Abstract
Genome-wide gene expression studies have indicated that the eukaryotic genome contains many gene pairs showing overlapping sense and antisense transcription. Regulation of these coding and/or noncoding gene pairs involves intricate regulatory mechanisms. In the present study, we utilized an enhanced green fluorescent protein (EGFP)-tagged reporter plasmid cis linked to a doxycycline-inducible antisense promoter, generating antisense transcription that fully overlaps EGFP, to study the mechanism and dynamics of gene silencing after induction of noncoding antisense transcription in undifferentiated and differentiating mouse embryonic stem cells (ESCs). We found that EGFP silencing is reversible in ESCs but is locked into a stable state upon ESC differentiation. Reversible silencing in ESCs is chromatin dependent and is associated with accumulation of trimethylated lysine 36 on histone H3 (H3K36me3) at the EGFP promoter region. In differentiating ESCs, antisense transcription-induced accumulation of H3K36me3 was associated with an increase in CpG methylation at the EGFP promoter. Repression of the sense promoter was affected by small-molecule inhibitors which interfere with DNA methylation and histone demethylation pathways. Our results indicate a general mechanism for silencing of fully overlapping sense-antisense gene pairs involving antisense transcription-induced accumulation of H3K36me3 at the sense promoter, resulting in reversible silencing of the sense partner, which is stabilized during ESC differentiation by CpG methylation.
INTRODUCTION
Strand-specific RNA sequencing analysis of the mammalian transcriptome has indicated that more than 20% of the sequenced transcripts belong to sense-antisense gene pairs (1). Many of these gene pairs show full overlap of at least one template or antisense transcription through the sense promoter and may consist of coding genes, noncoding genes, or a combination of coding and noncoding genes. Sense-antisense gene pairs are frequently found in imprinted gene clusters involved in setting up and maintaining parent-specific gene expression profiles. Imprinted gene loci are regulated by differentially methylated imprinting control regions (ICRs), which often direct parent-specific transcription of noncoding RNA (ncRNA) transcripts. Studies involving knockout alleles and alleles with introduced transcriptional stop sequences have indicated that these antisense noncoding genes play a crucial role in the regulation of the coding sense partner. For instance, the imprinted noncoding antisense genes Kcnq1ot1, Ube3a-ATS, Nespas, and Airn are the master regulators of the Kcnq1, Prader-Willi/Angelman syndrome, Gnas, and Igf2r clusters, respectively, and regulate these clusters by regulating the sense protein-coding partner. Kcnq1ot1-mediated repression of Kcnq1 and silencing of Igf2r by Airn does not depend on double-stranded RNA (dsRNA) molecules but has been attributed to the act of transcription involving transcription through the promoters of Kcnq1 and Igf2r (2, 3). This repression may involve the transcriptional interference mechanisms of the sense partner but may also include recruitment of chromatin remodeling complexes, leading to the local accumulation of histone modifications and DNA methylation, as was found for Nespas and Airn, respectively (4, 5). In addition, for Kcnq1ot1 and Airn it has been shown that recruitment of chromatin remodeling complexes is involved in the cis spreading of silencing toward nonoverlapping genes, leading to parent-specific inhibition of expression of flanking genes over long distances in cis (6, 7).
The Xist/Tsix gene pair represents one of the best-studied mammalian sense-antisense gene loci. In contrast to most imprinted gene loci, both Xist and Tsix are noncoding and the respective transcriptional activities or the transcribed ncRNAs are involved in mutually repressive mechanisms. Xist and Tsix, which fully overlaps Xist, are the main players in the X-chromosome inactivation (XCI) process. Random XCI occurs and can be studied in differentiating female mouse embryonic stem cells (ESCs) with two X chromosomes. Xist is upregulated on the future inactive X chromosome, and cis spreading and ncRNA-mediated recruitment of chromatin remodeling complexes, including PRC2, lead to inactivation of that one X chromosome. Tsix-mediated repression of Xist on the active X chromosome does not involve dsRNA or RNA interference mechanisms (8) but is dependent on Tsix antisense transcription through the Xist promoter, which leads to Xist promoter-associated changes in histone modifications and CpG methylation (9, 10). Whether this local recruitment of chromatin remodelers is ncRNA mediated or is dependent on transcription or transcriptional interference mechanisms is unknown.
The loss or gain of expression of a noncoding antisense partner of a sense gene has often been implicated in disease. For instance, in fragile X syndrome (FXS), a repeat expansion of a CGG repeat in the 5′ untranslated region of the human FMR1 gene results in induction of antisense transcription through the FMR1 promoter (11), initiating at the expanded repeat and producing an unstable noncoding transcript. This antisense transcription results in epigenetic silencing of FMR1, which involves CpG methylation of the expanded repeat. This silencing of FMR1 happens during a defined window of neuronal development (12). One form of alpha-thalassemia has been associated with the juxtaposition of LUC7L to HBA2, resulting in antisense transcription through HBA2. This aberrant antisense transcription leads to silencing of HBA2 and DNA methylation of its associated CpG island (CGI) by an unknown mechanism during a specific developmental time window (13). These examples highlight the close relationship between transcription, ncRNAs, and gene regulation with human disease in a developmental context.
For all these examples, the exact mechanisms involved in silencing of the sense partner by antisense transcription remain elusive, as the effects of the act of transcription and the biological activity of the respective ncRNA product cannot be separated. Therefore, a general question is whether transcriptional interference (e.g., collision of RNA polymerase II [Pol II] complexes, torsional strain, or displacement/occlusion of transcription factors/regulatory elements) or chromatin-mediated mechanisms are responsible for silencing of overlapping genes. Direct transcriptional interference has mostly been studied in prokaryotes and Saccharomyces cerevisiae, and it has been shown that collision of Pol II complexes and displacement/occlusion of transcription factors/regulatory elements do influence the activity of overlapping genes to some extent (reviewed in reference 14). In the case of Airn, transcriptional overlap with the Igf2r promoter alone is necessary and sufficient for silencing of Igf2r, thus precluding a role for the RNA molecule itself (3). However, chromatin modulation based on the process of transcription per se might still be involved. This model is supported by the observed association of Pol II with chromatin remodelers like Set2 (15), which catalyzes the deposition of trimethylated lysine 36 on histone H3 (H3K36me3) in transcribed regions. H3K36me3 in turn has been implicated in the recruitment of chromatin factors correlating with transcriptional repression, like histone deacetylases, histone demethylases, and DNA methyltransferases (16–18). In contrast, Airn, Kcnq1ot1, Xist, and other ncRNAs have been shown to recruit histone-modifying complexes like G9A and PRC2 in cis (6, 7, 19), suggesting the requirement of the RNA molecule itself. To be able to exclusively study the effects of antisense transcription, we have utilized an artificial sense-antisense gene pair consisting of an enhanced green fluorescent protein (EGFP) reporter and a fully overlapping inducible antisense transcription unit. Our studies indicate that antisense-mediated silencing of the EGFP gene is reversible in ESCs and is dependent on modifications of the chromatin environment. Interestingly, silencing is locked into a stable state upon ESC differentiation concomitantly with the accumulation of EGFP promoter-associated CpG methylation. Antisense transcription-induced silencing is augmented by blocking JARID1/JMJD2 family histone demethylases, suggesting that the transcription-coupled histone modification H3K36me3 provides a repressive environment for sense transcription initiation, which is locked into a stable state by CpG methylation upon ESC differentiation.
MATERIALS AND METHODS
Plasmids, reagents, and antibodies.
The plasmids used for the generation of transgenic lines and transient transfections were pTRE-Tight-BI-DsRed2 (Clontech) and pCAG-EGFP-N1, which was generated by replacing the cytomegalovirus promoter in pEGFP-N1 (Clontech) with the CAG promoter from pCAG-Rnf12-Flag (20). The reagents used were doxycycline (DOX), 5-aza-dC, suberoylanilide hydroxamic acid (SAHA), pyridine-2,4-dicarboxylic acid (2,4-PDCA), trans-2-phenylcyclopropylamine (2-PCPA), curcumin (CUR), pargyline (PAR), and JQ1 (all from Sigma).
Cell lines.
