Abstract
The histone H3-lysine-4 methyltransferase mixed-lineage leukemia 3 (MLL3) and its closest homolog, MLL4 (aka KMT2D), belong to two homologous transcriptional coactivator complexes, named MLL3 and MLL4 complexes, respectively. MLL3 plays crucial roles in multiple metabolic processes. However, the physiological roles of MLL4 in metabolism and the relationship between MLL3 and MLL4 in metabolic gene regulation are unclear. To address these issues, we analyzed the phenotypes of newly generated MLL4 mutant mice, along with MLL3 mutant and MLL3;MLL4 compound mutant mice. We also performed comparative genome-wide transcriptome analyses in livers of MLL3, MLL4, and MLL3;MLL4 mutant mice. These analyses revealed that MLL3 and MLL4 complexes are key epigenetic regulators of common metabolic processes and the hepatic circadian clock. Subsequent mechanistic analyses uncovered that MLL3/4 complexes function as pivotal coactivators of the circadian transcription factors (TFs), retinoid-related orphan receptor (ROR)-α and -γ, in the hepatic circadian clock. Consistent with disturbed hepatic clock gene expression in MLL4 mutant mice, we found that rhythmic fluctuation of hepatic and serum bile acid (BA) levels over the circadian cycle is abolished in MLL4 mutant mice. Our analyses also demonstrate that MLL4 primarily impinges on hepatic BA production among several regulatory pathways to control BA homeostasis. Together, our results provide strong in vivo support for important roles of both MLL3 and MLL4 in similar metabolic pathways. Conclusion: Both MLL3 and MLL4 complexes act as major epigenetic regulators of diverse metabolic processes (including circadian control of bile acid homeostasis) and as critical transcriptional coactivators of the circadian TFs, RORs.
Trimethylation of histone H3-lysine 4 (H3K4me3) marks active chromatin and counters the repressive chromatin milieu imposed by H3K9/H3K27 methylation in higher eukaryotes.1 The vertebrate H3K4 methyltransferases (H3K4MTs) performing H3K4 trimethylation, SET1A/B and mixed-lineage leukemia 1-4 (MLL1-4), form a family of six SET1-like complexes.1 Each complex contains an H3K4MT, complex-specific subunits, and a common subcomplex of retinoblastoma-binding protein 5 (RBBP5), ASH2L, WDR5, and DPY30, which facilitates the H3K4MT activity of SET1A/B/MLL1-4.1 We previously purified the first mammalian SET1-like complexes containing MLL3 or MLL4 (aka KMT2D).2,3 Their unique subunits include activating signal cointegrator 2 (ASC-2), PTIP/PA1, and the H3K27-demethylase, ubiquitously transcribed tetratricopeptide repeat, X chromosome (UTX).1-5 ASC-2 links several nuclear receptors (NRs) to MLL3/4 complexes using its two LXXLL motifs that recognize activated conformation of NRs.3,6-9
We reported on various metabolic phenotypes in the mouse line, MLL3Δ/Δ, which expresses a catalytically inactive mutant form of MLL3,3 including decreased white fat mass, impaired ability of high-fat diet (HFD) to induce obesity and lipid accumulation in the brown adipose tissue and liver, improved glucose tolerance and insulin sensitivity, hyperactivity, and increased energy expenditure and bile acid (BA) levels.7-10 However, it remains unknown whether MLL4 also regulates metabolic processes and, if so, whether MLL3 and MLL4 target the same or distinct metabolic pathways.
In the liver, BA synthesis is a major mechanism for elimination of cholesterol. Cholesterol-7α-hydroxylase (CYP7A1) is the rate-limiting enzyme in BA production and shows circadian oscillation in its messenger RNA (mRNA) expression and enzymatic activity, although BA synthesis peaks during the daytime and nighttime in humans and rodents, respectively.11,12 Accordingly, bile flow and biliary secretion of BAs display precise daily rhythms.13 Expression of CYP7A1 is regulated by an intricate gene network with feedback loops that operate in response to BA levels. The BA-activated NR, farnesoid X receptor (FXR), orchestrates the expression of many genes involved in BA homeostasis, including small heterodimer partner (SHP).14 SHP restrains BA synthesis by suppressing CYP7A1 expression.14 The tumor-suppressor, p53, also controls BA homeostasis by up-regulating SHP.10,15 Retinoid-related orphan receptors (RORα-γ) up-regulate the expression of Rev-erbα and Rev-erbα, which negatively regulate SHP.14 MLL3 was shown to serve as a coactivator of FXR and p53,7,10,16 providing some molecular mechanisms underlying disturbed BA homeostasis inMLL3Δ/Δ mice. Recently, MLL3 was found to show circadian oscillation in its expression and modulate many circadian genes in the liver.17 FXR and SHP showed distinct circadian expression patterns.18,19 These results suggest that MLL3 may regulate circadian oscillation of BA levels as a coactivator of FXR. However, this function of MLL3/4 complexes may involve a wider range of transcription factors (TFs), given the multipartite actions of MLL3/4 complexes.1
Circadian rhythms administer diverse metabolic and physiological functions.20 In mammals, the hypothalamic suprachiasmatic nucleus central clock is integrated with peripheral clocks throughout the rest of the body.20 In response to light, the central clock directs the 24-hour period length throughout the body in synchrony with the peripheral clocks. The circadian clock consists of an integral gene network of several feedback loops. The oscillator is activated by a heterodimer of brain and muscle ARNT-like (Bmal1) and circadian locomotor output cycles kaput (Clock) or its paralog, neuronal PAS domain protein 2, or inhibited by two cryptochrome and three period proteins.20 The Rev-erbs and RORα-γ also participate in the core loops by regulating the expression of circadian genes negatively and positively, respectively, through a common ROR-response element (RORE).20
In this study, we demonstrate that MLL3 and MLL4 play important roles in various metabolic processes. We also show that RORs are the major TFs that enable the circadian clock function of MLL3/4 in the liver, and that MLL3/4 complexes regulate circadian rhythmic control of BA homeostasis primarily through suppressing hepatic BA synthesis.
Materials and Methods
Animals
All mice were housed in a pathogen-free animal facility under standard 12-hour light-dark cycle. Mice were allowed to ad libitum access to water and regular rodent chow. For gene expression and chromatin immunoprecipitation (ChIP) analyses, mice were killed by cervical dislocation at indicated zeitgeber time (ZT) for snap freezing of liver tissues. To measure body-weight changes of MLL4+/− upon HFD, mice were fed HFD (42% fat; Harlan Laboratories, Indianapolis, IN) or normal diet for up to 3 months. Body weights of each group of mice were measured once a week. All animals received humane care according to the criteria outlined in the Guide for the Care and Use of Laboratory Animals according to National Institutes of Health publication 86-23 (revised 1985).
Measurements of Body Fat Mass and BA Levels
Fat mass was measured with EchoMRI-4in1 (EchoMRI LLC, Houston, TX). Blood samples were taken at indicated time points. The liver, the entire small intestine, and feces were weighed and homogenized in 75% ethanol, and the resulting homogenate was incubated at 50°C for 2 hours to extract BAs and centrifuged at 6,000g for 10 minutes at 4°C. BA content of serum or the supernatant of centrifuged samples was then measured using a Colormetric Total Bile acid Assay kit (Diazyme Laboratories, Poway, CA).
