Abstract
Assessment of virus neutralization (VN) activity in 176 pigs infected with porcine reproductive and respiratory syndrome virus (PRRSV) identified one pig with broadly neutralizing activity. A Tyr-10 deletion in the matrix protein provided escape from broad neutralization without affecting homologous neutralizing activity. The role of the Tyr-10 deletion was confirmed through an infectious clone with a Tyr-10 deletion. The results demonstrate differences in the properties and specificities of VN responses elicited during PRRSV infection.
TEXT
Porcine reproductive and respiratory syndrome virus (PRRSV) causes respiratory disease in young pigs, reduced performance in growing pigs, and reproductive failure in gilts and sows (1). Generally, vaccination with modified live virus (MLV) or previous natural infection results in only homologous protection against closely related isolates (2). Under some circumstances, protection is expanded to include more genetically distant viruses (3). The paucity of heterologous protection is attributed to a number of factors, such as antigenic drift in B and T cell epitopes (4, 5) and glycan shielding of conserved epitopes (6).
The enveloped virion surface contains at least six proteins. The major proteins, GP5 and M, encoded by open reading frames (ORFs) 5 and 6, respectively, form a disulfide-linked heterodimer (7). The minor surface glycoproteins, GP2, GP3, and GP4, encoded by ORFs 2, 3, and 4, respectively, form a noncovalent heterotrimer (8). Finally, there are two small nonglycosylated proteins, E and 5a, encoded by ORFs 2b and 5a, respectively (9–11). As summarized in Fig. S1 in the supplemental material, several previous studies have identified multiple neutralizing epitopes distributed among the major and minor surface proteins. For example, a PEPSCAN analysis of Lelystad virus (LV), a type 1 virus, identified a short peptide sequence in GP4 as the epitope linked with virus neutralization (VN) by a monoclonal antibody (MAb) prepared against purified virions (12) (epitope c in Fig. S1D in the supplemental material). The same region in GP4 was identified as a target for neutralizing antibodies derived from experimentally infected pigs (13). Furthermore, Costers et al. (14) recovered neutralization-resistant viruses propagated in the presence of an anti-GP4 MAb. Using peptide-specific antibodies, Vanhee et al. (15) characterized additional neutralizing epitopes in GP2 and GP3 (epitopes a and b in Fig. S1A in the supplemental material and epitopes a and b in Fig. S1C in the supplemental material). Ostrowski et al. (16) and Plagemann et al. (17) described an epitope in GP5 in a type 2 genotype virus located in the vicinity of two conserved glycosylation sites in the ectodomain region (epitopes a and b in Fig. S1F in the supplemental material). A similar epitope is found in GP5 of a closely related arterivirus, lactate dehydrogenase-elevating virus (LDV) (18). In an effort to understand the role of envelope-associated proteins in the cross-neutralization of genetically distinct PRRSV isolates, Kim and Yoon (19) reacted neutralizing swine serum with a panel of chimeric viruses constructed of structural genes derived from neutralization-sensitive and neutralization-resistant viruses. When individual ORFs were replaced, the largest increase in VN resistance or susceptibility was obtained following the exchange of GP3 or GP5. The search for additional epitopes has become more complicated by a recent report describing nsp2 as a virus-associated protein (20).
One explanation for the absence of agreement in the characterization of PRRSV neutralizing epitopes is a lack of understanding regarding the homologous versus heterologous nature of the different antibody reagents used in experiments. We hypothesize that homologous versus heterologous neutralization outcomes are the product of the recognition of different epitopes on the PRRSV proteome. Furthermore, we predict the existence of a new class of heterologous PRRSV antibody, referred to as broadly neutralizing antibody (BNAb). This hypothesis of BNAb for PRRSV is based on HIV studies, in which the screening sera from thousands of patients, or populations of HIV-specific B cells from individual patients, resulted in the identification of antibodies with the capacity to neutralize a wide range of HIV isolates (21–23). Several linear and conformational broadly neutralizing epitopes located in GP120 and GP41 have been identified (24). Similar results have been described for hepatitis C virus, dengue virus, West Nile virus, and influenza virus (reviewed in reference 39).
