Abstract
The groundbreaking technologies of induced pluripotency and lineage conversion have generated a genuine opportunity to address fundamental aspects of the diseases that affect the nervous system. These approaches have granted us unrestricted access to the brain and spinal cord of patients and have allowed for the study of disease in the context of human cells, expressing physiological levels of proteins and under each patient's unique genetic constellation. Along with this unprecedented opportunity have come significant challenges, particularly in relation to patient variability, experimental design and data interpretation. Nevertheless, significant progress has been achieved over the past few years both in our ability to create the various neural subtypes that comprise the nervous system and in our efforts to develop cellular models of disease that recapitulate clinical findings identified in patients. In this Review, we present tables listing the various human neural cell types that can be generated and the neurological disease modeling studies that have been reported, describe the current state of the field, highlight important breakthroughs and discuss the next steps and future challenges.
Keywords: directed differentiation, disease modeling, neurologic disorder, neuronal development
Introduction
Diseases of the nervous system represent an enormous burden for society in terms of human suffering and financial cost. While significant advancements have been achieved over the last few decades particularly in terms of genetic linkage, clinical classification and patient care, effective treatments are lacking. The inaccessibility of the relevant tissues and cell types in the central nervous system (CNS) and the complex multifactorial nature of most neurological disorders have hampered research progress. While animal models have been crucial in the investigation of disease mechanisms, fundamental developmental, biochemical and physiological differences exist between animals and humans. The importance of utilizing human cells for these purposes is evident by the large number of drugs that show efficacy and safety in rodent models of diseases but subsequently fail in human clinical trials, which are partly attributed to these species differences (Rubin, 2008). Furthermore, the overwhelming majority of neurological disease is of a sporadic nature, rendering animal modeling ineffective, while it is unclear whether the relatively rare monogenic forms of disease truly represent the vast majority of sporadic cases.
The simultaneous development of methods for reprogramming adult cells into induced pluripotent stem cells (iPSCs; Takahashi et al, 2007; Yu et al, 2007; Park et al, 2008) and the directed differentiation of pluripotent stem cells into distinct neuronal subtypes (Williams et al, 2012) suggested an attractive route to a novel model system for the study of neurological disorders. Patient-specific iPSCs can be generated by epigenetic reprogramming of various adult cell types such as skin fibroblasts and blood mononuclear cells and just like embryonic stem cells (ESCs), self-renew indefinitely and retain the potential to give rise to all cell types in the human body (Takahashi et al, 2007). More recently, sophisticated lineage conversion approaches have allowed for the direct generation of neurons and neural cell types from adult cells by means of overexpressing key transcription factors (for a detailed description see Tsunemoto et al, 2015). These methods have overcome some of the limitations of directed differentiation and have enabled for the generation of cell types that in many cases were previously unattainable.
The overwhelming advantages of using iPSCs and lineage conversion to develop models of diseases of the nervous system are that they allow one to study disease mechanisms in the context of human neurons and in the context of each patient's own unique genetic constellation. In many cases, established differentiation protocols allow for the generation of the particular neuronal subtype that is most vulnerable to the particular disease, such as spinal motor neurons (Davis-Dusenbery et al, 2014) and dopaminergic neurons (Kriks et al, 2011). These neurons can be produced in abundance from variable genetic backgrounds and could provide useful platforms for drug discovery.
The concept of using iPSCs and lineage conversion to study neurological disease appears straightforward: Both of these approaches allow for the generation of patient-specific neurons, which are relevant to the disease of interest, and when these are compared to neurons generated from healthy controls, any differences identified could be related to the disease. In practice, this approach has been proven to be more challenging than initially believed. What is the right cell type to make and study? How should quality control of neurons be performed? What are the right controls to use when assessing a disease-related phenotype? How do phenotypes identified in vitro relate to the clinical presentation of patients? These are just some of the questions that the community has struggled with, since the initial description of iPSCs and the onset of the development of in vitro patient-specific disease models. Perhaps the seemingly biggest advantage of this approach—the ability to study disease in the genetic background of the patient—has created the biggest challenge, as genetic background contributes to high variability in the properties of the patient-derived cells. This variability is a reality that neurologists have been facing for years, as often, two patients diagnosed with the same condition might present with very different clinical profiles. The technology of cellular reprogramming has brought this reality of clinical heterogeneity seen in patients from the bedside to the lab bench.
Since the initial description of reprogramming technologies, neuroscientists, neurologists and stem cell researchers have generated and characterized hundreds of patient-specific stem cell lines as well as neuronal cells derived from them (Table1). The first “wave” of disease modeling studies focused on generating patient-specific human neurons and confirming previously described pathologies (Dimos et al, 2008; Ebert et al, 2009; Marchetto et al, 2010; Brennand et al, 2011; Seibler et al, 2011; Bilican et al, 2012; Israel et al, 2012). More recent studies have revealed novel insights into disease mechanisms and employed gene editing approaches to clearly demonstrate the association of identified phenotypes with known genetic variants that contribute to disease (An et al, 2012; Corti et al, 2012; Fong et al, 2013; Reinhardt et al, 2013; Kiskinis et al, 2014; Wainger et al, 2014; Wen et al, 2014b). At the same time, there has been tremendous progress in our ability to generate neuronal subtypes both via directed differentiation and through the exogenous expression of transcription factors. Here, we review the current state of disease modeling and neuronal differentiation approaches, highlight breakthrough studies and discuss the shift in focus that is expected over the next few years.
Table 1.
List of published studies modeling human neurological diseases with iPSCs
| Disease | References | Patient genotype | Cell type analyzed | Identified phenotype | Notable |
|---|---|---|---|---|---|
| Alzheimer's Disease | Yagi et al (2011) | PSEN1, PSEN2 mutations | Neurons | Increased amyloid β42 secretion | |
| Alzheimer's Disease | Israel et al (2012) | APP mutations, sporadic cases | Neurons | Increased amyloid β40, Tau and GSK3β phosphorylation, accumulation of endosomes | One of two sporadic patients exhibited phenotypes |
| Alzheimer's Disease | Kondo et al (2013) | APP mutations, sporadic cases | Cortical neurons, astrocytes | Accumulated Aβ oligomers, ER & oxidative stress | One of two sporadic patients exhibited phenotypes |
| Alzheimer's Disease | Muratore et al (2014) | APP mutation | Forebrain neuron | Increase in Aβ42, Aβ38, pTAU | Aβ-antibodies reduce pTAU |
| Alzheimer's Disease | Sproul et al (2014) | PSEN1 mutation | Neural progenitors | Higher Aβ42/Aβ40 ratio, gene expression differences | Verification of gene expression differences in human AD brains |
| Alzheimer's Disease | Duan et al (2014) | Sporadic ApoE3/E4 | Basal forebrain cholinergic neurons | Higher Aβ42/Aβ40 ratio, increased vulnerability to glutamate-stress | |
| Alzheimer's Disease | Hossini et al (2015) | Sporadic | Neurons | Gene expression analysis | |
| Amyotrophic Lateral Sclerosis (ALS) | Dimos et al (2008) | SOD1 mutations | Motor neurons | N.D. | First report of patient-specific neurons |
| Amyotrophic Lateral Sclerosis (ALS) | Mitne-Neto et al (2011) | VAPB mutations | Fibroblasts, iPSCs, motor neurons | Reduced VAPB protein levels | Although VAPB levels were highest in neurons, the reduction was not specific to neurons |
| Amyotrophic Lateral Sclerosis (ALS) | Bilican et al (2012) | TDP43 mutations | Motor neurons | Cell death | Real-time survival analysis of HB9+ neurons |
| Amyotrophic Lateral Sclerosis (ALS) | Egawa et al (2012) | TDP43 mutations | Motor neurons | Expression differences, TDP43 pathology, shorter neurites | Rescue by anacardic acid, multiple clones per patient used |
| Amyotrophic Lateral Sclerosis (ALS) | Sareen et al (2013) | C9orf72 expansion | Motor neurons | RNA foci, hypoexcitability, gene expression differences | Repeat-containing RNA foci colocalized with hnRNPA1 and Pur-α, rescue of gene expression by ASO treatment |
| Amyotrophic Lateral Sclerosis (ALS) | Donnelly et al (2013) | C9orf72 expansion | Neurons | RNA foci, irregular interaction with ADARB2, susceptibility to glutamate excitotoxicity | Colocalization of repeat with ADARB2 validated in patient motor cortex. Rescue of gene expression by ASO treatment |
| Amyotrophic Lateral Sclerosis (ALS) | Yang et al (2013b) | SOD1, TDP43 mutations | Motor neurons | Sensitivity to growth factor withdrawal | Rescue by kenpaullone |
| Amyotrophic Lateral Sclerosis (ALS) | Serio et al (2013) | TDP43 mutations | Astrocytes | Cell death, TDP43 mislocalization | |
| Amyotrophic Lateral Sclerosis (ALS) | Wainger et al (2014) | SOD1, C9orf72, FUS mutations | Motor neurons | Hyperexcitability | Phenotype rescued by gene correction in SOD1, and by treatment with a Kv7 agonist |
| Amyotrophic Lateral Sclerosis (ALS) | Kiskinis et al (2014) | SOD1, C9orf72 mutations | Motor neurons | Cell death, reduced soma size, ER stress, mitochondrial abnormalities, gene expression changes | Phenotypes rescued by gene correction in SOD1 |
| Amyotrophic Lateral Sclerosis (ALS) | Chen et al (2014) | SOD1 mutations | Motor neurons | Neurofilament aggregation, cell death | Phenotype rescued by gene correction |
| Amyotrophic Lateral Sclerosis (ALS) | Barmada et al (2014) | TDP43 mutations | Neurons, astrocytes | Sensitivity to TDP43 accumulation | Autophagy stimulation increases survival |
| Amyotrophic Lateral Sclerosis (ALS) | Devlin et al (2015) | TDP43 and C9orf72 mutants | Neurons | Electrophysiological dysfunction | Hyperexcitability followed by loss of action potential output |
| Angelman & Prader–Willi Syndrome | Chamberlain et al (2010) | 15q11-q13 deletions | Neurons | UBE3A expression | Genomic imprint is maintained in iPSC neurons |
| Ataxia Telangiectasia | Lee et al (2013) | ATM mutations | NPCs & neurons | Defective DNA damage response | SMRT compounds rescue phenotype |
| Best Disease | Singh et al (2013) | BEST1 mutations | RPE cells | Delayed RHODOPSIN degradation, defective Ca2+ responses, oxidative stress | |
| Dravet Syndrome | Higurashi et al (2013) | SCN1A mutation | Neurons (mostly GABA+) | Reduced AP firing | |
| Dravet Syndrome | Liu et al (2013b) | SCN1A mutation | Neurons (GABA & Glutamate+) | Increase Na+ current density, altered excitability | |
| Dravet Syndrome | Jiao et al (2013) | SCN1A mutation | Neurons | Abnormal Na+ currents, increased firing | |
| Familial Dysautonomia | Lee et al (2009) | IKBKAP mutation | Peripheral neurons, neural crest precursors | Mis-splicing & IKBKAP expression, neurogenesis & migration defects | Phenotypes are tissue specific |
| Familial Dysautonomia | Lee et al (2012) | IKBKAP mutation | Neural crest precursors | IKBKAP expression levels | First large-scale drug screening approach, first follow-up study |
| Fragile X Syndrome | Sheridan et al (2011) | FMR1 expansion | NPCs & neurons | FMR1 promoter methylation & reduced expression, reduced length of processes | |
| Fragile X Syndrome | Liu et al (2012b) | FMR1 expansion | Neurons | Decreased PSD95 expression & density, neurite length, electrophysiological defects | |
| Fragile X Syndrome | Doers et al (2014) | FMR1 expansion | Neurons | Neurite extension & initiation defects | |
| Friedreich's Ataxia | Liu et al (2011) | FXN expansion | Peripheral neurons, cardiomyocytes | FXN expression, repeat instability | |
| Friedreich's Ataxia | Hick et al (2013) | FXN expansion | Neurons, cardiomyocytes | FXN expression, mitochondrial dysfunction | |
| Friedreich's Ataxia | Eigentler et al (2013) | FXN expansion | Peripheral neurons | FXN expression | |
| Frontotemporal Dementia | Almeida et al (2013) | C9orf72 expansion | Neurons | RNA foci, RAN products, sensitivity to autophagy inhibitors | |
| Frontotemporal Dementia (Bv) | Gascon et al (2014) | Sporadic patients | Neurons | Alterations in miR-124 & AMPAR levels | Confirmation of mouse model findings in iPSC neurons & patients |
| Frontotemporal Dementia | Raitano et al (2015) | PGRN mutation | Cortical & motor neurons | Cortical differentiation defects | Rescue by PGRN expression |
| Gaucher's Disease | Mazzulli et al (2011) | GBA1 mutations | Dopaminergic neurons | Declined proteolysis, increased α-synuclein | Provides links between GD & PD |
| Gaucher's Disease | Tiscornia et al (2013) | GBA1 mutations | Neurons & macrophages | Reduction in acid-β-glucosidase activity | Identification of two small molecules |
| Gyrate Atrophy | Meyer et al (2011) | OAT mutation | RPE cells | Decreased OAT activity | Rescued by BAC-mediated introduction of OAT |
| Hereditary Spastic Paraplegia | Denton et al (2014) | SPAST mutation | Glutamatergic neurons | Axonal swelling, increased levels of acetylated tubulin | |
| Hereditary Spastic Paraplegia | Zhu et al (2014) | ATL1 mutation | Forebrain neurons | Impaired axonal growth, defects in mitochondrial motility | |
| Huntington's Disease | Camnasio et al (2012) | HTT expansion | Neurons | Altered lysosomal activity | |
| Huntington's Disease | Juopperi et al (2012) | HTT expansion | Astrocytes | Cytoplasmic vacuolization | |
| Huntington's Disease | HD Consortium (2012) | HTT expansion | NPCs & GABA+ neurons | Altered gene expression, morphological alterations, survival deficit, sensitivity to stressors | Correlation between repeat length & vulnerability to cell stress |
| Huntington's Disease | An et al (2012) | HTT expansion | NPCs, neurons | Cell death, gene expression, mitochondrial dysfunction | Genetic correction rescued phenotypes |
| Huntington's Disease | Guo et al (2013) | HTT expansion | Neurons (GABA+) | Mitochondrial damage | |
| Huntington's Disease | Yao et al (2015) | HTT expansion | Striatal neurons | Cell death, caspase-3 activation | Identified Gpr52 as a stabilizer of HTT |
| Lesch–Nyhan Syndrome | Mekhoubad et al (2012) | HPRT1 mutation | Neurons | Neuronal differentiation efficiency and neurite number defects | Demonstrate that X-inactivation erodes in culture & could affects modeling of X-linked disease |
| Microcephaly | Lancaster et al (2013) | CDK5RAP2 mutation | Cerebral organoids | Smaller neuroepithelial regions & RGs, premature neurogenesis, RG spindle disarray | Generated 3-dimensional brain structures |