Construction of MLL4+/− mice, glucose and insulin tolerance tests, reverse transcription/quantitative reverse-transcriptase polymerase chain reaction (RT/qRT-PCR) analysis, RNA sequencing (RNA-Seq) and data analysis, ChIP, coimmunoprecipitation (CoIP), and all primer and small interfering RNA (siRNA) sequence info are detailed in the Supporting Information.
Statistical Analysis
Statistical differences were determined by two-tailed Student t test. Statistical significance is displayed as P < 0.05 (*), P < 0.01 (**), or P < 0.001 (***).
Results
Metabolic Phenotypes of MLL4+/− Mice
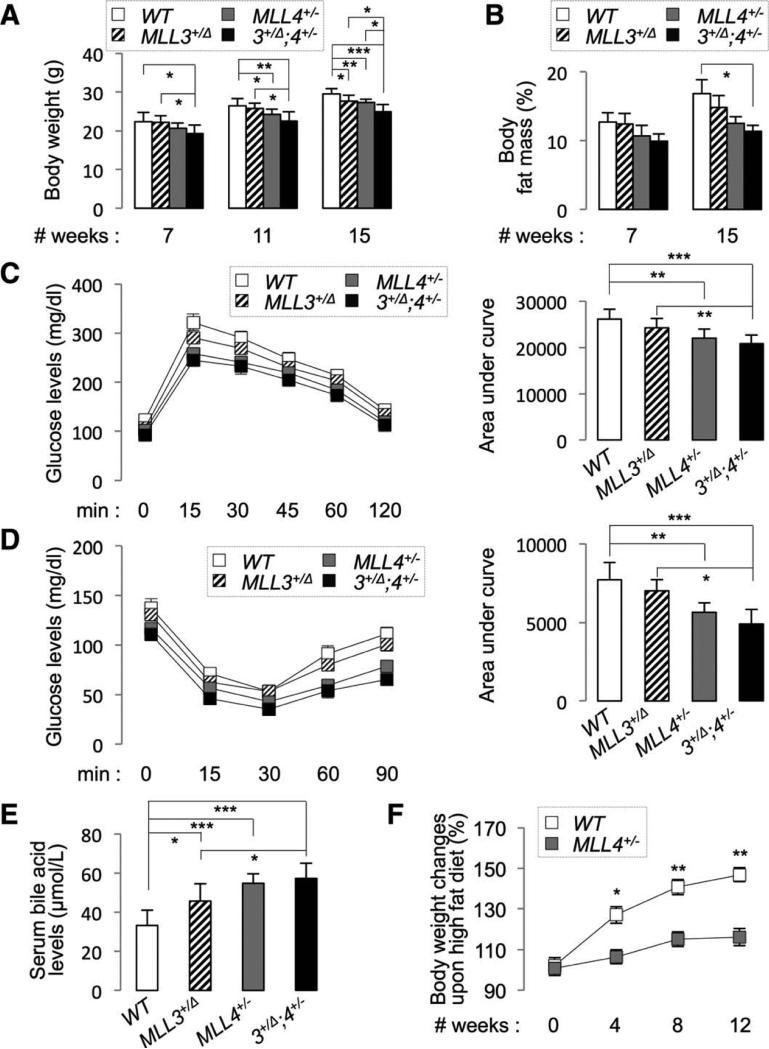
To elucidate the physiological roles of MLL4, we developed mutant mice carrying an MLL4-null allele, MLL4− (Supporting Fig. 1A-D). MLL4−/− embryos died before embryonic day 9.5 (data not shown), indicating the requirement for MLL4 in embryonic development. The MLL4+/− mice survived into adulthood and were significantly lighter than wild-type (WT) mice after 11 weeks of age (Fig. 1A). Consistently, MLL4+/− mice showed low levels of body fat mass (Fig. 1B). Relative to WT mice, MLL4+/− mice showed enhanced glucose tolerance and insulin sensitivity (Fig. 1C,D), increased serum and feces BAs (Fig. 1E and Supporting Fig. 2A), and resistance to HFD-induced obesity (Fig. 1F). These phenotypes were similar to those of MLL3Δ/Δ mice.7-10 MLL3+/Δ mice exhibited similar phenotypes, although the severity of the phenotypes was weaker than those in MLL4+/− mice (Fig. 1A-E), and the MLL3+/Δ;MLL4+/− compound mice showed the tendency of stronger phenotypes than MLL3+/Δ or MLL4+/− mice (Fig. 1A-E). These results indicate that MLL4 and MLL3 play critical roles in regulating common metabolic pathways.
Fig 1.
MLL4+/− mice show strong metabolic phenotypes. (A-E) Body weight (A), body fat mass (B), glucose tolerance (C), insulin sensitivity (D), and serum BA levels (E) were measured for 16-week-old male WT, MLL3+/Δ, MLL4+/−, and MLL3+/Δ;MLL4+/− mice (n = 6~7). Similar results were also obtained with female animals. Serum BA levels were measured at ZT10 (E). (F) Starting with 8-week-old male WT and MLL4+/− mice (n = 4), body-weight changes were measured for the next 12 weeks of HFD feeding as indicated and the results were plotted as percent increase in body weight relative to the beginning of HFD (0 week). P values are for differences between WT and MLL4+/− mice (F).
Critical Roles of MLL3/4 in the Liver
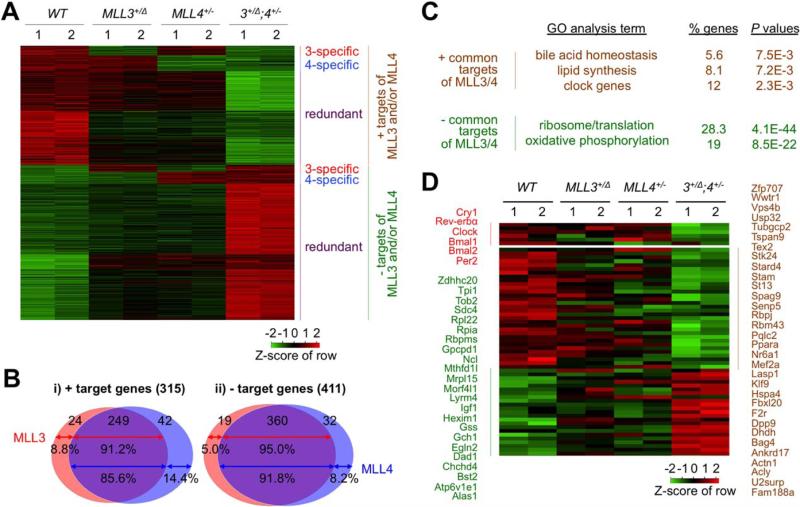
Because MLL3 plays important metabolic roles in the liver7-10 and liver is an ideal tissue for transcriptome studies because of its relative homogeneity, we chose the liver as a model organ to investigate the gene-regulatory pathways controlled by MLL3/4. To identify the target genes of MLL3/4 in an unbiased genome-wide scale, we determined the transcriptome changes in the liver of 5-month-old WT, MLL3+/Δ, MLL4+/−, and MLL3+/Δ;MLL4+/− male mice using RNA-Seq. Our analyses identified 726 genes, whose expression was significantly changed by mutation in MLL3 and/or MLL4 (more than 1.5-fold changes, false discovery rate of <10%; Fig. 2A; Supporting Table 1). Among these, 315 genes (~43%) were down-regulated in the MLL3/4 mutants (positive targets of MLL3/4), whereas 411 genes (~57%) were up-regulated (negative targets of MLL3/4; Fig. 2A,B). The majority of these targets (~83%) were similarly changed in MLL3+/Δ and MLL4+/− mice and were more severely affected in MLL3+/Δ;MLL4+/− livers than livers with a single gene mutation (Fig. 2A,B), suggesting that they are commonly regulated by MLL3/4. This is consistent with the similarities we observed in the metabolic phenotypes between MLL3+/Δ and MLL4+/− mice. Interestingly, a small fraction of the target genes were changed specifically by the mutation of MLL3 or MLL4 alone, and their expression levels were not further affected by the compound mutation of both genes (Fig. 2A,B), suggesting that these genes represent MLL3- or MLL4-specific targets. Collectively, our systemic gene expression analyses uncovered that MLL3/4 share a majority of their target genes in the liver.