In this study, BNAb for PRRSV by evaluating the virus neutralization (VN) response of pigs experimentally infected with PRRSV. All experiments involving animals and viruses were approved by the Kansas State University institutional animal care and biosafety committee. As part of a study on host genetics associated with PRRS (25–28), 200 pigs, 7 weeks of age, were experimentally infected with a type 2 PRRSV isolate, KS62 (GenBank accession number KM035798), and porcine circovirus 2b (PCV2b) (GenBank accession number JQ692110) (42). Four weeks prior, half of the pigs were vaccinated with a commercial MLV vaccine (Ingelvac PRRS MLV; Boehringer Ingelheim) according to the label instructions (GenBank accession number AF159149). VR2332 (GenBank number AY150564) is the parent for the vaccine strain. Forty-two days after virus challenge, serum samples from 89 vaccinated and 87 nonvaccinated pigs were assayed for the presence of VN activity against the homologous isolate and six genetically diverse isolates, including a type 1 virus. The percent identities for the seven viruses are described in Table 1. For ORFs 2 through 6, the nucleotide identity ranged from 64.5% (LV versus NVSL) to 97.8% (KS62 versus NVSL). The GP5 amino acid sequence identity was 55% for LV versus KS06, compared to 96.5% for KS62 versus NVSL (data not shown). A comparison of all the envelope-associated structural proteins, including GP2, E, GP3, GP4, 5a, GP5, and M, for the seven isolates is shown in Fig. S1 in the supplemental material. Consistent with previous reports (29), the most conserved protein was M, ranging between 78.9 and 99.4% amino acid sequence identity, compared to the least conserved, 5a (40.4 to 96.2% amino acid sequence identity). The selected isolates were different in regions containing previously described neutralizing epitopes. A phylogenetic analysis supported by 75 ORF 2 to 6 nucleotide sequences showed that the seven isolates are well distributed within the tree and are well separated from each other (Fig. 1). Even though it is not currently possible to specify the minimum number of isolates needed to represent the entire genetic diversity of PRRSV, the seven selected provide a good representation.
TABLE 1.
Identity at the nucleotide level of envelope-associated structural genes of viruses used for screening VN activity
| Virusa | % Nucleotide sequence identity for ORFs 2 through 6 |
|||||
|---|---|---|---|---|---|---|
| KS62 | NVSL | P129 | VR | SDSU | KS06 | |
| KS62 | ||||||
| NVSL | 97.8 | |||||
| P129 | 94.2 | 95.5 | ||||
| VR | 92.2 | 92.9 | 93.5 | |||
| SDSU | 91.4 | 92.3 | 92.8 | 96.0 | ||
| KS06 | 91.7 | 92.1 | 91.4 | 91.1 | 90.1 | |
| LV | 64.7 | 64.5 | 65.1 | 65.1 | 64.8 | 65.4 |
FIG 1.
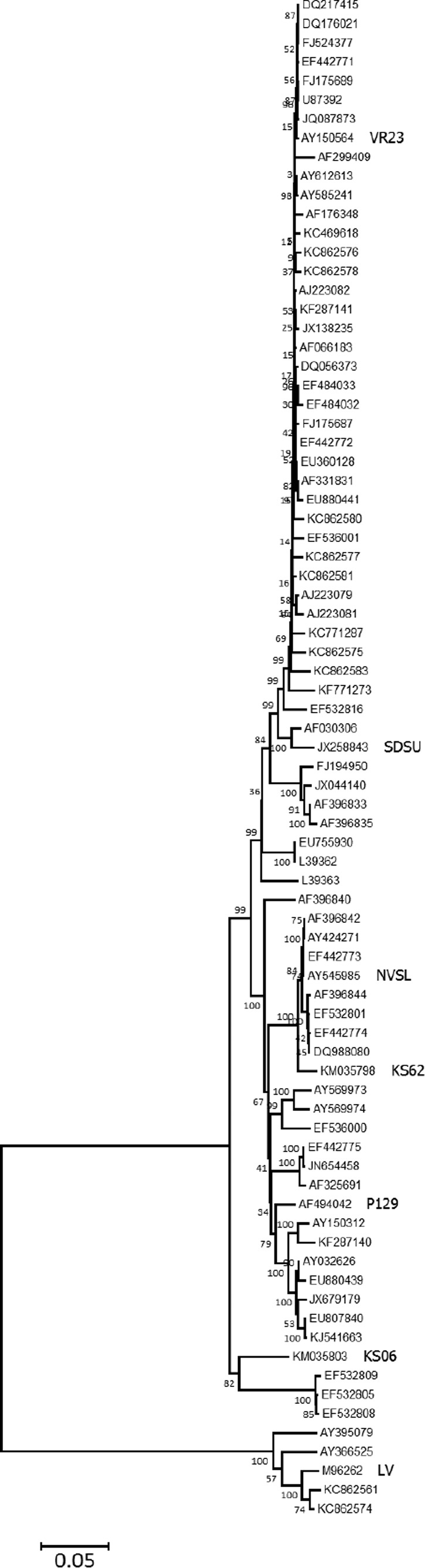
Locations of isolates within a phylogenetic tree constructed from ORFs 2 to 6. The seven isolates incorporated in neutralization assays (Table 1) are identified in large type. The remaining 73 nucleotide sequences were obtained from GenBank. The phylogenetic analysis was performed using MEGA5 software (35). The tree was constructed by the neighbor-joining method, and the percentages of replicate trees in which the associated isolates clustered together in the bootstrap test (1,000 replicates) are shown next to the branches (36). The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances (p), which were computed using the Kimura 2-parameter method (37).