The culture media and conditions for ESC culture and differentiation have been described previously (21). The final concentration of doxycycline was 2 μg/ml. The final concentrations of the small-molecule inhibitors were 20 nM for 5-aza-dC, 400 nM for SAHA, 5 mM for 2,4-PDCA, 200 μM for 2-PCPA, 10 μM for CUR, 1.5 mM for PAR, and 150 nM for JQ1. Transgenic ESC lines were generated using a polymorphic male 129/Sv-Cast/Ei cell line harboring an M2rtTA transcriptional activator in the ROSA26 locus (22), as follows. A tetracycline-responsive ptet promoter excised from pTRE-Tight-BI-DsRed2 was inserted downstream and in the antisense direction of the EGFP in pCAG-EGFP-N1 (pCAG-EGFP-as-ptet). This construct was transfected into the M2 ESC line by electroporation (Gene Pulser Xcell; Bio-Rad), and stable clones with random integrations were obtained by 1 week of selection in 350 μg/ml G-418 (Gibco). Clones were screened for expression of EGFP and responsiveness to doxycycline. For transient transfections, M2 ESCs were cotransfected with pCAG-EGFP-as-ptet and pTRE-Tight-BI-DsRed2 using an Amaxa Nucleofector device and a mouse ESC Nucleofector kit (Lonza). After 18 h, EGFP-positive cells were sorted and used for experiments.
Fluorescence-activated cell sorter (FACS) analysis.
Single-cell suspensions were prepared by treatment with Tris-EDTA (TE) for 7 min at 37°C and 30 min of preplating to remove feeder cells, if necessary. Duplets were excluded by appropriate gating, and dead/dying cells were detected by staining with Hoechst 33258 (1 μg/ml; Molecular Probes). Relative fluorescence intensities (FIs) for EGFP and mCherry were determined. Cell analysis was performed on an LSRFortessa cell analyzer, and cell sorting was performed on a FACSAria III cell sorter (BD Biosciences) with FACSDiva software. Statistical analysis was performed with FlowJo software.
Strand-specific expression analysis.
RNA was isolated using the TRIzol reagent (Invitrogen) according to the manufacturer's instructions. DNase I treatment was performed to remove genomic DNA, and cDNA was prepared using SuperScript II reverse transcriptase (Invitrogen) with strand-specific primers for the target and control in the same reaction mixture. Quantitative reverse transcription-PCR (RT-PCR) was performed on a CFX384 real-time PCR detection system (Bio-Rad) using Fast SYBR green master mix (Applied Biosystems). The primers used are listed in Table S2 in the supplemental material. Results were normalized to those for actin using the change-in-threshold-cycle (ΔCT) method (23) and are mostly presented as the fold change in expression versus the levels of expression for the undifferentiated no-doxycycline-treated control.
Bisulfite sequencing.
Phenol-chloroform-extracted DNA was converted using an EpiTect bisulfite kit (Qiagen) following the manufacturer's instructions. Part of the CAG promoter was amplified from bisulfite-converted DNA with Platinum Taq polymerase (Invitrogen) using primers 204 and 207. The PCR product was gel purified and subcloned into the pGEM-T Easy vector (Promega), and the vector was transformed into bacteria. Single bacterial clones were isolated, and the fragment of the CAG promoter was amplified by colony PCR using primers 208 and 209, followed by Sanger sequencing using primer 302. Sequence reads were analyzed using the quantification tool for methylation analysis (24).
ChIP.
In short, for chromatin immunoprecipitation (ChIP), approximately 5 × 106 cells were cross-linked in a dish for 10 min at room temperature by adding 1/10 volume 11% buffered formaldehyde solution (50 mM HEPES-KOH, pH 7.6, 100 mM NaCl, 1 mM EDTA, pH 8, 0.5 mM EGTA, pH 8, 11% [vol/vol] formaldehyde) and cross-linking was quenched for 10 min at room temperature with 125 mM glycine. The cells were washed twice in ice-cold phosphate-buffered saline with protease inhibitors and resuspended in SDS lysis buffer (50 mM Tris-HCl, pH 8, 10 mM EDTA, pH 8, 1% SDS), followed by sonication until fragments of ca. 500 bp were obtained. Chromatin was diluted 1:10 in ChIP dilution buffer (16.7 mM Tris-HCl, pH 8, 167 mM NaCl, 1.2 mM EDTA, pH 8, 1.1% Triton X-100, 0.01% SDS) and incubated with antibodies overnight at 4°C. Chromatin was then incubated with preblocked protein G Dynabeads (Novex) for 1 h at 4°C. The beads were washed thrice in low-salt buffer (20 mM Tris-HCl, pH 8, 150 mM NaCl, 2 mM EDTA, pH 8, 1% Triton X-100, 0.1% SDS), once in high-salt buffer (20 mM Tris-HCl, pH 8, 500 mM NaCl, 2 mM EDTA, pH 8, 1% Triton X-100, 0.1% SDS), once in LiCl buffer (10 mM Tris-HCl, pH 8, 250 mM LiCl, 1 mM EDTA, 0.5% deoxycholate, 0.5% NP-40), and once in TE buffer (10 mM Tris-HCl, pH 8, 50 mM NaCl, 1 mM EDTA, pH 8). Complexes were eluted in elution buffer (10 mM Tris-HCl, pH 8, 150 mM NaCl, 1 mM EDTA, pH 8, 5 mM dithiothreitol, 1% SDS) for 15 min at 65°C, and cross-links were reversed by incubation overnight at 65°C. DNA fragments were recovered by proteinase K treatment followed by phenol-chloroform extraction and analyzed by quantitative RT-PCR. Enrichment was estimated by determining the original amount of template in the pulldown and input fractions as 2−CT(pulldown)/2−CT(input).
RESULTS
Inducible antisense transcription reversibly silences an EGFP reporter.
To be able to study the general effect of antisense transcription on gene regulation during development, we generated transgenic mouse ESC lines containing an EGFP reporter cassette and a doxycycline-responsive promoter in the antisense direction downstream of the reporter. This antisense ptet promoter was intended to initiate antisense transcription that fully overlaps the sense EGFP reporter. To this end, the tetracycline-responsive ptet promoter was inserted downstream and in the antisense direction of a CAG promoter-driven (25) EGFP (Fig. 1A). This construct was randomly integrated into previously established male ESCs originally generated from a cross of a male Cast/Ei mouse and a female 129/Sv mouse carrying an M2rtTA transcriptional activator in the ROSA26 locus (22), allowing induction of doxycycline (DOX)-dependent antisense transcription through the EGFP-coding sequence and its promoter. EGFP-positive clones were isolated and expanded, and reactivity to DOX was tested. Most clones showed a reduction in EGFP intensity upon DOX addition, and six clones, denominated M2-3, M2-4, M2-5, M2-8, M2-20, and M2-29, were used for further analysis (an example is shown in Fig. 1B).
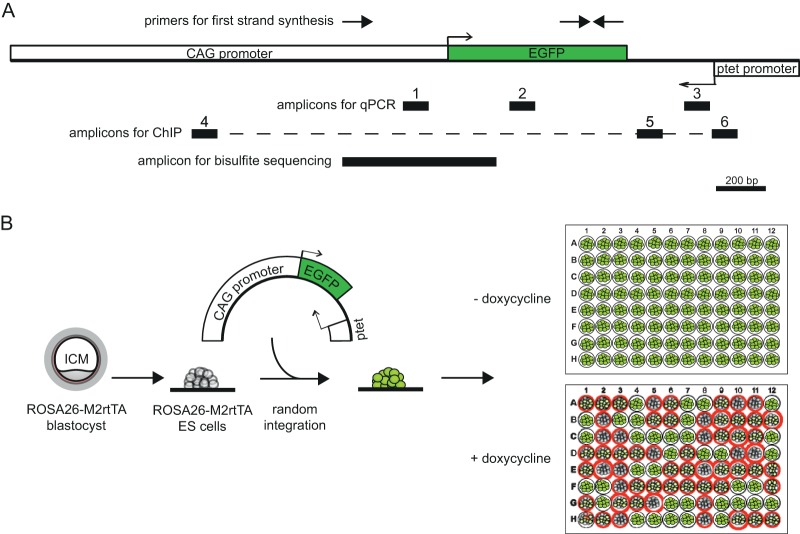
FIG 1.
Map of EGFP-antisense gene-ptet construct and transfection strategy. (A) Map (to scale) showing the construct used for generation of transgenic cell lines. Promoters and the EGFP open reading frame are depicted as white and green boxes, respectively. The location of the primers used for first-strand synthesis for strand-specific expression analysis is indicated on top, and the black boxes numbered 1 to 6 mark the locations of amplicons for expression analysis and ChIP analysis. The area amplified from bisulfite-treated DNA for bisulfite sequencing is shown on the bottom. (B) Cartoon depicting the transfection and screening strategy used to identify clones for further analysis. ESCs harboring an M2rtTA transcriptional activator in the ROSA26 locus were transfected, and stable random integration was forced by G-418 selection. EGFP-positive clones were expanded and plated in duplicate, and the responsiveness to doxycycline was tested in 96-well plates (as an example, red circles denote clones with a decreased level of EGFP expression).
