Abstract
Cronobacter species are opportunistic pathogens commonly found in the environment. Among the seven Cronobacter species, Cronobacter sakazakii sequence type 4 (ST-4) is predominantly associated with recorded cases of infantile meningitis. This study reports on a 26-month powdered infant formula (PIF) surveillance program in four production facilities located in distinct geographic regions. The objective was to identify the ST(s) in PIF production environments and to investigate the phenotypic features that support their survival. Of all 168 Cronobacter isolates, 133 were recovered from a PIF production environment, 31 were of clinical origin, and 4 were laboratory type strains. Sequence type 1 (n = 84 isolates; 63.9%) was the dominant type in PIF production environments. The majority of these isolates clustered with an indistinguishable pulsotype and persisted for at least an 18-month period. Moreover, DNA microarray results identified two phylogenetic lineages among ST-4 strains tested. Thereafter, the ST-1 and -4 isolates were phenotypically compared. Differences were noted based on the phenotypes expressed by these isolates. The ST-1 PIF isolates produced stronger biofilms at both 28°C and 37°C, while the ST-4 clinical isolates exhibited greater swimming activity and increased binding to Congo red dye. Given the fact that PIF is a low-moisture environment and that the clinical environment provides for an interaction between the pathogen and its host, these differences may be consistent with a form of pathoadaptation. These findings help to extend our current understanding of the epidemiology and ecology of Cronobacter species in PIF production environments.
INTRODUCTION
Cronobacter (formerly known as Enterobacter sakazakii) was accepted as a new bacterial genus in 2007 (1). It consists of seven species, including C. sakazakii, C. malonaticus, C. turicensis, C. muytjensii, C. dublinensis, C. universalis, and C. condimenti (2, 3). The epidemiological link between Cronobacter infection in neonates and contaminated powdered infant formula (PIF) has been previously established (4, 5), with C. sakazakii sequence type 4 (ST-4) being linked to cases of meningitis (6). Outbreaks have been associated with contaminated food products and the presence of this bacterium in PIF production environments (5, 7).
In order to rapidly and accurately characterize Cronobacter species in PIF and its associated environments, several molecular-based protocols have been developed, which include direct target gene detection and subtyping methods (8–20). Pulsed-field gel electrophoresis (PFGE) is an accepted method for tracking isolates across the food chain, and this approach is generally considered suitable for epidemiological studies (12, 21–28). A multilocus sequence typing (MLST) scheme for Cronobacter species was developed, which focuses on single nucleotide polymorphisms associated with seven housekeeping genes (including atpD, fusA, glnS, gltB, gyrB, infB, and pps) and identifies their associated alleles (29). This protocol has been used to describe some of the diversity related to the genus (6, 29–31). Both PFGE and MLST have been widely applied to study the genomic diversity of Cronobacter isolated from manufacturing facilities, commercial PIF, and follow-up formula, as well as clinical isolates (20, 32, 33). These reports highlight the dominance of C. sakazakii and the importance of the ST-4 clonal complex as the etiological agent in meningitis cases. However, there is a lack of data comparing the phenotypes of isolates within these clusters. Cruz-Cordova et al. (34) investigated the role of flagella from C. sakazakii ST-1 and -4. No significant difference was observed during proinflammatory cytokine activation in macrophages when ST-1 and -4 strains were compared.
This study reports on a 26-month PIF surveillance program in four production facilities geographically located in distinct regions. The study was designed to describe the genotypes and phenotypes of the Cronobacter species recovered (summarized in Fig. S1 in the supplemental material). The objective was to characterize the ST(s) in these PIF manufacturing environments and to investigate the phenotypes of the isolates.
MATERIALS AND METHODS
Bacterial isolates.
A total of 133 Cronobacter isolates were cultured from finished products (FP), semifinished products (BP), and environmental swabs (Env) at PIF facilities in four different geographical regions between April 2011 and May 2013. Thirty-one clinical isolates and four laboratory type strains were included for comparison (see Table S1 in the supplemental material). All bacteria were grown on tryptone soy agar (TSA; Oxoid Ltd., Basingstoke, United Kingdom) at 37°C overnight and stored at −80°C on cryo-beads (Technical Service Consultants Ltd., Lancashire, United Kingdom).
Purification of DNA and PCR amplification of target genes.
Template DNA was extracted by using a simple boiling procedure (which yielded approximately 50 ng from a 50-μl PCR mixture). Real-time PCR was used to confirm the bacterial genus (9, 35). The species were identified using rpoB as the gene target for the PCR amplification, as described previously (13, 17). Serotyping was conducted using PCR amplification of targeted genes within the species, as originally described (11, 15, 16, 19). All amplicons were analyzed in 1% [wt/vol] agarose gels in 1× Tris-borate-EDTA buffer (TBE; Sigma-Aldrich, Gillingham, United Kingdom) that were stained with SYBR Safe (Life Technologies, CA, USA), visualized, and photographed with a Kodak Gel Logic 1500 imaging system (Carestream Health, Inc., NY, USA).
PFGE subtyping of Cronobacter.
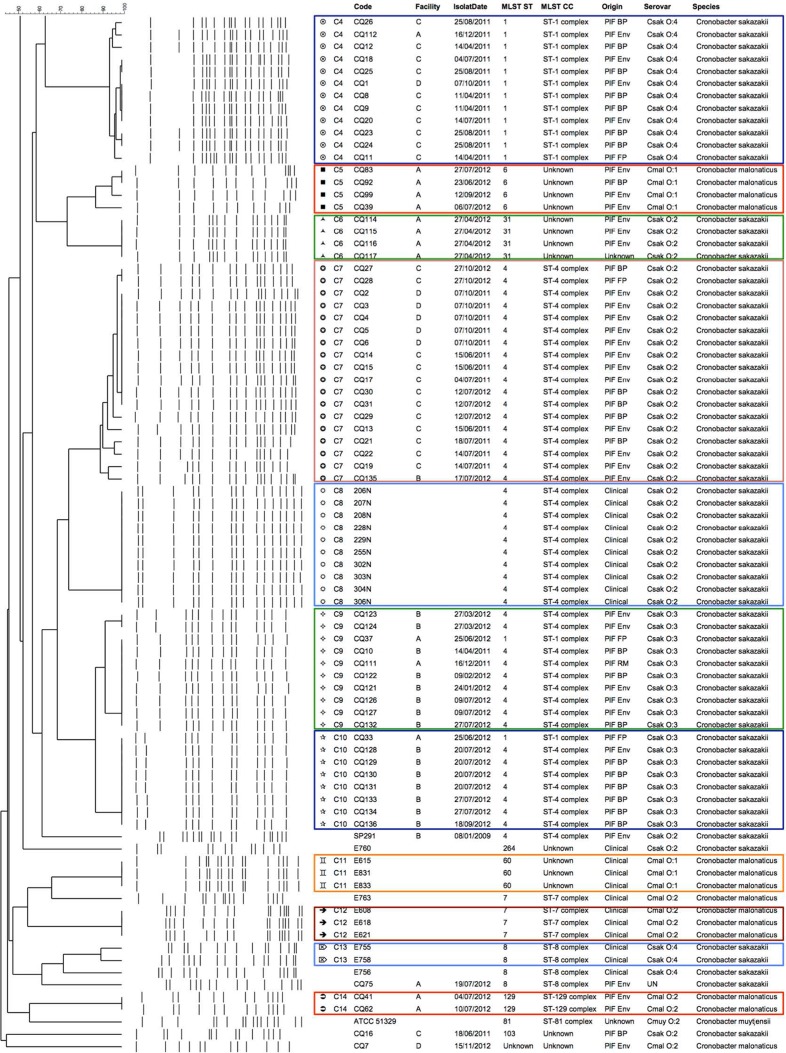
PFGE analysis was performed on all 168 bacterial isolates (Fig. 1; see also Table S1 in the supplemental material). A modified version of the standard PulseNet protocol for Cronobacter (36) was used. Briefly, pulsed-field certified agarose (Bio-Rad, Hercules, CA) was selected for preparation of agarose plugs. Each plug was lysed with 0.1 mg/ml proteinase K at 54°C for 90 min, followed by two 10-min washes with 18 MΩ water and a further three 15-min washes in Tris-HCl (pH 8.0)-EDTA [TE] buffer. The restriction digestion was carried out at 37°C for 3 h using 50 U XbaI. Treated plugs were cast into a 1% (wt/vol) agarose gel and electrophoresed in 0.5× TBE buffer with a CHEF Mapper XA system (Bio-Rad, Hercules, CA). The running conditions used were as follows: initial switch time, 2.16 s; final switch time, 63.8 s; voltage. 6 V; angle of 120°; run time of 20 h at a constant temperature of 14°C. The resulting gel was then stained with 0.01% (wt/vol) SYBR Safe staining buffer for 30 min and destained with 500 ml 18 MΩ water for a further 30 min. Tiff images were then acquired and uploaded to BioNumerics version 7.1 (Applied Maths, Saint-Martens-Latem, Belgium) for analysis using the DICE coefficient and unweighted pair group method with arithmetic mean (UPGMA). Both the optimization and band-matching tolerance were 1.0%. When comparing the DNA fingerprint patterns, a cutoff value of 90% similarity was applied.
FIG 1.
Dendrogram showing the 14 clusters of the 168 strains, denoted clusters C1 to C14, generated using BioNumerics v7.1. The similarity cutoff value was 90%. Both the optimization and tolerance values were 1.0%.
MLST characterization of Cronobacter.
