchta K, Rombauts S, Nishiyama T, Hasebe M, Van de Peer Y, Rensing SA, Reski R. 2013. Reannotation and extended community resources for the genome of the non-seed plant Physcomitrella patens provide insights into the evolution of plant gene structures and functions. BMC Genomics, 14, 498. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.