When undifferentiated M2-3 ESCs were grown for 2 days in the presence of DOX and analyzed by FACS, the EGFP relative fluorescence intensity (FI) level was reduced to 40% of that of control cells. This demonstrates that antisense transcription can reduce the transcriptional activity of a sense partner in this experimental context, where sense and antisense partners are biologically unrelated (Fig. 2A and B; see Fig. S1 in the supplemental material). This silencing appeared to be very dynamic, as DOX removal within 1 day resulted in the almost complete recovery of the EGFP FI to the levels measured without the induction of antisense transcription (Fig. 2A and B; see Fig. S1 in the supplemental material). Comparable results were found with several other M2 clones. Due to heterogeneity between independent experiments, statistical significance for the difference between recovery and repression (with DOX treatment) was not reached for all clones. However, the trend was highly similar in all independent experiments. This is also reflected by the statistical significance for the difference when the data for all clones are pooled (Fig. 2B). To evaluate the time course of synthesis of both EGFP mRNA and the transcript originating from the ptet promoter, RNA was isolated from these pulse-chase experiments and strand-specific quantitative RT-PCR was performed. Two different sets of primers, one for amplification of a ptet proximal product and one for a ptet distal product, were used (Fig. 1A). As expected, quantification of the proximal transcript demonstrated reversible induction of the ptet promoter by doxycycline, which resulted in a concomitant reduction of the EGFP mRNA level. After DOX washout, the EGFP mRNA level recovered. As for fluorescence levels, heterogeneity between independent experiments was observed, but these independent experiments always showed the same trend. Therefore, not all clones showed statistically significant differences, whereas pooled data for all clones did (Fig. 2C). The same pattern was observed for the distal ptet amplicon, confirming that ptet-induced transcription fully overlaps the EGFP reporter and runs through the CAG promoter (Fig. 2D). These results show that, in the present system using undifferentiated mouse ESCs, repression of a coding gene by antisense transcription is completely reversible, so that the repression is dependent on continuous antisense transcription.
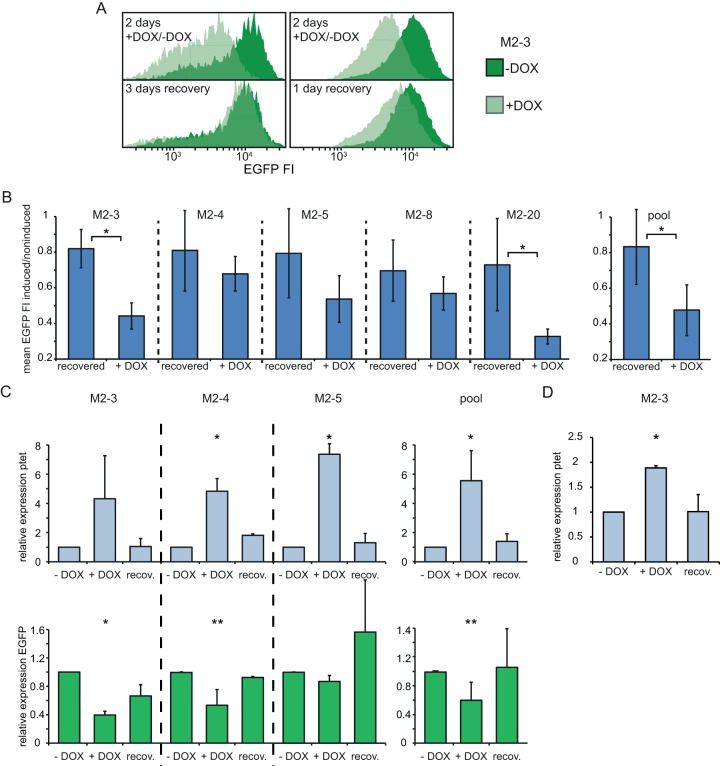
FIG 2.
Antisense transcription and EGFP reporter expression in undifferentiated ESCs. (A) Histograms of the EGFP FI distribution in clone M2-3 determined by FACS analysis. −Dox, uninduced cells; +Dox, cells treated with doxycycline. (Top left) Repression after 2 days of doxycycline treatment compared to that for the untreated control; (bottom left) the situation 3 days later, after doxycycline has been washed out; (top and bottom right) results of another experiment with the same experimental setup as that for the panels on the left, but with only 1 day of recovery from doxycycline induction. (B) The mean EGFP FI of induced and noninduced cells is shown after 2 days of doxycycline treatment (+DOX) and after 2 days of doxycycline treatment followed by 1 day of recovery (recovered). (C) Strand-specific expression analysis in noninduced (no DOX treatment [−DOX]), induced (+DOX), and recovered (recov.) cells, as outlined in the legend to panel B and the text. (Top) for the ptet proximal amplicon (amplicon 3 in Fig. 1A); (bottom) results for the EGFP amplicon (amplicon 2). Quantification is depicted as the fold change (relative expression) compared to the level of expression in noninduced cells. (D) Same as the top of panel C, but the results for the ptet distal amplicon are shown (amplicon 1). The mean and SD from at least two independent experiments for each clone and for all clones pooled are shown in panels B to D. *, P < 0.05 by a two-sample Student's t test (B) or single-factor analysis of variance (C, D); **, P < 0.1 by a two-sample Student's t test (B) or single-factor analysis of variance (C, D).
Antisense transcription changes the chromatin structure of the CAG promoter.
For several specific examples of sense-antisense gene pairs, it has been described that silencing of the sense partner is accompanied by changes in the promoter chromatin structure. In some cases, transcriptional overlap was found to be sufficient for silencing (3), but for other such gene pairs, a requirement for antisense ncRNA to recruit chromatin-modifying complexes has been reported (6). Thus, mechanisms of regulation appear to vary, and it is not fully understood how antisense transcription is converted into a repressive chromatin environment. Our ptet-EGFP system provides an experimental tool to study the effect of antisense transcription on the chromatin structure for specific genes and RNA sequences which are not taking part in a biological context in a sense-antisense regulation system. Hence, we anticipated that the present experiments would provide information regarding the more general aspects of such regulatory mechanisms.
Elongating Pol II itself interacts with a plethora of histone-modifying proteins, and we hypothesized that transcriptional read-through per se might be sufficient to create a specific repression-instructive chromatin signature in promoters. We were particularly interested in methylation of histone H3 at residues K4 and K36, which is catalyzed by two Pol II-associated proteins, SET1 (26) and SET2 (15), respectively. ChIP analysis of the CAG promoter in undifferentiated, uninduced M2-3 cells showed a strong enrichment of trimethylation of histone H3 at residue K4 (H3K4me3), while H3K36me3 levels were close to background levels (Fig. 3A). The H3K36me3 signal between the EGFP cassette and the ptet promoter (amplicon 5 in Fig. 1A) found in uninduced cells is most likely caused by read-through transcription derived from the EGFP cassette. Upon DOX addition, however, the H3K36me3 signal at the CAG promoter increased approximately 3-fold, with a concomitant decrease in H3K4me3 being detected (Fig. 3A; see Fig. 5C), thereby creating a specific chromatin environment that corresponded to the repressed expression of the EGFP reporter. The gain of H3K36me3 just downstream of the ptet promoter also demonstrates effective transcriptional elongation, while enrichment at the ptet promoter itself most likely reflects a lack of resolution or initiation of transcription slightly upstream of the amplicon tested by quantitative PCR (qPCR). This might also explain the lack of induction of the H3K4me3 signal at the ptet promoter (amplicon 6 in Fig. 1A) itself. Analogously to fluorescence and mRNA abundance measurements, this effect was almost completely reversible after DOX washout (Fig. 3A).