| Neuronal ceroid lipofuscinosis | Lojewski et al (2014) | CNL2, CNL3 mutations | NPCs, neurons | Morphological abnormalities in ER, Golgi, mitochondria & lysosomes | Rescue by expression of NCL proteins |
| Niemann–Pick type C1 disease | Trilck et al (2013) | NPC1 mutation | NPCs & neurons | Accumulation of cholesterol | |
| Parkinson's Disease | Byers et al (2011) | SCNA triplication | Dopaminergic neurons | Oxidative stress, α-synuclein accumulation | |
| Parkinson's Disease | Nguyen et al (2011) | LRRK2 mutations | Dopaminergic neurons | Oxidative stress, α-synuclein accumulation, sensitivity to stress reagents | |
| Parkinson's Disease | Seibler et al (2011) | PINK1 mutations | Dopaminergic neurons | Increased mitochondrial copy number, PGC1a upregulation | Rescue by PINK1 overexpression |
| Parkinson's Disease | Devine et al (2011) | SNCA triplication | Dopaminergic neurons | Upregulation of α-synuclein | |
| Parkinson's Disease | Sanchez-Danes et al (2012) | Sporadic & LRRK2 mutations | Dopaminergic neurons | Reduction in neurite number & density, vacuolization, sensitivity to lysosomal inhibition | A total of 15 patients examined, long-term culture ∽75 DIV |
| Parkinson's Disease | Cooper et al (2012) | PINK1 & LRRK2 mutations | Dopaminergic neurons | Mitochondrial dysfunction in response to stressors | Pharmacological rescue of phenotypes |
| Parkinson's Disease | Imaizumi et al (2012) | PARK2 mutations | Dopaminergic neurons | Oxidative stress, mitochondrial dysfunction, Nrf2 induction, α-synuclein accumulation | |
| Parkinson's Disease | Liu et al (2012a) | LRRK2 mutation | Neural stem cells | Susceptibility to proteosomal stress, differentiation & clonal expansion deficiencies | Genetic correction rescued phenotypes |
| Parkinson's Disease | Reinhardt et al (2013) | LRRK2 mutation | Dopaminergic neurons | Gene expression differences, ERK phosphorylation & activity | Genetic correction rescued phenotypes |
| Parkinson's Disease | Su and Qi (2013) | LRRK2 mutation | Dopaminergic neurons | Mitochondrial damage, shorter neuritis, lysosomal hyperactivity | Pharmacological rescue |
| Parkinson's Disease | Chung et al (2013) | SNCA mutation | Cortical neurons | Nitrosative & ER stress | Pharmacological rescue, combination between a yeast and an iPSC platform |
| Parkinson's Disease | Miller et al (2013) | PINK1 & PARKIN mutations | Dopaminergic neurons | TH reduction, dendritic degeneration | Phenotypes induced only after overexpressing progerin |
| Parkinson's Disease | Ryan et al (2013) | SNCA mutation | Dopaminergic neurons | Nitrosative stress, gene expression alterations, mitochondrial stress | Genetic & pharmacological rescue of phenotypes |
| Parkinson's Disease | Flierl et al (2014) | SNCA triplication | NPCs | Viability, metabolism & stress resistance defects | Rescue by SNCA knockdown |
| Parkinson's Disease | Sanders et al (2014) | LRRK2 mutations | NPCs & neurons | Mitochondrial DNA damage | Genetic correction rescued phenotypes |
| Phelan–McDermid Syndrome | Shcheglovitov et al (2013) | 22q13.3 deletion | Forebrain neurons | Defective excitatory synaptic transmission | Rescue by SHANK3 expression or IGF1 treatment |
| Retinitis Pigmentosa | Jin et al (2011) | RP1, RP9, PRPH2, RHO mutations | Rod photoreceptors | Cell death, oxidative & ER stress | Differential response to treatment with α-Tocopherol |
| Retinitis Pigmentosa | Tucker et al (2011) | MAK mutations | Retinal precursors | Defective MAK mRNA splicing | |
| Retinitis Pigmentosa | Jin et al (2012) | RHO mutations | RPE cells | Cell death & ER stress | |
| Retinitis Pigmentosa | Tucker et al (2013) | USH2A mutations | Retinal precursors | USH2A transcript defects, ER stress | |
| Rett Syndrome | Marchetto et al (2010) | MeCP2 mutations | Neurons | MeCP2 expression, reduced synapses, spine density, soma size, altered calcium signaling | |
| Rett Syndrome | Ananiev et al (2011) | MeCP2 mutations | Neurons | Reduced nuclear size | |
| Rett Syndrome | Cheung et al (2011) | MeCP2 deletion | Neurons | MeCP2 expression, reduced soma size | |
| Rett Syndrome | Kim et al (2011c) | MeCP2 mutations | Neurons | Lower TUJ1 & Na+ channel expression | |
| Rett Syndrome | Amenduni et al (2011) | CDKL5 mutations | Neurons | No phenotype described | |
| Rett Syndrome | Ricciardi et al (2012) | CDKL5 mutations | Neurons | Aberrant dendritic spines | |
| Rett Syndrome | Larimore et al (2013) | MeCP2 mutations | Neurons | Reduced expression of PLDN | |
| Rett Syndrome | Griesi-Oliveira et al (2014) | TRPC6 mutation | NPCs & cortical neurons | Gene expression differences, Ca2+ influx defects, decreased axonal length & arborization | Overlap in molecular pathways between TRPC6 & MeCPT2 |
| Rett Syndrome | Williams et al (2014) | MeCP2 mutations | Astrocytes | Mutant astrocytes cause morphological and firing defects in healthy neurons | Demonstrates non-cell autonomous contribution of astrocytes in Rett Syndrome |
| Rett Syndrome | Djuric et al (2015) | MeCP2e1 mutation | Cortical neurons | Reduced soma size, dendritic density, capacitance & firing defects | Rescue of phenotypes by overexpression of MeCP2e1 |
| Rett Syndrome | Livide et al (2015) | MeCP2 & CDKL5 mutations | NPCs & neurons | Gene expression differences | Identified GRID1 as a common target in two distinct genetic classes of RTT |
| Schizophrenia | Brennand et al (2011) | Familial & sporadic SCZD patients | NPCs & neurons | Decreased connectivity, neurite number, PSD95 protein, gene expression changes | Recovery after treatment with loxapine |
| Schizophrenia | Pedrosa et al (2011) | 22q11.2 deletion & sporadic SCZD | Glutamatergic neurons | No phenotype described | |
| Schizophrenia | Paulsen Bda et al (2012) | SCZD patient | NPCs | Elevated ROS, extramitochondrial consumption | Treatment with valproic acid reduced ROS |
| Schizophrenia | Robicsek et al (2013) | SCZD patients | NPCs, dopaminergic, glutamatergic neurons | Differentiation & maturation deficiencies, mitochondrial defects | |
| Schizophrenia | Yoon et al (2014) | 15q11.2 microdeletion | NPCs | Deficits in adherent junctions & apical polarity | Identified haploinsufficiency of CYFIP1 as a potential contributor to neuropsychiatric disorders |
| Schizophrenia | Hook et al (2014) | SCZD patients | Neurons | Increased secretion of catecholamines, higher numbers of TH+ neurons | |
| Schizophrenia | Wen et al (2014b) | DISC1 mutations | Forebrain neurons | Synaptic vesicle release deficits, gene expression changes | Isogenic controls included in this study |
| Schizophrenia | Brennand et al (2015) | Familial & sporadic SCZD patients | NPCs & neurons | RNA & protein-level differences related to cytoskeleton & oxidative stress, aberrant migration | |
| Spinal Muscular Atrophy | Ebert et al (2009) | Type 1 SMA | Motor neurons | Cell death, soma size, reduced SMN levels | First study of iPSC-based approach to report a disease-associated phenotype |
| Spinal Muscular Atrophy | Sareen et al (2012) | Type 1 SMA | Motor neurons | Cell death, increased caspase-8 & 3 activation | Rescue by apoptotic inhibitors |
| Spinal Muscular Atrophy | Corti et al (2012) | Type 1 SMA | Motor neurons | Cell death, smaller soma size, reduced axonal length, gene expression and RNA splicing defects | Gene correction, transplantation of iPSC motor neurons extends lifespan of SMA mouse model |
| Tauopathy | Fong et al (2013) | TAU mutation | Neurons | TAU fragmentation & phosphorylation, axonal degeneration | Gene editing to correct the mutation & generate a homozygous mutant used as controls |
| Timothy Syndrome | Pasca et al (2011) | CACNA1C mutations | NPCs & cortical neurons | Ca2+ signaling, activity-dependent gene expression | Rescue by roscovitine treatment |
| Timothy Syndrome | Krey et al (2013) | CACNA1C mutations | Cortical neurons | Activity-dependent dendrite retraction | Rescue by GTPase Gem |
NPCs, neural progenitor cells; RPE, retinal pigment epithelium; ND, not determined; ASO, allele-specific oligonucleotide; GD, Gaucher's disease; PD, Parkinson's disease; AP, action potential.
The table includes neurodevelopmental and neurodegenerative diseases for which patient-specific iPSCs have been generated and neuronal cells differentiated to develop a cell-based model of disease.
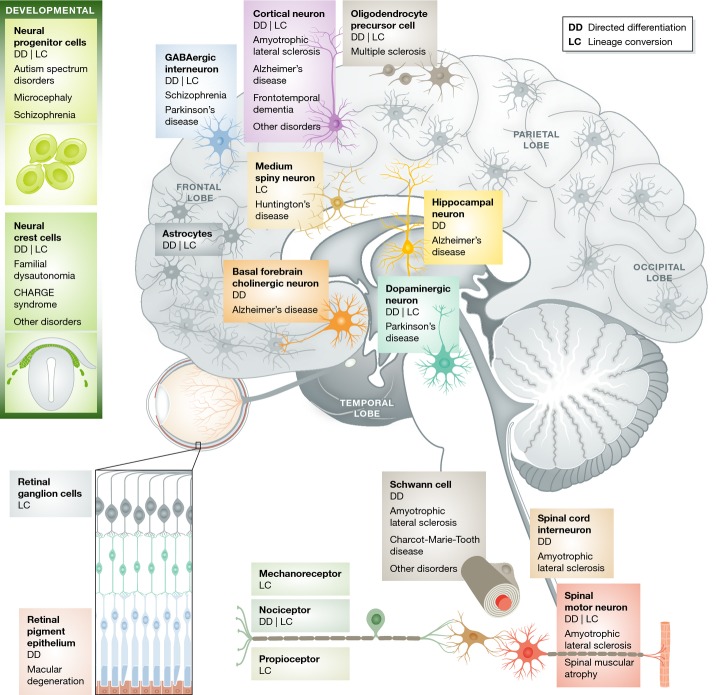
You can study only what you can make
With an eye on modeling neurological disease, stem cell scientists have steadily developed protocols for generating relevant human neural subtypes in vitro (Fig1 and Table2). Many directed differentiation and lineage conversion studies have focused on cell types that are selectively vulnerable in neurodegenerative or neurological diseases such as spinal motor neurons (amyotrophic lateral sclerosis, ALS), midbrain dopaminergic neurons (Parkinson's disease, PD) and striatal medium spiny neurons (Huntington's disease, HD). Their selective vulnerability in patients provides confidence that the phenotypes identified in iPSC-derived or lineage-converted cells in vitro represent relevant disease processes. In addition, it provides the opportunity to sift out phenotypes that may be disease non-relevant by using neuronal subtypes that are not affected in vivo as negative controls.
Figure 1. You can model only what you can make.
A number of different human neural cells can be efficiently generated by directed differentiation (DD) from pluripotent stem cells, or by lineage conversion (LC) from somatic cell types.
Table 2.
List of neural cells generated by directed differentiation of stem cells and lineage conversion of somatic cells
| Initial cell population | Target cell type | Morphogens/Small molecules | Reprogramming factors | References |
|---|---|---|---|---|
| Lineage conversion | ||||
| Fibroblasts | Neural crest cells | SOX10 | Kim et al (2011a) | |
| Fibroblasts | Neural stem cells | SOX2 | Ring et al (2012) | |
| Fibroblasts | Neurons | CHIR99021, SB431542 | ASCL1, NGN2 | Ladewig et al (2012) |
| Fibroblasts | Neurons | ASCL1 | Chanda et al (2014) | |
| Pericyte-derived cells | Neurons | SOX2, ASCL1 | Karow et al (2012) | |
| Fibroblasts | Dopaminergic neurons | ASCL1, BRN2, MYT1L, LMX1A, FOXA2 | Pfisterer et al (2011) | |
| Fibroblasts | Dopaminergic neurons | ASCL1, LMX1A, NURRL | Caiazzo et al (2011) | |
| Fibroblasts | Dopaminergic neurons | Lmx1a, Foxa2, Ascl1, Brn2 | Sheng et al (2012a) | |
| Fibroblasts | Dopaminergic neurons | Ascl1, Pitx3, Lmx1a, Nurr1, Foxa2, EN1 | Kim et al (2011b) | |
| Fibroblasts | Dopaminergic neurons | ASCL1, NGN2, SOX2, NURR1, PITX3 | Liu et al (2012c) | |
| Fibroblasts | Glutamatergic Neurons | ASCL1, BRN2, MYT1L, NEUROD1 | Pang et al (2011) | |
| Fibroblasts | Glutamatergic neurons | BRN2, MYT1L, miR-124 | Ambasudhan et al (2011) | |
| Fibroblasts | Glutamatergic neurons | Forskolin, Dorsomorphin | NGN2 | Liu et al (2013a) |
| Fibroblasts | Glutamatergic and GABAergic neurons | ASCL1, MYT1L, NEUROD2, miR-9/9*, miR-124 | Yoo et al (2011) | |
| Fibroblasts | Medium spiny neurons | DLX1, DLX2, MYT1L, CTIP2, miR-9/9*, miR-124 | Victor et al (2014) | |
| Fibroblasts | Nociceptor, mechanoreceptor, proprioceptor neurons | Brn3a, Ngn1/2 | Blanchard et al (2015) | |
| Fibroblasts | Nociceptor Neurons | ASCL1, MYT1L, ISL2, KLF7, NGN1 | Wainger et al (2015) | |
| Fibroblasts | Oligodendrocyte progenitor cells | Sox10, Olig2, Zfp536 | Yang et al (2013a) | |
| Fibroblasts | Oligodendrocyte progenitor cells | Olig1, Olig2, Nkx2.2, Nkx6.2, Sox10, ST18, Gm98, Myt1 | Najm et al (2013) | |
| Fibroblasts | Spinal motor neurons | ASCL1, BRN2, MYT1L, NGN2, ISL1, LHX3, NEUROD1 | Son et al (2011) | |
| Fibroblasts | Astrocytes | Nfia, Nfib, Sox9 | Caiazzo et al (2015) | |
| Fibroblasts | Neural precursor cells | Brn2, Sox2, FoxG1 | Lujan et al (2012) | |
| Fibroblasts | Neural progenitor cells | VPA, CHIR99021, RepSox | Cheng et al (2014) | |
| Fibroblasts | Neural stem cells | Brn4, Sox2, Klf4, c-Myc, E47 | Han et al (2012) | |
| Fibroblasts | Neural stem cells | Sox2, Klf4,c-Myc, Oct4 | Thier et al (2012) | |
| Sertoli cells | Neural stem cells | Ascl1, Ngn2, Hes1, Id1, Pax6, Brn2, Sox2, c-Myc, Klf4 | Sheng et al (2012b) | |
| Astrocytes | Neuroblasts | Sox2 | Niu et al (2013) | |
| Hepatocytes | Neurons | Ascl1, Brn2, Myt1l | Marro et al (2011) | |
| Fibroblasts | Neurons | PTB repression | Xue et al (2013) | |
| Astrocytes | GABAergic neurons | Ascl1, Dlx2 | Heinrich et al (2010) | |
| Directed differentiation | ||||
| Pluripotent stem cells | Forebrain neuronal precursors | SB431542, LDN189193, N2, B27 | Chambers et al (2009) | |
| Pluripotent stem cells | Forebrain neurons | SB431542, LDN189193, N2, B27 | Chambers et al (2009) | |
| Pluripotent stem cells | Telencephalic neurons | N2, B27, IGF1, Heparin, SHH, DKK1, WNT3A, BDNF, GDNF | Li et al (2009) | |
| Pluripotent stem cells | Forebrain neural cells | Heparin, N2, B27, BDNF, GDNF, IGF1 | Zeng et al (2010) | |
| Pluripotent stem cells | Cortical neurons | B27, N2, BSA, sodium pyruvate, 2-mercaptoethanol, Noggin, Y27632 | Espuny-Camacho et al (2013) | |
| Pluripotent stem cells | Granule cerebellar neurons | FGF2, heparin, N2, Glutamax, FGF8, retinoic acid, ITS, FGF4, WNT1, WNT3A, B27, BMP7, BMP6, GDF7, SHH, NT3, JAG1 | Erceg et al (2012) | |
| Pluripotent stem cells | Hypothalamic neurons | Neurobasal-A, Glutamax, N2, B27, sodium bicarbonate, dibutyryl cyclic AMP, GDNF, BDNF, CNTF | Merkle et al (2015) | |
| Pluripotent stem cells | Dopaminergic neurons | Heparin, N2, serum replacer, cAMP, ascorbic acid, BDNF, GDNF, SHH, FGF8 | Yan et al (2005) | |
| Pluripotent stem cells | Dopaminergic neurons | LDN193189, SB431542, SHH C25II, purmorphamine, FGF8, CHIR99021, N2, B27, L-Glut, BDNF, ascorbic acid, GDNF, TGFβ3, dibutyryl cAMP, DAPT | Kriks et al (2011) | |
| Pluripotent stem cells | Spinal motor neurons | SB431542, LDN189193, N2, B27, retinoic acid, smoothened agonist | Amoroso et al (2013) | |
| Pluripotent stem cells | Astrocytes | B27, BMP2, BMP4, LIF | Gupta et al (2012) | |
| Pluripotent stem cells | Astrocytes | EGF, FGF, Glutamax, N2, CNTF | Krencik and Zhang (2011) | |
| Pluripotent stem cells | Oligodendrocytes | N2, N1, cAMP, biotin, heparin, retinoic acid, SHH, purmorphamine, FGF2, B27, PDGF, IGF, NT3 | Hu et al (2009) | |
| Pluripotent stem cells | Hippocampal neurons | DKK1, SB431542, Noggin, cyclopamine, N2, B27, Wnt3a, BDNF, FGF2, ascorbic acid, cyclic AMP, fetal bovine serum | Yu et al (2014) | |
| Pluripotent stem cells | Astrocytes (ventralized) | SB431542, LDN189193, RA, SHH, N2, B27, FGF1, FGF2 | Roybon et al (2013) | |
| Pluripotent stem cells | Basal forebrain cholinergic neurons | RA/SSH/FGF8/BMP9 | Lhx8 & Gbx1 | Bissonnette et al (2011) |
| Pluripotent stem cells | Cortical interneurons | SB431542, LDN189193, XAV939, SHH, purmorphamine, N2, B27 | Maroof et al (2013) | |
One important area requiring further development of in vitro protocols is region-specific cortical differentiation. Many diseases affect specific regions of the cortex, such as frontotemporal dementia (FTD), which affects the anterior cingulate, orbitofrontal cortex, and temporal lobes, or ALS, which affects layer V neurons in the motor cortex. Thus, region-specific attributes play a large role in the disease vulnerability of neuronal subtypes. While protocols exist to generate neurons from both deep and upper layers of the cortex (Shi et al, 2012b; Kadoshima et al, 2013), they have not shown to be specific for a given region of the cortex. The identification of marker genes and neuronal projection patterns specific to neurons in different cortical regions will greatly facilitate the development and validation of region-specific cortical neuron protocols.