VN assays incorporating the seven selected isolates were performed as previously described by us (5). Serum dilutions were incubated with 200 tissue culture infective doses (TCID50) of virus in a 200-μl volume of minimal essential medium (MEM) with 7% fetal bovine serum (FBS) and antibiotics (MEM-FBS) at 37°C and 5% CO2. After 1 h of incubation, the well contents were transferred to a 96-well plate of confluent MARC-145 cells and incubated for 4 days. For the initial screening, the absence of cytopathic effect (CPE) at a 1:16 dilution was considered positive for the presence of VN activity. Although a 1:8 VN titer cutoff is considered the standard for diagnostic purposes, cell toxicity was frequently seen at this dilution, making it difficult to interpret the results. Osorio et al. (30) demonstrated that 1:16 is the lowest VN titer sufficient to protect pregnant sows from experimental challenge. For selected sera that possessed only homologous neutralization of KS62 or showed broadly neutralizing activity, a second round of VN assays was performed to calculate the log2 50% neutralizing activity (NA50)/ml (31). Using the VN results, presented in Fig. 2, pigs could be placed into distinct categories or groups based on the number of virus isolates neutralized. Approximately 13% of pigs in both the vaccine and nonvaccine groups possessed no detectable VN activity at the 1:16 cutoff (group 1 in Fig. 2). Fifty-two percent of pigs in the nonvaccine group possessed homologous VN activity, compared to 38% in the vaccine group (group 2 in Fig. 2). All sera that possessed only homologous VN activity were directed against KS62, the virus used for challenge, and not VR-2332, the parent virus of the vaccine strain. The percentage of pigs in the nonvaccine group that neutralized more than one virus, but not all viruses, was 35%, compared to 47% for the vaccine group (group 3 in Fig. 2). For the vaccine group, the increase in the percentage of sera in group 3 was largely the result of sera that neutralized both KS62 and VR-2332. Group 4 (Fig. 2) is comprised of serum from a single pig, 16-45, that was able to neutralize all viruses tested, including the type 1 virus, LV. The 16-45 serum was further found to neutralize four additional PRRSV isolates (Table 2). Therefore, 16-45 was described as possessing broadly neutralizing activity and was selected for further study.
FIG 2.
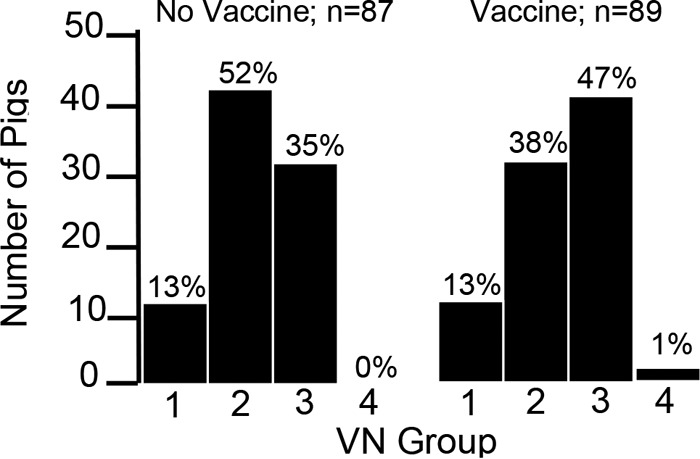
Placement of 87 nonvaccinated and 89 vaccinated pigs challenged with KS62 into different VN groups. VN assays were performed by reacting a single replicate of each vaccinated or nonvaccinated pig serum sample against the seven viruses listed in Table 1. With the exception of pigs placed in group 1, which showed no detectable VN activity, the results show the percentage of pigs in each group with a VN titer greater than or equal to 1:16. Assays were performed 42 days after virus challenge or 70 days after vaccination. The numbers of pigs in each category for vaccinated and nonvaccinated groups were not statistically different based on the chi-square test. Group 1, no detectable VN activity; group 2, homologous VN against KS62 only; group 3, VN activity against the homologous isolate and 1 to 5 additional isolates; group 4, reactivity against all seven isolates.
TABLE 2.