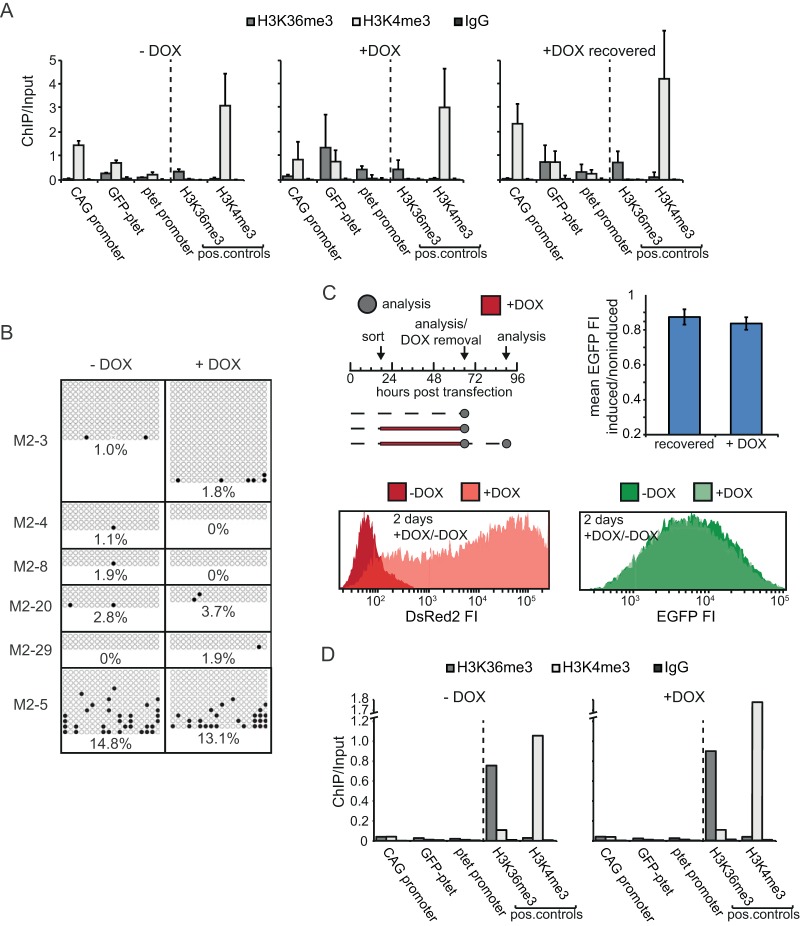
FIG 3.
Chromatin structure in the CAG and ptet promoter regions in undifferentiated ESCs. (A) ChIP-qPCR analysis of the region encompassing CAG and ptet promoters (the CAG promoter is amplicon 4 in Fig. 1A, GFP-ptet is amplicon 5, and the ptet promoter is amplicon 6) in the M2-3 clone for noninduced (−DOX), induced (+DOX), and recovered (+DOX recovered) conditions, as outlined in the text. Antibodies against H3K36me and H3K4me3 and against whole IgG as a mock-treated control were used, as indicated. The loci analyzed, including H3K36me3- and H3K4me3-positive controls, are indicated. Values are plotted as the ratio of the original amount of template DNA for pulldown and input fractions. The mean and SD from three independent experiments are shown. (B) Bisulfite sequencing of the CAG promoter in several clones treated with doxycycline (+DOX) or not treated with doxycycline (−DOX) for 2 days. For independent clones represented in the separate panels, filled circles represent methylated CpGs and empty circles represent unmethylated CpGs. Average percentages for each condition and clone are indicated. (C) (Top left) Timing of transient-transfection experiments. Arrows, time points of sorting, analysis, and doxycycline removal; dashed lines, times that cells were grown in the absence of doxycycline; red lines, times that cells were grown in the presence of doxycycline. (Bottom) Histograms of FACS analysis of the DsRed2 and EGFP FI distribution for conditions without and with doxycycline. (Top right) Mean EGFP FI of induced and noninduced cells after 2 days of doxycycline treatment and after 2 days of doxycycline treatment followed by 1 day of recovery (recovered). Error bars represent SDs from three independent experiments. (D) Exactly as described in the legend to panel A, but for transient transfections. pos., positive.
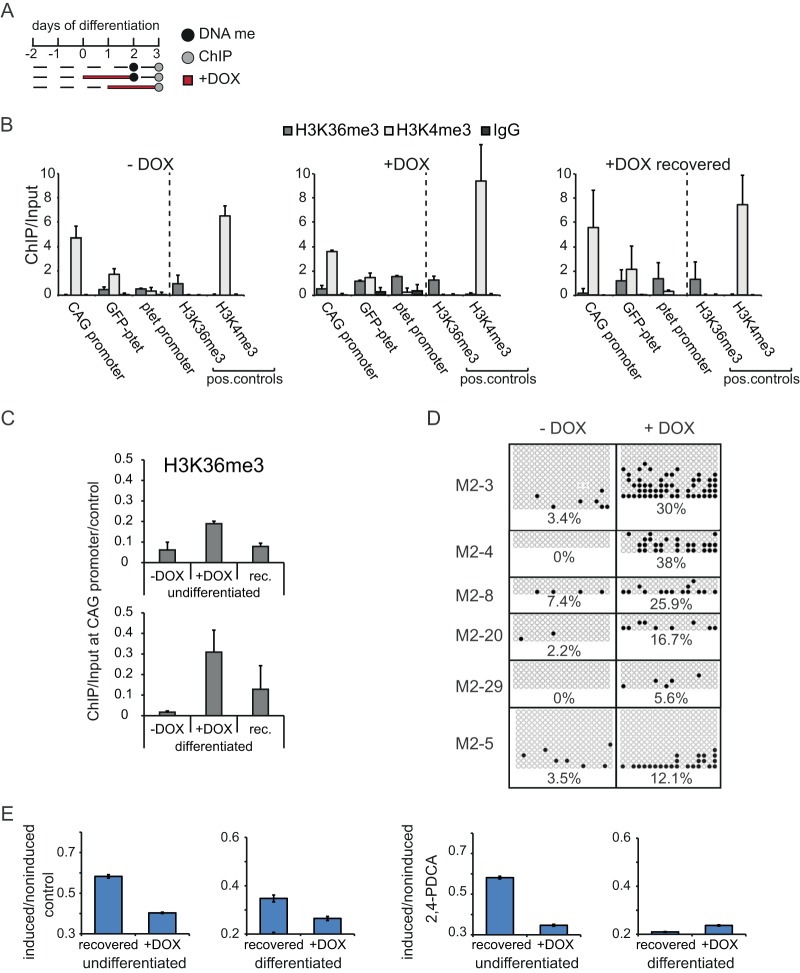
FIG 5.
Epigenetic modifications of the CAG promoter in differentiating ESCs. (A) Time schedule of induction experiments in differentiating ESCs. Dashed lines, times that cells were grown in the absence of doxycycline; red lines, times that cells were grown in the presence of doxycycline; gray dots, time points of ChIP analysis; black dots, time points of bisulfite sequencing. (B) ChIP-qPCR analysis of the region encompassing the CAG and ptet promoters in differentiating clone M2-3 under noninduced (−DOX), induced (+DOX), and recovered (+DOX recovered) conditions, as outlined in panel A and the text. Antibodies against H3K36me and H3K4me3 and against whole IgG as a mock-treated control were used, as indicated. The loci analyzed, including the H3K36me3- and H3K4me3-positive controls, are indicated. Values are plotted as the ratio of the original amount of template DNA for the pulldown and input fractions. The mean and SD from two independent experiments are shown. (C) Direct comparison of H3K36me3 levels at the CAG promoter between undifferentiated and differentiating cells. Values for undifferentiated cells (Fig. 3A) and differentiating cells (B) were divided by the corresponding values for the H3K36me3-positive controls for normalization. The mean and SD from two (differentiated) or three (undifferentiated) independent experiments are shown. rec., recovered. (D) Bisulfite sequencing of the CAG promoter in several differentiating clones treated with doxycycline or not treated with doxycycline for 2 days, as outlined in panel A. For the independent clones represented in the separate panels, filled circles represent methylated CpGs and empty circles represent unmethylated CpGs. Average percentages for each condition and clone are indicated. (E) Mean EGFP FI of induced and noninduced M2-3 cells (undifferentiated and differentiating) after 2 days of doxycycline treatment and after 2 days of doxycycline treatment followed by 1 day of recovery (recovered). All cells were treated with 2,4-PDCA. Error bars represent the SDs from three independent experiments.
CpG islands (CGIs) are CG-rich genomic regions which frequently initiate transcription and constitute more than 50% of all annotated promoters in vertebrates (27). Most CGIs remain unmethylated, but DNA methylation of CpG residues is correlated with the stable repression of transcription. Several promoters of developmentally regulated genes acquire DNA methylation during development (28, 29). To test if DOX-induced repression of the CAG promoter that drives EGFP expression and contains a CGI involves DNA methylation, we performed bisulfite sequencing on undifferentiated ESCs grown in the absence and presence of DOX. In most clones, the CAG promoter was found to be completely devoid of DNA methylation, regardless of the induction of antisense transcription (Fig. 3B). Only clone M2-5 showed higher levels of DNA methylation, but this was unresponsive to the induction of antisense transcription, meaning that this higher level most likely is related to a position effect. Thus, antisense transcription generates a special chromatin state at an unrelated promoter that is located nearby and transcribed in the sense direction. This sense partner is reversibly repressed in cis. Importantly, this effect is not dependent on any specific RNA sequences or locus requirements.