Adult neural stem cells of the dentate gyrus play a key role in memory formation and pattern separation tasks and could be an important therapeutic target for Alzheimer's disease (AD). Although it is possible to generate human neural stem or progenitor cells in vitro, these are likely more embryonic and it is not clear how closely these mimic adult stem cells of the dentate gyrus (Chambers et al, 2009, 2011; Shi et al, 2012a,b). One reason is that until recently, rigorous molecular characterization of these cells was missing. We are now starting to get a clearer picture. There seem to be several adult neural stem cell populations or states that can be distinguished by markers such as Ascl1 or Gli1, and single-cell RNA sequencing data have been generated (Bonaguidi et al, 2011). This new information will serve as a template for generating adult neural stem cells in vitro.
A third cell type that has not been produced on a patient-specific level in vitro is microglia. Microglia perform inflammatory and non-inflammatory tasks that enable normal neuronal function. Through these roles, they are known to regulate the progression of ALS and AD (Zhong et al, 2009; de Boer et al, 2014; Johansson et al, 2015), and potentially other neurodegenerative diseases. Mouse studies showed that microglia from SOD1G93A ALS mice express higher levels of the prostaglandin E2 receptor (Di Giorgio et al, 2008; de Boer et al, 2014). Similarly, microglia from an AD model sharply upregulate the prostaglandin E2 receptor in response to amyloid-β (Ab) exposure in an age-dependent manner (Johansson et al, 2015). Higher prostaglandin E2 signaling in microglia caused reduced microglial cytokine generation, chemotaxis, clearance of Aboligomers, resolution of inflammatory responses to Ab42 and trophic factor release (Johansson et al, 2015). In both the ALS and AD models, deletion of the prostaglandin E2 receptor significantly slowed disease progression (de Boer et al, 2014; Johansson et al, 2015). Under normal conditions, microglia are derived from the embryonic yolk sac and go through a maturation process after they enter the nervous system (Nayak et al, 2014). The signaling and gene expression changes that occur during this process are not well understood and will need to be characterized further to enable the production of patient-specific microglia.
Specificity of phenotypes: the importance of controls
Significant technical advancements achieved over that last few years currently allow for the generation of patient-specific iPSCs that are free from genomic integration of the reprogramming factors (Malik & Rao, 2013). The essential quality of any newly derived iPSC can be easily assessed by (i) immunocytochemistry for pluripotency markers (e.g. NANOG/SSEA3), (ii) a quantitative pluripotency assay such as the Scorecard or the Pluritest and (iii) analysis of genomic integrity (karyotype, array CGH).
Disease modeling studies based on iPSC technology have relied on the use of diseased cells derived from patients as a model for disease, and cells derived from healthy individuals as controls. However, genetic and potentially epigenetic heterogeneity of iPSC lines contributes to functional variability of differentiated somatic cells, confounding evaluation of disease modeling experiments (Sandoe & Eggan, 2013). Such variability can be introduced at multiple different levels including generation of stem cell lines, continuous in vitro culture, variation in cell culture reagents, differential efficiencies of neural generation and genetic background. There are different approaches to overcoming this variation. One approach is through the use of targeted gene editing that results in the generation of a control stem cell line that is isogenic to the patient one, except for the disease-causing mutation. Such an approach effectively minimizes line-to-line differences and is a very important tool for iPSC-based disease modeling.
CRISPR/Cas9, a recent technology that has emerged, allows for the efficient generation of such isogenic stem cell lines (Hsu et al, 2014). The system contains two essential components, an enzyme that can cleave DNA such that a double-strand break or a single nick is generated and a guide RNA that targets the enzyme to a specific genomic location. By simultaneously introducing either a single-stranded oligodeoxynucleotide (ssODN) containing the desired edit or a targeting plasmid with larger desired sequence alterations, the genomic sequence can be precisely edited via the cells' own endogenous repair mechanism, homologous recombination. Given the incredible versatility of the CRISPR/Cas9 system and the continuous evolvement of the technical aspects of this approach, it should be expected that every iPSC study that focuses on genetic forms of disease should include an isogenic control cell line. The rescue of a phenotype by genetic correction can lead to the conclusion that the genetic lesion is necessary for the onset of the phenotype. The same technique can be used to introduce a disease-associated mutation in a healthy iPSC line in order to assess whether the mutation in itself is sufficient for the onset of particular phenotypes.
An alternative approach to the concern of variation would be to utilize multiple stem cell clones from each individual patient and compare the desired measurement against multiple healthy individuals. The use of multiple patient clones would ensure that the phenotype is not an artifact of a defective clonal cell line, while the use of multiple healthy controls should encapsulate sufficient technical and genetic variation, so that the measured cellular properties neuronal firing, dendritic density, etc. will represent a true average. This approach will be important in studies of sporadic disease.
Additionally, approaches that are complementary to the iPSC method should also be considered for the verification of identified phenotypes. These could include the generation of neurons via direct conversion as well as the investigation of human patient material such as postmortem CNS tissue and cerebrospinal fluid (CSF). Other non-invasive techniques such as transcranial magnetic stimulation (TMS) (Fox et al, 2014), which allows in vivo neurostimulation and neuromodulation, and an electroencephalogram (EGG), can also be used to examine changes associated with electrical excitability of neurons.
An important point to consider when assessing the specificity of an identified phenotype is whether it is only apparent in the cell type known to be most vulnerable to the disease being modeled. In ALS patients for example, it is the upper and lower motor neurons that are initially targeted by disease mechanisms and gradually lost, while sensory neurons remain relatively unaffected. It would therefore be predicted that a phenotype that is truly relevant to disease would not be evident in a sensory neuron generated from the same individual. Although this could be a valuable approach, it should be taken with caution for two reasons: firstly because a sensory neuron might simply be resistant to a phenotype, and therefore, it is the effect of the phenotype on the sensory cell that should be considered and not simply the presence of the phenotype in itself, and secondly because it might be the in vivo microenvironment of a sensory neuron that confers resistance and not a cell autonomous trait. Nevertheless, studies have demonstrated neuronal-type specificity of a phenotype including the sensitivity of mutant PD tyrosine hydroxylase (TH)-positive neurons but not TH-negative neurons to H2O2-induced toxicity (Nguyen et al, 2011), and morphometric deficiencies of mutant ALS, ISL-positive motor neurons but not ISL-negative neurons grown in the same culture dishes (Kiskinis et al, 2014).
A major advantage of using reprogramming approaches to study a neurological disease is the ability to assess the biological variation associated with a specific neuronal defect. Consider that a phenotype, for example, defective lysosomal function, has been identified in neurons derived from a patient cell line and that this phenotype is mutation dependent (i.e. it is corrected in an isogenic control line). The first level of biological variation can be addressed by examining neurons derived from a different individual that harbors the exact same mutation in the same gene. If the phenotype is not present, then additional genetic or epigenetic factors might be necessary for the onset of the defect. The next level of biological variability can be addressed by examining neurons from a patient with a different mutation in the same gene. Lastly, the broader relevance of the identified phenotype for the disease can be assessed by examining the lysosomal function of neurons from patients with mutations in different disease-causing genes as well as in a large number of sporadic cases.
A more direct route to the CNS?
Lineage conversion provides a progenitor-free approach for generating various neural types. Lineage conversion relies on the overexpression of transcription factors to internally drive differentiation programs. The forced expression of these factors replaces external developmental morphogens utilized in iPSC differentiation by directly activating downstream genes. Additionally, either purified external cues or other cell types normally present in vivo are sometimes added to further guide the developmental trajectory and maturation of various cell types (Son et al, 2011; Meyer et al, 2014).
An advantage of this approach is that it simplifies the identification of protocols for generating new neural subtypes because it only requires knowledge of transcription factor expression during the terminal stages of development, as opposed to requiring a deep understanding of morphogen signaling dynamics starting from the pluripotent state through the terminally differentiated state. In the same way that identifying the signals that produce the target cell type is simpler for lineage conversion, optimizing the efficiency of their production is more complicated for iPSC-directed differentiation because one must optimize the efficiency of each progenitor step as opposed to one step as in lineage conversion. Due to these advantages, reprogramming biologists have rapidly developed lineage conversion protocols for almost all neural subtypes attainable by directed differentiation just a few years after the initial demonstration of iPSC reprogramming (Takahashi et al, 2007), which showed that dramatic changes in cell fate are possible (see Fig1).
Several groups have taken advantage of the modular nature of transcriptional networks to generate distinct neuronal subtypes. Genetic neuralization through introduction of BRN2, ASCL1 and MYT1L (BAM) to fibroblasts generates induced neurons (iN; Pang et al, 2011). The transcription factor ASCL1 is sufficient in generating iNeurons alone, indicating that it is the key driver in this reprogramming approach (Chanda et al, 2014). The addition of NEUROD1 further enhances this conversion (Pang et al, 2011). From iNs, a secondary layer of transcription factors guide cells to particular neurons. Spinal motor neurons have been generated by adding ISL1, LHX3, NGN2 and HB9 to the BAM factors (Son et al, 2011). Addition of LMX1A and FOXA2 to the BAM cocktail results in dopaminergic neurons (Pfisterer et al, 2011). Striatal medium spiny neurons can be generated using a microRNA-based neuralization platform (Yoo et al, 2011) supplemented with CTIP2, DLX1, DLX3 and MYT1L (Victor et al, 2014). Neuronal induction has also been achieved through the repression of polypyrimidine tract binding (PTB), a single RNA binding protein (Xue et al, 2013). Oligodendrocyte precursor cells follow a separate glial lineage that is independent of BAM-mediated neuralization. Induced oligodendrocyte precursor cells can be made by overexpressing either SOX10, OLIG2 and ZFP536 (Yang et al, 2013a) or OLIG1, OLIG2, NKX2.2, NKX6.2, SOX10, ST18, GM98 and MYT1 (Najm et al, 2013). Just like in iPSC differentiation, inductive signals are added during lineage conversion protocols to further guide cells to mature fates.
During development, early neural progenitors produce neurons whereas late progenitors differentiate into astrocytes (Stiles & Jernigan, 2010). iPSC-directed differentiation recapitulates this developmental process. As a result, while the production of neurons from human iPSCs occurs within 30 days, astrocytes only emerge after 3 months (Krencik & Zhang, 2011). Recently, Broccoli and colleagues reported that three transcription factors, NFIA, NFIB and SOX9, convert fibroblasts into astrocytes (Caiazzo et al, 2015). A major advantage of this approach is that it requires < 3 weeks to generate functional astrocytes (Caiazzo et al, 2015).
The key consideration in evaluating the utility of lineage-converted cells is how similar they are to their primary counterparts and whether they reliably recapitulate disease phenotypes. We and others have shown that lineage-converted cells such as motor neurons (Son et al, 2011), dopaminergic neurons (Kim et al, 2011b) and pancreatic beta cells (Zhou et al, 2008; Li et al, 2014) express transcriptional profiles and DNA methylation patterns (Li et al, 2014) very similar to their primary targets. Although bulk analysis of lineage-converted cultures suggested that these cells retained more residual gene expression from the starting somatic cells than iPSC-derived cells (Cahan et al, 2014), single-cell studies suggest that this reflects heterogeneous cultures of converted and non-converted cells rather than “confused” or mixed-property cells (Li et al, 2014). Detailed epigenetic and single-cell analysis for more lineage-converted cell types will be required to rigorously assess the quality of these cells.
Recent studies have shown that lineage-converted cells are able to recapitulate disease phenotypes and provide insight into pathogenic mechanisms. Induced motor neurons derived from patients with C9orf72 ALS degenerated rapidly in cell culture relative to control neurons (Wen et al, 2014a). In addition, they expressed dipeptide repeat proteins specific to the C9orf72 form of the disease, indicating that they reproduce the phenotypes observed in vivo (Wen et al, 2014a). The authors used this model to determine that dipeptide repeat proteins induce toxicity in C9orf72 ALS (Wen et al, 2014a). Meyer and colleagues used lineage conversion to generate astrocytes from sporadic ALS patients (Meyer et al, 2014). Astrocytes from the familial SOD1 form of the disease induce the degeneration of motor neurons (Di Giorgio et al, 2008; Marchetto et al, 2008; Meyer et al, 2014), and the authors used this approach to assess the neurotoxicity of sporadic ALS patient astrocytes. They found that lineage-converted astrocytes from sporadic ALS patients consistently induced neurodegeneration, suggesting that an inherent disease mechanism is maintained in most sporadic patients. These studies demonstrate that lineage-converted cells are effective tools for studying CNS diseases.
Which approach would be better for disease modeling experiments—lineage conversion or iPSC-directed differentiation? It depends on several considerations. Has the disease affected cell type been generated in vitro previously? If not, how much is known about their developmental signaling or transcriptional profile? This would dictate which approach would be more effective to pursue. If both developmental signaling and transcriptional profiling are known, then lineage conversion might be a faster route to disease studies.
How many cells are required for the designed study? If a large number of cells are needed, for example for biochemical or epigenetic studies, iPSC-directed differentiation would be more suitable because the number of differentiated cells gets amplified at each progenitor step, whereas lineage conversion does not amplify the number of differentiated cells.
Are lineage-related cell types desirable or undesirable for the specific model? For example, non-cell autonomous neurotoxic stimuli from astrocytes are a key aspect of ALS (Di Giorgio et al, 2008; Marchetto et al, 2008; Meyer et al, 2014) and perhaps AD disease processes. It therefore would be informative to have patient-derived astrocytes included in these disease models. Most iPSC-directed differentiation protocols result in the production of multiple cell types within the same developmental lineage. In addition, several groups have started to develop three-dimensional protocols that produce several cell types from the same tissue that self-organize into structures that mimic the primary tissue (Eiraku et al, 2011; Nakano et al, 2012; Koehler et al, 2013; Koehler & Hashino, 2014). For certain tissues, such as the inner ear, this may enable more relevant disease models.