Fig. 2.
RNA-Seq assay identifies target genes of MLL3 and MLL4 in metabolism. (A) A heatmap shows that target genes of MLL3 and/or MLL4 uncovered by RNA-Seq analysis of liver mRNAs of 5-month-old WT, MLL3+/Δ, MLL4+/−, and MLL3+/Δ;MLL4+/− (n = 2) are grouped into six different clusters. (B) Venn diagram showing that a large portion of target genes of MLL3 and MLL4 (85.6%-95.0%) is commonly regulated by MLL3 and MLL4, whereas a much smaller portion of target genes of MLL3 and MLL4 (5.0%-14.4%) is regulated by MLL3 and MLL4 alone. (C) GO categories of the genes commonly regulated by MLL3 and MLL4. (D) A heatmap showing a subgroup of the target genes of MLL3 and MLL4, which have been previously shown to be regulated by MLL3 and to fluctuate in a circadian clock-dependent manner.16
Roles of MLL3/4 in the Hepatic Circadian Clock
To identify the hepatic gene pathways controlled by MLL3/4, we performed Gene Ontology (GO) analyses on the MLL3/4 targets discovered from our RNA-Seq analyses and found several interesting features of MLL3/4 targets linked to metabolism (Supporting Table 2). First, the common negative targets of MLL3/ 4 were highly enriched for the categories of genes involved in ribosomal biogenesis/protein synthesis and oxidative phosphorylation, both of which represent primary energy-consuming processes in cells (Fig. 2C), consistent with our report that MLL3Δ/Δ mice show increased energy expenditure.9 Second, the positive common targets of MLL3/4 included genes implicated in bile synthesis/secretion, lipid synthesis, and circa-dian clock (Fig. 2C). These findings provide a molecular basis for our reports for the disturbance of lipid and BA homeostasis in MLL3Δ/Δ mice.7-10
Recently, MLL3 was suggested as an important epi-genetic modifier of 119 circadian genes in the liver.17 Among these genes,17 55 were identified as common targets of MLL3/4 in our RNA-Seq results (genes in green and brown; Fig. 2D). Given that our RNA-Seq was performed with liver samples collected at ZT10 (4 pm), the circadian genes targeted by MLL3/4 at other time points were likely missed in our RNA-Seq analyses. Our analyses also identified several core clock genes, such as cryptochrome 1 (Cry1), Rev-erbα, Clock, Bmal1, Bmal2, and period 2 (Per2), as common positive targets of MLL3/4 (genes in red; Fig. 2D).
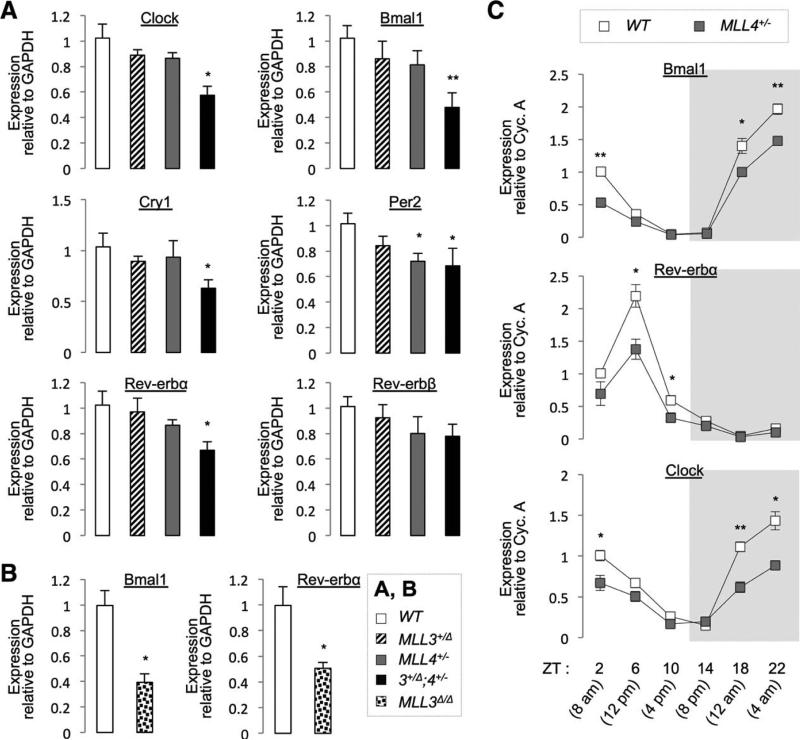
To validate our RNA-Seq results for the core clock genes, we performed qRT-PCR on liver of 5-month-old WT, MLL3+/Δ, MLL4+/−, and MLL3+/Δ;MLL4+/− male mice sacrificed at ZT10 (4 pm). Expression of Clock, Bmal1, Cry1, Per2, and Rev-erbα/β tended to be lower in MLL3+/Δ and MLL4+/− livers, relative to WT livers, and their level was further down-regulated in MLL3+/Δ;MLL4+/− livers (Fig. 3A). Likewise, expression of Bmal1 and Rev-erbα was significantly lower in livers of 2-month-old MLL3Δ/Δ male mice, relative to WT mice, sacrificed at ZT4 (10 am; Fig. 3B). These results prompted us to ask whether the rhythmic fluctuation of core clock genes throughout a day is disturbed in MLL4+/− mice. Thus, we collected liver samples of WT and MLL4+/− mice at six time points over the circadian cycle and examined the expression pattern of Bmal1, Rev-erbα, and Clock using qRT-PCR. Their overall expression levels were reduced in MLL4+/− livers, compared to WT livers (most prominently at the highest expression time points), resulting in the substantially dampened oscillation in core clock gene expression in MLL4+/− livers (Fig. 3C).
Fig. 3.
Validation of hepatic targets of MLL3/4 using qRT-PCR. (A) Livers of 5-month-old male WT, MLL3+/Δ, MLL4+/−, and MLL3+/Δ;MLL4+/− mice (N = 6-7) were harvested at ZT10 (4 pm). (B) Livers of 2-month-old male WT and MLL3Δ/Δ mice (n = 4) were harvested at ZT4 (10 AM). (C) Livers of 5-month-old female WT or MLL4+/− (n = 4) were harvested at indicated ZT. P values are for differences between WT and MLL3/4 mutant mice (A-C). Abbreviation: GAPDH, glyceraldehyde 3-phosphate dehydrogenase.
These results suggest that MLL3/4 control a wide range of circadian genes, including core clock genes, establishing MLL3/4 as crucial epigenetic regulators of the hepatic circadian clock.