Antisense transcription-mediated repression requires an intact chromatin template.
ChIP analysis of the CAG promoter suggested that antisense transcription induces a specific chromatin signature over promoters. Next, we asked whether chromatin modifications are important for silencing by antisense transcription. We therefore exploited transient transfection as a system in which the regular chromatin structure is perturbed (30, 31). The same sense-EGFP-antisense-ptet construct that was used for the generation of M2 cell clones was transiently transfected into M2rtTA-ROSA26 male ESCs, and EGFP-positive cells were sorted after 18 h into medium containing DOX or no DOX. As a control for DOX induction, a ptet-DsRed construct was cotransfected. After 2 days of either DOX or no DOX treatment, cells were analyzed by FACS. For another set of cells, DOX was removed after 48 h and cells were left to recover for an additional 24 h before FACS analysis. Surprisingly, addition of DOX almost completely failed to repress EGFP transcription from the transiently transfected plasmid, even though DOX induction of ptet transcription per se was functional, as demonstrated by the expression of DsRed (Fig. 3C). Thus, recovery from DOX treatment did not significantly increase EGFP FI levels compared to those obtained under the induced condition (Fig. 3C). To study if a perturbed chromatin arrangement on a transiently transfected template carries chromatin modifications as they are laid down by the transcription machinery and that are thus involved in the specific chromatin state induced by antisense transcription, ChIP was performed on transiently transfected cells. Intriguingly, neither H3K4me3 nor H3K36me3 was found to reside on the transiently transfected plasmid (Fig. 3D). Thus, even though EGFP expression and ptet induction are not hampered on a transiently transfected template devoid of the histone modifications H3K4me3 and H3K36me3, repression of EGFP mediated by antisense transcription does not occur in that situation.
Antisense transcription-mediated repression during ESC differentiation.
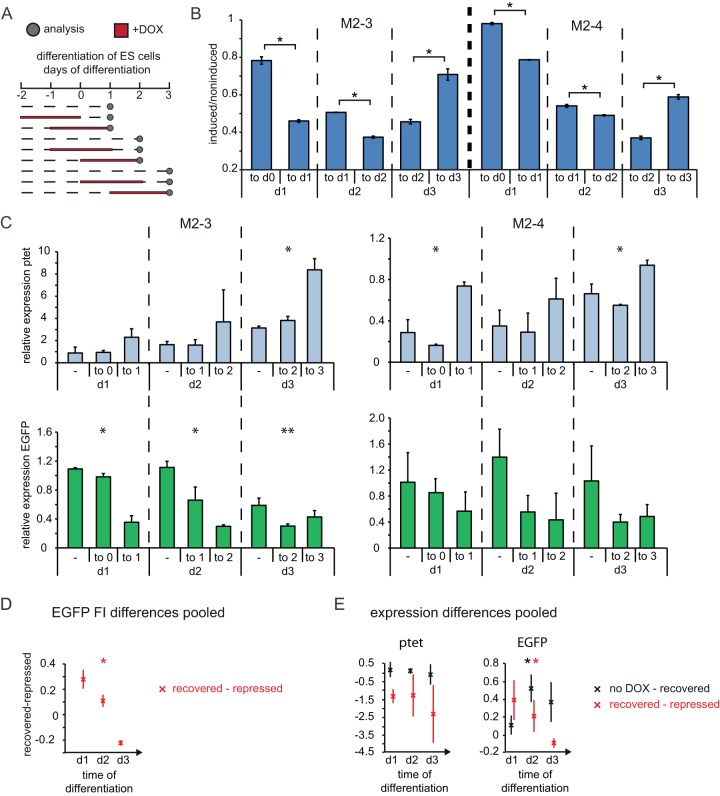
To investigate the effect of antisense transcription on expression of the EGFP reporter during differentiation, we performed pulse-chase-type time course experiments. Cells were differentiated by removal of feeders and leukemia inhibitory factor (LIF) and kept in culture until day 3 of differentiation. We opted for this window of time because, even in the absence of DOX, the EGFP reporter was increasingly silenced at later time points in all clones analyzed. Moreover, in the present system, 50% of female ESCs initiated XCI during the first 3 days of differentiation, demonstrating that during this developmental time window, major epigenetic and gene regulatory changes occur. The different conditions for days 1, 2, and 3 of differentiation were (i) no DOX treatment, (ii) 2 days of DOX treatment followed by washout and 1 day of recovery, and (iii) addition of DOX for 2 days before analysis (Fig. 4A). FACS analysis showed that while DOX treatment until the start of differentiation did not interfere with the recovery of FI levels, the DOX treatment exerted a stronger inhibitory effect on recovery if DOX was administered during differentiation (Fig. 4B). Plotting of the difference in the EGFP FI ratios between recovered and repressed cells for all clones revealed significant differences over the time of differentiation (Fig. 4D). This difference trended toward zero, demonstrating that at later time points of differentiation cells are not able to recover from antisense transcription-induced EGFP repression (Fig. 4D). In addition, when ptet antisense transcription through the EGFP cassette was induced from days 1 to 3 of differentiation, repression of the EGFP was attenuated. This suggests either that antisense transcription at the onset of differentiation is important for proper silencing of the antisense partner or that at later time points during differentiation any kind of forced expression helps to maintain the locus in an open conformation. To test whether the loss of reversible silencing during differentiation is caused by general repression of the EGFP reporter and to verify that induction of transcription from the ptet promoter is working under differentiation conditions, RNA was isolated from cells in the same time course experiments and transcripts emanating from the ptet and CAG promoters were quantified by strand-specific qPCR. The abundance of ptet transcripts increased 1.5- to 3-fold upon DOX addition at all time points analyzed, and this expression reverted to levels similar to those obtained under the noninduced condition after DOX removal (Fig. 4C), demonstrating that the inducible system functions normally in differentiating cells. Of note, the ptet promoter appeared to become increasingly derepressed while differentiation lasted. Upon addition or washout of DOX, EGFP transcripts displayed the expected anticorrelation with ptet-derived transcription, mirroring the data obtained by FACS analysis (Fig. 4C). Plotting the differences in expression levels for all clones pooled, we found that ptet levels do not differ significantly over the time of differentiation. The difference between uninduced and recovered cells clustered around zero, showing that ptet expression is effectively terminated upon DOX washout, whereas the difference between recovered and repressed cells was negative, showing that ptet expression can be induced at all time points during differentiation (Fig. 4E, left). In contrast, the levels of EGFP expression change significantly over the time of differentiation. The difference between uninduced and recovered cells increased, whereas the difference between recovered and repressed cells decreased, demonstrating that EGFP expression levels do not recover from DOX-induced repression at later time points of differentiation (Fig. 4E, right). Importantly, the EGFP mRNA abundance in the absence of DOX did not substantially decrease during differentiation but stayed at levels comparable to those in undifferentiated cells. Taken together, these data indicate that antisense transcription during differentiation might lead to stable repression of a gene on the opposite strand, in contrast to the reversible silencing observed in undifferentiated cells.
FIG 4.
Stable repression of CAG promoter by antisense transcription in differentiating ESCs. (A) Time schedule of induction experiments in differentiating ESCs. Dashed lines, times that cells were grown in the absence of doxycycline; red lines, times that cells were grown in the presence of doxycycline; dots, time points of analysis. (B) The mean EGFP FI of induced and noninduced cells is shown for cells treated for 2 days with doxycycline until the day of analysis and for cells treated for 2 days followed by 1 day of recovery, as outlined in panel A. The upper label on the x axis gives the time period of doxycycline treatment, and the lower label indicates the day of analysis. (C) Strand-specific expression analysis in noninduced, recovered, and induced cells, as outlined in panel A and the text. The upper label on the x axis gives the time period of doxycycline treatment (−, noninduced), and the lower label indicates the day of analysis. (Top) ptet proximal amplicon (amplicon 3 in Fig. 1a); (bottom) EGFP amplicon (amplicon 2). Quantification is depicted as the fold change compared to the level of expression of noninduced, undifferentiated cells. (D) Difference in EGFP FI ratios from panel B between cells after doxycycline washout (recovered) and under doxycycline treatment (repressed). (E) Difference in relative expression of ptet and EGFP from panel C between cells with no doxycycline treatment (no DOX) and recovered cells and between recovered and repressed cells. The mean and SD from two to three independent experiments for each clone are shown in panels B and C, and the mean and SD for all clones pooled are shown in panels D and E. *, P < 0.05 by a two-sample Student's t test (B) or single-factor analysis of variance (C to E); **, P < 0.1 by a two-sample Student's t test (B) or single-factor analysis of variance (C to E). d0, d1, d2, and d3, days 0, 1, 2, and 3, respectively.