MLST analysis was carried out on all 168 strains (strain information is listed in Fig. 1; see also Table S1 in the supplemental material). All PCRs mixtures included Taq DNA polymerase with ThermoPol buffer (New England BioLabs Inc., USA) and were assembled into mixtures following the manufacturer's instructions. Primers for all seven housekeeping genes were those used previously (29). Amplicons were dispatched for commercial Sanger sequencing (MWG Eurofins, Ebersberg, Germany). The nucleotide sequence trace files were uploaded to BioNumerics version 7.1 (Applied Maths, Saint-Martens-Latem, Belgium) and mapped against information on the MLST Cronobacter website (http://pubmlst.org/cronobacter/) for assignment of each allele, ST profile, and clonal cluster. A minimum spanning tree was generated to analyze the relatedness of all isolates studied.
DNA microarray analysis.
A custom-designed multigenome DNA microarray was developed by the U.S. FDA for the identification and characterization of Cronobacter species (37). This array contains over 21,402 unique genes, representing the pan-genomes of all seven currently recognized Cronobacter species. The probe development and optimization have been described in detail by Tall et al. (37). For this study, 59 representatives of the 168 isolates were selected, based on their PFGE clusters and ST(s) along with 14 isolates representing other Cronobacter species and nearest neighbors, such as Salmonella enterica serovar Typhimurium, Klebsiella pneumoniae, Citrobacter freundii, Siccibacter turicensis, Franconibacter helveticus, and Franconibacter pulveris, which were used as controls for this custom-designed pan-genome microarray (Table 1).
TABLE 1.
Pan-genome DNA microarray analysis of a subset of isolates selected from the PFGE and MLST studies as well as other isolates representing other species of Cronobacter and their nearest neighbors
| Isolate group and ID | Species | Serotype | MLST ST no. | ST clonal complex |
|---|---|---|---|---|
| Cronobacter isolates selected from PFGE and MLST assays | ||||
| 206N | Cronobacter sakazakii | O:2 | 4 | ST-4 |
| 207N | Cronobacter sakazakii | O:2 | 4 | ST-4 |
| 208N | Cronobacter sakazakii | O:2 | 4 | ST-4 |
| 255N | Cronobacter sakazakii | O:2 | 4 | ST-4 |
| 306N | Cronobacter sakazakii | O:2 | 4 | ST-4 |
| CQ2 | Cronobacter sakazakii | O:2 | 4 | ST-4 |
| CQ3 | Cronobacter sakazakii | O:2 | 4 | ST-4 |
| CQ4 | Cronobacter sakazakii | O:2 | 4 | ST-4 |
| CQ5 | Cronobacter sakazakii | O:2 | 4 | ST-4 |
| CQ6 | Cronobacter sakazakii | O:2 | 4 | ST-4 |
| CQ13 | Cronobacter sakazakii | O:2 | 4 | ST-4 |
| CQ14 | Cronobacter sakazakii | O:2 | 4 | ST-4 |
| CQ15 | Cronobacter sakazakii | O:2 | 4 | ST-4 |
| CQ17 | Cronobacter sakazakii | O:2 | 4 | ST-4 |
| CQ19 | Cronobacter sakazakii | O:2 | 4 | ST-4 |
| CQ24 | Cronobacter sakazakii | O:4 | 1 | ST-1 |
| CQ25 | Cronobacter sakazakii | O:4 | 1 | ST-1 |
| CQ26 | Cronobacter sakazakii | O:4 | 1 | ST-1 |
| CQ31 | Cronobacter sakazakii | O:2 | 4 | ST-4 |
| CQ32 | Cronobacter sakazakii | O:1 | 1 | ST-1 |
| CQ34 | Cronobacter sakazakii | O:1 | 1 | ST-1 |
| CQ35 | Cronobacter sakazakii | O:1 | 1 | ST-1 |
| CQ36 | Cronobacter sakazakii | O:1 | 1 | ST-1 |
| CQ37 | Cronobacter sakazakii | O:3 | 1 | ST-1 |
| CQ38 | Cronobacter sakazakii | O:1 | 1 | ST-1 |
| CQ40 | Cronobacter sakazakii | O:1 | 1 | ST-1 |
| CQ42 | Cronobacter sakazakii | O:1 | 1 | ST-1 |
| CQ43 | Cronobacter sakazakii | O:1 | 1 | ST-1 |
| CQ44 | Cronobacter sakazakii | O:1 | 1 | ST-1 |
| CQ45 | Cronobacter sakazakii | O:1 | 1 | ST-1 |
| CQ46 | Cronobacter sakazakii | O:1 | 1 | ST-1 |
| CQ61 | Cronobacter sakazakii | O:1 | 1 | ST-1 |
| CQ68 | Cronobacter sakazakii | O:1 | 57 | ST-1 |
| CQ75 | Cronobacter sakazakii | O:3 | 8 | ST-8 |
| CQ83 | Cronobacter sakazakii | O:4 | 6 | Unknown |
| CQ92 | Cronobacter sakazakii | O:4 | 6 | Unknown |
| CQ99 | Cronobacter sakazakii | O:4 | 6 | Unknown |
| CQ111 | Cronobacter sakazakii | O:3 | 4 | ST-4 |
| CQ112 | Cronobacter sakazakii | O:4 | 1 | ST-1 |
| CQ114 | Cronobacter sakazakii | O:2 | 31 | Unknown |
| CQ115 | Cronobacter sakazakii | O:2 | 31 | Unknown |
| CQ116 | Cronobacter sakazakii | O:2 | 31 | Unknown |
| CQ121 | Cronobacter sakazakii | O:3 | 4 | ST-4 |
| CQ122 | Cronobacter sakazakii | O:3 | 4 | ST-4 |
| CQ123 | Cronobacter sakazakii | O:3 | 4 | ST-4 |
| CQ124 | Cronobacter sakazakii | O:3 | 4 | ST-4 |
| CQ126 | Cronobacter sakazakii | O:3 | 4 | ST-4 |
| CQ127 | Cronobacter sakazakii | O:3 | 4 | ST-4 |
| CQ128 | Cronobacter sakazakii | O:3 | 4 | ST-4 |
| E654 | Cronobacter sakazakii | O:1 | 1 | ST-1 |
| E657 | Cronobacter sakazakii | O:1 | 1 | ST-1 |
| E755 | Cronobacter sakazakii | O:4 | 8 | ST-8 |
| E758 | Cronobacter sakazakii | O:4 | 8 | ST-8 |
| E760 | Cronobacter sakazakii | O:2 | 264 | Unknown |
| E788 | Cronobacter sakazakii | O:3 | 4 | ST-4 |
| ATCC BAA-894 | Cronobacter sakazakii | O:1 | 1 | ST-1 |
| CQ41 | Cronobacter malonaticus | O:2 | 129 | ST-129 |
| CQ62 | Cronobacter malonaticus | O:2 | 129 | ST-129 |
| ATCC 51329 | Cronobacter muytjensii | O:2 | 81 | ST-81 |
| Other Cronobacter species included as internal controls | ||||
| CI825 | Cronobacter malonaticus | O:2 | 7 | ST-7 |
| 464 | Cronobacter dublinensis | O:1 | 79 | |
| CFS237 | Cronobacter dublinensis | O:1 | 106 | |
| E515 | Cronobacter dublinensis | O:2 | 80 | ST-80 |
| z3032 | Cronobacter turicensis | O:1 | 19 | ST-24 |
| 797-2 | Cronobacter universalis | O:1 | 54 | |
| LMG26250 | Cronobacter condimenti | Unknown | 98 | |
| Nearest neighbors used as controls | ||||
| STM | Salmonella enterica serovar Typhimurium | |||
| 214 | Klebsiella pneumoniae | |||
| 508 | Siccibacter turicensis | |||
| z1159 | Franconibacter helveticus | |||
| z513 | Franconibacter helveticus | |||
| 1160 | Franconibacter pulveris | |||
| 601 | Franconibacter pulveris |
Briefly, genomic DNA was purified from all of these bacterial isolates, fragmented by DNase I, and labeled as reported previously (37, 38). Hybridization was performed according to the Affymetrix GeneChip expression analysis technical manual for the 49-format array. Following hybridization, washing and staining procedures were carried out on an Affymetrix FS-450 fluidics station (Affymetrix, CA, USA) using the mini_prok2v1_450 fluidics script. Reagents for washing and staining were prepared according to the GeneChip expression analysis technical manual. Arrays were then scanned using an Affymetrix GeneChip scanner 3000 running AGCC software (Affymetrix, CA, USA). For each gene represented on the microarray, probe set intensities were summarized using the Robust multiarray averaging (RMA) method (Bioconductor Affy package and Affymetric power tools) and compared across all strains investigated. If the same gene in different strains had an RMA intensity difference greater than 8-fold (log2 = 3), this gene was considered to be “different” between the two isolates. Thereafter, the absence/presence gene calls similar to binary nucleotide calls for each isolate were generated into Fasta-formatted files, which were then directly uploaded to the MEGA5 software package. Phylogenetic trees were generated using the maximum likelihood method as described by Jackson et al. (38).
Bacterial motility assays.
Swim and swarm assays were performed with all 168 isolates as described previously (39). Briefly, agar plates were freshly prepared using Luria-Bertani (LB) broth (Becton Dickinson, MD, USA) supplemented with 0.3% (wt/vol) agar (Sigma-Aldrich, Gillingham, United Kingdom) for the swim assay or with 0.6% (wt/vol) agar along with 0.5% (wt/vol) glucose for the swarm assay. Overnight cultures were stabbed into the center of the swim plates and spotted onto the swarm plates, and these plates were subsequently incubated at 37°C for 8 and 24 h, respectively. The diameter of the colony growth on each plate was measured using a standard ruler, recorded, and imaged using a Kodak Gel Logic 1500 imaging system (Carestream, Dublin, Ireland) and a Nikon D3100 camera (Nikon, Japan). Salmonella enterica serovar Typhimurium DT104 13348, a strain that expresses a high-motility phenotype, was included as the positive control for these assays. All assays were performed in duplicate.
Biofilm formation assay.