Rhythmic Expression of MLL3/4
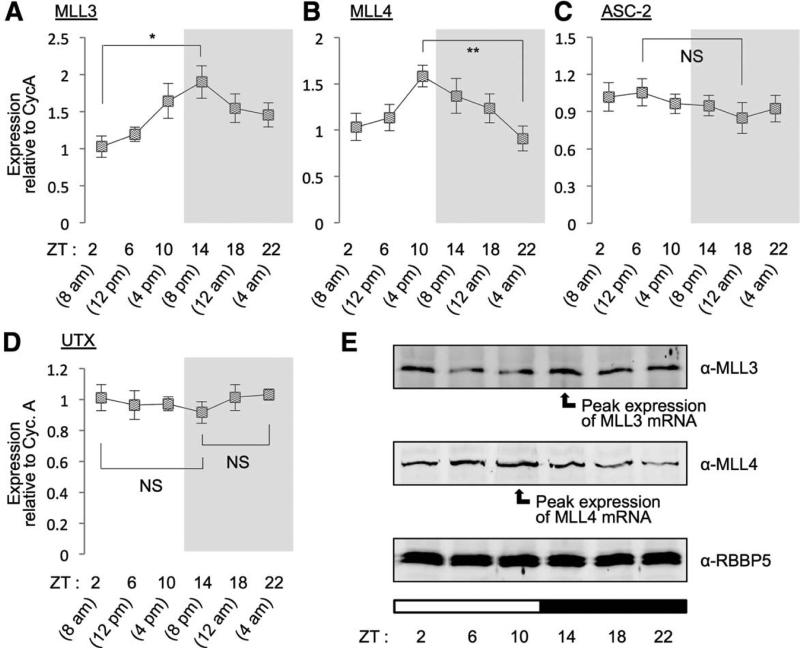
The important role of MLL3/4 in the hepatic clock raises the possibility that expression of key components of the MLL3/4 complexes may show the circadian rhythmic oscillation in the liver, similarly to MLL3.17 Therefore, we monitored expression of MLL3, MLL4, ASC-2, and UTX in WT liver at various time points using qRT-PCR. Expression of MLL3 and MLL4 underwent rhythmic changes over the circadian cycle, with peak expression at ZT14 (8 pm) and ZT10 (4 pm), respectively, whereas expression levels of ASC-2 and UTX did not (Fig. 4A-D). Similar results were also obtained in protein levels (Fig. 4E). Notably, the rhythmic fluctuation pattern of MLL3/4 levels is highly correlated with the cycling of the H3K4me3 mark over the clock at whole-genome level in the liver,17 suggesting that the circadian rhythmic oscillation of MLL3/4 expression likely contributes to epigenetic control of circadian gene expression in the liver.
Fig. 4.
Circadian regulation of MLL3 and MLL4. Measurement of mRNA levels in livers of 9-week-old WT male mice (n = 5) for MLL3 (A), MLL4 (B), ASC-2 (C), and UTX (D) throughout different time points using qRT-PCR reveals that mRNA levels of both MLL3 and MLL4, but neither ASC-2 nor UTX, fluctuate over the circadian day and night. (E) Immunoblotting analysis of liver lysates pooled from all 5 animals at each time point, which were used in the above mRNA measurements, reveals that protein levels of MLL3 and MLL4, but not those of RBBP5, fluctuate and closely match the circadian pattern of mRNA levels. White bars indicate circadian daytime; black bars signify circadian night.
MLL3/4 as Coactivators of the Clock TFs RORs
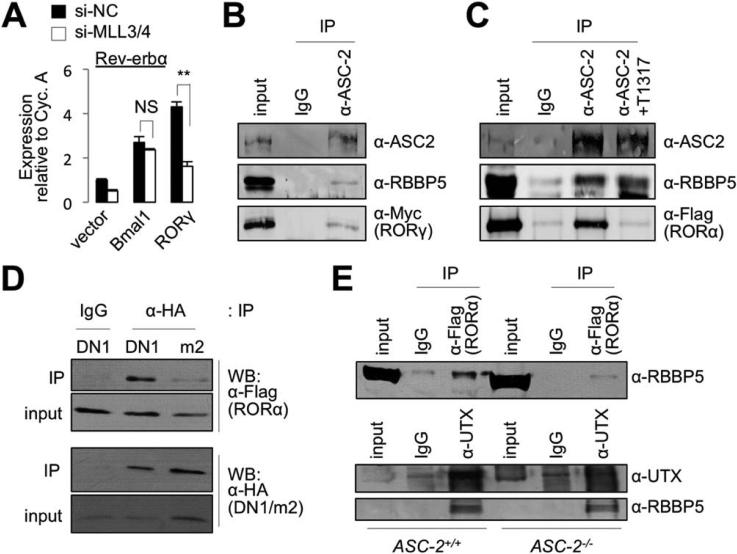
Given the crucial roles of MLL3/4 in circa-dian gene regulation and the well-established function of MLL3/4 in transcriptional activation,1 we considered the possibility that MLL3/4 complexes function as hepatic coactivators for the core clock TFs, such as Bmal1/Clock heterodimer and the RORs. Rev-erbα, another core clock TF, functions as a transcriptional repressor in circadian clock gene regulation.20 In circadian gene networks, Bmal1/Clock and RORs trigger transcription of the Rev-erbα gene by binding to its promoter.20 To test whether MLL3/4 are important for Bmal1/Clock- or ROR-dependent transactivation, we expressed Bmal1 or RORγ with short hairpin RNAs (shRNAs) targeting MLL3/4 or control shRNA in the human hepatocarcinoma cell line, HepG2, and monitored induction levels of Rev-erbα using qRT-PCR. Expression of Rev-erbα was up-regulated by Bmal1 or RORγ (Fig. 5A), as previously reported.20 Knockdown of MLL3/4 impaired induction of Rev-erbα by RORγ, but not induction of Rev-erbα by Bmal1 (Fig. 5A), suggesting that MLL3/4-complexes function as coactivators of RORs in regulating hepatic circadian clock genes.
Fig. 5.
MLL3/4 complexes as coactivators of RORs. (A) Expression of Rev-erbα in HepG2 cells transfected with expression vector for Bmal1 or RORγ, along with control siRNA or siRNA against MLL3 and MLL4. (B and C) CoIP experiments in HEK293 cells transfected with expression vector for Myc-tagged RORγ (B) or Flag-tagged RORα (C). Endogenous ASC-2 and RBBP5 in HEK293 cells were detected using Abs against ASC-2 and RBBP5. The antagonist of RORs, T0901317 (T1317), was used in 20 lM (C). (D) CoIP experiments with HEK293 cells transfected with expression vectors for Flag-tagged RORα and either DN1 or DN1/m2. (E) CoIP experiments with WT and ASC-2−/− mouse embryo fibroblast cells transfected with expression vector for Flag-tagged RORα. Abbreviation: IgG, immunoglobulin G.
RORs Bind to ASC-2 in MLL3/4 Complexes
In the MLL3/4 complexes, ASC-2 serves as an adaptor that recruits MLL3/4 complexes to several NRs in a ligand-dependent manner.6 To test whether ASC-2 mediates the association of the MLL3/4 complexes with RORs, we performed CoIP experiments in HEK293 cells transfected with an expression vector for RORγ or RORα. ASC-2 coimmunoprecipitated RORs and RBBP5, a core subunit of MLL3/4 complexes (Fig. 5B,C), suggesting that the MLL3/4 complexes associate with RORs. Supporting our results in Fig. 5A, ASC-2 failed to coimmunoprecipitate Bmal1 (data not shown).