Chromatin structure induced by antisense transcription during differentiation.
Since we observed that transcriptional antisense read-through resulted in a specific chromatin signature at the CAG promoter without altering DNA methylation levels in undifferentiated ESCs, we next asked what effect antisense transcription would have on chromatin structure during differentiation. Therefore, differentiating cells were grown in the absence or presence of DOX or were allowed to recover for 1 day after DOX removal, and ChIP analysis of H3K4me3 and H3K36me3 was performed at day 3 of differentiation (Fig. 5A to C). Similar to the findings for undifferentiated cells without DOX induction in the absence of ptet transcription, H3K4me3 was found to be strongly enriched at the CAG promoter, while H3K36me3 levels were close to the background levels. In DOX-induced cells, H3K36me3 became enriched at the CAG promoter, while H3K4me3 levels decreased slightly, suggesting that antisense transcription through the CAG promoter creates a specific chromatin environment also during differentiation. However, at 1 day after DOX washout, reversal of chromatin marks to the noninduced state was less complete than it was in undifferentiated cells, even though the absolute drop was equally prominent (Fig. 5C). This might be attributed either to the enhanced levels of DOX-induced ptet transcription during later phases of differentiation (Fig. 4C) or to a more stable silencing of EGFP. DNA methylation, which is believed to be important for stable repression of the inactivated X chromosome (32) and several other genes (29), is highly dynamic during embryonic development and is essential for embryonic development (33, 34). We therefore tested if DNA methylation is involved in the repression of the EGFP reporter by bisulfite sequencing of cells differentiated for 2 days. Strikingly, all clones analyzed displayed a marked increase in DNA methylation in the CAG promoter which was strictly dependent on antisense transcription (Fig. 5D). The values ranged from 0% to 8% methylated CpGs without DOX addition to up to 40% CpG methylation after 2 days of DOX treatment. These findings indicate that antisense transcription generates a particular chromatin signature and, contrary to the situation in undifferentiated ESCs, is capable of inducing promoter-associated CGI DNA methylation only in the specific context of differentiation. These events might lead, in turn, to stable gene repression.
To follow up on the observations that properly assembled chromatin and specific combinations of chromatin modifications may have a role in the antisense transcription-mediated repression of the EGFP reporter, we used several small-molecule inhibitors (see Table S1 in the supplemental material) to interfere with enzymes that catalyze DNA methylation or the addition or removal of histone modifications. As described above, pulse-chase-type experiments were performed on undifferentiated and differentiating M2-3 cells, but this time the experiments were performed in the presence of these inhibitors. Most inhibitors had only minimal effects on repression and recovery in undifferentiated and differentiating cells (see Fig. S2 in the supplemental material). However, in contrast to all other small-molecule inhibitors used in this study, 2,4-PDCA, a histone demethylase inhibitor with high specificity for JARID1 and JMJD2 family demethylases, which are responsible for H3K4me3 and H3K36me3 demethylation, respectively (35), strongly enhanced both the direct repressive effect and stable silencing by antisense transcription during differentiation (Fig. 5E). Taken together, these data point to a general role for H3K4me3 and/or H3K36me3 in antisense transcription-mediated repression and the establishment of silent chromatin at the CAG promoter driving EGFP. In particular, maintenance of the silent state, which is put into place only during differentiation, appears to be influenced by the H3K4me3 and H3K36me3 histone modifications.
DISCUSSION
Transcriptional interference mechanisms have been thoroughly studied in prokaryotes and yeast (reviewed in reference 14). These studies indicate transcriptional inhibition of the sense gene of a sense-antisense gene pair, which might involve transcription-invoked torsional effects or transcriptional collision. In addition, studies of the SRG1/SER3 system in yeast have implicated SRG1 transcription-dependent nucleosome occupancy in the SER3 promoter in the repression of SER3 (36, 37). Torsional or topological effects have also been implicated in transcriptional interference in higher eukaryotes (38), and for the Xist/Tsix locus, chromatin remodeling of the Xist promoter by overlapping Tsix transcription has been shown to be involved in Xist silencing (9, 10). Our findings indicate that antisense-mediated repression of a sense gene is absent on transiently transfected templates, precluding an important role for direct transcriptional interference in silencing of the reporter. Our ChIP experiments revealed the absence of chromatin modifications that are normally found on templates randomly integrated at different positions in the genome, suggesting that histone modifications play a key role in antisense-mediated repression of fully overlapping sense-antisense gene pairs.
We found that silencing of a stably integrated reporter plasmid is reversible in ESCs and is accompanied by an increase in H3K36me3 and a reduction of H3K4me3 in the CAG promoter region driving EGFP transcription. Interestingly, during the ESC differentiation process repression of EGFP is stabilized concomitantly with partial maintenance of accumulated H3K36me3, the loss of H3K4me3, and a significant increase in CpG methylation at the EGFP promoter. Addition of inhibitors interfering with specific epigenetic pathways had little effect on green fluorescent protein (GFP) expression during and after recovery of antisense transcription for most compounds tested. However, addition of 2,4-PCDA had a pronounced effect on EGFP expression in differentiating ESCs both during doxycycline-induced antisense transcription and after recovery from this antisense transcription. The compound 2,4-PCDA inhibits H3K4me3 and H3K36me3 demethylases, so the results suggest that accumulation of these modifications leads to silencing of the CAG promoter, which might also be the case for any other gene promoter with overlapping antisense transcription. The observed effect of 2,4-PCDA was also present, but less pronounced, in undifferentiated ESCs and was possibly related to the reversibility of the silencing process. Interestingly, treatment of differentiating ESCs with the DNA methylation inhibitor 5-aza-dC revealed the opposite effect, resulting in an increase in EGFP expression, pointing to a specific role for DNA methylation in silencing of antisense promoters in a developmental context.
Although we cannot formally exclude the possibility of a role for the noncoding antisense RNA in this process, the synthetic nature of our reporter construct favors a model where the act of transcription and Pol II-associated chromatin remodelers play a crucial role in the regulation of sense-antisense gene pairs where at least one of the genes initiates transcription through the promoter of the other gene. In yeast, the methyltransferase Set2 associates with Pol II, and the resulting accumulation of H3K36me3 in the gene body is important for recruitment of the histone deacetylase Rpd3 (15, 16, 39). In mammals, H3K36me3 is involved in recruitment of the H3K4me3 demethylase KDM5B (17) and the DNA de novo methyltransferase DNMT3A (18), implicating a role for H3K36me3 in the repression of transcription from cryptic intragenic promoters. Our data are consistent with these findings and suggest that promoter-associated H3K36me3 reversibly represses transcription initiation in ESCs, which might be dependent on the recruitment of KDM5B, whose function is associated with ESC self-renewal. Upon ESC differentiation, H3K36me3-enriched gene bodies, including our antisense-transcribed EGFP reporter, might be targeted specifically by DNMT3A, which is upregulated during this differentiation process. Whether silencing of the EGFP promoter requires only H3K36me3 or is also dependent on H3K4me3 needs further investigation. Similar observations have been made for Xist and Tsix, two endogenous overlapping gene loci. The loss of Tsix antisense transcription through the Xist promoter has been shown to result in promoter-associated chromatin changes, allowing aberrant initiation of Xist transcription (9, 10, 40). In addition, forced Tsix expression during development results in Xist promoter methylation (41). Xist promoter methylation is also required to stably repress Xist at later stages of development in the absence of ongoing Tsix transcription, which is shut down in differentiated cells (42). Also for the imprinted Igf2r/Airn locus, Airn antisense transcription through the Igf2r promoter is required for silencing of Igf2r (5). Studies with an inducible Airn promoter indicate that antisense Airn transcription leads to CpG methylation of the Igf2r promoter, stabilizing the silent state, which can then be maintained in the absence of Airn transcriptional read-through (43). The present findings obtained for an experimental sense-antisense gene pair in undifferentiated and differentiating ESCs, taken together with the above-described findings on physiological gene pairs, clearly demarcate a developmental time window where irreversible silencing is established. Our experimental system provides a powerful tool to study the respective regulatory mechanisms.