Virus neutralization properties of pig sera used in this study
| Pig | Neutralization (log2 NA50/ml) of virusa: |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| KS62 | NVSL | VR | P129 | KS06 | SD | AZ25 | CO84 | CO90 | WY27 | LV | |
| 16-21 | 10.7 | <5.0 | <5.0 | <5.0 | <5.0 | <5.0 | ND3 | ND | ND | ND | <5.0 |
| 16-45 | 9.0 | 6.0 | 6.0 | 6.4 | 6.0 | 6.4 | 6.0 | 6.4 | 6.7 | 6.0 | 7.7 |
| 16-63 | <5.0 | <5.0 | <5.0 | <5.0 | <5.0 | <5.0 | ND | ND | ND | ND | <5.0 |
| 15-4 | 7.7 | 8.0 | 6.0 | 6.4 | 6.0 | 6.0 | ND | ND | ND | ND | 7.0 |
GenBank accession numbers for PRRSV isolates KS62, NVSL, VR, P129, KS06, SD, and LV are listed in footnote a of Table 1. Accession numbers for additional isolates: AZ25, KM035800; CO84, KM03502; CO90, KM035799; and WY27, KM035801. Values are based on reacting serum samples in triplicate with viruses. ND, not determined.
In order to characterize the epitopes recognized by sample 16-45, KS62 was propagated in the presence of 16-45 serum as a means to select for VN resistance. A 24-well plate that contained 16-45 serum diluted 1:2 going across the plate and virus diluted 1:10 going down the plate was prepared. After 1 h at 37°C and 5% CO2, the well contents were transferred to a 24-well plate of confluent MARC-145 cells. After 4 days, the lowest dilution of virus in each row showing CPE was pooled and 100 μl transferred to a T-25 flask of MARC-145 cells containing 5 ml MEM-FBS. After 3 days of incubation in the absence of serum, the contents of the flask were harvested and the selection protocol repeated on a new 24-well plate. After six rounds of antibody selection all wells showed CPE, demonstrating that the virus had acquired resistance to the 16-45 serum. As described in Fig. 3, the resulting virus, KS62-R4, was resistant to serum 16-45 and to a second broadly neutralizing serum, 15-4, obtained from a pig experimentally challenged with a different PRRSV isolate, NVSL 97-7895 (NVSL) (Table 2 shows the sequence comparison between KS62 and NVSL). However, KS62-R4 retained sensitivity to neutralization with 16-21, a serum with homologous VN activity against KS62 (Table 2).
FIG 3.
VN assay results for KS62 parent, neutralization-resistant, and EGFP recombinant viruses. (A) Viruses were assayed against serum samples, in triplicate, possessing broad VN activity (16-45 and 15-4) or homologous VN activity to KS62 (serum 16-21) in MARC-145 cells. The log2NA50/ml was calculated using the method of Reed and Muench (38). (B) VN results for PAM cells are presented as the log2 neutralizing titer, which was calculated as the log2 of the inverse of the highest dilution of serum showing a 90% reduction in FFU. For PAM cell-based assays, serum samples were reacted, in duplicate, with viruses in three independent assays. Table 2 shows the VN properties of sera used in the experiments.
Amino acid substitutions in KS62-R4 responsible for escape from neutralization were identified by amplifying the 3.1-kb region covering the structural genes, ORFs 2 to 7, using the forward primer 5′-GAGGACTGGGAGGATTAC and reverse primer 5′-CCATTCACCACACATTCTTCC. Reverse transcription-PCR (RT-PCR) was performed using SuperScript III one-step RT-PCR system (Life Technologies) according to the manufacturer's directions. The RT-PCR product was sequenced directly, as well as individual cDNA fragments cloned into pcr2.1 TOPO (Life Technologies). The only sequence difference between KS62 and KS62-R4 was a tyrosine codon deletion, TAT, in ORF 6 at amino acid position 10 (Y10) in the M protein (Fig. 4). For KS62-R4, the sequencing chromatogram from the RT-PCR product showed only single peaks after the Y10 deletion, indicating that sequences with the Y-10 deletion were the predominant species within the population of sequences. All cloned PCR products had the Y10 deletion.
FIG 4.
M protein peptide sequence alignment of PRRSV isolates incorporated in this study. The alignment of M protein is shown for the seven viruses listed in Table 1. Peptide sequences were aligned using CLC Main Workbench version 6.6.2 (CLC bio). The location of the deletion in M of KS62 associated with escape from broadly neutralizing serum 16-45 is shown in the gray box.