In contrast, lineage conversion strategies would not be expected to produce developmentally related cell types at a substantial rate nor 3D structures (unless a progenitor was formed that gives rise to self-organizing structures). But this would be desirable for screening applications where pure cultures of one neuronal subtype simplify high-throughput scaling and assay interpretation.
Overall, there are advantages and disadvantages to both approaches for the production of in vitro patient-derived cells depending on the disease and application. However, the emergence of the same phenotype in cells derived by both methods would certainly enhance confidence in the results.
A shift in focus: from developing neurons to maturing and aging them
A critical area that deserves further investigation is the maturity and aging of in vitro derived cells (Fig2). We like to think that there are three stages we need to consider when setting up in vitro models of disease: the development, the maturation and the natural aging process of a neural cell type. While significant advancements have been achieved in generating and maturing neural cell types—either by directed differentiation or lineage conversion—little has been done in terms of affecting the aging of cells. For late onset diseases such as ALS, FTD, HD, PD and AD, it is possible that changes elicited by aging are required to induce the disease process. Age is the strongest risk factor for neurodegenerative diseases, and although there are rare cases with early onset presentation, the overwhelming majority of patients develop clinical symptoms in the later stages of their lives. The nature of the age-related risk remains largely unknown, and whether it arises from cell autonomous mechanisms or as a result of a systemic dysfunction remains to be determined. A number of studies support the notion that cellular epigenetic changes in the CNS correlate with aging. For example, recent work has demonstrated that profound changes in DNA methylation levels occur in the brains of mice with age (Lister et al, 2013), while aging oligodendrocytes lose their ability to effectively remyelinate damaged nerves (Ruckh et al, 2012). Importantly, under conditions of heterochronic parabiosis in mice, the effects on oligodendrocytes were reversible, implicating some aspect of epigenetic regulation.
Figure 2. Developing stem cell-based models of neurological disorders.
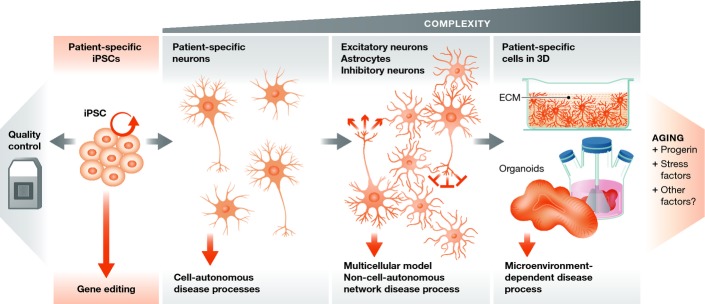
Patient-specific iPSCs should be properly quality controlled for genomic integrity and pluripotent potential, while gene editing techniques allow for the generation of isogenic controls in cases where the disease-causing allele is known. Simple cell autonomous or more sophisticated multi-cellular and 3D disease models can be developed depending on the hypothesis being addressed. Neuronal maturity increases with the complexity of the cellular system, while methods for effectively aging neurons are lacking.
Current studies suggest that the transcriptional and electrophysiological properties of both iPSC-derived and lineage-converted neurons are more similar to fetal neurons than adult (Son et al, 2011; Takazawa et al, 2012). It is likely that extrinsic factors present during normal development or aging are required to activate the maturation process. We and others have shown, for example, that the addition of primary astrocytes to lineage conversion cultures significantly improves the maturation of induced neurons (Son et al, 2011; Chanda et al, 2013; Wainger et al, 2015). Additional progress in generating more mature and aged cells will require a better understanding of the gene expression and functional changes associated with maturation and aging. This has been difficult to obtain for specific neuronal subtypes because of the scarcity of available human tissue. Efforts such as those of the Allen Brain Institute have shed some light on these markers, but future studies will need to analyze specific neuronal subtypes in order to be sure that differences between aged neurons and young neurons are truly due to aging and not different neuronal subtypes.
In addition to glial-derived factors, Rubin and colleagues recently showed that circulatory factors also contribute to the aging process in the CNS (Katsimpardi et al, 2014). They were able to identify a single factor, GDF11, which normally declines in expression with age. Interestingly, restoring GDF11 levels in old mice rejuvenated the proliferative and neurogenic properties of neural stem cells in the mouse (Katsimpardi et al, 2014). This raises the notion that there may be other factors that control the aging of neurons and could be exploited to regulate this process in vitro.
Studer and colleagues took a more intrinsic approach to inducing aging in iPSC-derived neurons by expressing Progerin, which is a mutant form of the Lamin A protein that causes accelerated aging phenotypes in humans (Miller et al, 2013). Expression of Progerin induced higher levels of DNA damage and mitochondrial reactive oxygen species in dopaminergic neurons derived from PD patients, which enabled the detection of PD-associated disease phenotypes such as dendrite degeneration, mitochondrial enlargement, Lewy body precursor inclusions and suppression of tyrosine hydroxylase expression (Miller et al, 2013). It remains unclear whether this approach induces the recapitulation of bona fide disease processes, but it represents a new line of targeted aging procedures.
From cell autonomy to more sophisticated systems
Neurons do not exist in isolation in the human nervous system. They form elaborate and functional networks with other neurons and also rely on a sophisticated microenvironment that is created by the interactions with other neural and non-neural cell types, which provide structural, metabolic and functional support as well as effective communication (Abbott et al, 2006). Glial cells, astrocytes, oligodendrocytes, microglia and endothelial cells exist in abundance in the nervous system and play vital functional roles. Glial cells buffer harmful ions, astrocytes provide nutrients and circulate neurotransmitters around synapses, oligodendrocytes form myelin sheaths around axons, microglia scavenge and degrade dead cells, and endothelial cells are important in maintaining the blood–brain barrier. Cell–cell interactions and the microenvironment as a whole might mediate important neuroprotective or neurotoxic activities in response to disease or injury. In fact, a number of studies over the last few years have clearly demonstrated that non-cell autonomous processes involving astrocytes, oligodendrocytes and microglia play a critical role in mediating disease progression and potentially onset in neurodegeneration including in ALS, HD, PD, prion disease, the spinal cerebellar ataxias (SCAs) and AD in vivo (Ilieva et al, 2009). The strength of utilizing iPSCs to study neurological disease is in their ability to generate a range of different cell types from the same genetic background (Fig2). This allows for the assessment of how a specific genetic lesion, for example, might differentially impact neuronal subtypes. It also allows for a rational step-by step approach to assess how cellular interactions might contribute toward the evolvement of a disease-associated phenotype or a cellular response to stress.
The co-culture of spinal motor neurons with cortical astrocytes has previously been utilized in one of the first stem cell-based models of ALS to demonstrate how mutant or healthy astrocytes significantly compromised or maintained, respectively, the health of a pure population of motor neurons (Di Giorgio et al, 2008; Marchetto et al, 2008). The co-culture of cortical excitatory with cortical inhibitory neurons and the establishment of functional circuitry might be beneficial when studying epileptic syndromes. The clinical presentation of epileptic patients is the result of the functional control—or lack thereof—of a network of neurons, and recapitulating such a network could be an essential step toward the development of a cellular disease model. The importance of the local microenvironment in neuronal function and potentially dysfunction during disease is also relevant in the context of the three dimensionality that it creates. Neither the brain nor the spinal cord hosts isolated neurons surrounded by an entirely liquid trophic support (akin to culture media) in which nutrients, molecules and proteins can freely diffuse and float around. Recently, Kim, Tanzi and colleagues were able to successfully recapitulate amyloid-β plaques, and tau neurofibrillary tangles—the two pathological hallmarks of AD—in a single 3D human neural cell culture system (Choi et al, 2014). Although this system is not based on iPSCs and their cell lines expressed slightly elevated protein levels of PSEN1 and APP, they designed a simple but innovative cell culture system with neurons grown embedded within a 0.3-mm layer of an extracellular matrix composed of BD Matrigel. This viscous layer reduced the diffusion of secreted amyloid-β and led to the accumulation of aggregated plaques. This is the first time this has been achieved in a cell-based in vitro system and demonstrates the importance of a 3D environment for disease modeling assays.
The recent description of cerebral organoids generated from human pluripotent stem cells and resembling the three-dimensional regional organization of a developing brain has created an exciting opportunity for iPSC-based disease modeling approaches (Lancaster et al, 2013). These brain-like structures, formed by the combination of external growth factor patterning and intrinsic and environmental cues, exhibit distinct regional identities that functionally interact and importantly recapitulate human cortical organization. The authors utilized this method to study microcephaly and demonstrate that patient-specific organoids show premature neuronal differentiation and are only capable of developing to a smaller size. Importantly, mouse models have failed to effectively recapitulate these disease phenotypes for microcephaly, probably due to the dramatic differences in the development and regional organization of the brain as mice do not have an outer subventricular zone (SVZ). This system may be suitable for the study of other neurodevelopmental and neuropsychiatric syndromes in which there are moderate but crucial defects in cortical organization and function. This approach may also be useful in recapitulating human neurodegenerative models that primarily affect brain function as it may allow for the establishment of neuronal circuitry as well as biochemical networks.
Patient stratification based on molecular pathways affected
Neurological disorders including schizophrenia, ALS, PD, FTD and epilepsy are often characterized by a profound clinical and genetic heterogeneity, suggesting that they might represent a syndrome rather than a single nosological entity (Fanous & Kendler, 2005; Tremblay et al, 2013; Jeste & Geschwind, 2014). The variable combination of positive and negative symptoms in schizophrenia, the variable degree of upper and lower motor neuron dysfunction in ALS, the heterogeneity of cognitive symptoms in PD, the variable rate of progression in FTD and the differential response to anti-epileptic treatments in epileptic syndromes are some examples of the clinical diversity in neurological disorders. In addition, genetic studies in ALS, for example, have demonstrated that the disease can be caused by mutations in genes that encode proteins involved in diverse cellular functions ranging from RNA metabolism, vesicle transport, cytoskeletal homeostasis and the processing of unfolded proteins (Cleveland & Rothstein, 2001; Pasinelli & Brown, 2006; Sreedharan & Brown, 2013). While progress has been achieved in terms of genetic taxonomy, pathological stratification and the classification of patients based on their clinical presentation, little is known about how similar or different patients are, in terms of the molecular pathways that mediate their disease processes. Reprogramming technologies can be used to develop in vitro models of genetic and sporadic disease cases and effectively stratify patients, based on (i) the neuronal subtype that exhibits a disease-associated phenotype and (ii) the pathway that leads to this phenotype in each case (Fig3). This approach may lead to the identification of overlapping disease mechanisms that will be broadly relevant and represent the best therapeutic opportunities, or toward a personalized approach to clinical trials and therapeutic treatments.
Figure 3. Patient stratification based on the molecular pathways that are affected.
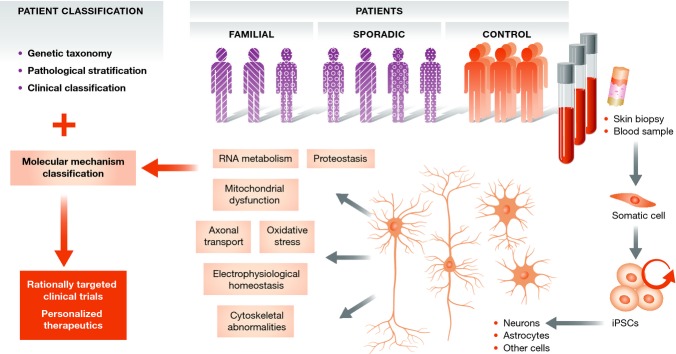
Reprogramming and stem cell-based disease modeling can be utilized to address the level of heterogeneity by defining the molecular mechanisms that lead to disease in different patients. This novel classification of patients could lead to rationally targeted clinical trials and personalized therapeutic approaches.
Concluding remarks
Tremendous progress has been achieved in our efforts to develop cellular models of neurological disease since 2007 and the initial description of induced pluripotency and the concept of cellular reprogramming (Takahashi et al, 2007). We are now able to generate a wealth of different neural subtypes, have created and characterized hundreds of patient-specific iPSCs and their neural derivatives, have developed efficient gene editing approaches and are continuously establishing elaborate methods for the functional analysis of neurons. During the next phase in the field, it is imperative that the research community offers unrestricted access to cell lines, human samples and differentiation protocols, maintains close communication and attempts to further establish standards for the quality control of pluripotent stem cells and the neuronal subtypes that are utilized for disease modeling and drug screening experiments. It is also worth pointing out that broad collaborative efforts with substantial financial support need to take center stage in order to address important outstanding questions. What is the variation in the properties of neurons generated from a significant number of healthy individuals? Can we assess the broader relevance of phenotypes identified in genetic types of disease by monitoring hundreds of sporadic cases? Can we predict how patients will respond to a potential therapeutic treatment by studying their stem cell-derived neurons? Can we match an in vivo clinical trial with an in vitro iPSC-based clinical trial to monitor the correlation of outcome measures? The answers to these questions will help us conclude what are the capabilities and limitations of this promising technological tool. Despite the challenges that have arisen over the last few years, the community has responded with sustained effort and is steadily moving forward toward the development of systems that will have an impact in our efforts to understand and treat diseases that affect the nervous system.
Acknowledgments
JI is supported by the Donald E. and Delia B. Baxter Foundation, SC-CTSI, an NIH K99 award R00-NS07743 and the Tau Consortium. EK is supported by the Les Turner ALS Foundation and Target ALS.
Conflict of interest
The authors declare that they have no conflict of interest.