Biofilm formation in standard 96-well microtiter plates for all 168 isolates was performed using minimal medium M9 (6 g/liter Na2HPO4, 3 g/liter KH2PO4, 0.5 g/liter NaCl, 1 g/liter NH4Cl, 2 mM MgSO4, 0.1% glucose, and 0.1 mM CaCl2). After overnight growth in TSB medium, a 1:100 dilution was prepared, and 200 μl of cell suspension was inoculated in each of the 96-well microtiter plate wells. These inoculated plates were then incubated at either 28 or 37°C for 72 h. A crystal violet (CV) staining assay was carried out, which comprised three brief washes with 200 μl of phosphate-buffered saline (PBS) solution, followed by a 20-min fixation step with 200 μl methanol. Plates were allowed to air dry for 15 min. After the latter step, all plates were then stained with 200 μl 0.4% (wt/vol) CV for a period of 15 min and washed with 200 μl PBS, followed by air drying for another 15 min. The formed biofilm was then dissolved with 200 μl of 33% (vol/vol) acetic acid for 30 min. The biofilm formed was then measured at an optical density of 570 nm in a microtiter plate reader (Tecan, Männedorf, Switzerland) and analyzed as described previously (39). Salmonella Typhimurium ATCC 14028, a strong biofilm-forming strain, was selected as the positive control for the biofilm formation assays. These biofilm assays were performed in triplicate that included biological duplicates.
Detection of the Congo red dye binding.
The colony morphology of Cronobacter species on Congo red agar was examined for the binding of the Congo red dye as described previously (40). Congo red agar plates were prepared using LB agar without salt as a base and supplemented with 40 μg/ml Congo red dye (Sigma-Aldrich, Gillingham, United Kingdom) as well as 20 μg/ml of Coomassie brilliant blue (ThermoFisher Scientific, Waltham, MA). Three microliters of an overnight culture was inoculated in the center of the Congo red plates and plates were incubated at 28°C for 72 h. The morphology of each colony was photographed using a Nikon D3100 camera (Nikon, Japan). Salmonella Typhimurium ATCC 14028 was included as the positive control. The experiment was performed in duplicate.
Examination of cellulose production.
The production of cellulose by Cronobacter species was examined on calcofluor agar plates as described previously (40). A concentration of 50 μg/ml fluorescent brightener 28 (Sigma-Aldrich, Gillingham, United Kingdom) was added into the LB agar without salt. Three microliters of an overnight culture grown in TSB medium was inoculated into the center of the plate, and plates were incubated at 28°C for 72 h. The morphology and color of each colony were photographed under a 366-nm UV light using a Nikon D3100 camera (Nikon, Japan). Binding of any cellulose produced by the bacterial isolate was observed based on the presence of a blue colony under UV light. Salmonella Typhimurium ATCC 14028, a strong biofilm-forming strain, was included as the positive control. Each isolate was tested in duplicate.
Statistical analysis.
All data were analyzed using Microsoft Excel 2010 and IBM SPSS statistics version 20 unless indicated specifically. The Student t test was performed with the null hypothesis to understand the statistical significance among isolates of different groups. Bivariate correlation analysis was carried out using a Spearman's rho coefficient test among different phenotype traits in various groups. Correlation was considered significant at the level of 0.01 and 0.05.
RESULTS
Cronobacter species and serotypes identified in the PIF production sites.
Using species-specific PCR amplification of the rpoB gene, as described by Stoop et al. (13) and Lehner et al. (17), C. sakazakii was identified among 126 (94.7%) of the 133 isolates cultured from PIF production sites, thus constituting the dominant species (Table 2). Seven isolates (5.3%) were identified as C. malonaticus. No other Cronobacter species were cultured from either PIF or its production environments in this study. Serotypes of all PIF isolates were determined by using PCR assays as described by Mullane et al. (11) and Jarvis et al. (15, 19) (Fig. 1). Of the recovered 126 Cronobacter sakazakii isolates, the following serotypes were identified (Table 2): C. sakazakii O:1 (71 isolates, 53.4%), C. sakazakii O:2 (24 isolates, 18.0%), C. sakazakii O:3 (19 isolates, 14.3%), and C. sakazakii O:4 (15 isolates, 9.0%). Four isolates were identified as C. malonaticus O:1 (3.0%), and three others were identified as C. malonaticus O:2 (2.2%) (Table 2).
TABLE 2.
Species and serotype distribution of all 168 Cronobacter isolates studieda
| Species and serotype | No. of isolates cultured from PIF/PIF facilities | No. of clinical strains | No. of ATCC type strains |
|---|---|---|---|
| C. sakazakii O:1 | 71 | 6 | 2 |
| C. sakazakii O:2 | 24 | 14 | 1 |
| C. sakazakii O:3 | 19 | 3 | 0 |
| C. sakazakii O:4 | 12 | 0 | 0 |
| C. malonaticus O:1 | 4 | 3 | 0 |
| C. malonaticus O:2 | 3 | 5 | 0 |
| C. muytjensii O:2 | 0 | 0 | 1 |
| Total | 133 | 31 | 4 |
The isolates studied included 133 PIF isolates cultured from raw materials, swabs from the production environment, semifinished products, or finished products in PIF production facilities, and also 31 clinical isolates and 4 laboratory type strains.
PFGE subtyping analysis.
Following molecular subtyping by PFGE, and based on the analysis of the pulsotypes obtained, 14 clusters (with similarities above 90%) were identified comprising 133 PIF isolates along with 35 clinical and type strains, which were included for comparison (Fig. 1). Four clustered pulsotypes (denoted as C1, C5, C6, and C14) were identified in one of the four facilities analyzed. The pulsotype designated cluster C1 consisted of 70 isolates cultured from facility A, and all of these were identified as C. sakazakii serotype O:1 (Fig. 1). These isolates were recovered from finished product (denoted as FP) or the production environments (denoted as Env), and these were isolated on three separate occasions in December 2011, May through September 2012, and again in May 2013. Cluster C5 included four isolates of C. malonaticus serotype O:1, which were cultured on three occasions in June, July, and September 2012 from either base powder or the environment of facility A (Fig. 1). Cluster C6 consisted of four isolates cultured from the environment of facility A in April 2012 and which were identified as C. sakazakii serotype O:2 (Fig. 1). Cluster C14 included two C. malonaticus serotype O:2 isolates which were recovered from the environment of facility A in July 2012 (Fig. 1).
Of note, four clusters (including C4, C7, C9, and C10) were recovered from facilities in different geographical regions. Cluster C4 contained 12 C. sakazakii isolates, which were cultured from either finished product, base powder, or the environment during April, July, August, October, and December 2011 in three production locations (facilities A, C, and D) (Fig. 1). All were identified as C. sakazakii serotype O:4. Similarly, cluster C7 included 18 C. sakazakii isolates of serotype O:2, and these were cultured from base powder, finished product, or the environments in three of the facilities (facilities B, C, and D) (Fig. 1) during June, July, and October 2011 and July and October 2012. Cluster C9 included 10 C. sakazakii of serotype O:3. These were cultured from two facilities (facilities A and B) (Fig. 1) on a number of occasions during April and December 2011 and January through March, June, and July 2012. Finally, cluster C10 consisted of eight C. sakazakii serotype O:3 isolates, which were isolated in June, July, and September 2012 from base powder, finished product, or environments at facilities A and B (Fig. 1).
Cronobacter isolates of clinical origin clustered independently into six groups, which included cluster C2 (2 isolates) and C3 (2 isolates) of C. sakazakii serotype O:1, C8 (10 isolates) of C. sakazakii serotype O:2, C11 (3 isolates) of C. malonaticus serotype O:1, C12 (3 isolates) of C. malonaticus serotype O:2, and C13 (2 isolates) of C. sakazakii serotype O:4. In addition, five PIF isolates (including one C. malonaticus and four C. sakazakii isolates) and nine clinical isolates, along with four type strains, did not group into any of the previously described clusters.
MLST profiling.
MLST subtyping was performed with all 133 PIF isolates, along with the 31 clinical isolates and four type strains (Fig. 1). Sequence trace files were uploaded to BioNumerics v7.1 to aid with the generation of a minimum spanning tree (Fig. 2). The cross-link among isolates of PIF origin is highlighted in the orange circle. Of the 133 PIF isolates, ST-1 (n = 84 isolates, 63.2%) was identified as the dominant sequence type, followed by 35 isolates (26.3%) that were identified as ST-4 (Fig. 2a). Other STs identified included ST-6 (3.0%), ST-8 (0.8%), ST-31 (3.0%), ST-57 (0.8%), ST-103 (0.8%), ST-129 (1.5%), and a single unknown type (0.8%) (Sequence information for the unknown type is described in Text S1 in the supplemental material). When these STs were grouped by bacterial species, C. sakazakii was identified among ST-1, -4, -6, -8, -31, -57, and -103, while C. malonaticus was represented by ST-6 and -129 (Fig. 2a). A single unknown ST was identified as C. malonaticus serotype O:2. Additionally, the 31 clinical strains studied consisted of 23 C. sakazakii and 8 C. malonaticus isolates. ST-4 was the dominant type among these clinical isolates, comprising 45.2% of these. Lastly, five ST-7 C. malonaticus isolates and five ST-8 C. sakazakii isolates together comprised 32.2% of the collection.
FIG 2.
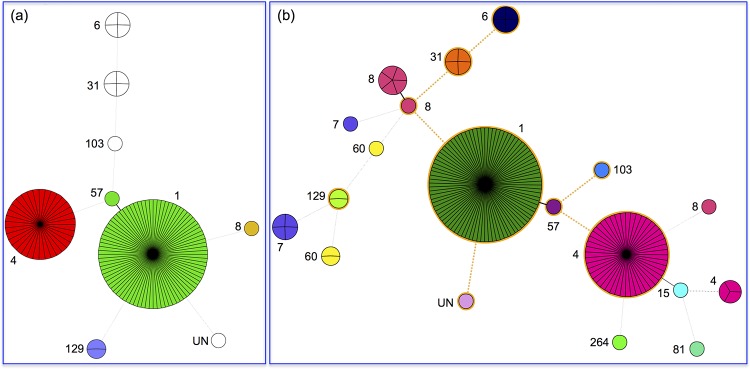
MLST distributions of 133 PIF and 31 clinical isolates. (a) PIF isolates; (b) all 164 PIF and clinical isolates studied. Distinct sequence types are shown in different colors.