In prokaryotes and yeast, it has been found that genes with a clear on-off switch show an enrichment for antisense expression from a neighboring locus (44). This antisense transcription has been implicated in providing thresholds that need to be overcome for sense genes to be expressed (45). Also in higher eukaryotes, the best-studied sense-antisense fully overlapping gene pairs, including Xist/Tsix and Igf2r/Airn, show such a binary switch pattern in gene expression during development, where the antisense partner provides a threshold for initiation of transcription of the sense gene. Our findings with an engineered reporter indicate that fully overlapping sense-antisense and, possibly, sense-sense gene pairs might be subject to a general silencing mechanism which does not involve transcriptional interference but at least partially relies on transcription-mediated accumulation of histone modifications in promoters, leading to gene silencing (Fig. 6). Our model does not exclude alternative models like nucleosome occupancy-mediated repression of overlapping promoters, as was observed for the SRG1/SER3 locus in yeast (36, 37), or ncRNA-mediated recruitment of chromatin modifiers at specific loci, as proposed for Airn, Kcnq1ot1, and Xist (6, 7, 19). Rather, several complementary mechanisms might act cooperatively to ensure the faithful regulation of overlapping gene pairs. Genome-wide strand-specific transcriptome sequencing and ChIP sequencing studies will be required to determine whether such a general surveillance mechanism is indeed active in mammalian systems.
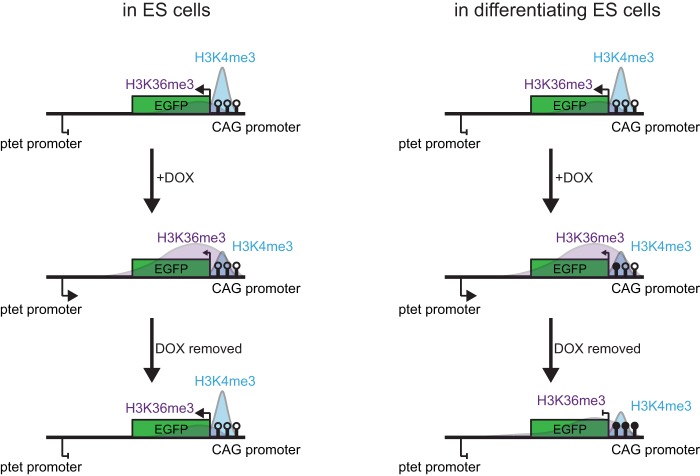
FIG 6.
Model of chromatin-mediated repression of sense-antisense gene pairs. In undifferentiated ES cells (left), transcription through the CAG promoter driving EGFP expression results in a specific chromatin signature at and repression of this promoter. This effect is completely dependent on antisense transcription and is thus reversible upon doxycycline washout. (Right) In contrast, antisense transcription in differentiating ES cells leads to accumulation of DNA methylation (filled circles) in the CAG promoter and stable silencing of EGFP, even if antisense transcription is stopped at later stages of differentiation. Only histone modifications in the CAG promoter and downstream region are displayed.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by NWO VICI (project no. 865.10.003) and ERC grants (project no. 260587).
We thank all members of the Department of Developmental Biology, Erasmus MC, for helpful discussions.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.00029-15.
REFERENCES
- 1.Katayama S, Tomaru Y, Kasukawa T, Waki K, Nakanishi M, Nakamura M, Nishida H, Yap CC, Suzuki M, Kawai J, Suzuki H, Carninci P, Hayashizaki Y, Wells C, Frith M, Ravasi T, Pang KC, Hallinan J, Mattick J, Hume DA, Lipovich L, Batalov S, Engstrom PG, Mizuno Y, Faghihi MA, Sandelin A, Chalk AM, Mottagui-Tabar S, Liang Z, Lenhard B, Wahlestedt C, RIKEN Genome Exploration Research Group, Genome Science Group (Genome Network Project Core Group), FANTOM Consortium. 2005. Antisense transcription in the mammalian transcriptome. Science 309:1564–1566. doi: 10.1126/science.1112009. [DOI] [PubMed] [Google Scholar]
- 2.Thakur N, Tiwari VK, Thomassin H, Pandey RR, Kanduri M, Gondor A, Grange T, Ohlsson R, Kanduri C. 2004. An antisense RNA regulates the bidirectional silencing property of the Kcnq1 imprinting control region. Mol Cell Biol 24:7855–7862. doi: 10.1128/MCB.24.18.7855-7862.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Latos PA, Pauler FM, Koerner MV, Senergin HB, Hudson QJ, Stocsits RR, Allhoff W, Stricker SH, Klement RM, Warczok KE, Aumayr K, Pasierbek P, Barlow DP. 2012. Airn transcriptional overlap, but not its lncRNA products, induces imprinted Igf2r silencing. Science 338:1469–1472. doi: 10.1126/science.1228110. [DOI] [PubMed] [Google Scholar]
- 4.Williamson CM, Ball ST, Dawson C, Mehta S, Beechey CV, Fray M, Teboul L, Dear TN, Kelsey G, Peters J. 2011. Uncoupling antisense-mediated silencing and DNA methylation in the imprinted gnas cluster. PLoS Genet 7:e1001347. doi: 10.1371/journal.pgen.1001347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sleutels F, Zwart R, Barlow DP. 2002. The non-coding Air RNA is required for silencing autosomal imprinted genes. Nature 415:810–813. doi: 10.1038/415810a. [DOI] [PubMed] [Google Scholar]
- 6.Pandey R, Mondal T, Mohammad F, Enroth S, Redrup L, Komorowski J, Nagano T, Mancini-Dinardo D, Kanduri C. 2008. Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol Cell 32:232–246. doi: 10.1016/j.molcel.2008.08.022. [DOI] [PubMed] [Google Scholar]
- 7.Nagano T, Mitchell JA, Sanz LA, Pauler FM, Ferguson-Smith AC, Feil R, Fraser P. 2008. The Air noncoding RNA epigenetically silences transcription by targeting G9a to chromatin. Science 322:1717–1720. doi: 10.1126/science.1163802. [DOI] [PubMed] [Google Scholar]
- 8.Nesterova TB, Popova BC, Cobb BS, Norton S, Senner CE, Tang YA, Spruce T, Rodriguez TA, Sado T, Merkenschlager M, Brockdorff N. 2008. Dicer regulates Xist promoter methylation in ES cells indirectly through transcriptional control of Dnmt3a. Epigenetics Chromatin 1:2. doi: 10.1186/1756-8935-1-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sado T, Hoki Y, Sasaki H. 2005. Tsix silences Xist through modification of chromatin structure. Dev Cell 9:159–165. doi: 10.1016/j.devcel.2005.05.015. [DOI] [PubMed] [Google Scholar]
- 10.Ohhata T, Hoki Y, Sasaki H, Sado T. 2008. Crucial role of antisense transcription across the Xist promoter in Tsix-mediated Xist chromatin modification. Development 135:227–235. doi: 10.1242/dev.008490. [DOI] [PubMed] [Google Scholar]
- 11.Ladd PD, Smith LE, Rabaia NA, Moore JM, Georges SA, Hansen RS, Hagerman RJ, Tassone F, Tapscott SJ, Filippova GN. 2007. An antisense transcript spanning the CGG repeat region of FMR1 is upregulated in premutation carriers but silenced in full mutation individuals. Hum Mol Genet 16:3174–3187. doi: 10.1093/hmg/ddm293. [DOI] [PubMed] [Google Scholar]
- 12.Brouwer JR, Willemsen R, Oostra BA. 2009. The FMR1 gene and fragile X-associated tremor/ataxia syndrome. Am J Med Genet B Neuropsychiatr Genet 150B:782–798. doi: 10.1002/ajmg.b.30910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tufarelli C, Stanley JA, Garrick D, Sharpe JA, Ayyub H, Wood WG, Higgs DR. 2003. Transcription of antisense RNA leading to gene silencing and methylation as a novel cause of human genetic disease. Nat Genet 34:157–165. doi: 10.1038/ng1157. [DOI] [PubMed] [Google Scholar]
- 14.Shearwin KE, Callen BP, Egan JB. 2005. Transcriptional interference—a crash course. Trends Genet 21:339–345. doi: 10.1016/j.tig.2005.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li J, Moazed D, Gygi SP. 2002. Association of the histone methyltransferase Set2 with RNA polymerase II plays a role in transcription elongation. J Biol Chem 277:49383–49388. doi: 10.1074/jbc.M209294200. [DOI] [PubMed] [Google Scholar]
- 16.Carrozza MJ, Li B, Florens L, Suganuma T, Swanson SK, Lee KK, Shia WJ, Anderson S, Yates J, Washburn MP, Workman JL. 2005. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell 123:581–592. doi: 10.1016/j.cell.2005.10.023. [DOI] [PubMed] [Google Scholar]