Since 16-45 is a polyclonal serum, the mutation(s) responsible for escape from neutralization could have been located outside the structural gene region. Therefore, an infectious clone with the Y-10 deletion was constructed by cloning ORF 3 to 7 cDNA of the KS62 viruses into a green fluorescent protein (GFP)-expressing DNA-launched infectious clone, PCMV-S-P129 (32). The GFP cDNA was replaced with the red-shifted mammalian adapted variant, enhanced GFP (EGFP) (Clontech; GenBank accession number U55762). EGFP provided an easy means for detecting virus replication in culture. A shuttle vector was created by cloning a BstBI (position 13590) and SpeI (16240) fragment, which covered ORF 3 through the poly(A) tail from the P129-EGFP plasmid, into pcr2.1. The region between BstEII (15059) and BsmI (16029), which covered ORFs 6 and 7, was replaced with the corresponding KS62 restriction fragment. This was followed in a second step by replacing the BstBI-BstEII fragment from KS62, which covered ORFs 3 to 5. The Y10 mutation was inserted into the KS62 shuttle vector by replacing the region between BstEII and XmaI (15580) from KS62-R4. The shuttle fragment was excised with BstBI-SpeI and inserted back into the infectious clone. After sequencing to confirm the correct products, plasmids (1.1 μg) were transfected into HEK 293T cells on a 24-well plate using Fugene HD (Promega) according to the product recommendations. Viruses were propagated by two additional passages on MARC-145 cells and ORFs 2 to 7 sequenced to confirm the presence of the Y10 deletion and rule out other amino acid substitutions. The VN properties of the recombinant viruses are summarized in Fig. 3. In addition to recording CPE, the VN titration endpoints for the recombinant viruses were determined using an EGFP fluorescence focus-forming unit (FFU) reduction assay. Wells were examined using an inverted fluorescence microscope and VN titers calculated as the last serum dilution showing at least 90% reduction in FFU. Compared to their respective parent viruses, P129-EGFP-KS62 and P129-EGFP-KS62-R4 possessed similar VN properties, including susceptibility to neutralization by 16-21 and escape from neutralization by sera 16-45 and 15-4 for P129-EGFP-KS62-R4. As a control, serum 16-63 (Table 2), which was negative for detectable VN activity, failed to neutralize any of the viruses (data not shown). The VN titration endpoints determined using EGFP fluorescence were in agreement with the VN results obtained by examining for CPE (data not shown).
To further determine the relevance of the Y10 deletion, VN assays were performed using primary cultures of porcine alveolar macrophage (PAM) cells. PAM cells were obtained by lung lavage from a healthy 4-week-old PRRSV-negative pig. Briefly, lungs were removed and PAM cells were collected in phosphate-buffered saline (PBS). Following washing, PAM cells were counted and stored in liquid nitrogen. Approximately 5 × 104 PAM cells in RPMI 1640 supplemented with 10% FBS were seeded into each well of a 96-well plate and incubated at 37°C and 5% CO2 for 24 h prior to infection. Similar to the MARC-145 cell assay, serial 1:2 dilutions of sera were mixed with approximately 200 FFU of recombinant virus. Following a 1-h incubation, the contents were transferred to plates containing PAM cells and fluorescence recorded 24 h later. Overall, the maximum VN titers for 16-21 and 16-45 were lower in PAM cells than in MARC-145 cells (compare Fig. 3B and A). However, the patterns for neutralization were the same. Serum 16-21 neutralized P129-EGFP-KS62 and P129-EGFP-KS62-R4, while 16-45 neutralized the EGFP-KS-62 virus but not the Y10 deletion virus. Together, the results demonstrate that the Y-10 deletion in the M protein is sufficient to escape from neutralization by 16-45.
In the absence of the availability of a PRRSV MAb with broadly neutralization activity, this study demonstrates the utility of screening populations of infected pigs to identify naturally occurring antibodies as tools to probe the nature of the immune response to PRRSV. In this study, reverse genetics of an infectious cDNA clone was used to confirm that a single amino acid deletion at position 10 in M of KS62 resulted in escape from broadly neutralizing sera, 16-45 and 15-4, while retaining sensitivity to serum with homologous neutralizing activity. The results suggest that epitopes recognized by sera with different neutralization properties are distinct, which can explain the large number of neutralizing epitopes previously described in the literature (12–17). The results also point to the importance of thoroughly characterizing the neutralization properties of polyclonal and monoclonal antibodies used for epitope mapping experiments.