RORs activate transcription of their target genes without exogenous ligand, but this transcriptional activity is suppressed by an inverse agonist of RORs, T0901317.21 To test whether ASC-2 associates with a transcriptionally active form of RORs, we monitored whether T0901317 affects interaction between ASC-2 and RORα (Fig. 5C). T0901317 blocked this interaction, but not the association between ASC-2 and RBBP5 (Fig. 5C). These results suggest that ASC-2 binds to the transcriptionally active conformation of RORs.
The two LXXLL motifs of ASC-2 directly recognize the activated conformation of several NRs.6 The first LXXLL motif interacts with multiple NRs, whereas the second LXXLL motif is highly specific to the liver X receptors.6 To test whether ASC-2 binds to RORs by its LXXLL motifs, we performed CoIP experiments in HEK293 cells expressing an hemagglutinin (HA)-tagged fragment of ASC-2 containing the first LXXLL motif, named DN1.22 The DN1 fragment readily coimmuno-precipitated RORα (Fig. 5D). A mutation in the first LXXLL motif within DN1 disrupted the association between DN1 and RORα (Fig. 5D), indicating that RORs bind to the first LXXLL motif in ASC-2.
To test whether ASC-2 recruits the MLL3/4 complexes to RORs, we performed CoIP experiments between RORs and MLL3/4-complexes in WT and ASC-2-null mouse embryonic fibroblast cells.3 RBBP5, a core subunit of SET1-like complexes, was coimmunoprecipitated by α-Flag antibody (Ab; against Flag-RORα; Fig. 5E, upper panel), establishing the interaction between MLL3/4 complexes and RORα. However, this interaction was reduced in ASC-2-null cells, relative to WT cells (Fig. 5E, upper panel), suggesting that ASC-2 is required for RORα to recruit MLL3/4 complexes. In contrast, UTX associated with RBBP5 equally well in both WT and ASC-2-null cells (Fig. 5E, lower panels), indicating that ASC-2 is not essential for the assembly of core MLL3/4 complexes.
These results demonstrate that ASC-2 senses the transcriptionally active conformation of RORs and tethers the MLL3/4 complexes to RORs.
MLL3/4 Complexes as an Epigenetic Switch to Activate the Hepatic Circadian Target Genes of RORs
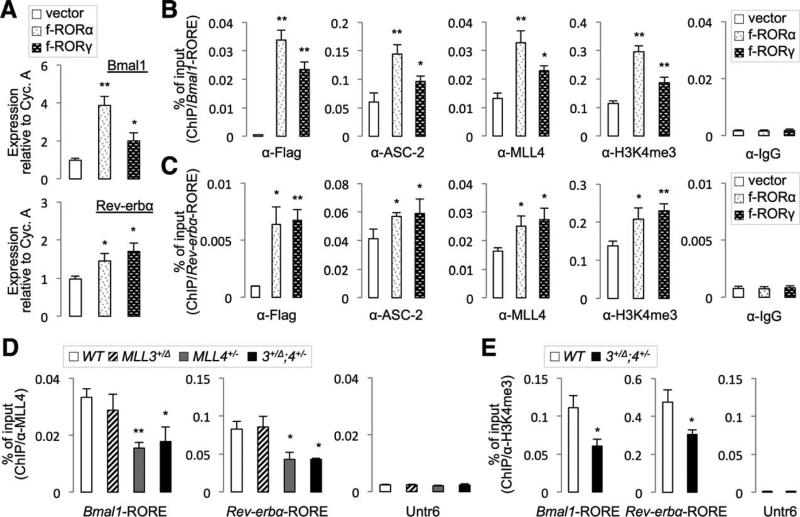
Induction of Bmal1 and Rev-erbα genes by RORs represents a crucial pathway in the regulation of circadian gene networks.20 Combined with our results that Bmal1 and Rev-erbα are down-regulated in MLL3/4 mutant livers (Figs. 2 and 3), this notion suggests that the MLL3/4 complexes serve as an epigenetic switch in the hepatic circadian gene networks by enabling ROR-mediated transactivation of target genes, such as Bmal1 and Rev-erbα, by their activity to trigger an open chromatin marker H3K4me3.
To test this possibility, we examined whether RORs recruit the MLL4 complex to the promoters of Bmal1 and Rev-erbα and increase their H3K4me3 mark.
Expression of RORα or RORγ led to induction of expression of Bmal1 and Rev-erbα in the murine hepatoma cell line, Hepa1c1c7 (Fig. 6A). As reported previously,23,24 RORα and RORγ bound to the ROREs in the promoters of Bmal1 and Rev-erbα genes (Fig. 6B,C). Binding of RORs coincided with the increased recruitment of ASC-2 and MLL4 to ROREs as well as elevated levels of H3K4me3 in the same genomic regions (Fig. 6B,C). These results suggest that RORs recruit the MLL4 complex, which triggers the transcriptionally active chromatin in Bmal1 and Rev-erbα genes.
Fig. 6.
Binding of MLL4 to the promoter regions of clock genes. (A) qRT-PCR for expression of Bmal1 and Rev-erbα in Hepa1c1c7 cells transfected with Flag-tagged RORα or RORγ. (B and C) ChIP for RORs, ASC-2, MLL4, and H3K4me3 using Hepa1c1c7 cells transfected with Flag-tagged RORα or RORγ on Bmal1-RORE (B) and Rev-erbα-RORE (C). (D and E) ChIP for MLL4 (D) and H3K4me3 (E) using liver lysates of indicated mouse lines (n = 5-6 per each genotype). P values are for differences between vector alone and ROR-expression vectors (A-C) or between WT and MLL3/4 mutant mice (D and E). Abbreviation: IgG, immunoglobulin G.
To validate these results in vivo, we pooled the liver lysates of 5-month-old WT, MLL3+/Δ, MLL4+/−, and MLL3+/Δ;MLL4+/− male mice sacrificed at ZT10 (4 PM) and performed three separate sets of ChIP experiments with biological replicate samples. MLL4 was robustly recruited to both Bmal1- and Rev-erbα-ROREs in the liver, whereas it did not bind to a negative control genomic region untranslated region on chromosome 6 (Untr6)25 (Fig. 6D). The MLL4 binding to ROREs was significantly dampened in MLL4+/− and MLL3+/Δ;MLL4+/− livers, but not in MLL3+/Δ livers (Fig. 6D), consistent with the finding that MLL4 protein levels are reduced by ~50% in MLL4+/− mice (Supporting Fig. 1D). We also monitored H3K4me3 levels of these RORE regions in WT and MLL3+/Δ;MLL4+/− livers using ChIP with α-H3K4me3 Abs. In MLL3+/Δ;MLL4+/− livers, H3K4me3 levels on Bmal1- and Rev-erbα-ROREs were substantially diminished, compared to levels in WT tissues, although they were still higher than the Untr6 control region (Fig. 6E).
These results suggest that the MLL3/4 complexes manifest their function as an epigenetic switch of the hepatic circadian clock, at least in part, by triggering open chromatin formation in the circadian target genes of RORs.