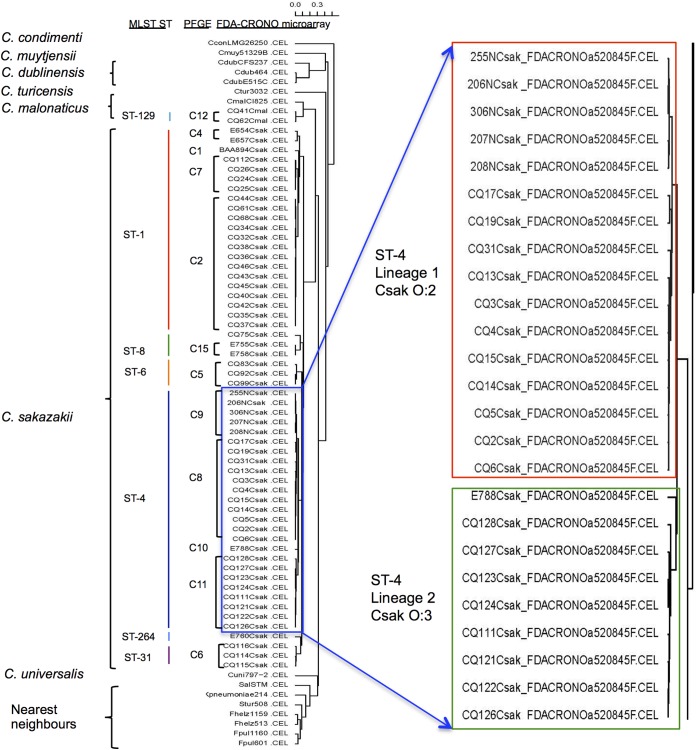
Phylogenetic analysis of Cronobacter species based on a custom-designed multigenome DNA microarray.
Microarray analysis aptly distinguished the seven Cronobacter species from one another and from non-Cronobacter species, which were used as controls for this custom-designed pan-genome microarray. Within each species, the isolates grouped into various distinct subclusters based on their pan-genomic diversity (Fig. 3). In addition, the microarray analysis separated C. sakazakii isolates into six clusters, and the strains clearly segregated according to sequence type. Interestingly the microarray also placed 25 ST-4 isolates into two distinct subclusters. Microarray gene differences noted between the two lineages are shown in Tables S2 and S3 in the supplemental material. Strains from lineage 1 differed from those in lineage 2 by 24 genes, of which 7 were phage related. In contrast, isolates within lineage 2 differed from those in lineage 1 across 71 genes, of which 17 were associated with the pESA3-harbored type 6 secretion system (T6SS) gene cluster (41).
FIG 3.
The multigenome microarray identification of Cronobacter species and the comparison with MLST and PFGE subtyping method results.
To better understand this diversity, PCR analysis of these 25 isolates was carried out using primers to detect plasmid pESA3 and the presence or absence of four regions within the T6SS gene cluster, as described by Franco et al. (41). Table S4 in the supplemental material summarizes the results of this PCR analysis. All 25 isolates were PCR positive for the single plasmid IncFIB incompatibility group replication protein gene repA (ESA_pESA3; location 115 to 588). Further PCR analysis of the T6SS in the 25 isolates revealed that most of the isolates representing each lineage possessed the 5′ end of the T6SS gene cluster. However, none of the ST4 lineage 1 isolates possessed the vgrG gene, a known T6SS effector protein (42), while eight out of nine ST-4 lineage 2 isolates did. This lineage-specific pattern was repeated for the 3′ targets associated with this end of the T6SS gene cluster.
Bacterial motility assays.
Bacterial motility is a phenotype that can support the survival of an organism in a given ecological niche. Cronobacter species are by definition a motile genus; nonetheless, few studies have explored this phenotype in a collection of isolates cultured from the PIF production environment. Salmonella Typhimurium DT104 13348 was included for comparison purposes, as it demonstrated a suitable swim and swarm phenotype (39, 40). After 8 h of incubation at 37°C, 34 isolates (20.2%) were observed to have an increased swim phenotype, with 1 (0.6%) exhibiting no change in its swimming ability and 133 others (79.2%) displaying reduced swim activity compared to the reference strain (Fig. 4; see also Table S1 in the supplemental material). By extending the incubation time to a total of 24 h at 37°C, 25 isolates (14.9%) were observed to have reduced swim activity, with 143 of the remaining isolates (85.1%) able to spread across the entire plate, indicating that these isolates possessed the same swim activities as the reference. The reference strain Salmonella Typhimurium DT104 13348 demonstrated good swarming activity; however, in comparison with the reference, all Cronobacter isolates exhibited a reduced swarming activity at both 8 and 24 h when incubated at 37°C (see Table S1).
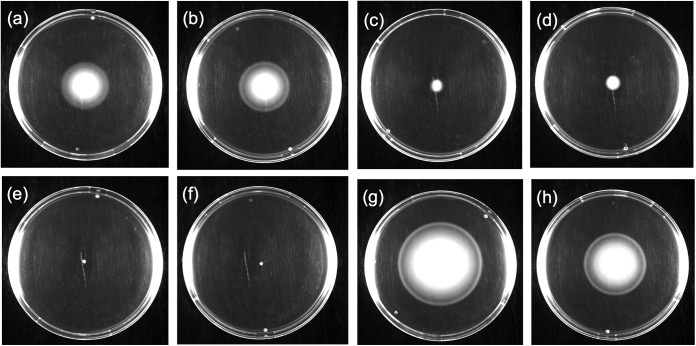
FIG 4.

Swimming ability of Cronobacter isolates compared to that of the reference strain S. Typhimurium DT104 13348. The reference strain showed an average swimming activity of 60 mm after 8 h of incubation at 37°C. (a) E657, C. sakazakii serotype O:1, clinical origin; (b) CQ8, C. sakazakii serotype O:4, PIF origin; (c) ATCC BAA-894, C. sakazakii serotype O:1, PIF origin; (d) 206N, C. sakazakii serotype O:2, clinical origin.
Biofilm formation under defined substrate growth conditions.
The ability to form a biofilm under laboratory-defined conditions for all Cronobacter isolates was studied. This was investigated by incubating bacterial cultures in M9 minimal medium at 28 and 37°C in standard microtiter plates (see Table S1 in the supplemental material). Salmonella Typhimurium ATCC 14028, a strong biofilm-forming strain (39), was included as the positive control. When incubated at 28°C, the formation of a strong biofilm was observed with 115 isolates (68.5%), whereas 35 isolates (20.8%) were defined as moderate biofilm formers and 18 isolates (10.7%) demonstrated weak biofilm formation. Similarly, when exposed to a temperature of 37°C, 119 isolates (70.8%) formed strong biofilms, with 32 isolates (19.1%) producing moderate biofilms and 17 isolates (10.1%) forming weak biofilms. Interestingly, temperature-dependent biofilm formation was observed among 36 isolates. Fifteen isolates produced strong biofilms at 28°C and formed moderate biofilms when incubated at 37°C. Twenty strong biofilm formers at 37°C produced moderate or weak biofilms at 28°C, while one moderate biofilm former at 37°C showed weak biofilm formation when incubated at 28°C.
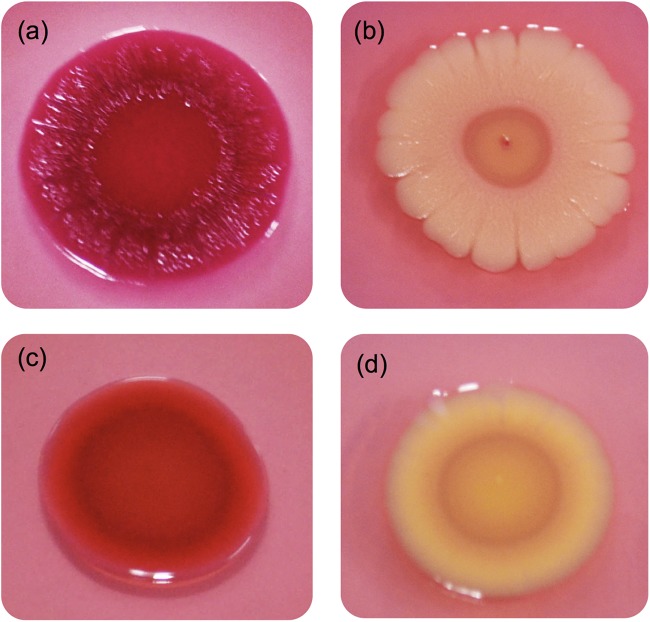
Morphotypes of Cronobacter species.
All Cronobacter isolates were incubated separately on LB agar plates supplemented with either Congo red or calcofluor. The colony color and morphology were recorded as an indication of the binding of the Congo red dye (Fig. 5) and cellulose production (see Table S1 in the supplemental material). Salmonella Typhimurium ATCC 14028 was selected as the reference strain (40). Four morphotypes were noted, as shown in Fig. 5, and included red, dry, and rough (RDAR) (Fig. 5a), brown, dry, and rough (BDAR) (Fig. 5b), red and smooth (RAS) (Fig. 5c), and brown and smooth (BAS) (Fig. 5d) types. The reference strain was characterized as RDAR (Fig. 5a). The most common morphotype among the Cronobacter species studied was BAS, as noted for 95 isolates (56.5%), followed by 43 isolates (25.6%) that were identified as BDAR, 21 isolates (12.5%) with the RAS morphotype, and 9 isolates (5.4%) with the RDAR morphotype. Only isolates defined by either the RDAR or BDAR morphotypes were considered positive for the Congo red dye binding assay, and these accounted for 31.0% of the tested isolates. Isolates that exhibited a RAS morphotype (12.5%) were considered to exhibit reduced binding of the Congo red dye, while those with the BAS morphotype (56.5%) did not show any binding of the Congo red dye.