- 17.Xie L, Pelz C, Wang W, Bashar A, Varlamova O, Shadle S, Impey S. 2011. KDM5B regulates embryonic stem cell self-renewal and represses cryptic intragenic transcription. EMBO J 30:1473–1484. doi: 10.1038/emboj.2011.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dhayalan A, Rajavelu A, Rathert P, Tamas R, Jurkowska RZ, Ragozin S, Jeltsch A. 2010. The Dnmt3a PWWP domain reads histone 3 lysine 36 trimethylation and guides DNA methylation. J Biol Chem 285:26114–26120. doi: 10.1074/jbc.M109.089433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Plath K, Fang J, Mlynarczyk-Evans SK, Cao R, Worringer KA, Wang H, de la Cruz CC, Otte AP, Panning B, Zhang Y. 2003. Role of histone H3 lysine 27 methylation in X inactivation. Science 300:131–135. doi: 10.1126/science.1084274. [DOI] [PubMed] [Google Scholar]
- 20.Gontan C, Achame EM, Demmers J, Barakat TS, Rentmeester E, van IW, Grootegoed JA, Gribnau J. 2012. RNF12 initiates X-chromosome inactivation by targeting REX1 for degradation. Nature 485:386–390. doi: 10.1038/nature11070. [DOI] [PubMed] [Google Scholar]
- 21.Barakat TS, Gunhanlar N, Gontan Pardo C, Achame EM, Ghazvini M, Boers R, Kenter A, Rentmeester E, Grootegoed JA, Gribnau J. 2011. RNF12 activates Xist and is essential for X chromosome inactivation. PLoS Genet 7:e1002001. doi: 10.1371/journal.pgen.1002001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Friedrich G, Soriano P. 1991. Promoter traps in embryonic stem cells: a genetic screen to identify and mutate developmental genes in mice. Genes Dev 5:1513–1523. doi: 10.1101/gad.5.9.1513. [DOI] [PubMed] [Google Scholar]
- 23.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−delta delta C(T)) method. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 24.Kumaki Y, Oda M, Okano M. 2008. QUMA: quantification tool for methylation analysis. Nucleic Acids Res 36:W170–W175. doi: 10.1093/nar/gkn294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alexopoulou AN, Couchman JR, Whiteford JR. 2008. The CMV early enhancer/chicken beta actin (CAG) promoter can be used to drive transgene expression during the differentiation of murine embryonic stem cells into vascular progenitors. BMC Cell Biol 9:2. doi: 10.1186/1471-2121-9-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ng HH, Robert F, Young RA, Struhl K. 2003. Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol Cell 11:709–719. doi: 10.1016/S1097-2765(03)00092-3. [DOI] [PubMed] [Google Scholar]
- 27.Saxonov S, Berg P, Brutlag DL. 2006. A genome-wide analysis of CpG dinucleotides in the human genome distinguishes two distinct classes of promoters. Proc Natl Acad Sci U S A 103:1412–1417. doi: 10.1073/pnas.0510310103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A, Zhang X, Bernstein BE, Nusbaum C, Jaffe DB, Gnirke A, Jaenisch R, Lander ES. 2008. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature 454:766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mohn F, Weber M, Rebhan M, Roloff TC, Richter J, Stadler MB, Bibel M, Schubeler D. 2008. Lineage-specific polycomb targets and de novo DNA methylation define restriction and potential of neuronal progenitors. Mol Cell 30:755–766. doi: 10.1016/j.molcel.2008.05.007. [DOI] [PubMed] [Google Scholar]
- 30.Jeong S, Stein A. 1994. Micrococcal nuclease digestion of nuclei reveals extended nucleosome ladders having anomalous DNA lengths for chromatin assembled on non-replicating plasmids in transfected cells. Nucleic Acids Res 22:370–375. doi: 10.1093/nar/22.3.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hebbar PB, Archer TK. 2008. Altered histone H1 stoichiometry and an absence of nucleosome positioning on transfected DNA. J Biol Chem 283:4595–4601. doi: 10.1074/jbc.M709121200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sado T, Fenner MH, Tan SS, Tam P, Shioda T, Li E. 2000. X inactivation in the mouse embryo deficient for Dnmt1: distinct effect of hypomethylation on imprinted and random X inactivation. Dev Biol 225:294–303. doi: 10.1006/dbio.2000.9823. [DOI] [PubMed] [Google Scholar]
- 33.Borgel J, Guibert S, Li Y, Chiba H, Schubeler D, Sasaki H, Forne T, Weber M. 2010. Targets and dynamics of promoter DNA methylation during early mouse development. Nat Genet 42:1093–1100. doi: 10.1038/ng.708. [DOI] [PubMed] [Google Scholar]
- 34.Okano M, Bell DW, Haber DA, Li E. 1999. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 99:247–257. doi: 10.1016/S0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 35.Kristensen LH, Nielsen AL, Helgstrand C, Lees M, Cloos P, Kastrup JS, Helin K, Olsen L, Gajhede M. 2012. Studies of H3K4me3 demethylation by KDM5B/Jarid1B/PLU1 reveals strong substrate recognition in vitro and identifies 2,4-pyridine-dicarboxylic acid as an in vitro and in cell inhibitor. FEBS J 279:1905–1914. doi: 10.1111/j.1742-4658.2012.08567.x. [DOI] [PubMed] [Google Scholar]
- 36.Hainer SJ, Pruneski JA, Mitchell RD, Monteverde RM, Martens JA. 2011. Intergenic transcription causes repression by directing nucleosome assembly. Genes Dev 25:29–40. doi: 10.1101/gad.1975011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thebault P, Boutin G, Bhat W, Rufiange A, Martens JA, Nourani A. 2011. Transcription regulation by the noncoding RNA SRG1 requires Spt2-dependent chromatin deposition in the wake of RNA polymerase II. Mol Cell Biol 31:1288–1300. doi: 10.1128/MCB.01083-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Eszterhas SK, Bouhassira EE, Martin DI, Fiering S. 2002. Transcriptional interference by independently regulated genes occurs in any relative arrangement of the genes and is influenced by chromosomal integration position. Mol Cell Biol 22:469–479. doi: 10.1128/MCB.22.2.469-479.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Houseley J, Rubbi L, Grunstein M, Tollervey D, Vogelauer M. 2008. A ncRNA modulates histone modification and mRNA induction in the yeast GAL gene cluster. Mol Cell 32:685–695. doi: 10.1016/j.molcel.2008.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Navarro P, Page DR, Avner P, Rougeulle C. 2006. Tsix-mediated epigenetic switch of a CTCF-flanked region of the Xist promoter determines the Xist transcription program. Genes Dev 20:2787–2792. doi: 10.1101/gad.389006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ohhata T, Senner CE, Hemberger M, Wutz A. 2011. Lineage-specific function of the noncoding Tsix RNA for Xist repression and Xi reactivation in mice. Genes Dev 25:1702–1715. doi: 10.1101/gad.16997911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Beard C, Li E, Jaenisch R. 1995. Loss of methylation activates Xist in somatic but not in embryonic cells. Genes Dev 9:2325–2334. doi: 10.1101/gad.9.19.2325. [DOI] [PubMed] [Google Scholar]
- 43.Santoro F, Mayer D, Klement RM, Warczok KE, Stukalov A, Barlow DP, Pauler FM. 2013. Imprinted Igf2r silencing depends on continuous Airn lncRNA expression and is not restricted to a developmental window. Development 140:1184–1195. doi: 10.1242/dev.088849. [DOI] [PubMed] [Google Scholar]
- 44.Duhring U, Axmann IM, Hess WR, Wilde A. 2006. An internal antisense RNA regulates expression of the photosynthesis gene isiA. Proc Natl Acad Sci U S A 103:7054–7058. doi: 10.1073/pnas.0600927103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu Z, Wei W, Gagneur J, Clauder-Munster S, Smolik M, Huber W, Steinmetz LM. 2011. Antisense expression increases gene expression variability and locus interdependency. Mol Syst Biol 7:468. doi: 10.1038/msb.2011.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.