The deletion of a tyrosine at position 10 in M suggests that the loss of neutralization in the presence of 16-45 is the result of the deletion of a critical contact amino acid residue. Even though the M protein is relatively conserved, the analysis of 573 type 2 PRRSV M protein peptide sequences from GenBank (see Table S1 in the supplemental material for list of these GenBank numbers) showed several amino acid substitutions at position 10, including the substitution of asparagine (87.4%), histidine (9.2%), tyrosine (2.6%), or arginine (0.7%). No deletions were identified within the group of sequences. In this study, the viruses incorporated in the neutralization assays possessed histidine, tyrosine, and asparagine substitutions at position 10 (Fig. 4). Therefore, the presence of different amino acids at position 10 suggests that the peptide sequence variation is not sufficient to escape from broadly neutralizing serum, 16-45. An alternative explanation can be based on the structure formed by the GP5-M heterodimer molecule and its interaction with the receptor on macrophages. Previous models identified the interaction between CD169 and GP5 as being responsible for the initial uptake and internalization of the virus, which is followed by uncoating via an interaction between GP2, -3, and -4 and CD163 (40). However, productive infection of CD169 knockout pigs with PRRSV has brought into question the role of GP5/M as a receptor-binding molecule (41). With this in mind, the Tyr-10 in the M protein of KS62 is immediately adjacent to a highly conserved cysteine at position 9 that forms the disulfide bond with Cys-54 in GP5 (33, 34). One possibility is that the Y-10 deletion creates a conformational change sufficient to block the interaction of broadly neutralizing antibody with a previously characterized linear epitope located between amino acids 43 and 51 in GP5 (16) (see Fig. S1F in the supplemental material). Alternatively, the target of broadly neutralizing antibody could be a much larger conformational epitope formed by the interaction of GP5, M, and other viral structural proteins.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported wholly or in part by National Pork Board grant 12-120 and USDA-NIFA award 2013-68004-20362.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.03287-14.
REFERENCES
- 1.Snijder EJ, Spaan WJM. 2007. Arteriviruses, p 1337–1355. In Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE (ed), Fields virology, 5th ed Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 2.Huang YW, Meng XJ. 2010. Novel strategies and approaches to develop the next generation of vaccines against porcine reproductive and respiratory syndrome virus (PRRSV). Virus Res 154:141–149. doi: 10.1016/j.virusres.2010.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Darwich L, Díaz I, Mateu E. 2010. Certainties, doubts and hypotheses in porcine reproductive and respiratory syndrome virus immunobiology. Virus Res 154:123–132. doi: 10.1016/j.virusres.2010.07.017. [DOI] [PubMed] [Google Scholar]
- 4.Fang Y, Schneider P, Zhang WP, Faaberg KS, Nelson EA, Rowland RRR. 2007. Diversity and evolution of a newly emerged North American type 1 porcine arterivirus: analysis of isolates collected between 1999 and 2004. Arch Virol 152:1009–1017. doi: 10.1007/s00705-007-0936-y. [DOI] [PubMed] [Google Scholar]
- 5.Rowland RRR, Steffen M, Ackerman T, Benfield DA. 1999. The evolution of porcine reproductive and respiratory syndrome virus: quasispecies and emergence of a virus subpopulation during infection of pigs with VR-2332. Virology 259:262–266. doi: 10.1006/viro.1999.9789. [DOI] [PubMed] [Google Scholar]
- 6.Ansari IH, Kwon B, Osorio FA, Pattnaik AK. 2006. Influence of N-linked glycosylation of porcine reproductive and respiratory syndrome virus GP5 on virus infectivity, antigenicity, and ability to induce neutralizing antibodies. J Virol 80:3994–4004. doi: 10.1128/JVI.80.8.3994-4004.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Snijder EJ, Dobbe JC, Spaan WJM. 2003. Heterodimerization of the two major envelope proteins is essential for arterivirus infectivity. J Virol 77:97–104. doi: 10.1128/JVI.77.1.97-104.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wissink EHJ, Kroese MV, van Wijk HAR, Rijsewijk FAM, Meulenberg JJM, Rottier PJM. 2005. Envelope protein requirements for the assembly of infectious virions of porcine reproductive and respiratory syndrome virus. J Virol 79:12495–12506. doi: 10.1128/JVI.79.19.12495-12506.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu W-H, Fang Y, Rowland RRR, Lawson SR, Christopher-Hennings J, Yoon K-J, Nelson EA. 2005. The 2b protein as a minor structural component of PRRSV. Virus Res 114:177–181. doi: 10.1016/j.virusres.2005.