FIG 5.
Four types of cell morphology observed with Congo red dye binding. (a) red, dry, and rough (RDAR); (b) brown, dry, and rough (BDAR); (c) red and smooth (RAS); (d) brown and smooth (BAS).
Production of cellulose was detected through monitoring the fluorescent signal observed at 366 nm under UV light. The reference strain generated a strong fluorescent signal. Most Cronobacter isolates (118 isolates, 70.2%) showed weak cellulose production, while 50 isolates (29.8%) were negative for this phenotype (see Table S1 in the supplemental material).
Comparative analysis of selected phenotypes expressed by Cronobacter ST-1 and -4 when recovered from various sources.
In order to determine whether or not the origin of a Cronobacter isolate would reflect its phenotype, we sought to evaluate a number of correlations among the isolates in the various groups, as shown in Table 3. When all ST-1 isolates were compared against those of ST-4 regardless of origins, significant differences in the ability to swim (when measured for 8 and 24 h) and to bind Congo red dye (P = 0.000) were noted (Table 3). Analysis of these phenotypic comparisons showed that STs had significant negative correlations with swim activities at both 8 h (r = −0.414, P = 0.000) and 24 h (r = −0.527, P = 0.000), as well as with the ability to form biofilms under laboratory-defined conditions at 37°C (r = −0.353, P = 0.000) (Table 3). In contrast, correlations that were significantly positive were noted for biofilm formation at 28°C (r = 0.173, P = 0.044) and Congo red dye binding (r = 0.310, P = 0.000) (Table 3). Similar correlations were observed between ST-1/PIF and ST-4/PIF isolates; however, a significant difference was only observed in the ability to swarm after 24 h of incubation between ST-1/clinical and ST-4/clinical isolates (Table 3).
TABLE 3.
Bivariate correlation analysis results for all phenotypic traits investigateda
| Comparison | Swimmingb |
Swarmingb |
Biofilm formation atc: |
Cellulose productiond | Congo red dye bindinge | |||
|---|---|---|---|---|---|---|---|---|
| 8 h | 24 h | 8 h | 24 h | 28°C | 37°C | |||
| Overall ST-1 vs ST-4 | −0.414** | −0.527** | 0.015 | 0.125 | 0.173* | −0.353** | 0.048 | 0.310* |
| ST-1/PIF vs ST-4/PIF | −0.651** | −0.667** | 0.055 | 0.106 | 0.346** | −0.231** | 0.030 | 0.292** |
| ST-1/clinical vs ST-4/clinical | 0.000 | 0.000 | −0.415 | 0.562* | 0.451 | 0.041 | −0.182 | 0.162 |
| ST-1/PIF vs ST-1/clinical | −0.009 | 0.000 | 0.147 | −0.232* | −0.195 | −0.222* | 0.099 | 0.061 |
| ST-4/PIF vs ST-4/clinical | 0.717** | 0.480** | −0.181 | 0.180 | −0.592** | −0.510** | 0.053 | −0.016 |
| ST-1/PIF vs ST-4/clinical | 0.199* | 0.000 | −0.056 | 0.102 | −0.237* | −0.456** | 0.067 | 0.212* |
| ST-4/PIF vs ST-1/clinical | 0.258 | 0.226 | 0.269 | −0.377* | −0.374* | −0.245 | 0.140 | −0.137 |
The phenotypic traits investigated in the various isolate groups were analyzed on the basis of STs and sample origins by using Spearman's rho coefficient test (data shown are the resulting r values). Statistical significance is indicated by asterisks: **, P ≤ 0.01; *, P ≤ 0.05.
In the motility assays (including swimming and swarming tests), the diameter of each colony was measured after incubation and normalized against that of the reference strain.
Optical densities (at 570 nm) for each isolate were normalized to that of the reference strain.
Cellulose production, when lacking (negative), was assigned a score of 1, while weak production was assigned a score of 2.
Congo red dye binding results were interpreted as follows: the RAS group was assigned a score of 1; the BAS group score was 2 (both RAS and BAS results were considered negative for Congo red dye binding); the RDAR group was assigned a score of 3; the BDAR group received a score of 4 (both the RDAR and BDAR groups were considered positive for Congo red dye binding).
When the bacterial isolates defined as ST-1/PIF and ST-1/clinical were compared, a significantly different phenotype (P = 0.000) was observed for cellulose production. The origin of an isolate (PIF or clinical, as shown here) demonstrated a significant negative correlation with the ability to swarm after 24 h of incubation (r = −0.232, P = 0.023) and to form a biofilm at 37°C (r = −0.222, P = 0.038) (Table 3). Similarly, when ST-4/PIF and ST-4/clinical were compared, phenotypic differences were observed in the ability to swim (when measured at both 8 and 24 h; P = 0.000) and to form a biofilm at both 28°C (P = 0.000) and 37°C (P = 0.009). In this case, there was a significantly positive correlation with the ability to swim after incubation for 8 h (r = 0.717, P = 0.000) or 24 h (r = 0.480, P = 0.001) and a negative ability to form biofilms at both 28°C (r = −0.592, P = 0.000) and 37°C (r = −0.510, P = 0.000) (Table 3).
When ST-1/PIF isolates were compared against the recognized ST-4/clinical isolates, statistically significant differences in biofilm formation at both 28°C (P = 0.001) and 37°C (P = 0.000), as well as the Congo red dye binding (P = 0.014) were noted. Furthermore, some of these results correlated positively between the two groups, specifically in the case of their swim phenotype (r = 0.199, P = 0.047) and the Congo red dye binding (r = 0.212, P = 0.034) (Table 3). In contrast, negative correlations were observed when comparing biofilm formation at both 28°C (r = −0.237, P = 0.018) and 37°C (r = −0.456, P = 0.000) (Table 3). Finally, when ST-4/PIF and ST-1/clinical groups were compared, the statistically significant differences observed included the ability to swim (P = 0.000) and the production of cellulose (P = 0.002). ST-4/PIF isolates appeared to have a greater ability to swarm (r = −0.377, P = 0.024) and to form stronger biofilms at 28°C (r = −0.374, P = 0.025) than ST-1/clinical isolates (Table 3).
DISCUSSION
Molecular subtyping methods have been recognized for their utility in supporting epidemiology investigations involving bacteria of importance to public health. Data presented in this study were obtained via several techniques, including targeted gene-based PCR, PFGE, MLST, and a recently described pan-genomic-based DNA microarray, to molecularly characterize a collection of 168 Cronobacter isolates. These isolates were cultured from PIF and the production environment, along with others obtained from clinical cases. Following molecular subtyping, phenotypic features related to the survival of Cronobacter in limited nutrient environments, such as clinical and PIF production environments, were investigated.
The predominant Cronobacter species cultured from the four PIF production environments studied was C. sakazakii. This finding is in agreement with others, including Müller et al. (32), who reported on the microbial ecology of a Swiss PIF facility, Mozrová et al. (43), who studied a dairy farm and its environment in the Czech Republic, and Pan et al. (20), who reported on Cronobacter species found in commercially available PIF products.
Cronobacter sakazakii O:1 (53.4%) was identified as a common serotype detected in PIF and its manufacturing environment, followed by C. sakazakii O:2 (18.0%) in this study. In contrast, in the Swiss investigation, Müller et al. (32) reported C. sakazakii O:2 (62.4%) was the most common serotype, followed by C. sakazakii O:7 (17.0%). These differences may reflect the diversity among the genus serotypes associated with facilities located in these regions.
The 133 Cronobacter isolates cultured from PIF and its associated production environment, along with the additional 35 clinical and type strains, were further investigated using PFGE and MLST (Fig. 1 and 2). The PFGE data revealed that these contaminations occurred within a facility or among different facilities. The latter observation could be attributed to the transfer of ingredients between these locations, as part of the formulation steps involved in the production of PIF. Notably, the isolates in cluster C1 appeared to persist in a PIF production facility for a period of at least 18 months, and the contaminations occurred in the production environment and the finished product within this facility. Isolates in cluster C4 appeared to persist for a period of over 9 months and contaminated the production environment. Cronobacer isolates in cluster C7 persisted for a period of 17 months in these facilities. Cluster C9 isolates were cultured from finished product, base powder, and the production environments and appeared to persist for a period of at least 16 months.
We were able to identify 14 different pulsotypes by using PFGE, while MLST identified 13 STs, one of which was a new sequence type. Interestingly, the PFGE profiles showed differences between isolates of PIF and those of clinical origin by clustering these separately, while MLST did not differentiate between them (Fig. 2b). Most of the isolates within a given pulsotype were associated with a single ST or clonal complex. Some exceptions to this included isolates within clusters C9 and -10, each of which included one isolate designated ST-1, while all remaining isolates within the cluster belonged to ST-4. This observation has been reported previously, and our data support these earlier studies, which stated that combining both PFGE and MLST as a subtyping approach would improve accuracy (20).
Two earlier studies have been reported that entailed microarray-based protocols to investigate the genomic diversity within the genus Cronobacter. Healy et al. (44) used a microarray design platform based on 276 open reading frames, which were selected from C. sakazakii ATCC BAA-894, to determine the gene differences among five of the six Cronobacter species initially described by Iversen et al. (2). Kucerova et al. (45) constructed a 387,000-probe oligonucleotide microarray covering the whole genome of C. sakazakii ATCC BAA-894, in an effort to identify the pan-genome of Cronobacter using five of the seven recognized species.