MLL4 Is Crucial for Circadian Regulation of BA
Homeostasis
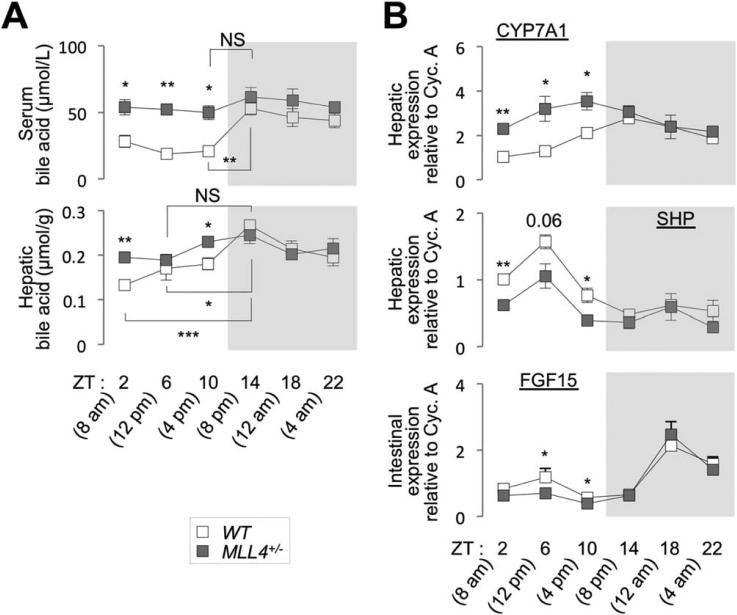
Production and excretion of BAs in murine liver exhibit a synchronous circadian rhythm.11,13 Thus, we examined whether dysregulation of the hepatic circadian genes in MLL4+/− mice leads to compromised circadian fluctuation of BA levels. Serum and hepatic BA levels showed a clear circadian rhythmic pattern with peak levels during nighttime feeding period in WT mice (Fig. 7A). Both serum and hepatic BA levels were substantially increased during daytime in MLL4+/− mice, compared to WT mice, resulting in a loss of rhythmic fluctuation of BA levels over the circadian cycle (Fig. 7A).
Fig. 7.
Circadian control of BA homeostasis by MLL4. (A and B) Serum and hepatic BA levels (A) and expression levels of CYP7A1 and SHP in liver and FGF15 in the intestine (B) of 5-month-old female WT and MLL4+/− mice (n = 4) were measured at multiple time points throughout the circadian clock. P values are for differences between WT and MLL4+/− mice unless they are accompanied with brackets. Abbreviation: NS, not significant.
The key contributing factor in establishing the synchronous rhythmic pattern of BA levels during circadian cycle is BA synthesis in the liver, for which CYP7A1 serves as a rate-limiting enzyme.14 Expression and enzymatic activity of CYP7A1 exhibit circadian oscillation.13 Also, transport of BAs from hepatocytes to bile, the reabsorption of BA in the intestine, and the process to transport reabsorbed BAs back to hepatocytes contribute to setting up circadian clock regulation of BA levels.13,14 To test whether any of these pathways is interrupted in MLL4+/− mice, we collected livers and intestines from 5-month-old WT and MLL4+/− male mice at six distinct time points over the circadian cycle and then monitored the expression pattern of genes controlling hepatic synthesis (CYP7A1), intestinal reabsorption (OSTalpha, OSTbeta, FABP6 and ASBT), and hepatic transport (NTCP and BSEP) of BA using qRT-PCR. In WT tissues, the genes of all pathways exhibited unambiguous circadian oscillation pattern except NTCP (Fig. 7B and Supporting Fig. 2B,C). CYP7A1 expression was markedly increased in MLL4+/− liver, relative to WT liver, during daytime, but not during the nighttime feeding period, greatly dampening the circadian oscillation pattern of CYP7A1 expression (Fig. 7B). Notably, the weakened rhythmic pattern of CYP7A1 mRNA expression in MLL4+/− liver mirrored the loss of circadian fluctuation of BA levels in MLL4+/− mice. The circa-dian rhythmic expression of genes involved in intestinal reabsorption and hepatic transport of BA was largely intact in MLL4+/− mice (Supporting Fig. 2B,C).
These results suggest that MLL4 plays crucial roles in establishing circadian rhythmic BA levels mainly by controlling the BA production driven by CYP7A1.
Impaired Negative Feedback Loops in BA Homeostasis in MLL4+/− Mice
Our findings indicate that CYP7A1 is a major regulatory point for MLL4 to establish BA homeostasis over the circadian cycle. Expression of CYP7A1 is tightly controlled by intricate feedback-regulatory pathways that respond to BA levels in the body (Fig. 8A). The aberrantly up-regulated CYP7A1 expression in MLL4+/− liver during daytime (Fig. 7B) suggests that the regulatory feedback loops for CYP7A1 expression are impaired in MLL4+/− mice. A key pathway to suppress CYP7A1 gene in the liver is the FXR-dependent induction of two genes, SHP in the liver and fibroblast growth factor 15 (FGF15) in the intestine, in response to increased levels of BAs that function as an agonist of FXR.14 SHP and FGF15 cooperatively antagonize the hepatic expression of CYP7A1, suppressing BA production.14,26 NRs, such as RORs, Rev-erbs and FXR, and p53 control the expression of SHP and FGF15 in regulating BA levels.14,15 Given the essential roles of SHP and FGF15 in directing the rhythmic fluctuation of BA levels,14 we monitored the expression pattern of hepatic SHP and intestinal FGF15 in 5-month-old WT and MLL4+/− mice over the circadian cycle. Expression of SHP and FGF15 in MLL4+/− mice was substantially reduced specifically during daytime, when compared to WT liver (Fig. 7B). These results also identify FGF15 as a novel target of MLL4 complex in BA homeostasis. The down-regulation of SHP and FGF15 in MLL4+/− mice during daytime likely leads to the derepression of CYP7A1 and increased BA synthesis, resulting in disruption of circadian cycling of BA levels. Collectively, our data suggest that MLL3/4 complexes establish circadian rhythms of BA levels, in part, by participating in gene networks that control the expression of SHP and FGF15, two critical negative regulators of the CYP7A1 gene.
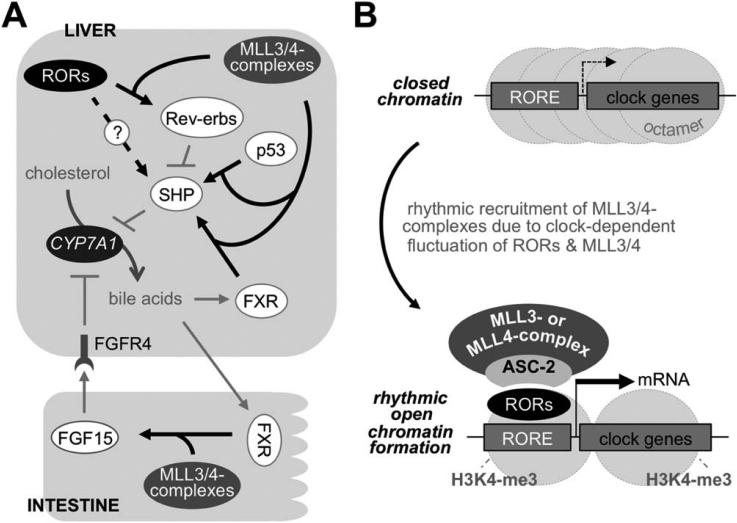
Fig. 8.