References
- Abbott NJ, Ronnback L, Hansson E. Astrocyte-endothelial interactions at the blood-brain barrier. Nat Rev Neurosci. 2006;7:41–53. doi: 10.1038/nrn1824. [DOI] [PubMed] [Google Scholar]
- Almeida S, Gascon E, Tran H, Chou HJ, Gendron TF, Degroot S, Tapper AR, Sellier C, Charlet-Berguerand N, Karydas A, Seeley WW, Boxer AL, Petrucelli L, Miller BL, Gao FB. Modeling key pathological features of frontotemporal dementia with C9ORF72 repeat expansion in iPSC-derived human neurons. Acta Neuropathol. 2013;126:385–399. doi: 10.1007/s00401-013-1149-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambasudhan R, Talantova M, Coleman R, Yuan X, Zhu S, Lipton SA, Ding S. Direct reprogramming of adult human fibroblasts to functional neurons under defined conditions. Cell Stem Cell. 2011;9:113–118. doi: 10.1016/j.stem.2011.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amenduni M, De Filippis R, Cheung AY, Disciglio V, Epistolato MC, Ariani F, Mari F, Mencarelli MA, Hayek Y, Renieri A, Ellis J, Meloni I. iPS cells to model CDKL5-related disorders. Eur J Hum Genet. 2011;19:1246–1255. doi: 10.1038/ejhg.2011.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amoroso MW, Croft GF, Williams DJ, O'Keeffe S, Carrasco MA, Davis AR, Roybon L, Oakley DH, Maniatis T, Henderson CE, Wichterle H. Accelerated high-yield generation of limb-innervating motor neurons from human stem cells. J Neurosci. 2013;33:574–586. doi: 10.1523/JNEUROSCI.0906-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An MC, Zhang N, Scott G, Montoro D, Wittkop T, Mooney S, Melov S, Ellerby LM. Genetic correction of Huntington's disease phenotypes in induced pluripotent stem cells. Cell Stem Cell. 2012;11:253–263. doi: 10.1016/j.stem.2012.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ananiev G, Williams EC, Li H, Chang Q. Isogenic pairs of wild type and mutant induced pluripotent stem cell (iPSC) lines from Rett syndrome patients as in vitro disease model. PLoS ONE. 2011;6:e25255. doi: 10.1371/journal.pone.0025255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barmada SJ, Serio A, Arjun A, Bilican B, Daub A, Ando DM, Tsvetkov A, Pleiss M, Li X, Peisach D, Shaw C, Chandran S, Finkbeiner S. Autophagy induction enhances TDP43 turnover and survival in neuronal ALS models. Nat Chem Biol. 2014;10:677–685. doi: 10.1038/nchembio.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilican B, Serio A, Barmada SJ, Nishimura AL, Sullivan GJ, Carrasco M, Phatnani HP, Puddifoot CA, Story D, Fletcher J, Park IH, Friedman BA, Daley GQ, Wyllie DJ, Hardingham GE, Wilmut I, Finkbeiner S, Maniatis T, Shaw CE, Chandran S. Mutant induced pluripotent stem cell lines recapitulate aspects of TDP-43 proteinopathies and reveal cell-specific vulnerability. Proc Natl Acad Sci USA. 2012;109:5803–5808. doi: 10.1073/pnas.1202922109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bissonnette CJ, Lyass L, Bhattacharyya BJ, Belmadani A, Miller RJ, Kessler JA. The controlled generation of functional basal forebrain cholinergic neurons from human embryonic stem cells. Stem Cells. 2011;29:802–811. doi: 10.1002/stem.626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchard JW, Eade KT, Szucs A, Lo Sardo V, Tsunemoto RK, Williams D, Sanna PP, Baldwin KK. Selective conversion of fibroblasts into peripheral sensory neurons. Nat Neurosci. 2015;18:25–35. doi: 10.1038/nn.3887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Boer AS, Koszka K, Kiskinis E, Suzuki N, Davis-Dusenbery BN, Eggan K. Genetic validation of a therapeutic target in a mouse model of ALS. Sci Transl Med. 2014;6:248ra104. doi: 10.1126/scitranslmed.3009351. [DOI] [PubMed] [Google Scholar]
- Bonaguidi MA, Wheeler MA, Shapiro JS, Stadel RP, Sun GJ, Ming GL, Song H. In vivo clonal analysis reveals self-renewing and multipotent adult neural stem cell characteristics. Cell. 2011;145:1142–1155. doi: 10.1016/j.cell.2011.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennand KJ, Simone A, Jou J, Gelboin-Burkhart C, Tran N, Sangar S, Li Y, Mu Y, Chen G, Yu D, McCarthy S, Sebat J, Gage FH. Modelling schizophrenia using human induced pluripotent stem cells. Nature. 2011;473:221–225. doi: 10.1038/nature09915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennand K, Savas JN, Kim Y, Tran N, Simone A, Hashimoto-Torii K, Beaumont KG, Kim HJ, Topol A, Ladran I, Abdelrahim M, Matikainen-Ankney B, Chao SH, Mrksich M, Rakic P, Fang G, Zhang B, Yates JR, III, Gage FH. Phenotypic differences in hiPSC NPCs derived from patients with schizophrenia. Mol Psychiatry. 2015;20:361–368. doi: 10.1038/mp.2014.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byers B, Cord B, Nguyen HN, Schule B, Fenno L, Lee PC, Deisseroth K, Langston JW, Pera RR, Palmer TD. SNCA triplication Parkinson's patient's iPSC-derived DA neurons accumulate alpha-synuclein and are susceptible to oxidative stress. PLoS ONE. 2011;6:e26159. doi: 10.1371/journal.pone.0026159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahan P, Li H, Morris SA, Lummertz da Rocha E, Daley GQ, Collins JJ. CellNet: network biology applied to stem cell engineering. Cell. 2014;158:903–915. doi: 10.1016/j.cell.2014.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caiazzo M, Dell'Anno MT, Dvoretskova E, Lazarevic D, Taverna S, Leo D, Sotnikova TD, Menegon A, Roncaglia P, Colciago G, Russo G, Carninci P, Pezzoli G, Gainetdinov RR, Gustincich S, Dityatev A, Broccoli V. Direct generation of functional dopaminergic neurons from mouse and human fibroblasts. Nature. 2011;476:224–227. doi: 10.1038/nature10284. [DOI] [PubMed] [Google Scholar]
- Caiazzo M, Giannelli S, Valente P, Lignani G, Carissimo A, Sessa A, Colasante G, Bartolomeo R, Massimino L, Ferroni S, Settembre C, Benfenati F, Broccoli V. Direct conversion of fibroblasts into functional astrocytes by defined transcription factors. Stem Cell Rep. 2015;4:25–36. doi: 10.1016/j.stemcr.2014.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camnasio S, Delli Carri A, Lombardo A, Grad I, Mariotti C, Castucci A, Rozell B, Lo Riso P, Castiglioni V, Zuccato C, Rochon C, Takashima Y, Diaferia G, Biunno I, Gellera C, Jaconi M, Smith A, Hovatta O, Naldini L, Di Donato S, et al. The first reported generation of several induced pluripotent stem cell lines from homozygous and heterozygous Huntington's disease patients demonstrates mutation related enhanced lysosomal activity. Neurobiol Dis. 2012;46:41–51. doi: 10.1016/j.nbd.2011.12.042. [DOI] [PubMed] [Google Scholar]
- Chamberlain SJ, Chen PF, Ng KY, Bourgois-Rocha F, Lemtiri-Chlieh F, Levine ES, Lalande M. Induced pluripotent stem cell models of the genomic imprinting disorders Angelman and Prader-Willi syndromes. Proc Natl Acad Sci USA. 2010;107:17668–17673. doi: 10.1073/pnas.1004487107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers SM, Fasano CA, Papapetrou EP, Tomishima M, Sadelain M, Studer L. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat Biotechnol. 2009;27:275–280. doi: 10.1038/nbt.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers SM, Mica Y, Studer L, Tomishima MJ. Converting human pluripotent stem cells to neural tissue and neurons to model neurodegeneration. Methods Mol Biol. 2011;793:87–97. doi: 10.1007/978-1-61779-328-8_6. [DOI] [PubMed] [Google Scholar]
- Chanda S, Marro S, Wernig M, Sudhof TC. Neurons generated by direct conversion of fibroblasts reproduce synaptic phenotype caused by autism-associated neuroligin-3 mutation. Proc Natl Acad Sci USA. 2013;110:16622–16627. doi: 10.1073/pnas.1316240110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanda S, Ang CE, Davila J, Pak C, Mall M, Lee QY, Ahlenius H, Jung SW, Sudhof TC, Wernig M. Generation of induced neuronal cells by the single reprogramming factor ASCL1. Stem Cell Rep. 2014;3:282–296. doi: 10.1016/j.stemcr.2014.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Qian K, Du Z, Cao J, Petersen A, Liu H, Blackbourn LWT, Huang CL, Errigo A, Yin Y, Lu J, Ayala M, Zhang SC. Modeling ALS with iPSCs reveals that mutant SOD1 misregulates neurofilament balance in motor neurons. Cell Stem Cell. 2014;14:796–809. doi: 10.1016/j.stem.2014.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng L, Hu W, Qiu B, Zhao J, Yu Y, Guan W, Wang M, Yang W, Pei G. Generation of neural progenitor cells by chemical cocktails and hypoxia. Cell Res. 2014;24:665–679. doi: 10.1038/cr.2014.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung AY, Horvath LM, Grafodatskaya D, Pasceri P, Weksberg R, Hotta A, Carrel L, Ellis J. Isolation of MECP2-null Rett Syndrome patient hiPS cells and isogenic controls through X-chromosome inactivation. Hum Mol Genet. 2011;20:2103–2115. doi: 10.1093/hmg/ddr093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi SH, Kim YH, Hebisch M, Sliwinski C, Lee S, D'Avanzo C, Chen H, Hooli B, Asselin C, Muffat J, Klee JB, Zhang C, Wainger BJ, Peitz M, Kovacs DM, Woolf CJ, Wagner SL, Tanzi RE, Kim DY. A three-dimensional human neural cell culture model of Alzheimer's disease. Nature. 2014;515:274–278. doi: 10.1038/nature13800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung CY, Khurana V, Auluck PK, Tardiff DF, Mazzulli JR, Soldner F, Baru V, Lou Y, Freyzon Y, Cho S, Mungenast AE, Muffat J, Mitalipova M, Pluth MD, Jui NT, Schule B, Lippard SJ, Tsai LH, Krainc D, Buchwald SL, et al. Identification and rescue of alpha-synuclein toxicity in Parkinson patient-derived neurons. Science. 2013;342:983–987. doi: 10.1126/science.1245296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleveland DW, Rothstein JD. From Charcot to Lou Gehrig: deciphering selective motor neuron death in ALS. Nat Rev Neurosci. 2001;2:806–819. doi: 10.1038/35097565. [DOI] [PubMed] [Google Scholar]
- Cooper O, Seo H, Andrabi S, Guardia-Laguarta C, Graziotto J, Sundberg M, McLean JR, Carrillo-Reid L, Xie Z, Osborn T, Hargus G, Deleidi M, Lawson T, Bogetofte H, Perez-Torres E, Clark L, Moskowitz C, Mazzulli J, Chen L, Volpicelli-Daley L, et al. Pharmacological rescue of mitochondrial deficits in iPSC-derived neural cells from patients with familial Parkinson's disease. Sci Transl Med. 2012;4:141ra190. doi: 10.1126/scitranslmed.3003985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corti S, Nizzardo M, Simone C, Falcone M, Nardini M, Ronchi D, Donadoni C, Salani S, Riboldi G, Magri F, Menozzi G, Bonaglia C, Rizzo F, Bresolin N, Comi GP. Genetic correction of human induced pluripotent stem cells from patients with spinal muscular atrophy. Sci Transl Med. 2012;4:165ra162. doi: 10.1126/scitranslmed.3004108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis-Dusenbery BN, Williams LA, Klim JR, Eggan K. How to make spinal motor neurons. Development. 2014;141:491–501. doi: 10.1242/dev.097410. [DOI] [PubMed] [Google Scholar]
- Denton KR, Lei L, Grenier J, Rodionov V, Blackstone C, Li XJ. Loss of spastin function results in disease-specific axonal defects in human pluripotent stem cell-based models of hereditary spastic paraplegia. Stem Cells. 2014;32:414–423. doi: 10.1002/stem.1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devine MJ, Ryten M, Vodicka P, Thomson AJ, Burdon T, Houlden H, Cavaleri F, Nagano M, Drummond NJ, Taanman JW, Schapira AH, Gwinn K, Hardy J, Lewis PA, Kunath T. Parkinson's disease induced pluripotent stem cells with triplication of the alpha-synuclein locus. Nat Commun. 2011;2:440. doi: 10.1038/ncomms1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin AC, Burr K, Borooah S, Foster JD, Cleary EM, Geti I, Vallier L, Shaw CE, Chandran S, Miles GB. Human iPSC-derived motoneurons harbouring TARDBP or C9ORF72 ALS mutations are dysfunctional despite maintaining viability. Nat Commun. 2015;6:5999. doi: 10.1038/ncomms6999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Giorgio FP, Boulting GL, Bobrowicz S, Eggan KC. Human embryonic stem cell-derived motor neurons are sensitive to the toxic effect of glial cells carrying an ALS-causing mutation. Cell Stem Cell. 2008;3:637–648. doi: 10.1016/j.stem.2008.09.017. [DOI] [PubMed] [Google Scholar]
- Dimos JT, Rodolfa KT, Niakan KK, Weisenthal LM, Mitsumoto H, Chung W, Croft GF, Saphier G, Leibel R, Goland R, Wichterle H, Henderson CE, Eggan K. Induced pluripotent stem cells generated from patients with ALS can be differentiated into motor neurons. Science. 2008;321:1218–1221. doi: 10.1126/science.1158799. [DOI] [PubMed] [Google Scholar]
- Djuric U, Cheung AY, Zhang W, Mok RS, Lai W, Piekna A, Hendry JA, Ross PJ, Pasceri P, Kim DS, Salter MW, Ellis J. MECP2e1 isoform mutation affects the form and function of neurons derived from Rett syndrome patient iPS cells. Neurobiol Dis. 2015;76:37–45. doi: 10.1016/j.nbd.2015.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doers ME, Musser MT, Nichol R, Berndt ER, Baker M, Gomez TM, Zhang SC, Abbeduto L, Bhattacharyya A. iPSC-derived forebrain neurons from FXS individuals show defects in initial neurite outgrowth. Stem Cells Dev. 2014;23:1777–1787. doi: 10.1089/scd.2014.0030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly CJ, Zhang PW, Pham JT, Heusler AR, Mistry NA, Vidensky S, Daley EL, Poth EM, Hoover B, Fines DM, Maragakis N, Tienari PJ, Petrucelli L, Traynor BJ, Wang J, Rigo F, Bennett CF, Blackshaw S, Sattler R, Rothstein JD. RNA toxicity from the ALS/FTD C9ORF72 expansion is mitigated by antisense intervention. Neuron. 2013;80:415–428. doi: 10.1016/j.neuron.2013.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan L, Bhattacharyya BJ, Belmadani A, Pan L, Miller RJ, Kessler JA. Stem cell derived basal forebrain cholinergic neurons from Alzheimer's disease patients are more susceptible to cell death. Mol Neurodegener. 2014;9:3. doi: 10.1186/1750-1326-9-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert AD, Yu J, Rose FF, Jr, Mattis VB, Lorson CL, Thomson JA, Svendsen CN. Induced pluripotent stem cells from a spinal muscular atrophy patient. Nature. 2009;457:277–280. doi: 10.1038/nature07677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egawa N, Kitaoka S, Tsukita K, Naitoh M, Takahashi K, Yamamoto T, Adachi F, Kondo T, Okita K, Asaka I, Aoi T, Watanabe A, Yamada Y, Morizane A, Takahashi J, Ayaki T, Ito H, Yoshikawa K, Yamawaki S, Suzuki S, et al. Drug screening for ALS using patient-specific induced pluripotent stem cells. Sci Transl Med. 2012;4:145ra104. doi: 10.1126/scitranslmed.3004052. [DOI] [PubMed] [Google Scholar]
- Eigentler A, Boesch S, Schneider R, Dechant G, Nat R. Induced pluripotent stem cells from friedreich ataxia patients fail to upregulate frataxin during in vitro differentiation to peripheral sensory neurons. Stem Cells Dev. 2013;22:3271–3282. doi: 10.1089/scd.2013.0126. [DOI] [PubMed] [Google Scholar]
- Eiraku M, Takata N, Ishibashi H, Kawada M, Sakakura E, Okuda S, Sekiguchi K, Adachi T, Sasai Y. Self-organizing optic-cup morphogenesis in three-dimensional culture. Nature. 2011;472:51–56. doi: 10.1038/nature09941. [DOI] [PubMed] [Google Scholar]