The microarray approach reported here was developed for the molecular characterization of Cronobacter from foods, primarily to address source attribution in trace-back investigations and to investigate the genomic diversity and evolutionary history of Cronobacter species. Fifty-nine isolates selected from this study were compared directly with five nearest neighbors and seven type strains (Fig. 3). The microarray was able to accurately assess each strain's identity and could differentiate Cronobacter species from the nearest neighbors. Furthermore, the microarray results supported the rpoB-based identities of Cronobacter species as described by Stoop et al. (13) and Lehner et al. (17). The results also concurred with recently published studies that have discussed the phylogenetic divergence of the genus from the most recent common ancestral species into two major clusters, one consisting of C. dublinensis and C. muytjensii, and the other comprised of C. sakazakii, C. malonaticus, C. universalis, and C. turicensis as postulated by Grim et al. (46). C. condimenti was a distant outlier of these two clusters. Of note, these results also offer a more in-depth analysis to the recent proposal to include Enterobacter pulveris, E. helveticus, and E. turicensis as members of this genus (36), and the results support their reclassification as proposed by Stephan et al. (47). These data agree with the genome sequence information reported previously (48–50).
In addition, our microarray data suggest the existence of some evolving lineages at the nucleotide level for C. sakazakii compared with other members of the genus. This is consistent with the fact that C. sakazakii accounts for approximately 84.8% of the isolates studied using this protocol. As an example, ST-4 strains separated into two clades comprising distinct lineages, which differed in the presence or absence of genes associated with the pESA3-harbored T6SS. As is known for C. sakazakii ATCC BAA-894, the T6SS is a recently characterized protein secretion system that consists of 16 ORFs (ESA_pESA3p 05491 to -5506) (41). T6SSs have been studied primarily in the context of pathogenic bacterium-host interactions (51). Recent data suggest, however, that these versatile protein secretion systems may also function to promote commensal or mutualistic relationships between bacteria and eukaryotes, or to mediate cooperative or competitive interactions between bacteria (52). One hypothesis is that T6SSs may be involved in overall cell fitness, promulgating ecology-driven selection processes as cells interact with other cells present within their environment. Our data showed the presence of a repA gene in all of the isolates studied, which signified that these isolates possess the pESA3 common virulence plasmid. The vgrG (valine-glycine repeat G protein gene) codes for a T6SS effector protein and is a single-copy gene in this cluster. The VgrG protein also has related sequences that are distributed on the chromosome, but most of these are not associated with any other T6SS gene cluster (41). Furthermore, the chromosomal T6SS genes in C. sakazakii ATCC BAA-894 do not share significant homology at the nucleotide level with the pESA3 T6SS gene locus (45). These results suggest that this region of the virulence plasmid, in these strains, may be in “genetic flux,” with either insertions or deletions most likely occurring in the 3′ region of the gene locus. These observations further support the perceived changes in gene content measured via PCR positive and negative controls with relevance for this region, as described by Franco et al. (41). However, the reasons for these changes remain unknown.
Microarray analysis showed that within the C. sakazakii cluster, six STs were identified, while 11 PFGE pulsotypes could also be grouped (Fig. 3). Interestingly, the ST-4 isolates, which were divided into two subclusters denoted lineage 1 and lineage 2, also differentiated according to serotypes: C. sakazakii O:2 and C. sakazakii O:3, respectively. Previous results reported by Hariri et al. (6) suggested that ST-4 strains form a distinct cluster with related STs, such as ST-110, -107, and -108; this cluster has been defined as the ST-4 clonal complex. The finding of two closely related lineages among ST-4 strains further defines and improves the phylogenetic resolution of this important meningitis-causing group.
Phenotype correlation analysis showed significant differences (P < 0.01) with respect to swim phenotype exhibited after a 24-h incubation period, along with an ability to swarm at both 8 and 24 h. Isolates in lineage 1 demonstrated an increased ability to swarm at both 8 h (r = −0.650, P = 0.000) and 24 h (r = −0.442, P = 0.027), while isolates in lineage 2 had better swimming ability at 24 h (r = 0.579, P = 0.002) (data not shown).
Previously, MLST studies highlighted the importance of ST-4 isolates linked to serious cases of meningitis (6). The same ST has also been identified among nonclinical isolates, including those cultured from PIF and follow-up formula (20, 30, 32). An example of the latter is C. sakazakii SP291, which was included in this study but did not cluster with any of the 14 pulsotypes. This isolate was originally cultured from an environmental sample obtained from facility B and is historically known to persist in PIF production environments for a period of at least 30 months (53). In this study, ST-1 is identified as the most frequent sequence type recovered, following the screening of all four PIF production sites from different geographical regions (Fig. 2a), a feature reported previously by Pan et al. (20). Furthermore, an ST-1 isolate, C. sakazakii ATCC BAA-894, was cultured from a PIF source and had a pulsotype profile that matched that of a clinical isolate. This isolate was linked to the death of an infant who had consumed a portion of a contaminated batch of PIF in Tennessee, USA, in 2001 (5). These observations raise questions as to the nature of the phenotypic differences between ST-1 and ST-4, which may in part account for the dominance of each ST in different niche settings.
The phenotypes related to the survival of ST-1 and ST-4 isolates have not been compared previously, and moreover, such a comparison is warranted, based on the findings of this study. Phenotypic experiments designed to compare both sequence types were performed on 168 isolates. These experiments included comparisons between cell motility (swimming and swarming), biofilm formation, Congo red dye binding, and cellulose production (see Table S1 in the supplemental material). In general, Cronobacter isolates of ST-1 exhibited a greater ability to swim and to form biofilms at 37°C compared to ST-4 isolates, while the latter formed a stronger biofilm at 28°C and exhibited greater Congo red dye binding than the former (Table 3). Similarly, ST-1/PIF isolates demonstrated greater swarming activity and formed stronger biofilms at 37°C than did ST-1/clinical isolates; while among ST-4 the clinical isolates exhibited better swimming activity than PIF isolates (Fig. 6); however, the latter forms stronger biofilms at both 28 and 37°C compared to the former. More importantly, ST-1/PIF isolates formed a stronger biofilm at both 28 and 37°C than did ST-4/clinical isolates, while the latter exhibited a more active swimming ability and greater binding of Congo red dye. As demonstrated in the present study, ST-1 is a common sequence type that is cultured from PIF and the associated manufacturing environments investigated. The abilities of a bacterium to swim and to bind Congo red dye are known to be determinants related to virulence. The phenotypic differences between these two Cronobacter sequence types may in part explain why ST-4 isolates of clinical origin are more often linked to cases of meningitis. It is tempting to speculate that these observations may represent a type of pathoadaptation. Further, our findings are consistent with phenotypes described by Yan et al. (54) for C. sakazakii SP291, an ST-4 isolate whose origin was a PIF production environment.
FIG 6.
Comparisons among ST-1 and ST-4 isolates, of PIF or clinical origin, for the ability to swim. (a) CQ74, ST-1/PIF; (b) CQ113, ST-1/PIF; (c) E654, ST-1/clinical; (d) E657, ST-1/clinical; (e) CQ14, ST-4/PIF; (f) CQ27, ST-4/PIF; (g) 206N, ST-4/clinical; (h) E788, ST-4/clinical.
In conclusion, C. sakazakii O:1 ST-1 was found to be the most common sequence type cultured from four geographically distinct PIF production facilities. Seventy of 84 ST-1 C. sakazakii isolates clustered as a distinct pulsotype, and these could be recovered over an 18-month period. Phenotypic differences were noted when comparisons were made between ST-1 and -4 isolates, including differences in bacterial motility, biofilm formation, Congo red dye binding, and cellulose production, all of which are considered to be relevant for bacterial survival in these environments. These characteristics may contribute to the pathoadaptation of this pathogen as it becomes disseminated along the food chain, and they account for the epidemiological observations in both cases. The adaptation of this opportunistic pathogen to the PIF manufacturing environment could lead to the survival of these organisms in finished product and increase thereafter the risk of causing infections once the contaminated food is consumed. Further in-depth phenotypic characterizations may provide clues as to how this phenotype is controlled in response to critical signals and subsequently expressed. Moreover, this type of approach may highlight bacterial targets that could be useful in the development of biomarkers for use in control protocols.
Supplementary Material
ACKNOWLEDGMENTS
We thank Matthew McCusker and Sarah Finn for their assistance with the phenotype assays.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00359-15.
