Abstract
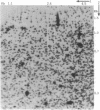
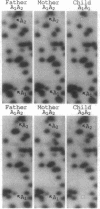
The continuing efforts to evaluate specific human populations for altered germinal mutation rates would profit from more efficient and more specific approaches than those of the past. To this end, we have explored the potential usefulness of two-dimensional electrophoresis of DNA fragments obtained from restriction-enzyme-digested genomic DNA. This permits the analysis, on a single preparation, of approximately 2000 DNA fragments varying in size from 1.0 to 5.0 kb in the first dimension and from 0.3 to 2.0 kb in the second dimension. To enter into a genetic analysis, these fragments must exhibit positional and quantitative stability. With respect to the latter, if spots that are the product of two homologous DNA fragments are to be distinguished with the requisite accuracy from spots that are the product of only one fragment, the coefficient of variation of spot intensity should be approximately < or = 0.12. At present, 482 of the spots in our preparations meet these standards. In an examination of preparations based on three Japanese mother/father/child trios, 43 of these 482 spots were found to exhibit variation that segregated within families according to Mendelian principles. We have established the feasibility of cloning a variant fragment from such gels and establishing its nucleotide sequence. This technology should be highly efficient in monitoring for mutations resulting in loss/gain/rearrangement events in DNA fragments distributed throughout the genome.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Antequera F., Bird A. Number of CpG islands and genes in human and mouse. Proc Natl Acad Sci U S A. 1993 Dec 15;90(24):11995–11999. doi: 10.1073/pnas.90.24.11995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bickmore W. A., Sumner A. T. Mammalian chromosome banding--an expression of genome organization. Trends Genet. 1989 May;5(5):144–148. doi: 10.1016/0168-9525(89)90055-3. [DOI] [PubMed] [Google Scholar]
- Bird A., Taggart M., Frommer M., Miller O. J., Macleod D. A fraction of the mouse genome that is derived from islands of nonmethylated, CpG-rich DNA. Cell. 1985 Jan;40(1):91–99. doi: 10.1016/0092-8674(85)90312-5. [DOI] [PubMed] [Google Scholar]
- Gardiner-Garden M., Frommer M. CpG islands in vertebrate genomes. J Mol Biol. 1987 Jul 20;196(2):261–282. doi: 10.1016/0022-2836(87)90689-9. [DOI] [PubMed] [Google Scholar]
- Hatada I., Hayashizaki Y., Hirotsune S., Komatsubara H., Mukai T. A genomic scanning method for higher organisms using restriction sites as landmarks. Proc Natl Acad Sci U S A. 1991 Nov 1;88(21):9523–9527. doi: 10.1073/pnas.88.21.9523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashizaki Y., Shibata H., Hirotsune S., Sugino H., Okazaki Y., Sasaki N., Hirose K., Imoto H., Okuizumi H., Muramatsu M. Identification of an imprinted U2af binding protein related sequence on mouse chromosome 11 using the RLGS method. Nat Genet. 1994 Jan;6(1):33–40. doi: 10.1038/ng0194-33. [DOI] [PubMed] [Google Scholar]
- Kuick R. D., Skolnick M. M., Hanash S. M., Neel J. V. A two-dimensional electrophoresis-related laboratory information processing system: spot matching. Electrophoresis. 1991 Oct;12(10):736–746. doi: 10.1002/elps.1150121007. [DOI] [PubMed] [Google Scholar]
- Larsen F., Gundersen G., Lopez R., Prydz H. CpG islands as gene markers in the human genome. Genomics. 1992 Aug;13(4):1095–1107. doi: 10.1016/0888-7543(92)90024-m. [DOI] [PubMed] [Google Scholar]
- Neel J. V., Satoh C., Myers R. International Commission for Protection against Environmental Mutagens and Carcinogens. Report of a workshop on the application of molecular genetics to the study of mutation in the children of atomic bomb survivors. Mutat Res. 1993 Feb;291(1):1–20. doi: 10.1016/0165-1161(93)90012-o. [DOI] [PubMed] [Google Scholar]
- Skolnick M. M., Sternberg S. R., Neel J. V. Computer programs for adapting two-dimensional gels to the study of mutation. Clin Chem. 1982 Apr;28(4 Pt 2):969–978. [PubMed] [Google Scholar]
- Uitterlinden A. G., Slagboom P. E., Knook D. L., Vijg J. Two-dimensional DNA fingerprinting of human individuals. Proc Natl Acad Sci U S A. 1989 Apr;86(8):2742–2746. doi: 10.1073/pnas.86.8.2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi M., Au L. C., Ichikawa N., Ts'o P. O. Enhanced resolution of DNA restriction fragments: a procedure by two-dimensional electrophoresis and double-labeling. Proc Natl Acad Sci U S A. 1990 May;87(10):3919–3923. doi: 10.1073/pnas.87.10.3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- te Meerman G. J., Mullaart E., van der Meulen M. A., den Daas J. H., Morolli B., Uitterlinden A. G., Vijg J. Linkage analysis by two-dimensional DNA typing. Am J Hum Genet. 1993 Dec;53(6):1289–1297. [PMC free article] [PubMed] [Google Scholar]