- Erceg S, Lukovic D, Moreno-Manzano V, Stojkovic M, Bhattacharya SS. Derivation of cerebellar neurons from human pluripotent stem cells. Curr Protoc Stem Cell Biol. 2012 doi: 10.1002/9780470151808.sc01h05s20. Chapter 1: Unit 1H 5. [DOI] [PubMed] [Google Scholar]
- Espuny-Camacho I, Michelsen KA, Gall D, Linaro D, Hasche A, Bonnefont J, Bali C, Orduz D, Bilheu A, Herpoel A, Lambert N, Gaspard N, Peron S, Schiffmann SN, Giugliano M, Gaillard A, Vanderhaeghen P. Pyramidal neurons derived from human pluripotent stem cells integrate efficiently into mouse brain circuits in vivo. Neuron. 2013;77:440–456. doi: 10.1016/j.neuron.2012.12.011. [DOI] [PubMed] [Google Scholar]
- Fanous AH, Kendler KS. Genetic heterogeneity, modifier genes, and quantitative phenotypes in psychiatric illness: searching for a framework. Mol Psychiatry. 2005;10:6–13. doi: 10.1038/sj.mp.4001571. [DOI] [PubMed] [Google Scholar]
- Flierl A, Oliveira LM, Falomir-Lockhart LJ, Mak SK, Hesley J, Soldner F, Arndt-Jovin DJ, Jaenisch R, Langston JW, Jovin TM, Schule B. Higher vulnerability and stress sensitivity of neuronal precursor cells carrying an alpha-synuclein gene triplication. PLoS ONE. 2014;9:e112413. doi: 10.1371/journal.pone.0112413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong H, Wang C, Knoferle J, Walker D, Balestra ME, Tong LM, Leung L, Ring KL, Seeley WW, Karydas A, Kshirsagar MA, Boxer AL, Kosik KS, Miller BL, Huang Y. Genetic correction of tauopathy phenotypes in neurons derived from human induced pluripotent stem cells. Stem Cell Rep. 2013;1:226–234. doi: 10.1016/j.stemcr.2013.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox MD, Buckner RL, Liu H, Chakravarty MM, Lozano AM, Pascual-Leone A. Resting-state networks link invasive and noninvasive brain stimulation across diverse psychiatric and neurological diseases. Proc Natl Acad Sci USA. 2014;111:E4367–E4375. doi: 10.1073/pnas.1405003111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gascon E, Lynch K, Ruan H, Almeida S, Verheyden JM, Seeley WW, Dickson DW, Petrucelli L, Sun D, Jiao J, Zhou H, Jakovcevski M, Akbarian S, Yao WD, Gao FB. Alterations in microRNA-124 and AMPA receptors contribute to social behavioral deficits in frontotemporal dementia. Nat Med. 2014;20:1444–1451. doi: 10.1038/nm.3717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griesi-Oliveira K, Acab A, Gupta AR, Sunaga DY, Chailangkarn T, Nicol X, Nunez Y, Walker MF, Murdoch JD, Sanders SJ, Fernandez TV, Ji W, Lifton RP, Vadasz E, Dietrich A, Pradhan D, Song H, Ming GL, Gu X, Haddad G, et al. Modeling non-syndromic autism and the impact of TRPC6 disruption in human neurons. Mol Psychiatry. 2014 doi: 10.1038/mp.2014.141. doi: 10.1038/mp.2014.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X, Disatnik MH, Monbureau M, Shamloo M, Mochly-Rosen D, Qi X. Inhibition of mitochondrial fragmentation diminishes Huntington's disease-associated neurodegeneration. J Clin Invest. 2013;123:5371–5388. doi: 10.1172/JCI70911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta N, Henry RG, Strober J, Kang SM, Lim DA, Bucci M, Caverzasi E, Gaetano L, Mandelli ML, Ryan T, Perry R, Farrell J, Jeremy RJ, Ulman M, Huhn SL, Barkovich AJ, Rowitch DH. Neural stem cell engraftment and myelination in the human brain. Sci Transl Med. 2012;4:155ra137. doi: 10.1126/scitranslmed.3004373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han DW, Tapia N, Hermann A, Hemmer K, Hoing S, Arauzo-Bravo MJ, Zaehres H, Wu G, Frank S, Moritz S, Greber B, Yang JH, Lee HT, Schwamborn JC, Storch A, Scholer HR. Direct reprogramming of fibroblasts into neural stem cells by defined factors. Cell Stem Cell. 2012;10:465–472. doi: 10.1016/j.stem.2012.02.021. [DOI] [PubMed] [Google Scholar]
- HD iPSC Consortium. Induced pluripotent stem cells from patients with Huntington's disease show CAG-repeat-expansion-associated phenotypes. Cell Stem Cell. 2012;11:264–278. doi: 10.1016/j.stem.2012.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinrich C, Blum R, Gascon S, Masserdotti G, Tripathi P, Sanchez R, Tiedt S, Schroeder T, Gotz M, Berninger B. Directing astroglia from the cerebral cortex into subtype specific functional neurons. PLoS Biol. 2010;8:e1000373. doi: 10.1371/journal.pbio.1000373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hick A, Wattenhofer-Donze M, Chintawar S, Tropel P, Simard JP, Vaucamps N, Gall D, Lambot L, Andre C, Reutenauer L, Rai M, Teletin M, Messaddeq N, Schiffmann SN, Viville S, Pearson CE, Pandolfo M, Puccio H. Neurons and cardiomyocytes derived from induced pluripotent stem cells as a model for mitochondrial defects in Friedreich's ataxia. Dis Model Mech. 2013;6:608–621. doi: 10.1242/dmm.010900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higurashi N, Uchida T, Lossin C, Misumi Y, Okada Y, Akamatsu W, Imaizumi Y, Zhang B, Nabeshima K, Mori MX, Katsurabayashi S, Shirasaka Y, Okano H, Hirose S. A human Dravet syndrome model from patient induced pluripotent stem cells. Mol Brain. 2013;6:19. doi: 10.1186/1756-6606-6-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hook V, Brennand KJ, Kim Y, Toneff T, Funkelstein L, Lee KC, Ziegler M, Gage FH. Human iPSC neurons display activity-dependent neurotransmitter secretion: aberrant catecholamine levels in schizophrenia neurons. Stem Cell Rep. 2014;3:531–538. doi: 10.1016/j.stemcr.2014.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossini AM, Megges M, Prigione A, Lichtner B, Toliat MR, Wruck W, Schroter F, Nuernberg P, Kroll H, Makrantonaki E, Zoubouliss CC, Adjaye J. Induced pluripotent stem cell-derived neuronal cells from a sporadic Alzheimer's disease donor as a model for investigating AD-associated gene regulatory networks. BMC Genom. 2015;16:84. doi: 10.1186/s12864-015-1262-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu BY, Du ZW, Zhang SC. Differentiation of human oligodendrocytes from pluripotent stem cells. Nat Protoc. 2009;4:1614–1622. doi: 10.1038/nprot.2009.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilieva H, Polymenidou M, Cleveland DW. Non-cell autonomous toxicity in neurodegenerative disorders: ALS and beyond. J Cell Biol. 2009;187:761–772. doi: 10.1083/jcb.200908164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imaizumi Y, Okada Y, Akamatsu W, Koike M, Kuzumaki N, Hayakawa H, Nihira T, Kobayashi T, Ohyama M, Sato S, Takanashi M, Funayama M, Hirayama A, Soga T, Hishiki T, Suematsu M, Yagi T, Ito D, Kosakai A, Hayashi K, et al. Mitochondrial dysfunction associated with increased oxidative stress and alpha-synuclein accumulation in PARK2 iPSC-derived neurons and postmortem brain tissue. Mol Brain. 2012;5:35. doi: 10.1186/1756-6606-5-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Israel MA, Yuan SH, Bardy C, Reyna SM, Mu Y, Herrera C, Hefferan MP, Van Gorp S, Nazor KL, Boscolo FS, Carson CT, Laurent LC, Marsala M, Gage FH, Remes AM, Koo EH, Goldstein LS. Probing sporadic and familial Alzheimer's disease using induced pluripotent stem cells. Nature. 2012;482:216–220. doi: 10.1038/nature10821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeste SS, Geschwind DH. Disentangling the heterogeneity of autism spectrum disorder through genetic findings. Nat Rev Neurol. 2014;10:74–81. doi: 10.1038/nrneurol.2013.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao J, Yang Y, Shi Y, Chen J, Gao R, Fan Y, Yao H, Liao W, Sun XF, Gao S. Modeling Dravet syndrome using induced pluripotent stem cells (iPSCs) and directly converted neurons. Hum Mol Genet. 2013;22:4241–4252. doi: 10.1093/hmg/ddt275. [DOI] [PubMed] [Google Scholar]
- Jin ZB, Okamoto S, Osakada F, Homma K, Assawachananont J, Hirami Y, Iwata T, Takahashi M. Modeling retinal degeneration using patient-specific induced pluripotent stem cells. PLoS ONE. 2011;6:e17084. doi: 10.1371/journal.pone.0017084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin ZB, Okamoto S, Xiang P, Takahashi M. Integration-free induced pluripotent stem cells derived from retinitis pigmentosa patient for disease modeling. Stem Cells Transl Med. 2012;1:503–509. doi: 10.5966/sctm.2012-0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson JU, Woodling NS, Wang Q, Panchal M, Liang X, Trueba-Saiz A, Brown HD, Mhatre SD, Loui T, Andreasson KI. Prostaglandin signaling suppresses beneficial microglial function in Alzheimer's disease models. J Clin Invest. 2015;125:350–364. doi: 10.1172/JCI77487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juopperi TA, Kim WR, Chiang CH, Yu H, Margolis RL, Ross CA, Ming GL, Song H. Astrocytes generated from patient induced pluripotent stem cells recapitulate features of Huntington's disease patient cells. Mol Brain. 2012;5:17. doi: 10.1186/1756-6606-5-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadoshima T, Sakaguchi H, Nakano T, Soen M, Ando S, Eiraku M, Sasai Y. Self-organization of axial polarity, inside-out layer pattern, and species-specific progenitor dynamics in human ES cell-derived neocortex. Proc Natl Acad Sci USA. 2013;110:20284–20289. doi: 10.1073/pnas.1315710110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karow M, Sanchez R, Schichor C, Masserdotti G, Ortega F, Heinrich C, Gascon S, Khan MA, Lie DC, Dellavalle A, Cossu G, Goldbrunner R, Gotz M, Berninger B. Reprogramming of pericyte-derived cells of the adult human brain into induced neuronal cells. Cell Stem Cell. 2012;11:471–476. doi: 10.1016/j.stem.2012.07.007. [DOI] [PubMed] [Google Scholar]
- Katsimpardi L, Litterman NK, Schein PA, Miller CM, Loffredo FS, Wojtkiewicz GR, Chen JW, Lee RT, Wagers AJ, Rubin LL. Vascular and neurogenic rejuvenation of the aging mouse brain by young systemic factors. Science. 2014;344:630–634. doi: 10.1126/science.1251141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Efe JA, Zhu S, Talantova M, Yuan X, Wang S, Lipton SA, Zhang K, Ding S. Direct reprogramming of mouse fibroblasts to neural progenitors. Proc Natl Acad Sci USA. 2011a;108:7838–7843. doi: 10.1073/pnas.1103113108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Su SC, Wang H, Cheng AW, Cassady JP, Lodato MA, Lengner CJ, Chung CY, Dawlaty MM, Tsai LH, Jaenisch R. Functional integration of dopaminergic neurons directly converted from mouse fibroblasts. Cell Stem Cell. 2011b;9:413–419. doi: 10.1016/j.stem.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KY, Hysolli E, Park IH. Neuronal maturation defect in induced pluripotent stem cells from patients with Rett syndrome. Proc Natl Acad Sci USA. 2011c;108:14169–14174. doi: 10.1073/pnas.1018979108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiskinis E, Sandoe J, Williams LA, Boulting GL, Moccia R, Wainger BJ, Han S, Peng T, Thams S, Mikkilineni S, Mellin C, Merkle FT, Davis-Dusenbery BN, Ziller M, Oakley D, Ichida J, Di Costanzo S, Atwater N, Maeder ML, Goodwin MJ, et al. Pathways disrupted in human ALS motor neurons identified through genetic correction of mutant SOD1. Cell Stem Cell. 2014;14:781–795. doi: 10.1016/j.stem.2014.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koehler KR, Mikosz AM, Molosh AI, Patel D, Hashino E. Generation of inner ear sensory epithelia from pluripotent stem cells in 3D culture. Nature. 2013;500:217–221. doi: 10.1038/nature12298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koehler KR, Hashino E. 3D mouse embryonic stem cell culture for generating inner ear organoids. Nat Protoc. 2014;9:1229–1244. doi: 10.1038/nprot.2014.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo T, Asai M, Tsukita K, Kutoku Y, Ohsawa Y, Sunada Y, Imamura K, Egawa N, Yahata N, Okita K, Takahashi K, Asaka I, Aoi T, Watanabe A, Watanabe K, Kadoya C, Nakano R, Watanabe D, Maruyama K, Hori O, et al. Modeling Alzheimer's disease with iPSCs reveals stress phenotypes associated with intracellular Abeta and differential drug responsiveness. Cell Stem Cell. 2013;12:487–496. doi: 10.1016/j.stem.2013.01.009. [DOI] [PubMed] [Google Scholar]
- Krencik R, Zhang SC. Directed differentiation of functional astroglial subtypes from human pluripotent stem cells. Nat Protoc. 2011;6:1710–1717. doi: 10.1038/nprot.2011.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krey JF, Pasca SP, Shcheglovitov A, Yazawa M, Schwemberger R, Rasmusson R, Dolmetsch RE. Timothy syndrome is associated with activity-dependent dendritic retraction in rodent and human neurons. Nat Neurosci. 2013;16:201–209. doi: 10.1038/nn.3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriks S, Shim JW, Piao J, Ganat YM, Wakeman DR, Xie Z, Carrillo-Reid L, Auyeung G, Antonacci C, Buch A, Yang L, Beal MF, Surmeier DJ, Kordower JH, Tabar V, Studer L. Dopamine neurons derived from human ES cells efficiently engraft in animal models of Parkinson's disease. Nature. 2011;480:547–551. doi: 10.1038/nature10648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladewig J, Mertens J, Kesavan J, Doerr J, Poppe D, Glaue F, Herms S, Wernet P, Kogler G, Muller FJ, Koch P, Brustle O. Small molecules enable highly efficient neuronal conversion of human fibroblasts. Nat Methods. 2012;9:575–578. doi: 10.1038/nmeth.1972. [DOI] [PubMed] [Google Scholar]
- Lancaster MA, Renner M, Martin CA, Wenzel D, Bicknell LS, Hurles ME, Homfray T, Penninger JM, Jackson AP, Knoblich JA. Cerebral organoids model human brain development and microcephaly. Nature. 2013;501:373–379. doi: 10.1038/nature12517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larimore J, Ryder PV, Kim KY, Ambrose LA, Chapleau C, Calfa G, Gross C, Bassell GJ, Pozzo-Miller L, Smith Y, Talbot K, Park IH, Faundez V. MeCP2 regulates the synaptic expression of a Dysbindin-BLOC-1 network component in mouse brain and human induced pluripotent stem cell-derived neurons. PLoS ONE. 2013;8:e65069. doi: 10.1371/journal.pone.0065069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee G, Papapetrou EP, Kim H, Chambers SM, Tomishima MJ, Fasano CA, Ganat YM, Menon J, Shimizu F, Viale A, Tabar V, Sadelain M, Studer L. Modelling pathogenesis and treatment of familial dysautonomia using patient-specific iPSCs. Nature. 2009;461:402–406. doi: 10.1038/nature08320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee G, Ramirez CN, Kim H, Zeltner N, Liu B, Radu C, Bhinder B, Kim YJ, Choi IY, Mukherjee-Clavin B, Djaballah H, Studer L. Large-scale screening using familial dysautonomia induced pluripotent stem cells identifies compounds that rescue IKBKAP expression. Nat Biotechnol. 2012;30:1244–1248. doi: 10.1038/nbt.2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee P, Martin NT, Nakamura K, Azghadi S, Amiri M, Ben-David U, Perlman S, Gatti RA, Hu H, Lowry WE. SMRT compounds abrogate cellular phenotypes of ataxia telangiectasia in neural derivatives of patient-specific hiPSCs. Nat Commun. 2013;4:1824. doi: 10.1038/ncomms2824. [DOI] [PubMed] [Google Scholar]