REFERENCES
- 1.Iversen C, Lehner A, Mullane N, Bidlas E, Cleenwerck I, Marugg J, Fanning S, Stephan R, Joosten H. 2007. The taxonomy of Enterobacter sakazakii: proposal of a new genus Cronobacter gen. nov. and descriptions of Cronobacter sakazakii comb. nov. Cronobacter sakazakii subsp. sakazakii, comb. nov., Cronobacter sakazakii subsp. malonaticus subsp. nov., Cronobacter turicensis sp. nov., Cronobacter muytjensii sp. nov., Cronobacter dublinensis sp. nov. and Cronobacter genomospecies 1. BMC Evol Biol 7:64. doi: 10.1186/1471-2148-7-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Iversen C, Mullane N, McCardell B, Tall BD, Lehner A, Fanning S, Stephan R, Joosten H. 2008. Cronobacter gen. nov., a new genus to accommodate the biogroups of Enterobacter sakazakii, and proposal of Cronobacter sakazakii gen. nov., comb. nov., Cronobacter malonaticus sp. nov., Cronobacter turicensis sp. nov., Cronobacter muytjensii sp. nov., Cronobacter dublinensis sp. nov., Cronobacter genomospecies 1, and of three subspecies, Cronobacter dublinensis subsp. dublinensis subsp. nov., Cronobacter dublinensis subsp. lausannensis subsp. nov. and Cronobacter dublinensis subsp. lactaridi subsp. nov. Int J Syst Evol Microbiol 58:1442–1447. doi: 10.1099/ijs.0.65577-0. [DOI] [PubMed] [Google Scholar]
- 3.Joseph S, Cetinkaya E, Drahovska H, Levican A, Figueras MJ, Forsythe SJ. 2012. Cronobacter condimenti sp. nov., isolated from spiced meat, and Cronobacter universalis sp. nov., a species designation for Cronobacter sp. genomospecies 1, recovered from a leg infection, water and food ingredients. Int J Syst Evol Microbiol 62:1277–1283. doi: 10.1099/ijs.0.032292-0. [DOI] [PubMed] [Google Scholar]
- 4.Hunter CJ, Bean JF. 2013. Cronobacter: an emerging opportunistic pathogen associated with neonatal meningitis, sepsis and necrotizing enterocolitis. J Perinatol 33:581–585. doi: 10.1038/jp.2013.26. [DOI] [PubMed] [Google Scholar]
- 5.Himelright I, Harris E, Lorch V, Anderson M, Jones T, Craig A, Kuehnert M, Forster T, Arduino M, Jensen B, Jernigan D. 2002. Enterobacter sakazakii infections associated with the use of powdered infant formula—Tennessee, 2001. MMWR Morb Mortal Wkly Rep 51:297–300. [PubMed] [Google Scholar]
- 6.Hariri S, Joseph S, Forsythe SJ. 2013. Cronobacter sakazakii ST4 strains and neonatal meningitis, United States. Emerg Infect Dis 19:175–177. doi: 10.3201/eid1901.120649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stephan R, Lehner A, Tischler P, Rattei T. 2011. Complete genome sequence of Cronobacter turicensis LMG 23827, a food-borne pathogen causing deaths in neonates. J Bacteriol 193:309–310. doi: 10.1128/JB.01162-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Derzelle S, Dilasser F. 2006. A robotic DNA purification protocol and real-time PCR for the detection of Enterobacter sakazakii in powdered infant formulae. BMC Microbiol 6:100. doi: 10.1186/1471-2180-6-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Drudy D, O'Rourke M, Murphy M, Mullane NR, O'Mahony R, Kelly L, Fischer M, Sanjaq S, Shannon P, Wall P, O'Mahony M, Whyte P, Fanning S. 2006. Characterization of a collection of Enterobacter sakazakii isolates from environmental and food sources. Int J Food Microbiol 110:127–134. doi: 10.1016/j.ijfoodmicro.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 10.Liu Y, Cai X, Zhang X, Gao Q, Yang X, Zheng Z, Luo M, Huang X. 2006. Real time PCR using TaqMan and SYBR Green for detection of Enterobacter sakazakii in infant formula. J Microbiol Methods 65:21–31. doi: 10.1016/j.mimet.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 11.Mullane N, O'Gaora P, Nally JE, Iversen C, Whyte P, Wall PG, Fanning S. 2008. Molecular analysis of the Enterobacter sakazakii O-antigen gene locus. Appl Environ Microbiol 74:3783–3794. doi: 10.1128/AEM.02302-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Molloy C, Cagney C, O'Brien S, Iversen C, Fanning S, Duffy G. 2009. Surveillance and characterisation by pulsed-field gel electrophoresis of Cronobacter spp. in farming and domestic environments, food production animals and retail foods. Int J Food Microbiol 136:198–203. doi: 10.1016/j.ijfoodmicro.2009.07.007. [DOI] [PubMed] [Google Scholar]
- 13.Stoop B, Lehner A, Iversen C, Fanning S, Stephan R. 2009. Development and evaluation of rpoB based PCR systems to differentiate the six proposed species within the genus Cronobacter. Int J Food Microbiol 136:165–168. doi: 10.1016/j.ijfoodmicro.2009.04.023. [DOI] [PubMed] [Google Scholar]
- 14.Fricker-Feer C, Cernela N, Bolzan S, Lehner A, Stephan R. 2011. Evaluation of three commercially available real-time PCR based systems for detection of Cronobacter species. Int J Food Microbiol 146:200–202. doi: 10.1016/j.ijfoodmicro.2011.02.006. [DOI] [PubMed] [Google Scholar]
- 15.Jarvis KG, Grim CJ, Franco AA, Gopinath G, Sathyamoorthy V, Hu L, Sadowski JA, Lee CS, Tall BD. 2011. Molecular characterization of Cronobacter lipopolysaccharide O-antigen gene clusters and development of serotype-specific PCR assays. Appl Environ Microbiol 77:4017–4026. doi: 10.1128/AEM.00162-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun Y, Wang M, Liu H, Wang J, He X, Zeng J, Guo X, Li K, Cao B, Wang L. 2011. Development of an O-antigen serotyping scheme for Cronobacter sakazakii. Appl Environ Microbiol 77:2209–2214. doi: 10.1128/AEM.02229-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lehner A, Fricker-Feer C, Stephan R. 2012. Identification of the recently described Cronobacter condimenti by a rpoB-gene-based PCR system. J Med Microbiol 61:1034–1035. doi: 10.1099/jmm.0.042903-0. [DOI] [PubMed] [Google Scholar]
- 18.Cai XQ, Yu HQ, Ruan ZX, Yang LL, Bai JS, Qiu DY, Jian ZH, Xiao YQ, Yang JY, Le TH, Zhu XQ. 2013. Rapid detection and simultaneous genotyping of Cronobacter spp. (formerly Enterobacter sakazakii) in powdered infant formula using real-time PCR and high resolution melting (HRM) analysis. PLoS One 8:e67082. doi: 10.1371/journal.pone.0067082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jarvis KG, Yan QQ, Grim CJ, Power KA, Franco AA, Hu L, Gopinath G, Sathyamoorthy V, Kotewicz ML, Kothary MH, Lee C, Sadowski J, Fanning S, Tall BD. 2013. Identification and characterization of five new molecular serogroups of Cronobacter spp. Foodborne Pathog Dis 10:343–352. doi: 10.1089/fpd.2012.1344. [DOI] [PubMed] [Google Scholar]
- 20.Pan Z, Cui J, Lyu G, Du X, Qin L, Guo Y, Xu B, Li W, Cui Z, Zhao C. 2014. Isolation and molecular typing of Cronobacter spp. in commercial powdered infant formula and follow-up formula. Foodborne Pathog Dis 11:456–461. doi: 10.1089/fpd.2013.1691. [DOI] [PubMed] [Google Scholar]
- 21.Mullane NR, Whyte P, Wall PG, Quinn T, Fanning S. 2007. Application of pulsed-field gel electrophoresis to characterise and trace the prevalence of Enterobacter sakazakii in an infant formula processing facility. Int J Food Microbiol 116:73–81. doi: 10.1016/j.ijfoodmicro.2006.12.036. [DOI] [PubMed] [Google Scholar]
- 22.Mullane N, Healy B, Meade J, Whyte P, Wall PG, Fanning S. 2008. Dissemination of Cronobacter spp. (Enterobacter sakazakii) in a powdered milk protein manufacturing facility. Appl Environ Microbiol 74:5913–5917. doi: 10.1128/AEM.00745-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.El-Sharoud WM, O'Brien S, Negredo C, Iversen C, Fanning S, Healy B. 2009. Characterization of Cronobacter recovered from dried milk and related products. BMC Microbiol 9:24. doi: 10.1186/1471-2180-9-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Terragno R, Salve A, Pichel M, Epszteyn S, Brengi S, Binsztein N. 2009. Characterization and subtyping of Cronobacter spp. from imported powdered infant formulae in Argentina. Int J Food Microbiol 136:193–197. doi: 10.1016/j.ijfoodmicro.2009.10.013. [DOI] [PubMed] [Google Scholar]
- 25.Craven HM, McAuley CM, Duffy LL, Fegan N. 2010. Distribution, prevalence and persistence of Cronobacter (Enterobacter sakazakii) in the nonprocessing and processing environments of five milk powder factories. J Appl Microbiol 109:1044–1052. doi: 10.1111/j.1365-2672.2010.04733.x. [DOI] [PubMed] [Google Scholar]
- 26.Miled-Bennour R, Ells TC, Pagotto FJ, Farber JM, Kerouanton A, Meheut T, Colin P, Joosten H, Leclercq A, Besse NG. 2010. Genotypic and phenotypic characterisation of a collection of Cronobacter (Enterobacter sakazakii) isolates. Int J Food Microbiol 139:116–125. doi: 10.1016/j.ijfoodmicro.2010.01.045. [DOI] [PubMed] [Google Scholar]
- 27.Brengi SP, O'Brien SB, Pichel M, Iversen C, Arduino M, Binsztein N, Jensen B, Pagotto F, Ribot EM, Stephan R, Cernela N, Cooper K, Fanning S. 2012. Development and validation of a PulseNet standardized protocol for subtyping isolates of Cronobacter species. Foodborne Pathog Dis 9:861–867. doi: 10.1089/fpd.2012.1161. [DOI] [PubMed] [Google Scholar]
- 28.Yan QQ, Fanning S. 2015. Pulsed-field gel electrophoresis (PFGE) for pathogenic Cronobacter species, p 55–69. In Jordan K, Marion D (ed), Pulsed field gel electrophoresis: methods and protocols. Springer Science, New York, NY. [DOI] [PubMed] [Google Scholar]