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Firth AE, Zevenhoven-Dobbe JC, Wills NM, Go YY, Balasuriya UBR, Atkins JF, Snijder EJ, Posthuma CC. 2011. Discovery of a small arterivirus gene that overlaps the GP5 coding sequence and is important for virus production. J Gen Virol 92:1097–1106. doi: 10.1099/vir.0.029264-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johnson CR, Griggs TF, Gnanandarajah J, Murtaugh MP. 2011. Novel structural protein in porcine reproductive and respiratory syndrome virus encoded by an alternative ORF5 present in all arteriviruses. J Gen Virol 92:1107–1116. doi: 10.1099/vir.0.030213-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meulenberg JJ, van Nieuwstadt AP, van Essen-Zandbergen A, Langeveld JP. 1997. Posttranslational processing and identification of a neutralization domain of the GP4 protein encoded by ORF4 of Lelystad virus. J Virol 71:6061–6067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vanhee M, Costers S, Van Breedam W, Geldhof MF, Van Doorsselaere J, Nauwynck HJ. 2010. A variable region in GP4 of European-type porcine reproductive and respiratory syndrome virus induces neutralizing antibodies against homologous but not heterologous virus strains. Viral Immunol 23:403–413. doi: 10.1089/vim.2010.0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Costers S, Lefebvre DJ, Van Doorsselaere J, Vanhee M, Delputte PL, Nauwynck HJ. 2010. GP4 of porcine reproductive and respiratory syndrome virus contains a neutralizing epitope that is susceptible to immunoselection in vitro. Arch Virol 155:371–378. doi: 10.1007/s00705-009-0582-7. [DOI] [PubMed] [Google Scholar]
- 15.Vanhee M, Van Breedam W, Costers S, Geldhof M, Noppe Y, Nauwynck H. 2011. Characterization of antigenic regions in the porcine reproductive and respiratory syndrome virus by the use of peptide-specific serum antibodies. Vaccine 29:4794–4804. doi: 10.1016/j.vaccine.2011.04.071. [DOI] [PubMed] [Google Scholar]
- 16.Ostrowski M, Galeota JA, Jar AM, Platt KB, Osorio FA, Lopez OJ. 2002. Identification of neutralizing and nonneutralizing epitopes in the porcine reproductive and respiratory syndrome virus GP5 ectodomain. J Virol 76:4241–4250. doi: 10.1128/JVI.76.9.4241-4250.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Plagemann PGW, Rowland RRR, Faaberg KS. 2002. The primary neutralization epitope of porcine respiratory and reproductive syndrome virus strain VR-2332 is located in the middle of the GP5 ectodomain. Arch Virol 147:2327–2347. doi: 10.1007/s00705-002-0887-2. [DOI] [PubMed] [Google Scholar]
- 18.Li K, Chen Z, Plagemann P. 1998. The neutralization epitope of lactate dehydrogenase-elevating virus is located on the short ectodomain of the primary envelope glycoprotein. Virology 242:239–245. doi: 10.1006/viro.1997.9014. [DOI] [PubMed] [Google Scholar]
- 19.Kim W-I, Yoon K-J. 2008. Molecular assessment of the role of envelope-associated structural proteins in cross neutralization among different PRRS viruses. Virus Genes 37:380–391. doi: 10.1007/s11262-008-0278-1. [DOI] [PubMed] [Google Scholar]
- 20.Kappes MA, Miller CL, Faaberg KS. 2013. Highly divergent strains of porcine reproductive and respiratory syndrome virus incorporate multiple isoforms of nonstructural protein 2 into virions. J Virol 87:13456–13465. doi: 10.1128/JVI.02435-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Simek MD, Rida W, Priddy FH, Pung P, Carrow E, Laufer DS, Lehrman JK, Boaz M, Tarragona-Fiol T, Miiro G, Birungi J, Pozniak A, McPhee DA, Manigart O, Karita E, Inwoley A, Jaoko W, Dehovitz J, Bekker L-G, Pitisuttithum P, Paris R, Walker LM, Poignard P, Wrin T, Fast PE, Burton DR, Koff WC. 2009. Human immunodeficiency virus type 1 elite neutralizers: individuals with broad and potent neutralizing activity identified by using a high-throughput neutralization assay together with an analytical selection algorithm. J Virol 83:7337–7348. doi: 10.1128/JVI.00110-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Doria-Rose NA, Klein RM, Manion MM, O'Dell S, Phogat A, Chakrabarti B, Hallahan CW, Migueles SA, Wrammert J, Ahmed R, Nason M, Wyatt RT, Mascola JR, Connors M. 2009. Frequency and phenotype of human immunodeficiency virus envelope-specific B cells from patients with broadly cross-neutralizing antibodies. J Virol 83:188–199. doi: 10.1128/JVI.01583-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Euler Z, van Gils MJ, Bunnik EM, Phung P, Schweighardt B, Wrin T, Schuitemaker H. 2010. Cross-reactive neutralizing humoral immunity does not protect from HIV type 1 disease progression. J Infect Dis 201:1045–1053. doi: 10.1086/651144. [DOI] [PubMed] [Google Scholar]