Working models for roles of MLL3/4 complexes. (A and B) Roles of MLL3/4 complexes in BA homeostasis (A) and as critical epigenetic circadian coactivators of RORs (B).
Discussion
In this study, we made several major findings in epigenetic regulation of metabolism. First, we demonstrated that MLL3/4 complexes coregulate genes involved in the similar metabolic pathways. MLL3 and MLL4 mutant mice exhibited similar metabolic phenotypes, and their phenotypes were more severe in the compound mutants in both genes (Fig. 1). The idea that MLL3/4 complexes control common metabolic processes is reinforced by our comparative genome-wide gene expression analyses in the liver (Fig. 2). Among six mammalian SET1-like complexes, MLL3/4 complexes have been closely associated with regulation of metabolic genes.7-10,16 We speculate that this is a result of the presence of ASC-2, an adaptor that directly binds to NRs, in MLL3/4 complexes, given that many NRs are the central regulators of diverse metabolic pathways.27
Second, our study revealed that MLL3/4 are key epigenetic regulators of the hepatic circadian clock. Whereas MLL3 has recently been linked to the hepatic circadian clock,17 this idea has not been tested in MLL3 mutant animals. This study provides the missing physiological support for the roles of MLL3 in the hepatic circadian clock and underlines an important role for MLL4 in the same process.
Third, our results identified RORs as the core circa-dian TFs that convey the hepatic circadian action of MLL3/4 complexes (Figs. 5 and 6). ASC-2 senses the transcriptionally active conformation of RORs through at least its first LXXLL motif (involvement of the second LXXLL motif was not tested) and brings MLL3/4 complexes to the ROREs of the circadian genes regulated by RORs, such as Bmal1 and Rev-erbα, decorating them with the transcriptionally active chromatin marker, H3K4me3 (Fig. 8B). Given that RORs are integral players for the central and peripheral clocks,20 our findings would be widely applicable to the circa-dian regulation in diverse biological processes in multiple organs.
Fourth, our study established that MLL3/4 complexes direct circadian oscillation of BA levels in the body. BA homeostasis is maintained by elaborate regulatory pathways involving multiple tissues that control BA production in the liver, excretion to bile and the intestine, reabsorption in the ileum, and retransport to the liver.14 Among these complex regulatory steps, BA production was most significantly altered in MLL4 mutant mice, resulting in an increase in the whole BA pool size, as reflected in the increase in serum and feces BA levels (Fig. 1E and Supporting Fig. 2A). At a molecular level, MLL3/4 complexes impinge on the gene networks driving rhythmic expression of CYP7A1, which involve the orchestrated actions of NRs, such as FXR and SHP, and p53.14,15 Rev-erbα directly represses expression of SHP by SHP-RORE, thereby increasing CYP7A levels.28 This opens the possibility that RORs may indirectly up-regulate CYP7A1 by up-regulating Rev-erbα expression (Fig. 8A). Because Rev-erbα and RORs share the same response element, it is also possible that RORs directly activate expression of SHP,24 particularly when Rev-erbα levels are lower (Fig. 8A). These two opposing forces of RORs and Rev-erbs in regulating SHP may contribute to the rhythmic expression pattern of SHP.18,19 Our studies revealed that MLL3/4 complexes function as important coactivators of FXR, RORs, and p53 (this study and previously published works7,10). Thus, the reduced activities of MLL3/4 in MLL3/4 mutant mice would result in dysregulation of SHP from weaker transcriptional activity of FXR, RORs, and p53, attributing to the loss of CYP7A1 mRNA rhythmicity (Fig. 8A). Overall, our results suggest that the circadian regulation of BA homeostasis by MLL3/4 complexes involves their coactivator roles for multiple TFs and pathways, which primarily converge on the control of CYP7A1 levels (Fig. 8A).
Finally, a subset of genes appears to be regulated by MLL3 or MLL4 alone in the liver (Fig. 2A). Given that the overall homology between MLL3 and MLL4 is less than 30%, direct, but differential, recognition of transcription factors by MLL3 and MLL4 may result in MLL3- or MLL4-specific activation of genes in a cell-context–dependent manner.
Supplementary Material
Acknowledgment
The authors thank Ashly Brown for critically reading the manuscript and Prof. Robert Roeder for sharing his MLL4 antibody.
This research was supported by grants from the Basic Science Research Program (2012R1A1A1001749) and the Bio & Medical Technology Development Program (2012M3A9C6050508) of the National Research Foundation (NRF) funded by the Korean government (MEST) and the National R&D Program for Cancer Control, Ministry of Health & Welfare, Republic of Korea (1220120), and Aspiring Researcher Program of Seoul National University in 2014 (to S.L.) as well as the National Institutes of Health (NIH)/National Institute of Neurological Disorders and Stroke (R01 NS054941; to S.K.L.), and the NIH/National Institute of Diabetes and Digestive and Kidney Diseases (R01 DK064678 and R01 DK103664; to J.W.L.).
Abbreviations
- Abs
antibodies
- ASC-2
activating signal cointegrator 2
- BA
bile acid
- Bmal1
brain and muscle ARNT-like
- ChIP
chromatin immunoprecipitation
- Clock
circadian locomotor output cycles kaput
- CoIP
coimmunoprecipitation
- Cry1
cryptochrome 1
- CYP7A1
cholesterol-7a hydroxylase
- FGF15
fibroblast growth factor 15
- FXR
farnesoid X receptor
- GO
Gene Ontology
- HA
hemagglutinin
- H3K4me3
histone H3-lysine 4 trimethylation
- H3K4MT
H3K4 methyltransferase
- HFD
high-fat diet
- IP
immunoprecipitation
- MLL
mixed-lineage leukemia
- mRNA
messenger RNA
- NRs
nuclear receptors
- Per2
period 2
- qRT-PCR
quantitative reverse-trasncription polymerase chain reaction
- RBBP5
retinoblastoma-binding protein 5
- RNA-Seq
RNA sequencing
- ROR
retinoid-related orphan receptor
- RORE
ROR-response element
- SHP
small heterodimer partner
- shRNAs
short hairpin RNAs
- siRNA
small interfering RNA
- TFs
transcription factors
- Untr6
untranslated region on chromosome 6
- UTX
ubiquitously transcribed tetratricopeptide repeat, X chromosome
- WT
wild-type
- ZT
zeitgeber time
Footnotes
Additional Supporting Information may be found at onlinelibrary.wiley.com/doi/10.1002/hep.27578/suppinfo.
Potential conflict on interest: Nothing to report.