- Li XJ, Zhang X, Johnson MA, Wang ZB, Lavaute T, Zhang SC. Coordination of sonic hedgehog and Wnt signaling determines ventral and dorsal telencephalic neuron types from human embryonic stem cells. Development. 2009;136:4055–4063. doi: 10.1242/dev.036624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Cavelti-Weder C, Zhang Y, Clement K, Donovan S, Gonzalez G, Zhu J, Stemann M, Xu K, Hashimoto T, Yamada T, Nakanishi M, Zhang Y, Zeng S, Gifford D, Meissner A, Weir G, Zhou Q. Long-term persistence and development of induced pancreatic beta cells generated by lineage conversion of acinar cells. Nat Biotechnol. 2014;32:1223–1230. doi: 10.1038/nbt.3082. [DOI] [PubMed] [Google Scholar]
- Lister R, Mukamel EA, Nery JR, Urich M, Puddifoot CA, Johnson ND, Lucero J, Huang Y, Dwork AJ, Schultz MD, Yu M, Tonti-Filippini J, Heyn H, Hu S, Wu JC, Rao A, Esteller M, He C, Haghighi FG, Sejnowski TJ, et al. Global epigenomic reconfiguration during mammalian brain development. Science. 2013;341:1237905. doi: 10.1126/science.1237905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Verma PJ, Evans-Galea MV, Delatycki MB, Michalska A, Leung J, Crombie D, Sarsero JP, Williamson R, Dottori M, Pebay A. Generation of induced pluripotent stem cell lines from Friedreich ataxia patients. Stem Cell Rev. 2011;7:703–713. doi: 10.1007/s12015-010-9210-x. [DOI] [PubMed] [Google Scholar]
- Liu GH, Qu J, Suzuki K, Nivet E, Li M, Montserrat N, Yi F, Xu X, Ruiz S, Zhang W, Wagner U, Kim A, Ren B, Li Y, Goebl A, Kim J, Soligalla RD, Dubova I, Thompson J, Yates J, III, et al. Progressive degeneration of human neural stem cells caused by pathogenic LRRK2. Nature. 2012a;491:603–607. doi: 10.1038/nature11557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Koscielska KA, Cao Z, Hulsizer S, Grace N, Mitchell G, Nacey C, Githinji J, McGee J, Garcia-Arocena D, Hagerman RJ, Nolta J, Pessah IN, Hagerman PJ. Signaling defects in iPSC-derived fragile X premutation neurons. Hum Mol Genet. 2012b;21:3795–3805. doi: 10.1093/hmg/dds207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Li F, Stubblefield EA, Blanchard B, Richards TL, Larson GA, He Y, Huang Q, Tan AC, Zhang D, Benke TA, Sladek JR, Zahniser NR, Li CY. Direct reprogramming of human fibroblasts into dopaminergic neuron-like cells. Cell Res. 2012c;22:321–332. doi: 10.1038/cr.2011.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu ML, Zang T, Zou Y, Chang JC, Gibson JR, Huber KM, Zhang CL. Small molecules enable neurogenin 2 to efficiently convert human fibroblasts into cholinergic neurons. Nat Commun. 2013a;4:2183. doi: 10.1038/ncomms3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Lopez-Santiago LF, Yuan Y, Jones JM, Zhang H, O'Malley HA, Patino GA, O'Brien JE, Rusconi R, Gupta A, Thompson RC, Natowicz MR, Meisler MH, Isom LL, Parent JM. Dravet syndrome patient-derived neurons suggest a novel epilepsy mechanism. Ann Neurol. 2013b;74:128–139. doi: 10.1002/ana.23897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livide G, Patriarchi T, Amenduni M, Amabile S, Yasui D, Calcagno E, Lo Rizzo C, De Falco G, Ulivieri C, Ariani F, Mari F, Mencarelli MA, Hell JW, Renieri A, Meloni I. GluD1 is a common altered player in neuronal differentiation from both MECP2-mutated and CDKL5-mutated iPS cells. Eur J Hum Genet. 2015;23:195–201. doi: 10.1038/ejhg.2014.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lojewski X, Staropoli JF, Biswas-Legrand S, Simas AM, Haliw L, Selig MK, Coppel SH, Goss KA, Petcherski A, Chandrachud U, Sheridan SD, Lucente D, Sims KB, Gusella JF, Sondhi D, Crystal RG, Reinhardt P, Sterneckert J, Scholer H, Haggarty SJ, et al. Human iPSC models of neuronal ceroid lipofuscinosis capture distinct effects of TPP1 and CLN3 mutations on the endocytic pathway. Hum Mol Genet. 2014;23:2005–2022. doi: 10.1093/hmg/ddt596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lujan E, Chanda S, Ahlenius H, Sudhof TC, Wernig M. Direct conversion of mouse fibroblasts to self-renewing, tripotent neural precursor cells. Proc Natl Acad Sci USA. 2012;109:2527–2532. doi: 10.1073/pnas.1121003109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik N, Rao MS. A review of the methods for human iPSC derivation. Methods Mol Biol. 2013;997:23–33. doi: 10.1007/978-1-62703-348-0_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchetto MC, Muotri AR, Mu Y, Smith AM, Cezar GG, Gage FH. Non-cell-autonomous effect of human SOD1 G37R astrocytes on motor neurons derived from human embryonic stem cells. Cell Stem Cell. 2008;3:649–657. doi: 10.1016/j.stem.2008.10.001. [DOI] [PubMed] [Google Scholar]
- Marchetto MC, Carromeu C, Acab A, Yu D, Yeo GW, Mu Y, Chen G, Gage FH, Muotri AR. A model for neural development and treatment of Rett syndrome using human induced pluripotent stem cells. Cell. 2010;143:527–539. doi: 10.1016/j.cell.2010.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maroof AM, Keros S, Tyson JA, Ying SW, Ganat YM, Merkle FT, Liu B, Goulburn A, Stanley EG, Elefanty AG, Widmer HR, Eggan K, Goldstein PA, Anderson SA, Studer L. Directed differentiation and functional maturation of cortical interneurons from human embryonic stem cells. Cell Stem Cell. 2013;12:559–572. doi: 10.1016/j.stem.2013.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marro S, Pang ZP, Yang N, Tsai MC, Qu K, Chang HY, Sudhof TC, Wernig M. Direct lineage conversion of terminally differentiated hepatocytes to functional neurons. Cell Stem Cell. 2011;9:374–382. doi: 10.1016/j.stem.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzulli JR, Xu YH, Sun Y, Knight AL, McLean PJ, Caldwell GA, Sidransky E, Grabowski GA, Krainc D. Gaucher disease glucocerebrosidase and alpha-synuclein form a bidirectional pathogenic loop in synucleinopathies. Cell. 2011;146:37–52. doi: 10.1016/j.cell.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mekhoubad S, Bock C, de Boer AS, Kiskinis E, Meissner A, Eggan K. Erosion of dosage compensation impacts human iPSC disease modeling. Cell Stem Cell. 2012;10:595–609. doi: 10.1016/j.stem.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkle FT, Maroof A, Wataya T, Sasai Y, Studer L, Eggan K, Schier AF. Generation of neuropeptidergic hypothalamic neurons from human pluripotent stem cells. Development. 2015;142:633–643. doi: 10.1242/dev.117978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer JS, Howden SE, Wallace KA, Verhoeven AD, Wright LS, Capowski EE, Pinilla I, Martin JM, Tian S, Stewart R, Pattnaik B, Thomson JA, Gamm DM. Optic vesicle-like structures derived from human pluripotent stem cells facilitate a customized approach to retinal disease treatment. Stem Cells. 2011;29:1206–1218. doi: 10.1002/stem.674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer K, Ferraiuolo L, Miranda CJ, Likhite S, McElroy S, Renusch S, Ditsworth D, Lagier-Tourenne C, Smith RA, Ravits J, Burghes AH, Shaw PJ, Cleveland DW, Kolb SJ, Kaspar BK. Direct conversion of patient fibroblasts demonstrates non-cell autonomous toxicity of astrocytes to motor neurons in familial and sporadic ALS. Proc Natl Acad Sci USA. 2014;111:829–832. doi: 10.1073/pnas.1314085111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JD, Ganat YM, Kishinevsky S, Bowman RL, Liu B, Tu EY, Mandal PK, Vera E, Shim JW, Kriks S, Taldone T, Fusaki N, Tomishima MJ, Krainc D, Milner TA, Rossi DJ, Studer L. Human iPSC-based modeling of late-onset disease via progerin-induced aging. Cell Stem Cell. 2013;13:691–705. doi: 10.1016/j.stem.2013.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitne-Neto M, Machado-Costa M, Marchetto MC, Bengtson MH, Joazeiro CA, Tsuda H, Bellen HJ, Silva HC, Oliveira AS, Lazar M, Muotri AR, Zatz M. Downregulation of VAPB expression in motor neurons derived from induced pluripotent stem cells of ALS8 patients. Hum Mol Genet. 2011;20:3642–3652. doi: 10.1093/hmg/ddr284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muratore CR, Rice HC, Srikanth P, Callahan DG, Shin T, Benjamin LN, Walsh DM, Selkoe DJ, Young-Pearse TL. The familial Alzheimer's disease APPV717I mutation alters APP processing and Tau expression in iPSC-derived neurons. Hum Mol Genet. 2014;23:3523–3536. doi: 10.1093/hmg/ddu064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Najm FJ, Lager AM, Zaremba A, Wyatt K, Caprariello AV, Factor DC, Karl RT, Maeda T, Miller RH, Tesar PJ. Transcription factor-mediated reprogramming of fibroblasts to expandable, myelinogenic oligodendrocyte progenitor cells. Nat Biotechnol. 2013;31:426–433. doi: 10.1038/nbt.2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano T, Ando S, Takata N, Kawada M, Muguruma K, Sekiguchi K, Saito K, Yonemura S, Eiraku M, Sasai Y. Self-formation of optic cups and storable stratified neural retina from human ESCs. Cell Stem Cell. 2012;10:771–785. doi: 10.1016/j.stem.2012.05.009. [DOI] [PubMed] [Google Scholar]
- Nayak D, Roth TL, McGavern DB. Microglia development and function. Annu Rev Immunol. 2014;32:367–402. doi: 10.1146/annurev-immunol-032713-120240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen HN, Byers B, Cord B, Shcheglovitov A, Byrne J, Gujar P, Kee K, Schule B, Dolmetsch RE, Langston W, Palmer TD, Pera RR. LRRK2 mutant iPSC-derived DA neurons demonstrate increased susceptibility to oxidative stress. Cell Stem Cell. 2011;8:267–280. doi: 10.1016/j.stem.2011.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu W, Zang T, Zou Y, Fang S, Smith DK, Bachoo R, Zhang CL. In vivo reprogramming of astrocytes to neuroblasts in the adult brain. Nat Cell Biol. 2013;15:1164–1175. doi: 10.1038/ncb2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang ZP, Yang N, Vierbuchen T, Ostermeier A, Fuentes DR, Yang TQ, Citri A, Sebastiano V, Marro S, Sudhof TC, Wernig M. Induction of human neuronal cells by defined transcription factors. Nature. 2011;476:220–223. doi: 10.1038/nature10202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park IH, Arora N, Huo H, Maherali N, Ahfeldt T, Shimamura A, Lensch MW, Cowan C, Hochedlinger K, Daley GQ. Disease-specific induced pluripotent stem cells. Cell. 2008;134:877–886. doi: 10.1016/j.cell.2008.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasca SP, Portmann T, Voineagu I, Yazawa M, Shcheglovitov A, Pasca AM, Cord B, Palmer TD, Chikahisa S, Nishino S, Bernstein JA, Hallmayer J, Geschwind DH, Dolmetsch RE. Using iPSC-derived neurons to uncover cellular phenotypes associated with Timothy syndrome. Nat Med. 2011;17:1657–1662. doi: 10.1038/nm.2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasinelli P, Brown RH. Molecular biology of amyotrophic lateral sclerosis: insights from genetics. Nat Rev Neurosci. 2006;7:710–723. doi: 10.1038/nrn1971. [DOI] [PubMed] [Google Scholar]
- Paulsen Bda S, de Moraes Maciel R, Galina A, Souza da Silveira M, dos Santos Souza C, Drummond H, Nascimento Pozzatto E, Silva H, Jr, Chicaybam L, Massuda R, Setti-Perdigao P, Bonamino M, Belmonte-de-Abreu PS, Castro NG, Brentani H, Rehen SK. Altered oxygen metabolism associated to neurogenesis of induced pluripotent stem cells derived from a schizophrenic patient. Cell Transplant. 2012;21:1547–1559. doi: 10.3727/096368911X600957. [DOI] [PubMed] [Google Scholar]
- Pedrosa E, Sandler V, Shah A, Carroll R, Chang C, Rockowitz S, Guo X, Zheng D, Lachman HM. Development of patient-specific neurons in schizophrenia using induced pluripotent stem cells. J Neurogenet. 2011;25:88–103. doi: 10.3109/01677063.2011.597908. [DOI] [PubMed] [Google Scholar]
- Pfisterer U, Kirkeby A, Torper O, Wood J, Nelander J, Dufour A, Bjorklund A, Lindvall O, Jakobsson J, Parmar M. Direct conversion of human fibroblasts to dopaminergic neurons. Proc Natl Acad Sci USA. 2011;108:10343–10348. doi: 10.1073/pnas.1105135108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raitano S, Ordovas L, De Muynck L, Guo W, Espuny-Camacho I, Geraerts M, Khurana S, Vanuytsel K, Toth BI, Voets T, Vandenberghe R, Cathomen T, Van Den Bosch L, Vanderhaeghen P, Van Damme P, Verfaillie CM. Restoration of progranulin expression rescues cortical neuron generation in an induced pluripotent stem cell model of frontotemporal dementia. Stem Cell Rep. 2015;4:16–24. doi: 10.1016/j.stemcr.2014.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt P, Schmid B, Burbulla LF, Schondorf DC, Wagner L, Glatza M, Hoing S, Hargus G, Heck SA, Dhingra A, Wu G, Muller S, Brockmann K, Kluba T, Maisel M, Kruger R, Berg D, Tsytsyura Y, Thiel CS, Psathaki OE, et al. Genetic correction of a LRRK2 mutation in human iPSCs links parkinsonian neurodegeneration to ERK-dependent changes in gene expression. Cell Stem Cell. 2013;12:354–367. doi: 10.1016/j.stem.2013.01.008. [DOI] [PubMed] [Google Scholar]
- Ricciardi S, Ungaro F, Hambrock M, Rademacher N, Stefanelli G, Brambilla D, Sessa A, Magagnotti C, Bachi A, Giarda E, Verpelli C, Kilstrup-Nielsen C, Sala C, Kalscheuer VM, Broccoli V. CDKL5 ensures excitatory synapse stability by reinforcing NGL-1-PSD95 interaction in the postsynaptic compartment and is impaired in patient iPSC-derived neurons. Nat Cell Biol. 2012;14:911–923. doi: 10.1038/ncb2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ring KL, Tong LM, Balestra ME, Javier R, Andrews-Zwilling Y, Li G, Walker D, Zhang WR, Kreitzer AC, Huang Y. Direct reprogramming of mouse and human fibroblasts into multipotent neural stem cells with a single factor. Cell Stem Cell. 2012;11:100–109. doi: 10.1016/j.stem.2012.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robicsek O, Karry R, Petit I, Salman-Kesner N, Muller FJ, Klein E, Aberdam D, Ben-Shachar D. Abnormal neuronal differentiation and mitochondrial dysfunction in hair follicle-derived induced pluripotent stem cells of schizophrenia patients. Mol Psychiatry. 2013;18:1067–1076. doi: 10.1038/mp.2013.67. [DOI] [PubMed] [Google Scholar]
- Roybon L, Lamas NJ, Garcia-Diaz A, Yang EJ, Sattler R, Jackson-Lewis V, Kim YA, Kachel CA, Rothstein JD, Przedborski S, Wichterle H, Henderson CE. Human stem cell-derived spinal cord astrocytes with defined mature or reactive phenotypes. Cell Rep. 2013;4:1035–1048. doi: 10.1016/j.celrep.2013.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin LL. Stem cells and drug discovery: the beginning of a new era? Cell. 2008;132:549–552. doi: 10.1016/j.cell.2008.02.010. [DOI] [PubMed] [Google Scholar]