- 24.Pejchal R, Wilson IA. 2010. Structure-based vaccine design in HIV: blind men and the elephant? Curr Pharm Des 16:3744–3753. doi: 10.2174/138161210794079173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boddicker N, Waide EH, Rowland RRR, Lunney JK, Garrick DJ, Reecy JM, Dekkers JCM. 2012. Evidence for a major QTL associated with host response to porcine reproductive and respiratory syndrome virus challenge. J Anim Sci 90:1733–1746. doi: 10.2527/jas.2011-4464. [DOI] [PubMed] [Google Scholar]
- 26.Boddicker NJ, Garrick DJ, Rowland RRR, Lunney JK, Reecy JM, Dekkers JCM. 2014. Validation and further characterization of a major quantitative trait locus associated with host response to experimental infection with porcine reproductive and respiratory syndrome virus. Anim Genet 45:48–58. doi: 10.1111/age.12079. [DOI] [PubMed] [Google Scholar]
- 27.Boddicker NJ, Bjorkquist A, Rowland RRR, Lunney JK, Reecy JM, Dekkers JCM. 2014. Genome-wide association and genomic prediction for host response to porcine reproductive and respiratory syndrome virus infection. Genet Sel Evol 46:18. doi: 10.1186/1297-9686-46-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lunney JK, Steibel JP, Reecy JM, Fritz E, Rothschild MF, Kerrigan M, Trible B, Rowland RR. 2011. Probing genetic control of swine responses to PRRSV infection: current progress of the PRRS host genetics consortium. BMC Proc 5(Suppl 4):S30. doi: 10.1186/1753-6561-5-S4-S30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meng XJ, Paul PS, Halbur PG, Lum MA. 1995. Phylogenetic analyses of the putative M (ORF 6) and N (ORF 7) genes of porcine reproductive and respiratory syndrome virus (PRRSV): implication for the existence of two genotypes of PRRSV in the USA and Europe. Arch Virol 140:745–755. doi: 10.1007/BF01309962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Osorio FA, Galeota JA, Nelson E, Brodersen B, Doster A, Wills R, Zuckermann F, Laegreid WW. 2002. Passive transfer of virus-specific antibodies confers protection against reproductive failure induced by a virulent strain of porcine reproductive and respiratory syndrome virus and establishes sterilizing immunity. Virology 302:9–20. doi: 10.1006/viro.2002.1612. [DOI] [PubMed] [Google Scholar]
- 31.Finney DJ. 1964. The Spearman-Karber method, p 524–530. In Finney DJ. (ed), Statistical methods in biological assay, 2nd ed Charles Griffin, London, United Kingdom. [Google Scholar]
- 32.Lee C, Calvert JG, Welch S-KW, Yoo D. 2005. A DNA-launched reverse genetics system for porcine reproductive and respiratory syndrome virus reveals that homodimerization of the nucleocapsid protein is essential for virus infectivity. Virology 331:47–62. doi: 10.1016/j.virol.2004.10.026. [DOI] [PubMed] [Google Scholar]
- 33.Mardassi H, Massie B, Dea S. 1996. Intracellular synthesis, processing, and transport of proteins encoded by ORFs 5 to 7 of porcine reproductive and respiratory syndrome virus. Virology 221:98–112. doi: 10.1006/viro.1996.0356. [DOI] [PubMed] [Google Scholar]
- 34.Dokland T. 2010. The structural biology of PRRSV. Virus Res 154:86–97. doi: 10.1016/j.virusres.2010.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Felsenstein J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791. doi: 10.2307/2408678. [DOI] [PubMed] [Google Scholar]
- 37.Kimura M. 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 38.Reed JL, Muench H. 1938. A simple method of estimating fifty percent endpoints. Am J Hyg 27:493–497. [Google Scholar]
- 39.Corti D, Lanzavecchia A. 2013. Broadly neutralizing antiviral antibodies. Annu Rev Immunol 31:705–742. doi: 10.1146/annurev-immunol-032712-095916. [DOI] [PubMed] [Google Scholar]
- 40.Van Breedam W, Delputte PL, Van Gorp H, Misinzo G, Vanderheijden N, Duan X, Nauwynck HJ. 2010. Porcine reproductive and respiratory syndrome virus entry into the porcine macrophage. J Gen Virol 91:1659–1667. doi: 10.1099/vir.0.020503-0. [DOI] [PubMed] [Google Scholar]
- 41.Prather RS, Rowland RRR, Ewen C, Trible B, Kerrigan M, Bawa B, Teson JM, Mao J, Lee K, Samuel MS, Whitworth KM, Murphy CN, Egen T, Green JA. 2013. An intact sialoadhesin (Sn/SIGLEC1/CD169) is not required for attachment/internalization of the porcine reproductive and respiratory syndrome virus. J Virol 87:9538–9546. doi: 10.1128/JVI.00177-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Trible BR, Kerrigan M, Crossland N, Potter M, Faaberg K, Hesse R, Rowland RRR. 2011. Antibody recognition of porcine circovirus type 2 capsid protein epitopes after vaccination, infection, and disease. Clin Vaccine Immunol 18:749–757. doi: 10.1128/CVI.00418-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.