References
- 1.Gu B, Lee MG. Histone H3 lysine 4 methyltransferases and demethylases in self-renewal and differentiation of stem cells. Cell Biosci. 2013;3:39. doi: 10.1186/2045-3701-3-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goo YH, Sohn YC, Kim DH, Kim SW, Kang MJ, Jung DJ, et al. Activating signal cointegrator 2 belongs to a novel steady-state complex that contains a subset of trithorax group proteins. Mol Cell Biol. 2003;23:140–149. doi: 10.1128/MCB.23.1.140-149.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee S, Lee DK, Dou Y, Lee J, Lee B, Kwak E, et al. Coactivator as a target gene specificity determinant for histone H3 lysine 4 methyltransferases. Proc Natl Acad Sci U S A. 2006;103:15392–15397. doi: 10.1073/pnas.0607313103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cho YW, Hong T, Hong S, Guo H, Yu H, Kim D, et al. PTIP associates with MLL3- and MLL4-containing histone H3 lysine 4 methyltransferase complex. J Biol Chem. 2007;282:20395–20406. doi: 10.1074/jbc.M701574200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Issaeva I, Zonis Y, Rozovskaia T, Orlovsky K, Croce CM, Nakamura T, et al. Knockdown of ALR reveals ALR target genes and leads to alterations in cell adhesion and growth. Mol Cell Biol. 2007;27:1889–1903. doi: 10.1128/MCB.01506-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee SK, Jung SY, Kim YS, Na SY, Lee YC, Lee JW. Two distinct nuclear receptor-interaction domains and CREB-binding protein-dependent transactivation function of activating signal cointegrator-2. Mol Endocrinol. 2001;15:241–254. doi: 10.1210/mend.15.2.0595. [DOI] [PubMed] [Google Scholar]
- 7.Kim DH, Lee J, Lee B, Lee JW. ASCOM controls farnesoid X receptor transactivation through its associated histone H3 lysine 4 methyltransferase activity. Mol Endocrinol. 2009;23:1556–1562. doi: 10.1210/me.2009-0099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee S, Lee J, Lee SK, Lee JW. Activating signal cointegrator-2 is an essential adaptor to recruit histone H3 lysine 4 methyltransferases MLL3 and MLL4 to the liver X receptors. Mol Endocrinol. 2008;22:1312–1319. doi: 10.1210/me.2008-0012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee J, Saha PK, Yang QH, Lee S, Park JY, Suh Y, et al. Targeted inactivation of MLL3 histone H3-Lys-4 methyltransferase activity in the mouse reveals vital roles for MLL3 in adipogenesis. Proc Natl Acad Sci U S A. 2008;105:19229–19234. doi: 10.1073/pnas.0810100105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim DH, Kim J, Lee JW. Requirement for MLL3 in p53 regulation of hepatic expression of small heterodimer partner and bile acid homeostasis. Mol Endocrinol. 2011;25:2076–2083. doi: 10.1210/me.2011-1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kai M, Eto T, Kondo K, Setoguchi Y, Higashi S, Mayeda Y, et al. Synchronous circadian rhythms of mRNA levels and activities of cholesterol-7-alpha-hydroxylase in the rabbit and rat. J Lipid Res. 1995;36:367–374. [PubMed] [Google Scholar]
- 12.Duane WC, Levitt DG, Mueller SM, Behrens JC. Regulation of bile acid synthesis in man. Presence of a diurnal rhythm. J Clin Invest. 1983;72:1930–1936. doi: 10.1172/JCI111157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nakano A, Tietz PS, LaRusso NF. Circadian rhythms of biliary protein and lipid excretion in rats. Am J Physiol. 1990;258:G653–G659. doi: 10.1152/ajpgi.1990.258.5.G653. [DOI] [PubMed] [Google Scholar]
- 14.de Aguiar Vallim TQ, Tarling EJ, Edwards PA. Pleiotropic roles of bile acids in metabolism. Cell Metab. 2013;17:657–669. doi: 10.1016/j.cmet.2013.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim DH, Lee JW. Tumor suppressor p53 regulates bile acid homeostasis via small heterodimer partner. Proc Natl Acad Sci U S A. 2011;108:12266–12270. doi: 10.1073/pnas.1019678108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ananthanarayanan M, Li Y, Surapureddi S, Balasubramaniyan N, Ahn J, Goldstein JA, et al. Histone H3K4-trimethylation by MLL3 as part of ASCOM complex is critical for NR activation of bile acid transporter genes and is downregulated in cholestasis. Am J Physiol Gastrointest Liver Physiol. 2011;300:G771–G781. doi: 10.1152/ajpgi.00499.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Valekunja UK, Edgar RS, Oklejewicz M, van der Horst GT, O'Neill JS, Tamanini F, et al. Histone methyltransferase MLL3 contributes to genome-scale circadian transcription. Proc Natl Acad Sci UK S A. 2013;110:1554–1559. doi: 10.1073/pnas.1214168110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bookout AL, Jeong Y, Downes M, Yu RT, Evans RM, Mangelsdorf DJ. Anatomical profiling of nuclear receptor expression reveals a hierarchical transcriptional network. Cell. 2006;126:789–799. doi: 10.1016/j.cell.2006.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang X, Downes M, Yu RT, Bookout AL, He W, Straume M, et al. Nuclear receptor expression links the circadian clock to metabolism. Cell. 2006;126:801–810. doi: 10.1016/j.cell.2006.06.050. [DOI] [PubMed] [Google Scholar]
- 20.Asher G, Schibler U. Crosstalk between components of circadian and metabolic cycles in mammals. Cell Metab. 2011;13:125–137. doi: 10.1016/j.cmet.2011.01.006. [DOI] [PubMed] [Google Scholar]
- 21.Kumar N, Solt LA, Conkright JJ, Wang Y, Istrate MA, Busby SA, et al. The benzenesulfoamide T0901317 [N-(2,2,2-trifluoroethyl)-N-[4-[2,2,2-trifluoro-1-hydroxy-1-(trifluoromethyl)ethyl]phenyl]-benzene-sulfonamide] is a novel retinoic acid receptor-related orphan receptor-alpha/gamma inverse agonist. Mol Pharmacol. 2010;77:228–236. doi: 10.1124/mol.109.060905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim SW, Cheong C, Sohn YC, Goo YH, Oh WJ, Park JH, et al. Multiple developmental defects derived from impaired recruitment of ASC-2 to nuclear receptors in mice: implication for posterior lenticonus with cataract. Mol Cell Biol. 2002;22:8409–8414. doi: 10.1128/MCB.22.24.8409-8414.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guillaumond F, Dardente H, Giguère V, Cermakian N. Differential control of Bmal1 circadian transcription by REV-ERB and ROR nuclear receptors. J Biol Rhythms. 2005;20:391–403. doi: 10.1177/0748730405277232. [DOI] [PubMed] [Google Scholar]
- 24.Takeda Y, Jothi R, Birault V, Jetten AM. RORc directly regulates the circadian expression of clock genes and downstream targets in vivo. Nucleic Acids Res. 2012;40:8519–8535. doi: 10.1093/nar/gks630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Quintana FJ, Basso AS, Iglesias AH, Korn T, Farez MF, Bettelli E, et al. Control of Treg and TH17 cell differentiation by the aryl hydro-carbon receptor. Nature. 2008;453:65–71. doi: 10.1038/nature06880. [DOI] [PubMed] [Google Scholar]
- 26.Inagaki T, Choi M, Moschetta A, Peng L, Cummins CL, McDonald JG, et al. Fibroblast growth factor 15 functions as an enterohepatic signal to regulate bile acid homeostasis. Cell Metab. 2005;2:217–225. doi: 10.1016/j.cmet.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 27.Hong SH, Ahmadian M, Yu RT, Atkins AR, Downes M, Evans RM. Nuclear receptors and metabolism: from feast to famine. Diabetologia. 2014;57:860–867. doi: 10.1007/s00125-014-3209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Duez H, van der Veen JN, Duhem C, Pourcet B, Touvier T, Fontaine C, et al. Regulation of bile acid synthesis by the nuclear receptor Reverbalpha. Gastroenterology. 2008;135:689–698. doi: 10.1053/j.gastro.2008.05.035. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.