- 29.Joseph S, Sonbol H, Hariri S, Desai P, McClelland M, Forsythe SJ. 2012. Diversity of the Cronobacter genus as revealed by multilocus sequence typing. J Clin Microbiol 50:3031–3039. doi: 10.1128/JCM.00905-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baldwin A, Loughlin M, Caubilla-Barron J, Kucerova E, Manning G, Dowson C, Forsythe S. 2009. Multilocus sequence typing of Cronobacter sakazakii and Cronobacter malonaticus reveals stable clonal structures with clinical significance which do not correlate with biotypes. BMC Microbiol 9:223. doi: 10.1186/1471-2180-9-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Joseph S, Forsythe SJ. 2012. Insights into the emergent bacterial pathogen Cronobacter spp., generated by multilocus sequence typing and analysis. Front Microbiol 3:397. doi: 10.3389/fmicb.2012.00397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Müller A, Stephan R, Fricker-Feer C, Lehner A. 2013. Genetic diversity of Cronobacter sakazakii isolates collected from a Swiss infant formula production facility. J Food Prot 76:883–887. doi: 10.4315/0362-028X.JFP-12-521. [DOI] [PubMed] [Google Scholar]
- 33.Gičová A, Oriešková M, Oslanecová L, Drahovská H, Kaclíková E. 2014. Identification and characterization of Cronobacter strains isolated from powdered infant foods. Lett Appl Microbiol 58:242–247. doi: 10.1111/lam.12179. [DOI] [PubMed] [Google Scholar]
- 34.Cruz-Cordova A, Rocha-Ramirez LM, Ochoa SA, Gonzalez-Pedrajo B, Espinosa N, Eslava C, Hernandez-Chinas U, Mendoza-Hernandez G, Rodriguez-Leviz A, Valencia-Mayoral P, Sadowinski-Pine S, Hernandez-Castro R, Estrada-Garcia I, Munoz-Hernandez O, Rosas I, Xicohtencatl-Cortes J. 2012. Flagella from five Cronobacter species induce pro-inflammatory cytokines in macrophage derivatives from human monocytes. PLoS One 7:e52091. doi: 10.1371/journal.pone.0052091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Seo KH, Brackett RE. 2005. Rapid, specific detection of Enterobacter sakazakii in infant formula using a real-time PCR assay. J Food Prot 68:59–63. [DOI] [PubMed] [Google Scholar]
- 36.Brady C, Cleenwerck I, Venter S, Coutinho T, De Vos P. 2013. Taxonomic evaluation of the genus Enterobacter based on multilocus sequence analysis (MLSA): proposal to reclassify E. nimipressuralis and E. amnigenus into Lelliottia gen. nov. as Lelliottia nimipressuralis comb. nov. and Lelliottia amnigena comb. nov., respectively, E. gergoviae and E. pyrinus into Pluralibacter gen. nov. as Pluralibacter gergoviae comb. nov. and Pluralibacter pyrinus comb. nov., respectively, E. cowanii, E. radicincitans, E. oryzae and E. arachidis into Kosakonia gen. nov. as Kosakonia cowanii comb. nov., Kosakonia radicincitans comb. nov., Kosakonia oryzae comb. nov. and Kosakonia arachidis comb. nov., respectively, and E. turicensis, E. helveticus and E. pulveris into Cronobacter as Cronobacter zurichensis nom. nov., Cronobacter helveticus comb. nov. and Cronobacter pulveris comb. nov., respectively, and emended description of the genera Enterobacter and Cronobacter. Syst Appl Microbiol 36:309–319. doi: 10.1016/j.syapm.2013.03.005. [DOI] [PubMed] [Google Scholar]
- 37.Tall BD, Gangiredla J, Gopinath GR, Yan Q, Chase HR, Lee B, Hwang S, Trach L, Park E, Yoo Y, Chung T, Jackson SA, Patel IR, Sathyamoorthy V, Pava-Ripoll M, Kotewicz ML, Carter L, Iversen C, Pagotto F, Stephan R, Lehner A, Fanning S, Grim CJ. 2015. Development of a custom-designed, pan genomic DNA microarray to characterize strain-level diversity among Cronobacter spp. Front Pediatr 3:1–11. doi: 10.3389/fped.2015.00036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jackson SA, Patel IR, Barnaba T, LeClerc JE, Cebula TA. 2011. Investigation the global genomic diversity of Escherichia coli using a multi-genome DNA microarray platform with novel gene prediction strategies. BMC Genomics 12:349. doi: 10.1186/1471-2164-12-349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martins M, McCusker MP, McCabe EM, O'Leary D, Duffy G, Fanning S. 2013. Evidence of metabolic switching and implications for food safety from the phenome(s) of Salmonella enterica serovar Typhimurium DT104 cultured at selected points across the pork production food chain. Appl Environ Microbiol 79:5437–5449. doi: 10.1128/AEM.01041-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Finn S, Hinton JC, McClure P, Amezquita A, Martins M, Fanning S. 2013. Phenotypic characterization of Salmonella isolated from food production environments associated with low-water activity foods. J Food Prot 76:1488–1499. doi: 10.4315/0362-028X.JFP-13-088. [DOI] [PubMed] [Google Scholar]
- 41.Franco AA, Kothary MH, Gopinath G, Jarvis KG, Grim CJ, Hu L, Datta AR, McCardell BA, Tall BD. 2011. Cpa, the outer membrane protease of Cronobacter sakazakii, activates plasminogen and mediates resistance to serum bactericidal activity. Infect Immun 79:1578–1587. doi: 10.1128/IAI.01165-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hachani A, Allsopp LP, Oduko Y, Filloux A. 2014. The VgrG proteins are “a la carte” delivery systems for bacterial type VI effectors. J Biol Chem 289:17872–17884. doi: 10.1074/jbc.M114.563429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mozrová V, Bøeňová N, Mrázek J, Lukešová D, Marounek M. 2014. Surveillance and characterisation of Cronobacter spp. in Czech retail food and environmental samples. Folia Microbiol (Praha) 59:63–38. doi: 10.1007/s12223-013-0266-2. [DOI] [PubMed] [Google Scholar]
- 44.Healy B, Huynh S, Mullane N, O'Brien S, Iversen C, Lehner A, Stephan R, Parker CT, Fanning S. 2009. Microarray-based comparative genomic indexing of the Cronobacter genus (Enterobacter sakazakii). Int J Food Microbiol 136:159–164. doi: 10.1016/j.ijfoodmicro.2009.07.008. [DOI] [PubMed] [Google Scholar]
- 45.Kucerova E, Clifton SW, Xia XQ, Long F, Porwollik S, Fulton L, Fronick C, Minx P, Kyung K, Warren W, Fulton R, Feng DY, Wollam A, Shah N, Bhonagiri V, Nash WE, Pepin KH, Wilson RK, McClelland M, Forsythe SJ. 2010. Genome sequence of Cronobacter sakazakii BAA-894 and comparative genomic hybridization analysis with other Cronobacter species. PLoS One 5:e9556. doi: 10.1371/journal.pone.0009556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Grim CJ, Kotewicz ML, Power K, Pagotto F, Gopinath G, Mammel MK, Jarvis KG, Yan QQ, Kothary MH, Franco AA, Patel IR, Jackson SA, Hu L, Sathyamoorthy V, Iversen C, Lehner A, Stephan R, Farber JM, Fanning S, Tall BD. 2013. Pan genome analysis of the emerging foodborne pathogen Cronobacter spp. suggests a species-level bidirectional divergence driven by niche adaption. BMC Genomics 14:366. doi: 10.1186/1471-2164-14-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stephan R, Grim CJ, Gopinath GR, Mammel MK, Sathyamoorthy V, Trach LH, Chase HR, Fanning S, Tall BD. 2014. Re-examination of the taxonomic status of Enterobacter helveticus, Enterobacter pulveris, and Enterobacter turicensis as members of the genus Cronobacter and their reclassification in the genera Franconibacter gen. nov. and Siccibacter gen. nov. as Franconibacter helveticus comb. nov., Franconibacter pulveris comb. nov. and Siccibacter turicensis comb. nov., respectively. Int J Syst Evol Microbiol 64:3402–3410. doi: 10.1099/ijs.0.059832-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stephan R, Grim CJ, Gopinath GR, Mammel MK, Sathyamoorthy V, Trach LH, Chase HR, Fanning S, Tall BD. 2013. Genome sequence of Enterobacter turicensis strain 610/05 (LMG 23731), isolated from fruit powder. Genome Announc 1:e00996-13. doi: 10.1128/genomeA.00996-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gopinath GR, Grim CJ, Tall BD, Mammel MK, Sathyamoorthy V, Trach LH, Chase HR, Fanning S, Stephan R. 2013. Genome sequences of two Enterobacter pulveris strains, 601/05T (=LMG 24057T =DSM 19144T) and 1160/04 (=LMG 24058 =DSM 19146), isolated from fruit powder. Genome Announc 1:e00991-13. doi: 10.1128/genomeA.00991-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Grim CJ, Gopinath GR, Mammel MK, Sathyamoorthy V, Trach LH, Chase HR, Tall BD, Fanning S, Stephan R. 2013. Genome sequence of an Enterobacter helveticus strain, 1159/04 (LMG 23733), isolated from fruit powder. Genome Announc 1:e01038-13. doi: 10.1128/genomeA.01038-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ma AT, McAuley S, Pukatzki S, Mekalanos JJ. 2009. Translocation of a Vibrio cholerae type VI secretion effector requires bacterial endocytosis by host cells. Cell Host Microbe 5:234–243. doi: 10.1016/j.chom.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jani AJ, Cotter PA. 2010. Type VI secretion: not just for pathogenesis anymore. Cell Host Microbe 8:2–6. doi: 10.1016/j.chom.2010.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cooney S. 2012. A study on the epidemiology and behavior of selected bacteria colonizing a powdered infant formula (PIF) low moisture production environment. Ph.D. dissertation University College Dublin, Dublin, Ireland. [Google Scholar]
- 54.Yan QQ, Power KA, Cooney S, Fox E, Gopinath GR, Grim CJ, Tall BD, McCusker MP, Fanning S. 2013. Complete genome sequence and phenotype microarray analysis of Cronobacter sakazakii SP291: a persistent isolate cultured from a powdered infant formula production facility. Front Microbiol 4:256. doi: 10.3389/fmicb.2013.00256. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.