- Ruckh JM, Zhao JW, Shadrach JL, van Wijngaarden P, Rao TN, Wagers AJ, Franklin RJ. Rejuvenation of regeneration in the aging central nervous system. Cell Stem Cell. 2012;10:96–103. doi: 10.1016/j.stem.2011.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan SD, Dolatabadi N, Chan SF, Zhang X, Akhtar MW, Parker J, Soldner F, Sunico CR, Nagar S, Talantova M, Lee B, Lopez K, Nutter A, Shan B, Molokanova E, Zhang Y, Han X, Nakamura T, Masliah E, Yates JR, III, et al. Isogenic human iPSC Parkinson's model shows nitrosative stress-induced dysfunction in MEF2-PGC1alpha transcription. Cell. 2013;155:1351–1364. doi: 10.1016/j.cell.2013.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Danes A, Richaud-Patin Y, Carballo-Carbajal I, Jimenez-Delgado S, Caig C, Mora S, Di Guglielmo C, Ezquerra M, Patel B, Giralt A, Canals JM, Memo M, Alberch J, Lopez-Barneo J, Vila M, Cuervo AM, Tolosa E, Consiglio A, Raya A. Disease-specific phenotypes in dopamine neurons from human iPS-based models of genetic and sporadic Parkinson's disease. EMBO Mol Med. 2012;4:380–395. doi: 10.1002/emmm.201200215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders LH, Laganiere J, Cooper O, Mak SK, Vu BJ, Huang YA, Paschon DE, Vangipuram M, Sundararajan R, Urnov FD, Langston JW, Gregory PD, Zhang HS, Greenamyre JT, Isacson O, Schule B. LRRK2 mutations cause mitochondrial DNA damage in iPSC-derived neural cells from Parkinson's disease patients: reversal by gene correction. Neurobiol Dis. 2014;62:381–386. doi: 10.1016/j.nbd.2013.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandoe J, Eggan K. Opportunities and challenges of pluripotent stem cell neurodegenerative disease models. Nat Neurosci. 2013;16:780–789. doi: 10.1038/nn.3425. [DOI] [PubMed] [Google Scholar]
- Sareen D, Ebert AD, Heins BM, McGivern JV, Ornelas L, Svendsen CN. Inhibition of apoptosis blocks human motor neuron cell death in a stem cell model of spinal muscular atrophy. PLoS ONE. 2012;7:e39113. doi: 10.1371/journal.pone.0039113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sareen D, O'Rourke JG, Meera P, Muhammad AK, Grant S, Simpkinson M, Bell S, Carmona S, Ornelas L, Sahabian A, Gendron T, Petrucelli L, Baughn M, Ravits J, Harms MB, Rigo F, Bennett CF, Otis TS, Svendsen CN, Baloh RH. Targeting RNA foci in iPSC-derived motor neurons from ALS patients with a C9ORF72 repeat expansion. Sci Transl Med. 2013;5:208ra149. doi: 10.1126/scitranslmed.3007529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seibler P, Graziotto J, Jeong H, Simunovic F, Klein C, Krainc D. Mitochondrial Parkin recruitment is impaired in neurons derived from mutant PINK1 induced pluripotent stem cells. J Neurosci. 2011;31:5970–5976. doi: 10.1523/JNEUROSCI.4441-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serio A, Bilican B, Barmada SJ, Ando DM, Zhao C, Siller R, Burr K, Haghi G, Story D, Nishimura AL, Carrasco MA, Phatnani HP, Shum C, Wilmut I, Maniatis T, Shaw CE, Finkbeiner S, Chandran S. Astrocyte pathology and the absence of non-cell autonomy in an induced pluripotent stem cell model of TDP-43 proteinopathy. Proc Natl Acad Sci USA. 2013;110:4697–4702. doi: 10.1073/pnas.1300398110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shcheglovitov A, Shcheglovitova O, Yazawa M, Portmann T, Shu R, Sebastiano V, Krawisz A, Froehlich W, Bernstein JA, Hallmayer JF, Dolmetsch RE. SHANK3 and IGF1 restore synaptic deficits in neurons from 22q13 deletion syndrome patients. Nature. 2013;503:267–271. doi: 10.1038/nature12618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng C, Zheng Q, Wu J, Xu Z, Sang L, Wang L, Guo C, Zhu W, Tong M, Liu L, Li W, Liu ZH, Zhao XY, Wang L, Chen Z, Zhou Q. Generation of dopaminergic neurons directly from mouse fibroblasts and fibroblast-derived neural progenitors. Cell Res. 2012a;22:769–772. doi: 10.1038/cr.2012.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng C, Zheng Q, Wu J, Xu Z, Wang L, Li W, Zhang H, Zhao XY, Liu L, Wang Z, Guo C, Wu HJ, Liu Z, Wang L, He S, Wang XJ, Chen Z, Zhou Q. Direct reprogramming of Sertoli cells into multipotent neural stem cells by defined factors. Cell Res. 2012b;22:208–218. doi: 10.1038/cr.2011.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheridan SD, Theriault KM, Reis SA, Zhou F, Madison JM, Daheron L, Loring JF, Haggarty SJ. Epigenetic characterization of the FMR1 gene and aberrant neurodevelopment in human induced pluripotent stem cell models of fragile X syndrome. PLoS ONE. 2011;6:e26203. doi: 10.1371/journal.pone.0026203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Kirwan P, Livesey FJ. Directed differentiation of human pluripotent stem cells to cerebral cortex neurons and neural networks. Nat Protoc. 2012a;7:1836–1846. doi: 10.1038/nprot.2012.116. [DOI] [PubMed] [Google Scholar]
- Shi Y, Kirwan P, Smith J, Robinson HP, Livesey FJ. Human cerebral cortex development from pluripotent stem cells to functional excitatory synapses. Nat Neurosci. 2012b;15:477–486. doi: 10.1038/nn.3041. s471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R, Shen W, Kuai D, Martin JM, Guo X, Smith MA, Perez ET, Phillips MJ, Simonett JM, Wallace KA, Verhoeven AD, Capowski EE, Zhang X, Yin Y, Halbach PJ, Fishman GA, Wright LS, Pattnaik BR, Gamm DM. iPS cell modeling of Best disease: insights into the pathophysiology of an inherited macular degeneration. Hum Mol Genet. 2013;22:593–607. doi: 10.1093/hmg/dds469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son EY, Ichida JK, Wainger BJ, Toma JS, Rafuse VF, Woolf CJ, Eggan K. Conversion of mouse and human fibroblasts into functional spinal motor neurons. Cell Stem Cell. 2011;9:205–218. doi: 10.1016/j.stem.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sproul AA, Jacob S, Pre D, Kim SH, Nestor MW, Navarro-Sobrino M, Santa-Maria I, Zimmer M, Aubry S, Steele JW, Kahler DJ, Dranovsky A, Arancio O, Crary JF, Gandy S, Noggle SA. Characterization and molecular profiling of PSEN1 familial Alzheimer's disease iPSC-derived neural progenitors. PLoS ONE. 2014;9:e84547. doi: 10.1371/journal.pone.0084547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreedharan J, Brown RH., Jr Amyotrophic lateral sclerosis: problems and prospects. Ann Neurol. 2013;74:309–316. doi: 10.1002/ana.24012. [DOI] [PubMed] [Google Scholar]
- Stiles J, Jernigan TL. The basics of brain development. Neuropsychol Rev. 2010;20:327–348. doi: 10.1007/s11065-010-9148-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su YC, Qi X. Inhibition of excessive mitochondrial fission reduced aberrant autophagy and neuronal damage caused by LRRK2 G2019S mutation. Hum Mol Genet. 2013;22:4545–4561. doi: 10.1093/hmg/ddt301. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Takazawa T, Croft GF, Amoroso MW, Studer L, Wichterle H, Macdermott AB. Maturation of spinal motor neurons derived from human embryonic stem cells. PLoS ONE. 2012;7:e40154. doi: 10.1371/journal.pone.0040154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thier M, Worsdorfer P, Lakes YB, Gorris R, Herms S, Opitz T, Seiferling D, Quandel T, Hoffmann P, Nothen MM, Brustle O, Edenhofer F. Direct conversion of fibroblasts into stably expandable neural stem cells. Cell Stem Cell. 2012;10:473–479. doi: 10.1016/j.stem.2012.03.003. [DOI] [PubMed] [Google Scholar]
- Tiscornia G, Vivas EL, Matalonga L, Berniakovich I, Barragan Monasterio M, Eguizabal C, Gort L, Gonzalez F, Ortiz Mellet C, Garcia Fernandez JM, Ribes A, Veiga A, Izpisua Belmonte JC. Neuronopathic Gaucher's disease: induced pluripotent stem cells for disease modelling and testing chaperone activity of small compounds. Hum Mol Genet. 2013;22:633–645. doi: 10.1093/hmg/dds471. [DOI] [PubMed] [Google Scholar]
- Tremblay C, Achim AM, Macoir J, Monetta L. The heterogeneity of cognitive symptoms in Parkinson's disease: a meta-analysis. J Neurol Neurosurg Psychiatry. 2013;84:1265–1272. doi: 10.1136/jnnp-2013-305021. [DOI] [PubMed] [Google Scholar]
- Trilck M, Hubner R, Seibler P, Klein C, Rolfs A, Frech MJ. Niemann-Pick type C1 patient-specific induced pluripotent stem cells display disease specific hallmarks. Orphanet J Rare Dis. 2013;8:144. doi: 10.1186/1750-1172-8-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsunemoto RK, Eade KT, Blanchard JW, Baldwin KK. Forward engineering neuronal diversity using direct reprogramming. EMBO J. 2015;34:1445–1455. doi: 10.15252/embj.201591402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker BA, Scheetz TE, Mullins RF, DeLuca AP, Hoffmann JM, Johnston RM, Jacobson SG, Sheffield VC, Stone EM. Exome sequencing and analysis of induced pluripotent stem cells identify the cilia-related gene male germ cell-associated kinase (MAK) as a cause of retinitis pigmentosa. Proc Natl Acad Sci USA. 2011;108:E569–E576. doi: 10.1073/pnas.1108918108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker BA, Mullins RF, Streb LM, Anfinson K, Eyestone ME, Kaalberg E, Riker MJ, Drack AV, Braun TA, Stone EM. Patient-specific iPSC-derived photoreceptor precursor cells as a means to investigate retinitis pigmentosa. eLife. 2013;2:e00824. doi: 10.7554/eLife.00824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Victor MB, Richner M, Hermanstyne TO, Ransdell JL, Sobieski C, Deng PY, Klyachko VA, Nerbonne JM, Yoo AS. Generation of human striatal neurons by microRNA-dependent direct conversion of fibroblasts. Neuron. 2014;84:311–323. doi: 10.1016/j.neuron.2014.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wainger BJ, Kiskinis E, Mellin C, Wiskow O, Han SS, Sandoe J, Perez NP, Williams LA, Lee S, Boulting G, Berry JD, Brown RH, Jr, Cudkowicz ME, Bean BP, Eggan K, Woolf CJ. Intrinsic membrane hyperexcitability of amyotrophic lateral sclerosis patient-derived motor neurons. Cell Rep. 2014;7:1–11. doi: 10.1016/j.celrep.2014.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wainger BJ, Buttermore ED, Oliveira JT, Mellin C, Lee S, Saber WA, Wang AJ, Ichida JK, Chiu IM, Barrett L, Huebner EA, Bilgin C, Tsujimoto N, Brenneis C, Kapur K, Rubin LL, Eggan K, Woolf CJ. Modeling pain in vitro using nociceptor neurons reprogrammed from fibroblasts. Nat Neurosci. 2015;18:17–24. doi: 10.1038/nn.3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen X, Tan W, Westergard T, Krishnamurthy K, Markandaiah SS, Shi Y, Lin S, Shneider NA, Monaghan J, Pandey UB, Pasinelli P, Ichida JK, Trotti D. Antisense proline-arginine RAN dipeptides linked to C9ORF72-ALS/FTD form toxic nuclear aggregates that initiate in vitro and in vivo neuronal death. Neuron. 2014a;84:1213–1225. doi: 10.1016/j.neuron.2014.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen Z, Nguyen HN, Guo Z, Lalli MA, Wang X, Su Y, Kim NS, Yoon KJ, Shin J, Zhang C, Makri G, Nauen D, Yu H, Guzman E, Chiang CH, Yoritomo N, Kaibuchi K, Zou J, Christian KM, Cheng L, et al. Synaptic dysregulation in a human iPS cell model of mental disorders. Nature. 2014b;515:414–418. doi: 10.1038/nature13716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams LA, Davis-Dusenbery BN, Eggan KC. SnapShot: directed differentiation of pluripotent stem cells. Cell. 2012;149:1174–1174.e1171. doi: 10.1016/j.cell.2012.05.015. [DOI] [PubMed] [Google Scholar]
- Williams EC, Zhong X, Mohamed A, Li R, Liu Y, Dong Q, Ananiev GE, Mok JC, Lin BR, Lu J, Chiao C, Cherney R, Li H, Zhang SC, Chang Q. Mutant astrocytes differentiated from Rett syndrome patients-specific iPSCs have adverse effects on wild-type neurons. Hum Mol Genet. 2014;23:2968–2980. doi: 10.1093/hmg/ddu008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y, Ouyang K, Huang J, Zhou Y, Ouyang H, Li H, Wang G, Wu Q, Wei C, Bi Y, Jiang L, Cai Z, Sun H, Zhang K, Zhang Y, Chen J, Fu XD. Direct conversion of fibroblasts to neurons by reprogramming PTB-regulated microRNA circuits. Cell. 2013;152:82–96. doi: 10.1016/j.cell.2012.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi T, Ito D, Okada Y, Akamatsu W, Nihei Y, Yoshizaki T, Yamanaka S, Okano H, Suzuki N. Modeling familial Alzheimer's disease with induced pluripotent stem cells. Hum Mol Genet. 2011;20:4530–4539. doi: 10.1093/hmg/ddr394. [DOI] [PubMed] [Google Scholar]
- Yan Y, Yang D, Zarnowska ED, Du Z, Werbel B, Valliere C, Pearce RA, Thomson JA, Zhang SC. Directed differentiation of dopaminergic neuronal subtypes from human embryonic stem cells. Stem Cells. 2005;23:781–790. doi: 10.1634/stemcells.2004-0365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang N, Zuchero JB, Ahlenius H, Marro S, Ng YH, Vierbuchen T, Hawkins JS, Geissler R, Barres BA, Wernig M. Generation of oligodendroglial cells by direct lineage conversion. Nat Biotechnol. 2013a;31:434–439. doi: 10.1038/nbt.2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YM, Gupta SK, Kim KJ, Powers BE, Cerqueira A, Wainger BJ, Ngo HD, Rosowski KA, Schein PA, Ackeifi CA, Arvanites AC, Davidow LS, Woolf CJ, Rubin LL. A small molecule screen in stem-cell-derived motor neurons identifies a kinase inhibitor as a candidate therapeutic for ALS. Cell Stem Cell. 2013b;12:713–726. doi: 10.1016/j.stem.2013.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Y, Cui X, Al-Ramahi I, Sun X, Li B, Hou J, Difiglia M, Palacino J, Wu ZY, Ma L, Botas J, Lu B. A striatal-enriched intronic GPCR modulates huntingtin levels and toxicity. eLife. 2015;4:e05449. doi: 10.7554/eLife.05449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo AS, Sun AX, Li L, Shcheglovitov A, Portmann T, Li Y, Lee-Messer C, Dolmetsch RE, Tsien RW, Crabtree GR. MicroRNA-mediated conversion of human fibroblasts to neurons. Nature. 2011;476:228–231. doi: 10.1038/nature10323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon KJ, Nguyen HN, Ursini G, Zhang F, Kim NS, Wen Z, Makri G, Nauen D, Shin JH, Park Y, Chung R, Pekle E, Zhang C, Towe M, Hussaini SM, Lee Y, Rujescu D, St Clair D, Kleinman JE, Hyde TM, et al. Modeling a genetic risk for schizophrenia in iPSCs and mice reveals neural stem cell deficits associated with adherens junctions and polarity. Cell Stem Cell. 2014;15:79–91. doi: 10.1016/j.stem.2014.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, Slukvin II, Thomson JA. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- Yu DX, Di Giorgio FP, Yao J, Marchetto MC, Brennand K, Wright R, Mei A, McHenry L, Lisuk D, Grasmick JM, Silberman P, Silberman G, Jappelli R, Gage FH. Modeling hippocampal neurogenesis using human pluripotent stem cells. Stem Cell Rep. 2014;2:295–310. doi: 10.1016/j.stemcr.2014.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng H, Guo M, Martins-Taylor K, Wang X, Zhang Z, Park JW, Zhan S, Kronenberg MS, Lichtler A, Liu HX, Chen FP, Yue L, Li XJ, Xu RH. Specification of region-specific neurons including forebrain glutamatergic neurons from human induced pluripotent stem cells. PLoS ONE. 2010;5:e11853. doi: 10.1371/journal.pone.0011853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Z, Ilieva H, Hallagan L, Bell R, Singh I, Paquette N, Thiyagarajan M, Deane R, Fernandez JA, Lane S, Zlokovic AB, Liu T, Griffin JH, Chow N, Castellino FJ, Stojanovic K, Cleveland DW, Zlokovic BV. Activated protein C therapy slows ALS-like disease in mice by transcriptionally inhibiting SOD1 in motor neurons and microglia cells. J Clin Invest. 2009;119:3437–3449. doi: 10.1172/JCI38476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Brown J, Kanarek A, Rajagopal J, Melton DA. In vivo reprogramming of adult pancreatic exocrine cells to beta-cells. Nature. 2008;455:627–632. doi: 10.1038/nature07314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu PP, Denton KR, Pierson TM, Li XJ, Blackstone C. Pharmacologic rescue of axon growth defects in a human iPSC model of hereditary spastic paraplegia SPG3A. Hum Mol Genet. 2014;23:5638–5648. doi: 10.1093/hmg/ddu280. [DOI] [PMC free article] [PubMed] [Google Scholar]